Abstract
For stem cell-based therapies, the fate and distribution of stem cells should be traced using non-invasive or histological methods and a nanomaterial-based labelling agent. However, evaluation of the biophysical effects and related biological functions of nanomaterials in stem cells remains challenging. Here, we aimed to investigate the biophysical effects of nanomaterials on stem cells, including those on membrane fluidity, using total internal reflection fluorescence microscopy, and traction force, using micropillars of human bone marrow-derived mesenchymal stem cells (hBM-MSCs) labelled with silica-coated magnetic nanoparticles incorporating rhodamine B isothiocyanate (MNPs@SiO2(RITC)). Furthermore, to evaluate the biological functions related to these biophysical changes, we assessed the cell viability, reactive oxygen species (ROS) generation, intracellular cytoskeleton, and the migratory activity of MNPs@SiO2(RITC)-treated hBM-MSCs. Compared to that in the control, cell viability decreased by 10% and intracellular ROS increased by 2-fold due to the induction of 20% higher peroxidized lipid in hBM-MSCs treated with 1.0 µg/µL MNPs@SiO2(RITC). Membrane fluidity was reduced by MNPs@SiO2(RITC)-induced lipid oxidation in a concentration-dependent manner. In addition, cell shrinkage with abnormal formation of focal adhesions and ~30% decreased total traction force were observed in cells treated with 1.0 µg/µL MNPs@SiO2(RITC) without specific interaction between MNPs@SiO2(RITC) and cytoskeletal proteins. Furthermore, the migratory activity of hBM-MSCs, which was highly related to membrane fluidity and cytoskeletal abnormality, decreased significantly after MNPs@SiO2(RITC) treatment. These observations indicated that the migratory activity of hBM-MSCs was impaired by MNPs@SiO2(RITC) treatment due to changes in stem-cell biophysical properties and related biological functions, highlighting the important mechanisms via which nanoparticles impair migration of hBM-MSCs. Our findings indicate that nanoparticles used for stem cell trafficking or clinical applications should be labelled using optimal nanoparticle concentrations to preserve hBM-MSC migratory activity and ensure successful outcomes following stem cell localisation.
Keywords: magnetic nanoparticles, human bone marrow-derived mesenchymal stem cells, membrane fluidity, focal adhesion, cytoskeletal abnormality
1. Introduction
Nanoparticles are being increasingly used for disease diagnosis and therapy and cell tracing [1,2,3]. Among nanoparticles, magnetic nanoparticles (MNPs) and MNPs coated with biocompatible polymers and silica for safety are used for in vitro cell labelling, fluorescence-based in vivo cell tracking, and magnetic resonance imaging (MRI)-based stem cell-labelled in vivo tracing [4,5,6,7]. However, detailed information regarding the biophysical effects of nanoparticles at the cellular level is still limited.
Mesenchymal stem cells (MSCs) are used in biomedical applications (cytotherapy) for multiple sclerosis and for cardiovascular, ischemic, and neurodegenerative disorders [8,9,10,11,12]. In particular, bone marrow-derived MSCs (BM-MSCs) possess useful characteristics, including high degree of plasticity, trophic factor secretion, and immune response suppression capability [13,14,15]. Thus, human BM-MSCs (hBM-MSCs) are considered promising therapeutic candidates for clinical application [12,16,17,18]. For successful cytotherapeutic outcomes using stem cells [19,20], nanoparticle-based methods for tracking the localization of transplanted cells in the body are essential to ensure their distribution in the impaired tissue [21]. However, nanoparticle-induced biophysical disturbances caused by reactive oxygen species (ROS) generation, which result in changes in normal physiological redox-regulated functions and cellular alteration, are matters of concern [22]. For example, silica-coated magnetic nanoparticles incorporating rhodamine B isothiocyanate (MNPs@SiO2(RITC)) induce ROS production, leading to endoplasmic reticulum (ER) stress, reduced proteasome activity, and altered cellular metabolism [23,24,25].
Nanoparticle-induced ROS oxidize proteins to generate protein radicals [22] and induce lipid peroxidation [26], which impairs the functions of the plasma membrane [27,28]. These oxidative cleavage events deplete unsaturated phospholipids and cholesterol in the cell membrane, leading to loss of the fluidity and permeability of the membrane and, thereby, affecting its physiological functions [29,30,31]. Several studies have reported that lipid peroxidation decreases membrane fluidity, indicating that the membrane fluidity of nanoparticle treated-cells can change due to oxidative stress-induced membrane damage [30,32]. However, the relationship between nanoparticle-induced lipid peroxidation and membrane fluidity remains unclear.
The membrane and the cytoskeleton are tightly linked via phosphoinositides, especially via focal adhesion and actin assembly [33,34,35]. Therefore, membrane damage, caused by nanoparticle-induced oxidative stress, is highly related to the cytoskeleton. Cell morphology is determined by the balance between adhesion and tension. Nanoparticles disrupt the cytoskeleton to affect focal adhesion proteins and, thus, adhesion [36] initiated by the lamellipodia (branched actin filaments) and filopodia (extended finger-like protrusions) as focal complexes [37]. During cell death; membrane repair; and osmotic-, oxidative-, and heat stress; these structures are abolished, such that cells display a rounded and shrunken morphology [38,39]. However, studies on the effect of nanoparticles on cell adhesion and tension are limited.
The focal adhesion of hBM-MSCs is strongly associated with changes in cellular traction forces [40]. Elastomeric pillar arrays are considered excellent for measuring cellular traction force by calculating the nanometric level of pillar deflection [41,42]. In addition, sub-micron pillar arrays have been shown to mimic continuous substrates of specific rigidity [43]. Thus, biophysical changes in nanoparticle-treated cells have been quantitatively studied using elastomeric submicron pillars [41,43].
In this study, we aimed to investigate the biophysical properties of MNPs@SiO2(RITC)-treated hBM-MSCs, such as membrane fluidity (using total internal reflection fluorescence microscopy (TIRFM)), traction force (using micropillars), cytoskeletal characteristics, and migratory activity.
2. Materials and Methods
2.1. MNPs@SiO2(RITC) and Silica Nanoparticles (NPs)
MNPs@SiO2(RITC) particles, composed of a ~9 nm cobalt ferrite core (CoFe2O3) chemically bonded to rhodamine isothiocyanate dye (RITC) and coated by a silica shell [4], were purchased from BITERIALS (Seoul, Korea). Previously, these nanoparticles have been characterized for confirming their quality [44]. Size distribution and morphology are important factors determining the uniformity of nanoparticles and were analyzed using electron and atomic microscopy [44]. Hydrodynamic size, polydispersity, and surface charge were determined using dynamic light scattering [45]. The purity and contents of nanoparticles are usually analyzed using an X-ray based technique [44]. In this study, X-ray diffraction (XRD) analysis using a high-power X-Ray diffractometer (Ultima III, Rigaku, Japan) confirmed the structure of MNPs@SiO2(RITC) (data not shown). The silica NPs were composed of identical materials and were of a similar size as the MNPs@SiO2(RITC) shell, and their biological effects were similar to those of MNPs@SiO2(RITC) [23,24,46,47]. The diameters of the MNPs@SiO2(RITC) and silica NPs were 50 nm, and the zeta potential of MNPs@SiO2(RITC) was between −40 to −30 mV [4,46]. A previous study determined ~105 particles of MNPs@SiO2(RITC) per cell in MNPs@SiO2(RITC)-treated MCF-7 cells using inductively coupled plasma atomic emission spectrometry [4]. Furthermore, in previous reports, the dosage was determined by measuring the fluorescence intensity of HEK293 cells treated with MNPs@SiO2(RITC) at concentrations ranging from 0.01 to 2.0 μg/μL for 12 h. The optimal concentration of MNPs@SiO2(RITC) was 0.1 µg/µL for in vitro use, whereas 1.0 µg/µL was the plateau concentration for cellular uptake [24]. Furthermore, MNPs@SiO2(RITC) concentrations ranging from 0 to 1.0 μg/μL have been used for MRI contrasting without toxicological effects on human cord blood-derived MSCs [48], and caused changes in gene expression and metabolic profiles similar to those of the control HEK293 cells at 0.1 µg/µL [24]. In addition, the uptake efficiency of MNPs@SiO2(RITC) almost plateaued at 1.0 µg/µL in HEK293 cells [24,25]. The dose-dependent fluorescence intensity of MNPs@SiO2(RITC)-labelled hBM-MSCs was similar to those of labelled HEK293 cells. In addition, the viability of human cord blood-derived MSCs was determined to assess the cytotoxic effect of MNPs@SiO2(RITC) after 24, 48, and 72 h of treatment with 0–1.0 µg/µL MNPs@SiO2(RITC); compared to the control group, no significant cytotoxic effect was observed [48]. Therefore, in this study, hBM-MSCs were treated with 0.1 µg/µL (low dose) MNPs@SiO2(RITC)or 1.0 µg/µL (high dose), similarly to previous reports [23,24,47].
2.2. Cell Culture
hBM-MSCs were purchased from PromoCell (Heidelberg, Germany) and were cultured as described in previous studies [49,50]. Briefly, the cells were rinsed with phosphate buffered saline (PBS), resuspended, cultured in Dulbecco’s low-glucose modified Eagle’s medium (DMEM, Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA), 100 units/mL penicillin, and 100 μg/mL streptomycin (Gibco, USA), and incubated in a 5% humidified CO2 chamber at 37 °C. The hBM-MSC surface markers, CD73 and CD105, and negative markers of hBM-MSCs, namely, CD34 and CD45, were analyzed and maintained (data not shown).
2.3. Morphological Analysis of hBM-MSCs
To evaluate the MNPs@SiO2(RITC)-induced morphological changes, hBM-MSCs were treated with 0.1 and 1.0 µg/µL of MNPs@SiO2(RITC) for 12 h. Images were acquired with an Axio Vert 200M fluorescence microscope (Zeiss, Jena, Germany). The excitation wavelength for MNPs@SiO2(RITC) was 530 nm.
2.4. Cell Viability Assay
For analysis of cell viability, the CellTiter 96-cell proliferation assay kit (MTS, Promega, Madison, WI, USA) was used, according to the manufacturer’s instructions. Briefly, 2 × 104 hBM-MSCs were seeded on 96-well assay plates. After 16 h, the hBM-MSCs were washed with PBS and treated with MNPs@SiO2(RITC) for 12 h. The hBM-MSCs were then washed with PBS to remove excess MNPs@SiO2(RITC), and MTS solution was added to each well (1/10 volume of media). Subsequently, the plate was incubated for 1 h in a 5% CO2 chamber maintained at 37 °C. The absorbance of the soluble formazan was measured using a plate reader (Molecular Devices, San Jose, CA, USA) at 490 nm. Values were normalized relative to the protein absorbance value for each corresponding group.
2.5. Evaluation of Intracellular ROS Levels in hBM-MSCs
Intracellular ROS levels in hBM-MSCs were evaluated using DCFH-DA staining (Cell Biolabs, San Diego, CA, USA) according to the manufacturer’s protocol. Briefly, control and MNPs@SiO2(RITC)-treated hBM-MSCs were resuspended in 10 µM DCFH-DA and incubated in a 5% CO2, 37 °C chamber for 1 h. The hBM-MSCs were washed twice with PBS, and DCF fluorescence was measured using a fluorescence microplate reader (Gemini EM, Molecular Devices, Sunnyvale, CA, USA) at 480 nm excitation and 530 nm emission wavelengths.
2.6. Evaluation of Lipid Peroxidation
Peroxidized lipids in control and MNPs@SiO2(RITC)-treated hBM-MSCs were quantified using a lipid peroxidation kit (Cayman Chemical, Ann Arbor, MI, USA) according to the manufacturer’s instructions. Briefly, hBM-MSCs were treated with MNPs@SiO2(RITC) for 12 h. Subsequently, hBM-MSCs were trypsinized and washed twice with PBS. The hBM-MSCs were transferred to glass tubes, and the lipids were extracted in crystalline solid-saturated methanol and cold chloroform. The mixture was centrifuged (1500× g, 0 °C, 5 times) to form two layers, and the bottom chloroform layer was collected. The samples were mixed with 2.25 mM ferrous sulphate, 0.1 M hydrochloric acid, and 1.5% ammonium thiocyanate in methanol at a 9:1 ratio. Next, the mixtures were incubated at room temperature for 5 min. Ferric ions were produced in the reaction, and the level of peroxidized unsaturated lipids was evaluated using thiocyanate as a chromogen. Absorbance was measured at 500 nm using a quartz cuvette and microplate reader (Molecular Devices, San Jose, CA, USA).
2.7. Measurement of Membrane Fluidity
Membrane fluidity of hBM-MSCs was measured using a homemade combined differential interference contrast (DIC)-total internal reflection fluorescence microscopy (TIRFM) experimental system [49,51]. The procedure was based on a previously described protocol [52,53]. Briefly, hBM-MSCs were cultured on 0.13–0.16 mm thick cover slips and treated with MNPs@SiO2(RITC) for 12 h. The hBM-MSCs were incubated with media containing 10 µM Laurdan in a 5% CO2, 37 °C chamber for 2 h. The hBM-MSCs were washed twice with PBS and incubated with fixation buffer (Cytofix; BD, San Jose, CA, USA). Subsequently, the hBM-MSC-containing cover slips were mounted onto 0.13–0.16 mm thick cover slips with mounting medium (Prolong Gold antifade; Molecular Probes, Eugene, OR, USA). Laurdan fluorescence was observed with a 100× objective lens (oil-type, Olympus UPLFL 1.3 N.A., W.D. 0.1 mm) and an A CCD camera (QuantEM 512SC, Photometrics, Tucson, AZ, USA). The fluorescence intensity was measured using an excitation wavelength of 405 nm, and emission fluorescence was detected using 420 nm and 473 nm bandpass filter (resolution: ±5 nm). As a parameter of membrane fluidity, the generalized polarizations (GP) = (fluorescence intensity at 420 nm—fluorescence intensity at 473 nm)/(fluorescence intensity at 420 nm + fluorescence intensity at 473 nm) was calculated, and pseudo-colored GP value images were generated using the Image J 1.48v software (NIH, Bethesda, MD, USA) [54]. Gauss distributions were generated using the nonlinear fitting algorithm in Sigma Plot 10.0 (Systat Software Inc., San Jose, CA, USA).
2.8. Immunocytochemistry
hBM-MSCs were seeded on cover slips and treated with 0.1 μg/μL and 1.0 μg/μL MNPs@SiO2(RITC) for 12 h. The cells were, then, fixed in Cytofix buffer (BD, San Jose, CA, USA). To reduce non-specific binding, the cover slips were blocked with PBS containing 2% bovine serum albumin (BSA) and 0.1% Triton-X100 (Sigma-Aldrich, St Louis, MO, USA). For actin labelling, hBM-MSCs were then incubated with Alexa Fluor 488-conjugated phalloidin (Molecular Probe, Carlsbad, CA, USA, 1:200), diluted in blocking buffer, for 1 h at room temperature. For β-tubulin and vinculin labelling, hBM-MSCs were incubated with anti-β-tubulin mouse polyclonal antibody (Molecular Probes, Carlsbad, CA, USA, 1:200) or anti-vinculin rabbit monoclonal antibody (Becton Dickinson, Franklin Lakes, NJ, USA, 1:200) diluted in blocking buffer, for 12 h at 4 °C. Following three times rinsing in PBS containing 0.1% Triton-X100, the cells were incubated with Alexa Fluor 488-conjugated anti-mouse goat polyclonal antibody or Alexa Fluor 647-conjugated anti-rabbit goat polyclonal antibody for 1 h at room temperature. The labelled cells were washed thrice with PBS containing 0.1% Triton-X100 and incubated with PBS containing 10 µg/mL Hoechst 33342 for 10 min at room temperature to label the nuclei. After washing three times with PBS, cover slips were mounted onto slides using Prolong Gold antifade mounting medium (Molecular Probes). Fluorescent images were acquired using confocal laser scanning microscopy (LSM710, Carl Zeiss Microscopy GmbH, Jena, Germany). The excitation wavelengths for Alexa Fluor 488, Hoechst 33342, and MNPs@SiO2(RITC) were 488, 405, and 530 nm, respectively. The attached hBM-MSCs areas were analyzed using the Image J software.
2.9. Western Blotting
hBM-MSCs were seeded at a density of 2 × 105 cells/well in 6-well plates and cultured for 36 h. hBM-MSCs were treated with 0.1 or 1.0 µg/µL MNPs@SiO2(RITC) for 12 h and lysed in radioimmunoprecipitation assay (RIPA) buffer. The lysates were vortexed and incubated at 4 °C for 1 h. Then, the lysates were centrifuged at 14,000× g for 15 min at 4 °C, and the supernatants were collected. Next, 40 µg protein was subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE) and transferred onto nitrocellulose membranes. The membranes were blocked with 3% non-fat milk for 1 h at room temperature and incubated with primary antibody overnight at 4 °C. The following primary antibodies were used: t-c-SRC (1:2000, Santa Cruz Technologies, Santa Cruz, CA, USA), p-c-SRC (1:2000, Santa Cruz Technologies, Santa Cruz, CA, USA), t-FAK (1:2000, Santa Cruz Technologies, Santa Cruz, CA, USA), p-FAK (1:2000, Santa Cruz Technologies, Santa Cruz, CA, USA), and β-actin (1:5000, Cell Signaling, La Jolla, CA, USA). Secondary antibodies were used at a dilution of 1:2000 (Santa Cruz Technologies, USA). The blots were developed using enhanced chemiluminescence solution (ECL, Thermo Scientific, Waltham, MA, USA), and luminescence was captured on medical blue X-ray film (AGFA, Mortsel, Belgium) in a dark room.
2.10. Microfabrication of Pillar Arrays
A standard photolithograph was used to fabricate a mold with arrays of holes over a silicon wafer [55]. To fabricate pillar arrays, Polydimethylsiloxane (PDMS) was mixed at 10:1 ratio with its curing agent (Sylgard 184; Dow Corning, Midland, MI, USA) and degassed for 15 min. Next, it was spin-coated over the mold, degassed again for 30 min to remove trapped air bubbles within the mixture, and cured at 80 °C for 4 h and 30 min until the Young modulus of the PDMS reached 2 ± 0.1 MPa. Subsequently, the cured pillar array was carefully removed from the mold. In the case of micron scale pillars, cellular contractions occurred around individual pillars (diameter 2 µm) [43,56]. Thus, we generated linearly arranged pillar arrays of 900 nm diameter, 1 µm height, and a pillar diameter two times the center-to-center distance between pillars. The bending stiffness of the pillar was calculated based on Euler-Bernoulli beam theory [57]:
where D is the diameter, L is the length, and E is the Young modulus of the pillar [43]. The bending stiffness (k) of the pillar arrays used in this study was 28.8 nN/µm.
2.11. Measurement of Traction Force
Images of pillars in hBM-MSCs were captured using a live cell chamber at 1 Hz in a fluorescence microscope (Deltavision, GE Healthcare, Chicago, IL, USA) equipped with a camera (CoolSNAP HQ2, Photometrics) at 37 °C and 5% humidity. The place of each pillar in each frame was determined using the pillar tracking plugin (PillarTracker 1.1.3 version) of the Image J software. PillarTracker uses the pillar reconstruction algorithm to establish an exact grid of the pillar arrays, thus allowing users to automatically detect and track the locations of the pillars. Throughout this study, pillars with no cell contact were used as reference pillars. To account for stage drift, the average displacement of the reference pillars was deducted from the displacement data of pillars deflected by hBM-MSCs. To avoid unwanted displacement of pillars by MNPs@SiO2(RITC), MNPs@SiO2(RITC)-treated cells were washed five times using Dulbecco’s phosphate buffered saline (DPBS) before seeding on a pillar array. The displacement of each pillar was multiplied by its bending stiffness to calculate the traction force.
2.12. Wound Healing Assay
hBM-MSCs were seeded and grown to 100% confluence in 6-well plates, followed by washing in PBS. Cell monolayers were wounded with a 200-µL micropipette tip in two different places in each well, treated with MNPs@SiO2(RITC) in serum-free media, and allowed to migrate for 16 h. Images of the wounded areas were captured under an Axio Vert 200M fluorescence microscope (Zeiss, Jena, Germany) at 0 and 16 h. The excitation wavelength for MNPs@SiO2(RITC) was 530 nm. Migration activity was quantified by analyzing the cell number.
2.13. Invasion Assay
Invasion assays of hBM-MSCs were performed using an 8-µm pore size transwell polycarbonate membrane (Corning, Corning, NY, USA). The upper side of the membrane was coated with Matrigel (1:10 dilution in 0.01 M Tris pH 8.0, 0.7% NaCl) for 2 h at 37 °C. hBM-MSCs were treated with MNPs@SiO2(RITC) for 12 h. Next, 2.5 × 104 hBM-MSCs were transferred to the upper chamber of the transwell in serum-free media, and 10% FBS containing medium was added to the lower chamber as a chemoattractant. The cells were incubated for 12 h at 37 °C. Subsequently, the cells on the upper side of the membrane were removed with a cotton swab, and the invading cells on the lower side of the membrane were fixed in Cytofix buffer (BD, San Jose, CA, USA) and stained with Hoechst 33342. Images were acquired using an Axio Vert 200M fluorescence microscope (Zeiss, Jena, Germany). The excitation wavelengths for MNPs@SiO2(RITC) and Hoechst 33342 were 530 nm and 405 nm, respectively. The number of invading cells was counted using the Image J software.
2.14. Statistical Analysis and Error Correction
The results were analyzed using one-way analysis of variance (ANOVA) with Bonferroni’s multiple-comparison test of the IBM-SPSS software (IBM Corp., Armonk, NY, USA). Differences were considered significant for p values < 0.05. In the micropillar experiments, errors of the pillar deflections were corrected by reducing the average deflection of pillars outside the cell.
3. Results
3.1. Decrease in Cell Viability and ROS Generation of MNPs@SiO2(RITC)-Treated hBM-MSCs
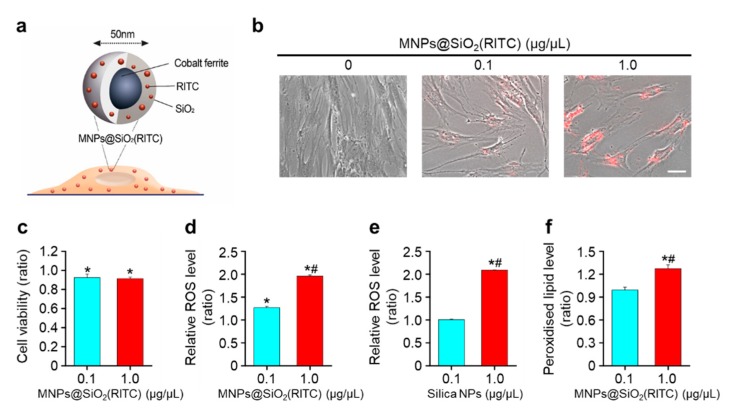
To evaluate the viability of and ROS generation in MNPs@SiO2(RITC)-treated hBM-MSCs, hBM-MSCs were treated with MNPs@SiO2(RITC) for 12 h before analysis (Figure 1a). A monolayer of hBM-MSCs was clearly observed for the non-treated control cells, while the monolayer was disintegrated for the MNPs@SiO2(RITC)-treated hBM-MSCs (Figure 1b). Furthermore, compared to that in the non-treated control, the viability of hBM-MSCs decreased by ~10% upon treatment with 0.1 and 1.0 µg/µL MNPs@SiO2(RITC) (Figure 1c). However, the viability of cells treated with 0.1 µg/µL MNPs@SiO2(RITC) was not statistically significantly different from that of cells treated with 1.0 µg/µL MNPs@SiO2(RITC). Intracellular ROS generation was evaluated using 2′,7′-dichlorodihydrofluorescin diacetate (DCFH-DA) staining in MNPs@SiO2(RITC)-treated hBM-MSCs. Treatment with MNPs@SiO2(RITC) and 50 nm-sized silica NPs increased intracellular ROS levels (Figure 1d,e). In particular, compared to that in non-treated control cells and cells treated with 0.1 µg/µL NPs, intracellular ROS levels increased by more than 50% in cells treated with 1.0 µg/µL MNPs@SiO2(RITC) and silica-NPs. Furthermore, the level of ROS-induced peroxidized lipids increased by ~30% in hBM-MSCs treated with 1.0 µg/µL MNPs@SiO2(RITC) (Figure 1f).
Figure 1.
Intracellular reactive oxygen species (ROS) generation and lipid peroxidation in MNPs@SiO2(RITC)-treated hBM-MSCs. (a) Schematic showing MNPs@SiO2(RITC) composition. (b) Morphological analysis of non-treated control and MNPs@SiO2(RITC)-treated hBM-MSCs. Scale bar = 50 µm (c) Cell viability assay with hBM-MSCs treated with MNPs@SiO2(RITC) for 12 h. Evaluation of intracellular ROS generation using DCFH-DA for 12 h in HEK293 cells treated with (d) MNPs@SiO2(RITC) and (e) silica NPs. The non-oxidized DCFH-DA was used as the blank. (f) Evaluation of peroxidized lipids using ferrous thiocyanate. Ferrous thiocyanate was used as the blank. Data represent mean ± SD of three independent experiments. * p < 0.05 vs. non-treated control, # p < 0.05 for the comparison between 0.1 and 1.0 µg/µL MNPs@SiO2(RITC) or silica NP-treated cells. Data represent mean ± SD of three independent experiments.
3.2. Reduction in Membrane Fluidity of hBM-MSCs after MNPs@SiO2(RITC) Treatment
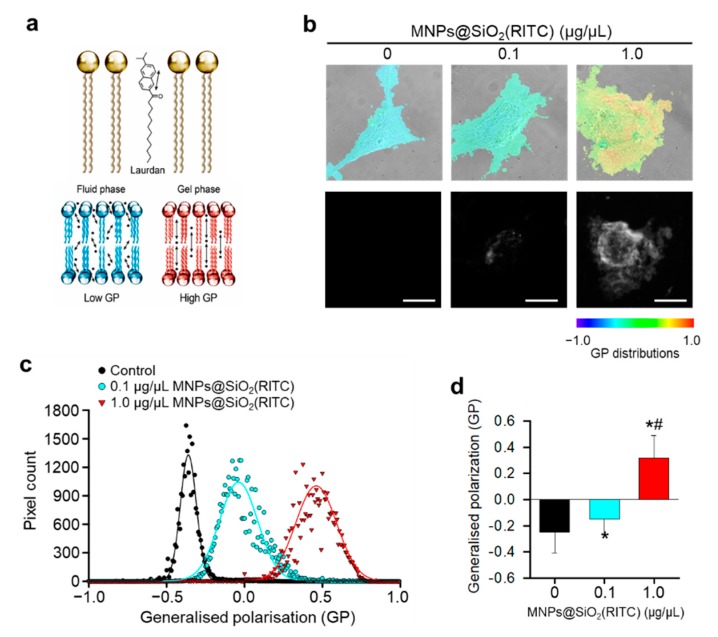
To analyze changes in membrane fluidity due to peroxidation of lipids, the MNPs@SiO2(RITC)-treated hBM-MSCs were stained with Laurdan and generalized polarization (GP) values were calculated using TIRFM (Figure 2a). The number of high-GP areas on the hBM-MSC surface—Corresponding to rigid domains—Increased upon MNPs@SiO2(RITC) treatment. In particular, the abundantly distributed region of MNPs@SiO2(RITC) majorly co-localized with the high GP-distribution region in a GP scale of −1.0 to 1.0 (Figure 2b). GP frequency distribution values of MNPs@SiO2(RITC) treated-hBM-MSCs were subtracted from the corresponding values of the non-treated control hBM-MSCs to obtain frequency difference curves (Figure 2c) and total mean GP values (Figure 2d).
Figure 2.
Laurdan generalized polarizations (GP) images and GP frequency distributions of hBM-MSCs treated with MNPs@SiO2(RITC) for 12 h. (a) Schematic of Laurdan for measuring membrane GP value. (b) Merged differential interference contrast (DIC) and total internal reflection fluorescence microscopy (TIRFM) images of human bone marrow-derived mesenchymal stem cells (hBM-MSCs) in each upper panel. Distributions of magnetic nanoparticles incorporating rhodamine B isothiocyanate (MNPs@SiO2(RITC)) are indicated in each lower panel. GP distributions ranged from −1.0 to 1.0. Scale bar = 2.5 µm. (c) GP frequency distributions of cells. GP values of each pixel are represented as dots and were fitted to Gaussian distributions. (d) Total GP values. Data represent mean ± SD of three independent experiments (n = 10). * p < 0.05 vs. non-treated control, # p < 0.05 for the comparison between cells treated with 0.1 and 1.0 µg/µL of MNPs@SiO2(RITC).
3.3. Cytoskeletal Abnormality in MNPs@SiO2(RITC)-Treated hBM-MSCs
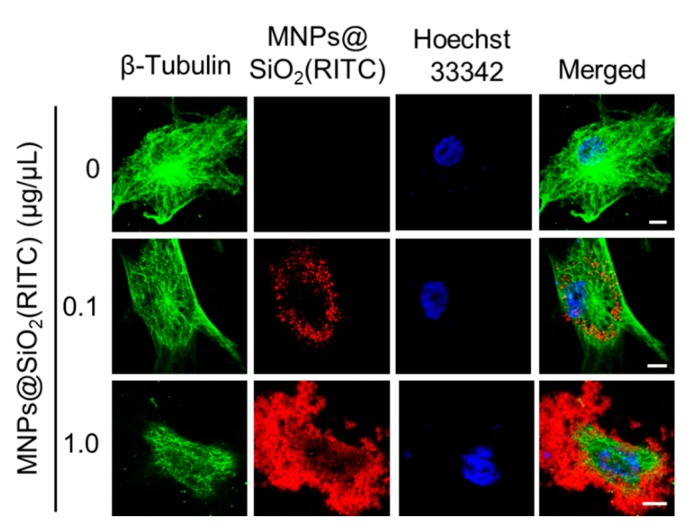
We investigated the changes in cell morphology and focal adhesion in MNPs@SiO2(RITC)-treated hBM-MSCs. As shown in Figure 2, cell shrinkage was observed with reduction in the attached area of MNPs@SiO2(RITC)-treated hBM-MSCs, indicating cytoskeletal changes and abnormal adhesion, which are followed by reduced membrane fluidity due to lipid peroxidation. Furthermore, immunocytochemical analysis of β-tubulin, a major cytoskeletal protein, showed shrinkage of the cell and β-tubulin structure, although interaction between MNPs@SiO2(RITC) and β-tubulin was not detected (Figure 3).
Figure 3.
Tubulin-based analysis of the cytoskeleton of hBM-MSCs treated with MNPs@SiO2(RITC) for 12 h. β-Tubulin stained images of hBM-MSCs treated with MNPs@SiO2(RITC). Blue, Hoechst 33342; Green, β-tubulin; red, MNPs@SiO2(RITC). Scale bar = 10 μm.
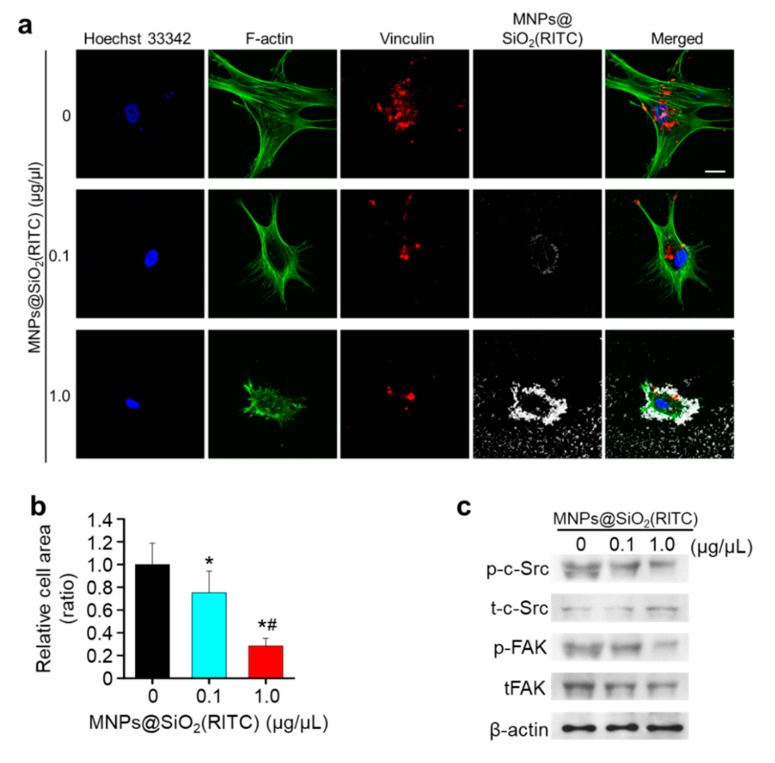
Immunocytochemical analysis of F-actin, a major cytoskeletal protein, co-stained with vinculin, a focal adhesion marker, showed that unlike that in non-treated control hBM-MSCs, lamellipodia and filopodia disappeared, and vinculin was abnormally congregated in the central region of hBM-MSCs treated with 1.0 µg/µL MNPs@SiO2(RITC) (Figure 4a). However, specific interaction between MNPs@SiO2(RITC) and F-actin or vinculin was not detected. Furthermore, compared to that in the non-treated control, the attached relative cell area was reduced in 0.1 and 1.0 µg/µL MNPs@SiO2(RITC)-treated hBM-MSCs (Figure 4b). Levels of phosphorylated proto-oncogene tyrosine-protein kinase SRC (c-SRC) and focal adhesion kinase (FAK), which are activated forms of the focal adhesion proteins SRC and FAK, respectively, were reduced in cells treated with 1.0 µg/µL MNPs@SiO2(RITC) (Figure 4c).
Figure 4.
Shrinkage and abnormal focal adhesion of hBM-MSCs treated with MNPs@SiO2(RITC). (a) Images of F-actin based cytoskeleton and focal adhesion in MNPs@SiO2(RITC)-treated HEK293 cells. Green, F-actin; red, vinculin; white, MNPs@SiO2(RITC); blue, Hoechst 33342. Scale bar = 20 µm. (b) Relative attached area of MNPs@SiO2(RITC)-treated hBM-MSCs compared to non-treated control. Data represent mean ± SD (n > 30). * p < 0.05 vs. non-treated control. # p < 0.05 for the comparison between 0.1 and 1.0 µg/µL MNPs@SiO2(RITC)-treated cells. (c) Immunoblotting analysis associated with focal adhesion. p-, phosphorylated protein; t, total protein. β-Actin was used as an internal control.
3.4. Reduction in Traction Force of hBM-MSCs after MNPs@SiO2(RITC) Treatment
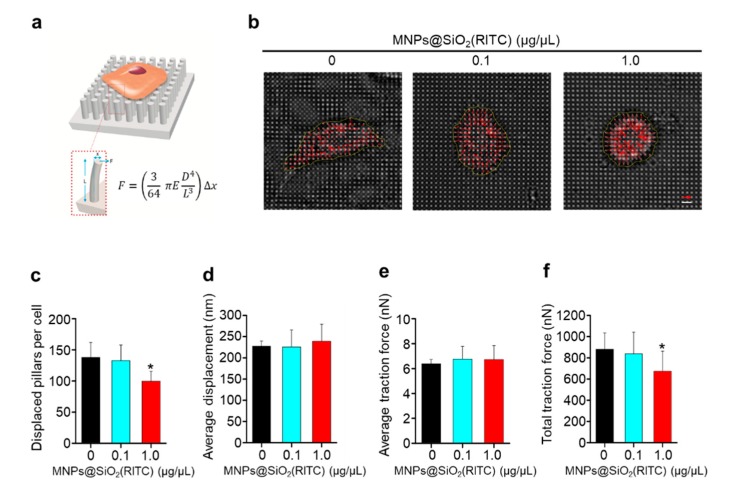
To analyze, in detail, the parameters of cell adhesion, cell spread areas were measured using images of MNPs@SiO2(RITC)-treated hBM-MSCs and submicron pillars at 12 h after cell seeding (Figure 5a). Compared to that in the non-treated control cells, cell spreading of hBM-MSCs treated with 0.1 and 1.0 µg/µL MNPs@SiO2(RITC) decreased (Figure 5b). Furthermore, the spread areas of hBM-MSCs treated with 0.1 and 1.0 µg/µL MNPs@SiO2(RITC) were significantly smaller than those of the non-treated control cells (Figure 5c).
Figure 5.
Change in pillar deflection, traction force, aspect ratio, and surface area of hBM-MSCs treated with MNPs@SiO2(RITC). (a) Schematic of traction force measurement using a micropillar. F = traction force; E = Young modulus of the pillar; D = diameter of pillar; L = length of pillar; Δx = pillar displacement. (b) Representative images showing the concentration of MNPs@SiO2(RITC) inside the cell, pillar deflections, and magnified pillar deflections at the edge of the cell (left to right). The red arrow represents 356 nm of deflection and the white bar represents 8 µm. The yellow line indicates the approximate cell boundary. The direction and length of the red arrow indicate the magnitude and direction of pillar deflection, respectively. (c) Displaced pillar number of pillar array (d) Average displacement of each pillar under the cell. (e) Average traction force of each pillar under the cell and (f) total traction force of pillars beneath the cell. Data represent mean ± SD (n = 21). * p < 0.05 vs. non-treated control.
The pillar deflection in the magnified images was used to measure pillar displacement (Figure 5d) and calculate traction forces (Figure 5e,f). To calculate the traction force of a pillar, displacement of each pillar was multiplied by the bending stiffness of the pillar [43]. There were no significant changes in pillar displacement and the average traction force of MNPs@SiO2(RITC)-treated hBM-MSCs. However, the total traction force of hBM-MSCs treated with 1.0 µg/µL MNPs@SiO2(RITC) was significantly lower than those of hBM-MSCs treated with 0.1 µg/µL MNPs@SiO2(RITC) and non-treated control hBM-MSCs. The results showed that cell attachment was impaired after treatment with 1.0 µg/µL MNPs@SiO2(RITC). Taken together with the spread area and traction force data, this result implies that the decrease in total traction force of hBM-MSCs results from cell shrinkage upon MNPs@SiO2(RITC) treatment.
3.5. Reduction in Migratory Activity of hBM-MSCs after MNPs@SiO2(RITC) Treatment
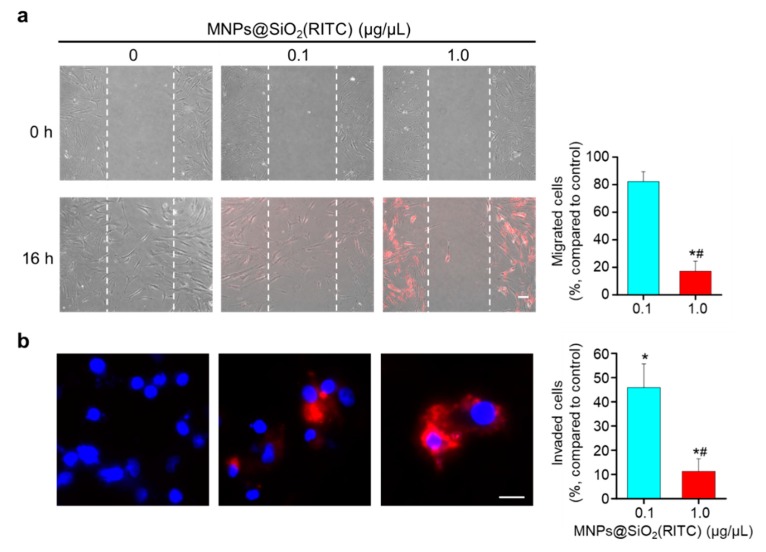
To evaluate membrane fluidity and adhesion-related biological functions, we assessed the effect of treatment with 0.1 or 1.0 µg/µL MNPs@SiO2(RITC) on the migratory activity of hBM-MSCs using wound healing and invasion assays. Compared to those of non-treated controls and cells treated with 0.1 μg/μL MNPs@SiO2(RITC), the migratory ability of hBM-MSCs treated with 1.0 μg/μL MNPs@SiO2(RITC) was seen to be significantly impaired through the wound healing assay (Figure 6a). Results of the invasion assay showed that compared to that of the non-treated controls, the invasion ability of hBM-MSCs was significantly impaired by MNPs@SiO2(RITC) treatment in a dose-dependent manner (Figure 6b).
Figure 6.
Migratory activity of hBM-MSCs treated with MNPs@SiO2(RITC). (a) Representative images of wound healing assay and quantitative image analysis. Images of the initial wounded (0 h) layer are shown the upper panels. Images of cells after MNPs@SiO2(RITC) treatment for 16 h are shown in the lower panels. Scale bar = 100 μm. Quantitative image analysis of migrated cells in MNPs@SiO2(RITC)-treated hBM-MSCs are shown in the bar graph. (b) Representative images of hBM-MSCs and quantitative image analysis of the invasion assay results after MNPs@SiO2(RITC) treatment for 12 h. Red, MNPs@SiO2(RITC); blue, Hoechst 33342. Scale bar = 20 μm. Quantitative image analysis of invaded cells in MNPs@SiO2(RITC)-treated hBM-MSCs are shown in the bar graph. Data represent mean ± SD of three independent experiments. Data represent mean ± SD of three independent experiments. * p < 0.05 vs. non-treated control, # p < 0.05 for the comparison between cells treated with 0.1 and 1.0 µg/µL of MNPs@SiO2(RITC).
4. Discussion
This study used molecular cellular biology tests, TIRFM measurement of membrane fluidity, micropillar measurement of traction force, and invasion and migration analyses to evaluate the biophysical effects of MNPs@SiO2(RITC) treatment in hBM-MSCs. Our results indicated that MNPs@SiO2(RITC) usage should be minimized in cell labelling to preserve the biophysical effect of hBM-MSCs.
Reduction in cell viability and ROS generation have been reported in MSCs treated with nanoparticles of various types and sizes [58,59,60]. In this study, cell viability decreased slightly by about 10% in hBM-MSCs treated with both 0.1 and 1.0 µg/µL MNPs@SiO2(RITC) and silica nanoparticles. This result is consistent with that of a previous study on 0–1.0 µg/µL MNPs@SiO2(RITC)-treated human cord blood–derived MSCs [48]. However, the viability of HEK293 cells treated with MNPs@SiO2(RITC) has been shown to decrease by about 1%–3% [24]. These discrepancies may be related to the cell-specific characteristics, as MSCs are more sensitive to excessive ROS induced by nanoparticles than differentiated cell lines [61].
We analyzed the biophysical changes and membrane fluidity in MNPs@SiO2(RITC)-treated hBM-MSCs, the effects of lipid peroxidation by MNPs@SiO2(RITC)-induced ROS, and the physical interaction between the cell membrane and MNPs@SiO2(RITC). Our results suggested that cell membrane damage induced by MNPs@SiO2(RITC) can occur via direct interaction between membrane lipids and nanoparticles [62] and that lipid peroxidation in nanoparticle-treated hBM-MSCs can induce oxidative membrane damage, resulting in biological alterations [63,64]. In addition, the biological effects of MNPs@SiO2(RITC) were caused by their silica shell rather than the cobalt ferrite core compounds, as reported previously [23,24].
We also observed that MNPs@SiO2(RITC)-treated hBM-MSCs showed a rounded and shrunken morphology with disrupted cytoskeletal structure. However, specific interactions between MNPs@SiO2(RITC) and actin or tubulin were not observed. Generally, the cytoskeleton is tightly linked to the membrane via phosphoinositides and linker proteins, such as spectrin, Ezrin/radixin/moesin (ERM), and myosin-I [33,34,65]. Thus, biophysical changes in the membrane are reflected as cytoskeletal changes in hMSCs [65]. Based on the relationship between the cytoskeleton and ROS, high ROS levels have been shown to induce microtubule dysfunction and sever F-actin structure [35]. Furthermore, endocytosis is another potential mechanism underlying cytoskeletal changes. During endocytosis, cells undergo reorganization of the cytoskeleton and the membrane [66]. Thus, the cytoskeletal rearrangement may be caused by ROS generation and bulk endocytosis post-MNPs@SiO2(RITC) treatment.
Previous studies have suggested that the major mechanism underlying nanoparticle-induced cell shrinkage and abnormal formation of focal adhesions involves cytoskeletal depolymerization and an increase in cellular traction force [67,68,69]. We observed that the lamellipodia and filopodia structures of MNPs@SiO2(RITC)-treated hBM-MSCs were disrupted and the phosphorylation of FAK and c-SRC, which are markers of focal adhesion formation, was reduced. However, there were no changes in the traction forces, although the total traction force was reduced. These results indicated that MNPs@SiO2(RITC)-induced reduction of FAK and c-SRC phosphorylation corresponded with the reduction in the attached area of cells and the number of focal adhesions during cell shrinkage and cytoskeletal structure disruption after MNPs@SiO2(RITC) treatment.
We observed that the changes in biophysical properties, such as reduced membrane fluidity, cell shrinkage with disrupted cytoskeletal structure, and reduced total traction force in MNPs@SiO2(RITC)-treated hBM-MSCs, were tightly linked to each other and contributed to the reduction in the migratory activity of hBM-MSCs. These findings are supported by two previous findings: (i) Actin assembly-based protrusions and generation of traction forces are key processes in cell migration [70]. Furthermore, migratory activity is highly related to cellular biophysical properties, such as membrane fluidity and traction force [71,72]. (ii) Condensation of the cytoskeleton due to MNPs@SiO2(RITC) treatment increases cell stiffness, which impedes cellular movement in the contractile machinery (shrinkage) [71].
Evaluation of the migratory activity is a major requirement for hBM-MSC applications and for successful outcome of stem cell therapy and tracking studies [66]. In addition, appropriate localization of hBM-MSCs in damaged tissues is important for the therapeutic effect of trophic factors and cytokines secreted by hBM-MSCs [73]. Previously, we have showed that PKH-26, a red fluorescent cell labelling dye, labelled hBM-MSCs in a localized, spotted-like pattern, implying secretion of the red dye outside the cell, in the border region of a lesion in a rat model of ischemic stroke [50]. However, MNPs@SiO2(RITC)-labelled human umbilical cord blood–derived MSCs in vivo rarely appeared as spotted particles in a mouse model, suggesting retention of MNPs@SiO2(RITC) in the cell for good tracking efficacy [48]. Hence, we believe that MNPs@SiO2(RITC) is better than the PKH-26 dye for hBM-MSC tracing. Further studies are required (i.e., in vivo experiment) for tracking MNPs@SiO2(RITC)-treated hBM-MSCs.
Based on our analysis of the effect of MNPs@SiO2(RITC) treatment on the biophysical and biological functions of hBM-MSCs, we suggest the mechanism of action of MNPs@SiO2(RITC) as follows: (i) MNPs@SiO2(RITC) are internalized into hBM-MSCs; (ii) intracellular ROS are generated by the internalized MNPs@SiO2(RITC); (iii) the ROS induces peroxidation of lipids, which are major components of the cell membrane; (iv) peroxidation decreases membrane fluidity; (v) cell shrinkage is induced by the reduction in membrane fluidity; (vi) owing to the cell shrinkage, abnormality in adhesion and reduction in total traction force are induced; (vii) owing to the reduction in membrane fluidity and abnormality in adhesion, fundamentally induced by intracellular ROS, the migratory activity of MNPs@SiO2(RITC)-treated hBM-MSCs decreases. The effect of this treatment on other cell functions will be addressed in future investigations.
In conclusion, our findings suggest that high-dose MNPs@SiO2(RITC) can alter biophysical properties and reduce the migratory activity of MNPs@SiO2(RITC)-treated hBM-MSCs. Thus, nanoparticles used for stem cell trafficking or clinical applications should be labelled using optimal nanoparticle concentrations to preserve hBM-MSC migratory activity and ensure successful outcomes following stem cell localization.
Acknowledgments
The authors thank the Ajou Core-Facility Center for technical support regarding cell image analysis.
Author Contributions
Conceptualization, T.H.S. and G.L.; methodology, B.M., S.B. and C.A.; software, T.H.S.; validation, S.B., T.H.S. and A.A.K.; formal analysis, D.Y.L. and S.L.; investigation, S.B. and C.A.; resources, S.P. and S.H.K.; data curation, T.H.S. and A.A.K.; writing—original draft preparation, T.H.S. and G.L.; writing—review and editing, T.H.S., S.P. and G.L.; visualization, T.H.S.; supervision, G.L.; project administration, G.L.; funding acquisition, G.L.
Funding
This work was supported by grants from the National Research Foundation (NRF) funded by the Ministry of Science and ICT (MSIT) in Korea [2018R1D1A1B07049494] and [2016M3C7A1904392], a grant from the Korea Basic Science Institute (KBSI) National Research Facilities & Equipment Center (NFEC) funded by the Korea government (Ministry of Education) [2019R1A6C1010003], and the BioNano Health-Guard Research Center as Global Frontier Project (NRF-2018M3A6B2057299) funded by the Ministry of Science and ICT (MSIT) in Korea.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Kim D., Kim J., Park Y.I., Lee N., Hyeon T. Recent Development of Inorganic Nanoparticles for Biomedical Imaging. ACS Cent. Sci. 2018;4:324–336. doi: 10.1021/acscentsci.7b00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parveen S., Misra R., Sahoo S.K. Nanoparticles: A boon to drug delivery, therapeutics, diagnostics and imaging. Nanomedicine. 2012;8:147–166. doi: 10.1016/j.nano.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 3.Kim J., Chhour P., Hsu J., Litt H.I., Ferrari V.A., Popovtzer R., Cormode D.P. Use of Nanoparticle Contrast Agents for Cell Tracking with Computed Tomography. Bioconjug. Chem. 2017;28:1581–1597. doi: 10.1021/acs.bioconjchem.7b00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoon T.J., Kim J.S., Kim B.G., Yu K.N., Cho M.H., Lee J.K. Multifunctional nanoparticles possessing a “magnetic motor effect” for drug or gene delivery. Angew. Chem. Int. Ed. 2005;44:1068–1071. doi: 10.1002/anie.200461910. [DOI] [PubMed] [Google Scholar]
- 5.Ding Y.F., Li S., Liang L., Huang Q., Yuwen L., Yang W., Wang R., Wang L.H. Highly Biocompatible Chlorin e6-Loaded Chitosan Nanoparticles for Improved Photodynamic Cancer Therapy. ACS Appl. Mater. Interfaces. 2018;10:9980–9987. doi: 10.1021/acsami.8b01522. [DOI] [PubMed] [Google Scholar]
- 6.Santoso M.R., Yang P.C. Magnetic Nanoparticles for Targeting and Imaging of Stem Cells in Myocardial Infarction. Stem Cells Int. 2016;2016:4198790. doi: 10.1155/2016/4198790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guldris N., Argibay B., Gallo J., Iglesias-Rey R., Carbo-Argibay E., Kolen’ko Y.V., Campos F., Sobrino T., Salonen L.M., Banobre-Lopez M., et al. Magnetite Nanoparticles for Stem Cell Labeling with High Efficiency and Long-Term in Vivo Tracking. Bioconjug. Chem. 2017;28:362–370. doi: 10.1021/acs.bioconjchem.6b00522. [DOI] [PubMed] [Google Scholar]
- 8.Lindvall O., Kokaia Z. Stem cells in human neurodegenerative disorders--time for clinical translation? J. Clin. Investig. 2010;120:29–40. doi: 10.1172/JCI40543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S., Wang X., Li J., Zhang J., Zhang F., Hu J., Qi Y., Yan B., Li Q. Advances in the Treatment of Ischemic Diseases by Mesenchymal Stem Cells. Stem Cells Int. 2016;2016:5896061. doi: 10.1155/2016/5896061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rivera F.J., de la Fuente A.G., Zhao C., Silva M.E., Gonzalez G.A., Wodnar R., Feichtner M., Lange S., Errea O., Priglinger E., et al. Aging restricts the ability of mesenchymal stem cells to promote the generation of oligodendrocytes during remyelination. Glia. 2019;67:1510–1525. doi: 10.1002/glia.23624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shim W.S., Jiang S., Wong P., Tan J., Chua Y.L., Tan Y.S., Sin Y.K., Lim C.H., Chua T., Teh M., et al. Ex vivo differentiation of human adult bone marrow stem cells into cardiomyocyte-like cells. Biochem. Biophys. Res. Commun. 2004;324:481–488. doi: 10.1016/j.bbrc.2004.09.087. [DOI] [PubMed] [Google Scholar]
- 12.Bang O.Y., Lee J.S., Lee P.H., Lee G. Autologous mesenchymal stem cell transplantation in stroke patients. Ann. Neurol. 2005;57:874–882. doi: 10.1002/ana.20501. [DOI] [PubMed] [Google Scholar]
- 13.Veronesi F., Giavaresi G., Tschon M., Borsari V., Nicoli Aldini N., Fini M. Clinical use of bone marrow, bone marrow concentrate, and expanded bone marrow mesenchymal stem cells in cartilage disease. Stem Cells Dev. 2013;22:181–192. doi: 10.1089/scd.2012.0373. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y., Tsai T.L., Li W.J. Strategies to retain properties of bone marrow-derived mesenchymal stem cells ex vivo. Ann. N. Y. Acad. Sci. 2017;1409:3–17. doi: 10.1111/nyas.13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuci S., Kuci Z., Schafer R., Spohn G., Winter S., Schwab M., Salzmann-Manrique E., Klingebiel T., Bader P. Molecular signature of human bone marrow-derived mesenchymal stromal cell subsets. Sci. Rep. 2019;9:1774. doi: 10.1038/s41598-019-38517-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee P.H., Kim J.W., Bang O.Y., Ahn Y.H., Joo I.S., Huh K. Autologous Mesenchymal Stem Cell Therapy Delays the Progression of Neurological Deficits in Patients with Multiple System Atrophy. Clin. Pharmacol. Ther. 2007;83:723–730. doi: 10.1038/sj.clpt.6100386. [DOI] [PubMed] [Google Scholar]
- 17.Suk K.T., Yoon J.H., Kim M.Y., Kim C.W., Kim J.K., Park H., Hwang S.G., Kim D.J., Lee B.S., Lee S.H., et al. Transplantation with Autologous Bone Marrow-Derived Mesenchymal Stem Cells for Alcoholic Cirrhosis: Phase 2 Trial. Hepatology. 2016;64:2185–2197. doi: 10.1002/hep.28693. [DOI] [PubMed] [Google Scholar]
- 18.Tse H.F., Kwong Y.L., Chan J.K., Lo G., Ho C.L., Lau C.P. Angiogenesis in ischaemic myocardium by intramyocardial autologous bone marrow mononuclear cell implantation. Lancet. 2003;361:47–49. doi: 10.1016/S0140-6736(03)12111-3. [DOI] [PubMed] [Google Scholar]
- 19.de Mel A., Murad F., Seifalian A.M. Nitric oxide: A guardian for vascular grafts? Chem. Rev. 2011;111:5742–5767. doi: 10.1021/cr200008n. [DOI] [PubMed] [Google Scholar]
- 20.Lee S., Yoon H.I., Na J.H., Jeon S., Lim S., Koo H., Han S.S., Kang S.W., Park S.J., Moon S.H., et al. In vivo stem cell tracking with imageable nanoparticles that bind bioorthogonal chemical receptors on the stem cell surface. Biomaterials. 2017;139:12–29. doi: 10.1016/j.biomaterials.2017.05.050. [DOI] [PubMed] [Google Scholar]
- 21.Pinho S., Macedo M.H., Rebelo C., Sarmento B., Ferreira L. Stem cells as vehicles and targets of nanoparticles. Drug Discov. Today. 2018;23:1071–1078. doi: 10.1016/j.drudis.2018.01.030. [DOI] [PubMed] [Google Scholar]
- 22.Fu P.P., Xia Q., Hwang H.M., Ray P.C., Yu H. Mechanisms of nanotoxicity: Generation of reactive oxygen species. J. Food Drug Anal. 2014;22:64–75. doi: 10.1016/j.jfda.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phukan G., Shin T.H., Shim J.S., Paik M.J., Lee J.K., Choi S., Kim Y.M., Kang S.H., Kim H.S., Kang Y., et al. Silica-coated magnetic nanoparticles impair proteasome activity and increase the formation of cytoplasmic inclusion bodies in vitro. Sci. Rep. 2016;6:29095. doi: 10.1038/srep29095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shim W., Paik M.J., Nguyen D.T., Lee J.K., Lee Y., Kim J.H., Shin E.H., Kang J.S., Jung H.S., Choi S., et al. Analysis of changes in gene expression and metabolic profiles induced by silica-coated magnetic nanoparticles. ACS Nano. 2012;6:7665–7680. doi: 10.1021/nn301113f. [DOI] [PubMed] [Google Scholar]
- 25.Shin T.H., Lee D.Y., Lee H.S., Park H.J., Jin M.S., Paik M.J., Manavalan B., Mo J.S., Lee G. Integration of metabolomics and transcriptomics in nanotoxicity studies. BMB Rep. 2018;51:14–20. doi: 10.5483/BMBRep.2018.51.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Potter T.M., Neun B.W., Stern S.T. Assay to detect lipid peroxidation upon exposure to nanoparticles. Methods Mol. Biol. 2011;697:181–189. doi: 10.1007/978-1-60327-198-1_19. [DOI] [PubMed] [Google Scholar]
- 27.Girotti A.W. Lipid hydroperoxide generation, turnover, and effector action in biological systems. J. Lipid Res. 1998;39:1529–1542. [PubMed] [Google Scholar]
- 28.Lehman S.E., Morris A.S., Mueller P.S., Salem A.K., Grassian V.H., Larsen S.C. Silica Nanoparticle-Generated ROS as a Predictor of Cellular Toxicity: Mechanistic Insights and Safety by Design. Environ. Sci. Nano. 2016;3:56–66. doi: 10.1039/C5EN00179J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Negre-Salvayre A., Coatrieux C., Ingueneau C., Salvayre R. Advanced lipid peroxidation end products in oxidative damage to proteins. Potential role in diseases and therapeutic prospects for the inhibitors. Br. J. Pharmacol. 2008;153:6–20. doi: 10.1038/sj.bjp.0707395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de la Haba C., Palacio J.R., Martinez P., Morros A. Effect of oxidative stress on plasma membrane fluidity of THP-1 induced macrophages. Biochimica et Biophysica Acta. 2013;1828:357–364. doi: 10.1016/j.bbamem.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 31.Blokhina O., Virolainen E., Fagerstedt K.V. Antioxidants, oxidative damage and oxygen deprivation stress: A review. Ann. Bot. 2003;91:179–194. doi: 10.1093/aob/mcf118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cazzola R., Rondanelli M., Russo-Volpe S., Ferrari E., Cestaro B. Decreased membrane fluidity and altered susceptibility to peroxidation and lipid composition in overweight and obese female erythrocytes. J. Lipid Res. 2004;45:1846–1851. doi: 10.1194/jlr.M300509-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Saarikangas J., Zhao H., Lappalainen P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol. Rev. 2010;90:259–289. doi: 10.1152/physrev.00036.2009. [DOI] [PubMed] [Google Scholar]
- 34.Geissler K.J., Jung M.J., Riecken L.B., Sperka T., Cui Y., Schacke S., Merkel U., Markwart R., Rubio I., Than M.E., et al. Regulation of Son of sevenless by the membrane-actin linker protein ezrin. Proc. Natl. Acad. Sci. USA. 2013;110:20587–20592. doi: 10.1073/pnas.1222078110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson C., Gonzalez-Billault C. Regulation of cytoskeletal dynamics by redox signaling and oxidative stress: Implications for neuronal development and trafficking. Front. Cell. Neurosci. 2015;9:381. doi: 10.3389/fncel.2015.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ibrahim M., Schoelermann J., Mustafa K., Cimpan M.R. TiO2 nanoparticles disrupt cell adhesion and the architecture of cytoskeletal networks of human osteoblast-like cells in a size dependent manner. J. Biomed. Mater. Res. A. 2018;106:2582–2593. doi: 10.1002/jbm.a.36448. [DOI] [PubMed] [Google Scholar]
- 37.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 38.Fulda S., Gorman A.M., Hori O., Samali A. Cellular stress responses: Cell survival and cell death. Int. J. Cell Biol. 2010;2010:214074. doi: 10.1155/2010/214074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Block E.R., Matela A.R., SundarRaj N., Iszkula E.R., Klarlund J.K. Wounding induces motility in sheets of corneal epithelial cells through loss of spatial constraints: Role of heparin-binding epidermal growth factor-like growth factor signaling. J. Biol. Chem. 2004;279:24307–24312. doi: 10.1074/jbc.M401058200. [DOI] [PubMed] [Google Scholar]
- 40.Notbohm J., Banerjee S., Utuje K.J.C., Gweon B., Jang H., Park Y., Shin J., Butler J.P., Fredberg J.J., Marchetti M.C. Cellular Contraction and Polarization Drive Collective Cellular Motion. Biophys. J. 2016;110:2729–2738. doi: 10.1016/j.bpj.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan J.L., Tien J., Pirone D.M., Gray D.S., Bhadriraju K., Chen C.S. Cells lying on a bed of microneedles: An approach to isolate mechanical force. Proc. Natl. Acad. Sci. USA. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.du Roure O., Saez A., Buguin A., Austin R.H., Chavrier P., Silberzan P., Ladoux B. Force mapping in epithelial cell migration. Proc. Natl. Acad. Sci. USA. 2005;102:2390–2395. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghassemi S., Meacci G., Liu S., Gondarenko A.A., Mathur A., Roca-Cusachs P., Sheetz M.P., Hone J. Cells test substrate rigidity by local contractions on submicrometer pillars. Proc. Natl. Acad. Sci. USA. 2012;109:5328–5333. doi: 10.1073/pnas.1119886109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mourdikoudis S., Pallares R.M., Thanh N.T.K. Characterization techniques for nanoparticles: Comparison and complementarity upon studying nanoparticle properties. Nanoscale. 2018;10:12871–12934. doi: 10.1039/C8NR02278J. [DOI] [PubMed] [Google Scholar]
- 45.Fornaguera C., Lazaro M.A., Brugada-Vila P., Porcar I., Morera I., Guerra-Rebollo M., Garrido C., Rubio N., Blanco J., Cascante A., et al. Application of an assay Cascade methodology for a deep preclinical characterization of polymeric nanoparticles as a treatment for gliomas. Drug Deliv. 2018;25:472–483. doi: 10.1080/10717544.2018.1436099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beck G.R., Jr., Ha S.W., Camalier C.E., Yamaguchi M., Li Y., Lee J.K., Weitzmann M.N. Bioactive silica-based nanoparticles stimulate bone-forming osteoblasts, suppress bone-resorbing osteoclasts, and enhance bone mineral density in vivo. Nanomedicine. 2012;8:793–803. doi: 10.1016/j.nano.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin T.H., Seo C., Lee D.Y., Ji M., Manavalan B., Basith S., Chakkarapani S.K., Kang S.H., Lee G., Paik M.J., et al. Silica-coated magnetic nanoparticles induce glucose metabolic dysfunction in vitro via the generation of reactive oxygen species. Arch. Toxicol. 2019;93:1201–1212. doi: 10.1007/s00204-019-02402-z. [DOI] [PubMed] [Google Scholar]
- 48.Park K.S., Tae J., Choi B., Kim Y.S., Moon C., Kim S.H., Lee H.S., Kim J., Kim J., Park J., et al. Characterization, in vitro cytotoxicity assessment, and in vivo visualization of multimodal, RITC-labeled, silica-coated magnetic nanoparticles for labeling human cord blood-derived mesenchymal stem cells. Nanomedicine. 2010;6:263–276. doi: 10.1016/j.nano.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 49.Shin T.H., Lee S., Choi K.R., Lee D.Y., Kim Y., Paik M.J., Seo C., Kang S., Jin M.S., Yoo T.H., et al. Quality and freshness of human bone marrow-derived mesenchymal stem cells decrease over time after trypsinization and storage in phosphate-buffered saline. Sci. Rep. 2017;7:1106. doi: 10.1038/s41598-017-01315-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shin T.H., Phukan G., Shim J.S., Nguyen D.T., Kim Y., Oh-Lee J.D., Lee H.S., Paik M.J., Lee G. Restoration of Polyamine Metabolic Patterns in In Vivo and In Vitro Model of Ischemic Stroke following Human Mesenchymal Stem Cell Treatment. Stem Cells Int. 2016;2016:4612531. doi: 10.1155/2016/4612531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee S., Cho N.P., Kim J.D., Jung H., Kang S.H. An ultra-sensitive nanoarray chip based on single-molecule sandwich immunoassay and TIRFM for protein detection in biologic fluids. Analyst. 2009;134:933–938. doi: 10.1039/b822094h. [DOI] [PubMed] [Google Scholar]
- 52.Owen D.M., Rentero C., Magenau A., Abu-Siniyeh A., Gaus K. Quantitative imaging of membrane lipid order in cells and organisms. Nat. Protoc. 2012;7:24–35. doi: 10.1038/nprot.2011.419. [DOI] [PubMed] [Google Scholar]
- 53.Schneckenburger H., Stock K., Strauss W.S., Eickholz J., Sailer R. Time-gated total internal reflection fluorescence spectroscopy (TG-TIRFS): Application to the membrane marker laurdan. J. Microsc. 2003;211:30–36. doi: 10.1046/j.1365-2818.2003.01201.x. [DOI] [PubMed] [Google Scholar]
- 54.Hansen J.S., Helix-Nielsen C. An epifluorescence microscopy method for generalized polarization imaging. Biochem. Biophys. Res. Commun. 2011;415:686–690. doi: 10.1016/j.bbrc.2011.10.138. [DOI] [PubMed] [Google Scholar]
- 55.Tan J.L., Liu W., Nelson C.M., Raghavan S., Chen C.S. Simple approach to micropattern cells on common culture substrates by tuning substrate wettability. Tissue Eng. 2004;10:865–872. doi: 10.1089/1076327041348365. [DOI] [PubMed] [Google Scholar]
- 56.Cui Y., Hameed F.M., Yang B., Lee K., Pan C.Q., Park S., Sheetz M. Cyclic stretching of soft substrates induces spreading and growth. Nat. Commun. 2015;6:6333. doi: 10.1038/ncomms7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Landau L.D., Lifshitz E.M., Kosevich A.M., Pitaevskii L.P. Theory of Elasticity. 3rd English ed. Butterworth-Heinemann; Oxford, UK: 1986. [DOI] [Google Scholar]
- 58.Rajanahalli P., Stucke C.J., Hong Y. The effects of silver nanoparticles on mouse embryonic stem cell self-renewal and proliferation. Toxicol. Rep. 2015;2:758–764. doi: 10.1016/j.toxrep.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ickrath P., Wagner M., Scherzad A., Gehrke T., Burghartz M., Hagen R., Radeloff K., Kleinsasser N., Hackenberg S. Time-Dependent Toxic and Genotoxic Effects of Zinc Oxide Nanoparticles after Long-Term and Repetitive Exposure to Human Mesenchymal Stem Cells. Int. J. Environ. Res. Public Health. 2017;14:1590. doi: 10.3390/ijerph14121590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Novotna B., Turnovcova K., Veverka P., Rossner P., Jr., Bagryantseva Y., Herynek V., Zvatora P., Vosmanska M., Klementova M., Sykova E., et al. The impact of silica encapsulated cobalt zinc ferrite nanoparticles on DNA, lipids and proteins of rat bone marrow mesenchymal stem cells. Nanotoxicology. 2016;10:662–670. doi: 10.3109/17435390.2015.1107144. [DOI] [PubMed] [Google Scholar]
- 61.Zhou D., Shao L., Spitz D.R. Reactive oxygen species in normal and tumor stem cells. Adv. Cancer Res. 2014;122:1–67. doi: 10.1016/B978-0-12-420117-0.00001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Lehn R.C., Ricci M., Silva P.H., Andreozzi P., Reguera J., Voitchovsky K., Stellacci F., Alexander-Katz A. Lipid tail protrusions mediate the insertion of nanoparticles into model cell membranes. Nat. Commun. 2014;5:4482. doi: 10.1038/ncomms5482. [DOI] [PubMed] [Google Scholar]
- 63.Chen F., Liu Y., Wong N.K., Xiao J., So K.F. Oxidative Stress in Stem Cell Aging. Cell Transplant. 2017;26:1483–1495. doi: 10.1177/0963689717735407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shaban S., El-Husseny M.W.A., Abushouk A.I., Salem A.M.A., Mamdouh M., Abdel-Daim M.M. Effects of Antioxidant Supplements on the Survival and Differentiation of Stem Cells. Oxid. Med. Cell. Longev. 2017;2017:5032102. doi: 10.1155/2017/5032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Titushkin I., Cho M. Regulation of cell cytoskeleton and membrane mechanics by electric field: Role of linker proteins. Biophys. J. 2009;96:717–728. doi: 10.1016/j.bpj.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Accomasso L., Gallina C., Turinetto V., Giachino C. Stem Cell Tracking with Nanoparticles for Regenerative Medicine Purposes: An Overview. Stem Cells Int. 2016;2016:7920358. doi: 10.1155/2016/7920358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhattacharya S., Ahir M., Patra P., Mukherjee S., Ghosh S., Mazumdar M., Chattopadhyay S., Das T., Chattopadhyay D., Adhikary A. PEGylated-thymoquinone-nanoparticle mediated retardation of breast cancer cell migration by deregulation of cytoskeletal actin polymerization through miR-34a. Biomaterials. 2015;51:91–107. doi: 10.1016/j.biomaterials.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 68.Rape A., Guo W.H., Wang Y.L. Microtubule depolymerization induces traction force increase through two distinct pathways. J. Cell Sci. 2011;124:4233–4240. doi: 10.1242/jcs.090563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tay C.Y., Cai P., Setyawati M.I., Fang W., Tan L.P., Hong C.H., Chen X., Leong D.T. Nanoparticles strengthen intracellular tension and retard cellular migration. Nano Lett. 2014;14:83–88. doi: 10.1021/nl4032549. [DOI] [PubMed] [Google Scholar]
- 70.Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., Horwitz A.R. Cell migration: Integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 71.Lange J.R., Fabry B. Cell and tissue mechanics in cell migration. Exp. Cell Res. 2013;319:2418–2423. doi: 10.1016/j.yexcr.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Matsuzaki T., Matsumoto S., Kasai T., Yoshizawa E., Okamoto S., Yoshikawa H.Y., Taniguchi H., Takebe T. Defining Lineage-Specific Membrane Fluidity Signatures that Regulate Adhesion Kinetics. Stem Cell Rep. 2018;11:852–860. doi: 10.1016/j.stemcr.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Lucas B., Perez L.M., Galvez B.G. Importance and regulation of adult stem cell migration. J. Cell. Mol. Med. 2018;22:746–754. doi: 10.1111/jcmm.13422. [DOI] [PMC free article] [PubMed] [Google Scholar]