Abstract
Background
Routine testing of baseline EGFR T790M mutation may have important clinical impact but many discordant data have been reported regarding the diagnostic, prognostic and predictive role of this marker. In this study we aimed to assess T790M frequency in 164 untreated EGFR-mutated NSCLCs using methods with different sensitivity as well as to analyze the relationship between baseline T790M mutation status, patient’s clinicopathologic features and tyrosine kinase inhibitors (TKI) treatment outcomes.
Methods
We compared the diagnostic performance, sensitivity and specificity of three methods, namely MALDI-TOF mass spectrometry (MS), Allele-Specific Real Time PCR (AS-PCR), droplet digital PCR (ddPCR). Ultra-deep next generation sequencing (NGS) validation of T790M-mutant NSCLCs was performed using SiRe® panel.
Results
Baseline T790M occurred in 17% of the tumors. Intermediately sensitive techniques such as MALDI-TOF MS (detection limit of T790M ≥5%) allow to detect T790M in 2% of cases exhibiting mutant-allele fractions ranging from 11.5% to 17%. Median overall survival (OS) in these patients was poor (7.3 months) and progression free survival (PFS) was of 3.3 months in patients treated with a 1st generation EGFR TKI. The remaining T790M-positive cases showed very low mutant-allele fractions ranging from 0.07% to 0.38% and required highly sensitive methods such as ddPCR and NGS to be identified. All these cases showed a concurrent sensitizing EGFR mutation (mainly exon 19 deletion), and clinicopathological features similar to those observed in EGFR mutant cancers. Median OS of these patients was 27 months while median PFS after TKI treatment was 20 months.
Conclusions
Routine test of baseline EGFR T790M may have an important role in the prediction to EGFR TKI therapy response and should be performed using highly sensitive and quantitative methods, such as ddPCR and NGS, in order to reliably distinguish NSCLCs with high or very low T790M mutant-allele fraction.
Keywords: EGFR T790M mutation, MALDI-TOF mass spectrometry, real time quantitative PCR (AS-PCR), droplet digital PCR (ddPCR), ultra-deep next generation sequencing (NGS)
Introduction
Acquired resistance to first- and second-generation tyrosine kinase inhibitors (TKIs) represents a hot topic in the personalized medicine of non-small cell lung cancer (NSCLC). The most common resistance mechanism is related to EGFR T790M mutation, which accounts for 50–60% of TKI relapses (1-3). T790M frequently co-exists with sensitizing mutations and several reports suggest that it may be selected during TKI treatment (4-6). The proportion of pre-treatment EGFR T790M-mutant alleles within a tumor may range from a small subclone to clonally dominant and the allelic frequency may have an important role in the prediction to EGFR TKI therapy response (7-11). Currently, the T790M mutation detection rate varies from 0.32% to 78.95% in EGFR-TKI-naïve patients, depending on the sensitivity of methods and on the quality of tumor DNA obtained from Formalin Fixed Paraffin Embedded (FFPE) samples. A recent systematic review of randomized controlled trials and observational studies reporting pretreatment T790M and EGFR-activating mutation, concluded that a careful stratified analysis based on the detection limits of the methods is mandatory to distinguish cases with higher T790M allelic fractions (mAF ≥5%) from those tested positive only using intermediately or highly sensitive techniques (detection limit of T790M <5% and ≥0.01%) (12). Although the clinical implication of baseline T790M mutations is still being debated (13,14), the identification of such EGFR double mutant NSCLCs stratifying cases according to T790M allelic fraction seems to be an important prerequisite to verify the potential predictive and prognostic value of this marker. Currently, the implementation of this test may be also valuable in treatment decision-making after the published results of the FLAURA trial showing efficacy of a third-generation EGFR inhibitors, also in basal setting (15).
In this current study, as a first endpoint, we aim to assess the rate of EGFR T790M in 164 EGFR exon 18–19 and 21 mutated NSCLCs tissue samples collected before TKI treatment, also defining the appropriate method to detect EGFR T790M in the routine clinical practice, comparing the results obtained from MALDI-TOF mass spectrometry (MS), real time quantitative PCR (AS-PCR), droplet digital PCR (ddPCR) and next-generation sequencing (NGS). In addition, as a secondary endpoint, the relationship among the pre-treatment EGFR T790M mutation status, tumor clinico-pathologic features and TKI treatment outcomes was also evaluated.
Methods
Study design
From January 2010 to March 2017 we had prospectively screened a multicenter series of advanced 1,354 NSCLCs to select patients eligible for first/second generation EGFR TKI inhibitors. The tissue specimens included 28% (n=379) surgically resected tumors, 56% (n=758) biopsies and 16% (n=216) cytological specimens. EGFR mutations were identified in 171 NSCLCs (12.6%) by using MassARRAY® System (Agena BioscienceTM, Hamburg, Germany) and Myriapod Lung kit (Diatech Pharmacogenetics, Jesi, Italy) (16). Input DNA for each reaction ranged from 20 to 40 ng. Type of mutations and number of cases showing a single or concurrent EGFR mutations were reported in Table 1. As reported in the study flow chart in Figure 1, patients inclusion criteria were: NSCLC from patients with locally advanced (stage IIIB) or metastatic (stage IV) disease; NSCLC with activating mutation in exons 18, 19, 21 and/or T790M; patients with histologic and/or cytological diagnosis of NSCLC; EGFR TKI naïve patients.
Table 1. EGFR mutations detected by MALDI-TOF MS.
| Mutation type | Cases (n) | Percent |
|---|---|---|
| Screened cases | 1,354 | |
| EGFR mutant cases | 171/1,354 | 12.6 |
| Activating mutations | 163/171 | 95.3 |
| Single mutation | ||
| E709K | 1 | 0.6 |
| G719C | 3 | 1.8 |
| Exon 19 del | 104 | 63.8 |
| L858R | 42 | 25.8 |
| L861Q | 6 | 3.7 |
| Concurrent mutations | ||
| G719A, L861Q | 1 | 0.6 |
| G719A, S768I | 1 | 0.6 |
| Exon 19 del, T790M | 1 | 0.6 |
| Exon 19 del, S768I | 1 | 0.6 |
| L858R, I744M | 1 | 0.6 |
| L858R, T790M | 1 | 0.6 |
| L858R, S768I | 1 | 0.6 |
| Exon 20 mutations | 8/171 | 4.8 |
| T790M | 1 | 0.6 |
| S768I | 1 | 0.6 |
| Ins exon 20 | 6 | 3.5 |
Figure 1.
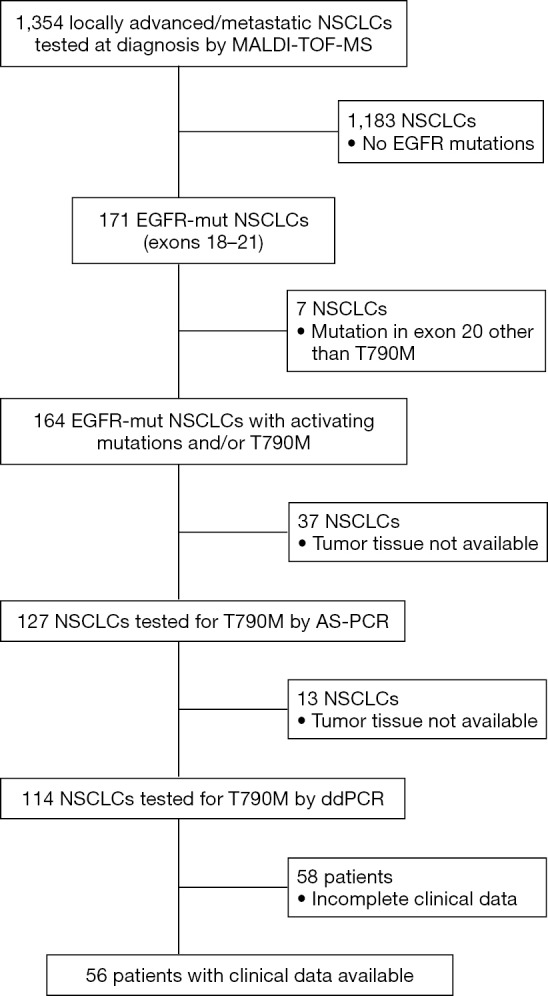
Flow diagram of the study population. NSCLCs, non-small cell lung cancer; AS-PCR, real time quantitative PCR; ddPCR, droplet digital PCR.
On the overall, 164 NSCLCs were included in this study and, prior to proceed with EGFR T790M mutation analysis, were histologically reviewed and classified according to the 2015 World Health Organisation criteria (17) by an experienced pathologist (FF). Clinicopathological features of all tumors are reported in Table 2. As expected, in line with literature data (18,19), our series encompassed more frequently females (68.9%); mean age was 67 years (range, 36–86 years) with only 7% of patients aged <50 years. Histological classification of lung adenocarcinomas was confirmed in all cases but 3 tumors that were diagnosed as squamous cell lung cancer (n=2) or as Mixed adenoneuroendocrine carcinomas (n=1). Grading and assessment of specific histological patterns was done only on surgically resected tumors (n=46, Table 2).
Table 2. Clinicopathological features of EGFR mutant non-small cell lung cancer (NSCLC) included in the study.
| Characteristics | Value |
|---|---|
| Patients | 164 |
| Gender, n (%) | |
| Female | 113 (68.9) |
| Male | 51 (31.1) |
| Age (years) | |
| Mean | 67 |
| Range | 36–86 |
| Specimen type, n (%) | |
| Surgical resection | 46 (28.0) |
| Biopsy | 92 (56.1) |
| Cytological sample | 26 (15.9) |
| Histology, n (%) | |
| Adenocarcinoma | 161 (98.2) |
| Squamous cell lung cancer | 2 (1.2) |
| Mixed adenoneuroendocrine carcinomas | 1 (0.6) |
| Pattern*, n (%) | |
| NOS | 70 (42.7) |
| Papillary | 39 (23.8) |
| Micropapillary | 7 (4.3) |
| Acinar | 32 (19.5) |
| Lepidic | 3 (1.8) |
| Solid | 13 (7.9) |
| Grading*, n (%) | |
| G1 | 8 (4.9) |
| G2 | 115 (70.1) |
| G3 | 41 (25.0) |
*, pattern and grading were evaluated only in surgical resections. NOS, not otherwise specified.
On the overall (n=164 NSCLCs), residual DNA sample or tissue was available to carry out AS-PCR and ddPCR analysis in 127 (77%) and 114 (70%) NSCLCs respectively (Figure 1). In addition, ultra-deep NGS analysis by using a previously validated panel (20,21) was mainly performed, to exclude false-positive results, on AS-PCR and ddPCR T790M-positive samples.
In 56 out of 164 patients, complete clinical data including type and lines of treatment, progression free survival (PFS), time to treatment failure (TTF) and overall survival (OS) were collected (Table S1). All these patients received a first/second generation TKI based therapy. In particular, 48 patients were treated with gefitinib, n=6 with erlotinib and n=2 with afatinib. Thirty-six patients (54%) were treated up-front with a standard EGFR-TKI, while the remaining 20 patients received an EGFR TKI as a second line after chemotherapy.
The study protocol was approved by the Ethics Committee of ASST-Sette Laghi Hospital (Protocol N°158/2016) and the work was undertaken in accordance with the 2013 Helsinki Declaration.
T790M mutation analysis by AS-PCR and ddPCR
AS-PCR was performed starting from 20 ng of input DNA using “Easy® EGFR” kit (Diatech Pharmacogenetics, Jesi, Italy) following the manufacturer’s instructions. To ensure an adequate input of tumor DNA, the minimum Ct of wild-type allele for each sample had to range from 19 to 24. Delta Ct (DCt) was calculated as the difference between the mutation assay Ct and control assay Ct from the same sample. A sample was defined as T790M-mutant if DCt value was equal or below 10.
ddPCR was performed in duplicate for each case on the QX200 droplet digital PCR system (BioRad, Hercules, CA) starting from 50 ng of input DNA/well. Validated assays for both the wild type and the T790M-mutated EGFR were purchased from BioRad and PCR reaction was performed with the ddPCR Supermix for Probes Reagents (BioRad Hercules, CA). Results were analyzed with the QuantaSoft software (Bio-Rad) (Supplementary material).
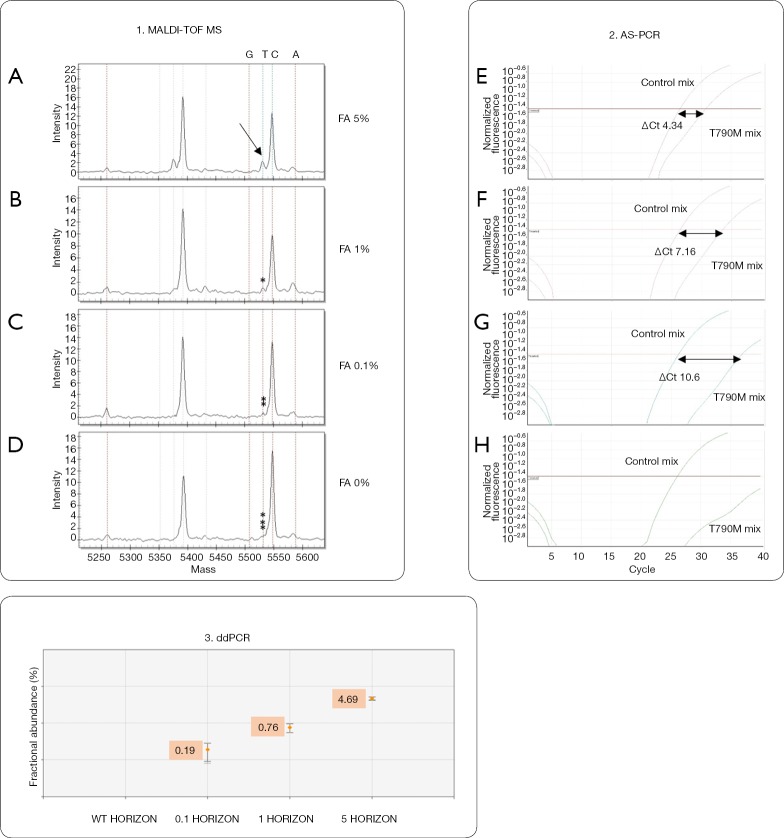
Sensitivity for T790M mutation of MALDI-TOF MS, AS-PCR and ddPCR approaches was previously assessed by determining the limit of detection (LOD) value using the Multiplex I cfDNA Reference Standard Set (Horizon) containing the mutant sequence (EGFR T790M) that was serially diluted with wild type DNA originating from a normal blood sample (mFA: 5%, 1%, 0.1% and 0%) (Figure S1).
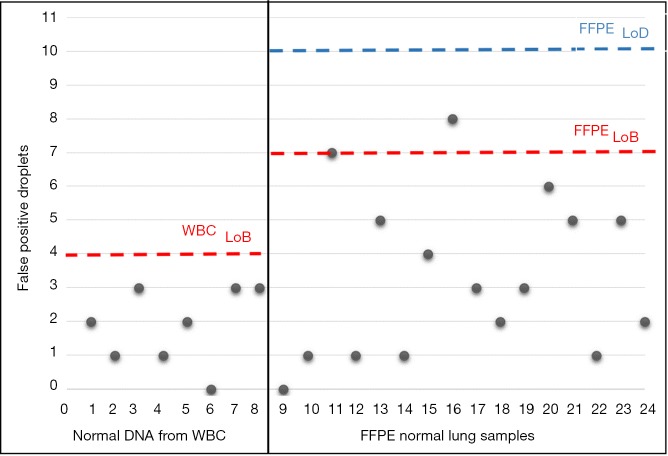
Specificity of AS-PCR and ddPCR was assessed by analyzing 24 mutation-negative controls and, only for ddPCR, determining the limit of blank (LoB) value. Additional details are described in Supplementary material.
NGS validation using SiRe® panel
T790M-mutant NSCLCs obtained with MALDI-TOF MS, AS-PCR and ddPCR were evaluated by SiRe® NGS panel (20,21). Briefly, a total of 10 ng of DNA for each sample were used to produce NGS libraries on the Ion Chef system (Thermofisher). Library was generated by adopting Ion AmpliSeq DL8 (Thermofisher) for amplification step, setting 22 cycles for DNA amplification and 6 cycles for library reamplification after barcoding, following the thermal conditions defined by the manufacturer. Purified libraries were diluted to 60 pM and combined to obtain 16 Ion Code pooled libraries. The two-pooled libraries were re-loaded into the Ion Chef instrument, and templates were prepared using the Ion 510™, 520™ & 530™ Kit Chef (Thermofisher). Finally, templates were loaded into the 510™ chip and sequenced on S5 instrument.
Signal processing and base calling were carried out basing analysis on the default base-caller parameters of Torrent Suite (v.5.0.2). BAM files were visually inspected by using the Golden Helix Genome Browser v.2.0.7 (Bozeman, MT, USA). Coverage analysis was performed using SiRe® designed bed files with coverage plug-in (v.5.0.2.0) and variants were automatically annotated adopting variant caller plug-in (v.5.0.2.1) at specific optimized parameters of the SiRe® panel as previously described. In particular, only variants with >5× allele coverage and a quality score >20, within an amplicon that covered at least 1,000× alleles, were called, and the frequency of each mutant allele was recorded.
Statistical analyses
Fisher exact test, ANOVA analysis, and the independent sample t-test were performed to analyze the relationship between the pretreatment T790M mutation and the patients’ clinico-pathologic features. Kaplan-Meier method and Cox proportional hazard regression were performed to analyze OS, PFS and TTF. PFS was measured from the first day of treatment to the date of first objective or clinical sign of PD, death (whichever came first) or to the date of last follow-up visit for patients censored without progression. TTF was measured from the time from the first day of treatment to treatment discontinuation for any reason, including disease progression, treatment toxicity, patient preference, or death. OS was measured from the first day of TKI treatment to the date of death or the date of last follow-up visit for patients who were still alive.
PFS and TTF were estimated at 24 months (6, 12, 18, 24 months) whereas OS was evaluated at 36 months (6, 12, 18, 24 and 36 months). A P value ≤0.05 was considered statistically significant.
Results
Diagnostic performance of MALDI-TOF MS, AS-PCR and ddPCR for EGFR T790M detection
The diagnostic performance of MALDI-TOF MS, Real Time PCR and ddPCR was assessed in order to identify sensitivity and specificity of each method. Titration experiments using Multiplex I cfDNA Reference Standard Set (Horizon) demonstrated high sensitivity of ddPCR that was able to identify mutated Allelic Fraction (mAF) of 0.1%. With a lower sensitivity, MALDI-TOF MS analysis revealed mutations with mAF of 5%, while Real Time PCR showed a sensitivity of 1% mutated allele (Figure S1).
Assay specificity was assessed for both AS-PCR and ddPCR. As regards AS-PCR we did not find any false-positive results in mutation-negative controls (data not shown). By contrast, ddPCR technique required a careful assay specificity assessment since false-positive droplets have been detected in mutation negative controls, especially with normal lung FFPE tissues (Figure S2). Based on the results obtained on mutation-negative controls, the lowest number of T790M-positive events to reliably distinguish mutant from wild-type samples was set to 10 positive droplets.
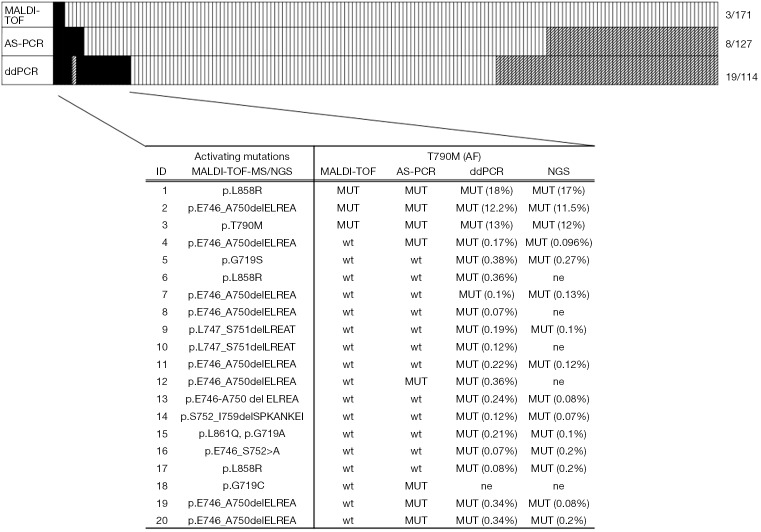
Comparison of the three methods was possible for a total of 113 tumor samples (Figure 2).
Figure 2.
Frequency of EGFR T790M comparing results obtained with MALDI-TOF MS, AS-PCR and ddPCR. T790M alone was observed only in case 3 (AF>10%) while the remaining 19 NSCLCs showed the coexistence of T790M with an activating mutation in exon 18, 19 or 21. T790M allelic fraction ranged from 0.07% to 0.38% in the tumors tested negative for T790M using MALDI-TOF-MS. NGS confirmed all the sensitizing mutations detected by MALDI-TOF MS and validated the presence of T790M in 15 out of 16 mutation-positive NSCLCs, reporting mAFs very close to those quantified by ddPCR. ne: not evaluable.
EGFR T790M was detected in 2% (3/171) of NSCLCs by MALDI-TOF MS and no germline T790M mutation was observed. When using more sensitive methods such as AS-PCR and ddPCR the detection rate increased to 6% (8/127) and 17% (19/114), respectively (Figure 2). Only three cases, i.e., those identified also by MALDI-TOF MS, had mAFs greater than 10%, while 17 cases showed mutation frequencies ranging from 0.07% to 0.38% and were identified by AS-PCR or more often by ddPCR. T790M alone was observed in only one case, while the remaining 19 NSCLCs showed the coexistence of T790M with exon 19 deletion (13 cases) or more rarely with mutations in exon 21 (4 cases) or in exon 18 (2 cases).
NGS validation of T790M-mutant NSCLCs
As reported in Figure 2, NGS was possible in 15 out of 20 T790M mutation-positive NSCLCs after evaluation by ddPCR and/or by MALDI-TOF MS and AS-PCR. All NGS analyzed samples resulted adequate for data interpretation. In particular, NGS analysis showed an average of 464,240.14 reads per sample with a median read length of 155.20 bp. The mean number of mapped reads per sample was 218,492.41. with 92.40% reads on selected targets. Regarding the coverage, each amplicon showed an average read of 4,372.40 and a uniformity distribution on the 42 amplicons of 93.20%. NGS confirmed all the sensitizing mutations detected by MALDI-TOF MS and allowed to characterize the uncommon in-frame complex deletion p.S752_I759delSPKANKEI in case N°14. In addition, NGS confirmed the presence of T790M in all mutation-positive NSCLCs reporting mAFs very close to those quantified by ddPCR (Figure 2).
Correlation analysis with clinicopathologic features
Fisher exact test, ANOVA analysis, and the independent sample t-test were performed to analyze the relationship between the pretreatment T790M mutation and the patients’ clinicopathologic features. However, no association was found between T790M mutation status and any of the clinicopathological features examined (Table S1).
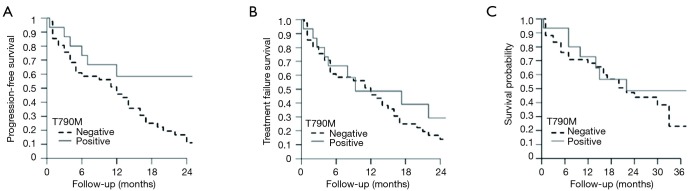
Kaplan-Meier method and Cox proportional hazard regression were performed to analyze the impact of pretreatment T790M mutation on OS, PFS, TTF of 56 patients that had been treated with 1st or 2nd generation TKIs. In these analyses two groups of patients were compared: 15 cases with T790M detected with at least one of the three methods used (Figure 2) and 41 patients with NSCLCs showing negativity for T790M with all the three tests.
As reported in Figure 3, T790M mutation was associated with longer PFS with a 60% decrease in the hazard of mortality after 24 months compared with absence of T790M (HR =0.40; 95% CI, 0.17–0.95, P=0.04). Similar results were obtained for TTF, although this finding was not statistically significant (HR =0.49; 95% CI, 0.22–1.11; P=0.09).
Figure 3.
Progression-free survival (A), time to treatment failure (B) and overall survival (C) at fixed times during follow-up according to EGFR T790M status.
Considering OS, T790M mutation was associated with a 30% decrease mortality at 36 months compared with absence of p.T790M, but this result was not statistically significant (HR =0.70; 95% CI, 0.30–1.63; P=0.4) (Figure 3).
Discussion
Routine testing of baseline EGFR T790M stratifying cases according to mAF may have important clinical implications and also clarify discordant data regarding the diagnostic, prognostic and predictive role of this marker. Currently, the needs are to define the frequency of baseline EGFR T790M, but also the most appropriate detection strategy in the clinical practice, considering accuracy, sensitivity and robustness.
Our study clearly demonstrates that a routine use of intermediately sensitive techniques such as MALDI-TOF MS (detection limit of T790M ≥5%) allows to detect T790M in about 2% (3 of 171 patients) of EGFR-TKI-naïve NSCLCs, a much lower frequency than that reported in Asiatic population (27 of 107 patients, 25.2%) using the same methodology (10). However, this frequency is very similar to those found in Caucasian population by means of low-intermediate sensitivity systems (12,13). NGS enabled to validate the T790M results detected by MALDI-TOF MS and to accurately quantify mAF in order to identify rare cases with germline T790M mutation associated with familial NSCLC (22) and also to dose mAF of both T790M and the concurrent EGFR sensitizing mutation. In our study, we did not identify any germline mutation since the tumors exhibited T790M mAF ranging from 11.5% to 17%. One case showed T790M alone while the other two NSCLCs had a concurrent exon 19 deletion (mAF: 40%) or L858R mutation (mAF: 29%). Overall survival in all these three patients was poor with median OS of 7.3 months. Moreover, the two patients with concurrent sensitizing mutations were treated with a first generation EGFR TKI but showed no response to the treatment, with a PFS of 3.3 months. These results are in line with several studies reporting a shorter PFS in advanced NSCLC patients harboring both the EGFR-activating and T790M mutations who were treated with first or second generation EGFR-TKI (8,10,11,23). Notably, in all these previous studies EGFR tests was performed using Sanger sequencing and/or MALDI-TOF MS. These data together with our results suggest that T790M mAF higher than 5% may be an appropriate threshold to identify TKI-naïve patients that may specifically benefit from a first-line treatment with third-generation EGFR inhibitors (24,25). This observation appears to be of great interest after the recent published results of the FLAURA trial showing efficacy of a third-generation EGFR inhibitors also in basal setting (15).
The remaining T790M-positive cases (17 out of 20) showed very low mAFs ranging from 0.07% to 0.38% and required highly sensitive methods such as ddPCR and NGS to be identified. As expected, ddPCR was confirmed to be both more sensitive and more reliable than AS-PCR. However, a careful specificity assessment of ddPCR assay was mandatory since the frequency of false-positive droplets was not negligible in normal controls, especially when using low quality DNA samples from FFPE tissues. Also in this subset of tumors, NGS was very useful to validate ddPCR results confirming a high correlation between the two methods considering both T790M detection and mAF quantitation.
Globally in this study, we demonstrated that before EGFR TKI treatment, T790M occurs in 17% of cases and this frequency is not significantly different from those reported in Caucasian population using NGS or ddPCR (4,26). By contrast it is much lower than that reported in Asiatic population by means of highly sensitive EGFR test (27), suggesting that not only the sensitivity of the diagnostic methods, but also patient population tested should be considered to understand many inconsistent data reported in literature about the frequency of baseline EGFR T790M.
In our series, NSCLCs with very low T790M mAFs had always a concurrent sensitizing EGFR mutation and the clinico-pathological characteristics of these NSCLCs are very similar to those observed in tumors harboring only sensitizing EGFR mutations, especially exon 19 deletion (28). Actually, we observed this mutation in 70% of cases, while only 14% and 11% of tumors exhibited L858R or G719C/S, respectively. Clinically, mean OS of these patients was 27 months while mean PFS after TKI treatment was 20 months. As these tumors represented the vast majority of the T790M positive cases (85% of cases), T790M mutation resulted associated with longer PFS with a 60% decrease in the hazard of mortality after 24 months compared with absence of T790M (P=0.04).
In conclusion, our study demonstrates that routine test of baseline EGFR T790M should be performed using highly sensitive and quantitative methods, such as ddPCR and NGS, in order to reliably distinguish NSCLCs with high (≥5%) or very low T790M mAF (<5% and ≥0.01%). The first group encompasses patients with a distinct clinical course, they should be addressed separately from other EGFR-mutant lung cancers since they may be more likely to gain profit from a third generation TKI as first-line regimen. This condition is rarely observed among Caucasian patients, seen in about 2% of pre-treatment EGFR positive NSCLCs.
By contrast, the clinicopathological characteristics of the second subset of NSCLCs are very similar to those observed in tumors harboring only sensitizing EGFR mutations. In our series, very low T790M allele content was mainly observed with exon 19 deletion. These results are interesting considering previous data reporting the same preferential association among patients with acquired resistance to first/second EGFR TKI and support the idea of a “clonal selection” of T790M during these treatments.
Acknowledgments
We thank Alice Ballerio, Micol Gilardoni and Remo Bedolis for their helpful support in clinical data management.
Funding: This study was supported by Astra Zeneca (ESR-16-11794 to D Furlan).
Supplementary
Droplet digital PCR analysis
ddPCR, measurement results of single PCR wells were excluded in case that the total number of accepted droplets was <10,000 and QuantaSoft Software returns “No Call” for wells with Quality Scores below 0.85 (Biorad ddPCR application guide https://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_6407.pdf). In our study, the starting DNA input for ddPCR analysis has been defined considering that the LOD is primarily determined by the number of wild-type molecules that are screened. As in an ideal assay (with a droplet false-positive rate of zero) a LOD of 0.1% is guaranteed using three times the number of expected wt molecules (i.e., 3,000 copies of haploid genomes that correspond to a minimum amount of 10 ng of DNA input) we adjusted the starting DNA input to 50 ng DNA/well and we loaded each case in duplicate.
Specificity of AS-PCR and ddPCR
To determine specificity level of AS-PCR and ddPCR we selected a total of 24 T790M-mutation negative controls. Eight were peripheral blood samples while 16 were FFPE histologically-normal lung tissue samples, which were eight from non-neoplastic patients and eight adjacent to lung neoplasia.
In AS-PCR we did not observe any positivity for T790M mutation in the tested control samples.
On the contrary, ddPCR required a careful evaluation of aspecific calls likely due to fixation artifacts. Thus, we evaluated the limit of blank (LoB), defined as the higher number of false-positive droplets measured in wild-type control DNA. Each sample was analyzed in eight replicates, using 50 ng of DNA input for each. The number of false-positive droplets was from 0 to 3 in the subset of blood samples while we recorded from 0 to 8 positive droplets in FFPE samples. Thus, we set the limit of detection for FFPE ddPCR assay at 10 positive droplets.
Figure S1.
Sensitivity of MALDI-TOF MS, AS-PCR and ddPCR evaluated by serial dilutions of control reference DNA. 1: MS of EGFR T790M assay at fractional abundance (FA) of 5% (A), 1% (B), 0.1% (C) and wild type (D). Only the 5% mutated sample was identified as positive (arrow in A). 2: the same analysis in real time PCR identified as positive only samples with 5% and 1% mutation (E and F, respectively) while samples with 0.1% (G) and 0% (H) were scored as negative. 3: ddPCR identified and quantified all the mutated samples, down to a FA of 0.1%.
Figure S2.
Scatter plot showing the number of false positive droplets for the EGFR T790M assay in ddPCR in wild type controls from WBC and from FFPE normal lung samples. Limit of blank (LoB) for whole blood cells (WBC) and Formalin Fixed Paraffin Embedded (FFPE) samples were respectively 4 and 7 droplets, as shown in red on the graph. We set the limit of detection (LOD) for FFPE ddPCR assay at 10 positive droplets.
Table S1. Non-small cell lung cancer (NSCLC) clinicopathological features of the 56 patients with clinical data available.
| Characteristics | T790M positive (n=15) | T790M negative (n=41) |
|---|---|---|
| Gender | ||
| Female | 6 (40%) | 28 (68%) |
| Male | 9 (60%) | 13 (32%) |
| Age (years) | ||
| Mean | 68 | 67 |
| Range | 52–77 | 36–84 |
| Specimen type | ||
| Surgical resection | 8 (53%) | 9 (22%) |
| Biopsy | 7 (47%) | 26 (63%) |
| Cytological sample | 0 | 6 (15%) |
| Histology | ||
| Adenocarcinoma | 15 (100%) | 41 (100%) |
| Squamous cell lung cancer | 0 | 0 |
| Mixed adenoneuroendocrine carcinomas | 0 | 0 |
| Pattern* | ||
| NOS | 3 (38%) | 0 |
| Papillary | 3 (38%) | 2 (22%) |
| Micropapillary | 0 | 1 (11%) |
| Acinar | 1 (12%) | 5 (56%) |
| Lepidic | 1 (12%) | 0 |
| Solid | 0 | 1 (11%) |
| Grading* | ||
| G1 | 1 (12%) | 1 (11%) |
| G2 | 7 (88%) | 3 (33%) |
| G3 | 0 | 5 (56%) |
*, pattern and grading were evaluated only in surgical resections.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study protocol was approved by the Ethics Committee of ASST-Sette Laghi Hospital (Protocol N°158/2016) and the work was undertaken in accordance with the 2013 Helsinki Declaration.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Arcila ME, Oxnard GR, Nafa K, et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res 2011;17:1169-80. 10.1158/1078-0432.CCR-10-2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. 10.1126/scitranslmed.3002003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. 10.1158/1078-0432.CCR-12-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosell R, Molina MA, Costa C, et al. Pretreatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin Cancer Res 2011;17:1160-8. 10.1158/1078-0432.CCR-10-2158 [DOI] [PubMed] [Google Scholar]
- 5.Sequist LV, Lynch TJ. EGFR tyrosine kinase inhibitors in lung cancer: an evolving story. Annu Rev Med 2008;59:429-42. 10.1146/annurev.med.59.090506.202405 [DOI] [PubMed] [Google Scholar]
- 6.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol 2008;26:2442-9. 10.1200/JCO.2007.14.8494 [DOI] [PubMed] [Google Scholar]
- 7.Costa C, Molina MA, Drozdowskyj A, et al. The impact of EGFR T790M mutations and BIM mRNA expression on outcome in patients with EGFR-mutant NSCLC treated with erlotinib or chemotherapy in the randomized phase III EURTAC trial. Clin Cancer Res 2014;20:2001-10. 10.1158/1078-0432.CCR-13-2233 [DOI] [PubMed] [Google Scholar]
- 8.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 2008;359:366-77. 10.1056/NEJMoa0800668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosell R, Dafni U, Felip E, et al. Erlotinib and bevacizumab in patients with advanced non-small-cell lung cancer and activating EGFR mutations (BELIEF): an international, multicentre, single-arm, phase 2 trial. Lancet Respir Med 2017;5:435-44. 10.1016/S2213-2600(17)30129-7 [DOI] [PubMed] [Google Scholar]
- 10.Su KY, Chen HY, Li KC, et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol 2012;30:433-40. 10.1200/JCO.2011.38.3224 [DOI] [PubMed] [Google Scholar]
- 11.Wu JY, Yu CJ, Chang YC, et al. Effectiveness of tyrosine kinase inhibitors on "uncommon" epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res 2011;17:3812-21. 10.1158/1078-0432.CCR-10-3408 [DOI] [PubMed] [Google Scholar]
- 12.Chen LY, Molina-Vila MA, Ruan SY, et al. Coexistence of EGFR T790M mutation and common activating mutations in pretreatment non-small cell lung cancer: A systematic review and meta-analysis. Lung Cancer 2016;94:46-53. 10.1016/j.lungcan.2016.01.019 [DOI] [PubMed] [Google Scholar]
- 13.Yu HA, Arcila ME, Hellmann MD, et al. Poor response to erlotinib in patients with tumors containing baseline EGFR T790M mutations found by routine clinical molecular testing. Ann Oncol 2014;25:423-8. 10.1093/annonc/mdt573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu HA, Riely GJ, Lovly CM. Therapeutic strategies utilized in the setting of acquired resistance to EGFR tyrosine kinase inhibitors. Clin Cancer Res 2014;20:5898-907. 10.1158/1078-0432.CCR-13-2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. 10.1056/NEJMoa1713137 [DOI] [PubMed] [Google Scholar]
- 16.Imperatori A, Sahnane N, Rotolo N, et al. LINE-1 hypomethylation is associated to specific clinico-pathological features in Stage I non-small cell lung cancer. Lung Cancer 2017;108:83-9. 10.1016/j.lungcan.2017.03.003 [DOI] [PubMed] [Google Scholar]
- 17.Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. 10.1097/JTO.0000000000000630 [DOI] [PubMed] [Google Scholar]
- 18.Pham D, Kris MG, Riely GJ, et al. Use of cigarette-smoking history to estimate the likelihood of mutations in epidermal growth factor receptor gene exons 19 and 21 in lung adenocarcinomas. J Clin Oncol 2006;24:1700-4. 10.1200/JCO.2005.04.3224 [DOI] [PubMed] [Google Scholar]
- 19.Tseng CH, Chiang CJ, Tseng JS, et al. EGFR mutation, smoking, and gender in advanced lung adenocarcinoma. Oncotarget 2017;8:98384-93. 10.18632/oncotarget.21842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malapelle U, Mayo de-Las-Casas C, Rocco D, et al. Development of a gene panel for next-generation sequencing of clinically relevant mutations in cell-free DNA from cancer patients. Br J Cancer 2017;116:802-10. 10.1038/bjc.2017.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pepe F, De Luca C, Smeraglio R, et al. Performance analysis of SiRe next-generation sequencing panel in diagnostic setting: focus on NSCLC routine samples. J Clin Pathol 2019;72:38-45. 10.1136/jclinpath-2018-205386 [DOI] [PubMed] [Google Scholar]
- 22.Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet 2005;37:1315-6. 10.1038/ng1671 [DOI] [PubMed] [Google Scholar]
- 23.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. 10.1016/S1470-2045(11)70393-X [DOI] [PubMed] [Google Scholar]
- 24.Liao BC, Lin CC, Lee JH, et al. Optimal management of EGFR-mutant non-small cell lung cancer with disease progression on first-line tyrosine kinase inhibitor therapy. Lung Cancer 2017;110:7-13. 10.1016/j.lungcan.2017.05.009 [DOI] [PubMed] [Google Scholar]
- 25.Ricciuti B, Chiari R. First line osimertinib for the treatment of patients with advanced EGFR-mutant NSCLC. Transl Lung Cancer Res 2018;7:S127-30. 10.21037/tlcr.2018.03.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prediletto EV, R. Bruno, E. Macerola, G. et al. Pre-treatment EGFR-T790M subclones in lung adenocarcinoma harboring activating mutation of EGFR: a positive prognostic factor for survival? Ann Oncol 2018;29:viii493-547.
- 27.Fujita Y, Suda K, Kimura H, et al. Highly sensitive detection of EGFR T790M mutation using colony hybridization predicts favorable prognosis of patients with lung cancer harboring activating EGFR mutation. J Thorac Oncol 2012;7:1640-4. 10.1097/JTO.0b013e3182653d7f [DOI] [PubMed] [Google Scholar]
- 28.Liang H, Pan Z, Wang W, et al. The alteration of T790M between 19 del and L858R in NSCLC in the course of EGFR-TKIs therapy: a literature-based pooled analysis. J Thorac Dis 2018;10:2311-20. 10.21037/jtd.2018.03.150 [DOI] [PMC free article] [PubMed] [Google Scholar]