Abstract
The Drosophila melanogaster sigma virus, a member of the Rhabdoviridae family, specifically propagates itself in D. melanogaster. It contains six genes in the order of 3′-N–P–X–M–G–L-5′. The sigma virus is the only arthropod-specific virus of the Rhabdoviridae family. Sigma-virus-infected Drosophila may suffer from irreversible paralysis when exposed to a high CO2 concentration, but generally, no other symptoms are reported. A recent study reported that host gene expression in immune pathways was not changed in sigma-virus-infected Drosophila, which does not necessarily suggest that they are not involved in virus–host interactions. The present study aimed to identify host genes associated with sigma virus replication. Immune pathways JAK-STAT and IMD were selected for detailed study. The results showed that the genome copy number of the sigma virus increased after knocking down the immune pathway genes domeless and PGRP-LC in Drosophila S2 cells. The knocking down of domeless and PGRP-LC significantly up-regulated the expression of the L gene compared to the other viral genes. We propose that the immune pathways respond to sigma virus infection by altering L expression, hence suppressing viral replication. This effect was further tested in vivo, when D. melanogaster individuals injected with dsdome and dsPGRP-LC showed not only an increase in sigma virus copy number, but also a reduced survival rate when treated with CO2. Our study proved that host immunity influences viral replication, even in persistent infection. Knocking down the key components of the immune process deactivates immune controls, thus facilitating viral expression and replication. We propose that the immunity system of D. melanogaster regulates the replication of the sigma virus by affecting the L gene expression. Studies have shown minimal host–virus interaction in persistent infection. However, our study demonstrated that the immunity continued to affect viral replication even in persistent infection because knocking down the key components of the immune process disabled the relevant immune controls and facilitated viral expression and replication.
Keywords: sigma virus, JAK-STAT pathway, IMD pathway, RNA interference
1. Introduction
Recent studies have used Drosophila melanogaster as a model for examining immune responses to viral infection, including responses to positive-sense RNA virus (Drosophila C virus, DCV) and negative-sense RNA virus (sigma virus) invasion [1,2]. The results showed that these viruses produced different symptoms depending on their host. Sigma virus is vertically transmitted and, in endemic areas, infects D. melanogaster, making it the only arthropod-specific member of the Rhabdoviridae family [2,3]. The RNA genome is similar to that described in Vesicular stomatitis Indiana, which comprises the genes N, P, M, G, and L (in order from 3′ to 5′); however, sigma virus differs due to the presence of the X gene between P and M (i.e., 3′-N–P–X–M–G–L-5′) [3], which is not typically observed in the rhabdovirus genome. Available information on the X gene is limited; it is only known to include a conserved region of reverse transcriptase. G and M produce structural proteins that are embedded in the lipid bilayer. N recognizes the viral RNA, and the N-RNA template binds to the RNA polymerase L (which is carried within the virions [4]) via the phosphoprotein P [5], forming a ribonucleoprotein complex that is released into the cytoplasm and initiates the processes of transcription and replication upon infection. In its natural course it does not produce marked symptoms [6], although it does cause irreversible paralysis in the presence of high CO2 concentrations [7,8]. Furthermore, it does not cause any immune response [9]. Despite this, it does have pathogenic effects (i.e., lower egg viability), which are strongly exhibited when infected female flies mate with uninfected males from a different population [10]. This observation suggests that the infected host can selectively control viral genome expression and subsequently viral replication.
The Drosophila model has facilitated the identification of numerous host genes up-regulated in response to pathogen infection. Defense of the host against viruses can be classified into inducible antiviral immunity and RNA interference (RNAi) [11]. To date, a number of major antiviral immune pathways, including Toll, Janus kinase signal transducer and activator of transcription (JAK-STAT) [12], and immune deficiency (IMD) pathways [13], have been recognized in D. melanogaster. These mechanisms regulate viral transcription and replication. The RNAi pathway affects viral replication by slicing/cleaving the viral genome [14,15,16,17]. The JAK-STAT pathways play a restricted role in only a few virus infections, whereas RNAi is active against most tested infections [12,18,19]. Genes associated with JAK-STAT and IMD pathways were reported to be silenced in sigma-virus-infected D. melanogaster [9], and not overexpressed, as occurs with other viruses and pathogens.
During viral infection, the host activates the immune mechanism by using pattern recognition receptors (PRRs) which detect foreign pathogen-associated molecular patterns, including viral nucleic acids and viral glycoproteins [20,21,22]. PRRs can be membrane bound, such as Toll, domeless (dome) [23], and peptidoglycan recognition protein (PGRP) [24]. Several key components instead of core RNAi mechanisms such as Dicer-2, r2d2, Argonaute-2, and piwi are responsible for initiating the antiviral response to Drosophila X virus (DXV), DCV, and other infectious viruses. However, host immunity may still interact with the virus and control its replication through other mechanisms. We analyzed the effect of suppressing the Toll signaling pathway on sigma virus infection, and therefore the present study aims to demonstrate individual and collective effects on viral replication in D. melanogaster when JAK-STAT and IMD pathways are knocked down.
2. Materials and Methods
2.1. Cells, Flies, Virus Stock, and CO2 Assay
Drosophila S2 cells were kept at 25 °C in Shields and Sang M3 insect medium (Sigma-Aldrich, St. Louis, MO, US) supplemented with yeast extract and bactopeptone according to the protocols of the Drosophila Genomics Resource Center (https://dgrc.bio.indiana.edu/Home), with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, US) [2]. Fly stocks were maintained in standard medium at 25 °C and 60% humidity under a 12:12 h (light:dark) photoperiod. The RC2 cell lines of D. melanogaster, which were or were not sigma-virus-infected, were used in this study. Df(2R)BSC22/SM6a, tub-Gal4, and UAS-mCD8-GFP flies were obtained from the Bloomington Drosophila Stock Center. A transgenic line (tub-Gal4>UAS-mCD8-GFP) with green fluorescent protein (GFP) gene was the progeny of tub-Gal4 males crossed to UAS-mCD8-GFP females, and this line was employed to demonstrate a successful knockdown of target gene after injection of dsgfp (dsRNA of GFP) [25]. In order to generate the virus stock, 3000 virus-infected flies were identified via a CO2 assay. This works on the principal that upon exposure to high concentrations of CO2, uninfected (or low viral titer) flies would recover, but those with a high viral titer would be paralyzed. Therefore, as per Tsai et al. [2], flies were exposed to pure CO2 for approximately 30 s and kept on ice for 10 min. After 30 min, the affected flies could be separated. To release the virus from the host, the flies were frozen using liquid nitrogen and ground using 250 mL of M3+ medium without FBS. Fly tissues were removed through viral crude extraction by centrifugation at 800× g for 10 min. The suspension of the crude extraction was filtered using an NML syringe with a 0.22-μm filter (Sartorius) [26]. The extraction product containing sigma virus was collected and stored at −80 °C. For the knockdown experiments, flies were injected with 50 nL of the sigma virus using a nanoinjector (InjectMan, Eppendorf, Hamburg, Germany) [27].
2.2. dsRNA Preparation
The dsRNAs for domeless and PGRP-LC were synthesized using a T7 Quick High Yield RNA Synthesis Kit (New England BioLabs, Ipswich, MA, US). Templates were generated by reverse-transcription polymerase chain reaction (RT-PCR) using the primers listed in Table 1, which include a gene-specific part and a T7 promoter overhang. The final products were purified with a total RNA purification kit (GeneMark, Taipei, TW).
Table 1.
Primer sequences for dsRNA synthesis.
| Gene Name | dsRNA Synthesis Primers |
|---|---|
| domeless (dome) | F: TAATACGACTCACTATAGGG TAACGGCAAGAGCGC |
| R: TAATACGACTCACTATAGGG AGGTTCTGGCCAGGT | |
| PGRP-LC | F: TAATACGACTCACTATAGGGG GCGGTT TCCATACGG |
| R: TAATACGACTCACTATAGGGG CCATTGCTGACGCTC | |
| GFP | F: GCTCGGGAGATCTCCTGCCTTTGGGTGTGTCTGGG |
| R: CTAGACTCGAGCGGCCAACGGATCCTTCGTAGCCC | |
| TBP | F: AATTAACCCTCACTAAAGGGAT GGACCAAATGCTAAGCCC |
| R: AATTAACCCTCACTAAAGGGTACTTTCTCGCTGCCAGTCT |
2.3. dsRNA Transfection, Virus Infection, and RNA Extraction
Approximately 2 × 105 cells/well of S2 cells were seeded on a 24-well plate. The transfection mix was prepared using 5 pmol of dsRNA (dsdome, dsPGRP-LC, or dsControl), 1.5 μL of RNAiMAX transfection reagent (Life Technology, Waltham, MA, US), and M3+ medium to a final volume of 50 μL. The mixture was incubated for 30 min to allow the formation of a dsRNA–lipid complex, which was then added in each well and incubated at 26 °C for 1 h. The transfected S2 cells were then infected with sigma virus using 100 μL of viral extraction product. After 48 h the cells were harvested in tubes, centrifuged at 800× g for 10 min to remove the supernatant, and lysed using a 2-mercaptoethanol solution. RNA was isolated and purified using a total RNA purification kit (GeneMark), treated with DNase I, and finally quantified using a NanoDrop spectrophotometer (Thermo Fisher, Waltham, MA, US).
2.4. Gene Expression Assay Using RT-qPCR
Total RNA was reverse-transcribed using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Waltham, MA, US) for RT-qPCR. A total of 500 ng of RNA sample and 0.5 µg of random primers was used.
For quantifying gene expression through RT-qPCR, a 20 μL mixture was used, containing 1 μL of cDNA, 0.5 μM forward and reverse primers, and 2× SYBR Green master mix (Applied Biosystems) under the following conditions: 94 °C for 15 s, 60 °C for 30 s, and 72°C for 30 s. The expression of actin was used as the control (Ct cutoff of 35). A list of primer sequences used in this study is given in Table 2. The relative fold changes of gene expression were calculated through the ΔΔCt method [28].
Table 2.
Primer sequences used in this study.
| Gene | qPCR Check Primers | Gene | qPCR Check Primers |
|---|---|---|---|
| domeless (dome) | F: ACAACAGGCGTCTTCGGATT | SV-NP | F: TAACTCGGGTGTGACAGCTC |
| R: ACCCTTCAGTTTTGCCATGGT | R: CTTCGTTCATCTTCCTGGGT | ||
| PGRP-LC | F: CGCAAGGCCGTCACAGTTAC | SV-N | F: CACATGAGAAAATGCAAACAGCTT |
| R: GGTTCAACGTCTTTCCGAAGAG | R: GAAAATGGAGCGAGGATCGA | ||
| Diptericin (Dpt) | F: CTATTCATTGGACTGGCTTGTGCC | SV-P | F: TCAAACCCAGAGCCAGAGATAGTAT |
| R: TGGAACTGGCGACGCACTCT | R: CGCTTTTATCTGACGCTCAGGTA | ||
| TEPS | F: AACTCCGCAAACACCAAGTTGG | SV-X | F: TGGCCCCAATATTTCCTGAA |
| R: CTTCAACGCTTCGTGTAACACCAC | R: GCGTCACTCCATCAGGGTTT | ||
| Actin | F: CAAAGCGCAAAAAGAACACA | SV-M | F: ACACACTCCACAGTTTACCACCAT |
| R: AGAGGAGAGGGCGAGGTTAG | R: CGCCCTCCTGTCAATGAATAG | ||
| GFP | F: GTGTTCAATGCTTTGCGAGA | SV-G | F: CCATGTTTCGTTGAGCTTTCC |
| R: AAAGGGCAGATTGTGTGGAC | R: CGCCTTCGTGTTCACTGAGTT | ||
| TBP | F: TAGTGGCCAATCCTGTGTACCA | SV-L | F: TTCCCTGAAGACGCCCATTA |
| R: TCAGCGGAACCTGGTGTGGC | R: TGCCGCCCTCATCCAA |
2.5. dsRNA Microinjection
Thirty virus-infected adult female flies per experiment were injected into the thorax with 30 nL of dsRNA (3 mg/mL) using a glass needle (Sutter instrument Co.) coupled to a nanoinjector (InjectMan; Eppendorf, Hamburg, Germany). Three days later, the whole bodies of flies were processed for total RNA extraction according to previous studies [29]. The obtained RNA was reverse-transcribed, and the cDNA was used for the analysis of gene expression using RT-qPCR as previously described.
2.6. Statistical Analysis
The Ct values of the target genes were normalized to the Ct values of actin (reference gene). The differences in expression levels of the target genes were analyzed in SPSS version using a Mann–Whitney U-test [30,31] considering p < 0.05 as the cutoff value for statistical significance.
3. Results
3.1. Viral Genome Replication Level Was Increased after the Knockdown of Immune Pathways in S2 Cells
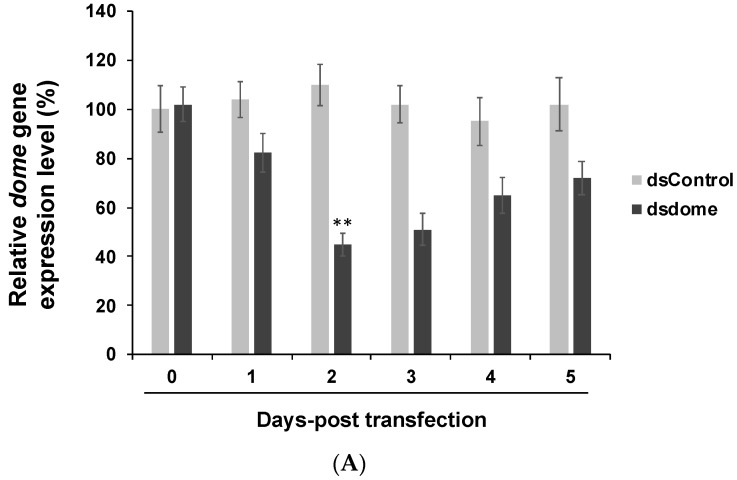
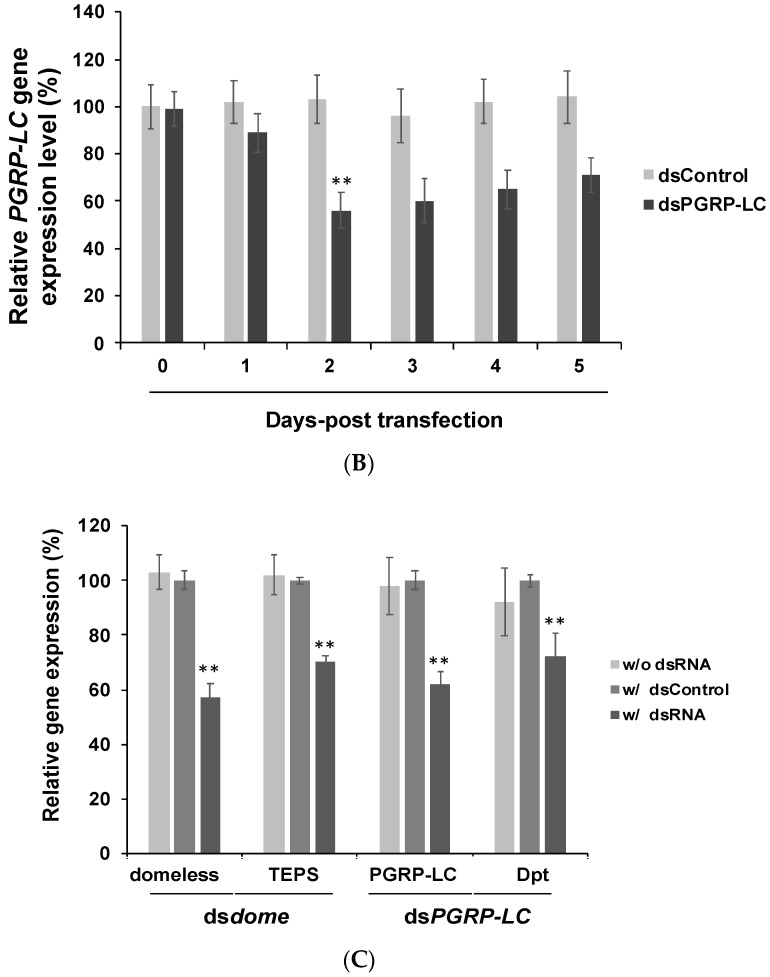
We first constructed a dsRNA that specifically suppresses the upstream genes of the JAK-STAT (domeless gene) and IMD (PGRP-LC gene) pathways in S2 cells. Drosophila S2 cells were transfected with the dsRNA and examined each day up to 5 days post-transfection (dpt) to find the optimal knockdown time point. The suppression of domeless (Figure 1A) and PGRP-LC (Figure 1B) expression reached a maximum at 2 dpt. The suppression of upstream genes thus efficiently silenced the expression of downstream genes such as antimicrobial peptide (AMP) genes (AMP genes; TEPS; Dpt, Diptericin) (Figure 1C).
Figure 1.
The expression of domeless and PGRP-LC decreased after being knocked down by dsRNA. (A) domeless and (B) PGRP-LC expression was at a minimum at 2 days post-transfection (dpt). Y-axis: relative domeless and PGRP-LC expression reading. X-axis: dpt. The maximum reading was set to 100, with other readings adjusted accordingly. Actin signals were used as a loading control. (C) The expression of antimicrobial peptide (AMP) genes detected by RT-qPCR. The downstream gene (AMP genes; TEPS; Dpt, Diptericin) expressions were decreased by silencing upstream genes. We set the dsControl group as 100% expression. Mean and SD shown, ** p < 0.005 one-sample t-test. All experiments were performed with three replicates.
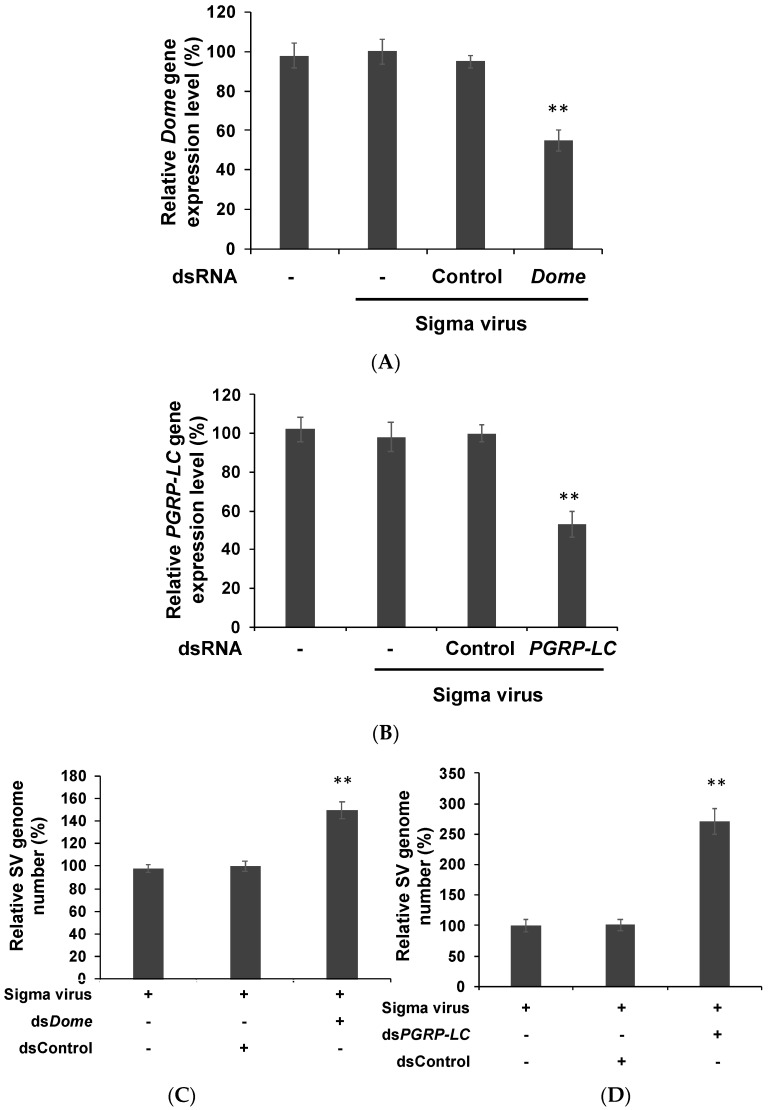
We knocked down the initial genes in these pathways using RNAi to examine whether there was any implication on sigma virus infectivity. As the knockdown efficiencies of dsRNA reached their maximum at 2 dpt (Figure 1), S2 cells were transfected with dsdomeless and dsPGRP-LC, followed by infection with sigma virus for 48 h. We confirmed the suppression of domeless and PGRP-LC expression by subjecting domeless and PGRP-LC knockdown to RT-qPCR (Figure 2A,B). Since a previous study showed that sigma virus infects and replicates in S2 cells [32], we further studied whether domeless and PGRP-LC suppression had any effect on sigma virus replication in S2 cells. Sigma virus contains six genes, ordered 3′-N–P–X–M–G–L-5′. Therefore, we designed primers that covered both N and P genes in order to avoid misjudging the outcome that single gene expression difference accounted for. The viral genome copy number of the domeless and PGRP-LC-knockdown cells significantly increased (Figure 2C,D), whereas dsControl (the negative control) revealed no increase in the viral genome copy number owing to dsRNA transfection (Figure 2). Cell morphology and the number before and after dsRNA transfection were approximately the same, indicating that transfection did not cause cell death (data not shown). Our results revealed that the aforementioned suppression caused a significant increase in viral replication. A previous study showed that the expression of immunity-related genes was not altered during persistent infection; however, further suppression of the basal expression of these genes by RNAi knockdown yielded results that suggest some regulatory roles of these genes in viral replication.
Figure 2.
Sigma virus (SV) genome copy number increased after knocking down PGRP-LC and dome. RT-qPCR showed that dsRNA can efficiently suppress domeless (A) or PGRP-LC (B) expression in S2 cells at 2 dpt. “-”: without dsRNA transfection; “Control”: S2 cells transfected with dsControl. The viral genome replication of the sigma virus was detectable by RT-qPCR after knocking down domeless (C) or PGRP-LC (D) using dsRNA. dsControl was used as the negative control. We set the dsControl group as 100% expression. A dsRNA targeting the GFP gene was used as the negative control and designated dsControl. The mean and SD shown are shown. ** p < 0.005, one-sample t-test. All experiments were performed with three biological replicates.
3.2. Viral Gene Expression Increased after the Knockdown of Immune Pathways in S2 Cells
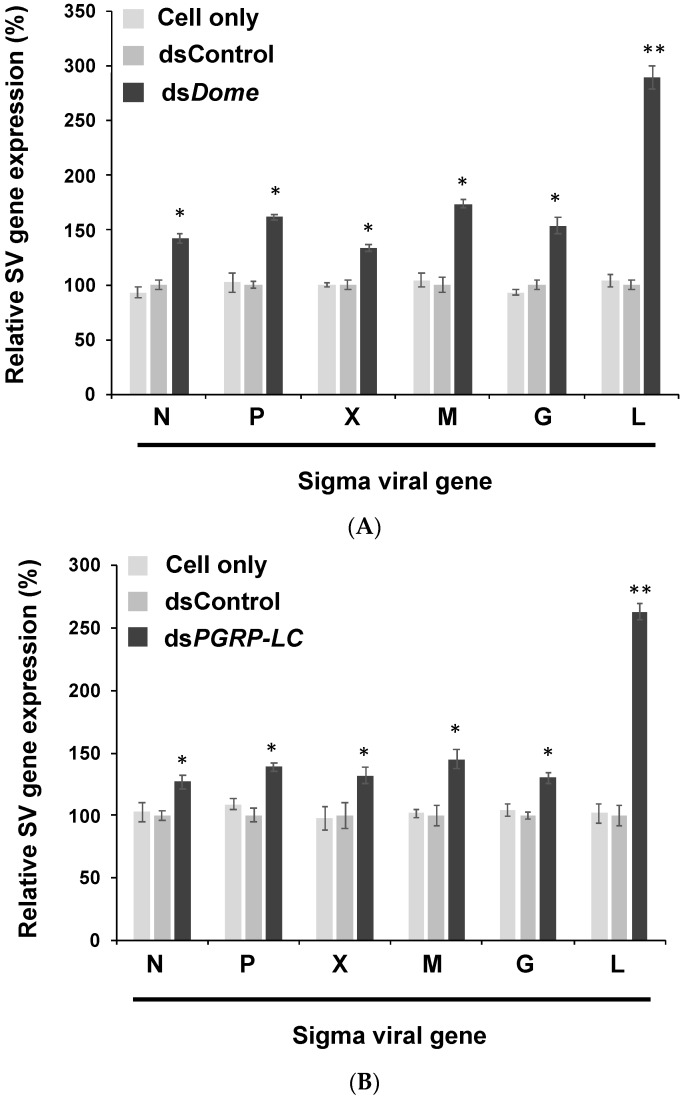
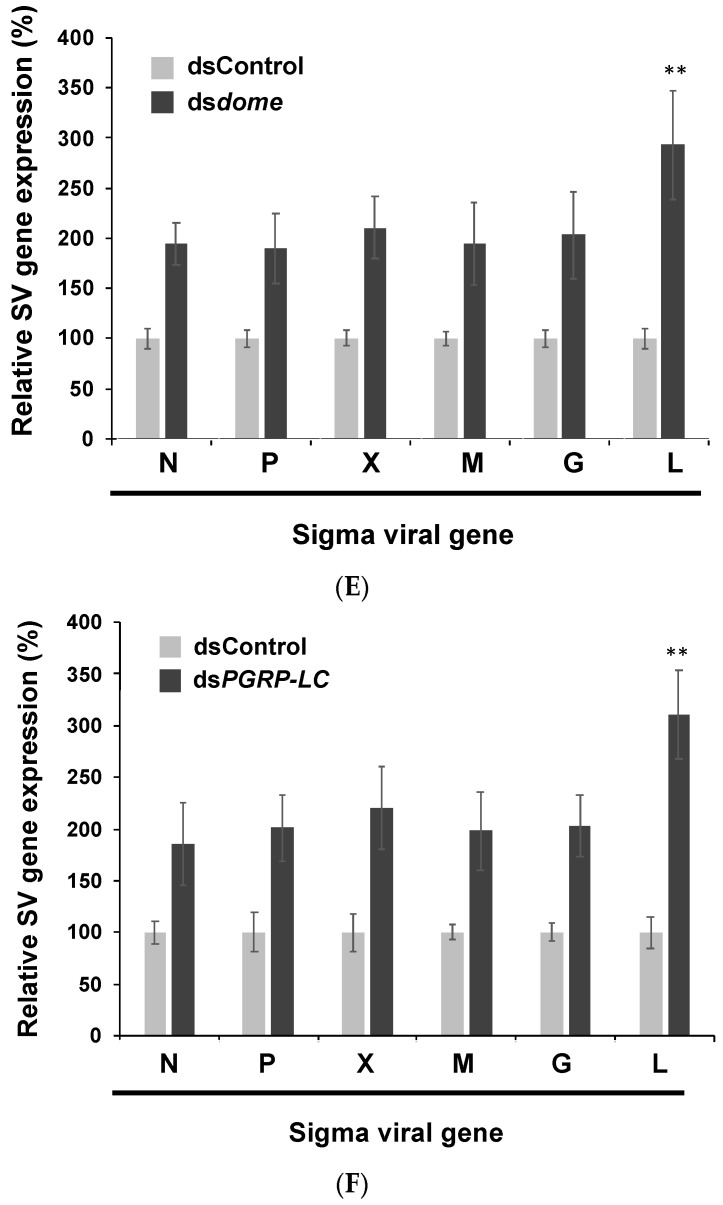
To investigate whether viral genes sufficiently promote viral genome replication, we transfected S2 cells with dsRNA in order to knock down immune signaling pathways; this was followed by viral infection. After suppressing the JAK-STAT and IMD pathways, we measured the expression of the six aforementioned genes. We observed that all genes were up-regulated; in particular, the L gene expression increased 2.5- to 3-fold (Figure 3A,B). The protein L is responsible for replication and transcription; therefore, we proposed that the defense mechanisms governed by the JAK-STAT and IMD pathways suppress the replication and transcription of sigma virus. The knockdown of immunity-related genes in these pathways consequently increased the viral genome copy number. Most viral genes were overexpressed, and expression of the L gene transcripts was significantly selectively elevated. This finding indicates that the host antiviral response may control viral replication and infection through L gene regulation.
Figure 3.
Blocking immune pathways resulted in a high level of viral gene expression. (A) The gene expression of the sigma virus in JAK-STAT pathway knockdown in S2 cells. The expression of all genes increased, and that of the L gene showed the highest increase (i.e., 4-fold). (B) Gene expression of sigma virus in the IMD pathway-knockdown cells. The mean and SD are shown. ** p < 0.005; * p < 0.05 one-sample t-test. All experiments were performed with three biological replicates.
3.3. In Vivo Experiments Confirmed the Findings of in Vitro Experiments
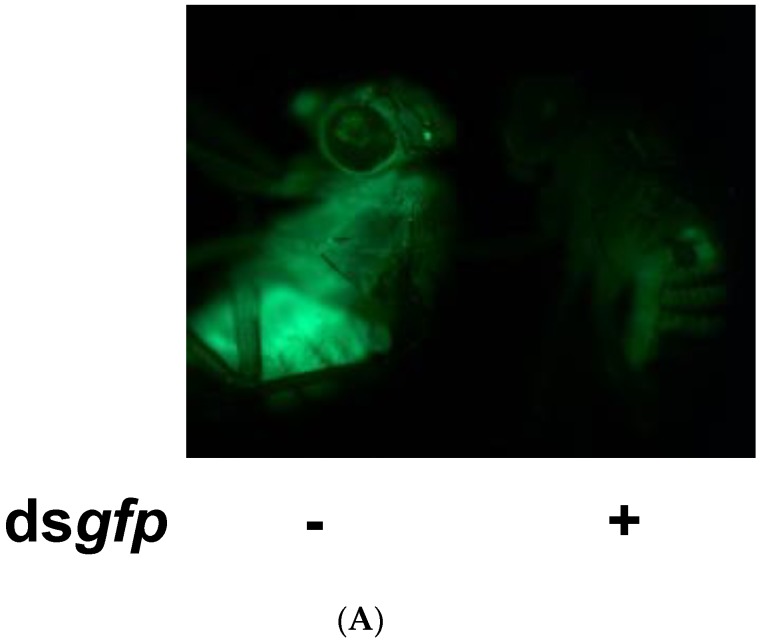
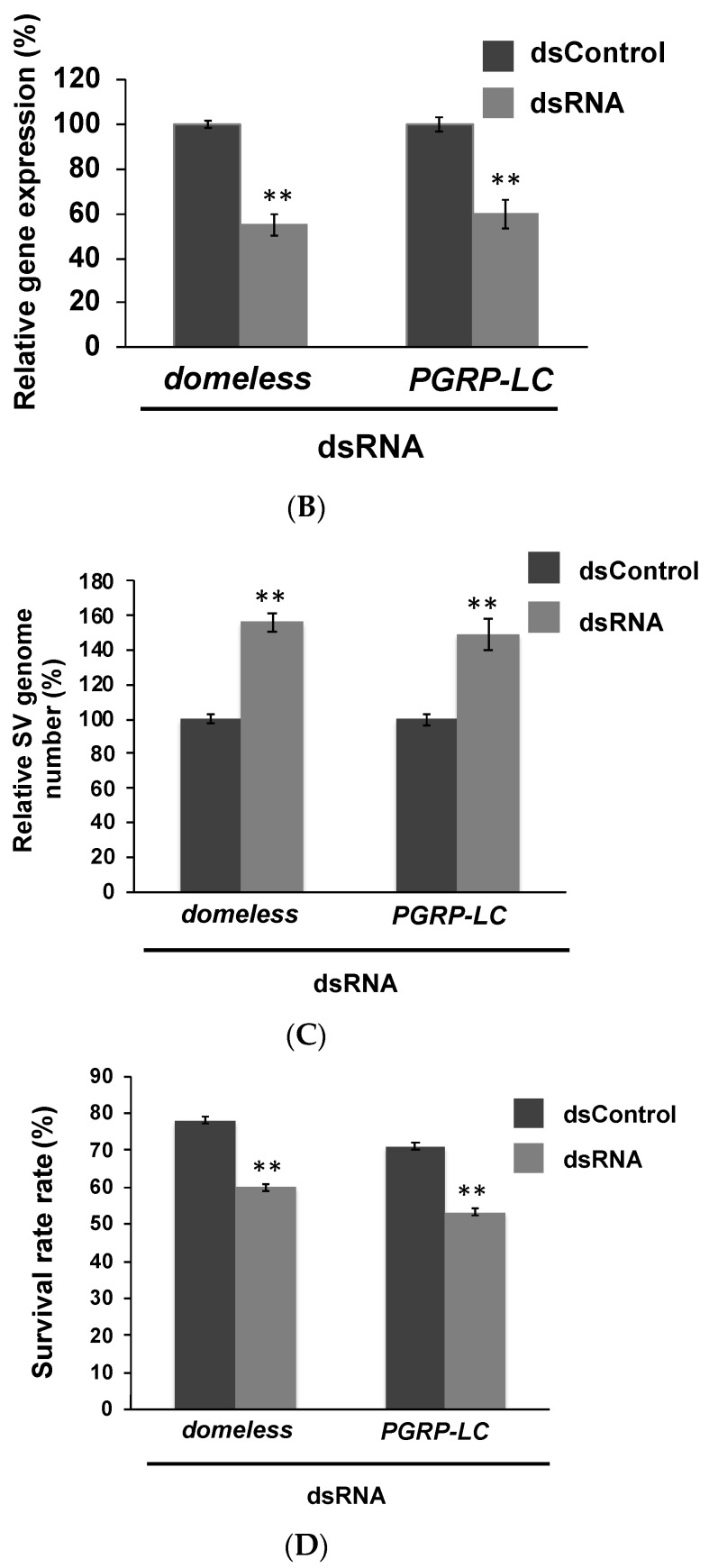
The knockdown of Drosophila immunity-related genes caused a high level of viral replication. Therefore, we conducted further experiments in Drosophila to explore whether the knockdown of host immunity genes has the same effect on persistent infection virus in vivo. dsdome and dsPGRP-LC were applied on sigma-virus-infected flies via injection. To confirm the successful knockdown of the target gene by using dsRNA, we injected dsgfp into GFP transgenic flies (tub-Gal4>UAS-mCD8-GFP fly) as the positive control [25]. The fluorescent signal in the flies decreased 3 days after injection, implying that the knockdown was achieved on day 3 (Figure 4A). We harvested sigma-virus-infected flies three days after dsdome or dsPGRP-LC injection. We examined the domeless and PGRP-LC gene expression in flies with or without dsRNA injection via RT-PCR (Figure 4B). The viral genome copy number increased approximately 1.5-fold, which was similar to the in vitro result (Figure 4C). Previous data have shown that sigma-virus-infected flies are permanently paralyzed after exposure to pure CO2 [7]. We therefore performed a CO2 assay on flies and compared the survival rate of sigma-virus-infected flies with and without domeless and PGRP-LC knockdown. The survival rate was found to be 20% lower in domeless and PGRP-LC-knockdown flies than in the wild-type flies (Figure 4D). The result indicated that the knockdown of immunity genes results in a higher replication and expression of the sigma virus and less resistance to CO2 treatment. We compared the transcriptional profiles of viral genes in buffer- or dsRNA-injected flies using RT-qPCR, and observed that viral gene expression was up-regulated, with the L gene expression significantly enhanced (Figure 4E,F). Our data suggest that immune genes protect D. melanogaster against sigma virus infection; this was found to be true and reproducible both in vitro and in vivo.
Figure 4.
Knockdown of domeless and PGRP-LC enhanced the sigma virus replication in flies. (A) Green fluorescent protein (GFP) transgenic flies were injected with dsgfp and then reared for 3 days. The fluorescence was quenched in dsgfp-injected flies, indicating successful knockdown of the target gene by dsRNA injection. (B) RT-qPCR analysis of domeless and PGRP-LC gene expression in dsRNA injected flies. (C) The RT-qPCR analysis of sigma virus copies in sigma-virus-infected domeless and PGRP-LC knockdown and untreated flies. (D) The survival rate of sigma-virus-infected flies in domeless and PGRP-LC-knockdown flies upon CO2 exposure. (E,F) RT-qPCR analysis of sigma virus copies in sigma-virus-infected domeless and PGRP-LC -knockdown and untreated flies. We set the dsControl group as 100% expression. The mean and SD are shown. ** p < 0.005, one-sample t-test. All experiments were performed with three replicates.
3.4. TATA-Binding Protein May Facilitate Viral Replication by Enhancing the RNA Polymerase Activity
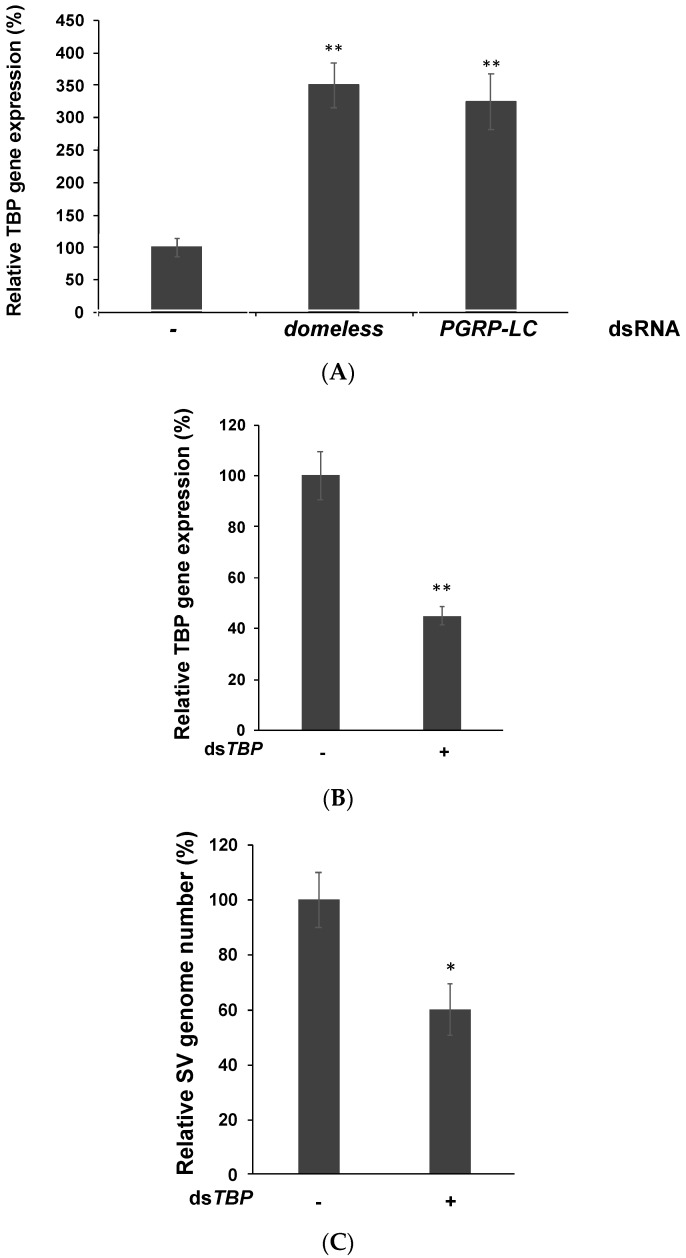
According to a recent study [33], in insects, TATA-binding protein (TBP) is associated with subunits of the viral RNA-dependent RNA polymerase complex (RdRp), which may indicate that TBP interacts with individual components of the viral RdRp complex for enhancing viral RNA replication. In sigma virus, the L gene encodes RdRp, whose main function is to regulate viral replication and translation. In our study, TBP expression markedly increased after the immune pathways were knocked down (Figure 5A). Thus, these immune pathways may suppress viral polymerase and the subsequent viral replication by reducing TBP expression. We also suppressed TBP using dsRNA and found sigma virus replication was significantly decreased (Figure 5B,C). This indicates TBP may play an important role in helping virus replicate. Currently, we are studying the relationship between TBP and sigma virus replication; these results will be published in further works.
Figure 5.
Gene expression of TATA-binding protein (TBP) increased after knocking down domeless and PGRP-LC upstream immune genes. (A) TBP expression increased after knocking down domeless and PGRP-LC. We set the untreated group as 100% expression. (B) TBP expression decreased with dsTBP transfected S2 cells at 2 dpt. (C) The RT-qPCR analysis of sigma virus copies in sigma-virus-infected TBP-knockdown and wild-type cells. Control treated cells were set to 100%. Mean and SD shown; ** p < 0.005; * p < 0.05; one-sample t-test. All experiments were performed with three replicates.
4. Discussion
Viruses infect hosts to complete their life cycles. Studies have reported that the expression of host genes in the JAK-STAT and IMD pathways are unaffected in sigma-virus-infected D. melanogaster [9]. In our study, the expression of sigma virus genes increased after knocking down the upstream genes in these two pathways. This showed that these pathways are indeed involved in virus–host interactions (Figure 2 and Figure 3).
In Drosophila, the JAK-STAT pathway has been considered to be triggered in bystander cells rather than in infected cells [12]. Carpenter et al. reported that there was no difference in the gene expression of the JAK-STAT pathway between flies infected and uninfected with sigma virus [9]. However, when domeless was selectively knocked down in S2 cells, the gene expression and genome copy number of sigma virus increased (Figure 2). This finding revealed that the pathway regulates the replication of sigma virus. Furthermore, the expression of the six sigma virus genes increased. Both findings reveal that in infected cells, the JAK-STAT pathway affected viral replication by affecting the synthesis of viral RNA genome and proteins. We hypothesize that even though no differential gene expression was detected in factors involved in the JAK-STAT pathway, it plays an essential role in suppressing L protein expression, which in turn affects the replication of sigma virus.
The IMD pathway employs an antibacterial mechanism because its receptor PGRPs are primarily activated by bacterial peptidoglycan (PG), which is present in the membrane of Gram-negative bacteria and some Gram-positive bacteria [13,24,34]. In contrast to its well-understood antibacterial mechanism, the antiviral mechanism of the IMD pathway has been much less studied. In our study, the gene upstream of the IMD pathway, PGRP-LC, was selectively knocked down in S2 cells; consequently, gene expression and genome copy number of sigma virus was increased, similar to when domeless was knocked down. These findings reveal that the IMD pathway also affects replication of this virus in a manner similar to the JAK-STAT pathway.
Consequently, these two immune pathways were involved in the regulation of the viral polymerase in sigma virus. The production of viral RNA and protein decreased when transcriptional repressor (DR1) was knocked down in mammalian cells, suggesting that DR1 is associated with an increased viral polymerase activity [35]. Further biochemical assays revealed that viral RNA replication was suppressed in DR1-knockdown cells. According to a recent study [36], in insects, TBP is associated with subunits of the viral RNA-dependent RNA polymerase complex (RdRp), which may indicate that TBP interacts with individual components of the viral RdRp complex for enhancing viral RNA replication. In our study, TBP expression markedly increased after the immune pathways were knocked down. Thus, these immune pathways may suppress viral polymerase and the subsequent viral replication by reducing TBP expression (Figure 5). However, inhibiting the RNAi pathway did not affect TBP expression, suggesting that immune pathways regulate viral replication via a route different from that of the RNAi pathway (Figure S1). Therefore, further research is required to reveal the association between these immune genes and TBP, if present, and the involved mechanism.
The knockdown of immune pathways increases the activity of viral polymerase and replication, possibly through an increased TBP production. To date, no efficient therapy exists against many viruses, particularly against RNA viruses (e.g., HIV), because of a high mutation rate, which complicates targeted antiviral therapy [37]. A cellular immune mechanism provides nonspecific protection against viruses. Therefore, administering drugs that enhance cellular mechanisms may be an efficient method that is less subjected to viral resistance. Although the expression of all six viral genes was up-regulated when the JAK-STAT and IMD pathways were knocked down, it is worth noting that the expression of L was increased more than the other genes. Considering the pivotal role of L during viral genome transcription and replication, it can be inferred that the Drosophila antiviral mechanisms will target this gene. Thus, we propose that the immune system of D. melanogaster regulates the replication of the sigma virus by affecting the gene expression of L.
5. Conclusions
Our results showed that the viral genome copy number increased after domeless and PGRP-LC were knocked down in S2 cells. Furthermore, the expression of all six viral genes, particularly that of the L gene, was up-regulated. Because the L gene plays a crucial role in genome transcription and replication, the Drosophila antiviral mechanism may be activated by suppressing this gene. Thus, we propose that the immunity system of D. melanogaster regulates the replication of the sigma virus by affecting the L gene expression. Studies have shown minimal host–virus interaction in persistent infection. However, our study demonstrated that the immunity continues to affect viral replication even in persistent infection because knocking down the key components of the immune process disabled the relevant immune controls and facilitated viral expression and replication.
Acknowledgments
We thank Alexander Barton for kindly revising the manuscript.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4450/10/10/339/s1, Figure S1: Gene expression of TBP increased after knocking down three upstream immune genes.
Author Contributions
Guarantors of integrity of entire study, study concepts, and manuscript preparation: J.-F.L., C.-P.W. and Y.-L.W. Study design, data acquisition/analysis, literature research and manuscript preparation: J.-F.L., C.-P.W., C.-K.T. and Y.-L.W. Data acquisition/analysis: J.-F.L., C.-P.W. and C.-K.T. Resources: C.-W.T. Manuscript editing, and revision: C.-P.W., L.R. and Y.-L.W. Project administration and unding acquisition: Y.-L.W. All authors reviewed the manuscript.
Funding
This research was funded by the grant MOST107-2311-B-002-024-MY3 to Y.L.W. from the Ministry of Science and Technology, Taiwan.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Chtarbanova S., Lamiable O., Lee K.Z., Galiana D., Troxler L., Meignin C., Hetru C., Hoffmann J.A., Daeffler L., Imler J.L. Drosophila C Virus Systemic Infection Leads to Intestinal Obstruction. J. Virol. 2014;88:14057–14069. doi: 10.1128/JVI.02320-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai C.W., McGraw E.A., Ammar E.D., Dietzgen R.G., Hogenhout S.A. Drosophila melanogaster mounts a unique immune response to the Rhabdovirus sigma virus. Appl. Environ. Microbiol. 2008;74:3251–3256. doi: 10.1128/AEM.02248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Longdon B., Obbard D.J., Jiggins F.M. Sigma viruses from three species of Drosophila form a major new clade in the rhabdovirus phylogeny. Proc. R. Soc. B. 2010;277:35–44. doi: 10.1098/rspb.2009.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang B., Li Z., Jenni S., Rahmeh A.A., Morin B.M., Grant T., Grigorieff N., Harrison S.C., Whelan S.P.J. Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy. Cell. 2015;162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green T.J., Luo M. Structure of the vesicular stomatitis virus nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P. Proc. Natl. Acad. Sci. USA. 2009;106:11713–11718. doi: 10.1073/pnas.0903228106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleuriet A. Evolution of the proportions of two sigma viral types in experimental populations of Drosophila melanogaster in the absence of the allele that is restrictive of viral multiplication. Genetics. 1999;153:1799–1808. doi: 10.1093/genetics/153.4.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosen L. Carbon dioxide sensitivity in mosquitoes infected with sigma, vesicular stomatitis, and other rhabdoviruses. Science. 1980;207:989–991. doi: 10.1126/science.6101512. [DOI] [PubMed] [Google Scholar]
- 8.Shroyer D.A., Rosen L. Extrachromosomal inheritance of carbon dioxide sensitivity in the mosquito Culex quinquefasciatus. Genetics. 1983;104:649–659. doi: 10.1093/genetics/104.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpenter J., Hutter S., Baines J.F., Roller J., Saminadin-Peter S.S., Parsch J., Jiggins F.M. The transcriptional response of Drosophila melanogaster to infection with the sigma virus (Rhabdoviridae) PLoS ONE. 2009;4:e6838. doi: 10.1371/journal.pone.0006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleuriet A., Sperlich D. Evolution of the Drosophila melanogaster-sigma virus system in a natural population from Tubingen. Theor. Appl. Genet. 1992;85:186–189. doi: 10.1007/BF00222858. [DOI] [PubMed] [Google Scholar]
- 11.Kingsolver M.B., Huang Z., Hardy R.W. Insect antiviral innate immunity: pathways, effectors, and connections. J. Mol. Biol. 2013;425:4921–4936. doi: 10.1016/j.jmb.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dostert C., Jouanguy E., Irving P., Troxler L., Galiana-Arnoux D., Hetru C., Hoffmann J.A., Imler J.L. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat. Immunol. 2005;6:946–953. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- 13.Kaneko T., Goldman W.E., Mellroth P., Steiner H., Fukase K., Kusumoto S., Harley W., Fox A., Golenbock D., Silverman N. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity. 2004;20:637–649. doi: 10.1016/S1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- 14.Mueller S., Gausson V., Vodovar N., Deddouche S., Troxler L., Perot J., Pfeffer S., Hoffmann J.A., Saleh M.C., Imler J.L. RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila. Proc. Natl. Acad. Sci. USA. 2010;107:19390–19395. doi: 10.1073/pnas.1014378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bronkhorst A.W., van Cleef K.W., Vodovar N., Ince I.A., Blanc H., Vlak J.M., Saleh M.C., van Rij R.P. The DNA virus Invertebrate iridescent virus 6 is a target of the Drosophila RNAi machinery. Proc. Natl. Acad. Sci. USA. 2012;109:E3604–E3613. doi: 10.1073/pnas.1207213109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabin L.R., Zheng Q., Thekkat P., Yang J., Hannon G.J., Gregory B.D., Tudor M., Cherry S. Dicer-2 processes diverse viral RNA species. PLoS ONE. 2013;8:e55458. doi: 10.1371/journal.pone.0055458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaines P.J., Olson K.E., Higgs S., Powers A.M., Beaty B.J., Blair C.D. Pathogen-derived resistance to dengue type 2 virus in mosquito cells by expression of the premembrane coding region of the viral genome. J. Virol. 1996;70:2132–2137. doi: 10.1128/jvi.70.4.2132-2137.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zambon R.A., Nandakumar M., Vakharia V.N., Wu L.P. The Toll pathway is important for an antiviral response in Drosophila. Proc. Natl. Acad. Sci. USA. 2005;102:7257–7262. doi: 10.1073/pnas.0409181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zambon R.A., Vakharia V.N., Wu L.P. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell Microbiol. 2006;8:880–889. doi: 10.1111/j.1462-5822.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- 20.Sabin L.R., Hanna S.L., Cherry S. Innate antiviral immunity in Drosophila. Curr. Opin. Immunol. 2010;22:4–9. doi: 10.1016/j.coi.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 22.Aliyari R., Wu Q., Li H.W., Wang X.H., Li F., Green L.D., Han C.S., Li W.X., Ding S.W. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belvin M.P., Anderson K.V. A conserved signaling pathway: the Drosophila toll-dorsal pathway. Annu. Rev. Cell Dev. Biol. 1996;12:393–416. doi: 10.1146/annurev.cellbio.12.1.393. [DOI] [PubMed] [Google Scholar]
- 24.Choe K.M., Lee H., Anderson K.V. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc. Natl. Acad. Sci. USA. 2005;102:1122–1126. doi: 10.1073/pnas.0404952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee T., Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/S0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 26.Teninges D., Bras-Herreng F. Rhabdovirus sigma, the hereditary CO2 sensitivity agent of Drosophila: nucleotide sequence of a cDNA clone encoding the glycoprotein. Pt 10J. Gen. Virol. 1987;68:2625–2638. doi: 10.1099/0022-1317-68-10-2625. [DOI] [PubMed] [Google Scholar]
- 27.Longdon B., Hadfield J.D., Webster C.L., Obbard D.J., Jiggins F.M. Host phylogeny determines viral persistence and replication in novel hosts. PLoS Pathogens. 2011;7:e1002260. doi: 10.1371/journal.ppat.1002260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y.W., Wu C.P., Wu T.C., Wu Y.L. Analyses of the transcriptome of Bombyx mori cells infected with either BmNPV or AcMNPV. J. Asia-Pac. Entomol. 2018;21:37–45. doi: 10.1016/j.aspen.2017.10.009. [DOI] [Google Scholar]
- 30.Dexter F. Wilcoxon-Mann-Whitney Test Used for Data That Are Not Normally Distributed. Anesth. Analg. 2013;117:537–538. doi: 10.1213/ANE.0b013e31829ed28f. [DOI] [PubMed] [Google Scholar]
- 31.Hu Y.T., Wu T.C., Yang E.C., Wu P.C., Lin P.T., Wu Y.L. Regulation of genes related to immune signaling and detoxification in Apis mellifera by an inhibitor of histone deacetylation. Sci. Rep-Uk. 2017;7 doi: 10.1038/srep41255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merkling S.H., van Rij R.P. Analysis of resistance and tolerance to virus infection in Drosophila. Nat. Protocols. 2015;10:1084–1097. doi: 10.1038/nprot.2015.071. [DOI] [PubMed] [Google Scholar]
- 33.Quadt I., Mainz D., Mans R., Kremer A., Knebel-Morsdorf D. Baculovirus infection raises the level of TATA-binding protein that colocalizes with viral DNA replication sites. J. Virol. 2002;76:11123–11127. doi: 10.1128/JVI.76.21.11123-11127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaneko T., Yano T., Aggarwal K., Lim J.H., Ueda K., Oshima Y., Peach C., Erturk-Hasdemir D., Goldman W.E., Oh B.H., et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the drosophila immune response to monomeric DAP-type peptidoglycan. Nat. Immunol. 2006;7:715–723. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- 35.Hsu S.F., Su W.C., Jeng K.S., Lai M.M. A host susceptibility gene, DR1, facilitates influenza A virus replication by suppressing host innate immunity and enhancing viral RNA replication. J. Virol. 2015;89:3671–3682. doi: 10.1128/JVI.03610-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H.D., Trivedi A., Johnson D.L. Regulation of RNA polymerase I-dependent promoters by the hepatitis B virus X protein via activated Ras and TATA-binding protein. Mol. Cell. Biol. 1998;18:7086–7094. doi: 10.1128/MCB.18.12.7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward D.M., Vaughn M.B., Shiflett S.L., White P.L., Pollock A.L., Hill J., Schnegelberger R., Sundquist W.I., Kaplan J. The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J. Biol. Chem. 2005;280:10548–10555. doi: 10.1074/jbc.M413734200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.