Abstract
Aim:
Cigarette smoking influences DNA methylation genome wide, in newborns from pregnancy exposure and in adults from personal smoking. Whether a unique methylation signature exists for in utero exposure in newborns is unknown.
Materials & methods:
We separately meta-analyzed newborn blood DNA methylation (assessed using Illumina450k Beadchip), in relation to sustained maternal smoking during pregnancy (9 cohorts, 5648 newborns, 897 exposed) and adult blood methylation and personal smoking (16 cohorts, 15907 participants, 2433 current smokers).
Results & conclusion:
Comparing meta-analyses, we identified numerous signatures specific to newborns along with many shared between newborns and adults. Unique smoking-associated genes in newborns were enriched in xenobiotic metabolism pathways. Our findings may provide insights into specific health impacts of prenatal exposure on offspring.
Keywords: : cigarette smoking, epigenetics, infant, maternal exposure, methylation
Cigarette smoking is the leading cause of preventable disease and death in the USA, despite decades of health advisories and progress in tobacco control [1]. Major causes of disability and death from personal smoking include various cancers, chronic obstructive pulmonary disease and cardiovascular disease [1]. Maternal smoking during pregnancy is associated with many adverse consequences on the offspring, including reduced birthweight, early respiratory illness, reduced pulmonary function, sudden infant death syndrome and neurobehavioral disorders [1].
The mechanisms underlying the diverse health impacts of smoking, both in adults from personal smoking and in infants from maternal smoking during pregnancy, remain largely unknown. Recent large meta-analyses in both adults and newborns have demonstrated that cigarette smoking has highly reproducible genome-wide impacts on DNA methylation, measured using the Illumina Infinium Human 450 Beadchip (Illumina450K, henceforth). In adults, a large-scale meta-analysis of the association between current smoking and adult blood DNA methylation in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium [2] identified numerous differentially methylated cytosine–phosphate–guanine sites (CpGs; false discovery rate [FDR] <0.05). In newborns, a meta-analysis from the Pregnancy And Childhood Epigenetics (PACE) consortium [3] identified widespread genome-wide differential methylation in relation to sustained maternal smoking across pregnancy. Although the statistically significant changes in methylation between the exposed and unexposed groups in both newborns and adults in relation to smoking exposures in whole blood, a mix of cell types are usually small, these are highly reproducible.
The exposures from tobacco smoke in an adult who smokes and in a fetus whose mother smokes during pregnancy are not identical. For example, smokers directly inhale tobacco smoke leading to irritative and inflammatory effects on the airways [1]. In contrast, fetal exposure is entirely blood borne. Further, although it has been shown that nicotine and some specific tobacco-related polyaromatic hydrocarbons and nitrosamines cross the placenta, this is not known for the other approximately 4000 compounds in tobacco smoke [4]. Nonetheless, potential differences in effects on DNA methylation between newborns from exposure to a mother who smoked during pregnancy and adults from their own smoking have not been well explored. Identifying differences in methylation patterns for the in utero and personal smoking exposure scenarios may shed light on the mechanisms that explain the numerous differential effects of tobacco smoke across the life course.
We aimed to identify potential unique methylation signatures of exposure to maternal smoking during pregnancy in newborns, that are not observed in adults from personal smoking. To this end, we analyzed the cohort-specific results for methylation in relation to smoking in the nine cohorts from the PACE newborn consortium [3] and in the 16 cohorts from the CHARGE adult consortium [2] using fixed-effects meta-analysis model. We compared the two sets of meta-analyzed results to identify CpGs differentially methylated in newborns in relation to maternal smoking exposure during pregnancy that were not observed for personal smoking exposure among the adults. After annotating the CpGs to genes, we identified genes with at least one CpG site significantly differentially methylated among the newborns, but none among the adults. We evaluated whether the genes unique to newborns showed enrichment for distinct pathways compared with genes shared between newborns and adults. We also estimated the associations between methylation levels of the CpGs unique to newborns and expression levels of nearby genes and compared those with associations with expression for CpGs shared between newborns and adults.
Materials & methods
Study participants
We identified differentially methylated CpGs related to smoking using existing results from the PACE and CHARGE consortia [2,3]. The details of these consortia have been previously described [2,3]. To estimate the association of maternal smoking with blood DNA methylation in newborns, we meta-analyzed the cohort-specific result files generated by the nine birth or pregnancy cohorts in the PACE consortium that contributed to the meta-analysis of sustained maternal smoking during pregnancy and genome-wide methylation reported by Joubert et al. [3]. These nine cohorts are the Avon Longitudinal Study of Parents and Children (ALSPAC), the GECKO Drenthe cohort, the Generation R Study, two cohorts participating in the Mechanisms of the Development of Allergy (MeDALL) study (EDEN [Etude des Determinants pre et post natals du developpement et de la sante de l’Enfant] and INMA [Infancia y Medio Ambiente]), three independent datasets from the Norwegian Mother and Child Cohort Study (MoBa1, MoBa2 and MoBa3), the Newborn Epigenetics Study (NEST) and the Norway Facial Clefts Study (NFCS).
To estimate the association of personal smoking on blood DNA methylation in adults we meta-analyzed the cohort-specific result files generated by the 16 cohorts that contributed to the CHARGE consortium meta-analysis of Joehanes et al. [2]. These 16 cohorts are the Atherosclerosis Risk in Communities (ARIC) study, Cardiovascular Health Study European Ancestry (CHS EA), Cardiovascular Health Study African Ancestry (CHS AA), European Prospective Investigation into Cancer (EPIC), European Prospective Investigation into Cancer and Nutrition-Norfolk (EPIC Norfolk), Framingham Heart Study (FHS), Genetic Epidemiology Network of Arteriopathy (GENOA), Genetics of Lipid Lowering Drugs and Diet Network (GOLDN), Grady Trauma Project (GTP), InCHIANTI, Cooperative health research in the Region of Augsburg follow-up survey 4 (KORA F4), Lothian Birth Cohorts of 1921 and 1936 (LBC 1921 and LBC 1936), the Multi Ethnic Study of Atherosclerosis (MESA), Normative Aging Study (NAS) and the Rotterdam Study (RS).
Details of the study populations and analyses preformed can be found in the earlier PACE [3] and CHARGE publications [2]. Ethical approval for study protocols was obtained by all cohorts.
DNA methylation measurements
In both PACE and CHARGE, DNA methylation was measured using the Illumina450K Beadchip (Illumina, Inc, CA, USA). In PACE, newborn DNA was extracted from umbilical cord blood in all cohorts except for NFCS, which used neonatal phlebotomy [3]. In CHARGE, DNA was extracted from whole blood in 14 cohorts, isolated CD4+ T cells in GOLDN, and monocytes (CD14+) in MESA [2]. In all PACE and CHARGE cohorts, DNA was subjected to bisulfite conversion using Zymo EZ DNA methylation (Zymo Research, CA, USA). Each cohort performed its own quality control removing low quality samples and low quality CpGs, and normalized their untransformed methylation β-values as previously described [2,3].
Smoking variables
In PACE, most cohorts ascertained sustained smoking by the mothers during pregnancy using questionnaires; two studies (MoBa1 and MoBa2) incorporated cotinine measurements, a biomarker of recent smoking, from maternal blood samples collected in mid pregnancy as part of the definition of sustained maternal smoking. We had previously found in the MoBa1 cohort, that significant associations between maternal smoking during pregnancy and DNA methylation in newborns were driven by sustained smoking during pregnancy and not by transient smoking that ended early in pregnancy [5]. Sustained smoking in PACE was defined as smoking at least one cigarette per day during most of the pregnancy and was compared with no smoking during pregnancy; newborns of mothers who reported quitting smoking early in pregnancy were excluded from the comparison group. In CHARGE, current smoking status of the adults was based on questionnaire reports. Current smoking was defined as smoking at least one cigarette per day within the prior 12 months. Current smokers were compared with never smokers; former smokers were excluded from analyses.
Cell-type proportion
In the earlier PACE consortium paper [3], each cohort had estimated cell-type proportions (CD8T, CD4T, Natural Killer cells, B cells, monocytes and granulocytes) using the reference-based Houseman method [6] implemented in minfi [7] with the Reinius et al. dataset as reference [8], the reference panel available when the cohort-specific analyses were performed for the paper of Joubert et al. [3]. In CHARGE, two cohorts (InCHIANTI and RS) had measured complete blood counts and used those cell proportions in the analysis. The two cohorts that used a specific cell type (CD4+ or CD14+) did not adjust for cell-type proportions. The other 12 cohorts that measured methylation in whole blood DNA estimated cell proportions using the same method as in PACE.
Cohort-specific analyses
In the earlier PACE consortium paper [3], each cohort used a robust linear regression model to estimate the association of sustained maternal smoking during pregnancy and newborn DNA methylation for each probe. The normalized methylation β-values were the outcome, and sustained maternal smoking status during pregnancy (versus no smoking during pregnancy) was the primary predictor. Models were adjusted for maternal age, maternal socioeconomic status (generally maternal education), parity and estimated cell proportion. Batch effects were adjusted by either using a batch correction method such as ComBat [9] or including the relevant batch covariates in the model. For cohorts that oversampled individuals based on a selection factor, the selection factor was also included in the model [3].
CHARGE cohorts had run a linear model to evaluate the association of current smoking (versus never smoking) and methylation at each probe [2]. The normalized methylation β-values were considered as outcome and current smoking status (versus never smoking) the predictor of interest. Covariates were age (continuous), sex and cell-type proportion. Batch effects were adjusted by using the relevant batch covariates in the model [2]. In both PACE and CHARGE, models were run in R [10].
Meta-analysis
Cohort-specific results for PACE cohorts were combined using inverse variance-weighted fixed-effects model with METAL [11]. The results presented in Joehanes et al. [2] for CHARGE were from a random-effects meta-analysis model. For the current work, we meta-analyzed the cohort-specific results from each consortium using fixed-effects meta-analysis, as recommended by Rice et al. [12], to increase comparability. The same probe exclusions were used as in the original publications [2,3]. FDR adjustment was performed using Benjamini–Hochberg method of multiplicity correction [13] and CpGs with FDR-adjusted p < 0.05 were considered significant. For all further analysis, we excluded CpGs that were not available in both CHARGE and PACE meta-analyses, leaving 464,547 CpGs.
Identification of CpGs differentially methylated in relation to smoking unique to newborns, shared between newborns & adults, & unique to adults
We compared the meta-analyzed results from PACE and CHARGE to identify CpGs that were significantly differentially methylated in response to the smoking exposure variable in newborns but were not significant in the adults (genome-wide FDR <0.05 in newborns but genome-wide FDR >0.05 among adults). Thus, these CpGs were potentially uniquely affected in newborns due to maternal smoking exposure during pregnancy but not due to personal smoking in adults. We also identified the CpGs that were significantly differentially methylated in relation to smoking in both newborns and adults (genome-wide FDR <0.05 in both newborns and adults) as well as those only in adults (genome-wide FDR <0.05 in adults but >0.05 in newborns). The genomic inflation factors (λ) were assessed using the Bayesian method based on the empirical null distribution [14] for the two meta-analyses.
Enrichment analysis for genomic features
We evaluated whether the significantly differentially methylated CpGs were enriched, relative to all the CpGs, for several genomic features (localization to CpG islands and CpG island shores, DNAse1 hypersensitivity sites, enhancers, promoters and Transcription Factor (TF) binding sites). Enrichment tests were performed by using the two-sided Fisher’s exact test for each feature. We performed enrichment tests separately for the three groups of CpGs significantly differentially methylated in relation to smoking – unique to newborns, shared between newborns and adults, and unique to adults.
Identification of differentially methylated genes unique in newborns, shared between newborns & adults, & unique to adults in relation to smoking
We annotated the CpGs to gene names using Illumina’s annotation file, enhanced by using the University of California Santa Cruz (UCSC) Genome Browser (including data from the RefSeq and Ensembl databases) to assign each gene to a single UCSC KnownGene. All the annotations were based on the human February 2009 (GRCh37/hg19) assembly. CpGs, not in a gene, were annotated to the closest gene in a 10-Mb window based on UCSC KnownGene annotation. Using this window, the maximum distance of a CpG site to the nearest gene in our data is 1.74 Mb and 99.96% of the CpGs are within a 1-Mb window.
To identify the genes uniquely differentially methylated among newborns, we identified all genes with at least one CpG site significantly differentially methylated among newborns, but no significantly differentially methylated CpG site related to personal smoking among adults. For a specific gene, different CpG(s) might be statistically significant in newborns and in adults. Therefore, we considered a gene unique to newborns if it did not have any significant CpG site in adults.
Similarly, we identified the set of genes that were shared between both newborns and adults; these had at least one CpG site significantly differentially methylated in relation to smoking in both groups (newborns and adults); the significant CpGs might be different in the two groups. For completeness, we also identified the genes unique to adults defined as those that had at least one significant CpG site in adults but none in newborns. However, we note that the adult meta-analysis is so much larger and thus, better powered, that we cannot conclude that in a larger study of newborns, these would not have been identified.
Pathway analysis
To further aid in interpretation of the gene-level results, gene set analysis was performed to identify putative pathways involved. Genes uniquely differentially methylated in relation to smoking exposure in newborns were used to identify relevant pathways with the missMethyl [15] package in R, which takes into account the differing number of probes per gene present on the Illumina450K array. For our pathway analysis, we used the Kyoto Encyclopedia of Genes and Genomes (KEGG) gene sets from the GSEA MSigDB (Gene Set Enrichment Analysis Molecular Signatures Database) [16–18]. The KEGG gene set was chosen for its extensive characterization for genes and genomes at the molecular and higher levels, which makes it particularly well suited for integrating data at multiple levels of biological function, incorporating evidence from molecular-level functions and higher-level functions (represented by networks of molecular interactions, reactions and relations) [19]. This pathway analysis was also done with the genes shared between newborns and adults, and we compared the two sets of results. All pathways with p < 0.05 were considered significant.
Correlation of methylation with transcription of nearby genes
We examined whether the significantly differentially methylated CpGs mapped to the genes that were unique to newborns and those CpGs that were shared between both groups (newborns and adults), had different associations with gene expression. We analyzed paired DNA methylation and gene expression data both measured in blood in age-specific datasets that were not included in the meta-analyses. We analyzed data from 157 cord blood samples from the Isle of Wight (IoW) 3rd Generation cohort. Methylation was measured using Illumina450K, and gene expression was measured using Agilent one-color microarray (Agilent Technologies, CA, USA) [20]. From another birth cohort (INMA), Illumina450K methylation data were available from 113 cord blood samples and 112 overlapping children with blood collected at age 4 years, comparing methylation at each timepoint to gene expression (Affymetrix Human Transcriptome Array [HTA 2.0], Affymetrix, Inc, CA, USA) measured in blood from the same children measured at age 4 years [21]. We also analyzed Illumina450K methylation and RNA-Seq gene expression data from the same 3075 adults from the Biobank-Based Integrative Omics Studies (BIOS) consortium [22]. In all datasets, for each significant CpG site differentially methylated in relation to smoking, we examined the association with expression of transcripts within a window of 500 kb (±250 kb) of the CpG site, by regressing methylation M-values on gene expression levels accounting for cohort-specific joint sources of variability (age, sex, differential cell counts, batch effects for both methylation and gene expression).
Results
Study participants
The nine cohorts from PACE, meta-analyzed to estimate the association of maternal smoking with blood DNA methylation in newborns, included 897 newborns exposed to sustained maternal smoking and 4097 newborns whose mothers did not smoke during pregnancy. The prevalence of exposure to sustained maternal smoking during pregnancy among newborns across the nine cohorts ranged from 10.1 to 27.6% (median = 14.6%) [3]. The participants of all the cohorts except NEST were of European ancestry; NEST had 49% European ancestry, 45% African ancestry and 6% ‘other’. The mean ages of the mothers across the nine cohorts ranged between 28.9 and 31.5 years [3].
The 16 cohorts meta-analyzed from CHARGE to estimate the association of personal smoking with blood DNA methylation in adults included 2433 current smokers whom we compared with 6956 never smokers [2]. Eleven cohorts (CHS EA, EPIC, EPIC Norfolk, FHS, GOLDN, InCHIANTI, KORA F4, LBC 1921, LBC 1936, NAS and RS) analyzed European ancestry individuals, four cohorts analyzed African-American ancestry individuals (ARIC, GTP, GENOA and CHS AA) and MESA had individuals of European, African and Hispanic ancestry; major ethnic groups were analyzed separately by the cohorts and then meta-analyzed together. The mean ages of the participants in the 16 cohorts ranged from 41.4 to 79.1 years. The percentage of current smokers ranged from 4 to 32.9 with a median across the 16 cohorts of 13.6 [2].
Differentially methylated CpGs in relation to smoking
In the original PACE publication, Joubert et al. [3] presented as the primary model analyses without cell-type adjustment. Because CHARGE presented results adjusting for cell-type proportions, we meta-analyzed the cell-type-adjusted results from PACE to increase consistency with CHARGE. At genome-wide FDR cutoff of 5%, we identified 5547 CpGs significantly differentially methylated in relation to sustained maternal smoking during pregnancy in newborns (Supplementary Table 1). In CHARGE, the original publication presented random effects meta-analysis results as the primary model. Here we used a fixed effects model for both consortia [12] and identified 34,541 CpGs significantly differentially methylated in relation to current smoking in adults (Supplementary Table 2). As in the original reports [2,3], CpGs significantly related to smoking during pregnancy in newborns were roughly equally split between higher (45%) and lower methylation (55%) with a similar distribution for the CpGs significant in adults (49% higher methylation). The magnitude of the effect estimates for differential methylation in relation to the smoking exposure variables was comparable for newborns and adults. In newborns, at the significant CpGs, methylation was on average 0.8% (standard deviation [SD] = 0.5%) higher in newborns exposed to sustained maternal smoking during pregnancy, compared with unexposed, for the CpGs with higher methylation and 0.6% (SD = 0.5%) lower for CpGs with lower methylation. In adults, at the significant CpGs, methylation was on average 0.4% (SD = 0.3%) higher in current smokers compared with never smokers for the CpGs with higher methylation and 0.4% (SD = 0.4%) lower for those with lower methylation. The inflation factors (λ) estimated using the Bayesian method based on the empirical null distribution [14] for the meta-analyses were 1.15 for PACE and 1.28 for CHARGE.
The 5547 CpGs significantly differentially methylated in relation to maternal smoking in pregnancy among newborns included 3838 CpGs that were not significantly differentially methylated in relation to smoking in adults (Supplementary Table 3). Among the 34,541 CpGs significantly differentially methylated in relation to smoking in adults, there were 32,832 CpGs not differentially methylated in newborns (Supplementary Table 4); this larger number of unique findings reflects the much greater sample size, and thus much higher power in the CHARGE analysis of adults than in the PACE newborn analysis. We found 1709 CpGs significantly differentially methylated in both newborns and adults (Supplementary Table 5). The proportions of higher and lower methylations were comparable for the significant CpGs that were unique to newborns (53% higher) and for those that were unique in adults (51% higher). The Spearman’s rank correlation coefficient between the effect estimates of these 1709 shared CpGs was 0.48 and the direction of the effect estimates matched for 79.4%. For newborns, the mean difference in methylation between exposed and unexposed groups was comparable for the CpGs unique to newborns (0.7% [SD = 0.4%] for CpGs with higher methylation and 0.5% [SD = 0.3%] for CpGs with lower methylation) and for those shared with adults (0.9% [SD = 0.7%] for CpGs with higher methylation and 0.7% [SD = 0.7%] for CpGs with lower methylation). Likewise, effect sizes were similar for CpGs unique in adults and those shared with newborns (Supplementary Tables 4 & 5).
Additionally, we evaluated the overlap between newborns and adults at a look-up level Bonferroni correction for significance in adults for all the 5547 CpGs that were genome-wide FDR significant in newborns. The 3838 CpGs, which were unique to newborns based on the genome-wide FDR correction in adults, remained uniquely significant based on the look-up level correction in adults (correction for 5547 tests).
Enrichment analysis
We observed similar patterns of significant enrichment for localization of CpGs differentially methylated in relation to smoking to CpG island shores, DNAse1 hypersensitivity sites, enhancers and TF binding sites for CpGs unique to newborns, those shared between newborns and adults, and those unique to adults (Supplementary Table 6). Conversely, we found significant depletion in CpG islands and promoters for all three groups of CpGs.
Genes uniquely differentially methylated in relation to smoking exposure in newborns & shared between newborns & adults
From the 3838 differentially methylated CpGs unique to newborns, we identified 743 genes annotated to at least one significant CpG site among the newborns but none among the adults (Supplementary Table 7). The gene SLC25A2 (OMIM: 608157) had the maximum number (six) of significant CpGs unique to newborns (cg25212131, cg07039560, cg07496545, cg14739664, cg21636683 and cg05845376). Among the 743 genes, there were 120 genes with more than one significant CpG site unique to newborns. We identified 2770 genes that had at least one significant CpG site in both newborns and adults. The gene, AHRR (OMIM: 606517), had the maximum number of significant CpGs in both groups (34 in newborns and 68 in adults) (Supplementary Table 8). There were 9894 genes that were unique to adults (no significant CpG site in newborns).
Pathway analysis
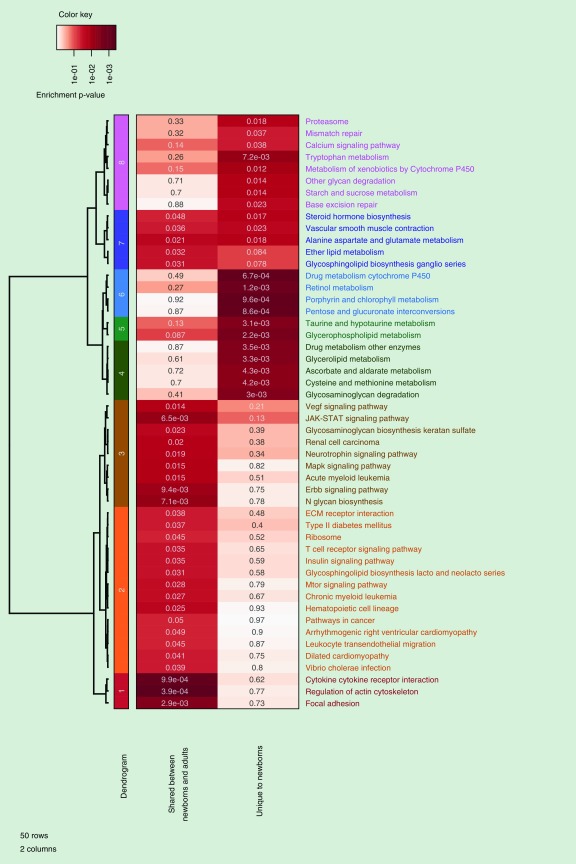
Pathway analysis yielded 22 enriched (p < 0.05) pathways (Supplementary Table 9) for the genes unique to newborns. For comparison, we also identified the enriched (p < 0.05) pathways (Supplementary Table 10) for the genes shared between newborns and adults. There were 31 enriched pathways for the genes shared between newborns and adults out of which 3 were also enriched among the genes unique to newborns (Figure 1). Several pathways (e.g., ‘drug metabolism cytochrome P450’, and ‘tryptophan metabolism’) were unique to newborn-only genes (Figure 1). Supplementary Figure 1 shows an example network of some of the pathways that were unique to newborn-only genes.
Figure 1. . Heatmap of biological pathways.
Heatmap of the pathways, significant at p-value cutoff of 0.05 in at least one of the two enrichment tests shown, functions enriched with newborn-specific genes, or functions enriched with genes shared between newborns and adults. For each pathway, the color coding is done to show the level of significance (based on p-values). Darker shades indicate higher level of significance.
Correlation of methylation with transcription of nearby genes
Among the 3838 significantly differentially methylated CpGs unique to newborns, there were 917 CpGs that mapped to the 743 genes unique to newborns. The remaining 2921 CpGs (out of the 3838 CpGs unique to newborns) were mapped to genes that were also mapped to at least one significant CpG site in adults. For the 917 CpGs that mapped to unique genes in newborns and for the 1709 significantly differentially methylated CpGs shared between newborns and adults, we assessed the association between paired levels of whole blood DNA methylation and whole blood gene expression for nearby transcripts.
In the Isle of Wight 3rd Generation cohort study [20], out of the 917 CpGs mapped to the genes unique to newborns, 770 CpGs were within ±250 kb of a gene transcript. In this dataset of modest size (N = 157), only a few CpGs were significantly associated with expression of a nearby transcript after adjusting for FDR at 0.05. At an arbitrary p-value cutoff of 0.01, 115 (15%) of the CpGs were associated with expression at one or more nearby transcripts (Supplementary Table 11). Of these associations (p < 0.01), for about 37%, higher methylation was related to higher gene expression and for 63% higher methylation was related to lower gene expression. For the 1709 CpGs shared between newborns and adults, 1540 were within ±250 kb of a gene transcript, out of which 277 (18%) were associated with expression of at least one nearby transcript at p < 0.01 (Supplementary Table 12). Among these associations (p < 0.01), for about 40% higher methylation was related to higher gene expression and for about 60% higher methylation was related to lower gene expression.
In the INMA study with newborn methylation (N = 113) [21], 905 of the 917 CpGs mapped to the genes unique to newborns were within ±250 kb of a gene transcript. At an arbitrary p-value cutoff of 0.01, 176 (19%) of the CpGs unique to newborns were associated with expression of at least one nearby transcript in blood collected at 4 years of age (Supplementary Table 13). Out of the 1709 CpGs shared between newborns and adults, 1707 CpGs were within ±250 kb of a gene transcript. Among these 1707 CpGs, 389 (23%) were associated with expression of one or more nearby transcripts at p < 0.01 (Supplementary Table 14). The results were similar for the INMA children with expression and methylation both measured at age four (N = 112) (Supplementary Table 15 & 16). Given the modest sample sizes, only a few CpGs were significantly associated with gene expression at FDR <0.05.
In the much larger study of adults with both gene expression and methylation (BIOS, N = 3075) [22], out of the 917 CpGs mapped to genes unique to newborns, 856 CpGs were within ±250 kb of a gene transcript. Among the 1709 CpGs shared between newborns and adults, 1664 CpGs were within ±250 kb of a gene transcript. The proportion of CpGs significantly related to gene expression was lower for those associated with smoking exposure uniquely in newborns compared with those shared between newborns and adults; at FDR <0.05, 48% of CpGs unique to newborns were associated with expression of at least one nearby transcript (Supplementary Table 17) compared with 78% of those shared between newborns and adults (Supplementary Table 18). The direction of significant associations between methylation and gene expression was similar for CpGs unique to newborns and for those shared between newborns and adults (55% higher methylation related to lower gene expression for those unique to newborns vs 57% for those shared with adults).
Discussion
We found a large number of differentially methylated CpGs (3838) that were unique to newborns in response to maternal smoking in pregnancy (Supplementary Table 3) but also a substantial number of differentially methylated CpGs (1709) in both newborns related to the in utero exposure and in adults related to their own smoking (Supplementary Table 5). We identified 743 genes unique to newborns with at least one significant CpG site in newborns but none among adults (Supplementary Table 7). For a specific gene, different CpG(s) might be significant in newborns and adults, therefore we considered a gene as unique to newborns if it did not have any CpG site significant in adults.
Overall, the CpGs related to smoking were enriched for localization to island shores, enhancers, DNase I hypersensitivity sites and TF binding sites. Methylation at these sites is dynamic and likely to have functional impact [23]. In addition, the pattern of enrichment for functional domains did not differ among CpGs unique to newborns, shared with adults and unique to adults.
In pathway analyses we observed several pathways uniquely enriched for the 743 genes unique to newborns but not for the 2770 genes implicated in both newborns and adults. These include xenobiotic-related pathways, such as ‘drug metabolism cytochrome P450’ (Supplementary Table 9). Most of the compounds in tobacco smoke are metabolized in two steps that include a generally activating step by the cytochrome P450 (CYP450) system and a subsequent detoxification process by enzymes such as glutathione-S-transferases (GSTs) [24] and uridine diphosphate-glucuronosyltransferases (UGTs). Nicotine, the primary alkaloid in tobacco smoke responsible for its addictive properties, readily crosses the placenta and has a much longer half-life in neonates than in adults; levels in fetal serum and amniotic fluid are higher than in the maternal serum [25]. Nicotine is primarily metabolized to cotinine via oxidation involving CYP450 enzymes. Both cotinine and nicotine can undergo glucuronidation, involving the UGTs, facilitating excretion [25]. In our data, differentially methylated genes unique to newborns that contribute to this drug metabolism pathway include the CYP450s CYP1A2 (OMIM: 124060), CYP2D6 (OMIM: 124030) and CYP2A7 (OMIM: 608054), the UGTs UGT2B4 (OMIM: 600067), UGT1A5 (OMIM: 606430) and UGT2A3 (OMIM: 616382), and the glutathione-S-transferase GSTA1 (OMIM: 138359). CYP1A2 and CYP2D6 can metabolize nicotine [26] and play key roles in the metabolism of many drugs and other xenobiotics [27]. CYP1A2 levels are also increased by smoking [28]. CYP2A7 has also a potential role in regulating nicotine metabolism rate [29]. This gene is 97% identical to the gene CYP2A6 (OMIM: 122720) [30], which is known to have a major role in nicotine metabolism [26,27]. The three UGTs we identified are not among those that have been implicated in nicotine glucuronidation, which has been primarily ascribed to UGT2B10 (OMIM: 600070) and UGT2B7 (OMIM: 600068) [31]. However, the three UGTs in this pathway are not as well studied, two (UGT1A5 and UGT2A3) having been much more recently characterized. Of interest substrate selectivity of UGT2B4 overlaps closely with that of UGT2B7 [32], and UGT1A5 has been shown to be polyaromatic hydrocarbon inducible, which are abundant in tobacco smoke [33]. The glutathione S-transferase GSTA1, which we identified in our data, has been previously found to mediate the effect of nicotine on lung cancer cell metastasis [34] and is induced in the placenta of smoking mothers [35] and is expressed during fetal development [36]. For five of the seven genes in these pathways (CYP1A2, CYP2D6, CYP2A7, UGT1A5 and UGT2A3), we found that methylation was correlated with expression of a nearby gene transcript in at least one of the newborn, child or adult datasets.
In general, CYP450s and UGTs are expressed in lower levels in the fetus than in adults [37]. Specifically, CYP2D6 [38] and the UGTs, we identified as differentially methylated in newborns but not in adults, have been documented to be expressed in fetal tissue, but at much lower levels than in adults [39]. Evolving data suggest that DNA methylation may play an important role in regulating low levels of drug metabolizing enzymes during fetal development [40]. CYP1A2, an enzyme induced by exposure to tobacco smoke in adults, has not been shown to be expressed in fetal tissue. There is evidence that CYP1A2 is not inducible in the fetus in response to prenatal tobacco smoke, which may make the fetus especially susceptible to deleterious effects of this exposure [41]. Of note, a recent study comparing the effects of smoking on gene expression in the frontal cortex of adults and fetuses found that smoking has more widespread effects in the fetus [42]. Our pathway analyses identified differentially methylated genes unique to newborns who have not been previously implicated in response to nicotine or tobacco and might be involved in in utero biological responses. The identification of drug metabolism pathways only in newborns resonates with the fact that nicotine is implicated as a causal exposure for some of the adverse consequences for the fetus, including preterm birth, stillbirth, interference with brain development, prenatal diaphragm movements and a modest contribution to growth restriction [1]. In contrast, nicotine is not clearly implicated in the primary health impacts of smoking in adults, such as cancer. Further, nicotine replacement therapy is regarded as a safe alternative to smoking in adults, whereas concern remains about use in pregnant women [1]. The fetus may be more vulnerable to some deleterious effects of nicotine than adults.
Although identifying the unique signatures in adults would be of great interest, given that the CHARGE meta-analysis is much larger than the PACE newborn analysis (both in the total number of samples as well as the proportion of samples exposed), we are very well powered to identify DNA methylation signatures in adults but not in newborns. Accordingly, we identified many more CpGs unique to adults, 32,832 in total. Failure to see these signals in newborns could easily be due to the lower power in the smaller newborn dataset. Thus, we are better powered to identify the unique DNA methylation signatures in newborns than those unique to adults.
A minority of the CpGs significantly differentially methylated in both newborns and adults had different directions of association with the smoking exposure variable. While we do not have a clear explanation for this finding, we note that in Joubert et al.’s meta-analysis, some of the associations for CpGs, differentially methylated in newborns, flipped directions in older children [3].
The primary model for each newborn cohort in PACE presented in the earlier publication [3] was not cell-type adjusted. Joubert et al. had performed the cell-type adjustment for all the cohorts, using the Reinius et al. dataset [8] of six adult men available at the time, as a sensitivity analysis, and found that this cell-type adjustment had minimal effect on the results [3]. Because the adult CHARGE meta-analysis results were cell-type adjusted [2], we used the cell-type adjusted results for PACE to increase consistency. We recognized that using an adult reference panel for cell-type adjustment in newborns is not ideal. Therefore, to address this issue, we estimated cell-type proportions in two of the larger newborn studies (MoBa1 and MoBa2) using a newer reference panel from newborn cord blood [43] and compared the results with those adjusted for cell type using the Reinius et al. adult reference panel [8]. Results were very similar. A slightly higher number of CpGs were associated with exposure at FDR <0.05 in the analyses using the newborn reference panel for cell-type adjustment but the regression coefficients were very highly correlated (>0.99 for both MoBa1 and MoBa2).
In the earlier CHARGE publication, results from an inverse variance-weighted random-effects model [2] were reported. Here we used the same meta-analysis model (inverse variance-weighted fixed effects) for both consortia, which increases comparability of the results. Recently, Rice et al. [12] showed that a fixed-effects model for meta-analysis of genomic experiments estimates a reasonable and interpretable parameter, even under the assumption that effect sizes differ, does not require the assumption of homogeneity, and is an appropriate model for discovery of novel loci in genomic analyses. In each consortium, data were combined across studies in large-scale epigenome-wide meta-analyses, and the results were robust to different modeling techniques [2,3]. Both meta-analyses [2,3] are the two largest studies with respect to the relevant smoking exposure and therefore, are well suited to identify the potential unique and contrasting methylation signatures.
While the unique methylation signatures among the newborns in relation to maternal smoking during pregnancy are of interest, we also identified many CpGs and genes shared between newborns and adults in response to smoking exposure. About 30% of the CpGs (1709 out of 5547) that were significant in newborns were also seen among the adults from personal smoking and 2770 genes were shared between newborns and adults. This similarity in results is especially notable because the pregnant women in PACE who smoke, smoked much less than the current smoking adults in CHARGE. The median number of cigarettes smoked per day during pregnancy was around 10 in most of the cohorts in PACE compared with an average of 20 cigarettes per day among the adults in CHARGE. In addition, the newborn exposure is entirely blood borne, whereas adults experience blood borne exposure plus direct effects of cigarette smoke inhalation to the lungs. It is remarkable that many associations with DNA methylation from a substantial history of personal smoking in adults can also be seen among the newborns who never smoked themselves, but due to maternal smoking exposure during pregnancy.
Conclusion
In summary, in addition to many overlapping exposure signatures, we identified genes differentially methylated in relation to maternal smoking that are not observed for personal smoking in adults. These genes implicate unique biologic pathways for effects of in utero exposure. These results can facilitate the development of biomarkers that are unique to smoking exposure during pregnancy. Our findings also may provide new insights about the impacts of tobacco smoking that are specific to newborns due to maternal cigarette use during pregnancy.
Future perspective
The methylation signatures identified in newborns from in utero exposure can facilitate the development of biomarkers unique to maternal smoking exposure during pregnancy and provide new insights about the mechanisms of health impacts of smoking that are specific to newborns.
Summary points.
We identified numerous methylation signatures in adults in relation to personal smoking.
We identified many methylation signatures in newborns in relation to maternal smoking exposure during pregnancy.
We are well powered to identify methylation signatures that are unique to newborns and are not seen for personal smoking in adults.
However, we are not very well powered to identify unique methylation signatures in adults because failure to see the signals in newborns could easily be due to the lower power in the smaller newborn dataset.
Although we identified important overlaps, the genome-wide effects from personal smoking in adults and from maternal smoking exposure during pregnancy in newborns are not identical.
We identified >3000 cytosine–phosphate–guanine sites (CpGs) that were unique to newborns.
Around 1700 cytosine–phosphate–guanine sites (CpGs) were differentially methylated in relation to smoking in both newborns and adults.
We identified 743 genes that were differentially methylated in newborns in relation to maternal smoking exposure during pregnancy that were not observed for personal smoking in adults.
We identified xenobiotics metabolism-related pathways that were uniquely enriched for the 743 genes unique to newborns.
Supplementary Material
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: https://www.futuremedicine.com/doi/suppl/10.2217/epi-2019-0066
Financial & competing interests disclosure
Funded by the Intramural Research Program of National Institutes of Health (National Institute of Environmental Health Sciences ZO1 ES49019) in addition to funding for individual authors and cohorts listed in the Supplementary Acknowledgements and Funding document. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
For each cohort participating in PACE and CHARGE, institutional ethical approval was obtained.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
Data & materials availability
The complete meta-analysis results files supporting the conclusions of this article will be deposited in database of Genotypes and Phenotypes (dbGAP) under acquisition number phs000930. Access to individual level methylation files for each of the participating CHARGE and PACE cohorts must be requested from the participating cohorts.
References
Papers of special note have been highlighted as: • of interest
- 1.National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health, Centers for Disease Control and Prevention. 2014 Surgeon General's Report: The Health Consequences of Smoking – 50 Years of Progress (2014). https://www.cdc.gov/tobacco/data_statistics/sgr/50th-anniversary/index.htm#report [Google Scholar]
- 2.Joehanes R, Just AC, Marioni RE. et al. Epigenetic signatures of cigarette smoking. Circ. Cardiovasc. Genet. 9(5), 436–447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Largest meta-analysis identifying the association between personal smoking and DNA methylation in adults.
- 3.Joubert BR, Felix JF, Yousefi P. et al. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am. J. Hum. Genet. 98(4), 680–696 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Largest meta-analysis identifying the association between maternal smoking exposure during pregnancy and DNA methylation in their newborns.
- 4.Shiverick KT. Cigarette smoking and reproductive and developmental toxicity. : Reproductive and Developmental Toxicology. Elsevier, London, UK, 319–331 (2011). [Google Scholar]
- 5.Joubert BR, Haberg SE, Bell DA. et al. Maternal smoking and DNA methylation in newborns: in utero effect or epigenetic inheritance? Cancer Epidemiol. Biomarkers Prev. 23(6), 1007–1017 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Shows that the significant methylation differences in newborns in relation to maternal smoking during pregnancy were driven by sustained maternal smoking but not when the mothers quit early in pregnancy.
- 6.Houseman EA, Accomando WP, Koestler DC. et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13, 86 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aryee MJ, Jaffe AE, Corrada-Bravo H. et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30(10), 1363–1369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinius LE, Acevedo N, Joerink M. et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE 7(7), e41361 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28(6), 882–883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: (2017). https://www.R-project.org/ [Google Scholar]
- 11.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26(17), 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rice K, Higgins JPT, Lumley T. A re-evaluation of fixed effect(s) meta-analysis. J. R. Stat. Soc. 181(1), 205–227 (2018). [Google Scholar]
- 13.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 289–300 (1995). [Google Scholar]
- 14.Van Iterson M, Van Zwet EW, Heijmans BT. Controlling bias and inflation in epigenome- and transcriptome-wide association studies using the empirical null distribution. Genome Biol. 18(1), 19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina's HumanMethylation450 platform. Bioinformatics 32(2), 286–288 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1), 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47(D1), D590–D595 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102(43), 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45(D1), D353–D361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arshad SH, Karmaus W, Zhang H, Holloway JW. Multigenerational cohorts in patients with asthma and allergy. J. Allergy Clin. Immunol. 139(2), 415–421 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guxens M, Ballester F, Espada M. et al. Cohort profile: the INMA – INfancia y Medio Ambiente – (Environment and Childhood) Project. Int. J. Epidemiol. 41(4), 930–940 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Bonder MJ, Luijk R, Zhernakova DV. et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat. Genet. 49(1), 131–138 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Ziller MJ, Gu H, Muller F. et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 500(7463), 477–481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang KH, Chou AK, Jeng SF. et al. The impacts of cord blood cotinine and glutathione-S-transferase gene polymorphisms on birth outcome. Pediatr. Neonatology 58(4), 362–369 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Benowitz NL, Hukkanen J, Jacob P III. Nicotine chemistry, metabolism, kinetics and biomarkers. Handb. Exp. Pharmacol. (2009). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2953858/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamazaki H, Inoue K, Hashimoto M, Shimada T. Roles of CYP2A6 and CYP2B6 in nicotine C-oxidation by human liver microsomes. Arch. Toxicol. 73(2), 65–70 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Puris E, Pasanen M, Gynther M. et al. A liquid chromatography-tandem mass spectrometry analysis of nine cytochrome P450 probe drugs and their corresponding metabolites in human serum and urine. Anal. Bioanal. Chem. 409(1), 251–268 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Hukkanen J, Jacob P, 3rd, Peng M, Dempsey D, Benowitz NL. Effect of nicotine on cytochrome P450 1A2 activity. Br. J. Clin. Pharmacol. 72(5), 836–838 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loukola A, Buchwald J, Gupta R. et al. A genome-wide association study of a biomarker of nicotine metabolism. PLoS Genet. 11(9), e1005498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raunio H, Rahnasto-Rilla M. CYP2A6: genetics, structure, regulation, and function. Drug Metabol. Drug Interact. 27(2), 73–88 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Mwenifumbo JC, Tyndale RF. Molecular genetics of nicotine metabolism. : Nicotine Psychopharmacology. Springer, 235–259 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Miners JO, Mackenzie PI, Knights KM. The prediction of drug-glucuronidation parameters in humans: UDP-glucuronosyltransferase enzyme-selective substrate and inhibitor probes for reaction phenotyping and in vitro–in vivo extrapolation of drug clearance and drug–drug interaction potential. Drug Metab. Rev. 42(1), 196–208 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Finel M, Li X, Gardner-Stephen D, Bratton S, Mackenzie PI, Radominska-Pandya A. Human UDP-glucuronosyltransferase 1A5: identification, expression, and activity. J. Pharmacol. Exp. Ther. 315(3), 1143–1149 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Wang W, Liu F, Wang C, Wang C, Tang Y, Jiang Z. Glutathione S-transferase A1 mediates nicotine-induced lung cancer cell metastasis by promoting epithelial–mesenchymal transition. Exp. Ther. Med. 14(2), 1783–1788 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huuskonen P, Storvik M, Reinisalo M. et al. Microarray analysis of the global alterations in the gene expression in the placentas from cigarette-smoking mothers. Clin. Pharmacol. Ther. 83(4), 542–550 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Raijmakers MT, Steegers EA, Peters WH. Glutathione S-transferases and thiol concentrations in embryonic and early fetal tissues. Hum. Reprod. 16(11), 2445–2450 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Isoherranen N, Thummel KE. Drug metabolism and transport during pregnancy: how does drug disposition change during pregnancy and what are the mechanisms that cause such changes? Drug Metab. Dispos. 41(2), 256–262 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevens JC, Marsh SA, Zaya MJ. et al. Developmental changes in human liver CYP2D6 expression. Drug Metab. Dispos. 36(8), 1587–1593 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Court MH, Zhang X, Ding X, Yee KK, Hesse LM, Finel M. Quantitative distribution of mRNAs encoding the 19 human UDP-glucuronosyltransferase enzymes in 26 adult and 3 fetal tissues. Xenobiotica 42(3), 266–277 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Habano W, Kawamura K, Iizuka N, Terashima J, Sugai T, Ozawa S. Analysis of DNA methylation landscape reveals the roles of DNA methylation in the regulation of drug metabolizing enzymes. Clin. Epigenetics 7, 105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Czekaj P, Wiaderkiewicz A, Florek E, Wiaderkiewicz R. Tobacco smoke-dependent changes in cytochrome P450 1A1, 1A2, and 2E1 protein expressions in fetuses, newborns, pregnant rats, and human placenta. Arch. Toxicol. 79(1), 13–24 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Semick SA, Collado-Torres L, Markunas CA. et al. Developmental effects of maternal smoking during pregnancy on the human frontal cortex transcriptome. Mol. Psychiatry (2018). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6438764/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bakulski KM, Feinberg JI, Andrews SV. et al. DNA methylation of cord blood cell types: applications for mixed cell birth studies. Epigenetics 11(5), 354–362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data & materials availability
The complete meta-analysis results files supporting the conclusions of this article will be deposited in database of Genotypes and Phenotypes (dbGAP) under acquisition number phs000930. Access to individual level methylation files for each of the participating CHARGE and PACE cohorts must be requested from the participating cohorts.