Highlights
-
•
Individuals with ANGPTL3 loss-of-function mutations have reduced cholesterol levels, triglyceride levels, and risk of coronary heart disease, making ANGPTL3 a potential therapeutic target.
-
•
An antisense oligonucleotide inhibitor of ANGPTL3 and a monoclonal antibody against ANGPTL3 have been advanced into clinical trials, with encouraging results to date.
-
•
A distinct approach to targeting ANGPTL3 would be therapeutic gene editing in patients to induce permanent loss of function mutations mimicking those in individuals with naturally occurring cardioprotective mutations.
Key Words: coronary artery disease, genetics, lipids
Abbreviations and Acronyms: ANGPTL3, angiopoietin-like 3; ASO, antisense oligonucleotide; CHD, coronary heart disease; HDL-C, high-density lipoprotein cholesterol; hoFH, homozygous familial hypercholesterolemia; LDL-C, low-density lipoprotein cholesterol; PCSK9, proprotein convertase subtilisin/kexin type 9
Summary
Hyperlipidemia is a major causal risk factor for atherosclerosis and coronary heart disease (CHD). Angiopoietin-like 3 (ANGPTL3) has emerged as a promising molecular target to reduce CHD risk due to its regulation of all 3 major lipid traits: low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, and triglycerides. Here, the authors review the discovery of ANGPTL3, the role of ANGPTL3 in lipoprotein metabolism, and the genetic association between naturally occurring ANGPTL3 loss-of-function mutations and CHD. In light of the favorable consequences of ANGPTL3 deficiency, various therapeutic strategies to target ANGPTL3 are currently in development, including a monoclonal antibody, an antisense oligonucleotide, and gene editing.
Central Illustration
Over the last few decades, tremendous progress has been made in understanding and decreasing the incidence of coronary heart disease (CHD). Low-density lipoprotein cholesterol (LDL-C) has been established as a major causal risk factor for atherosclerosis and CHD. Remarkable efforts have been made to develop LDL-lowering therapies, and statins have been proven to be an effective means of reducing the risk of CHD. However, even with the use of statin therapy, there remains a large residual risk of CHD, particularly in patients with familial hypercholesterolemia (1). In light of these observations, an intensive search for new molecular targets to further reduce CHD risk is ongoing. In 2015, the efficacy of alirocumab and evolocumab, 2 human monoclonal antibody-based drugs that target proprotein convertase subtilisin/kexin type 9 (PCSK9), was confirmed with respect to the reduction of LDL-C levels in patients 2, 3.
Despite advances in the development of lipid-lowering therapies, clinical trials have shown that a substantial risk of cardiovascular disease persists even when receiving currently recommended medical therapy. For example, in 1 trial involving patients who had an acute coronary syndrome, the lowering of LDL-C levels to a median of 54 mg/dl with the use of a statin plus ezetimibe was found to prevent only a slightly higher proportion of events than treatment with a statin alone; the difference in absolute risk associated with the 2 regimens was only 2 proportion points, and approximately one-third of both sets of patients had a major cardiovascular event within 7 years (4). A similarly small reduction in the absolute risk of coronary events was found among patients whose levels of LDL-C were reduced to 30 mg/dl with the use of a PCSK9 inhibitor (5). In addition, researchers have continued to search for additional molecular targets that are particularly relevant for the treatment of patients with homozygous familial hypercholesterolemia (hoFH) who have genetic deficiency of the LDL receptor. Since the efficacy of both statins and PCSK9 antibody therapies largely depends on functional LDL receptors, patients with hoFH show limited responses to both therapies.
One such promising molecular target is angiopoietin-like 3 (ANGPTL3). ANGPTL3 belongs to a subfamily of angiopoietin-like proteins involved in the regulation of plasma lipid metabolism. ANGPTL3 is secreted from liver and was first identified via positional cloning of a hypolipidemic mouse strain (6). ANGPTL3 is unique in that it regulates all 3 major lipid traits: LDL-C, high-density lipoprotein cholesterol (HDL-C), and triglycerides. The primary action of ANGPTL3 is to inhibit lipoprotein lipase (LPL), which hydrolyzes the triglycerides carried in triglyceride-rich lipoproteins in the circulation (7). ANGPTL3 also inhibits endothelial lipase to modulate HDL-C metabolism (8). The mechanism by which ANGPTL3 regulates LDL-C remains unclear (9).
Discovery of ANGPTL3
The Angptl3 gene was identified by positional cloning of the mutation responsible for the hypolipidemia phenotype in KK/San mice, a sub-strain of KK mice that have moderate obesity, abnormally high levels of plasma insulin (hyperinsulinemia), glucose (hyperglycemia), and lipids (hyperlipidemia). There is a 4-bp insertion in exon 6 of Angptl3 in KK/San mice, introducing a premature stop codon. KK/San mice have lower levels of triglycerides, total cholesterol, and non-esterified fatty acids in the circulation compared to KK mice. Using adenovirus expressing human or mouse ANGPTL3 rescued the lower levels of triglycerides, total cholesterol, and non-esterified fatty acids in KK/San mice. The same treatment also increased triglycerides and total cholesterol levels in wild-type (WT) C57BL/6 mice (6). These data proved that disruption of Angptl3 is responsible for the hypolipidemia in KK/San mice and that ANGPTL3 regulates circulating triglycerides and total cholesterol levels in mouse.
Angiopoietin-like proteins
ANGPTL3 belongs to a family of 8 angiopoietin-like proteins (ANGPTL1 to ANGPTL8) that share a similar structure and carry out related functions. Seven of 8 angiopoietin-like proteins (ANGPTL1 to ANGPTL7) contain a signal peptide, an N-terminal coiled-coil domain, a linker region, and a C-terminal fibrinogen-like domain (FLD) (10). ANGPTL8 differs from the other angiopoietin-like proteins in that it lacks a C-terminal FLD (11). Similar to other angiopoietin-like proteins, ANGPTL3 undergoes cleavage; the cleavage site is at amino acid residues 221-Arg-Ala-Pro-Arg-224, which yields separate fragments containing the coiled-coil domain and the FLD (12). ANGPTL3 is found in the plasma as full-length and truncated forms. ANGPTL3 is cleaved intracellularly by furin (also known as PCSK3) and extracellularly mainly by PACE4 (also known as PCSK6) (13). The truncated form of ANGPTL3 is more active, and cleavage enhances the ability of ANGPTL3 to inhibit lipoprotein lipase and regulate plasma levels of triglycerides both in vitro (11) and in vivo (12).
Similar to ANGPTL3, another 2 members of the angiopoietin-like protein family—ANGPTL4 and ANGPTL8—are involved in the regulation of plasma lipid metabolism. ANGPTL4 is highly expressed in the liver and adipose tissue and upregulated by fasting and hypoxia 14, 15. ANGPTL4 forms dimers and tetramers before secretion and undergoes cleavage at a canonical proprotein convertase cleavage site, 161-Arg-Arg-Lys-Arg-164, after secretion (16). The N-terminal fragment remains oligomerized after cleavage, binds transiently to LPL, and converts LPL from catalytically active dimers to inactive monomers to decrease its activity (17). Angptl4 knockout mice have lower triglyceride levels and modestly lower cholesterol levels (18). However, when Angptl4 knockout mice were fed a high-fat diet, they showed reduced viability associated with lipogranulomatous lesions, which raises a significant safety concern with respect to the proposed targeting of ANGPTL4 for the treatment of dyslipidemia and atherosclerosis 18, 19.
ANGPTL8 is an atypical member of the angiopoietin-like protein family because of its lack of a C-terminal FLD, but it does share structural homology with the N-terminal domains of ANGPTL3 and ANGPTL4 20, 21, and it can inhibit LPL and thereby regulate triglyceride metabolism (11).
ANGPTL3 genetics and plasma lipids
Genome-wide association studies and exome sequencing studies have identified associations between loss-of-function genetic variants in the ANGPTL3 gene and low levels of plasma LDL-C, HDL-C, and triglycerides 22, 23. The ANGPTL3 coding regions were sequenced in 3,551 individuals in the Dallas Heart Study, and a total of 35 nonsynonymous sequence variations (nonsense, missense, frameshift, and splice-site mutations) were identified. An excess of sequence variants in the lowest quartile for plasma triglyceride levels (14 vs. 5 variants) approached the nominal significance threshold (p = 0.06). In vitro functional studies revealed that all ANGPTL3 missense variants that were associated with low plasma triglyceride levels interfered either with the synthesis or secretion of the protein or with the ability of the ANGPTL3 protein to inhibit LPL activity (24).
Exome sequencing of 2 siblings with combined hypolipidemia, characterized by extremely low plasma levels of LDL-C, HDL-C, and triglycerides, led to the identification of 2 loss-of-function variants in ANGPTL3 as the cause. The siblings were compound heterozygotes for 2 distinct nonsense mutations (S17X and E129X) (23). Since the publication of this study, additional mutations in ANGPTL3 have been identified in combined hypolipidemia subjects without APOB gene mutations 25, 26, 27.
ANGPTL3 and coronary heart disease
Numerous animal models, such as inactivation of ANGPTL3 by an antibody (9), inhibition by an antisense oligonucleotide (ASO) (28), and genetic knockout of Angptl3 (29), support that ANGPTL3 regulates plasma lipid levels. In humans, individuals bearing 2 nonsense mutations in ANGPTL3 have familial combined hypolipidemia. The individuals with complete ANGPTL3 deficiency showed no evidence of coronary atherosclerotic plaque (30). Predicted loss-of-function (LOF) variants (nonsense, frameshift, and splice-site) in ANGPTL3 are rare in the general population, found in only ∼1 in 300 individuals in the United States, limiting the power of association analyses seeking to link LOF ANGPTL3 variants to coronary artery disease risk (30). Accordingly, a mouse model was used to functionally classify ANGPTL3 missense variants as LOF versus neutral. WT human ANGPTL3 and each ANGPTL3 missense variant were reconstituted in the livers of Angptl3 knockout mice by adenoviral expression. Missense variants were defined as LOF if they conferred <25% of WT activity as assessed by percent change in circulating triglyceride and cholesterol levels induced by heterologous gene expression. Eleven rare missense variants predicted to be damaging by 5 in silico prediction algorithms underwent functional validation in a mouse model, of which only 2 (p.Asp42Asn and p.Thr383Ser) were functionally validated as LOF. With the inclusion of nonsense, frameshift, and splice-site mutations along with the 2 LOF missense variants, there was a 34% lower risk of CHD among carriers of an ANGPTL3 LOF mutation compared with non-carriers in a meta-analysis of 21,980 patients with CHD and 158,200 control individuals (30). Subsequently, a different group reported that individuals carrying LOF variants in ANGPTL3 had a 41% lower risk of CHD than non-carriers (31).
ANGPTL3 and lipid-lowering therapy
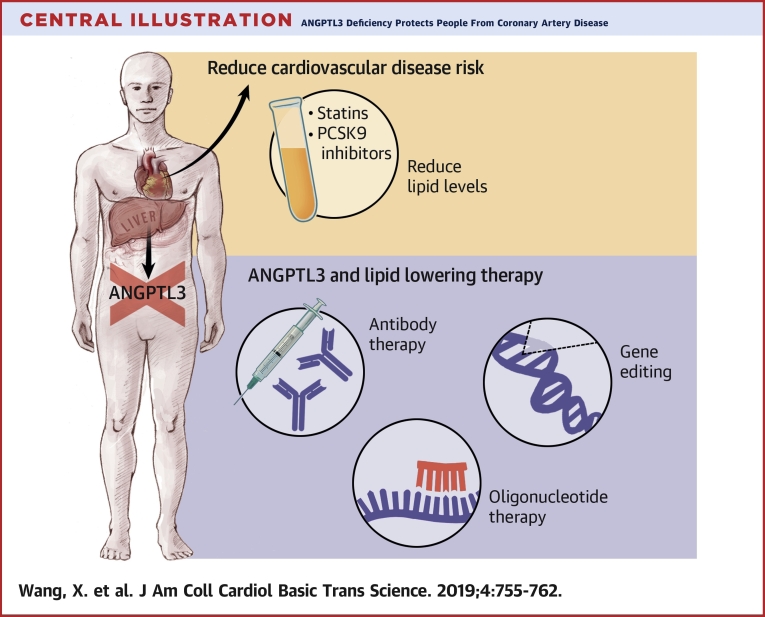
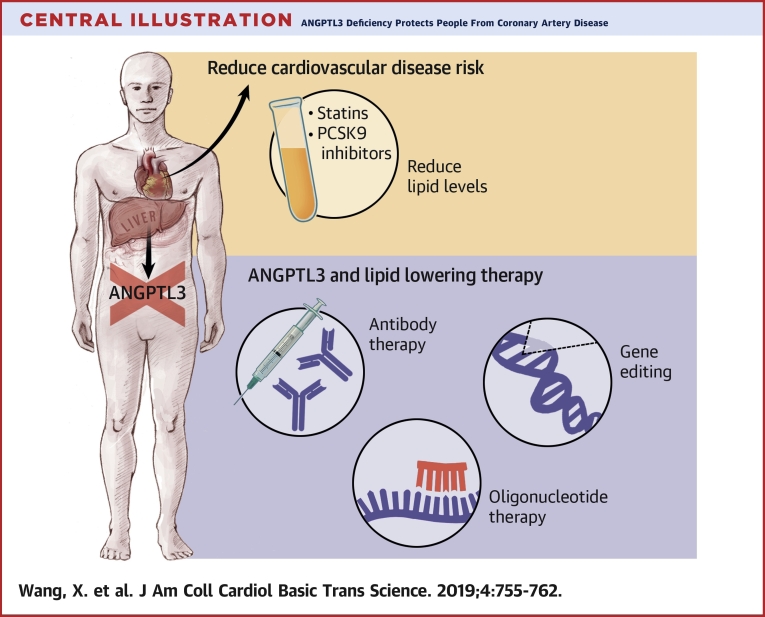
In light of the favorable consequences of ANGPTL3 deficiency, an ASO inhibitor of ANGPTL3 and a monoclonal antibody against ANGPTL3 have been advanced into clinical trials, with encouraging results so far 28, 31 (Central Illustration). The group that developed the ASO have performed studies in a variety of mouse models and humans. They found that after administration of the ASO, hepatic Angptl3 mRNA expression and plasma ANGPTL3 protein levels were significantly decreased in the mouse models they tested, including WT C57BL/6 mice, low-density lipoprotein receptor (LDLR) knockout mice(Ldlr−/−), double-knockout mice (Apoc3−/− and Ldlr−/−), heterozygous mice (Apoc3+/− and Ldlr−/−), mice with diet-induced obesity, and mice overexpressing human apolipoprotein C-III. The ASO decreased only triglyceride levels, not LDL-C or HDL-C, in C57BL/6 mice on standard chow, whereas in various hypercholesterolemic mouse models, the ASO resulted in reductions in levels of triglycerides (35% to 85%), LDL-C (7% to 64%), and HDL-C (3% to 23%). In addition, the ASO decreased liver triglyceride secretion and accumulation and improved insulin sensitivity, measured by means of intraperitoneal glucose tolerance and insulin tolerance testing in mice with diet-induced obesity. The ASO also slowed the progression of atherosclerosis in LDLR knockout mice.
Central Illustration.
ANGPTL3 Deficiency Protects People From Coronary Artery Disease
Despite the use of statins and PCSK9 inhibitors, there remains residual risk of coronary heart disease (CHD). ANGPTL3 has emerged as a promising molecular target to further reduce CHD risk due to its regulation of all 3 major lipid traits: low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, and triglycerides. In light of the favorable consequences of ANGPTL3 deficiency, various therapeutic strategies to target ANGPTL3 are currently in development, including a monoclonal antibody, an antisense oligonucleotide, and gene editing.
Subsequently, a randomized placebo-controlled phase 1 clinical trial of an ANGPTL3 ASO was conducted (28). Forty-four healthy human participants were randomly assigned to receive subcutaneous injections of the ASO or a placebo in a single dose or multiple doses. Among the single-dose groups at day 15, the investigators observed lower levels of ANGPTL3 protein, triglycerides, very low-density lipoprotein cholesterol, non-HDL cholesterol, and total cholesterol in the ASO group compared with placebo group. However, these differences were not significant, probably due to the small sizes of the groups (N = 3 for each group). Among the multiple-dose groups, they observed dose-dependent reductions of ANGPTL3 protein, triglycerides, non-HDL cholesterol, and apolipoprotein C-III in the ASO groups than in the placebo group.
Another group developed a fully human monoclonal antibody with high affinity for ANGPTL3. Intravenous administration of the antibody to normal C57BL/6 mice decreased plasma triglyceride levels by ≥50% and increased the LPL activity (9). Chronic administration of the antibody to dyslipidemic C57BL/6 mice reduced triglyceride, LDL-C, and HDL-C levels in the circulation without any changes in the triglyceride content of the liver, adipose, or heart. The investigators extended their observations to non-human primates by assessing the effect of the antibody on circulating lipid levels in cynomolgus monkeys (9). The antibody produced robust reductions in circulating triglycerides and non-HDL cholesterol in a dose-dependent way. It did not alter the plasma levels of LDL-C, perhaps due to those levels being low at baseline. The investigators also reported that inhibition of ANGPTL3 by the antibody could significantly decrease atherosclerotic lesion size compared to a control antibody in a mouse model of atherosclerosis (31).
A randomized placebo-controlled phase I clinical trial was performed to assess the safety, side effect profile, and pharmacodynamics of the antibody (31). A total of 83 healthy human volunteers with mildly to moderately elevated levels of triglycerides or LDL-C entered a single-ascending–dose trial. The antibody caused dose-dependent reductions in triglyceride levels of up to 76% and LDL-C levels of up to 23%. In addition, a single-group, open-label clinical trial of the antibody was conducted with 9 patients with homozygous familial hypercholesterolemia (hoFH). They received the antibody for 4 weeks, and their LDL-C levels were decreased by ∼50% at week 4, although the patients were already taking aggressive lipid-lowering therapy (32).
ANGPTL3 and therapeutic gene editing
Another approach to target ANGPTL3 is via gene editing to induce permanent LOF mutations in vivo (Central Illustration). Since the first report of successful use in mammalian cells in 2013, CRISPR-Cas9 technology has emerged as a promising gene editor for therapeutic applications due to its efficiency and ease of use (33).
CRISPR-Cas9 introduces a double-strand DNA break at a desired site in the genome. This activates endogenous DNA repair pathways, either non-homologous end-joining (NHEJ) or homology-directed repair (HDR), to fix the break (34). NHEJ is the default repair pathway in all cells, in which the free ends from the DNA break are reconnected. However, NHEJ is an error-prone process and results in introduction of insertions or deletions (indels) at the site of the break. NHEJ can introduce frameshift mutations into a targeted gene, thereby disrupting the gene. By contrast, the cell can accurately repair a DNA break through HDR, which occurs only in proliferating cells. HDR uses a template DNA strand to achieve high-fidelity repair, and if a repair template has matching sequence to the site of the break but also has a custom-made DNA mutation, the mutation can be introduced into the genome. However, introducing a specific mutation at the site with HDR is usually inefficient, and the efficiency in vivo in adult animals is very low (<1%), which limits its use to correct a disease-causing mutation (35). Another major concern about using a gene editor is that it might also cleave the genome at other sites and cause off-target mutations.
To circumvent these problems, CRISPR-Cas9 has been adapted so that it can directly alter specific nucleotides in the DNA sequence without generating double-strand breaks and without the need for a repair template—a phenomenon known as base editing 36, 37, 38, 39, 40. A fusion protein with a catalytically impaired Cas9 protein and a cytosine deaminase domain adapted from an RNA-editing or DNA-editing enzyme confers the ability to convert cytosine bases at the CRISPR-Cas9 target site into thymine bases.
The use of base editor 3 (BE3) successfully introduced LOF Angptl3 mutations into liver cells in mice (41). The investigators first screened potential sites of base-edited nonsense mutations in Angptl3 in Neuro-2a cells and identified high BE3 activity at the codon Gln-135 site. Then they produced an adenoviral vector expressing BE3 targeting Angptl3 Gln-135 and injected the vector into C57BL/6J mice. This resulted in significantly reduced plasma ANGPTL3, triglyceride, and total cholesterol levels (49%, 31%, and 19%, respectively). In hyperlipidemic LDLR knockout mice, even larger effects were observed. After treatment, triglycerides and cholesterol were reduced by 56% and 51%, respectively. Overall, this proof-of-concept study showed the ability to efficiently introduce LOF mutations in dyslipidemia-associated genes in vivo with significant lipid-lowering effects.
Several aspects make ANGPTL3 an attractive target for gene editing. First, naturally occurring LOF mutations in ANGPTL3 protect against coronary artery disease without causing serious adverse health consequences 30, 31, even in the homozygous or compound heterozygous state (i.e., full knockout of gene function) 23, 30. Second, ANGPTL3 is primarily expressed in hepatocytes and secreted into the bloodstream. The liver is an ideal organ for in vivo gene editing because of its accessibility for various delivery methods. Lastly, inhibition of ANGPTL3 requires only the introduction of inactivating mutations, which is easier to achieve than precisely correcting a specific gene mutation.
Will in vivo gene editing approaches translate into the clinic and provide a potential preventive strategy for CHD in humans? The primary benefit of in vivo gene editing is that it could ultimately yield a 1-shot, long-term therapy that would permanently modify a target gene, removing the need for repeated administration of drugs. Before consideration of translation into the clinic, ethical and regulatory challenges as well as technical issues must be addressed.
The first concern about in vivo gene editing from the scientific community has been the potential for off-target mutagenesis. It has the potential to introduce unanticipated mutations that cause oncogenesis, promoting a different disease than the 1 being treated. Second, unintended on-target mutagenesis requires more characterization before gene editing can be used as a therapy in humans because it causes irreversible genetic changes with possible serious adverse effects. Third, there might be the potential for toxicity or immune responses on delivery, and methods of safe delivery will need to be refined. Adeno-associated viruses are the preferred viral delivery vehicle compared with adenoviral vectors for use in humans because of safety concerns 42, 43. However, the size limitation of each adeno-associated virus vector makes it challenging to deliver a gene editor, especially a base editor. A nonviral solution would be the use of lipid nanoparticles to deliver gene editing tools into hepatocytes 44, 45.
Despite these challenges, ANGPTL3 is a promising gene editing target for potential clinical translation. The best candidates for first-in-human studies would be hoFH patients because they show limited response to current LDL-lowering therapies and the potential benefits might substantially outweigh the risks. With a demonstration of efficacy as well as safety in these selected patients, gene-editing therapies could then be considered for broader groups of patients at risk for CHD.
Footnotes
Dr. Musunuru is an unpaid scientific advisor to Verve Therapeutics. The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the JACC: Basic to Translational Scienceauthor instructions page.
References
- 1.Bonanni L., Cutolo A., Dalla Vestra M. Novel Approaches for the treatment of familial hypercholesterolemia. Exp Clin Endocrinol Diabetes. 2016;124:583–587. doi: 10.1055/s-0042-109063. [DOI] [PubMed] [Google Scholar]
- 2.Robinson J.G., Farnier M., Krempf M. Efficacy and safety of alirocumab in reducing lipids and cardiovascular events. N Engl J Med. 2015;372:1489–1499. doi: 10.1056/NEJMoa1501031. [DOI] [PubMed] [Google Scholar]
- 3.Sabatine M.S., Giugliano R.P., Wiviott S.D. Efficacy and safety of evolocumab in reducing lipids and cardiovascular events. N Engl J Med. 2015;372:1500–1509. doi: 10.1056/NEJMoa1500858. [DOI] [PubMed] [Google Scholar]
- 4.Cannon C.P., Blazing M.A., Giugliano R.P. Ezetimibe added to statin therapy after acute coronary syndromes. N Engl J Med. 2015;372:2387–2397. doi: 10.1056/NEJMoa1410489. [DOI] [PubMed] [Google Scholar]
- 5.Sabatine M.S., Giugliano R.P., Keech A.C. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med. 2017;376:1713–1722. doi: 10.1056/NEJMoa1615664. [DOI] [PubMed] [Google Scholar]
- 6.Koishi R., Ando Y., Ono M. Angptl3 regulates lipid metabolism in mice. Nat Genet. 2002;30:151–157. doi: 10.1038/ng814. [DOI] [PubMed] [Google Scholar]
- 7.Shimizugawa T., Ono M., Shimamura M. ANGPTL3 decreases very low density lipoprotein triglyceride clearance by inhibition of lipoprotein lipase. J Biol Chem. 2002;277:33742–33748. doi: 10.1074/jbc.M203215200. [DOI] [PubMed] [Google Scholar]
- 8.Shimamura M., Matsuda M., Yasumo H. Angiopoietin-like protein3 regulates plasma HDL cholesterol through suppression of endothelial lipase. Arterioscler Thromb Vasc Biol. 2007;27:366–372. doi: 10.1161/01.ATV.0000252827.51626.89. [DOI] [PubMed] [Google Scholar]
- 9.Gusarova V., Alexa C.A., Wang Y. ANGPTL3 blockade with a human monoclonal antibody reduces plasma lipids in dyslipidemic mice and monkeys. J Lipid Res. 2015;56:1308–1317. doi: 10.1194/jlr.M054890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattijssen F., Kersten S. Regulation of triglyceride metabolism by Angiopoietin-like proteins. Biochim Biophys Acta. 2012;1821:782–789. doi: 10.1016/j.bbalip.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Quagliarini F., Wang Y., Kozlitina J. Atypical angiopoietin-like protein that regulates ANGPTL3. Proc Natl Acad Sci U S A. 2012;109:19751–19756. doi: 10.1073/pnas.1217552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ono M., Shimizugawa T., Shimamura M. Protein region important for regulation of lipid metabolism in angiopoietin-like 3 (ANGPTL3): ANGPTL3 is cleaved and activated in vivo. J Biol Chem. 2003;278:41804–41809. doi: 10.1074/jbc.M302861200. [DOI] [PubMed] [Google Scholar]
- 13.Essalmani R., Susan-Resiga D., Chamberland A. Furin is the primary in vivo convertase of angiopoietin-like 3 and endothelial lipase in hepatocytes. J Biol Chem. 2013;288:26410–26418. doi: 10.1074/jbc.M113.501304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kersten S., Mandard S., Tan N.S. Characterization of the fasting-induced adipose factor FIAF, a novel peroxisome proliferator-activated receptor target gene. J Biol Chem. 2000;275:28488–28493. doi: 10.1074/jbc.M004029200. [DOI] [PubMed] [Google Scholar]
- 15.González-Muniesa P., de Oliveira C., Pérez de Heredia F., Thompson M.P., Trayhurn P. Fatty acids and hypoxia stimulate the expression and secretion of the adipokine ANGPTL4 (angiopoietin-like protein 4/fasting-induced adipose factor) by human adipocytes. J Nutrigenet Nutrigenomics. 2011;4:146–153. doi: 10.1159/000327774. [DOI] [PubMed] [Google Scholar]
- 16.Yin W., Romeo S., Chang S., Grishin N.V., Hobbs H.H., Cohen J.C. Genetic variation in ANGPTL4 provides insights into protein processing and function. J Biol Chem. 2009;284:13213–13222. doi: 10.1074/jbc.M900553200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sukonina V., Lookene A., Olivecrona T., Olivecrona G. Angiopoietin-like protein 4 converts lipoprotein lipase to inactive monomers and modulates lipase activity in adipose tissue. Proc Natl Acad Sci U S A. 2006;103:17450–17455. doi: 10.1073/pnas.0604026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai U., Lee E.C., Chung K. Lipid-lowering effects of anti-angiopoietin-like 4 antibody recapitulate the lipid phenotype found in angiopoietin-like 4 knockout mice. Proc Natl Acad Sci U S A. 2007;104:11766–11771. doi: 10.1073/pnas.0705041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lichtenstein L., Mattijssen F., de Wit N.J. Angptl4 protects against severe proinflammatory effects of saturated fat by inhibiting fatty acid uptake into mesenteric lymph node macrophages. Cell Metab. 2010;12:580–592. doi: 10.1016/j.cmet.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu Z., Yao F., Abou-Samra A.B., Zhang R. Lipasin, thermoregulated in brown fat, is a novel but atypical member of the angiopoietin-like protein family. Biochem Biophys Res Commun. 2013;430:1126–1131. doi: 10.1016/j.bbrc.2012.12.025. [DOI] [PubMed] [Google Scholar]
- 21.Zhang R. Lipasin, a novel nutritionally-regulated liver-enriched factor that regulates serum triglyceride levels. Biochem Biophys Res Commun. 2012;424:786–792. doi: 10.1016/j.bbrc.2012.07.038. [DOI] [PubMed] [Google Scholar]
- 22.Willer C.J., Sanna S., Jackson A.U. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Musunuru K., Pirruccello J.P., Do R. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010;363:2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romeo S., Yin W., Kozlitina J. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J Clin Invest. 2009;119:70–79. doi: 10.1172/JCI37118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martín-Campos J.M., Roig R., Mayoral C. Identification of a novel mutation in the ANGPTL3 gene in two families diagnosed of familial hypobetalipoproteinemia without APOB mutation. Clin Chim Acta. 2012;413:552–555. doi: 10.1016/j.cca.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 26.Noto D., Cefalù A.B., Valenti V. Prevalence of ANGPTL3 and APOB gene mutations in subjects with combined hypolipidemia. Arterioscler Thromb Vasc Biol. 2012;32:805–809. doi: 10.1161/ATVBAHA.111.238766. [DOI] [PubMed] [Google Scholar]
- 27.Pisciotta L., Favari E., Magnolo L. Characterization of three kindreds with familial combined hypolipidemia caused by loss-of-function mutations of ANGPTL3. Circ Cardiovasc Genet. 2012;5:42–50. doi: 10.1161/CIRCGENETICS.111.960674. [DOI] [PubMed] [Google Scholar]
- 28.Graham M.J., Lee R.G., Brandt T.A. Cardiovascular and metabolic effects of ANGPTL3 antisense oligonucleotides. N Engl J Med. 2017;377:222–232. doi: 10.1056/NEJMoa1701329. [DOI] [PubMed] [Google Scholar]
- 29.Fujimoto K., Koishi R., Shimizugawa T., Ando Y. Angptl3-null mice show low plasma lipid concentrations by enhanced lipoprotein lipase activity. Exp Anim. 2006;55:27–34. doi: 10.1538/expanim.55.27. [DOI] [PubMed] [Google Scholar]
- 30.Stitziel N.O., Khera A.V., Wang X. ANGPTL3 deficiency and protection against coronary artery disease. J Am Coll Cardiol. 2017;69:2054–2063. doi: 10.1016/j.jacc.2017.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dewey F.E., Gusarova V., Dunbar R.L. Genetic and pharmacologic inactivation of ANGPTL3 and cardiovascular disease. N Engl J Med. 2017;377:211–221. doi: 10.1056/NEJMoa1612790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaudet D., Gipe D.A., Pordy R. ANGPTL3 inhibition in homozygous familial hypercholesterolemia. N Engl J Med. 2017;377:296–297. doi: 10.1056/NEJMc1705994. [DOI] [PubMed] [Google Scholar]
- 33.Musunuru K. Genome editing: the recent history and perspective in cardiovascular diseases. J Am Coll Cardiol. 2017;70:2808–2821. doi: 10.1016/j.jacc.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.West S.C. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y., Wang L., Bell P. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishida K., Arazoe T., Yachie N. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353(6305) doi: 10.1126/science.aaf8729. pii:aaf8729. [DOI] [PubMed] [Google Scholar]
- 38.Ma Y., Zhang J., Yin W., Zhang Z., Song Y., Chang X. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods. 2016;13:1029–1035. doi: 10.1038/nmeth.4027. [DOI] [PubMed] [Google Scholar]
- 39.Hess G.T., Frésard L., Han K. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods. 2016;13:1036–1042. doi: 10.1038/nmeth.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim Y.B., Komor A.C., Levy J.M., Packer M.S., Zhao K.T., Liu D.R. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chadwick A.C., Evitt N.H., Lv W., Musunuru K. Reduced blood lipid levels with in vivo CRISPR-Cas9 base editing of ANGPTL3. Circulation. 2018;137:975–977. doi: 10.1161/CIRCULATIONAHA.117.031335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muruve D.A. The innate immune response to adenovirus vectors. Hum Gene Ther. 2004;15:1157–1166. doi: 10.1089/hum.2004.15.1157. [DOI] [PubMed] [Google Scholar]
- 43.Gregory S.M., Nazir S.A., Metcalf J.P. Implications of the innate immune response to adenovirus and adenoviral vectors. Future Virol. 2011;6:357–374. doi: 10.2217/fvl.11.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yin H., Song C.Q., Dorkin J.R. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang M., Zuris J.A., Meng F. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc Natl Acad Sci U S A. 2016;113:2868–2873. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]