Abstract
Rationale: A molecular test to distinguish between sepsis and systemic inflammation of noninfectious etiology could potentially have clinical utility.
Objectives: This study evaluated the diagnostic performance of a molecular host response assay (SeptiCyte LAB) designed to distinguish between sepsis and noninfectious systemic inflammation in critically ill adults.
Methods: The study employed a prospective, observational, noninterventional design and recruited a heterogeneous cohort of adult critical care patients from seven sites in the United States (n = 249). An additional group of 198 patients, recruited in the large MARS (Molecular Diagnosis and Risk Stratification of Sepsis) consortium trial in the Netherlands (www.clinicaltrials.gov identifier NCT01905033), was also tested and analyzed, making a grand total of 447 patients in our study. The performance of SeptiCyte LAB was compared with retrospective physician diagnosis by a panel of three experts.
Measurements and Main Results: In receiver operating characteristic curve analysis, SeptiCyte LAB had an estimated area under the curve of 0.82–0.89 for discriminating sepsis from noninfectious systemic inflammation. The relative likelihood of sepsis versus noninfectious systemic inflammation was found to increase with increasing test score (range, 0–10). In a forward logistic regression analysis, the diagnostic performance of the assay was improved only marginally when used in combination with other clinical and laboratory variables, including procalcitonin. The performance of the assay was not significantly affected by demographic variables, including age, sex, or race/ethnicity.
Conclusions: SeptiCyte LAB appears to be a promising diagnostic tool to complement physician assessment of infection likelihood in critically ill adult patients with systemic inflammation.
Clinical trial registered with www.clinicaltrials.gov (NCT01905033 and NCT02127502)
Keywords: inflammation, sepsis, infection, classifier, RT-qPCR
At a Glance Commentary
Scientific Knowledge on the Subject
Sepsis is a common and potentially lethal medical condition for which treatment is time sensitive. Distinguishing between sepsis and noninfectious systemic inflammatory response syndrome is clinically challenging, especially early in the course of illness when no site of infection or pathogen has been identified and when organ damage has not yet become evident. Molecular methods have increasingly been investigated as tools for diagnosing sepsis.
What This Study Adds to the Field
SeptiCyte LAB is the first host response gene expression assay cleared by the U.S. Food and Drug Administration as an aid for diagnosis of sepsis. SeptiCyte LAB was able to discriminate between systemic inflammatory response syndrome and sepsis with an area under the receiver operating characteristic curve of 0.82–0.89 and provided a probability of infection. The present study suggests that SeptiCyte LAB, especially when combined with commonly used demographic, clinical, or laboratory variables, potentially may contribute clinical utility for the diagnosis of sepsis.
Sepsis is a common and potentially lethal medical condition for which treatment is time sensitive. Distinguishing between sepsis and noninfectious systemic inflammatory response syndrome (SIRS) is clinically challenging, especially early in the course of illness when no site of infection or pathogen has been identified and when organ damage has not yet become evident. Uncertainty in diagnosis can lead to delays in therapy or unnecessary use of antibiotics. Cultures from blood and other body fluids are commonly ordered to identify a source or a pathogen but require at least 12 hours of incubation and are often either negative or inconclusive when positive for nonpathogenic organisms.
Molecular methods have increasingly been investigated as tools for diagnosing sepsis. Authors of several recent publications (1–10) have proposed diagnostic criteria for discriminating sepsis from SIRS on the basis of combinations of molecular (RNA or protein) biomarkers. Advantages of molecular testing for diagnosing sepsis include improved sensitivity and specificity beyond that provided by clinical or biochemical variables, as well as early detection of organ dysfunction.
Using a definition of sepsis as SIRS criteria plus infection (without requiring overt organ dysfunction), we previously reported the discovery and validation of SeptiCyte LAB (Immunexpress Inc.), a signature based on whole-blood expression levels of four genes (CEACAM4, LAMP1, PLAC8, PLA2G7) involved in the host response to infection (4, 7). SeptiCyte LAB is the first host response gene expression assay cleared by the U.S. Food and Drug Administration (FDA) for diagnosis of sepsis [510(k) premarket notification K163260 (11)]. SeptiCyte LAB was able to discriminate between SIRS and sepsis with an area under the receiver operating characteristic curve of 0.82–0.89, and it provided a probability of infection. In this paper, we report on the clinical performance of SeptiCyte LAB in a clinical trial involving 447 patients. We found that the test result (SeptiScore) directly correlates with the probability of sepsis, suggesting that the SeptiScore, especially when combined with commonly used demographic, clinical, or laboratory variables, could serve as a tool to aid clinicians in distinguishing sepsis from SIRS. Some of the results of this study were previously reported in the form of abstracts (12, 13).
Methods
Clinical Sites
This paper combines the results of three separate prospective, observational studies to achieve an evaluation of the clinical performance of SeptiCyte LAB. The first study involved the testing and analysis of 198 subjects from a much larger consortium trial, the MARS (Molecular Diagnosis and Risk Stratification of Sepsis) trial, which enrolled 7,500 patients at the Academic Medical Center (AMC) Amsterdam and the University Medical Center Utrecht in the Netherlands between January 2011 and December 2013 (www.clinicaltrials.gov identifier NCT01905033). The overall goal of the MARS consortium trial was to generate tools to provide rapid and accurate information about microorganisms responsible for infection and about the associated host immune responses. Besides the Amsterdam and Utrecht university hospitals, the MARS consortium project involved eight industrial partners, including Immunexpress. From among the large cohort of the MARS trial, we chose for examination in the present study a subset of 198 consecutive subjects who were enrolled at AMC between June 2013 and November 2013 and who satisfied a set of predefined inclusion and exclusion criteria (see below).
The second study, the VENUS (Validation of Septic Gene Expression Using SeptiCyte) trial, was conducted at Intermountain Medical Center (Murray, UT; n = 125) and Latter Day Saints Hospital (Salt Lake City, UT; n = 4) between May 2014 and April 2015 (www.clinicaltrials.gov identifier NCT02127502). The third study was a supplement to VENUS conducted between March 2016 and August 2016, which enrolled participants from five academic institutions in major U.S. metropolitan areas (Johns Hopkins Hospital, Baltimore, MD [n = 39]; Rush University Medical Center, Chicago, IL [n = 37]; Loyola University Medical Center, Maywood, IL [n = 11]; Northwell Healthcare, Long Island, NY [n = 26]; Grady Memorial Hospital, Atlanta, GA [n = 7]). The purpose of the VENUS Supplement was to increase ethnic diversity, as requested by the FDA.
Ethics Approval
Ethics approval was gained from the relevant medical ethics committee (MARS) or institutional review boards (VENUS): AMC, Amsterdam (10-056C); Intermountain Medical Center/Latter Day Saints Hospital (1024931); Johns Hopkins Hospital (IRB00087839); Rush University Medical Center (15111104-IRB01); Loyola University Medical Center (208291); Northwell Healthcare (16-02-42-03); and Grady Memorial Hospital (000-87806).
Study Objective and Inclusion/Exclusion Criteria
The study objective was to determine the diagnostic performance of SeptiCyte LAB in distinguishing sepsis from SIRS in adult critical care patients, either as a stand-alone test or in combination with other clinical variables and laboratory assessments used to confirm or exclude a diagnosis of sepsis. A “sepsis event” was defined operationally to have occurred when a patient displayed two or more signs of systemic inflammation and was given therapeutic systemic antibiotics by the attending clinician within 24 hours of ICU admission.
Adult critical care patients aged 18–89 years were considered qualified for enrollment if, upon admission to an ICU, they displayed two or more of the following signs: temperature above 38°C or less than 36°C, heart rate greater than 90 beats per minute, tachypnea greater than 20 breaths per minute or PaCO2 less than 32 mm Hg, and white blood cell (WBC) count greater than 12,000/mm3 or less than 4,000/mm3 or greater than 10% immature neutrophils (bands).
Patients were excluded if 1) clinical cultures or serologic tests were not obtained when sepsis was suspected, 2) they were admitted to ICU more than 24 hours before trial enrollment, 3) they had a delay of more than 24 hours between trial enrollment and sample draw, 4) they underwent elective cardiac surgery with an expected ICU length of stay less than 24 hours, 5) they were treated with antibiotics more than 24 hours before ICU admission for any reason other than surgical prophylaxis (VENUS and Venus Supplement only), or 6) they had a documented sepsis event that occurred more than 3 days before ICU admission or more than 2 days after ICU admission (MARS only). Exclusion criteria 5 and 6 were used to exclude patients who had experienced prior sepsis events. Additional details on study recruitment and patient exclusions are given in the online supplement, part 1.
Laboratory Procedures
SeptiCyte LAB
SeptiCyte LAB used a 2.5-ml sample of whole blood collected by venipuncture into a PAXgene Blood RNA Tube (catalog number 762165, FDA 510k number K042613; BD Biosciences) within 24 hours of admission to the ICU. Experiments in which increasing dilutions of WBC were spiked into leukocyte-depleted blood and assayed established that the limit of detection of SeptiCyte LAB was 270 WBC per cubic millimeter of whole blood (11). Therefore, a subject’s WBC count was required be at least 270 per cubic millimeter of whole blood to be successfully tested with SeptiCyte LAB. PAXgene tubes were handled per the manufacturer’s instructions and stored at less than or equal to −70°C until tested. Total RNA was isolated using the PAXgene Blood RNA Kit (catalog number 762164; Qiagen). Purified total RNA was required to have 2–50 ng/μl concentration (by A260 measurement) and adequate purity (as estimated by A260/A280 ratio ≥1.6). RNA was tested immediately after extraction or stored frozen in single-use portions at less than or equal to −70°C until tested.
SeptiCyte LAB is a real-time RT-qPCR assay that quantitates the relative expression levels of four genes (CEACAM4, LAMP1, PLAC8, PLA2G7) (4, 7). The format of the assay used in the present study was a boxed kit with turnaround time (from sample draw to report) of approximately 6 hours. The assay was performed on an Applied Biosystems 7500 Fast Dx Real-Time PCR instrument (Thermo Fisher Scientific) with kits provided to each site. Testing was performed in CLIA (Clinical Laboratory Improvement Amendments)-certified laboratories at Asuragen (Austin, TX), Providence Medical Center (Portland, OR), and Intermountain Medical Center (Murray, UT). All testing with SeptiCyte LAB was conducted on banked specimens. SeptiCyte LAB test results were not provided to the study investigators or clinicians.
SeptiCyte LAB generates a quantitative score (SeptiScore) falling into one of four ranges (bands) defined as follows: band 1 (0.0 ≤ SeptiScore ≤ 3.0), band 2 (3.1 ≤ SeptiScore ≤ 4.4), band 3 (4.5 ≤ SeptiScore ≤ 5.9), and band 4 (6.0 ≤ SeptiScore ≤ 10.0). The fractions of sepsis and SIRS subjects falling into each SeptiScore band, and the resultant sepsis/SIRS likelihood ratios, were calculated as described in the online supplement, part 2. For the goal of diagnosing sepsis, a SeptiScore value of 3.1 was operationally defined as a binary cutoff, with values greater than or equal to 3.1 considered to provide evidence for infection and values of 3.0 or less considered to provide evidence for noninfection. The cutoff of 3.1 had been preset on the basis of an entirely independent, nonoverlapping discovery dataset (4).
Procalcitonin
Procalcitonin (PCT) assays were performed at Providence Medical Center (Portland, OR) using a commercially available kit (VIDAS B.R.A.H.M.S PCT; bioMérieux). PCT concentrations greater than 2 mg/ml were considered to indicate a high risk of severe sepsis and/or septic shock, as per the VIDAS B.R.A.H.M.S PCT 510k Decision Summary (K071146).
Reference Method
Patient classification (sepsis, SIRS, or indeterminate) by an external three-member expert panel was used as the reference method (14) and is described fully in the online supplement, part 3. Some diagnostic discordance was expected, even for an expert panel, and especially for certain subpopulations (e.g., sepsis originating from pneumonia). Three different retrospective physician diagnosis (RPD) algorithms were used. Diagnosis was designated “consensus” if two panelists agreed (first algorithm; n = 180 sepsis, 230 SIRS, 37 indeterminate) or “unanimous” if all three panelists agreed with each other and with the consensus discharge evaluation of the site investigators at the hospital in question (second algorithm; n = 119 sepsis, 171 SIRS). If the panelists all disagreed, or if all deemed a subject indeterminate, then the diagnosis was considered indeterminate (37 of 447 [8.3%]). In the third RPD algorithm (“forced” approach), indeterminates underwent a second blinded independent case review and were forced into either the sepsis or SIRS category (n = 202 sepsis, 245 SIRS). The forced algorithm, by requiring a binary judgment (sepsis or SIRS) to be made about each indeterminate subject, was expected to result in some wrong classifications and therefore to produce a lower bound estimate of SeptiCyte LAB performance.
Data Analysis
Performance relative to the three reference algorithms (unanimous, consensus, forced RPD) using receiver operating characteristic (ROC) curve analyses and calculation of area under the curve (AUC) (15) was performed using the R package pROC (16) unless otherwise noted. Comparison of AUCs between correlated ROC curves was performed using DeLong’s method (17).
Single-variable and multivariable analyses were used to determine which variables were most predictive of sepsis versus SIRS. Four different multivariate analysis methods were used: 1) forward logistic regression with directed variable selection, 2) logistic regression with random variable selection, 3) ridge/lasso regression, and 4) random decision forests. The following 14 variables were examined by themselves and were also included in modeling: SeptiScore, PCT, maximum glucose concentration, minimum white blood cell count, maximum white blood cell count, maximum mean arterial pressure, minimum core temperature, maximum core temperature, minimum heart rate, maximum heart rate, number of SIRS criteria, age, sex, and race/ethnicity. Details are provided in the online supplement, parts 4–6.
Reporting
This article was composed according to the Standards for Reporting Diagnostic Accuracy (18). Descriptions of MARS and VENUS have been uploaded to www.clinicaltrials.gov under the identifiers NCT01905033 and NCT02127502, respectively.
Results
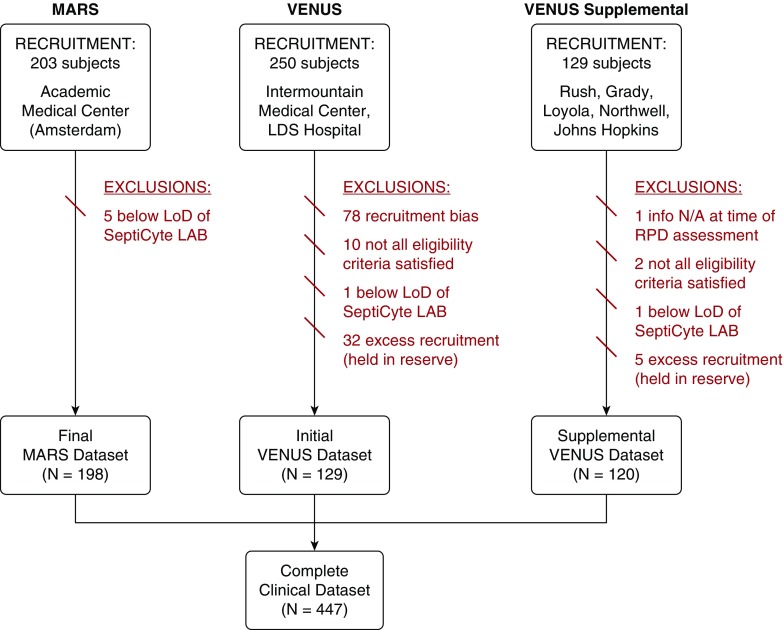
The complete clinical dataset consisted of 447 adult critical care patients from three different studies (Figure 1) across seven sites in the United States and one in the Netherlands. Although sources of admission to the ICU were diverse, about 64% of patients came to the ICU from the emergency department (online supplement, part 1). The racial and ethnic diversity of enrolled subjects varied by site; overall, white individuals (70.7%) and black individuals (19.7%) represented the largest subgroups (Table 1). SIRS diagnoses (51.4%) outnumbered sepsis diagnoses (40.3%). A unanimous classification was observed for 171 patients with SIRS (38.2%) and 119 patients with sepsis (26.6%). For 37 patients (8.3%), a consensus was not reached; therefore, they were classified as indeterminate (Table 1). Pneumonia was the most frequent infection source for patients with sepsis.
Figure 1.
CONSORT (Consolidated Standards of Reporting Trials) diagram for the complete clinical dataset (N = 447) used in validation of SeptiCyte LAB. Subject recruitment dates were as follows: MARS (Molecular Diagnosis and Risk Stratification of Sepsis), between June 2013 and November 2013; VENUS (Validation of Septic Gene Expression Using SeptiCyte), between May 2014 and April 2015; VENUS Supplement, between March 2016 and August 2016. LoD = limit of detection; N/A = not applicable; RPD = retrospective physician diagnosis.
Table 1.
Patient Demographics and Infection Source
| Characteristic | MARS (n = 198) | VENUS (n = 129) | VENUS Supplement (n = 120) | Total (N = 447) |
|---|---|---|---|---|
| Age, yr, median (IQR) | 59.5 (46–71) | 61.0 (48–70) | 57.0 (43.8–68) | 59.0 (46–70) |
| Sex, n (%) | ||||
| Female | 75 (16.8) | 62 (13.9) | 55 (12.3) | 192 (43.0) |
| Male | 123 (27.5) | 67 (15.0) | 65 (14.5) | 255 (57.0) |
| Race/ethnicity, n (%) | ||||
| White | 154 (34.4) | 117 (26.2) | 45 (10.1) | 316 (70.7) |
| Black | 26 (5.8) | 1 (0.2) | 61 (13.6) | 88 (19.7) |
| Asian | 14 (3.1) | 3 (0.7) | 7 (1.6) | 24 (5.4) |
| Hispanic | 0 (0.0) | 5 (1.1) | 7 (1.6) | 12 (2.7) |
| Other/unknown | 4 (0.9) | 3 (0.7) | 0 (0.0) | 7 (1.6) |
| Infection source, n (%) | ||||
| Pneumonia | 44 (9.8) | 17 (3.8) | 25 (5.6) | 86 (19.2) |
| UTI | 16 (3.6) | 10 (2.2) | 1 (0.2) | 27 (6.0) |
| Bloodstream | 0 (0.0) | 8 (1.8) | 10 (2.2) | 18 (4.0) |
| Abdominal | 25 (5.6) | 6 (1.3) | 6 (1.3) | 37 (8.3) |
| Other | 16 (3.6) | 12 (2.7) | 10 (2.2) | 38 (8.5) |
| Not identified initially | 97 (21.7) | 76 (17.0) | 68 (15.2) | 241 (53.9) |
| Classification: unanimous RPD, n (%) | ||||
| Sepsis | 50 (11.2) | 35 (7.8) | 34 (7.6) | 119 (26.6) |
| SIRS | 65 (14.5) | 49 (11.0) | 57 (12.8) | 171 (38.2) |
| Indeterminate | NA | NA | NA | NA |
| Classification: consensus RPD, n (%) | ||||
| Sepsis | 81 (18.1) | 55 (12.3) | 44 (9.8) | 180 (40.3) |
| SIRS | 100 (22.4) | 65 (14.5) | 65 (14.7) | 230 (51.4) |
| Indeterminate | 17 (3.8) | 9 (2.0) | 11 (2.4) | 37 (8.3) |
| Classification: forced RPD, n (%) | ||||
| Sepsis | 90 (20.1) | 59 (13.2) | 53 (11.9) | 202 (45.2) |
| SIRS | 108 (24.2) | 70 (15.7) | 67 (15.0) | 245 (54.8) |
| Indeterminate | NA | NA | NA | NA |
Definition of abbreviations: IQR = interquartile range; MARS = Molecular Diagnosis and Risk Stratification of Sepsis; NA = not applicable; RPD = retrospective physician diagnosis; SIRS = systemic inflammatory response syndrome; UTI = urinary tract infection; VENUS = Validation of Septic Gene Expression Using SeptiCyte.
Classification is by consensus RPD, unless indicated otherwise.
Clinical and laboratory characteristics for all patients are described in Table 2. Septic patients were sicker overall, with a longer length of ICU stay and total duration of hospitalization than those with SIRS or with indeterminate classifications. Common clinical variables used in SIRS criteria displayed small but significant differences between the SIRS and sepsis groups (P values in Table 2).
Table 2.
Patient Clinical Characteristics
|
P Value* (Sepsis vs. SIRS) |
|||||
|---|---|---|---|---|---|
| Characteristic | Sepsis (n = 180) | SIRS (n = 230) | Indeterminate (n = 37) | t Test | Mann-Whitney U Test |
| ICU LOS, d | 2.8 (1.7–5.8) | 1.9 (1.2–3.1) | 2.8 (1.3–6.8) | 7.0 × 10−6 | 3.2 × 10−7 |
| Hospital LOS, d | 7.4 (3.5–11.4) | 3.7 (2.6–6.6) | 6.4 (3.5–10.7) | 1.5 × 10−5 | 3.1 × 10−9 |
| SOFA score | 7.0 (4.8– 9.0) | 6.0 (4.0–8.0) | 5.0 (3.0–9.0) | 3.0 × 10−3 | 1.1 × 10−2 |
| WBC count, ×10−6 cells/ml, maximum | 15.0 (9.8–21.4) | 13.2 (8.8–17.4) | 13.7 (10.4–15.7) | 3.5 × 10−4 | 6.9 × 10−3 |
| Temperature, °C, maximum | 38 (37–39) | 37 (37–38) | 37 (37–38) | 1.2 × 10−8 | 9.0 × 10−8 |
| Heart rate, beats/min, maximum | 122 (109–136) | 113 (99–131) | 116 (103–129) | 6.8 × 10−4 | 1.5 × 10−4 |
| Respiratory rate, breaths/min | 24.0 (20.5–28.0) | 21.5 (18.5–25.0) | 24.4 (22.0–26.0) | <1.0 × 10−3 | 3.0 × 10−4 |
| PaO2, mm Hg | 75.0 (65.9–92.2) | 89.5 (68.0–119.2) | 80.5 (69.5–90.4) | 1.0 × 10−3 | 1.5 × 10−3 |
| Mean arterial pressure, mm Hg, minimum | 57 (49–64) | 64 (55–75) | 63 (58–73) | 3.7 × 10−7 | 1.2 × 10−7 |
| Lactate, mmol/L | 2.4 (1.6–4.0) | 2.1 (1.4–3.3) | 2.7 (1.6–3.5) | NS (0.57) | NS (0.22) |
| Creatinine, mg/dl | 5.4 (1.1–115.0) | 3.2 (1.0–81.8) | 3.7 (0.8–78.8) | NS (0.60) | NS (0.19) |
| Procalcitonin, ng/ml | 6.9 (1.6–24.0) | 0.3 (0.1–1.7) | 0.6 (0.1–5.0) | 8.4 × 10−7 | 6.1 × 10−20 |
| SeptiScore | 6.6 (5.2–7.7) | 3.7 (2.6–4.7) | 4.5 (2.9–6.0) | 4.1 × 10−40 | 2.2 × 10−34 |
| Mechanical ventilation, n (%) | 69 (15.4) | 102 (22.8) | 18 (4.0) | NS (0.06) (Z-score test) |
|
| Death, n (%) | 28 (6.3) | 27 (6.0) | 3 (0.7) | NS (0.90) (Z-score test) |
|
| SeptiScore band 1, n (%) | 10 (5.6) | 79 (34.3) | 10 (27.0) | ||
| SeptiScore band 2, n (%) | 20 (11.1) | 82 (35.6) | 6 (16.2) | ||
| SeptiScore band 3, n (%) | 45 (25.0) | 49 (21.3) | 11 (29.7) | ||
| SeptiScore band 4, n (%) | 105 (58.3) | 20 (8.7) | 10 (27.0) | ||
Definition of abbreviations: LOS = length of stay; NS = not significant; SIRS = systemic inflammatory response syndrome; SOFA = Sequential Organ Failure Assessment; WBC = white blood cell.
Consensus retrospective physician diagnosis was used to determine the patient numbers in the categories. Data are shown as median (interquartile range) unless otherwise noted.
For all parameters except mechanical ventilation and death, statistical significance was assessed with both Student’s t test (Microsoft Excel) and the Mann-Whitney U test (http://astatsa.com/WilcoxonTest/). For mechanical ventilation and death, a two-tailed Z-score test for difference in proportions was used (http://epitools.ausvet.com.au).
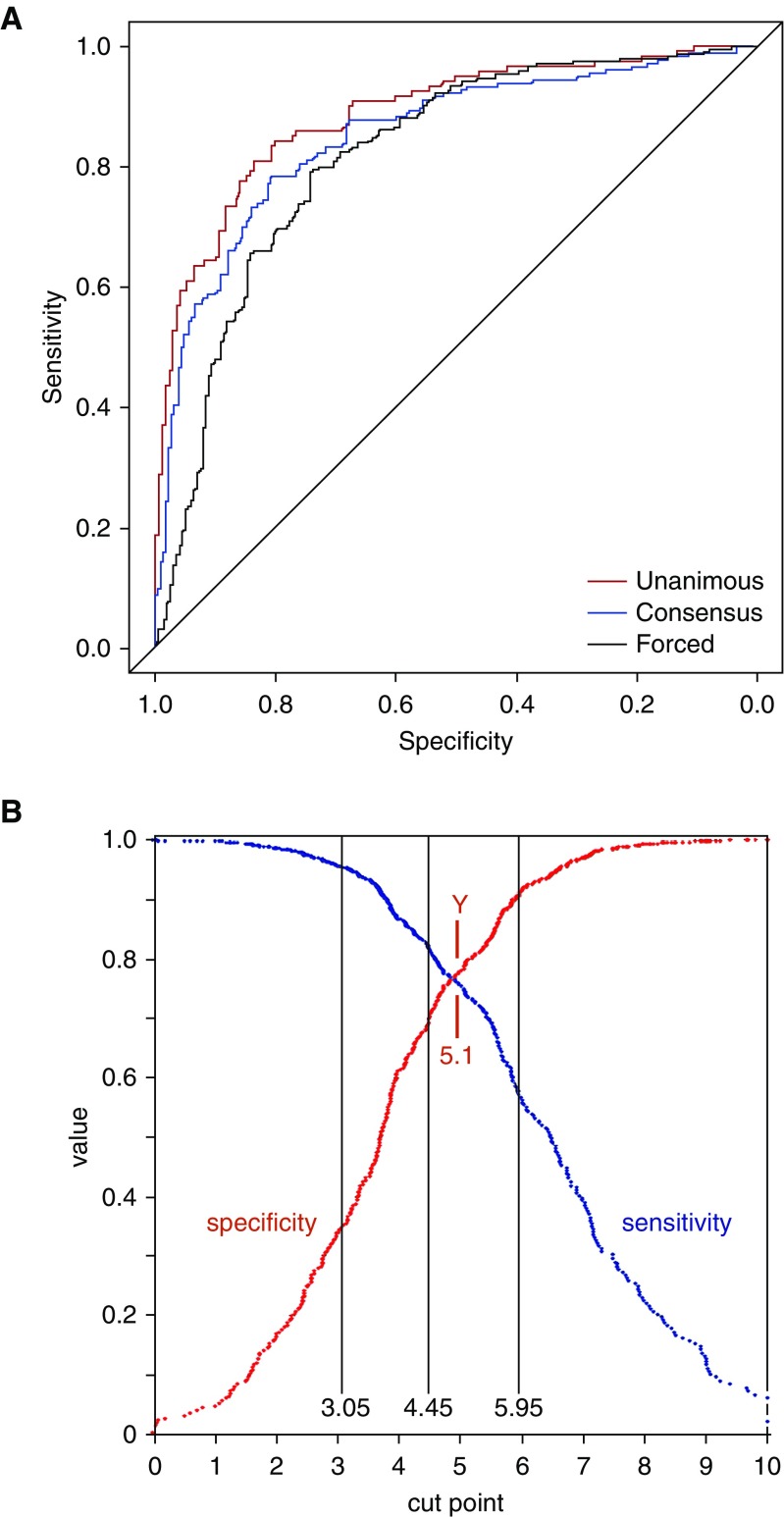
SeptiScore values were significantly higher in septic patients (Table 2), and ROC curve analysis produced AUC values of 0.89, 0.85, and 0.82 for unanimous, consensus, and forced RPD, respectively (Figure 2A). For the binary discrimination of sepsis versus SIRS in our dataset, the relationship between cut point, sensitivity, and specificity (derived from the ROC curve) is shown in Figure 2B. We used the SeptiScore value 3.1, previously chosen from an independent clinical dataset (4), as a binary cutoff. Values of 3.1 or greater provided evidence for sepsis, and values less than 3.1 provided evidence for SIRS. This choice of cutoff is biased toward sensitivity at the expense of specificity and was made because of asymmetry in the patient risk profile: a false-negative call (in which sepsis is missed and antibiotics potentially withheld) has greater deleterious consequence than a false-positive call (in which there is no systemic infection and antibiotics potentially given unnecessarily). Using a cutoff of 3.1, the sensitivity of SeptiScore for identifying septic patients was greatest for unanimous RPD cases (0.97) compared with consensus (0.94) or forced RPD (0.92); negative predictive values were 0.89 or greater (Table 3).
Figure 2.
Receiver operating characteristic curves for SeptiScore, calculated for the complete clinical dataset. (A) Receiver operating characteristic curves. Unanimous retrospective physician diagnosis (RPD) (nsepsis = 121; nSIRS = 171; nexcluded = 155; area under the curve [AUC], 0.89; 95% confidence interval [CI], 0.85–0.93), consensus RPD (nsepsis = 180; nSIRS = 230; nexcluded = 37; AUC, 0.85; 95% CI, 0.81–0.89), and forced RPD (nsepsis = 202; nSIRS = 245; AUC, 0.82; 95% CI, 0.78–0.86). (B) Sensitivity, specificity as a function of cut point. Comparator = consensus RPD. The following SeptiScore band boundaries are indicated: band 1/2 boundary at 3.05, band 2/3 boundary at 4.45, and band 3/4 boundary at 5.95, as well as the Youden index (Y) at 5.1 (when sensitivity + specificity is maximized). SIRS = systemic inflammatory response syndrome.
Table 3.
Summary of Results from Binary Analysis of Complete Clinical Dataset
| RPD | Description | Sepsis Prevalence | AUC | Sensitivity | Specificity | NPV | PPV |
|---|---|---|---|---|---|---|---|
| Unanimous, based on discharge evaluation (n = 290 of 447 [64.9%]) | All three panelists and site PI agree on SIRS (171 of 290 [59.0%]) or sepsis (119 of 290 [41.0%]) | 41.0% | 0.89 | 0.97 | 0.34 | 0.94 | 0.51 |
| Consensus (n = 410 of 447 [91.7%]) | Majority vote leads to exclusion of 37 indeterminates and classification of 230 of 410 (56.1%) as SIRS and 180 of 410 (43.9%) as sepsis | 43.9% | 0.85 | 0.94 | 0.35 | 0.89 | 0.53 |
| Forced (n = 447 of 447 [100.0%]) | All subjects classified as SIRS (245 of 447 [54.8%]) or sepsis (202 of 447 [45.2%]) | 45.2% | 0.82 | 0.92 | 0.65 | 0.91 | 0.69 |
Definition of abbreviations: AUC = area under the curve; NPV = negative predictive value; PI = principal investigator; PPV = positive predictive value; RPD = retrospective physician diagnosis; SIRS = systemic inflammatory response syndrome.
A SeptiCyte LAB cutoff value of 3.1 was used in the analysis. This value had been obtained previously from receiver operating curve analysis of an independent discovery dataset (4).
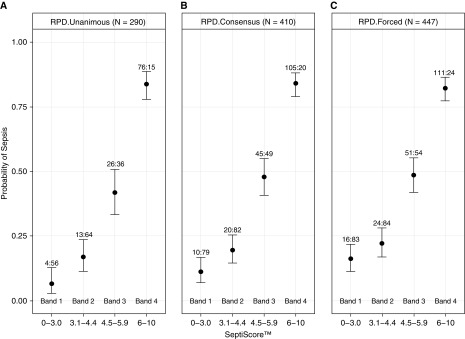
When SeptiScores were organized into numerical bands (band 1, 0–3.0; band 2, 3.1–4.4; band 3, 4.5–5.9; band 4, 6–10), a positive correlation between band number and probability of sepsis was observed (Figure 3). Unanimous sepsis patients more frequently had SeptiScore levels within bands 3 and 4 (86.0%) than patients with SIRS (29.8%) (Figure 3A). A similar distribution of sepsis by consensus and by forced classification was observed for each SeptiScore band, with 83.3% of consensus septic patients and 80.2% of forced sepsis classification falling within bands 3 and 4 (Figures 3B and 3C, respectively). Four patients (3.3%) unanimously determined to have sepsis had a SeptiScore within band 1 and did not have positive blood cultures. Sixty-nine patients with consensus RPD diagnoses of SIRS (30.0% of all patients with SIRS) had SeptiScores falling in bands 3 or 4. Heterogeneity within this subgroup is indicated by detailed analysis of clinical parameters (online supplement, part 7), with some patients showing clinical characteristics consistent with systemic infection or sepsis. SeptiScores were randomly distributed across the four bands for indeterminate cases. For blood culture–positive cases that were not suspected of contamination, no SeptiScores fell in band 1 (assay sensitivity, 1.00; 95% confidence interval, 0.93–1.00), and SeptiScores were heavily skewed toward bands 3 and 4. Additional information about the distribution of indeterminate and blood culture–positive subjects across SeptiScore bands is provided in the online supplement, part 7.
Figure 3.
Positive correlation between SeptiScore and the probability of sepsis. In each panel the probability of sepsis is plotted against SeptiScore for four different SeptiScore ranges (bands). Each box-and-whisker plot indicates the median and the upper and lower 80% confidence interval bounds on sepsis probability for a particular band. The number of sepsis:systemic inflammatory response syndrome (SIRS) subjects in each band is indicated. (A) Unanimous retrospective physician diagnosis (RPD) (171 SIRS, 119 sepsis; 155 excluded). (B) Consensus RPD (230 SIRS; 180 sepsis; 37 indeterminates excluded). (C) Forced RPD (245 SIRS; 202 sepsis).
We excluded subjects who received therapeutic antibiotics before ICU admission, and we reevaluated SeptiCyte LAB performance. The original set of 447 subjects gave an AUC of 0.852 with consensus RPD as the comparator. After removing 139 subjects who had received therapeutic antibiotics before ICU admission, the remaining set of 308 subjects gave an AUC of 0.848. DeLong’s test (17) for comparison of two ROC curves was performed, which demonstrated that there was no significant difference between the two ROC curves (D = 0.108, df = 569.54, P = 0.914).
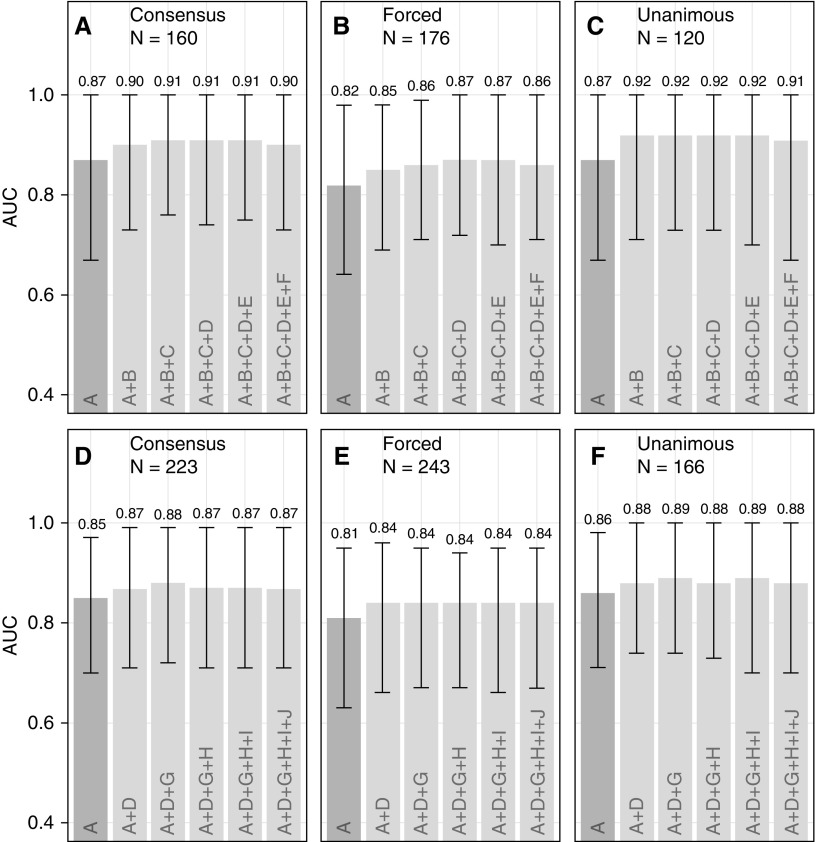
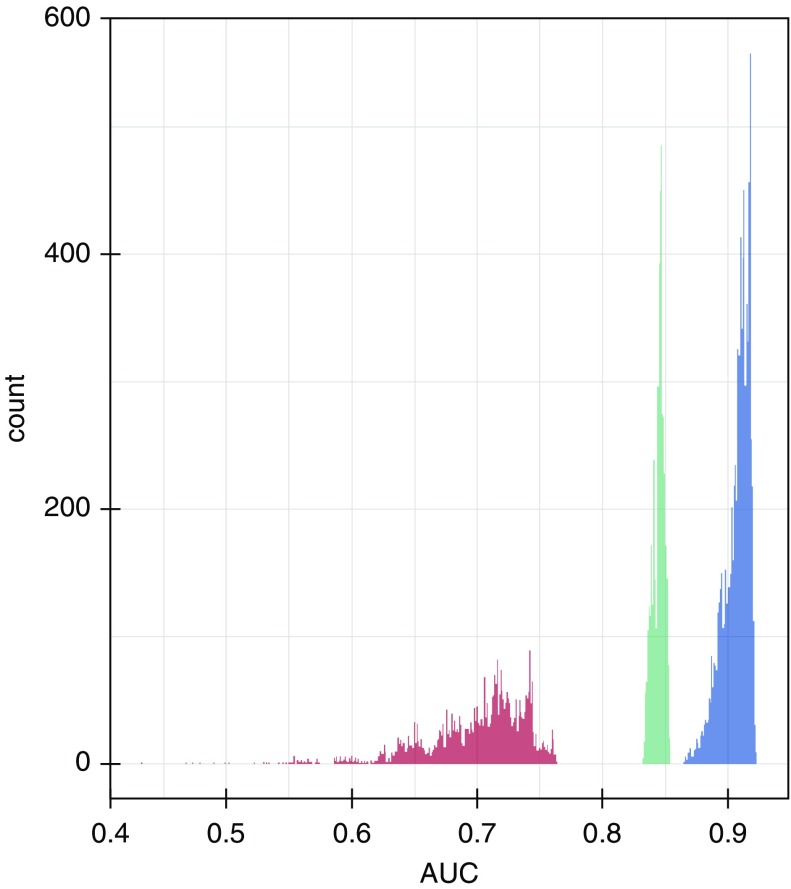
We evaluated SeptiCyte LAB in comparison to or in combination with other demographic, clinical, and laboratory variables, as well as with assessments available within the first 24 hours of ICU admission, for discriminating SIRS from sepsis. For these comparisons, the combined MARS + VENUS + VENUS Supplement cohort was used, with consensus RPD as the comparator. AUC values for individual variables all fell below 0.67, with the exception of PCT at 0.80, as compared with 0.85 for SeptiScore. Correlations between the individual variables and SeptiScore were relatively weak (r2 < 0.33), except for PCT (r2 = 0.46) (see online supplement, part 4). We stratified the data into the three separate cohorts (MARS, VENUS, VENUS Supplement), also by race/ethnicity or sex, and also by identified site of infection, without a significant effect on SeptiCyte LAB performance (online supplement, part 5). We next used forward logistic regression to combine variables in order of decreasing relative univariate contribution. The results from the forward logistic regression were in general agreement with three other multivariate analysis methods: logistic regression with random variable selection, ridge/lasso regression, and random decision forests. In all multivariate analyses, combining SeptiScore with clinical signs and laboratory variables, including PCT, did not significantly improve the overall performance of SeptiCyte LAB compared with its use alone (Figure 4). Taking this analysis further, Figure 5 presents the results from a complete permutation analysis of all 14 variables, resulting in 16,383 independent logistic models, using only complete datasets without missing values imputed (comparator = consensus RPD). Other methods of imputing missing values, and other methods of combining variables, were tested with essentially the same results (online supplement, part 6). In all analyses, adding SeptiScore as a variable significantly improved the discrimination of sepsis versus SIRS relative to classifiers without SeptiScore.
Figure 4.
Forward logistic regression models. No imputation of missing values was performed. (A) Consensus retrospective physician diagnosis (RPD) (n = 160), procalcitonin (PCT) included. (B) Forced RPD (n = 176), PCT included. (C) Unanimous RPD (n = 120), PCT included. For panels A–C, variables were added in the order A, B, C, D, E, F, and area under the curve (AUC) values were recalculated after each addition step. (D) Consensus RPD (n = 223), PCT excluded. (E) Forced RPD (n = 243), PCT excluded. (F) Unanimous RPD (n = 166), PCT excluded. For panels D–F, variables were added in the order A, D, G, H, I, J, and AUC values were recalculated after each addition step. Symbols, consistent with assignments in the online supplement, part 6, are as follows: A = SeptiScore; B = log2PCT; C = maximum white blood cell count; D = number of systemic inflammatory response syndrome criteria; E = minimum core temperature; F = minimum white blood cell count; G = maximum mean arterial pressure; H = race/ethnicity; I = maximum core temperature; J = sex.
Figure 5.
Area under the curve (AUC) distributions for logistic models. An exhaustive examination of all 16,383 possible logistic combinations of the following 14 variables was conducted: SeptiScore, procalcitonin, maximum glucose concentration, minimum white blood cell count, maximum white blood cell count, maximum mean arterial pressure, minimum core temperature, maximum core temperature, minimum heart rate, maximum heart rate, number of systemic inflammatory response syndrome criteria, age, sex, and race/ethnicity. No imputation of missing values was performed. The comparator was consensus retrospective physician diagnosis. Blue = models containing SeptiScore; green = models containing procalcitonin but not SeptiScore; red = models without SeptiScore or procalcitonin.
Discussion
Differential diagnosis of sepsis and SIRS continues to be a diagnostic challenge of practical importance (19–22). We report findings from a prospective, multicenter clinical evaluation of SeptiCyte LAB, an FDA-cleared and previously described four-gene host expression classifier designed to distinguish sepsis from SIRS in adult patients on the first day of ICU admission (4).
Our primary objective was to establish the performance characteristics of SeptiScore in a study with a racially and ethnically diverse patient population of 447 subjects from seven clinical sites in the United States and one clinical site in the Netherlands. We determined the AUC of SeptiCyte LAB to range from 0.82 to 0.89, depending on the degree of confidence in the clinical diagnosis. SeptiCyte LAB had the highest AUC when the clinical RPD was unanimous (0.89) and lowest with forced RPD (0.82). These two reference methods may thus represent upper and lower bounds, respectively, on SeptiCyte LAB performance independent of other clinical data. Performance characteristics in this study are similar to those of our previous discovery and validation cohorts, which were more racially homogeneous (mostly white) (4). Similar performance levels were observed for SeptiCyte LAB, regardless of stratification by study cohort/geographic origin (MARS vs. VENUS vs. VENUS Supplement), by race/ethnicity or sex, or by site of identified infection (online supplement, part 5).
Our secondary objective was to determine how SeptiCyte LAB performance compared with other clinical signs and laboratory variables often used to diagnose sepsis (including PCT) available within 24 hours of ICU admission. Individually, SeptiCyte LAB appeared to outperform these other variables (online supplement, part 4). Combining clinical variables with SeptiScores led to marginal increases in AUC score with or without inclusion of PCT. Performance of the multivariable predictor models varied on the basis of clinical confidence of the reference method. As expected, the highest AUCs were observed with the unanimous RPD and lowest with the forced RPD. We note that the FDA-cleared intended use of SeptiCyte LAB differs from and is complementary to that of PCT. The latter test is cleared for predicting progression from severe sepsis to septic shock, for predicting 28-day mortality, and for managing antibiotic de-escalation.
Our study included ICU patients who fulfilled at least two SIRS criteria. Our sepsis cohort presented with typically diverse sources of sepsis, with pneumonia being the most common infection site. SeptiCyte LAB had the highest AUC and SeptiScores in patients with bloodstream infection. In contrast, SeptiCyte LAB, as with other sepsis biomarkers (23, 24), appeared to perform less well in patients with pneumonia. Another challenging subgroup consisted of patients who had undergone mechanical ventilation. Difficulties in assessing these two overlapping patient subgroups may be due to inherent challenges in diagnosing pneumonia, particularly in ventilated patients (25, 26). In our study, this was indicated by relatively low values of interobserver concordance (Cohen’s kappa values for concordance between RPD panelists fell to 0.2–0.4 for pneumonia patients and 0.6 for ventilated patients, respectively). Of note, low interobserver concordance in the comparator method will lead to high apparent misclassification rates by the index test and thus to underestimation of the true performance of the index test relative to ground truth. With sufficiently low interobserver concordance, differences in apparent performance may be explained by this effect.
We believe our study offers a number of strengths. First, our population likely represents the diversity of patients with sepsis in ICUs encountered at most hospitals, because the study included patients from multiple geographical locations and medical facilities (online supplement, part 1). Second, patient status (sepsis, SIRS, or indeterminate) was assessed by three different methods (unanimous, consensus, or forced RPD). The best (unanimous) and worst (forced) scenarios establish the lower and upper performance bounds, respectively, for SeptiCyte LAB. To our knowledge, this approach to establishing performance bounds has not been described previously. Third, we applied a novel diagnostic method (host response gene expression assay) to a complex clinical problem.
Our study has several important limitations. First, we defined sepsis as SIRS criteria plus infection (without requiring overt organ dysfunction). Although this definition is sensitive to early septic events, it is not a perfect definition and could potentially miss cases of sepsis that occur in the absence of systemic inflammation (27). Second, the consensus RPD process was unable to assign a definitive classification to 8.3% (37 of 447) of patients (indeterminates). Thus, we bracketed the performance estimates by use of unanimous RPD (upper performance bound; many samples rejected) and forced RPD (lower performance bound; no samples rejected). Actual performance of SeptiCyte LAB (with consensus RPD as the comparator) likely will fall between these two bounds.
Third, a significant fraction of subjects (273 of 447 [61%]) had gaps in the clinical data record used in the multivariate analysis, as detailed in Table E13 of the online supplement, part 6. There was significant variability between sites with respect to the completeness of capture of clinical data in case report forms. To partially address the limitation, we conducted the multivariate analyses in two ways: 1) using only those patients with complete clinical data; and 2) using all patients, with imputation of the missing data values by various methods. Combining SeptiScore with other clinical variables (including or excluding PCT) did not significantly improve the performance of SeptiCyte LAB compared with its use alone, regardless of analysis method or exclusion or imputation of missing values.
Fourth, multivariate analysis may introduce or fail to limit bias. The RPD panelists considered all available clinical information in making their assessments, including the values of some of the variables used in the multivariate analysis (detailed in the online supplement, part 3). Thus, we would expect some correlation between the RPDs and the multivariate analysis results. The magnitude of such a correlation cannot be measured under our study design but would be expected to produce overestimates of AUC performance. We found in the multivariate analysis that little gain in AUC performance was produced by serial addition of variables, suggesting either that this type of bias was present or that the variables are truly not uniquely informative. Regardless, the test is not designed to be run independently of clinical variables or clinical judgment. Fifth, SeptiCyte LAB relies solely on host response characteristics, whereas the ideal sepsis biomarker strategy may require a combination of host response and direct pathogen detection (28–32). Finally, band cutoffs for SeptiCyte LAB were chosen to minimize the probability of false-negative results, which could lead to delayed antibiotic therapy and thus adverse consequences for patients. However, we acknowledge that the trade-off (increased false positivity) could lead to unnecessary antibiotic use while clinical workup progresses. Future clinical validation studies may lead to cutoff adjustments or to alternative cutoffs for different intended uses, such as patient stratification. A net benefit analysis (33, 34) of SeptiCyte LAB in comparison to clinical variables with or without PCT is presented in the online supplement, part 8. Additional analysis must await further clinical studies.
Conclusions
We demonstrated that SeptiCyte LAB successfully discriminated patients with sepsis from those with SIRS with good reliability. Performance predictably varied on the basis of degree of clinical diagnostic certainty. When commonly used clinical and laboratory information (including PCT) was added to SeptiCyte LAB, diagnostic accuracy improved only marginally. Future studies are warranted to determine how host gene expression could most effectively be integrated into clinical decision making to ensure that susceptible patients are accurately managed early in the course of disease.
Acknowledgments
Acknowledgment
The authors thank the clinical study coordinators of the VENUS study: Seth Pitts (Johns Hopkins University), Joyce D. Brown (Rush University), Margaret Gavor (Loyola University Medical Center), Ramona Ramdeo (Northwell Healthcare), Leona Wells (Grady Memorial Hospital), Stan Pies, Valerie Aston, and Heather Gallo (Intermountain Medical Center). The authors also thank the laboratory technical staff at Intermountain Medical Center (Murray, UT), Providence Portland Medical Center (Portland, OR), and Asuragen (Austin, TX).
Footnotes
Funded by Immunexpress and also in part by a Commercialisation Australia grant (CAU06263) from the Australian government to Immunexpress.
Author Contributions: Conceived of and designed the study: R.R.M., B.K.L., R.A.B., R.B.B., T.A.S., L.C.M., and A.R.; managed patient recruitment, initial clinical assessment, and clinical data capture: A.R., S.C., R.N., R.R.M., B.K.L., J.P.B., F.R.D., V.K.S., N.R.A., R.B., J.A.G., M.Y., G.P., E.G., M.A., J.P.P., G.S.M., A.M.E., J.A.K., M.N., A.T., S.H., P.M., T.v.d.P., M.J.S., and P.K.K.; generated data and contributed analytical tools: B.P.S., S.B., D.S., L.C.M., K.N., and B.A.F.; analyzed the data: R.R.M., B.K.L., M.L., S.O., R.E.R., B.P.S., P.K.K., A.R., T.A.S., L.C.M., T.D.Y., S.C., D.S., V.R., S.B., J.T.K., K.N., R.F.D., R.B.B., and B.A.F.; wrote the first draft of the manuscript: R.R.M., B.K.L., and T.D.Y.; contributed to the writing of the manuscript: R.R.M., B.K.L., T.D.Y., T.A.S., and R.B.B. All authors agree with the manuscript’s results and conclusions.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201712-2472OC on April 6, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Tsalik EL, Langley RJ, Dinwiddie DL, Miller NA, Yoo B, van Velkinburgh JC, et al. An integrated transcriptome and expressed variant analysis of sepsis survival and death. Genome Med. 2014;6:111. doi: 10.1186/s13073-014-0111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maslove DM, Wong HR. Gene expression profiling in sepsis: timing, tissue, and translational considerations. Trends Mol Med. 2014;20:204–213. doi: 10.1016/j.molmed.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sweeney TE, Shidham A, Wong HR, Khatri P. A comprehensive time-course–based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci Transl Med. 2015;7:287ra271. doi: 10.1126/scitranslmed.aaa5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McHugh L, Seldon TA, Brandon RA, Kirk JT, Rapisarda A, Sutherland AJ, et al. A molecular host response assay to discriminate between sepsis and infection-negative systemic inflammation in critically ill patients: discovery and validation in independent cohorts. PLoS Med. 2015;12:e1001916. doi: 10.1371/journal.pmed.1001916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oved K, Cohen A, Boico O, Navon R, Friedman T, Etshtein L, et al. A novel host-proteome signature for distinguishing between acute bacterial and viral infections. PLoS One. 2015;10:e0120012. doi: 10.1371/journal.pone.0120012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer M, Giamarellos-Bourboulis EJ, Kortgen A, Moller E, Felsmann K, Cavaillon JM, et al. A transcriptomic biomarker to quantify systemic inflammation in sepsis - a prospective multicenter phase II diagnostic study. EBioMedicine. 2016;6:114–125. doi: 10.1016/j.ebiom.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmerman JJ, Sullivan E, Yager TD, Cheng C, Permut L, Cermelli S, et al. Diagnostic accuracy of a host gene expression signature that discriminates clinical severe sepsis syndrome and infection-negative systemic inflammation among critically ill children. Crit Care Med. 2017;45:e418–e425. doi: 10.1097/CCM.0000000000002100. [DOI] [PubMed] [Google Scholar]
- 8.Mahajan P, Kuppermann N, Mejias A, Suarez N, Chaussabel D, Casper TC, et al. Association of RNA biosignatures with bacterial infections in febrile infants aged 60 days or younger. JAMA. 2016;316:846–857. doi: 10.1001/jama.2016.9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herberg JA, Kaforou M, Wright VJ, Shailes H, Eleftherohorinou H, Hoggart CJ, et al. Diagnostic test accuracy of a 2-transcript host RNA signature for discriminating bacterial vs viral infection in febrile children. JAMA. 2016;316:835–845. doi: 10.1001/jama.2016.11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burnham KL, Davenport EE, Radhakrishnan J, Humburg P, Gordon AC, Hutton P, et al. Shared and distinct aspects of the sepsis transcriptomic response to fecal peritonitis and pneumonia. Am J Respir Crit Care Med. 2017;196:328–339. doi: 10.1164/rccm.201608-1685OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.U.S. Food and Drug Administration (FDA) 510(k) Premarket notification K163260. Silver Spring, MD: FDA; 2017 Feb 17 [accessed 2018 Jul 12] Available from: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfPMN/pmn.cfm?ID=K163260.
- 12.Miller R, Lopansri B, McHugh L, Rapisarda A, Seldon T, Burke J. 1002: Validation of a novel host response assay to distinguish SIRS and sepsis in critically ill patients [abstract] Crit Care Med. 2015;43:252. [Google Scholar]
- 13.Miller RR, Lopansri BK, McHugh L, Seldon T, Burke JP. Evaluation of adjudicated sepsis diagnosis and a novel host response assay in the diagnosis of sepsis in critically ill patients [abstract] Am J Respir Crit Care Med. 2016;193:A7631. [Google Scholar]
- 14.Klein Klouwenberg PM, Ong DS, Bos LD, de Beer FM, van Hooijdonk RT, Huson MA, et al. Interobserver agreement of Centers for Disease Control and Prevention criteria for classifying infections in critically ill patients. Crit Care Med. 2013;41:2373–2378. doi: 10.1097/CCM.0b013e3182923712. [DOI] [PubMed] [Google Scholar]
- 15.Hajian-Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J Intern Med. 2013;4:627–635. [PMC free article] [PubMed] [Google Scholar]
- 16.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 18.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ. 2015;351:h5527. doi: 10.1136/bmj.h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ratzinger F, Schuardt M, Eichbichler K, Tsirkinidou I, Bauer M, Haslacher H, et al. Utility of sepsis biomarkers and the infection probability score to discriminate sepsis and systemic inflammatory response syndrome in standard care patients. PLoS One. 2013;8:e82946. doi: 10.1371/journal.pone.0082946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ratzinger F, Haslacher H, Perkmann T, Schmetterer KG, Poeppl W, Mitteregger D, et al. Sepsis biomarkers in neutropaenic systemic inflammatory response syndrome patients on standard care wards. Eur J Clin Invest. 2015;45:815–823. doi: 10.1111/eci.12476. [DOI] [PubMed] [Google Scholar]
- 21.Anand D, Das S, Bhargava S, Srivastava LM, Garg A, Tyagi N, et al. Procalcitonin as a rapid diagnostic biomarker to differentiate between culture-negative bacterial sepsis and systemic inflammatory response syndrome: a prospective, observational, cohort study. J Crit Care. 2015;30:218.e7–218.e12. doi: 10.1016/j.jcrc.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 22.Han JH, Nachamkin I, Coffin SE, Gerber JS, Fuchs B, Garrigan C, et al. Use of a combination biomarker algorithm to identify medical intensive care unit patients with suspected sepsis at very low likelihood of bacterial infection. Antimicrob Agents Chemother. 2015;59:6494–6500. doi: 10.1128/AAC.00958-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Müller F, Christ-Crain M, Bregenzer T, Krause M, Zimmerli W, Mueller B, et al. Procalcitonin levels predict bacteremia in patients with community-acquired pneumonia: a prospective cohort trial. Chest. 2010;138:121–129. doi: 10.1378/chest.09-2920. [DOI] [PubMed] [Google Scholar]
- 24.Pereira JM, Teixeira-Pinto A, Basílio C, Sousa-Dias C, Mergulhão P, Paiva JA. Can we predict pneumococcal bacteremia in patients with severe community-acquired pneumonia? J Crit Care. 2013;28:970–974. doi: 10.1016/j.jcrc.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 25.Craven DE, Hjalmarson KI. Ventilator-associated tracheobronchitis and pneumonia: thinking outside the box. Clin Infect Dis. 2010;51(Suppl 1):S59–S66. doi: 10.1086/653051. [DOI] [PubMed] [Google Scholar]
- 26.Grgurich PE, Hudcova J, Lei Y, Sarwar A, Craven DE. Diagnosis of ventilator-associated pneumonia: controversies and working toward a gold standard. Curr Opin Infect Dis. 2013;26:140–150. doi: 10.1097/QCO.0b013e32835ebbd0. [DOI] [PubMed] [Google Scholar]
- 27.Kaukonen KM, Bailey M, Bellomo R. Systemic inflammatory response syndrome criteria for severe sepsis. N Engl J Med. 2015;372:1629–1638. doi: 10.1056/NEJMoa1415236. [DOI] [PubMed] [Google Scholar]
- 28.Vincent JL. The clinical challenge of sepsis identification and monitoring. PLoS Med. 2016;13:e1002022. doi: 10.1371/journal.pmed.1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tissari P, Zumla A, Tarkka E, Mero S, Savolainen L, Vaara M. Accurate and rapid identification of bacterial species from positive blood cultures with a DNA-based microarray platform: an observational study. Lancet. 2010;375:224–230. doi: 10.1016/S0140-6736(09)61569-5. [DOI] [PubMed] [Google Scholar]
- 30.Yanagihara K, Kitagawa Y, Tomonaga M, Tsukasaki K, Kohno S, Seki M, et al. Evaluation of pathogen detection from clinical samples by real-time polymerase chain reaction using a sepsis pathogen DNA detection kit. Crit Care. 2010;14:R159. doi: 10.1186/cc9234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang SS. Multiplex PCR system for rapid detection of pathogens in patients with presumed sepsis – a systemic review and meta-analysis. PLoS One. 2013;8:e62323. doi: 10.1371/journal.pone.0062323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent JL. Rapid diagnosis of infection in the critically ill, a multicenter study of molecular detection in bloodstream infections, pneumonia, and sterile site infections. Crit Care Med. 2015;43:2283–2291. doi: 10.1097/CCM.0000000000001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rousson V, Zumbrunn T. Decision curve analysis revisited: overall net benefit, relationships to ROC curve analysis, and application to case-control studies. BMC Med Inform Decis Mak. 2011;11:45. doi: 10.1186/1472-6947-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vickers AJ, Van Calster B, Steyerberg EW. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ. 2016;352:i6. doi: 10.1136/bmj.i6. [DOI] [PMC free article] [PubMed] [Google Scholar]