SUMMARY
Nuclear glycogen was first documented in the early 1940s, but its role in cellular physiology remained elusive. In this study, we utilized pure nuclei preparations and stable isotope tracer to define the origin and metabolic fate of nuclear glycogen. Herein, we describe a key function for nuclear glycogen in epigenetic regulation through compartmentalized pyruvate production and histone acetylation. This pathway is altered in human non-small cell lung cancers, as surgical specimens accumulate glycogen in the nucleus. We demonstrate that the decreased abundance of malin, an E3 ubiquitin ligase, impaired nuclear glycogenolysis by preventing the nuclear translocation of glycogen phosphorylase and causing nuclear glycogen accumulation. Re-introduction of malin in lung cancer cells restored nuclear glycogenolysis, increased histone acetylation, and decreased growth of cancer cells transplanted into mice. This study uncovers a previously unknown role for glycogen metabolism in the nucleus and elucidates another mechanism by which cellular metabolites control epigenetic regulation.
Keywords: Glycogen, nuclear metabolism, histone acetylation, non-small cell lung cancer, ubiquitination, E3 ubiquitin ligase, malin, NHLRC1, EPM2B, glycogen phosphorylase
Graphical Abstract

eTOC:
Nuclear glycogen accumulation has been reported in multiple cancers. Sun et al. show that glycogen is de novo synthesized in the nucleus, and nuclear glycogenolysis provides a carbon pool for histone acetylation. Non-small cell lung cancers suppress nuclear glycogenolysis by down-regulating a key E3 ubiquitin ligase to drive cancer progression.
INTRODUCTION
Lung cancer is the most common cancer worldwide, accounting for approximately 1.8 million new cases and 1.6 million deaths every year (Bray et al., 2018). NSCLC accounts for approximately 85% of all lung cancer cases and is often caused by tobacco-induced genetic instability. The standard of care for non-small cell lung cancer (NSCLC) patients includes multi-agent chemotherapy to treat documented or potential metastatic disease, coupled with surgery and/or irradiation to treat the primary tumor. Although some incremental advances have been made in the last three decades through intensification of conventional chemotherapy agents, more significant improvements will likely depend on the identification of novel treatment strategies. Recent studies have identified major metabolic reprogramming in all types of NSCLC, suggesting that aberrant metabolism is an important feature in the transformation process and revealing potential novel therapeutic targets (Kerr et al., 2016; Kottakis et al., 2016; Shackelford and Shaw, 2009; Ying et al., 2012).
Glycogen is the primary source of storage carbohydrate in mammals; it is found in most tissues, including liver (Costill et al., 1973; Zois and Harris, 2016), muscle (Hultman and Nilsson, 1971), kidney (Krebs et al., 1963), brain (Brown and Ransom, 2007), white blood cells (Gibb and Stowell, 1949), and the lung (Bourbon and Jost, 1982). Glycogen synthesis and degradation either consumes or produces glucose-6-phosphate (G6P), a key metabolite essential for central carbon metabolism. Several studies have reported glycogen accumulation in specific sub-cellular organelles, suggesting that glycogen localization is not random. Nuclear glycogen was first reported in the 1940s in hepatocytes (Baird and Fisher, 1957; Bogoch et al., 1955; Chipps and Duff, 1942; Himes et al., 1956; Mori et al., 1970), and subsequent reports identified glycogen accumulation near the ER (Cardell Jr, 1977; De Man et al., 1966), and mitochondria (Ishikawa and Pei, 1965; Nielsen et al., 2010). Cumulatively, these data suggest compartment-specific roles for glycogen that have yet to be fully elucidated.
Elevated glycogen can be detected in multiple cancer cell lines, including lung, breast, kidney, uterus, bladder, ovary, skin, brain, and more recently colorectal cancer (Favaro et al., 2012; Rousset et al., 1979; Rousset et al., 1981; Sato et al., 2015; Staedel and Beck, 1978; Zhou et al., 2019). Hypoxia, a key characteristic of solid tumors, induces glycogen synthesis in certain cancer sub-types, although the exact mechanism of this phenotype has yet to be resolved (Iida et al., 2012; Pescador et al., 2010). Recently, hypoxia-induced glycogenolysis was shown to enhance tumorigenesis by suppressing reactive oxygen species levels and p53-dependent senescence in breast and colon cancer cells (Favaro et al., 2012). Several studies also suggest that some cancer cells accumulate glycogen as a stored energy source to enable survival and sustain metastases under adverse conditions (Chen et al., 2015; Liu et al., 2013; Zois and Harris, 2016). In the case of ovarian cancer, glycogen was recently shown to be a nutrient currency and exchanged between cancer cells and cancer associated fibroblast to sustain metastasis (Curtis et al., 2019). Thus, the connections between glycogen metabolism in tumorigenesis and cancer progression are beginning to emerge. However, potential roles of glycogen beyond a simple energy cache have yet to be identified.
The E3 ubiquitin ligase malin is a modulator of glycogen metabolism via an unknown mechanism(s) (Gentry et al., 2018; Nitschke et al., 2018; Verhalen et al., 2018). Malin is a RING-type E3 ubiquitin ligase that has been shown to ubiquitinate multiple proteins involved in glycogen metabolism in vitro, yet its in vivo function has not been elucidated (Cheng et al., 2007; Gentry et al., 2005; Solaz-Fuster et al., 2008; Vilchez et al., 2007; Worby et al., 2008). While the detailed mechanism of action for malin is unknown, it is clear that malin is required for coordinated glycogen metabolism as somatic loss-of-function mutations in malin results in aberrant glycogen with deleterious consequences (Gentry et al., 2018; Nitschke et al., 2018; Turnbull et al., 2016; Verhalen et al., 2018).
We report that aberrant accumulation of nuclear glycogen and decreased levels of malin are features of NSCLC. We employed ultra-pure nuclear preparations coupled with stable isotope technology to define the origin and biological destiny of nuclear glycogen. Malin is required to maintain normal nuclear glycogen metabolism, and loss of malin in NSCLC results in the accumulation of nuclear glycogen and reduced histone acetylation in the nucleus. Additionally, we demonstrate that nuclear glycogen is synthesized de novo and that NSCLC cells down-regulate nuclear glycogenolysis. Importantly, loss of malin promotes cellular proliferation while re-expression delays tumor growth in vivo. These data provide new insights for an alternative signaling pathway that regulates metabolism in the nucleus.
RESULTS
Glycogen Accumulates in Nuclei of Human NSCLC tissue
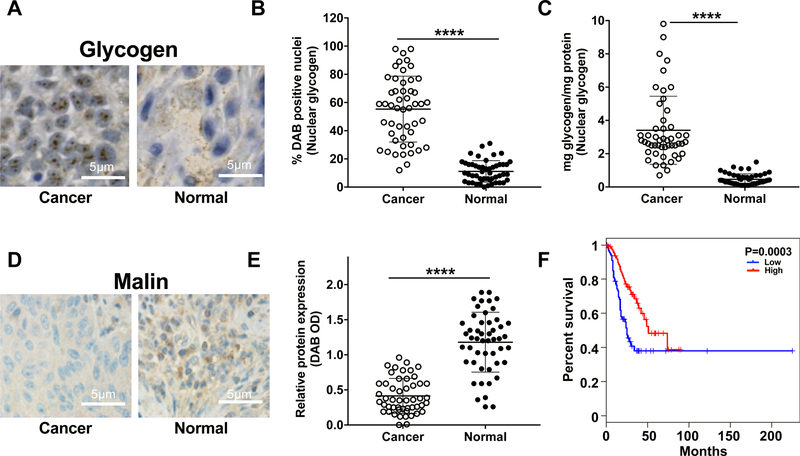
To define the function of malin in glycogen metabolism, we investigated the localization of both using immunohistochemical (IHC) analyses. Fifty paired NSCLC and tumor-distal benign lung tissues were analyzed using antibodies to glycogen (Oe et al., 2016) or malin (Figure 1A, 1D, and S1). This staining revealed punctate glycogen aggregates in the nuclei of cancer cells (Figure 1A and Figure S1A–C). Unbiased distribution analyses of whole sample slices using the multiplex IHC module of the HALO digital pathology software (Figure S1A) showed a greater than 5-fold increase in the number of glycogen-positive nuclei in cancer cells compared to normal cells (Figure 1B). To confirm the histology results, we isolated nuclei from NSCLC and tumor-distal benign tissue and biochemically quantified the amount of nuclear glycogen. Similar to the IHC results, cancer tissues displayed dramatically higher amounts of nuclear glycogen than normal tissues (Figure 1C). Nuclear glycogen content ranged from 1–10 mg/mg protein in patient cancer tissues, representing a 10–100-fold increase compared with normal tissue. Interestingly, normal tissues did contain measurable amounts of glycogen, suggesting that nuclear glycogen metabolism is a physiological event in normal cells. The increased nuclear glycogen in NSCLC tissue was accompanied by a reduction in the abundance of malin demonstrated by both IHC and immunoblotting (Figure 1D and Figure S1D–E). HALO analysis of the same cohort of fifty patients revealed a greater than 65% reduction of malin protein in NSCLC tissues compared to normal tissues (Figure 1E). Using TCGA survival data with matching RNAseq analysis, we found that high malin mRNA expression correlates with better survival, suggesting that malin is clinically important in NSCLC (Figure 1F).
Figure 1: Nuclear glycogen accumulation and loss of malin in NSCLC.
(A) Immunohistochemical staining for glycogen in paired NSCLC tumor and adjacent benign lung tissue (normal) resected at surgery. Scale bar = 5µm
(B) Quantification of the glycogen immunohistochemical staining detected with 3,3′-Diaminobenzidine (DAB) using the HALO digital pathology software. Tissue from 50 NSCLC patient tumor and adjacent benign lung tissue was used for this analysis and subsequent analyses.
(C) Biochemical quantification of nuclear glycogen from NSCLC patient tumor and adjacent benign (normal) lung tissue (n=50).
(D) Immunohistochemical staining for malin in paired NSCLC tumor and adjacent benign lung tissue (normal) resected at surgery. Scale bar = 5µm.
(E) Relative abundance of malin in patient tumor and normal samples (n=50), defined by average DAB intensity.
(F) Kaplan-Meier survival plot of NSCLC patient data from TCGA with high and low malin mRNA abundance separated by upper and lower quartile. Blue, low malin mRNA; red, high malin mRNA.
***P < 0.001, two-tailed t-test.
See also Figure S1
Glycogen is Synthesized De Novo in Nuclei
The identification of nuclear glycogen suggests the possibility of nuclear specific metabolic pathways. Since nuclear glycogen was identified in both normal and cancer cells, we sought to better elucidate nuclear metabolic processes. Traditional techniques for rapid purification of nuclei, such as a salt gradient followed by differential centrifugation, give rise to cytoplasmic contamination, so these protocols are not suitable for purifying nuclei from tissue. To overcome these issues, we utilized a method from a recent report for isolating nuclei using sucrose gradient centrifugation (Nagata et al., 2010) to develop parallel approaches for isolating highly pure nuclei from both cell lines and tissues. Cells or milled tissues were suspended in cell lysis buffer supplemented with collagenase. After gentle agitation, cells were centrifuged and washed, and the nuclei were isolated via sucrose gradient centrifugation. To avoid residual endoplasmic reticulum (ER) contamination and outer nuclear membrane proteins, isolated nuclei were further subjected to proteinase K digestion whereby only intranuclear proteins like Lamin A were protected (Besingi et al., 2013; Saurí et al., 2011; Selkrig et al., 2012). This methodology allowed sequential removal of ER proteins (e.g. GRP78), cytoplasmic proteins (e.g. actin), and outer nuclear membrane proteins (e.g. Nesprin-3) in each of the purification steps to obtain pure intact nuclei, which was monitored by immunoblotting (Figure 2A). Isolated nuclei were further analyzed for purity by bright field imaging of size distribution (Figure S2A) and immunoblotting for both the transcription factor USF1 and histones as nuclei markers (Figure S2B). Furthermore, we verified that the preparations were free of soluble cytosolic or mitochondrial contamination by immunoblotting for actin and succinate dehydrogenase (SDHA), respectively (Figure S2B). To confirm that the nuclei are intact and active, we traced the metabolic fate of 13C-pyruvate in both nuclear and cytosolic fractions (Figure S2C). In the cytosolic fraction, 13C-pyruvate was converted to 13C-citrate, 13C-malate, and 13C-succinate through both the oxidative and anaplerotic reactions of the TCA cycle. Conversely, we did not detect enrichment of citrate, malate, or succinate in the nuclear fraction (Figure S2C). Nuclei can utilize 13C-pyruvate for histone acetylation (Liu et al., 2018), and we observed robust histone acetylation in the nuclear fraction and not the cytosolic fraction, further confirming the purity of the nuclear preparations (Figure S2C). As an internal control, the cytosolic fraction was also subjected to protease digestion to demonstrate the removal of proteins and enzyme activity after treatment (Figure 2H, S2B–C).
Figure 2: De novo glycogen biosynthesis in the nucleus.
(A) Schematic of sequential purification steps to obtain pure nuclei from A549 cells (top), diagram of different markers used to assess nuclear purity (bottom left), and (bottom right) immunoblotting analysis of fractions from each purification step. Lysates from A549 cells were immunoblotted to assess contamination from ER proteins (GRP78), outer nuclear membrane proteins (Nesprin-3) and cytosolic proteins (actin). Lamin A is used as a control to confirm that nuclei are intact and nuclear proteins are protected from proteolysis. OP: osmotic pressure, SG: sucrose gradient, PK: Proteinase K digestion.
(B) Schematic of glycogen labeling and enrichment analysis.
(C)-(D) Glycogen synthesis assays were performed with purified intact nuclei from A549, H1299, and H2030 cells. Graphs represent combined results from each cell line with an n=3 for each cell line. 13C6-glucose (C) and 13C6-glucose-6-phosphate (D) were utilized as substrates for nuclear glycogen synthesis. A total of 10 µg of glycogen was hydrolyzed for each sample prior to analysis.
(E) Nuclear glycogen synthesis occurs in cultured HEK293 cells using 13C6-glucose as the substrate. 13C6-glucose was incubated with HEK293 cells, triplicate cells were lysed to purify nuclei, 10 µg of glycogen was hydrolyzed, and the sample was analyzed by GCMS.
(F) Purified nuclei from HEK293 cells were used to define the isotopic steady state for nuclear glycogen synthesis. 13C6-glucose-6-phosphate was incubated with nuclei and samples were removed every 30 minutes, 10µg of glycogen was hydrolyzed, and the sample was analyzed by GCMS.
(G) Diagram depicting that nuclear glycogen synthesis utilizes 13C6-glucose-6-phosphate.
(H) The nuclear and cytoplasmic fractions of A549 cells were probed by immunoblotting for glycogen synthase (GYS), UDP-glucose pyrophosphorylase (UGP2), hexokinase (HK), G6P-translocase (SLC37A4), and glucose transporters (Glut1 and Glut 4). Anti-actin was used as a cytoplasmic control and anti-histone as a nuclear control. Data in B and H are representative of three experiments. Data in C-G are from three experiments and are shown as mean ± SE.
*0.01 < P < 0.05; ** 0.001 < P < 0.01; *** P < 0.001, two-tailed t-test.
See also Figure S2
Nuclear glycogen was first identified in the 1940s and subsequent studies confirmed its presence (Caramia et al., 1968; Granzow et al., 1981; Himes et al., 1956), but its biological role has not been defined. The accumulation of nuclear glycogen in NSCLC created an ideal model to investigate the origin and biological consequences of nuclear glycogen. The molecular weight of glycogen is 5–20 million Daltons, a similar molecular weight as major enzyme complexes such as pyruvate dehydrogenase (PDH) (Linn et al., 1969) and a-ketoglutarate dehydrogenase complexes (Frank et al., 2007). Currently, there is no known mechanism for transporting glycogen across cell membranes. Additionally, glycogen synthase activity has been reported in the nucleus, suggesting that glycogen synthesis could occur de novo in the nucleus (Granzow et al., 1981; Zimmermann et al., 1976).
To test for de novo nuclear glycogen synthesis, A549 cell nuclei were purified and incubated with either 13C6-glucose or 13C6-glucose-6-phoshate in buffer supporting organelle metabolism to monitor 13C enrichment of glycogen through enzyme-mediated biosynthesis. After a six-hour incubation, the glycogen was purified and acid hydrolyzed followed by gas-chromatography mass spectrometry (GCMS) analysis (Figure 2B). The isolated nuclei were fully capable of generating glycogen, but 13C6-glucose was not incorporated into the glycogen (Figure 2C). Conversely, 13C6-glucose-6-phosphate (G6P) was readily incorporated into the nuclear glycogen (Figure 2D). To confirm this result, cultured cells and isolated nuclei were analyzed to determine their steady state 13C-nuclear glycogen synthesis from 13C6-glucose or 13C6-G6P, respectively. While cultured cells readily utilized 13C6-glucose to generate nuclear glycogen, the isolated nuclei required 13C6-G6P as the substrate for nuclear glycogen synthesis (Figure 2E–G).
Given that G6P is the substrate for nuclear glycogen synthesis, one would predict the presence of glycogen metabolic enzymes in the nucleus. To test this hypothesis, the nuclear and cytosolic fractions were immunoblotted for both glycogen synthesis enzymes and substrate transporters. Glycogen synthase (GYS), UDP-glucose pyrophosphorylase (UGP), and the glucose-6-phosphate transporter SLC37A4 were all identified in the nuclear and cytoplasmic fractions (Figure 2H). The glucose transporters Glut1 and Glut4 as well as hexokinase were only found in the cytoplasmic fractions, consistent with the finding that nuclear extracts could not utilize 13C6-glucose as a substrate for glycogen synthesis (Figure 2H). This finding is not surprising given that cytosolic hexokinase functions at Vmax even with low glucose concentrations in order to convert glucose to glucose-6-phosphate and prevent glucose efflux from the cytoplasm. (Engelking, 2015; Gots et al., 1972; Rose et al., 1974).
Nuclear Glycogenolysis Requires Malin-Directed Ubiquitination and Translocation of Glycogen Phosphorylase
Malin is comprised of a RING domain, a linker region, and six NHL protein-protein interaction repeats (Figure S3A) (Chan et al., 2003; Gentry et al., 2005). Ubiquitination can affect protein abundance or function by promoting proteasome-dependent degradation of the ubiquitinated protein or by altering its enzymatic activity, binding partners, or localization (Sun and Chen, 2004; Yau and Rape, 2016). Therefore, we hypothesized that the reduced abundance of malin in NSCLC contributes to the accumulation of nuclear glycogen by impacting one or more glycogen metabolic enzymes through at least one of these ubiquitin-mediated mechanisms of protein regulation.
To identify malin substrates that could modulate the abundance of nuclear glycogen, we purified recombinant malin, incubated the protein with cell extracts from HEK293 cells, and identified malin-bound proteins by mass spectrometry (Figure S3A). We identified known interacting proteins, including the glycogen phosphatase laforin (Gentry et al., 2005; Romá-Mateo et al., 2012), glycogen debranchin g enzyme (GDE) (Cheng et al., 2007), and heat shock protein 70 (HSP70) (Garyali et al., 2008), as well as several previously unreported malin-interacting partners that included glycogen branching enzyme (GBE) and glycogen phosphorylase brain isoform (GPBB) (Figure S3B). Using co-expression and co-immunoprecipitation experiments, we validated the known interactions and the interaction between malin and GPBB (Figure S3C–F) which, despite its name, is abundant in most tissues (Kim et al., 2014). Next, we performed reciprocal co-immunoprecipitation experiments from HEK293 lysates using antibodies against endogenous malin or GPBB to confirm the interaction between endogenous malin and GPBB (Figure S3G–H). Consistent with previous reports, malin overexpression decreased the amounts of laforin, HSP70, and GDE in HEK293 cells (Figure S3I) (Gentry et al., 2018; Gentry et al., 2005; Romá-Mateo et al., 2012). Conversely, the levels of GPBB did not decrease with malin overexpression, suggesting that malin does not target GPBB for degradation. To test if malin can ubiquitinate GP, we performed ubiquitination assays with purified proteins. GPBB ubiquitination was increased upon incubation with recombinant malin, ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), ATP, and ubiquitin (Figure S3J), indicating that GPBB is a substrate for malin. Furthermore, the A549 cell line overexpressing malin (OE) exhibits increased GPBB ubiquitination compared to cells expressing empty vector (EV) (Figure S3K). Similarly, alveolar type II (AT2) cells isolated from WT mice exhibit robust GP ubiquitination while AT2 cells from malin−/− show a dramatic reduction (Figure S3L).
Because GPBB is a previously unknown malin-interacting protein that could affect nuclear glycogen metabolism, we evaluated the effect of malin-mediated ubiquitination on its localization. In HEK293 cells, GPBB is mostly localized to the cytoplasm (Figure S3M). Co-expression of malin and GPBB in HEK293 cells resulted in GPBB translocation from the cytoplasm to the nucleus rather than proteasomal-directed degradation (Figure 3A). Conversely, co-expression with either malin containing a point mutation in the RING or NHL domain, mutations that compromise the E3 ligase activity of malin (Gentry et al., 2005), abolished the GPBB nuclear localization in HEK293, A549, H1299, and H2030 cells (Figure 3A and B and S3O–3Q). Moreover, exposing HEK293 cells to leptomycin, a nuclear exportin inhibitor (Kudo et al., 1998), resulted in the accumulation of endogenous GP in the nucleus (Figure S3M), suggesting that GPBB translocation is part of normal cellular physiology and is controlled through malin-mediated ubiquitination (Figure S3N).
Figure 3: Malin promotes nuclear localization of GP.
(A) Immunofluorescence localization of GP and malin in HEK293 cells co-expressing either wild-type malin (WT) or malin mutants C26S or Q302P. Data are representative of three independent experiments.
(B) Immunoblotting analysis of nuclear and cytosolic fractions for GPBB and malin from the A549, H2030, and H1299 cell lines stably overexpressing malin (OE) or empty vector (EV). Actin served as the loading control and indicator of cytosolic purity; histone served as the loading control and indicator of nuclear purity.
(C) Biochemical quantification of nuclear glycogen in the indicated cell lines stably overexpressing (OE) malin or empty vector (EV).
(D) IHC staining of GPBB in NSCLC tissue (cancer) and surrounding stroma cells (normal).
(E) The relative mRNA abundance for GPBB between lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) from TCGA database. Red = cancer; grey = normal.
(F) Quantification of IHC analysis for GPBB abundance in NSCLC patient tumor and normal samples (n=50).
(G) Quantification for the percent of GPBB-containing nuclei in NSCLC patient tumor and normal samples (n=50), quantified from IHC data.
B-C are from three experiments and shown as mean ±SE.
*0.01 < P < 0.05; ** 0.001 < P < 0.01; *** P < 0.001, two-tailed t-test.
See also Figure S3.
To validate the impact of malin on GPBB, we overexpressed malin in three different lung cancer cell lines and found that malin overexpression increased nuclear GP in each cell line (Figure 3B and S3R). As predicted, we also observed up to a 70% decrease in nuclear glycogen in the malin-overexpressing cell lines (Figure 3C). Thus, we propose that GP is targeted to the nucleus in a malin-dependent manner and reduced malin impairs the transport of GPBB into the nucleus, resulting in the accumulation of glycogen in the nucleus. To determine if cytosolic GPBB accumulation occurs in human patient specimens, we examined GPBB mRNA and protein abundance using the TCGA database and IHC analysis of patient samples. The same cohort of fifty patient samples were stained with anti-GPBB and analyzed using the HALO digital pathology platform to quantify both the intensity of GPBB staining and the percent of GPBB positive nuclei. GPBB mRNA and protein levels were both significantly increased in the cancer tissue compared to normal tissue (Figure 3D and E). Additionally, GPBB was largely cytosolic in cancerous tissues, whereas an average of 12% was localized to nuclei in the normal tissue (Figure 3F and G).
Nuclear Glycogen is a Substrate Pool for Histone Acetylation
We hypothesized that the nuclear glycogen represents a separate carbon pool from the cytosolic fraction, supplying metabolic substrates for histone modification independent of cytosolic metabolites. Multiple groups have demonstrated that individual glycolytic enzymes are detected in the nucleus, but many have undefined functions (Egea et al., 1992; Enzo et al., 2015; Funasaka et al., 2005; Hara et al., 2005; Matsuda et al., 2015; Sáez and Slebe, 2000). Moreover, pyruvate dehydroge nase translocates to the nucleus to regulate histone acetylation (Chen et al., 2018; Sutendra et al., 2014); however, the origin of nuclear pyruvate is currently ambiguous. Therefore, we investigated the functional role of GP and nuclear glycogen in pyruvate production and histone acetylation using isolated nuclei and stable isotope-labeling technology.
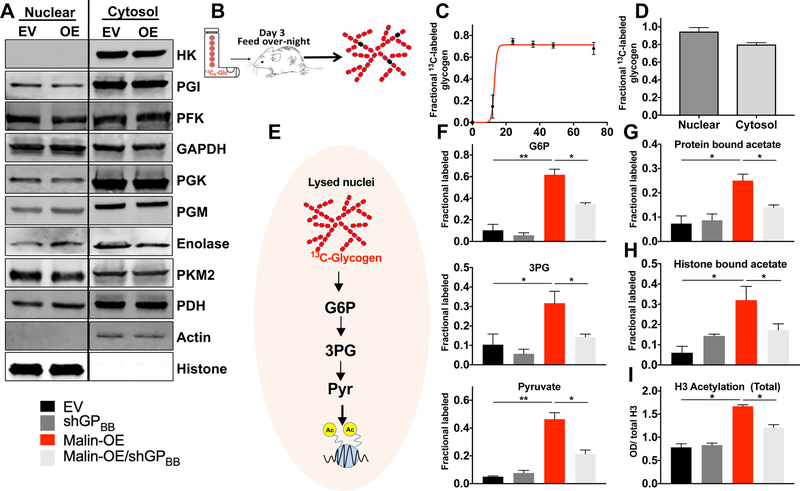
First, we tested whether glycolytic enzymes could perform glycolysis independent from the cytosolic fraction (Figure S4A). With the exception of hexokinase, we detected every glycolytic enzyme in the isolated nuclei, and increased levels of malin did not significantly alter the levels of these enzymes in the nuclear or cytosol fractions (Figure 4A). The intact nuclei were incubated with 13C6-glucose or 13C6-G6P, polar metabolites were isolated and derivatized, and each was analyzed by GCMS. The isolated nuclei metabolized G6P to glycolytic intermediates, including fructose-6-phosphate (F6P), 3-phosphoglycerate (3PG), and pyruvate, but they could not utilize glucose to generate intermediates (Figure S4B). These results are consistent with the production of glycogen by intact nuclei shown in Figure 2.
Figure 4: Glycogen provides a metabolic source for histone acetylation in isolated nuclei.
(A) Nuclei were isolated from A549 cells expressing either empty vector (EV) or malin over-expression (OE). Nuclear and cytosolic fractions were generated and immunoblotted for a panel of glycolytic enzymes. Data are representative of three experiments. HK, hexokinase; PGI, glucose-6-phosphate isomerase; PFK, phosphofructokinase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; PGK, phosphoglycerate kinase; PGM, phosphoglycerate mutase; PKM2, pyruvate kinase M2; PDH, pyruvate dehydrogenase E1 subunit.
(B) Schematic depicting the liquid diet using 13C6-glucose fed to mice to generate 13C-glycogen in the liver. The liver served as a source of labeled glycogen for subsequent experiments.
(C)-(D) 13C-glycogen enrichment in the liver reached steady state after 24-hours of ingesting the 13C-glucose liquid diet. Fractional label of cytosolic and nuclear glycogen isolated from mouse liver 24-hours after the 13C6-glucose liquid diet. Data are representative of mean ± SE from five animals.
(E) Schematic of the in organelle stable tracer experiment. 13C-glycogen was purified from the liver of mice fed a liquid diet enriched in 13C-glucose. This 13C-glycogen was used as substrate to trace its metabolic fate when incubated with the lysate of purified nuclei.
(F) Nuclei were purified from A549, H1299, and H2030 cells with empty vector (EV), GPBB-knockdown (shGPBB), malin overexpressing (OE), and malin-OE with shGPBB. The nuclei were lysed and lysates were incubated with 13C-glycogen. The production of 13C-enriched glucose-6-phosphate (G6P), 3-phosphoglycerate (3PG), and pyruvate generated by each lysate was quantified by GCMS.
(G) Following the experiment described in 4E, nuclei were lysed, proteins were purified, protein bound acetate was released by saponification, and the 13C-enriched acetate was quantified for each cell line.
(H) Following the experiment described in 4E, acid extraction of histone was performed, histone bound acetate was released by saponification, and the 13C-enriched acetate was quantified for each cell line.
(I) Total H3 histone acetylation was quantified for the indicated whole cell lysates. Acetylation was quantified by sandwich ELISA. The key applies to all graphs: EV, nuclei from or cells transfected with empty vector; shGPBB, nuclei from or cells with shGPBB; Malin-OE, nuclei or cells transfected with malin; Malin-OE/shGPBB, nuclei from or cells transfected with malin and shGPBB.
Unless otherwise stated, data are from three experiments and shown as mean ± SE. *0.01 < P < 0.05; ** 0.001 < P < 0.01; *** P < 0.001, two-tailed t-test.
To define the metabolic fate of nuclear glycogen in isolated nuclei, we first generated 13C-labeled glycogen using a liquid diet stable tracer enrichment method (Sun et al., 2017). We fed 8-week-old mice a liquid diet enriched in 13C6-glucose for 48-hours (Figure 4B) and purified liver glycogen by trichloroacetic acid extraction and ethanol precipitation (Bloom et al., 1951). Maximum 13C6-glucose incorporation occurred after 24-hours for both cytosolic and nuclear glycogen in the mouse liver (Figure 4C and D). We used this labeled glycogen as a substrate to quantify the metabolism of nuclei isolated from A549, H1299, and H2030 cells. For each cell line, we generated empty vector (EV) control cells, malin-overexpressing (OE) cells, GPBB knockdown (shGPBB) cells, and GPBB knockdown cells overexpressing malin (malin-OE/ shGPBB). After isolating nuclei in cell lysis buffer, we used a nuclear hypotonic lysis buffer to release nuclear enzymes while preserving enzyme integrity. The nuclear lysates were incubated for 6-hours with the isolated 13C-glycogen in buffer supporting organelle metabolism and polar metabolites and their isotopologues were analyzed by GCMS (Figure 4E). Nuclei from cells overexpressing malin metabolized 13C-glycogen, leading to the enrichment of labeled glycolytic metabolites G6P, 3PG, and pyruvate (Figure 4F). Importantly, the enrichment of labeled glycolytic metabolites by malin overexpression was significantly blunted in cells overexpressing malin that lacked GPBB (Figure 4F).
Pyruvate supplies a significant portion of intracellular acetate required for histone acetylation (Liu et al., 2018). Malin-directed glycogenolysis could provide an additional source for the nuclear pyruvate pool. Nuclei from cells overexpressing malin showed an increase in protein-bound 13C-acetate that was significantly blunted in the absence of GPBB (Figure 4G), suggesting that nuclei can use glycogen as a substrate source for protein acetylation. To confirm that malin overexpression increased histone acetylation in cells, we performed acid extraction of histones and observed increases in histone-bound 13C-acetate enrichment from 13C-glycogen (Figure 4H). We then quantified the amount of H4 and H3 histone acetylation in the cell lines with pan-acetylation H4 and H3 histone antibodies (Figure 4H–I and S6E). Cells overexpressing malin had increased acetylation of H4 and H3 histones, and the increase was ablated in malin-OE/shGPBB cells. In addition to pan-H4 and -H3 acetylation, we also probed the histone acetylation state for four specific H4 and four H3 residues. We observed similar changes in histone acetylation at these eight sites as we observed for total histone acetylation in the malin-overexpressing cells, shGPBB cells, and shGPBB/malin-overexpressing cells (Figure S5A–B). To further validate that malin driven cellular responses is dependent upon GPBB, we tested in vitro cell viability and in vivo xenograft growth of A549 cells with EV, malin-OE, GPBB knockdown (shGPBB), and malin-OE/ shGPBB. While malin overexpression significantly compromised tumor growth, this phenotype was rescued by GPBB knockdown in the malin-OE cell line (Figure S5C–E).
To validate that an increase in histone acetylation is a global event, we measured the relative abundance of an additional 26 histone acetylation residues by triple-quadrupole mass spectrometry. We observed increases in H1.4, H2A1, H2A3, H3, and H4 histone acetylation to further support the hypothesis that nuclear glycogen metabolism modulates histone acetylation (Table S1). Since histone acetylation was affected through malin/GPBB modulation of nuclear glycogen metabolism, we tested whether the malin-OE cell line would exhibit a combinatorial effect with the histone deacetylase inhibitor SAHA. Both A549 and H1299 malin-overexpression cell lines displayed additive effects with SAHA compared to the corresponding EV cells (Figure S5F). Conversely, H2030 malin-OE cells did not show further reduction in cell viability compared to H2030-EV cells. It should be noted that H2030 EV cells were already extremely sensitive to even the lowest SAHA concentrations tested.
Global changes in histone acetylation with malin overexpression suggests a shift in the transcriptional landscape. Therefore, we performed RNAseq analysis to define transcriptional changes after malin overexpression and found that all three cell lines showed similar changes with an average of 1761 genes that were downregulated and 1672 genes that were upregulated more than 1.5-fold (Figure S5G). Genes upregulated in malin-OE versus EV were enriched for pathways involved in smooth muscle contraction, DNA methylation/demethylation, and DNA alkylation (Figure S5H and S5J). Downregulated genes with malin-OE were enriched for chromosome segregation, cell proliferation, and cellular catabolic processes (Figure S5I and S5J). Next, we performed chromatin immunoprecipitation assays with high-throughput DNA sequencing (ChIPseq) using the acetylated-H4 antibody since increases in H4-acetylation were observed in the malin-OE cell line by both ELISA and immunoblotting (Figure S5K). A549 cells overexpressing malin exhibited increases in histone acetylation from nuclear glycogenolysis that directly led to epigenetic changes (Figure S5L). We observed a total of 200 genes/loci with greater than 2-fold changes after malin-OE and 198 of them are upregulated (Table S2), corroborating our findings at the molecular level. Cumulatively, these data support the model that malin controls nuclear glycogenolysis and histone acetylation by promoting the nuclear translocation of GPBB.
Nuclear Glycogenolysis is Involved in Normal Lung Physiology
Organoids are three-dimensional (3D) in vitro culture systems generated from self-assembling isolated epithelial progenitor cells with endothelial support cells (Zhang et al., 2017a). Over the past decade, they have become an indispensable tool to address the differentiation, proliferation, epigenetic regulation, and tumorigenesis of lung biology (Barkauskas et al., 2013; Lee et al., 2014; Rock et al., 2009). The adult mammalian lung contains several distinct epithelial cell populations with unique anatomical positions and specialized functions (Chen et al., 2014). The proximal airway is lined by pseudostratified columnar epithelial cells, in which the basal cells have been identified as important stem cells. Basal cells can be isolated from human and mouse based on their expression of nerve growth factor receptor (NGFR), integrin α6, CD166, and CD44 among other markers (Hegab et al., 2011; Rock et al., 2009). In the more distal lung, airways are lined with columnar epithelium composed of club, goblet, and ciliated cells. In the terminal bronchioles, airways open into the alveolar space, where surfactant-producing alveolar type II (AT2) cells and gas-exchanging alveolar type I (AT1) cells reside. Additionally, bronchioalveolar stem cells (BASCs) are bi-potent stem cells that reside at the bronchioalveolar duct junction and can give rise to both club and AT2 lineages. These 3D culture systems have emerged as an important method of characterizing lung stem cell properties including proliferation, differentiation, and self-renewal (Barkauskas et al., 2013; Lee et al., 2014; Rock et al., 2009).
We isolated BASCs and AT2 cells from wild-type (WT) and malin−/− mice to determine the impact of malin in lung 3D organoid physiology (Figure 5A). Malin−/− BASCs yielded 48% more lung organoids, suggesting increased stem cell function (Figure 5A and 5B). Immunoblotting analysis confirmed the lack of nuclear GPBB in malin−/− AT2 cells (Figure 5C). While WT lung organoids maintained a basal level of nuclear glycogen, malin−/−organoids accumulated 2.5-fold higher nuclear glycogen (Figure 5D), and significantly reduced histone acetylation compared to WT (Figure 5E–5F). We then either overexpressed (OE) malin or performed malin knockdown (KD) using a small-hairpin RNA (shRNA) non-cancerous human lung epithelial BEAS-2B cells and HEK293 cells (Figure 5G–5P). As predicted, malin overexpression resulted in increased nuclear GP while malin knockdown yielded decreased nuclear GP (Figure 5G and 5L). Malin knockdown cells showed increased nuclear glycogen accumulation (Figure 5H and 5M), decreased histone acetylation (Figure 5I, 5J 5N, and 5O), and increased cellular proliferation (Figure 5K and 5P). Further, as a control to confirm ectopic expression of malin is not cytotoxic in non-cancerous cells, we assessed in vitro cell growth of BEAS-2B and HEK293 cells overexpressing malin and did not observe a growth reduction (Figure 5K and 5P). These results demonstrate that malin regulation of nuclear glycogen is involved in normal lung physiology and a disruption in this process results in increased cellular proliferation, a notable phenotype similar to NSCLC.
Figure 5: Malin inhibition drives nuclear glycogen accumulation and growth in the normal lung.
(A) Representative bright field images of WT and malin−/− ex vivo organoids derived from BASCs isolated from WT and malin−/− mouse lungs.
(B) An equal number of WT and malin−/− BASCs were seeded and total colony numbers were quantified. Data are representative of mean ± SE from three separate animals per genotype.
(C) AT2 cells isolated from WT and malin−/− mouse lungs were probed by immunoblotting for malin, GPBB, histones as a nuclear marker, and actin as a cytoplasmic marker.
(D) Quantification of nuclear glycogen in WT and malin−/− BASC organoids.
(E)-(F) Analysis of total histone H4 and H3 acetylation in the indicated organoid cell lysates. Acetylation was quantified by sandwich ELISA.
(G) Nuclear and cytosolic fractions were generated from BEAS-2B cells overexpressing malin (OE), malin knockdown shRNA (shMalin) or empty vector (EV). The lysates were probed by immunoblotting for malin, GPBB, histones as a nuclear marker, and actin as a cytoplasmic marker.
(H) Quantification of nuclear glycogen in BEAS-2B cells: empty vector (EV), malin overexpression (OE), and malin knockdown (KD).
(I)-(J) Analysis of total H4 and H3 histone acetylation in the BEAS-2B cell line lysates as defined in H. Acetylation was quantified by sandwich ELISAs.
(K) Cell growth assay of the same BEAS-2B cell lines as indicated in H quantified by the neutral red assay over 4 days.
(L) Nuclear and cytosolic fractions were generated from HEK293 cells overexpressing malin (OE), malin knockdown shRNA (shMalin) or empty vector (EV). The lysates were probed by immunoblotting for malin, GPBB, histones as a nuclear marker, and actin as a cytoplasmic marker.
(M) Quantification of nuclear glycogen in HEK293 cells: empty vector (EV), malin overexpression (OE), and malin knockdown (KD).
(N)-(O) Analysis of total H4 and H3 histone acetylation from the HEK293 lysates of cell defined in M. Acetylation was quantified by sandwich ELISA.
(P) Cell growth assay of the same HEK293 cell lines as indicated in M quantified by the neutral red assay over 4 days.
Unless otherwise stated, all data are from three experiments and presented as mean ± SE.
*0.01 < P < 0.05; ** 0.001 < P < 0.01; *** P < 0.001, two-tailed t-test.
Re-introduction of Malin Suppresses Lung Cancer Proliferation In Vivo
Increased histone acetylation by the inhibition of histone deacetylases suppresses tumor growth and increases apoptosis in NSCLC cell lines (Bolden et al., 2006; Miyanaga et al., 2008). To test whether increased histone acetylation through activated glycogenolysis has the same effects, we injected A549, H2030, or H1299 control cells and malin-overexpressing cells as subcutaneous xenografts in nude mice and quantified tumor size over time. The overexpression of malin in any of the three NSCLC lines significantly reduced tumor growth when compared to growth of tumors from controls of the same cell line (Figure 6A). After confirming that malin was overexpressed in the isolated tumors (Figure 6B) and that GPBB was present in the nucleus of the malin-overexpressing tumors (Figure 6C), we measured nuclear glycogen (Figure 6D and S7A), H4 acetylation (Figure S6B), and H3 histone acetylation (Figure S6C) in the tumors. Accompanying the reduction in tumor growth, we observed a marked reduction in nuclear glycogen when malin was overexpressed and a concomitant increase in H3 and H4 acetylation for each model. Additionally, we observed a considerable reduced proliferative index, i.e. Ki67, in the malin overexpressing cells (Figure S6D). These data confirm that nuclear glycogen is associated with increased growth rates in NSCLC, which is inhibited by increasing malin abundance.
Figure 6: Malin modulation of nuclear glycogen and histone acetylation occurs in vivo.
(A) Tumor growth from xenografts of the indicated cell lines stably expressing empty vector (EV) or malin overexpression (OE).
(B) Relative abundance of malin in tumor xenograft extracts, defined by average DAB intensity.
(C) Percent of cells with nuclear GPBB in tumor xenografts, defined by DAB positive nuclei.
(D) Quantification of nuclear glycogen in nuclei purified from tumor xenografts. Tumor data are from 10 tumors in 5 mice per cell line.
Data are presented as mean ± SE. *0.01 < P < 0.05; ** 0.001 < P < 0.01; *** P < 0.001: ****P < 0.0001.
See also Figure S6.
DISCUSSION
The role of glycogen in different cancers is an intriguing and quickly evolving area of research. This study demonstrates the role of nuclear glycogen metabolism in providing substrates for histone acetylation and how lung cancer can perturb this pathway to favor cancer proliferation. We have shown that: 1) nuclear glycogen synthesis occurs de novo in the nucleus, 2) nuclear glycogen contributes to a compartmentalized pyruvate pool for histone acetylation, 3) nuclear glycogenolysis is dependent on ubiquitination and translocation of GP by the E3 ubiquitin ligase malin, and 4) lung cancer down-regulates malin-directed nuclear glycogenolysis, which results in nuclear glycogen accumulation. Finally, we validated these findings in multiple mouse xenograft models and in 50 NSCLC patients with matching normal tissue to demonstrate a direct clinical importance. Cumulatively, these data reveal a signaling role for nuclear glycogen that is beyond being a simple energy cache.
Compartmentalized metabolism is an area of intense investigation with rapid scientific progress. This work is being driven by improved isolation methods coupled to highly sensitive technologies and organelle-specific metabolisms are beginning to emerge (Abu-Remaileh et al., 2017; Chen et al., 2016). This study identifies nuclear glycogenolysis as a carbon source for histone acetylation. The pathway involves the E3 ubiquitin ligase malin and the ubiquitination of GP. This process represents a new concept in subcellular organelle communication and signaling by an E3 ligase through regulation of glycogen metabolism. Nuclear glycogenolysis downstream of malin-GP signaling contributes to the pyruvate pool and subsequently histone acetylation. In NSCLC, the downregulation of glycogenolysis due to reduced abundance of malin and the subsequent decrease in nuclear GP results in a lack of substrate for histone acetylation and contributes to the altered epigenetic landscape seen in this cancer.
These data also provide a context to understand emerging themes in cancer biology that connect metabolism with epigenetic regulation through undefined links. Mutations in metabolic enzymes, such as isocitrate dehydrogenase, fumarate hydratase, and succinate dehydrogenase, result in excess production of “oncometabolites,” such as 2-hydroxyglutarate (Lu et al., 2012), fumarate (Toro et al., 2003), and succinate (Xiao et al., 2012), respectively. Studies suggest that oncometabolites contribute to cancer malignancy through inhibition of histone and DNA demethylases. In addition to oncometabolites, pyruvate, acetate, and citrate are donors for histone acetylation, and serine and methionine are donors for histone methylation. Aberrant histone acetylation is a signature of multiple cancers including NSCLC (Komatsu et al., 2006). Indeed, histone deacetylases (HDAC) inhibitors that increase histone acetylation have anti-cancer effects in NSCLC and are in clinical trials (Reid et al., 2004; Vansteenkiste et al., 2008). Mammalian cells can promote histone acetylation through nuclear localization of pyruvate dehydrogenase (Sutendra et al., 2014), ATP citrate lyase (Gao et al., 2016; Wellen et al., 2009), and acetyl-CoA synthase (Bulusu et al., 2017; Mews et al., 2017); these enzymes utilize pyruvate and acetate to supply histone acetylation. While these mechanisms are elegantly described, the origin of the compartmentalized substrates for these enzymes was unknown. Recent work suggests that nuclear and cytosolic acetyl-CoA pools are maintained separately with limited equilibration between them, suggesting compartmentalized pyruvate and acetate production (Bulusu et al., 2017). We provide evidence that nuclear glycogen contributes to the nuclear pool of pyruvate and acetate, and downregulation of nuclear glycogenolysis results in decreased histone acetylation. The decreased histone acetylation leads directly to epigenetic changes within the genome as demonstrate by the H4-Ac ChIPseq results.
To date, aberrant histone acetylation has been attributed to the increased activity of histone deacetylases (HDACs) (Belinsky et al., 2003; Cedar and Bergman, 2009). Reductions in nuclear glycogenolysis or nuclear glycolysis represent additional mechanisms by which cancer can alter histone acetylation. Impairment of this nuclear metabolic program could coordinate synergistically with HDACs to change histone acetylation states and drive tumor proliferation. Indeed, malin overexpression in lung cancer cells exhibits additive effects with the HDAC inhibitor SAHA suggesting that malin-overexpression can improve the anti-proliferative effect of SAHA. Downregulation of malin promotes cellular proliferation in 3D lung organoid models, BEAS-2B cells, and HEK293 cells by preventing nuclear glycogen degradation and subsequent histone acetylation. Re-expression of malin in lung cancer cell lines results in down regulation in the set of genes responsible for chromosome segregation and nuclear division defined by gene set enrichment analysis. Since both processes are important during cell division, these data point towards possible mechanisms responsible for delayed cell growth after malin overexpression. Interestingly, malin overexpression increased gene signatures responsible for DNA methylation/demethylation and DNA alkylation, suggesting a feedforward loop for additional epigenetic regulations. These results support our RNAseq data of both up- and down-regulation of genes. While histone acetylation increases transcriptional activity (Struhl, 1998), additional chromosomal modifications are responsible for the overall transcriptional landscape seen by RNAseq analysis.
This study identifies a molecular mechanism for a more than 70-year-old observation and defines a role for nuclear glycogen beyond a simple energy cache to a substrate source for epigenetic transcriptional regulation by acetylation. Several recent reports suggested the use of GP inhibitors for the treatment of cancer (Curtis et al., 2019; Lee et al., 2004; Schnier et al., 2003). While short term reduction of xenograft is beneficial, our study suggests a further evaluation of GP inhibitors for long term use is needed to avoid unwanted epigenetic regulations. Given the significant reduction in mouse xenograft growth, the nuclear glycogen metabolic pathway represents a novel therapeutic target for NSCLC.
Limitation of The Study
This study employs isolated nuclei and stable isotope tracers to elucidate a previously unidentified nuclear glycogen metabolic pathway that is validated in 2D- and 3D-culture systems as well as xenografts. While this method is extremely powerful at dissecting organelle specific pathways without cytosolic interference, we cannot determine the fractional contributions of metabolites from other sources. For example, nuclear pyruvate production is likely a combination of nuclear glycogen breakdown, transport of cytosolic pyruvate into the nucleus, and acetate from glycolysis or fatty acid oxidation (Bulusu et al., 2017; Gao et al., 2016; Liu et al., 2018). Additionally, these pools are likely to shift subject to environmental stress or malignant transformation. Future studies should focus on developing better biological or informatics tools to elucidate compartmentalized metabolite pool generation, and identify environmental stimuli that alter nuclear metabolism. This study focuses on the role of nuclear glycogen in the lung, other tissues likely participate in nuclear glycogen metabolism as well, and their physiological roles remain to be identified.
STAR METHODS
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Matthew Gentry (matthew.gentry@uky.edu).
Experimental Model and Subject Details
Cell lines and animals
Cell lines H1299, H2030 and A549 were purchased from ATCC and maintained in high-glucose DMEM media supplemented with 10% FBS. BEAS-2B cell line was a gift Dr. Rebecca Dutch. For the generation of stable malin-OE cell lines, cells were infected with lentivirus carrying V5-tagged malin and selected by blasticidin (3µg/ml). V5-tagged malin lentiviral plasmid was purchased from Genecopoeia (EX-Y2418-LV242-B). Stable shGPBB cell lines were generated by transfection with lentivirus purchased from Santa Cruz Biotechnology (sc-105403-V) followed by puromycin selection (5µg/ml). Human tissue samples were obtained from the University of Kentucky Biospecimen Procurement and Translational Pathology Shared Resource Facility. Athymic Foxn1nu/Foxn1nu(nude) mice were purchased from Jackson Laboratory (Bar Harbor, ME). Mice were housed in a climate-controlled environment with a 1410 hours light/dark cycle (lights on at 0600 hours) with water and solid diet (except during tracer administration which received liquid diet, see below) provided ad libitum throughout the study. The institutional animal care and use committee at University of Kentucky has approved all of the animal procedures carried out in this study under PHS Assurance #A3336–01.
Method Details
Western blotting
Whole-cell extracts were generated in RIPA buffer (0.5% deoxycholate, 1% IGEPAL-CA630, 0.1% sodium dodecyl sulfate, 150 mM NaCl, 50 mM Tris-8.1), lysates were cleared by centrifugation and protein concentrations were quantified with the Pierce BCA Protein Assay Kit (Thermo). For Western blotting, 25µg of protein extract per sample was denatured with heat and reducing agents, separated on a 4–12% acrylamide gel (BioRad) and transferred to PVDF (BioRad). Antibodies used for western blotting were malin (Abcam, 1:1,000), acetyl-H3 (EMDmillipore, 1:1,000), pan-methylation (AbCAM, 1:1,000), myc (Abcam, 1:500), branching enzyme (Lsbio, 1:500), glycogen debranching enzyme (LSBio, 1:1,000), glycogen phosphorylase liver isoform (Cell Signaling Technology 4695S 1:2,000), glycogen phosphorylase brain isoform (LSBio, 1:1,000), GAPDH (Abcam 1:1,0000), Histone H3 (Abcam ab1791, 1:20,000), H3K27Ae (Abcam, 1:500), Flag-M2-HRP (Sigma, 1:5000), HSP70 (Abcam, 1:5000) and H3K27me3 (Millipore 07–449; 1:4,000). Each primary antibody was incubated overnight at 4 °C. Tubulin (Abcam, 1:20,000) was used as a loading control. All antibodies have detailed species validation available online from vendors. The secondary antibody anti-rabbit IgG, HRP-linked Antibody (Cell Signaling 7074, 1:5,000) or anti-mouse IgG, HRP-linked antibody (Cell Signaling 7076, 1:5,000) was incubated for 1 hour at room temperature. After washing, chemiluminescence images were acquired using the ChemiDocMP (BioRad). Western analyses were performed a minimum of three independent experiments, as detailed in figure legends, and the results are representative of a single experiment.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde and permeabilized with 10% normal donkey serum (Jackson Immunoresearch) and 0.25% Triton-X (Sigma), both in PBS pH 7.4. Primary antibodies, malin (Abcam) and glycogen phosphorylase liver isoform (LSBio) were incubated overnight at 1:100 dilution in PBS, 10% normal donkey serum. All antibodies have detailed species validation available online from vendors. Slides were washed three times and secondary antibodies, anti-mouse-AlexaFluor594 and anti-rabbit-AlexaFluor488 (Invitrogen) were incubated at 1:500 for 1 hour. After washing, cover slips were mounted using Vectashield with DAPI (Vector Labs). Imaging was performed with a Nikon 90i camera and NIS-Elements software and processed with NIS-Elements and Adobe Photoshop. All treatment groups were imaged with the same exposure time and equivalent processing.
Immunohistochemistry
De-identified human patient tissues were obtained from the University of Kentucky Biospecimen Procurement and Translation Pathology Shared Resource Facility. Cancer and N-distal (normal) tissues were fixed in neutral-buffered 10% formalin (NBF) then paraffin embedded and stored at the facility. Mice were sacrificed by spinal dislocation and tumors were dissected, fixed in 10% NBF. Fixed tumors and tissues were sectioned at 10 µ and immunohistochemistry was performed at the Biospecimen Procurement and Translation Pathology Shared Resource Facility using the method previously described (Zhang et al., 2017b). Antibodies used for other markers are malin (LSBio), Ki67 (Abcam), CC3 (Abcam), and IVS8B6 (glycogen, gift from Dr. Baba (Baba, 1993)). Digital images were acquired through the ZEISS Axio Scan.Z1 high resolution slide scanner. Quantitative image analysis was performed using the Halo software (Indica labs) using the multiplex IHC modules.
Glycogen Purification
Glycogen purification was performed based on the method described by Bloom et al. (Bloom et al., 1951). 10 ml of cold 10% trichloroacetic acid was added to 100 mg of frozen tissue ground to a fine suspension with SamplePrep freezer mill (SPEX) followed by incubation on ice for 30 minutes with periodic gentle mixing. After centrifugation at 300g for 10 minutes, the clear supernatant solution was decanted to a fresh prechilled 50 ml tube and equal volume of cold 95% ethanol was added. The mixture was incubated in the cold room overnight on a rotating platform. The following morning, a white flocculent precipitate (glycogen) was separated by centrifugation (300g for 15 minutes at 4°C) and washed twice with 95% ethanol, the glycogen was dried in a SpeedVac (Thermo), and re-suspended in water.
Glycogen Measurement
Cells were washed twice in PBS pH7.4 and then lysed in 20 mM NaAcetate pH 5.3, 150 mM NaCl, 10% (v/v) glycerol, 1% (v/v) NP-40. Lysates were centrifuged at 10,000 x g at 4 °C for ten minutes. Cleared lysates or purified g lycogen were dissolved in ddH2O, followed by acid hydrolysis with 2N HCL for 3 hours at 98°C. The reaction was quenched with 2N NaOH for subsequent experiments. Glycogen-derived glucose molecules were measured by either biochemical assay or gas chromatography-mass spectrometry (GC-MS). For the biochemical assay, a portion of the digested lysate was incubated for 30 minutes at room temperature with 100 U L−1 of both hexokinase and glucose-6-phosphate dehydrogenase (Sigma) in 50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 0.5 mM ATP, 0.5 mM NAD+. The absorbance was measured at 340 nm using glucose standards to determine glycogen concentration in lysates; undigested lysates were used for background subtraction. For GC-MS analysis, samples were first dried in a SpeedVac (Thermo), followed by sequential addition of 20 mg/ml methoxylamine hydrochloride in pyridine, and then the trimethylsilylating agent N-methyl-N-trimethylsilyl-trifluoroacetamide (MSTFA) was added. GC-MS settings are defined below. Glycogen content was normalized to protein concentration.
Kaplan-Meier Survival Analysis
Kaplan-Meier survival analysis was performed based on the method previously published (Tang et al., 2017). Briefly, RNA sequencing expression and survival data from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) project were used to perform the Kaplan-Meier survival analysis of malin in Lung adenocarcinoma patients. Disease-free survival was used as the end point and GAPDH for normalization. Upper quartile was considered as high expression and lower quartile were considered as low malin expression.
Affinity Purification-Mass Spectrometry
His6-tagged malin was purified using affinity purification with Ni-NTA resin. Lysates of HEK293 cells were subjected to centrifugation at 10,000 x g to remove insoluble debris. Malin was incubated with Ni-NTA-agarose beads either with or without the pre-cleared lysates from HEK293 cells for 2 hours. The samples were washed three times in wash buffer (Tris buffered saline (pH 7.4), 0.5% Triton X-100, and 2 mM dithiothreitol). Bound proteins were removed in the presence of Laemmli’s buffer and heated at 98°C for 10 minutes. Proteins were separated by denatured gel electrophoresis on NuPAGE 10% Bis-Tris gels, gels were stained with Coomassie, bands were excised from gels, digested with trypsin, desalted, and analyzed by MALDI TOF/TOF. The peptide fragments were searched with Protein Pilot software against Swiss-Prot database. The mass spectrometric analysis was performed at the University of Kentucky, Proteomics Core Facility.
Immunoprecipitations
Cells were collected on ice in lysis buffer [10 mM TrisHCl pH 8; 150 mM NaCl, 15 mM EDTA; 0.6 M sucrose, 0.5% nonidet P-40 (NP-40), protease inhibitor cocktail (Roche), 1 mM PMSF, 50 mM NaF and 5 mM Na2P2O7,]. Lysates were cleared by centrifugation at 10,000 x g. For immunoprecipitations in the absence of glycogen, 125 µl of 2 U/ml amyloglucosidase was added to lysates for 2 hrs at 4° C. The supernatants were mixed with anti-myc agarose (Sigma, A7470) or antibody of interest plus protein A-conjugated agarose beads (Invitrogen) and incubated for 2 hrs at 4°C. The beads were pelleted at 1,000 x g for 2 minutes and washed three times in lysis buffer. Samples were then mixed with Laemmli’s buffer and heated at 95°C for 10 minutes. Beads were removed by centrifugation and samples were subjected to Western blot analysis. Western analyses were performed a minimum of three independent experiments, and the results are representative of a single experiment.
In Vitro Ubiquitination
35S-labelled myc-GP was generated using the TNT Coupled transcription/translation System (Promega) per manufacturer’s protocol and probed for quality using gel electrophoresis and phosphorimaging using Typhoon FLA 9500 (GE). In a standard ubiquitin assay reaction, myc-GP was combined with GST-malin (2 µg), Mg-ATP (4 mM), His6-Ub (5 µg), His6-UbE1 (100 nM), GST-UbcH5a (200 nM) in assay buffer (50 mM Tris-HCl pH 7.5, 2.5 mM MgCl2, 0.2 mM DTT and 2 mM ATP) and was incubated for 1 hour at 37°C. myc-GP LL was immunoprecipitated from the reaction mixture using anti-myc agarose beads (Sigma), washed three times in assay buffer, and eluted in Laemmli’s buffer by heating samples at 95°C for 10 minutes. Proteins were separated using gel electrophoresis and probed by an anti-ubiquitin antibody (Biolegend).
13C-Glycogen Labeling in Mice
A liquid diet base containing casein, L-cystine, soy oil, cellulose, mineral mix (AIN-93G-MX), calcium phosphate, vitamin mix (AIN-93-VX), choline bitartrate, tert-butylhydroquinine (TBHQ), and Xanthan gum was purchased from Harlan Laboratories (Madison, WI). Ultrapure grade 13C6-glucose was obtained from Cambridge Isotope Laboratories (Tewkebury, MA). For the 13C-glycogen enrichment, unlabeled glucose and water were added to the diet base two days prior to the tracer study to give a final diet of 0.167 g glucose/g diet and a net protein content of 53 mg/g diet to provide sufficient carbon and nitrogen according to the vendor. 20 g mice were fed 13.6 g liquid diet (at 680 g diet/kg mouse). This pre-feeding of unlabeled liquid diet served to acclimate the mice to the liquid diet feeding. On the third day, 13C6-glucose at 0.173 g/g diet replaced the unlabeled glucose in the diet for each mouse and the mice were allowed to consume the diet ad libitum for 48 hours(Sun et al., 2017). At the end of the feeding period, mice were sacrificed by spinal dislocation and organs were harvested and snap frozen in liquid nitrogen. The frozen liver was pulverized into 10 µm particles in liquid N2 using a SamplePrep freezer mill (SPEX) and glycogen was extracted using TCA and ethanol precipitation via the method previously described(Sun et al., 2017).
Nuclear Purification
Nuclear extracts were prepared according to the method of Schreiber et al and Nagata et al (Nagata et al., 2010; Schreiber et al., 1990). For cell lines, after washing with PBS, 107 cells were suspended in cell lysis buffer (10 mM HEPES pH 7.5, 10 mM KCL, 0.1 mM EDTA, 1 mM DTT, 0.5% NP40 and protease/phosphatase inhibitor cocktail, Cell Signaling) for 20 minutes on ice with intermittent gentle mixing. At the end of incubation, tubes were vortexed, then nuclei were pelleted at 12,000 x g for 10 minutes at 4°C by centrifugation followed by two more washes with the lysis buffer. For tissue samples, frozen tissue was ground to a fine suspension (<10 µm particles) with a SamplePrep freezer mill (SPEX), and 20mg of tissue powder was suspended in tissue lysis buffer (10 mM HEPES pH 7.5, 10 mM KCL, 0.1 mM EDTA, 1 mM DTT, 0.5% NP40, 10mg/ml collagenase IV, Sigma and protease/phosphatase inhibitor cocktail, Cell Signaling) followed by centrifugation. The nuclei pellet was further resuspended in ice cold fractionation buffer (2M sucrose, 1mM MgCl2 and 10mM Tris-HCL pH7.4), mixed well, and then centrifuged at 16000 x g for 30 minutes at 4°C. After removal of supernatant, the ultrapure nuclei were further washed twice with cell lysis buffer, followed by proteinase K treatment for 30min on ice to removal outer nuclear membrane protein. PMSF was added at the end of incubation period then washed twice in cell lysis buffer for downstream processes. To study nuclear glycogen metabolism, pelleted nuclei were washed twice with cell lysis buffer, then resuspended in nuclear extraction buffer (20 mM HEPES, pH 7.5, 400 mM NaCl, 1 mM EDTA, and protease/phosphatase inhibitor cocktail, Cell Signaling) followed by the addition of 13C-glycogen.
Stable Isotope Labelling In Organella
Intact nuclei were resuspended in 500 µl of respiration buffer (125 mM KCL, 2 mM MgCl, 2.5 mM KH2PO4, 20 mM HEPES pH7.2, 1mM ATP, 0.01mM ADP, 1mM NAD+, 0.01mM NADH) supplemented with either 1 mM 13C-glycogen, 1 mM 13C6-glucose and 13C6-glucose-6-phosphate for 1–6 hour at 37 °C with periodic mixing. For lysed nuclei, 400 µl of respiration buffer supplemented with 1 mM 13C-glycogen were added to 100 µl of lysed nuclei and incubated for 3 hours at 37 °C with periodic mixing. At the end of incubation equal parts of pre-chilled 100% methanol was added to the mixture to precipitate out proteins and lipids. The polar fraction was transferred to a V-shaped GC-MS glass vial and dried using a SpeedVac (Thermo) followed by derivatization and GC-MS analysis. Dried polar samples were derivatized by sequential addition of 20 mg/ml methoxylamine hydrochloride in pyridine then the trimethylsilylating agent N-methyl-N-trimethylsilyl-trifluoroacetamide (MSTFA) with brief agitation in between at 80 °C. After cooling, the derivatized mixture analyzed by GC-MS.
Gas Chromatography-Mass Spectrometry (GC-MS) Analysis
GC-MS protocols were similar to those described previously (Jung and Oh, 2015; MacRae et al., 2012), except a modified temperature gradient was used for GC: Initial temperature was 130° C, held for 4 minutes, rising at 6° C/minutes to 243° C, rising at 60° C/minutes to 280° C, held for 2 minutes. The el ectron ionization (EI) energy was set to 70 eV. Scan (m/z:50–800) and selected ion monitoring mode were used for qualitative measurement and isotope monitoring, respectively. Ions used for metabolites that represent the entire carbon backbone are: glucose (554), glucose-6-phosphate (706), fructose-6-phosphate (706), 3PG (387), pyruvate (174) respectively. Batch data processing and natural 13C labeling correction were performed using the Data Extraction for Stable Isotope-labelled metabolites (DEXSI) software package (Dagley and McConville, 2018). Relative abundance for isotopic ions were corrected for recovery using the L-norvaline and adjusted to protein input.
Extraction of Total Acetate from Histones
Following in organello labelling from above, the protein pellet was further purified for histones by re-suspending in 0.4 N sulfuric acid followed by mixing and incubated on a rotating platform overnight. The following morning, the mixture was centrifuged at 16,000 x g for 10 minutes and the supernatant (containing histones) was mixed with TCA and incubated on ice for 30 minutes. Histones were pelleted at 16,000 x g for 10 minutes in a refrigerated centrifuge for acetate extraction. Bound acetate hydrolysis was performed by saponifying the histone pellet overnight by incubation with 200 µL of 10 M sodium hydroxide in a microfuge tube at 95° C. Each sample was then cooled on ice before adding 150 µL of concentrated hydrochloric acid, followed by drying by SpeedVac (Thermo). The dried samples were reconstituted in 200 µL of water and further derivatized as described below.
Chemical Derivatization of Acetate
200 µL of sample was added to a 2 mL microfuge tube, 50 µL of 1-propanol, and 50 µL of pyridine. The tube was then placed on ice for 5 minutes. 100 µL of 1 M sodium hydroxide was then added, immediately followed by 30 µL MCF and vigorous vortexing for 20 seconds. As gas built up in the microfuge tube during the derivatization reaction, the lid was kept closed with one finger and carefully opened after vortexing to relieve pressure (or the lid was kept open during vortexing). After vortexing, 300 µL of MTBE was added, the sample vortexed for another 20 seconds, and centrifuged at 10,000g for 5 minutes. 200 µL microliters of the resulting upper layer was transferred to a GC vial for analysis.
Acetate Quantification by GC-MS
The acetate samples were analyzed with an Agilent 7890B GC system coupled to a 5977B MSD GC-MS system. A DB-75 column was used (30 m × 0.25 mm × 0.25 m). Samples (2 µL) were injected using split mode (0.5 bar, 25 mL/minute split flow). The column gas flow was held at 1.0 mL of He per minutes. The temperature of the inlet was 280° C, the interface temperature 230° C, and the q uadrupole temperature 200° C. The column was equilibrated for 2 minutes before each analysis. The mass spectrometer was operated in scan mode between 2.2 and 2.7 minutes with a mass range of 30– 150 AMU at 1.47 scans/s. MNOVA software were employed for automated data processing using peak heights of m/z 61, 62 and 63 ions used to quantify 12C and 13C, respectively (the peak shapes were consistently highly symmetric, and using either peak area or peak heights gave equivalent results).
In Vitro Cell Viability Assay
For the assessment of cell viability, cells were plated in 96 well plates at a density of 3000 cells per well and 8 wells per group. At the indicated time points, cells were incubated for 3 hours with neutral red (30 g/ml) in fresh media, then washed with PBS, followed by the addition of lysis buffer (acetic acid/methanol, 80%/20%) and the absorbance at 540 nm was recorded. Results are expressed as mean ± S.D, calculations were performed using the Prism software package, ANOVA with Tukey posttest was applied and P < 0.05 was considered to be statistically significant. Experiments were performed at least three times and data presented are of one representative experiment.
Histone Extraction and Preparation for Protein Mass Spectrometry
Bulk histones were acid-extracted from cell pellets, propionylated and subjected to trypsin digestion as described previously (Zheng et al., 2013). Briefly, histones were extracted by incubating samples at room temperature for 1 hour in 0.2M sulfuric acid with intermittent vortexing. Histones were then precipitated by the addition of trichloroacetic acid (TCA) on ice, and recovered by centrifugation at 10,000 x g for 5 minutes at 4°C. The pellet was then washed once wit h 1mL cold acetone/0.1% HCl and twice with 100% acetone, and then air dried in a clean hood. The histones were propionylated by adding 1:3 v/v propionic anhydride/2-propanol and incrementally adding ammonium hydroxide to keep the pH around 8, and subsequently dried in a SpeedVac concentrator. The pellet was then resuspended in 100mM ammonium bicarbonate and adjusted to pH 7–8 with ammonium hydroxide. The histones were then digested with trypsin resuspended in 100mM ammonium bicarbonate overnight at 37°C, and dried in a SpeedVac concentrator. The pellet was resuspended in 100mM ammonium bicarbonate and propionylated a second time by adding 1:3 v/v propionic anhydride/2-propanol and incrementally adding ammonium hydroxide to keep the pH around 8, and subsequently dried in a SpeedVac concentrator. Histone peptides were resuspended in 0.1% TFA in H2O for mass spectrometry analysis.
Mass Spectrometry for Histone Acetylation
Samples were analyzed on a triple quadrupole (QqQ) mass spectrometer (Thermo Fisher Scientific TSQ Quantiva) directly coupled with an UltiMate 3000 Dionex nano-liquid chromatography system. Peptides were first loaded onto an in-house packed trapping column (3cm.150µm) and then separated on a New Objectives PicoChip analytical column (10 cm.75 µm). Both columns were packed with New Objectives ProntoSIL C18-AQ, 3µm, 200Å resin. The chromatography gradient was achi eved by increasing percentage of buffer B from 0 to 35% at a flow rate of 0.30 µl/minute over 45 minutes. Solvent A: 0.1% formic acid in water, and B: 0.1% formic acid in 95% acetonitrile. The QqQ settings were as follows: collision gas pressure of 1.5 mTorr; Q1 peak width of 0.7 (FWHM); cycle time of 2 s; skimmer offset of 10 V; electrospray voltage of 2.5 kV. Targeted analysis of unmodified and various modified histone peptides was performed. This entire process was repeated three separate times for each sample. Raw MS files were imported and analyzed in Skyline with Savitzky-Golay smoothing (MacLean et al., 2010). All Skyline peak area assignments for monitored peptide transitions were manually confirmed. Multiple peptide transitions were quantified for each modification. For each monitored amino acid residue, each modified (and unmodified) form was quantified by first calculating the sum of peak areas of corresponding peptide transitions; the sum of all modified forms was then calculated for each amino acid to represent the total pool of modifications for that residue. Finally, each modification is then represented as a percentage of the total pool of modifications. This process was carried out for each of the three separate mass spec runs, and the raw data provided in the data delivery spreadsheet corresponds to the mean and standard deviation of the resulting three values from this analysis for each modified and unmodified form of the corresponding amino acid residue.
Fluorescence Activated Cell Sorting of Murine Lung and Three-Dimensional Organoid Culture
Mouse cohorts of Malin KO and Malin WT mice (DePaoli-Roach et al., 2010) were all maintained in virus-free conditions on a c57bl/6 background. All care and treatment of experimental animals were in accordance with University of Kentucky Institute institutional animal care and use committee (IACUC) guidelines. Mice were euthanized via intraperitoneal avertin overdose and lungs were injected with 2ml of dispase (Corning). All five lung lobes were minced with scissors and incubated in PBS with 2mg/mL collagenase/dispase (Roche) for 45 minutes. Digested tissue was run through 100- and 40-micron filters. Next, cells were spun down and resuspended in 10% Fetal Bovine Serum in PBS (PF10), and subjected to red blood cell lysis buffer (0.15M NH4Cl, 10mM KHCO3, and 0.1mM Ethylenediaminetetraacetic acid) for 2 minutes. Resulting single cell suspensions were stained using rat-anti-mouse antibodies including anti-mouse-Sca1-APCCy7 (Fisher Scientific BDB560654), anti-mouse-EpCAM-PECy7 (BioLegend 118216), anti-mouse-CD31-APC (Fisher Scientific BDB551262) and anti-mouse-CD45-APC (Fisher Scientific BDB559864). Live cells were gated by exclusion of 4’,6-diamidino-2-phenylindole (DAPI) positive cells (SIGMA). All antibodies were incubated for 10–15 minutes at 1:100 dilutions. Cell sorting was performed with a Sony iCyt with a 100µm nozzle. Sorted BASC (Sca1+/EpCAM+/CD31-/CD45-/DAPI-) cells were seeded at 5000 cells per well with 100,000 low passage murine lung endothelial cells (as feeder cells) embedded in Matrigel (Corning, 50/50 v/v) in transwells with membrane pores of 0.4µm. Organoids that form in the culture have both bronchiolar and alveolar features (Lee et al., 2014; Zhang et al., 2017b). To the bottom chamber, 500µL of DME/F12 with 10% serum, 1x ITS supplement and 1x glutamax was added and refreshed every other day. Organoids grew over a period of 14 days at which point they were quantified and isolated by dispase for further analysis. AT2 (Sca1-/EpCAM+/CD31-/CD45-/DAPI-) cells were collected for immunoblotting analysis.
Xenograft
For in vivo tumor growth, H1299, H2030, and A549 cells were dissociated into single cells, counted, and resuspended at 5 × 106 cells pe r 100 µl of 1:1 media/matrigel (BD). Female 8–10-week-old Foxn1nu/Foxn1nu (Nude) mice (JAX Laboratories) were injected subcutaneously with 5 × 106 cells on both sides of the flanks. Tumor growth was measured twice a week by caliper in a non-blinded fashion. Tumors were allowed to grow to a maximum of 1000 mm3. All mouse experiments were approved by the BCH Animal Care and Use Committee and by the University of Kentucky Institutional Animal Care and Use Committee, both accredited by the Association for Assessment and Accreditation of Laboratory Animal Care and were performed in accordance with relevant institutional and national guidelines and regulations.
RNAseq GSEA analysis
RNA from cell lines were isolated using the RNeasy kit (Qiagen). RNA sequencing was performed using the IIIumina HiSeq 4000 system. The RNAseq data was processed using Cutadapt to trim adapters and STAR to align reads to the reference genome. For the differential expression (DE) analysis, HTSeq was used to count gene expression level, DESeq2 and EdgeR was used to identify differentially expressed genes between empty vector (EV) and malin-overexpression (OE). Gene ontology analysis was carried out by leading edge analysis, gene set enrichment analysis between A549-EV and A549-Malin cells using the biological processes molecular signature database. (GSEA, Broad Institute).
ChIP-Seq Sequencing and Data Processing
H4Ac (pan acetyl) (Active Motif, cat # 39925) ChIP-Seq of A549-EV and malin-OE cell lines has been performed at Active Motif according to proprietary methods. Libraries were sequenced on the Illumina HiSeq 2000 platform, yielding an average of 158 million (±41 million (s.d.)) 101 bp paired-end reads per sample. Genomic alignments were performed against the human reference genome (hg19, NCBI build 37.1) using BWA version 0.5.9-r16 and default parameters. Picard MarkDuplicates (version 1.61, http://picard.sourceforge.net) was used to remove putative PCR duplicates. To make the number of detected peaks comparable between samples, Picard DownsampleSam was applied to equalize the number properly paired reads per sample. Only the first-end was retained and files were converted to the Bed format (Quinlan and Hall, 2010). Peaks were identified using MACS version 1.4.1 using the non-default parameter -- llocal=100000. For downstream analyses, we generated whole-genome coverage tracks (WIG/bigWig files) of non-downstream paired-end reads normalized to all properly paired reads (RPM, paired-end reads/fragments per million). Integrated Genome Browser – version 9.0.2 and the default parameters was used for visualization.
Quantification and Statistical Analysis
Statistical analyses were carried out using GraphPad Prism. All numerical data are presented as mean ± SD except for xenograft tumor growth which is presented as mean ± S.E. Grouped analysis was performed using two-way ANOVA. Column analysis was performed using one-way ANOVA or t-test. A P-value less than 0.05 was considered statistically significant.
Data Availability
The RNA sequencing data that support the findings of this study is available online at the Gene Expression Omnibus (GEO): GSM3891766, National Center for Biotechnology Information (NCBI). All other plasmids and reagents will be available from the corresponding authors based on reasonable request to the University of Kentucky
Supplementary Material
Table S1: Mass spectrometry analysis if histone acetylation. Related to Figure 4.
Table S2: Most changed genes from H4Ac ChIPseq analysis. Related to Figure 4.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Malin | Abcam | ab225919 |
| GPBB | LSbio | LS-B4749 |
| LaminA | Abcam | ab26300 |
| Nesprin-3 | Abcam | ab74261 |
| Anti-Ubiquitin | Abcam | ab134953 |
| EPM2A | Abcam | ab129110 |
| Glycogen Synthase | Cell Signaling | 3886S |
| UAP1 | Abcam | ab155287 |
| GRP78 | Abcam | ab21685 |
| SLC37A4 | Sigma-Aldrich | HPA038939 |
| PGM | Sigma-Aldrich | HPA029760 |
| UGP2 | Sigma-Aldrich | HPA064836 |
| PKM2 | Abcam | ab137791 |
| PFKL | Abcam | ab37583 |
| Glucose 6 phosphate isomerase | Abcam | ab66340 |
| Hexokinase | Abcam | ab150423 |
| GLUT4 | Abcam | ab33780 |
| GLUT1 | Abcam | ab40084 |
| ATP citrate lyase | Abcam | ab40793 |
| hFAB™ Rhodamine Anti-Actin Primary Antibody | Bio-Rad | 12004163 |
| hFAB™ Rhodamine Anti-Tubulin Primary Antibody | Bio-Rad | 12004165 |
| StarBright™ Blue 700 Goat Anti-Rabbit IgG | Bio-Rad | 12004161 |
| StarBright Blue 520 Goat Anti-Mouse IgG | Bio-Rad | 12005866 |
| H4Ac (pan acetyl) | Active Motif | 39925 |
| Histone H3 | Abcam | ab1791 |
| Histone H3ac (pan acetyl) | Active Motif | 39139 |
| Bacterial and Virus Strains | ||
| Malin shRNA (h) Lentiviral Particles | Santa Cruz | sc-106193-V |
| Control shRNA Lentiviral Particles | Santa Cruz | sc-108080 |
| PYGB shRNA (h) Lentiviral Particles | Santa Cruz | sc-105403-V |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SAHA | Abcam | ab144480 |
| Proteinase K | New England Biolabs | P8107S |
| Collagenase | Sigma-Aldrich | C9407-500MG |
| MSTFA + 1 Percent TMCS | Sigma-Aldrich | 375934-10X1ML |
| Methanol | Sigma-Aldrich | 34860-1L-R |
| Leptomycin B | Cell Signaling Technology | 9676S |
| Methoxyamine hydrochloride | Sigma-Aldrich | 226904-25G |
| Pyridine | Thermo Fisher Scientific | TS-27530 |
| Matrigel Matrix Phenol Red Free | VWR | 47743-716 |
| U-13C6-glucose, 99% | Cambridge Isotope Laboratories | CLM-1396-MPT-PK |
| L-Norvaline | Sigma-Aldrich | N7627-5G |
| MG132 | Cayman Chemical Company | 10012628-5 |
| SODIUM PYRUVATE 13C3, 99% | Cambridge Isotope Laboratories | CLM-2440-1 |
| Protease/Phosphatase Inhibitor Cocktail (100X) | Cell Signaling Technology | 5872S |
| Neutral Red Cell Culture Tested | Sigma-Aldrich | N4638-1G |
| 4x Laemmli Sample Buffer | Bio-Rad | 1610747 |
| Precision Plus Protein All Blue Standards | Bio-Rad | 1610373 |
| Mini-PROTEAN® TGX Stain-Free™ Protein Gels | Bio-Rad | 4568024 |
| Fetal Bovine Serum | Sigma-Aldrich | F8067-500ML |
| Fetal Bovine Serum dialyzed | Thermo Fisher Scientific | A3382001 |
| DMEM (HG) With high-glucose, L-glutamine, and sodium pyruvate. | Caisson Labs | DML10-6X1000ML |
| Critical Commercial Assays | ||
| EpiQuik Total Histone H4 Acetylation Detection Kit | Epigentek | P-4032-96 |
| EpiQuik Total Histone Extraction Kit | Epigentek | OP-0006-100 |
| EpiQuik Total Histone H3 Acetylation Detection Fast Kit | Epigentek | P-4030-96 |
| EpiQuik Histone H4 Modification Multiplex Assay Kit | Epigentek | P-3102-96 |
| EpiQuik Histone H3 Modification Multiplex Assay Kit | Epigentek | P-3100-96 |
| Experimental Models: Cell Lines | ||
| A549 | ATCC | ATCC® CCL-185™ |
| H1299 | ATCC | ATCC® CRL-5803™ |
| H2030 | ATCC | ATCC® CRL-5914 |
| Experimental Models: Organisms/Strains | ||
| Athymic nude | The Jackson Laboratory | 002019 |
| Malin−/− | Gentry lab | N/A |
| Recombinant DNA | ||
| ORF expression clone for NHLRC1(NM_198586.2) | GeneCopoeia | EX-Y2418-Lv242-B |
| Empty control vector for pReceiver-Lv242 | GeneCopoeia | EX-NEG-Lv242 |
| Software and Algorithms | ||
Highlights:
De novo synthesized glycogen accumulates in the nucleus of non-small cell lung cancers
Nuclear glycogen provides a carbon pool for histone acetylation
Nuclear glycogenolysis is dependent on translocation of glycogen phosphorylase
Glycogen phosphorylase translocation is regulated by the E3-ubiquin ligase malin
Context and Significance:
Glycogen is the primary source of long-term sugar storage in mammals. Although glycogen is normally found in the cell’s cytoplasm, it has been detected in the nucleus, where our DNA resides. Using state-of-the art labelling techniques, investigators at the University of Kentucky report that non-small cell lung cancer cells (NSCLC) from clinical samples actively make and store glycogen in their nuclei and metabolize it to generate gene-modifying tags that promote cancerous cell growth. They show that nuclear glycogen metabolism is normally kept under check in the lung and that re-establishing the glycogen braking mechanism in tumor models reduces cancer cell growth. Nuclear glycogen metabolic pathways could be new therapeutic targets for NSCLC, a disease which claims 1.6 million deaths yearly.
ACKNOWLEDGMENTS
We would like to thank Dr. Otto Baba for providing the anti-glycogen antibody, Dr. Craig Vander Kooi and Gentry lab members for vigorous discussions regarding the work, Mrs. Dana Napier and Karrie Jones for performing immunohistochemistry on tissue slices and the Markey Cancer Center, University of Kentucky for providing human NSCLC tissues that made this study possible. We also would like to thank Dr. Nancy Gough at Bioserendipity for editing this manuscript.
Funding
This study was supported by National Institute of Health (NIH) grants R01 N070899 and P01 NS097197 to M.S.G., American Cancer Society grants in the form of an institutional research grant #16-182-28 to R.C.S., and a research scholar grant 133123-RSG-19-081-01-TBG to C.F.B.; NCI K22 CA201036 to C.F.B., NCI R01 CA237643 to C.F.B., and American Association for Cancer Research Innovation and Discovery Grant to C.F.B., AHA Great Rivers Affiliate Postdoctoral fellowship (12POST12030381) to V.V.D. This research was also supported by funding from the University of Kentucky Markey Cancer Center, NIH National Center for Advancing Translational Sciences UL1TR001998, the NIH-funded University of Kentucky Center for Cancer and Metabolism P20 GM121327, the Biospecimen Procurement & Translational Pathology Shared Resource Facility of the University of Kentucky Markey Cancer Center P30CA177558.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors declare no competing interests.
REFERENCE
- Abu-Remaileh M, Wyant GA, Kim C, Laqtom NN, Abbasi M, Chan SH, Freinkman E, and Sabatini DM (2017). Lysosomal metabolomics reveals V-ATPase-and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 358, 807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba O (1993). Production of monoclonal antibody that recognizes glycogen and its application for immunohistochemistry. Kokubyo Gakkai Zasshi 60, 264–287. [DOI] [PubMed] [Google Scholar]
- Baird WF, and Fisher ER (1957). Observations concerning vacuolation and deposition of glycogen in nuclei of hepatic cells. Lab Invest 6, 324–333. [PubMed] [Google Scholar]
- Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, and Hogan BLM (2013). Type 2 alveolar cells are stem cells in adult lung. The Journal of Clinical Investigation 123, 3025–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belinsky SA, Klinge DM, Stidley CA, Issa J-P, Herman JG, March TH, and Baylin SB (2003). Inhibition of DNA Methylation and Histone Deacetylation Prevents Murine Lung Cancer. Cancer Research 63, 7089. [PubMed] [Google Scholar]
- Besingi RN, Chaney JL, and Clark PL (2013). An alternative outer membrane secretion mechanism for an autotransporter protein lacking a C-terminal stable core. Mol Microbiol 90, 1028–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom WL, Lewis GT, Schumpert MZ, and Shen TM (1951). Glycogen fractions of liver and muscle. J Biol Chem 188, 631–636. [PubMed] [Google Scholar]
- Bogoch A, Casselman W, Kaplan A, and Bockus H (1955). Studies of hepatic function in diabetes mellitus, portal cirrhosis and other liver diseases: A correlation of clinical, biochemical and liver needle biopsy findings I. Diabetes mellitus. The American journal of medicine 18, 354–384. [DOI] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, and Johnstone RW (2006). Anticancer activities of histone deacetylase inhibitors. Nature Reviews Drug Discovery 5, 769. [DOI] [PubMed] [Google Scholar]
- Bourbon J, and Jost A (1982). Control of glycogen metabolism in the developing fetal lung. Pediatric research 16, 50. [DOI] [PubMed] [Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, and Jemal A (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians 68, 394–424. [DOI] [PubMed] [Google Scholar]
- Brown AM, and Ransom BR (2007). Astrocyte glycogen and brain energy metabolism. Glia 55, 1263–1271. [DOI] [PubMed] [Google Scholar]
- Bulusu V, Tumanov S, Michalopoulou E, van den Broek NJ, MacKay G, Nixon C, Dhayade S, Schug ZT, Vande Voorde J, Blyth K, et al. (2017). Acetate Recapturing by Nuclear Acetyl-CoA Synthetase 2 Prevents Loss of Histone Acetylation during Oxygen and Serum Limitation. Cell Reports 18, 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramia F, Ghergo FG, Branciari C, and Menghini G (1968). New aspect of hepatic nuclear glycogenosis in diabetes. J Clin Pathol 21, 19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardell RR Jr (1977). Smooth endoplasmic reticulum in rat hepatocytes during glycogen deposition and depletion. In International review of cytology (Elsevier; ), pp. 221–279. [DOI] [PubMed] [Google Scholar]
- Cedar H, and Bergman Y (2009). Linking DNA methylation and histone modification: patterns and paradigms. Nature Reviews Genetics 10, 295. [DOI] [PubMed] [Google Scholar]
- Chan EM, Young EJ, Ianzano L, Munteanu I, Zhao X, Christopoulos CC, Avanzini G, Elia M, Ackerley CA, and Jovic NJ (2003). Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nature genetics 35, 125. [DOI] [PubMed] [Google Scholar]
- Chen J, Guccini I, Mitri DD, Brina D, Revandkar A, Sarti M, Pasquini E, Alajati A, Pinton S, Losa M, et al. (2018). Compartmentalized activities of the pyruvate dehydrogenase complex sustain lipogenesis in prostate cancer. Nat Genet 50, 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lee H-J, Wu X, Huo L, Kim S-J, Xu L, Wang Y, He J, Bollu LR, Gao G, et al. (2015). Gain of Glucose-Independent Growth upon Metastasis of Breast Cancer Cells to the Brain. Cancer Research 75, 554–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WW, Freinkman E, Wang T, Birsoy K, and Sabatini DM (2016). Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism. Cell 166, 1324–1337. e1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Fillmore CM, Hammerman PS, Kim CF, and Wong K-K (2014). Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 14, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Zhang M, Gentry MS, Worby CA, Dixon JE, and Saltiel AR (2007). A role for AGL ubiquitination in the glycogen storage disorders of Lafora and Cori’s disease. Genes & development 21, 2399–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipps HD, and Duff GL (1942). Glycogen infiltration of the liver cell nuclei. The American journal of pathology 18, 645. [PMC free article] [PubMed] [Google Scholar]
- Costill DL, Gollnick PD, Jansson ED, Saltin B, and Stein EM (1973). Glycogen Depletion Pattern in Human Muscle Fibres During Distance Running. Acta Physiologica Scandinavica 89, 374–383. [DOI] [PubMed] [Google Scholar]
- Curtis M, Kenny HA, Ashcroft B, Mukherjee A, Johnson A, Zhang Y, Helou Y, Batlle R, Liu X, and Gutierrez N (2019). Fibroblasts mobilize tumor cell glycogen to promote proliferation and metastasis. Cell metabolism 29, 141–155. e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagley MJ, and McConville MJ (2018). DExSI: a new tool for the rapid quantitation of 13C-labelled metabolites detected by GC-MS. Bioinformatics 34, 1957–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Man J, Blok A, and Beens W (1966). Relationship between glycogen and agranular endoplasmic reticulum in rat hepatic cells. Journal of Histochemistry & Cytochemistry 14, 135–146. [DOI] [PubMed] [Google Scholar]
- DePaoli-Roach AA, Tagliabracci VS, Segvich DM, Meyer CM, Irimia JM, and Roach PJ (2010). Genetic depletion of the malin E3 ubiquitin ligase in mice leads to lafora bodies and the accumulation of insoluble laforin. Journal of Biological Chemistry 285, 25372–25381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea G, Ureña J, Graña X, Marsal J, Carreras J, and Climent F (1992). Nuclear location of phosphoglycerate mutase BB isozyme in rat tissues. Histochemistry 97, 269–275. [DOI] [PubMed] [Google Scholar]
- Engelking LR (2015). Chapter 22 - Glucose Trapping. In Textbook of Veterinary Physiological Chemistry (Third Edition). Engelking LR, ed. (Boston: Academic Press; ), pp. 141–146. [Google Scholar]
- Enzo E, Santinon G, Pocaterra A, Aragona M, Bresolin S, Forcato M, Grifoni D, Pession A, Zanconato F, and Guzzo G (2015). Aerobic glycolysis tunes YAP/TAZ transcriptional activity. The EMBO journal 34, 1349–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro E, Bensaad K, Chong Mei G., Tennant Daniel A., Ferguson David J.P., Snell C, Steers G, Turley H, Li J-L, Günther Ulrich L., et al. (2012). Glucose Utilization via Glycogen Phosphorylase Sustains Proliferation and Prevents Premature Senescence in Cancer Cells. Cell Metabolism 16, 751–764. [DOI] [PubMed] [Google Scholar]
- Frank RA, Price AJ, Northrop FD, Perham RN, and Luisi BF (2007). Crystal structure of the E1 component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex. Journal of molecular biology 368, 639–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funasaka T, Yanagawa T, Hogan V, and Raz A (2005). Regulation of phosphoglucose isomerase/autocrine motility factor expression by hypoxia. The FASEB journal 19, 1422–1430. [DOI] [PubMed] [Google Scholar]
- Gao X, Lin S-H, Ren F, Li J-T, Chen J-J, Yao C-B, Yang H-B, Jiang S-X, Yan G-Q, Wang D, et al. (2016). Acetate functions as an epigenetic metabolite to promote lipid synthesis under hypoxia. Nature Communications 7, 11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garyali P, Siwach P, Singh PK, Puri R, Mittal S, Sengupta S, Parihar R, and Ganesh S (2008). The malin–laforin complex suppresses the cellular toxicity of misfolded proteins by promoting their degradation through the ubiquitin–proteasome system. Human molecular genetics 18, 688–700. [DOI] [PubMed] [Google Scholar]
- Gentry MS, Guinovart JJ, Minassian BA, Roach PJ, and Serratosa JM (2018). Lafora disease offers a unique window into neuronal glycogen metabolism. Journal of Biological Chemistry 293, 7117–7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry MS, Worby CA, and Dixon JE (2005). Insights into Lafora disease: Malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Proceedings of the National Academy of Sciences of the United States of America 102, 8501–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb RP, and Stowell RE (1949). Glycogen in human blood cells. Blood 4, 569–579. [PubMed] [Google Scholar]
- Gots RE, Gorin FA, and Bessman SP (1972). Kinetic enhancement of bound hexokinase activity by mitochondrial respiration. Biochemical and biophysical research communications 49, 1249–1255. [DOI] [PubMed] [Google Scholar]
- Granzow C, Kopun M, and Zimmermann HP (1981). Role of nuclear glycogen synthase and cytoplasmic UDP glucose pyrophosphorylase in the biosynthesis of nuclear glycogen in HD33 Ehrlich-Lettré ascites tumor cells. The Journal of Cell Biology 89, 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, and Hester LD (2005). S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nature cell biology 7, 665. [DOI] [PubMed] [Google Scholar]
- Hegab AE, Ha VL, Gilbert JL, Zhang KX, Malkoski SP, Chon AT, Darmawan DO, Bisht B, Ooi AT, Pellegrini M, et al. (2011). Novel Stem/Progenitor Cell Population from Murine Tracheal Submucosal Gland Ducts with Multipotent Regenerative Potential. Stem Cells (Dayton, Ohio) 29, 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himes M, Pollister A, and Moore B (1956). The normal occurrence of hepatic intranuclear glycogen in larval and metamorphic stages of rana-pipiens. Journal of histochemistry & cytochemistry 4, 433–434. [Google Scholar]
- Hultman E, and Nilsson LH (1971). Liver Glycogen in Man. Effect of Different Diets and Muscular Exercise. In Muscle Metabolism During Exercise: Proceedings of a Karolinska Institutet Symposium held in Stockholm, Sweden, September 6–9, 1970 Honorary guest: E Hohwü Christensen Pernow B, and Saltin B, eds. (Boston, MA: Springer US; ), pp. 143–151. [Google Scholar]
- Iida Y, Aoki K, Asakura T, Ueda K, Yanaihara N, Takakura S, Yamada K, Okamoto A, Tanaka T, and Ohkawa K (2012). Hypoxia promotes glycogen synthesis and accumulation in human ovarian clear cell carcinoma. Int J Oncol 40, 2122–2130. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, and Pei YF (1965). Intramitochondrial glycogen particles in rat retinal receptor cells. The Journal of cell biology 25, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J-Y, and Oh M-K (2015). Isotope labeling pattern study of central carbon metabolites using GC/MS. Journal of Chromatography B 974, 101–108. [DOI] [PubMed] [Google Scholar]
- Kerr EM, Gaude E, Turrell FK, Frezza C, and Martins CP (2016). Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature 531, 110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M-S, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, et al. (2014). A draft map of the human proteome. Nature 509, 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu N, Kawamata N, Takeuchi S, Yin D, Chien W, Miller CW, and Koeffler HP (2006). SAHA, a HDAC inhibitor, has profound anti-growth activity against non-small cell lung cancer cells. Oncology reports 15, 187–191. [PubMed] [Google Scholar]
- Kottakis F, Nicolay BN, Roumane A, Karnik R, Gu H, Nagle JM, Boukhali M, Hayward MC, Li YY, Chen T, et al. (2016). LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature 539, 390–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs HA, Bennett DAH, De Gasquet P, Gascoyne T, and Yoshida T (1963). Renal gluconeogenesis. The effect of diet on the gluconeogenic capacity of rat-kidney-cortex slices. Biochemical Journal 86, 22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, Horinouchi S, and Yoshida M (1998). Leptomycin B Inhibition of Signal-Mediated Nuclear Export by Direct Binding to CRM1. Experimental Cell Research 242, 540–547. [DOI] [PubMed] [Google Scholar]
- Lee J-H, Bhang Dong H., Beede A, Huang Tian L., Stripp Barry R., Bloch Kenneth D., Wagers Amy J., Tseng Y-H, Ryeom S, and Kim Carla F. (2014). Lung Stem Cell Differentiation in Mice Directed by Endothelial Cells via a BMP4-NFATc1-Thrombospondin-1 Axis. Cell 156, 440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WP, Guo P, Lim S, Bassilian S, Lee S, Boren J, Cascante M, Go V, and Boros L (2004). Metabolic sensitivity of pancreatic tumour cell apoptosis to glycogen phosphorylase inhibitor treatment. British journal of cancer 91, 2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn TC, Pettit FH, and Reed LJ (1969). α-Keto acid dehydrogenase complexes, X. Regulation of the activity of the pyruvate dehydrogenase complex from beef kidney mitochondria by phosphorylation and dephosphorylation. Proceedings of the National Academy of Sciences 62, 234–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Gao L, Zhang H, Wang D, Wang M, Zhu J, Pang C, and Wang C (2013). Succinate Dehydrogenase 5 (SDH5) Regulates Glycogen Synthase Kinase 3β-β-Catenin-mediated Lung Cancer Metastasis. Journal of Biological Chemistry 288, 29965–29973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Cooper DE, Cluntun AA, Warmoes MO, Zhao S, Reid MA, Liu J, Wellen KE, Kirsch DG, and Locasale JW (2018). Acetate is generated de novo from glucose metabolism in mammals and is coupled to central carbon metabolism. bioRxiv
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. (2012). IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483, 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, and MacCoss MJ (2010). Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae James I., Sheiner L, Nahid A, Tonkin C, Striepen B, and McConville Malcolm J. (2012). Mitochondrial Metabolism of Glucose and Glutamine Is Required for Intracellular Growth of Toxoplasma gondii. Cell Host & Microbe 12, 682–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Adachi J, Ihara M, Tanuma N, Shima H, Kakizuka A, Ikura M, Ikura T, and Matsuda T (2015). Nuclear pyruvate kinase M2 complex serves as a transcriptional coactivator of arylhydrocarbon receptor. Nucleic acids research 44, 636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mews P, Donahue G, Drake AM, Luczak V, Abel T, and Berger SL (2017). Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature 546, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanaga A, Gemma A, Noro R, Kataoka K, Matsuda K, Nara M, Okano T, Seike M, Yoshimura A, Kawakami A, et al. (2008). Antitumor activity of histone deacetylase inhibitors in non-small cell lung cancer cells: development of a molecular predictive model. Molecular Cancer Therapeutics 7, 1923. [DOI] [PubMed] [Google Scholar]
- Mori M, DEMPO K, ABE M, and ONOE T (1970). Electron microscopic study of intranuclear glycogen. Microscopy 19, 163–169. [PubMed] [Google Scholar]
- Nagata T, Redman RS, and Lakshman R (2010). Isolation of intact nuclei of high purity from mouse liver. Analytical biochemistry 398, 178–184. [DOI] [PubMed] [Google Scholar]
- Nielsen J, Suetta C, Hvid LG, Schrøder HD, Aagaard P, and Ørtenblad N (2010). Subcellular localization dependent decrements in skeletal muscle glycogen and mitochondria content following short-term disuse in young and old men. American Journal of Physiology-Heart and Circulatory Physiology [DOI] [PubMed]
- Nitschke F, Ahonen SJ, Nitschke S, Mitra S, and Minassian BA (2018). Lafora disease—from pathogenesis to treatment strategies. Nature Reviews Neurology, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oe Y, Baba O, Ashida H, Nakamura KC, and Hirase H (2016). Glycogen distribution in the microwave-fixed mouse brain reveals heterogeneous astrocytic patterns. Glia 64, 1532–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pescador N, Villar D, Cifuentes D, Garcia-Rocha M, Ortiz-Barahona A, Vazquez S, Ordonez A, Cuevas Y, Saez-Morales D, Garcia-Bermejo ML, et al. (2010). Hypoxia promotes glycogen accumulation through hypoxia inducible factor (HIF)-mediated induction of glycogen synthase 1. PLoS One 5, e9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid T, Valone F, Lipera W, Irwin D, Paroly W, Natale R, Sreedharan S, Keer H, Lum B, and Scappaticci F (2004). Phase II trial of the histone deacetylase inhibitor pivaloyloxymethyl butyrate (Pivanex, AN-9) in advanced non-small cell lung cancer. Lung cancer 45, 381–386. [DOI] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, and Hogan BLM (2009). Basal cells as stem cells of the mouse trachea and human airway epithelium. Proceedings of the National Academy of Sciences 106, 12771–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romá-Mateo C, Sanz P, and Gentry MS (2012). Deciphering the role of malin in the Lafora progressive myoclonus epilepsy. IUBMB life 64, 801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose IA, O’Connell EL, Litwin S, and Tana JB (1974). Determination of the rate of hexokinase-glucose dissociation by the isotope-trapping method. Journal of Biological Chemistry 249, 5163–5168. [PubMed] [Google Scholar]
- Rousset M, Chevalier G, Rousset J-P, Dussaulx E, and Zweibaum A (1979). Presence and Cell Growth-related Variations of Glycogen in Human Colorectal Adenocarcinoma Cell Lines in Culture. Cancer Research 39, 531–534. [PubMed] [Google Scholar]
- Rousset M, Zweibaum A, and Fogh J (1981). Presence of Glycogen and Growth-related Variations in 58 Cultured Human Tumor Cell Lines of Various Tissue Origins. Cancer Research 41, 1165–1170. [PubMed] [Google Scholar]
- Sáez DE, and Slebe JC (2000). Subcellular loc alization of aldolase B. Journal of cellular biochemistry 78, 62–72. [DOI] [PubMed] [Google Scholar]
- Sato A, Kawasaki T, Kashiwaba M, Ishida K, Nagashima Y, Moritani S, Ichihara S, and Sugai T (2015). Glycogen-rich clear cell carcinoma of the breast showing carcinomatous lymphangiosis and extremely aggressive clinical behavior. Pathol Int 65, 674–676. [DOI] [PubMed] [Google Scholar]
- Saurí A, Oreshkova N, Soprova Z, Jong WS, Sani M, Peters PJ, Luirink J, and van Ulsen P (2011). Autotransporter β-domains have a specific function in protein secretion beyond outer-membrane targeting. Journal of molecular biology 412, 553–567. [DOI] [PubMed] [Google Scholar]
- Schnier JB, Nishi K, Monks A, Gorin FA, and Bradbury EM (2003). Inhibition of glycogen phosphorylase (GP) by CP-91,149 induces growth inhibition correlating with brain GP expression. Biochemical and biophysical research communications 309, 126–134. [DOI] [PubMed] [Google Scholar]
- Schreiber E, Harshman K, Kemler I, Malipiero U, Schaffner W, and Fontana A (1990). Astrocytes and glioblastoma cells express novel octamer-DNA binding proteins distinct from the ubiquitous Oct-1 and B cell type Oct-2 proteins. Nucleic Acids Research 18, 5495–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkrig J, Mosbahi K, Webb CT, Belousoff MJ, Perry AJ, Wells TJ, Morris F, Leyton DL, Totsika M, and Phan M-D (2012). Discovery of an archetypal protein transport system in bacterial outer membranes. Nature structural & molecular biology 19, 506. [DOI] [PubMed] [Google Scholar]
- Shackelford DB, and Shaw RJ (2009). The LKB1-AMPK pathway: metabolism and growth control in tumor suppression. Nature reviews. Cancer 9, 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solaz-Fuster MC, Gimeno-Alcaniz JV, Ros S, Fernandez-Sanchez ME, Garcia-Fojeda B, Criado Garcia O, Vilchez D, Dominguez J, Garcia-Rocha M, Sanchez-Piris M, et al. (2008). Regulation of glycogen synthesis by the laforin-malin complex is modulated by the AMP-activated protein kinase pathway. Hum Mol Genet 17, 667–678. [DOI] [PubMed] [Google Scholar]
- Staedel C, and Beck J-P (1978). Resurgence of glycogen synthesis and storage capacity in cultured hepatoma cells. Cell Differentiation 7, 61–71. [DOI] [PubMed] [Google Scholar]
- Struhl K (1998). Histone acetylation and transcriptional regulatory mechanisms. Genes & development 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Sun L, and Chen ZJ (2004). The novel functions of ubiquitination in signaling. Current Opinion in Cell Biology 16, 119–126. [DOI] [PubMed] [Google Scholar]
- Sun RC, Fan TWM, Deng P, Higashi RM, Lane AN, Le A-T, Scott TL, Sun Q, Warmoes MO, and Yang Y (2017). Noninvasive liquid diet delivery of stable isotopes into mouse models for deep metabolic network tracing. Nature Communications 8, 1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson Trevor H., Haromy A, Hashimoto K, Zhang N, Flaim E, and Michelakis Evangelos D. (2014). A Nuclear Pyruvate Dehydrogenase Complex Is Important for the Generation of Acetyl-CoA and Histone Acetylation. Cell 158, 84–97. [DOI] [PubMed] [Google Scholar]
- Tang Z, Li C, Kang B, Gao G, Li C, and Zhang Z (2017). GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Research 45, W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro JR, Nickerson ML, Wei M-H, Warren MB, Glenn GM, Turner ML, Stewart L, Duray P, Tourre O, Sharma N, et al. (2003). Mutations in the Fumarate Hydratase Gene Cause Hereditary Leiomyomatosis and Renal Cell Cancer in Families in North America. American Journal of Human Genetics 73, 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull J, Tiberia E, Striano P, Genton P, Carpenter S, Ackerley CA, and Minassian BA (2016). Lafora disease. Epileptic Disorders 18, S38–S62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vansteenkiste J, Van Cutsem E, Dumez H, Chen C, Ricker JL, Randolph SS, and Schöffski P (2008). Early phase II trial of oral vorinostat in relapsed or refractory breast, colorectal, or non-small cell lung cancer. Investigational new drugs 26, 483–488. [DOI] [PubMed] [Google Scholar]
- Verhalen B, Arnold S, and Minassian BA (2018). Lafora Disease: A Review of Molecular Mechanisms and Pathology. Neuropediatrics [DOI] [PMC free article] [PubMed]
- Vilchez D, Ros S, Cifuentes D, Pujadas L, Valles J, Garcia-Fojeda B, Criado-Garcia O, Fernandez-Sanchez E, Medrano-Fernandez I, Dominguez J, et al. (2007). Mechanism suppressing glycogen synthesis in neurons and its demise in progressive myoclonus epilepsy. Nat Neurosci 10, 1407–1413. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, and Thompson CB (2009). ATP-Citrate Lyase Links Cellular Metabolism to Histone Acetylation. Science 324, 1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby CA, Gentry MS, and Dixon JE (2008). Malin decreases glycogen accumulation by promoting the degradation of protein targeting to glycogen (PTG). The Journal of biological chemistry 283, 4069–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, et al. (2012). Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev 26, 1326–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau R, and Rape M (2016). The increasing complexity of the ubiquitin code. Nature Cell Biology 18, 579. [DOI] [PubMed] [Google Scholar]
- Ying H, Kimmelman Alec C., Lyssiotis Costas A., Hua S, Chu Gerald C., Fletcher-Sananikone E, Locasale Jason W., Son J, Zhang H, Coloff Jonathan L., et al. (2012). Oncogenic Kras Maintains Pancreatic Tumors through Regulation of Anabolic Glucose Metabolism. Cell 149, 656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Brainson CF, Koyama S, Redig AJ, Chen T, Li S, Gupta M, Garcia-de-Alba C, Paschini M, and Herter-Sprie GS (2017a). Lkb1 inactivation drives lung cancer lineage switching governed by Polycomb Repressive Complex 2. Nature communications 8, 14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Fillmore Brainson C, Koyama S, Redig AJ, Chen T, Li S, Gupta M, Garcia-de-Alba C, Paschini M, Herter-Sprie GS, et al. (2017b). Lkb1 inactivation drives lung cancer lineage switching governed by Polycomb Repressive Complex 2. Nat Commun 8, 14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Thomas PM, and Kelleher NL (2013). Measurement of acetylation turnover at distinct lysines in human histones identifies long-lived acetylation sites. Nature communications 4, 2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Kinslow CJ, Hibshoosh H, Guo H, Cheng SK, He C, Gentry MS, and Sun RC (2019). Clinical Features, Survival and Prognostic Factors of Glycogen-Rich Clear Cell Carcinoma (GRCC) of the Breast in the US Population. Journal of clinical medicine 8, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann H-P, Granzow V, and Granzow C (1976). Nuclear glycogen synthesis in ehrlich ascites cells. Journal of Ultrastructure Research 54, 115–123. [DOI] [PubMed] [Google Scholar]
- Zois CE, and Harris AL (2016). Glycogen metabolism has a key role in the cancer microenvironment and provides new targets for cancer therapy. Journal of Molecular Medicine 94, 137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Mass spectrometry analysis if histone acetylation. Related to Figure 4.
Table S2: Most changed genes from H4Ac ChIPseq analysis. Related to Figure 4.
Data Availability Statement
The RNA sequencing data that support the findings of this study is available online at the Gene Expression Omnibus (GEO): GSM3891766, National Center for Biotechnology Information (NCBI). All other plasmids and reagents will be available from the corresponding authors based on reasonable request to the University of Kentucky