SUMMARY
The small program Illustrate generates non-photorealistic images of biological molecules for use in dissemination, outreach and education. The method has been used as part of the “Molecule of the Month,” an ongoing educational column at the RCSB Protein Data Bank (http://rcsb.org). Insights from 20 years of application of the program are presented, and the program has been released both as open source Fortran at Github and through an interactive web-based interface.
eTOC Blurb
The small program Illustrate generates non-photorealistic images of biological molecules for use in dissemination, outreach and education. Insights from 20 years of application of the program at the RCSB Protein Data Bank are presented, and the program is available as open Fortran source and through an interactive web-based interface.
INTRODUCTION
Biomolecular visualization is a mature field, with many effective methods for interactive display and exploration (O’Donoghue et al., 2010; Olson, 2018). The majority of these tools use a look-and-feel that reflects the type of scene generation that historically has been relatively simple to generate, including models of basic primitive geometries with glossy shaded surfaces or rapidly-rendered lines with no shading at all. These representations are essential tools for the structural biology community and are used widely at all stages of structure determination, analysis and dissemination.
Over the years, we have explored less traditional approaches for rendering biomolecular structures, which take their lead from methods used by illustrators (Figure 1). In popular and professional publications, cartoons and schematics are often used to distill salient properties from complex objects, creating readily recognizable and comprehensible images. We began experimenting with non-photorealistic rendering the 1990s, when computer graphics was rapidly becoming an essential tool in structural biology. At the time, line-based images from ORTEP (Johnson, 1965) and other programs were common due to the widespread use of pen plotters. Use of outlines in raster images was beginning to be actively explored, for example, in the enhancement of virus images (Namba et al., 1989), postscript molecular images from MolScript (Kraulis, 1991), and more recently, brought together with ambient occlusion shading in QuteMol (Tarini et al., 2006) and cartoony images in ProteinShader (Weber, 2009) and VMD (https://www.ks.uiuc.edu/Research/vmd/minitutorials/glsloutline/).
Figure 1.
Selected biomolecules involved in protein synthesis, drawn at the same scale for easy comparison. These images were presented as part of a poster at PDB-101, available at http://pdb101.rcsb.org/learn/flyers-posters-and-other-resources/flyer/molecular-machinery-a-tour-of-the-protein-data-bank. PDB accession codes for the structures depicted in this article are included in the STAR Methods, and full citations are available on the RCSB PDB site in the structure browser pages, for example, https://www.rcsb.org/structure/1aon.
As part of a postdoctoral project, we modified a small Fortran program for generating raster molecular surfaces (Goodsell, 1988) to generate outlines that highlight the shape and form of spacefilling representations (Goodsell and Olson, 1992), as described below. We are lucky to have a visual record of development after that point: the software was used to illustrate the “Molecule of the Month” (Goodsell et al., 2015), a monthly column at the RCSB Protein Data Bank (Berman et al., 2000). From 2000 to the present, the column has been published as part of PDB-101 (http://pdb101.rcsb.org), the educational portal of the RCSB Protein Data Bank. Images from the column show improvements in antialiasing of outlines in 2005 and replacement of cast shadows with screen-space ambient occlusion in 2007. Over the years, we have released this software in an informal basis, but it was not made generally available because of difficulty of use and lack of interactivity. Recently, to celebrate a 20-year milestone the Molecule of the Month, we developed a web interface for Illustrate to make the method more generally available, and for adventurous users, we have also released the Fortran code on GitHub. In this manuscript, we present a short description and history of the method, some insights into creation of effective illustrations, and some prospects for future uses of the method.
RESULTS AND DISCUSSION
Illustrate is a Fortran program that reads coordinates in PDB format and creates a cartoon image composed of spacefilling spheres. The program relies on a simple but flexible selection and representation scheme that uses the typical punchcard-based approach of early Fortran programming. The user defines a series of atom specification cards, which include a range of residue numbers and a text block that is matched with the atom name and residue name fields of each PDB-format atom record. The cards also include red-green-blue values for the color and a radius. As atoms are read from the PDB file, they are compared with each specification card, and when there is a match, the atom is assigned those values.
The outlines are created using a feature detection technique that looks for large changes in the Z-buffer and other properties (Figure 2). We initially used first and second derivatives on depth, but later moved to a simple kernel that looks in a two-by-two neighborhood and counts the number of pixels that have large height differences relative to the central pixel. The initial version of the program also used cast shadows to heighten three-dimensionality of the images, but we moved to a direction-independent approach in 2007 using screen-space ambient occlusion to give a cleaner image. For large assemblies this calculation is considerably faster than cast shadow approaches, since it is performed on the z-buffer rather than on the list of atoms. The outline and shadowing methods are described in the STAR Methods section below.
Figure 2.
Outlines are generated by looking for differences in subunit numbers between chains, residue numbers within chains, and relative depth of pixels in the image. These are then combined to give the final image (right). The molecules are elongation factor Tu (blue) and transfer RNA (red).
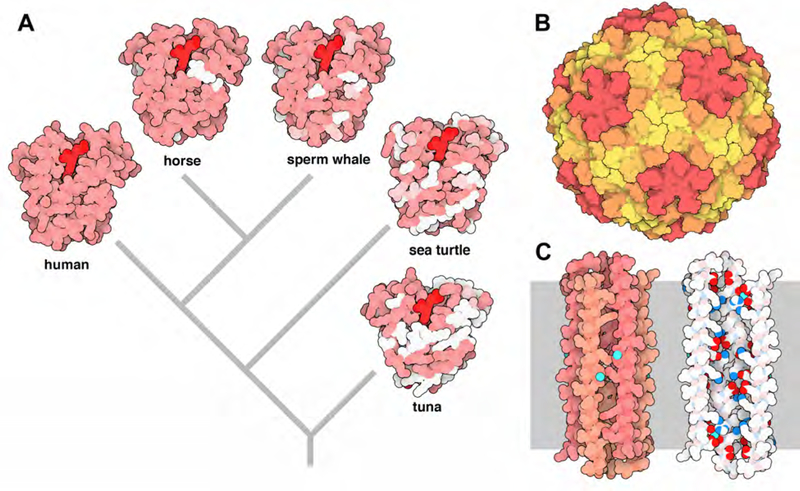
The new web-based interface provides a turnkey graphical user interface for Illustrate, allowing novice users to type in a PDB ID and quickly get an image. The interface uses NGL (Rose 2015) to provide a preview to help with orientation. Building on NGL, it also provides more facile access to several of the features available at the RCSB Protein Data Bank, including selection of biological assemblies and unit cells. Several curated styles are available in a pull-down menu (Figure 3), including a style that highlights proteins and nucleic acids, traditional coloring by atom type, and a subunit coloring scheme that uses IWantHue (http://tools.medialab.sciences-po.fr/iwanthue/) to generate an “optimally distinct” color palette. The interface also provides the ability to read user-specified coordinate files and Illustrate command files, for users who want to get their hands dirty and create custom illustrations.
Figure 3.
Illustrations from the web interface. (A) P-glycoprotein colored using the property-based atomic coloring. The gray membrane schematic was added in Photoshop. (B) Supercell packing of hemoglobin, using transformations provided by NGL. (C) Small subunit of the ribosome using subunit colors from IWantHue. (D) Four nucleosomes using a coarse representation, with DNA in orange and histones in blue.
Personal Best Practices
Scientific illustration is subjective: as long as the science is depicted accurately, the illustrator has much artistic freedom to customize the look and feel of an illustration. Looking back over 20 years of creating illustrations, we have identified a few general principles that have improved our approach. While largely based on personal preferences, we have received a few informal validations of these principles in practice, first through direct feedback from readers of the Molecule of the Month and other publications, and through adoption of the style in popular programs such as Chimera (https://www.cgl.ucsf.edu/chimera/ImageGallery/entries/clathrin/clathrin.html) and QuteMol (http://qutemol.sourceforge.net).
First and foremost is the incredible richness of the biostructural archive of knowledge, which calls for an improvisational approach to visualization. Defaults are a great place to start, but nearly every new subject requires some measure of customization. For example, we used a slightly modified version of atomic coloring for many of the early subjects, which is useful for viewers who are familiar with the basics of hydrophilicity and hydrophobicity. We gradually moved away from this approach, towards an approach that focuses on subunits and molecular interactions, which tells a higher-level story that is more appropriate for the non-expert audiences of the column. Defaults are a double-edged sword: traditional coloring schemes are instantly recognizable and comprehensible without much description, but they lock you into traditional storylines that may cause confusion when you wander into new areas.
This higher-level coloring and rendering approach also has the great advantage of simplifying the images. This embodies two principles that often make for more effective imagery: to formulate goals early in the process, and then simplify the image so that it’s not trying to address multiple goals simultaneously. As is often the case, less is more. However, this must be tempered by a healthy trust of viewers, so that we give them enough of the underlying complexity inherent in these remarkable structures to invite exploration.
To address these two best practices (the ability to customize and the desire to simplify), the Illustrate program provides a flexible approach to selection and coloring that streamlines focus at a high level, with illustrative rendering controls that allow us to balance complexity against comprehensibility. In the 20 years of work at the RCSB Protein Data Bank, we have experimented with a number of approaches, but it has all settled down to a style with a few basic principles that are applied in most cases (Figure 1). Solid colors are used to highlight subunits, and the carbon atoms in each subunit are given a slightly lighter shade. This slight color variation is enough to give a feeling for the complexity of the structure, while not distracting from the overall shape and form of the subunit. Similar shades of the color are used to distinguish subunits within an assembly, such as multiple subunits in an oligomeric enzyme or all the different protein chains in a ribosome. The difference is just enough to highlight that they are separate chains, but not enough to distract from the form of the overall assembly. Larger color differences are reserved for portions of the subject that have functional differences, for example, protein vs. DNA in a repressor complex or catalytic vs. regulatory subunits in an enzymatic complex. Finally, specific coloring schemes are layered on this general approach when faced with the many interesting twists of biomolecular structure and function (Figure 4).
Figure 4.
Specialized illustrations for specific topics presented in the “Molecule of the Month” at PDB-101. A. Color is used to show evolutionary conservation in myoglobin. B. Viral quasisymmetry is highlighted with color and outlines that surround hexamers and pentamers of subunits. C. A hydrophilic tunnel formed by dermcidin is shown with a modified atomic coloration scheme that has charged atoms in bright colors.
Sprites
These types of cartoony illustrations have another great advantage: they are quite modular, so compositing approaches are simple and effective. Scale levels and magnification are slippery parameters in most interactive molecular viewers, but Illustrate is designed to create illustrations using orthographic perspective with a user-defined scale level (in Angstroms/pixel). We have taken advantage of this over the years in many posters and reviews, displaying a panel of images, or even an entire book of images, drawn at the same scale to allow easy comparison (as in Figure 1). The simple style of the illustration also streamlines the combination of multiple images into a composite image, making it easy to create images of multidomain proteins or higher-order assemblies in popular programs like Photoshop. For example, illustrations of the DNA-binding proteins in Figure 1 were generated separately from entries in the PDB archive, and then composited, eliminating the need for a detailed modeling effort to generate an atomic model of the entire assembly. Taking this to the next step, we are currently developing CellPAINT, software that acts much like a digital painting program (Gardner et al., 2018). Sprites (two dimensional bitmaps that may be quickly integrated into larger scenes) of individual molecules are presented to the user in a palette, and then may be painted into a larger image (Figure 5). The current prototype allows creation of sprites with Illustrate, to populate the palette with structures of interest.
Figure 5.
Illustration of blood plasma, created interactively using CellPAINT with sprites from Illustrate. A poliovirus is included in blue. For several molecules, such as the star-shaped IgM at lower right, structure files included only alpha-carbon positions, so a coarse representation was used.
Invitation
We invite the community to use these programs in science dissemination, education and outreach, and welcome feedback. Information on free availability of the web-based interface and the source code is included in the STAR Methods section.
STAR METHODS
Lead Contact and Materials Availability
Software, documentation, and worked examples for Illustrate are freely available as described below in Data and Code Availability. We welcome feedback about use of the method to the Lead Contact: David S. Goodsell (goodsell@scripps.edu).
Method Details
Outlines
Illustrate visits each pixel and sums the number of pixels in a two-by-two neighborhood with height differences greater than a user-defined threshold. The user then specifies two contouring thresholds to define how this information is used to create outlines: typically, if this sum is less than about three, the pixel is not modified, if the sum is greater than eight, the pixel is colored black, and intermediate values are given antialiased shades of gray. Outlines that separate subunits are drawn similarly, by counting up the number of pixels in the neighborhood that are assigned to different subunits. Similarly, outlines that highlight different regions of a chain, highlighting the details of a protein fold, are drawn by using a user-defined threshold for differences in residue number in a chain, and then counting up the number of neighboring pixels that show differences greater than this threshold.
Screen-space Ambient Occulsion
Each pixel in the z-buffer casts a cone-shaped shadow away from the viewer, approximating a large, diffuse light source behind the viewer’s head. After all atoms are added to the z-buffer, each pixel casts a conical shadow, darkening neighboring pixels based on a user-defined angle and shadowing fraction. Two additional parameters allow the user to tune the quality of the shadows. First, a threshold for the minimum height difference is used to reduce shadowing of atoms by themselves. Second, a maximal value for the total shadowing fraction of each pixel may be defined, to produce softer shadows.
Quantification and Statistical Analysis
No tools for quantification or statistical analysis were used in this work.
Data and Code Availability
We invite the community to use these programs in science dissemination, education and outreach. The Fortran open-source code and documentation is available and free for use and reuse at Github (https://github.com/ccsb-scripps/Illustrate). The web-based interface is available with documentation and worked examples at http://ccsb.scripps.edu/illustrate.
HIGHLIGHTS.
A web-based and standalone program for molecular illustration is available for use.
Non-photorealistic rendering allows the creation of illustrations of biomolecules.
ACKNOWLEDGEMENTS
Development of Illustrate has been kindly supported by Damon Runyon-Walter Winchell Cancer Research Fund Fellowship DRG 972, the US National Institutes of Health R01-GM120604 and the RCSB Protein Data Bank (National Science Foundation DBI-1832184, the National Institutes of Health R01GM133198, and the US Department of Energy DE-SC0019749). This is manuscript 29865 from the Scripps Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, and Bourne PE (2000). The Protein Data Bank. Nucleic Acids Res 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner A, Autin L, Barbaro B, Olson AJ, and Goodsell DS (2018). CellPAINT: Interactive illustration of dynamic mesoscale cellular environments. IEEE Computer Graphics Appl 38, 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell DS (1988). RMS - Programs for Generating Raster Molecular-Surfaces. Journal of Molecular Graphics 6, 41–44. [Google Scholar]
- Goodsell DS, Dutta S, Zardecki C, Voigt M, Berman HM, and Burley SK (2015). The RCSB PDB “Molecule of the Month”: Inspiring a Molecular View of Biology. PLoS Biol 13, e1002140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell DS, and Olson AJ (1992). Molecular illustration in black and white. J Molec Graphics 10, 235–240. [DOI] [PubMed] [Google Scholar]
- Johnson CK (1965). ORTEP, A Fortran thermal-ellipsoid plot protein from crystal structure illustrations. In ONRL Report #3794 (Oak Ridge National Laboratory, TN: ). [Google Scholar]
- Kraulis P (1991). MolScript: a program to produce both detailed and schematic plots of protein structures. J Appl Cryst 24, 946–950. [Google Scholar]
- Namba K, Caspar DLD, and Stubbs G (1989). Enhancement and simplification of macromolecular images. Biophys J 53, 469–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donoghue SI, Goodsell DS, Frangakis AS, Jossinet F, Laskowski RA, Nilges M, Saibil HR, Schafferhans A, Wade RC, Westhof E, et al. (2010). Visualization of macromolecular structures. Nat Methods 7, S42–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson AJ (2018). Perspectives on Structural Molecular Biology Visualization: From Past to Present. J Mol Biol 430, 3997–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarini M, Cignoni P, and Montani C (2006). Ambient occlusion and edge cueing to enhance real time molecular visualization. Ieee Transactions on Visualization and Computer Graphics 12, 1237–1244. [DOI] [PubMed] [Google Scholar]
- Weber JR (2009). ProteinShader: illustrative rendering of macromolecules. BMC Struct Biol 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We invite the community to use these programs in science dissemination, education and outreach. The Fortran open-source code and documentation is available and free for use and reuse at Github (https://github.com/ccsb-scripps/Illustrate). The web-based interface is available with documentation and worked examples at http://ccsb.scripps.edu/illustrate.