Worldwide, approximately 1.1 billion people are smokers and more than 7 million people die from the negative effects of smoking every year (WHO report, 2017). One of the main natural ingredients causing dependence on tobacco is nicotine. Tobacco with a lowered nicotine content could help people to overcome their nicotine addiction. Nicotine‐free (or nicotine reduced) cigarettes may contribute to reduce the number of smokers and nicotine consumption, thus reducing the risk of death from tobacco use. Most genes involved in the nicotine biosynthesis in tobacco are known and well characterized (Dewey and Xie, 2013). This opens the possibility to employ genetic engineering approaches to alter the alkaloid content of the plant, and in particular to reduce the nicotine content. Nicotine itself is composed of a pyrrolidine and a pyridine ring, which are synthesized in independent pathways (Figure 1a). Recent approaches dealt with the silencing of upper pathway genes encoding the putrescine N‐methyltransferase (PMT) or A622, a phosphatidylinositol‐4‐phosphate (PIP)‐family member of NADPH reductases. The applied RNA silencing methods either resulted in the increased biosynthesis of other alkaloids like anatabine (i.e. Wang et al., 2009) or were only successful in hairy root cultures and BY‐2 cells, but not in whole plants (Kajikawa et al., 2009). The final oxidation step in the biosynthesis of nicotine, as well as anatabine and anabasine, is proposed to be catalysed by flavoproteins of the berberine bridge enzyme‐like (BBL) family (Kajikawa et al., 2011). The knockdown of the three most highly expressed BBL genes (BBLa–BBLc) by RNAi or the knockout with EMS‐induced mutations resulted in a reduction of the nicotine content without increasing the content of other alkaloids (Kajikawa et al., 2011; Lewis et al., 2015). Recently, the BBL gene family in tobacco was expanded by the identification of BBLd.2 and BBLe, leading to six known isoforms (Kajikawa et al., 2017). Thus, the simultaneous knockout of these BBL genes is a promising approach to generate a nicotine‐free tobacco plant.
Figure 1.
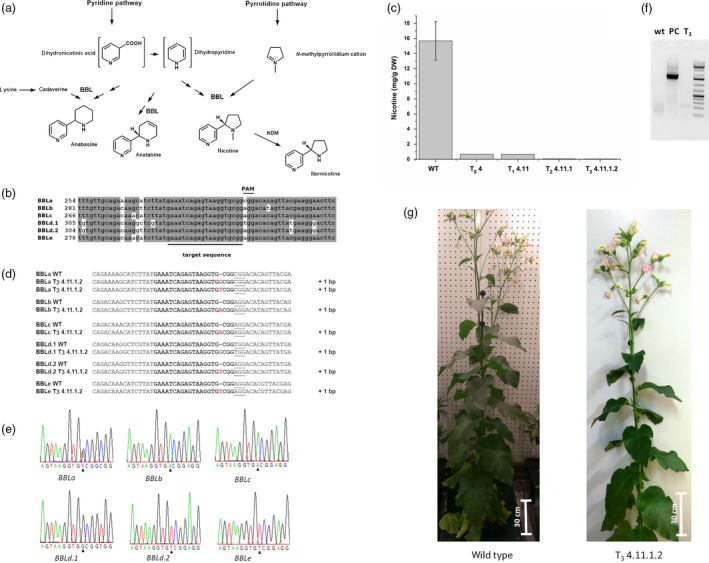
Nicotine‐free, nontransgenic Nicotiana tabacum l. edited by CRISPR‐Cas9. (a) Alkaloid biosynthesis pathway:Alkaloid biosynthesis consists of two independent pathways, the pyrrolidine and a pyridine pathway. Enzymes of the BBL gene family are proposed to be involved in the final oxidation step for the formation of tobacco alkaloids. (b) Alignment of all BBL gene family members: the six gene sequences of the BBL gene family were aligned and possible gene targets were evaluated resulting in a single target sequence identical in all six genes. (c) Comparison of nicotine content of plant 4 in all generations: The amount of nicotine was quantified with GC‐FID from 200 mg of dried and grounded leaf material of plants extracted with MTBE. Nicotine content was calculated as mg per gram dry weight (DW). (d) Genomic DNA was isolated from the plant T3 4.11.1.2 and a wild‐type plant for the amplification of fragments of the six BBL genes; fragments were cloned into a vector for easier sequencing. One base pair insertion was observed in all six genes; for BBLa, two different base pair insertions could be observed. (e) Sequencing of fragments of genomic DNA of plant T3 4.11.1.2 without cloning into a vector to verify the results obtained from vector sequencing. (f) Amplification of the T‐DNA cassette to test whether plant T3 4.11.1.2 (T3) is still transgenic. As a positive control (PC), a T0 plant was used, as a negative control the wild type (wt). (g) Phenotype of a wild‐type plant and the nicotine‐free plant (T3 4.11.1.2).
We aimed for a simple CRISPR Cas9‐based knockout strategy and searched for an identical target sequence present in all six published coding sequences (BBLa, BBLc, BBLd.2 originated from Nicotiana sylvestris; BBLb, BBLd.1, BBLe from Nicotiana tomentosiformis; Kajikawa et al., 2011, 2017) to enable the use of a single‐guide RNA. Except for the PAM sequence, the chosen target sequence is identical in all six BBL sequences (Figure 1b). Mismatches to other sequences of the N. tabacum genome were excluded by BLAST search. We cloned the 20 base pair target sequence between the ubiquitin 6‐26 promoter from Arabidopsis thaliana and the chimeric sgRNA. The gene cassette was subsequently transferred into the transformation vector pCas9‐TPC carrying a bar‐gene as selection marker (Fauser et al., 2014).
After transformation in Nicotiana tabacum l. plants ‘Virginia Smoking Tobacco’ (Strictly Medicinal Seeds LLC, United States), we regenerated ten plants, denoted them as T0 generation and analysed the plants with regard to their nicotine content. Extraction of alkaloids was done from grounded leaves, and nicotine levels were analysed with GC‐FID. The change in nicotine levels ranged from unchanged (T0 5) over a medium reduction of 65% (T0 3) to a reduction of around 95% (T0 1 and T0 4) compared to the wild type, indicating that not all BBL loci were knocked out. Sequencing the fragments of genomic DNA from T0 1 and T0 4 plants showed no editing of BBLe, whereas all other BBL genes were mutated. The T0 plants 1, 3 and 4 were chosen for further characterizations in following generations. Rooted plantlets were cultivated in a plant chamber for self‐pollination in order to produce T1 seeds. To enable further gene editing with CRISPR Cas9, we selected transgenic T1 plants with phosphinothricin (PPT). Transgenic T1 plants were cultivated until flowering and T2 plants were grown from collected seeds. This growing cycle was continued to obtain T3 plants. While the nicotine content in plant T1 1.2 did not decrease further, a decrease in nicotine content by 95% was observed for plant T1 3.1. The nicotine level of plant T1 4.11 was as low as in the T0 generation.
In order to identify nicotine‐free tobacco plants carrying knockouts in all six BBL genes, progenies of plant T0 4 up to generation T3 were screened initially with regard to their nicotine content. The GC analysis of the nicotine content of the analysed T2 and T3 plants resulted in minimal peaks with retention times identical to the nicotine standard. To ensure the correct identification of the peak as nicotine, a GC‐MS measurement was performed. A m/z of 162.23 identical to nicotine was detected with a signal‐to‐noise ratio intensity of nearly 1:1. Since the signal‐to‐noise ratio of the peaks was too low for an automated peak detection, a manual analysis of the peak area was performed for estimation of the residual nicotine content. It was calculated to 0.06 mg g per DW nicotine in the T2 4.11.1 and to 0.04 mg g per DW nicotine in the T3 4.11.1.2 plant (Figure 1c), which means a reduction of 99.6% and 99.7%, respectively, compared to the wild type. Based on these results, the plant T3 4.11.1.2 was considered as nicotine‐free.
Finally, to check whether all 12 loci were knocked out by our approach, a PCR‐based method was applied. The combination of a vector‐based cloning strategy for the amplicons followed by Sanger sequencing with the direct sequencing of the PCR product renders whole genome sequencing unnecessary. The precondition for that approach was an insertion of a single nucleotide three base pairs upstream of the PAM sequence resulting in a frameshift, that is a knockout. In contrast, in Arabidopsis and rice, insertions or deletions of several base pairs were observed after nonhomologous end joining (NHEJ) events (Jiang et al., 2013). However, for BBLa, sequencing results showed either the insertion of the base guanine or thymine, whereas sequencing results of the other BBL genes always showed the same base pair insertion (Figure 1d). To confirm these results, fragments of the BBL genes were amplified and sequenced directly. The previous results were confirmed by analysis of the sequencing trace of the samples (Figure 1e). Except from BBLa, the sequencing trace of the fragments showed a distinct signal peak for the appropriate base insertion. For BBLa, the sequencing trace showed a double peak for thymine and guanine, which is consistent with the results from the cloning experiments, in which both base pair insertions were detected.
To prove that the nicotine‐free plant is nontransgenic, leaf discs were cut out and transferred, after surface sterilization, to MS medium with PPT for selection. As control, a transgenic T1 generation plant was used. After 2 weeks, leaf discs of the T1 plant were still green and even started to grow, whereas the leaf discs of the nicotine‐free plant died. This result was additionally confirmed with PCR using primers that bind inside the transformation cassette thereby spanning the terminator of Cas9 and the PPT gene (Figure 1f). As a positive control, genomic DNA from a T0 plant was used. Neither for the wild type nor for the tested nicotine‐free plant a PCR product was obtained. Thus, the nicotine‐free plant was declared as nontransgenic.
Analysis of the GC measurements showed that the alkaloids anatabine, nornicotine and anabasine were present in wild‐type extracts, but the latter two at the detection limit. The chromatogram of the nicotine‐free plant showed again traces of anabasine and nornicotine, as well as a reduced peak area for anatabine. Additionally, 1H‐NMR measurements verified that no substantial changes in the primary metabolism occurred, showing that the complete knockout of the BBL gene family had no negative impact on other biosynthetic pathways using the tested growth conditions. Finally, no changes in the phenotype were observed (Figure 1g).
Conclusion
The generation of a nicotine‐free and nontransgenic tobacco plant enables the introgression of this plant into other tobacco varieties and the reduction of overall nicotine and alkaloid content. Furthermore, the use of a single gRNA for the complete knockout of all BBL‐relevant genes makes it easy to apply this method to other tobacco varieties and species, for example Nicotiana benthamiana that are commonly used as heterologous production platforms. This will expand the production spectrum of N. benthamiana to the biotechnological production of plant‐made pharmaceuticals beyond antibodies.
Author contributions
F.S. conceived the study. F.S and J.S designed the experiments and wrote the manuscript. J.S. performed the experiments. All authors read and approved the final manuscript.
Conflict of interest
The authors declare no competing financial interests.
Acknowledgements
We are especially grateful to Oliver Kayser for great encouragement, fruitful discussions and financial support. We thank Holger Puchta (University of Karlsruhe, Germany) for providing us with CRISPR‐Cas9 plasmids and Heribert Warzecha (University of Darmstadt, Germany) for providing the A. tumefaciens strain. We thank Gabriele Hardes and Michael Kubicki (INFU; TU Dortmund University, Germany) for assistance with GC‐MS measurements. We are also grateful to Chantale Zammarelli for excellent technical assistance with NMR sample preparation and GC measurements, as well as Angela Sester for helping with NMR data analysis and Patty Krabbe and Jan Volmer for their help with GC devices. The authors thank Magdalena Thoma and Fabio Alvarado for their assistance in revising the English text. We acknowledge financial support by Technische Universität Dortmund/TU Dortmund University for Open Access Publishing.
References
- Dewey, R.E. and Xie, J. (2013) Molecular genetics of alkaloid biosynthesis in Nicotiana tabacum . Phytochemistry, 94, 10–27. [DOI] [PubMed] [Google Scholar]
- Fauser, F. , Schiml, S. and Puchta, H. (2014) Both CRISPR/Cas‐based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana . Plant J. 79, 348–359. [DOI] [PubMed] [Google Scholar]
- Jiang, W. , Zhou, H. , Bi, H. , Fromm, M. , Yang, B. and Weeks, D.P. (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajikawa, M. , Hirai, N. and Hashimoto, T. (2009) A PIP‐family protein is required for biosynthesis of tobacco alkaloids. Plant Mol. Biol. 69, 287–298. [DOI] [PubMed] [Google Scholar]
- Kajikawa, M. , Shoji, T. , Kato, A. and Hashimoto, T. (2011) Vacuole‐localized berberine bridge enzyme‐like proteins are required for a late step of nicotine biosynthesis in tobacco. Plant Physiol. 155, 2010–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajikawa, M. , Sierro, N. , Kawaguchi, H. , Bakaher, N. , Ivanov, N.V. , Hashimoto, T. and Shoji, T. (2017) Genomic Insights into the Evolution of the Nicotine Biosynthesis Pathway in Tobacco. Plant Physiol. 174, 999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, R.S. , Lopez, H.O. , Bowen, S.W. , Andres, K.R. , Steede, W.T. and Dewey, R.E. (2015) Transgenic and mutation‐based suppression of a berberine bridge enzyme‐like (BBL) gene family reduces alkaloid content in field‐grown tobacco. PLoS ONE, 10, e0117273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Zeng, J. , Liang, Z. , Miao, Z. , Sun, X. and Tang, K. (2009) Silencing of PMT expression caused a surge of anatabine accumulation in tobacco. Mol. Biol. Rep. 36, 2285–2289. [DOI] [PubMed] [Google Scholar]
- World Health Organization (2017) WHO report on the Global Tobacco Epidemic 2017: Monitoring Tobacco use and Prevention Policies. Geneva: World Health Organization. Licence: CC BY‐NC‐SA 3.0 IGO [Google Scholar]


