Abstract
Epigenetic variation has the potential to influence environmentally dependent development and contribute to phenotypic responses to local environments. Environmental epigenetic studies of sexual organisms confirm the capacity to respond through epigenetic variation. An epigenetic response could be even more important in a population when genetic variation is lacking. A previous study of an asexual snail, Potamopyrgus antipodarum, demonstrated that different populations derived from a single clonal lineage differed in both shell phenotype and methylation signature when comparing lake versus river populations. Here, we examine methylation variation among lakes that differ in environmental disturbance and pollution histories. Snails were collected from a more pristine rural Lake 1 (Lake Lytle), and two urban lakes, Lake 2 (Capitol Lake) and Lake 3 (Lake Washington) on the Northwest Pacific coast. DNA methylation was assessed for each sample population using methylated DNA immunoprecipitation, MeDIP, followed by next-generation sequencing. The differential DNA methylation regions (DMRs) identified among the different lake comparisons suggested a higher number of DMRs and variation between rural Lake 1 and one urban Lake 2, and between the two urban Lakes 2 and 3, but limited variation between the rural Lake 1 and urban Lake 3. DMR genomic characteristics and gene associations were investigated. The presence of site-specific differences between each of these lake populations suggest an epigenetic response to varied environmental factors. The results do not support an effect of geographic distance in these populations. The role of dispersal distance among lakes, population history, environmental pollution and stably inherited methylation versus environmentally triggered methylation in producing the observed epigenetic variation are discussed. Observations support the proposal that epigenetic alterations may associate with phenotypic variation and environmental factors and history of the different lakes.
Keywords: epigenetics, DNA methylation, asexual reproduction, clonal animal, environment, ecology
Introduction
Ecological factors and environmental toxicants have a potential to alter developmental trajectories in organisms through epigenetic mechanisms [1, 2]. Epigenetics is defined as molecular factors or processes that regulate genome activity independent of DNA sequence and are mitotically stable [3]. Epigenetic factors and processes include DNA methylation, non-coding RNA, histone modifications, chromatin structure and RNA methylation [4]. All elements of genome activity are influenced by epigenetics including the activation or repression of gene expression, splicing, RNA stability and general gene expression. The ability to respond to environmental stressors through epigenetic mechanisms could be particularly important among asexual species [5]. Asexual species tend to lack significant genetic variation [6–8], which should result in a limited response to natural selection. The generation of phenotypic variation through alterations in the epigenome could explain the ability of asexual populations to persist in the context of environmental changes and stressors. Epigenetic responses may accelerate adaptation because epigenetic modifications occur on a much faster time scale than de novo genetic mutations [9]. Natural asexual populations provide excellent opportunities to examine the process of epigenetic response to environmental stressors in the absence of genetic variation [5]. The implications of this additional path to respond to environmental variation are potentially important in all natural populations with limited genetic variation, but may be especially relevant among asexual populations.
Invasive populations of the asexual Potamopyrgus antipodarum (New Zealand fresh water snail) exhibit rapid adaptation to diverse environments after establishment in the western USA [6, 7]. Genetic analysis of these invasive populations shows a paucity of genetic variation characterized by six allozyme loci, microsatellite loci and mtDNA haplotype [6]. All North American populations which have progressed beyond the initial invasion location exhibit the same multilocus allozyme and microsatellite genotype and the corresponding mitochondrial DNA haplotype [6]. Earlier studies showed that shell shape varies between sites which vary in water current speeds [7]. A demonstrated association between this shell shape divergence and epigenetic alterations between populations inhabiting diverse environmental conditions supports the potential for asexual species to respond to environmental cues through epigenetic mechanisms which result in adaptive phenotypic change [10].
The potential for epigenetic differentiation among populations, and for it to play a role in phenotypic divergence, will depend on the reproductive mode and epigenetic reprogramming. Sexual reproduction in vertebrates is associated with epigenetic reprogramming and resetting. The development of a stem cell requires reprogramming to reduce epigenetic regulation and allow pluripotency of the cells. This process has been established in sexual reproduction, early embryo development and generational cellular differentiation [2, 3]. Although similar processes are required in asexual species, the epigenetic reprogramming and stem cell biology is less defined. Interestingly, sexual species exhibit epigenetic divergence among populations. For example, an examination of three populations of Alligator mississippiens (American alligator) revealed site-specific epigenetic modifications in two contaminated lakes when compared to a pristine lake [11]. Comparison in differential DNA-methylated regions (DMRs) between each contaminated lake and the pristine lake suggests that epigenetic responses to environmental toxicants and factors can be the result of the specific environment experienced by each population and may be unique between isolated populations. Similarly, comparison of two species of Darwin’s finches (Geospiza fuliginosa and G. scandens) in the Galapagos in rural/wild and urban environments demonstrates phenotypic variation [12] and associated epigenetic variation with negligible genetic alterations observed [13]. The potential role of epigenetic variation as a response to the environment might be greater in asexual populations that lack meiosis and genetic recombination. Although the mechanisms of epigenetic reprogramming have not yet been explored in parthenogenetic animal species, it is well-known meiosis is aborted in the formation of apomictic plant lineages [14] and the same process is expected in apomictic animal lineages [5]. Regardless, the assumption is stem cell-like epigenetic reprogramming will be critical in the adaptation and development of asexually reproducing populations. Examinations of the epigenetic variation between populations of the asexual P. antipodarum opened the question that habitat-driven differences could play a role in controlling phenotypic responses when genetic variation is lacking. Divergent epialleles or epimutations may accumulate if they are stably inherited through asexual reproduction once populations become geographically isolated, or be driven by the divergent environments and environmental stressors encountered during development.
This study examined epigenetic variation among three isolated populations of a single clonal lineage residing in lakes which vary in level of ecological contamination and urban development (Fig. 1). The differences in DNA methylation are examined between these populations to see if there is a significant pattern associated with geographic distance between sites, differences in urban and rural location and presence of ecological contaminants. The initial hypothesis was that the two urban lakes may have more similarities than the rural pristine lake due to common urban environmental conditions. The accumulation of epigenetic alterations and correlated gene expression variation is a potential molecular mechanism for an organism to respond to environmental stress. Following analysis of the epigenetic responses for each population, an analysis of gene associations with these epigenetic alterations was performed.
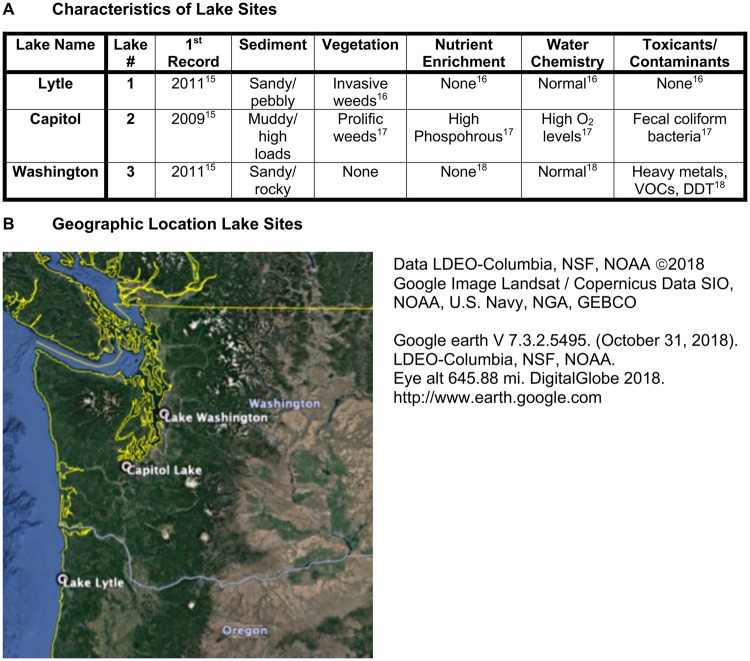
Figure 1:
lake characteristics. (A) Lake name, number, historic record sediment, vegetation, nutrition, water, toxicant information. (B) Regional map of lake locations in Washington and Oregon states USA
Results
Comparisons were made between three populations of an invasive and asexual freshwater snail, P. antipodarum inhabiting three different lakes with differing urban associations and levels of environmental exposures (e.g. toxicants), Fig. 1. Lake 1 (Lake Lytle) is located in a more rural and residential area with relatively low population compared to the urban lakes of Lake 2 (Capitol Lake) having the adjacent cities of Olympia and Lacy Washington and Lake 3 (Lake Washington) within Seattle, WA Previous analyses of these lakes have described differences in sediment and vegetation associated with differences in nutrient enrichment, water chemistry and contamination levels, Fig. 1 [15–18]. The local county population for the rural Lake 1, Tillamook county Oregon is 26 000. The urban Lake 2 within Thurston county Washington, which has a population of 280 000, and Lake 3 within King county Washington, which has a population of over 2 million. The geographic locations and distances are shown in Figs 1 and 2C. The collection of P. antipodarum from these sites has been previously described in [10]. The current investigation characterized epigenetic differences between populations inhabiting the same water flow rate but differing levels of urban association and environmental conditions.
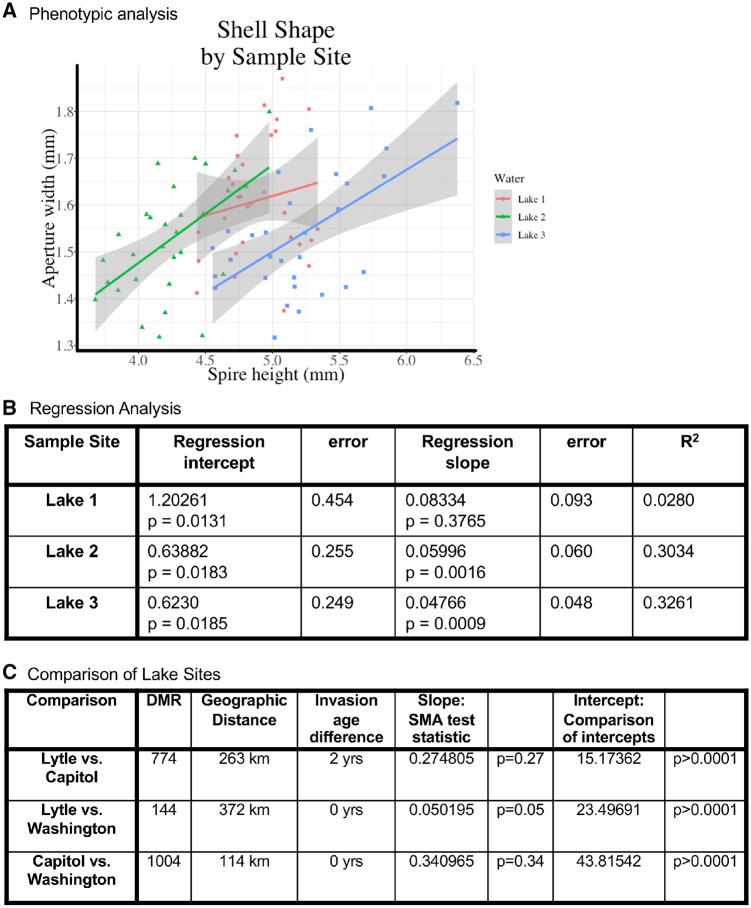
The shell shape of mature snails collected at the various sites (n = 30 individuals from each site) was characterized. The overall length of the shell is plotted against the size of the aperture (the opening through which the foot muscle extends) and it shows that populations differ in relative growth rates (allometry) of shell length versus aperture width (Fig. 2). The regression values for these phenotypic comparisons are shown in Fig. 2B. Lakes 2 and 3 populations show similar allometry, although the shell length of snails sampled from Lake 2 is shorter than those from Lake 3. The snails sampled from Lake 1 show a reduction in distribution of shell length, but a similar relationship to shell length and aperture size as those sampled in the urban lakes. Allometric differences were examined among the three sample sites using the R package smatr [19], which fits a standardized major axis (SMA) to bivariate lines. The slopes for Lakes 1 and three differ significantly (Fig. 2C), whereas the comparison between the other sites do not show a significant difference. The intercept for all three lakes is significantly different (Fig. 2C) showing site-specific allometric differences in shell shape.
Figure 2:
phenotypic differences. (A) Shell shape by sample site for aperture width (mm) versus spire height (mm) for each lake. (B) Regression analysis of slope and intercept for each lake site showing the relationship between snail spire height and snail aperture width. (C) Comparison between lakes, including geographic distance, age of invasion difference, DMR numbers for combination of cDNA DMR and de novo DMR, and SMA analysis
As previously described in [10], the foot pad of the snail was dissected from each individual and has been shown to primarily be a muscle cell population through histology analysis [10]. The total number of individuals for each lake was 30. This tissue was then used to create three different pools for each lake location and contain different individuals (n = 10) for each pool. The DNA was isolated from each pool. Equivalent amounts of DNA were used for each pool to shear into 300 bp fragments and used in a methylated DNA immunoprecipitation (MeDIP) procedure using a methylcytosine antibody, as described in the Methods section. The MeDIP DNA was then used to generate a sequencing library for MeDIP-Seq analysis [20], as previously described in the Methods section. A previously described cDNA library sequence of the snail species was used [21], and a de novo assembly of the MeDIP sequence obtained to assess potential DNA methylation changes [22]. For this study, the de novo analysis was used to identify DMRs that were associated with intergenic and gene regions, and then the cDNA library used for gene association analysis. Comparisons were made between Lakes 1 and 2 (Lytle vs. Capitol), Lakes 1 and 3 (Lytle vs. Washington) and Lakes 2 and 3 (Capitol vs. Washington) to determine the differential DMRs for each comparison.
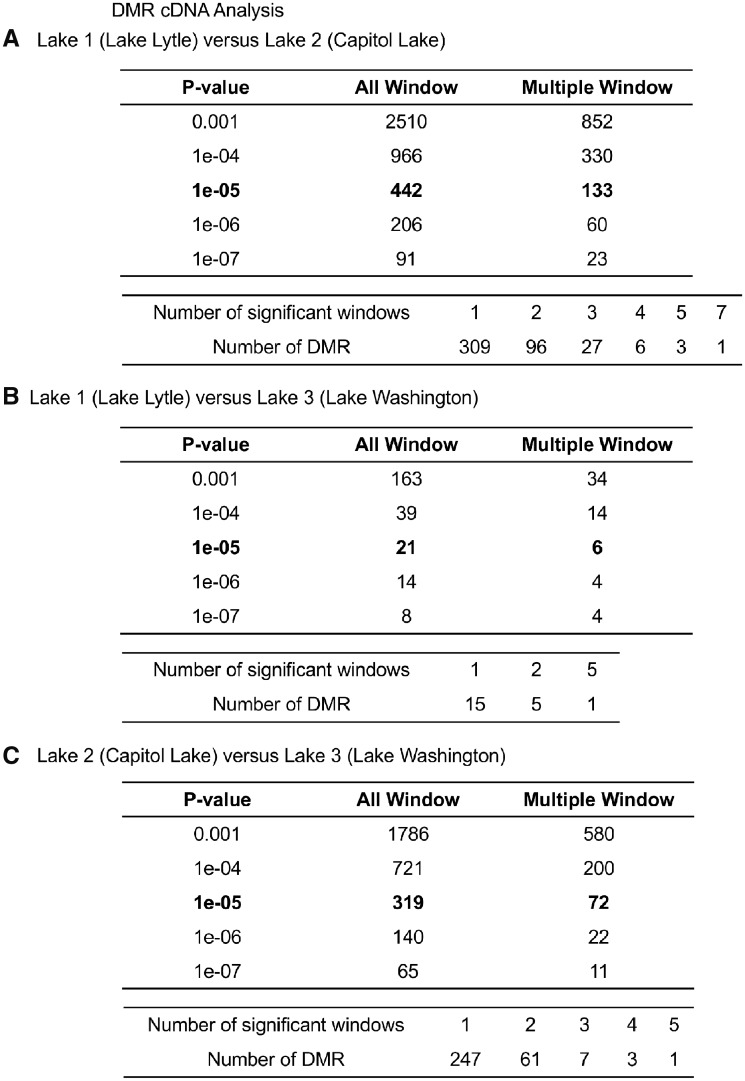
The DMRs from the cDNA library analysis [21] for a variety of P-values are presented in Fig. 3 for all windows (100 bp per window) and for multiple (≥2) windows for each DMR, as more thoroughly described in the Methods section. A P-value of 1e-05 was used for subsequent analysis, but DMRs were identified at reduced stringency P-values, Fig. 3. The Lakes 1 and 2 comparison identified 442 DMRs, Lakes 1 and 3 comparison 21 DMRs and Lakes 2 and 3 comparison 319 DMRs at 1e-05 P-value. The false discovery rate (FDR), a multiple testing correction, for all the comparisons was significant with P < 0.05. The multiple window DMRs are also listed for P < 1e-05 with the majority having a single 100 bp window at this P-value. Flanking 100 bp windows with P < 0.01 were merged with the DMR. For the cDNA DMR analysis the location, length and CpG density are presented for all DMRs in Supplementary Tables S1–S3.
Figure 3:
the number of cDNA DMRs found using different P-value cutoff thresholds. The All Window column shows all DMRs. The Multiple Window column shows the number of DMRs containing at least two significant windows. The number of DMR with each specific number of significant windows at a P-value threshold of 1e-05. (A) Lake 1 (Lytle Lake) versus Lake 2 (Capitol Lake). (B) Lake 1 (Lytle Lake) versus Lake 3 (Washington Lake). (C) Lake 2 (Capitol Lake) versus Lake 3 (Washington Lake)
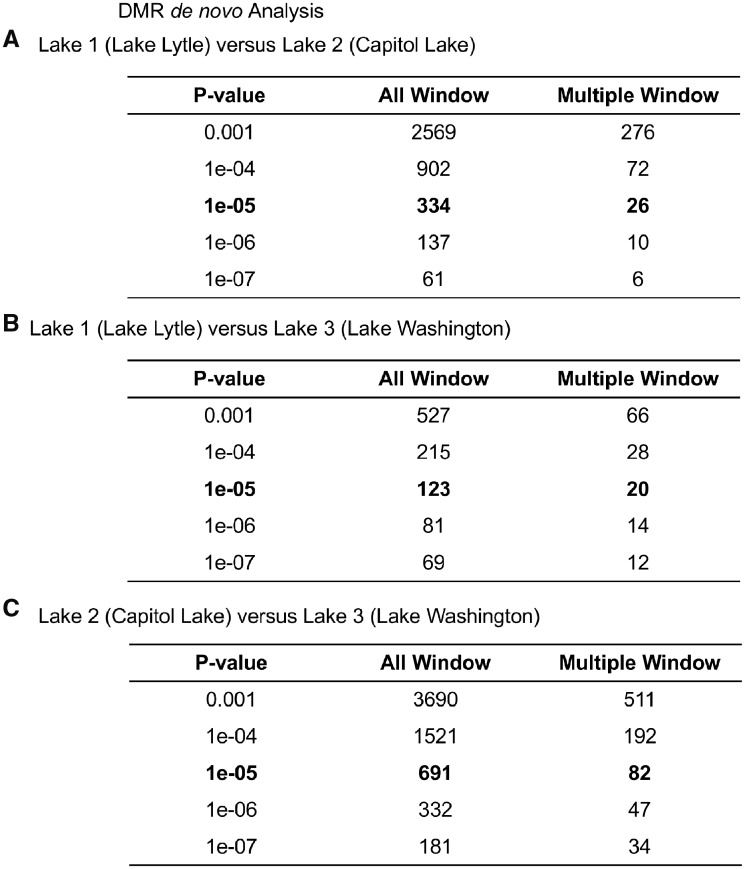
The DMRs from the de novo analysis, focused on intergenic regions, for a variety of P-values are presented in Fig. 4 for all windows (100 bp per window) and adjacent multiple windows, as more thoroughly discussed in the Methods section. A P-value of P < 1e-05 was used for subsequent analysis, but DMRs identified at various P-values presented, Fig. 4. The FDR for the de novo DMRs was also P < 0.05. The Lakes 1 and 2 de novo comparison identified 334 DMRs, Lakes 1 and 3 comparison identified 123 DMRs, and Lakes 2 and 3 comparison identified 691 DMRs at P < 1e-05, Fig. 4. The multiple adjacent window DMRs are also listed with the majority being a single 100 bp window. Since a full genome is not available, it was not possible to map the locations of the de novo DMRs or easily assess lengths or overlaps. As discussed below, the de novo DMRs are primarily intergenic and have comparable numbers as the cDNA DMR analysis.
Figure 4:
the number of de novo DMRs found using different P-value cutoff thresholds. The All Window column shows all DMRs. The Multiple Window column shows the number of DMRs containing at least two significant windows. (A) Lake 1 (Lytle Lake) versus Lake 2 (Capitol Lake). (B) Lake 1 (Lytle Lake) versus Lake 3 (Washington Lake). (C) Lake 2 (Capitol Lake) versus Lake 3 (Washington Lake)
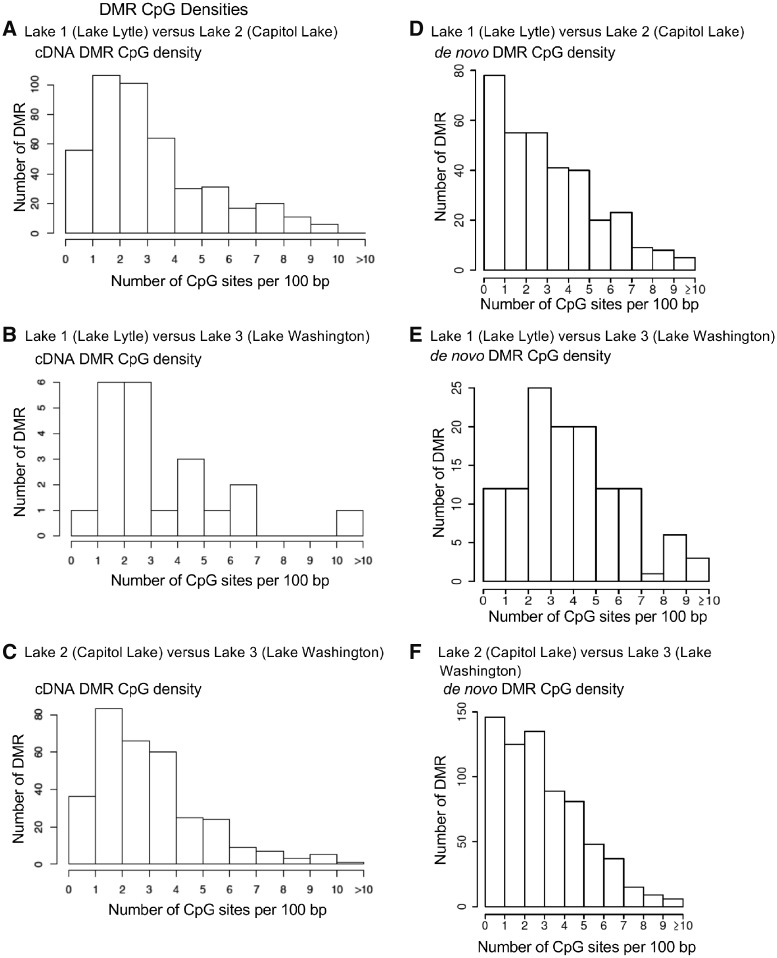
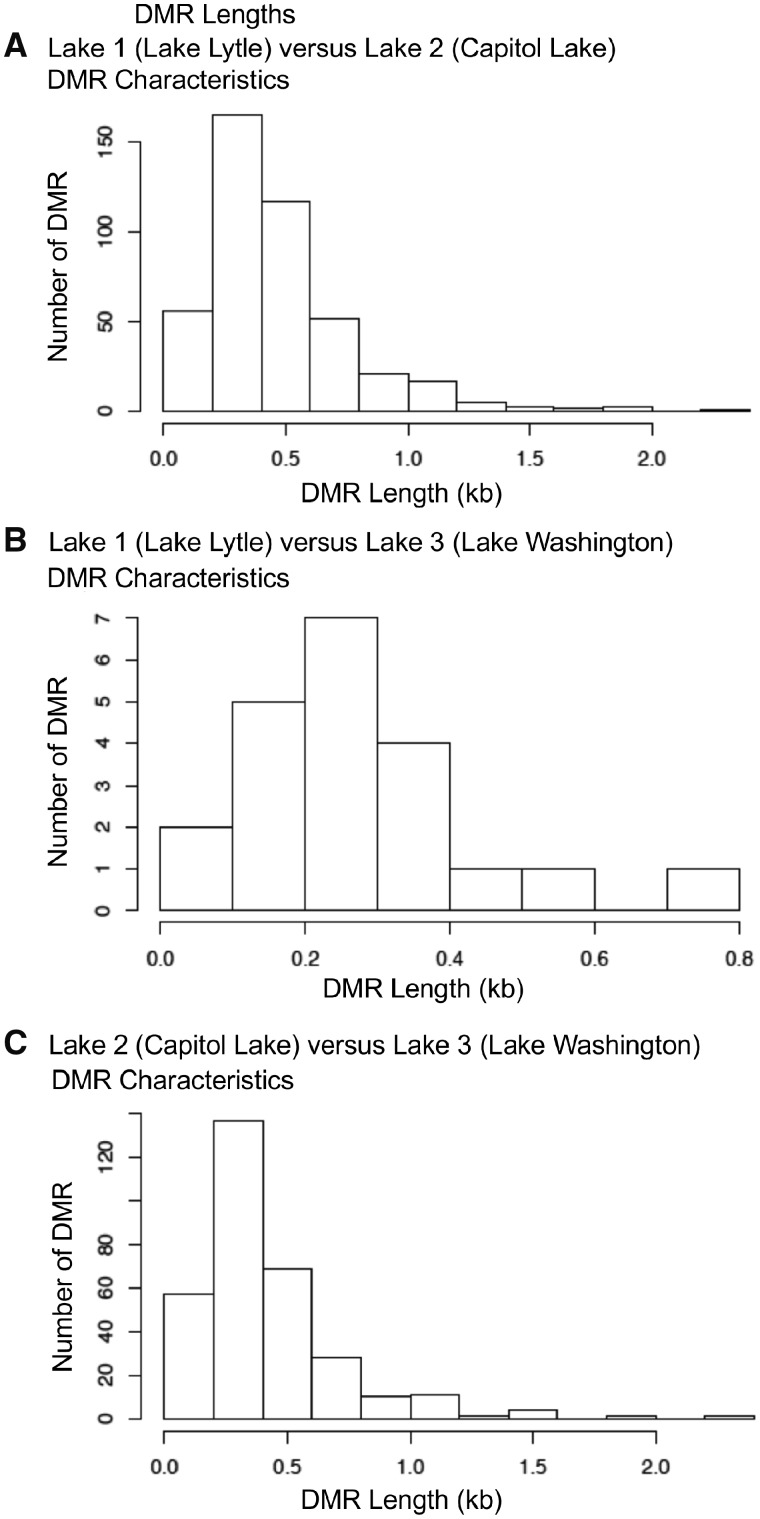
The genomic features of both the cDNA and de novo DMRs identified considered the CpG density to be associated with DNA methylation and the DMR length, as described in the Methods section. The CpG density for each lake comparison ranged between 1 and 10 CpG per 100 bp with some >10 CpGs per 100 bp, Fig. 5. The predominant CpG density for all the lake comparisons cDNA DMRs was 2–3 CpGs per 100 bp so are low density CpG deserts, as previously described in [23]. The CpG density of the de novo DMRs was similar to higher numbers in the 1 CpG/100 bp density than the cDNA DMRs. The cDNA DMR lengths ranged between 200 bp and 1 kb with the predominant size between 300 and 400 bp in size, Fig. 6. The de novo DMR fragmented reference does not allow the lengths to be determined. For all comparisons with both the cDNA and de novo DMRs there was ∼65 to 85% of DMRs with an increase in DNA methylation and 15–35% with a decrease in DNA methylation. For the de novo analysis, Lakes 1 and 2 82% of the DMRs had an increase in DNA methylation observed, Lakes 1 and 3 76% of the DMRs had an increase in DNA methylation, and Lakes 2 and 3 66% of the DMRs had an increase in DNA methylation. The cDNA analysis Lakes 1 and 2 51% of the DMRs had an increase in DNA methylation, Lakes 1 and 3 57% of the DMRs had an increase in DNA methylation and Lakes 2 and 3 58% of the DMRs had an increase in DNA methylation. The lack of a well-defined genome for the P. antipodarum influences this analysis. The specific details and characteristics for each cDNA DMR are presented in Supplementary Tables S1–S3.
Figure 5:
the number of DMRs at different CpG densities. All DMRs at a P-value threshold of P < 1e-5 are presented for cDNA DMRs (A–C) and de novo (D–F) CpG densities. (A and D) Lake 1 (Lytle Lake) versus Lake 2 (Capitol Lake). (B and E) Lake 1 (Lytle Lake) versus Lake 3 (Washington Lake). (C and F) Lake 2 (Capitol Lake) versus Lake 3 (Washington Lake)
Figure 6:
the cDNA DMR lengths. All cDNA DMRs at a P-value threshold of P < 1e-05. (A) Lake 1 (Lytle Lake) versus Lake 2 (Capitol Lake). (B) Lake 1 (Lytle Lake) versus Lake 3 (Washington Lake). (C) Lake 2 (Capitol Lake) versus Lake 3 (Washington Lake)
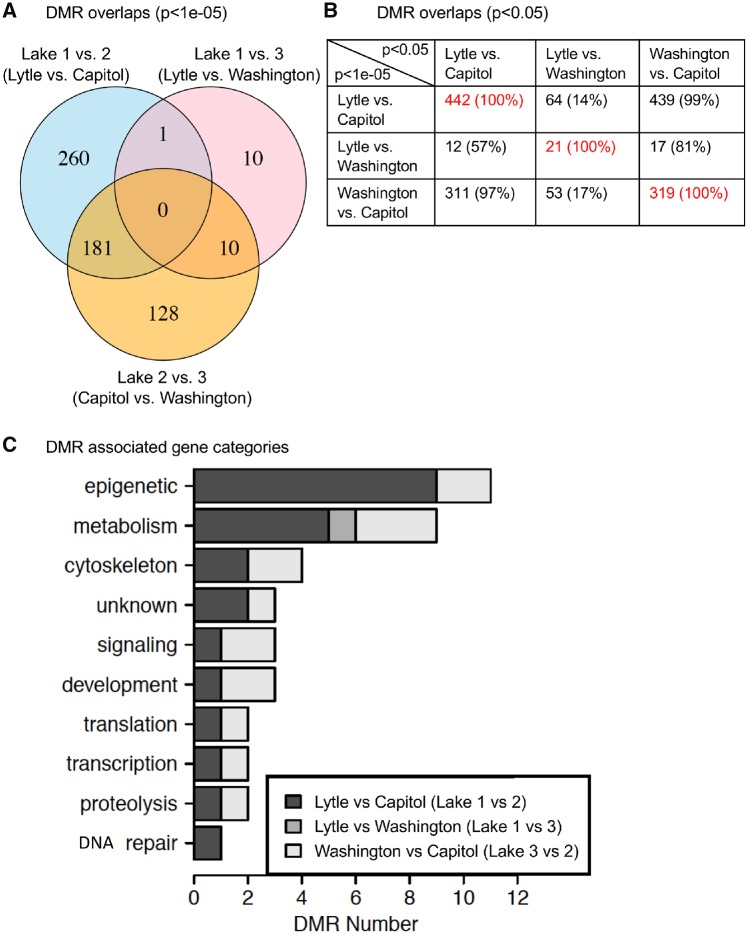
An analysis of the cDNA DMRs for each lake comparison was made to assess potential overlap of the specific DMRs. A Venn diagram of the DMRs for each lake comparison using a P < 1e-05 demonstrated negligible overlap between the Lakes 1 versus 2 and Lakes 1 versus 3 comparisons. Much more overlap was observed in the Lakes 1 versus 2 and Lakes 2 versus 3 DMRs, Fig. 7A. In fact, 40% of the DMRs between Lakes 1 and 2 are present in the comparison of Lakes 2 and 3. Interestingly, 56% of the cDNA DMRs between Lakes 2 and 3 are present in the comparison of Lakes 1 and 2. Since this was a stringent comparison with P < 1e-05, another analysis with reduced stringency was performed, Fig. 7B. When cDNA DMRs at P < 1e-05 are tested for overlap with DMRs at P < 0.05, the overlap between the cDNA DMRs in the Lakes 1 versus 2 and Lakes 2 versus 3 comparisons becomes more apparent. For example, 99% of DMRs identified between Lakes 2 and 3 are identified between Lakes 1 and 2, Fig. 7B. Therefore, while there are DMRs specific to each comparison of lakes, the most overlap was observed whenever comparisons included Lake 2. This could suggest that some factor of the environment in Lake 2 creates similar differences in methylation between snails inhabiting Lake 2 and snails inhabiting the other two lakes. The de novo DMR short fragments and lack of a full genome did not allow an efficient overlap analysis of the different lake comparison de novo DMRs.
Figure 7:
DMR overlap and gene associations. (A) Overlap each cDNA DMR comparison (Lakes 1 vs. 2; Lakes 1 vs. 3; Lakes 2 vs. 3) with cDNA DMR P < 1e-05. (B) Overlap cDNA DMR (P < 1e-05) (y-axis) with reduced P < 0.05 (x-axis) for comparison DMR data. (C) cDNA DMR-associated functional gene categories
The cDNA DMR-associated genes were identified and for each DMR are presented in Supplementary Tables S1–S3. Although the P. antipodarum genome is not assembled or fully annotated a limited cDNA library is available [21]. The de novo assembly of the MeDIP sequences also allowed intergenic regions with DNA methylation to be analysed. Blast search for similar sequences among all species was performed for all the DMRs to identify gene associations as described in the Methods section. The gene categories were assigned to each cDNA DMR-associated gene, when identified, and are presented in Fig. 7C. The majority of associations are associated with homologies of other genomic sites and not currently characterized with a specific gene, so are presented to allow future analysis of these sites. Since the full genome is not available nor gene sets for this species, a gene enrichment analysis is not possible. One of the most predominant gene functional categories for all comparisons was the metabolism pathway which is expected due to the large number of genes associated with metabolism. Interestingly, the epigenetics category (e.g. PHF5A) was predominant in the Lakes 1 and 2 comparison and also high in the Lakes 2 and 3 comparison. Other major pathways are the cytoskeleton, signalling and development pathways, Fig. 7C. The lack of gene associations in the Lakes 1 and 3 comparison is likely due to the low number of cDNA DMRs in that data set.
The overlap with the de novo DMRs and the cDNA DMRs only identified two overlaps for the Lakes 1 versus 2 comparison, no overlap with the Lakes 1 versus 3 comparison, and six overlap with the Lakes 2 versus 3 comparison. Due to the short fragments of the de novo analysis, an under representations of the actual DMR numbers are anticipated. Higher numbers likely exist if a full genome was available for analysis. A comparison of the sequencing reads between the cDNA analysis and de novo analysis demonstrated 50% of the transcriptome cDNA reads overlapped with the de novo reads, while 28% of the de novo reads overlapped with the cDNA reads. Therefore, the de novo analysis identified DMRs that appear predominantly in intergenic regions of the snail genome. Since the cDNA DMRs and de novo DMRs were similar in number and provided the same general trends in the analysis with the comparisons, the data are supportive. A combination of the two DMR sets identified 774 DMRs for the Lakes 1 versus 2 comparison, 144 DMRs for the Lakes 1 versus 3 comparison, and 1004 DMRs for the Lakes 2 versus 3 comparison for the P < 1e-05 threshold, Fig. 2C.
Discussion
Epigenetic responses can occur and accumulate relatively rapidly, particularly when compared with the accumulation of genetic mutations [24]. The rapid rate of response through epigenetic mechanisms can be important among range-expanding and invasive populations, which are encountering environmental heterogeneity and novel environmental stressors. In addition, the rapid response through epigenetics can be crucially important for asexually reproducing species, in which genetic variation will accumulate or spread through a population at a very low rate. We compared methylation in a clonal lineage of an asexual snail (P. antipodarum) and found differentially methylated regions between the pristine rural Lake 1 (Lake Lytle) and the urban, more contaminated Lake 2 (Capitol Lake) and Lake 3 (Lake Washington) populations. Analysis of the allometric growth patterns of each population shows differences among populations, while the relationship between shell length and aperture size differed significantly for all three populations. The phenotypic variation observed within a single clonal lineage among isolated lake populations, along with methylation differences, supports a role for epigenetic alterations in gene expression and development.
The DMR analysis provides evidence for alterations in methylation within this asexual invasive snail. This study used de novo assembly of the MeDIP sequence and the cDNA sequence previously identified [21], so provides the most complete genome sequence for the New Zealand snail (P. antipodarum) reported. The data are deposited in the NCBI GEO #GSE93836 and #GSE133502 as described in the Methods section. Therefore, both intergenic and gene-associated DMRs are identified. A relatively high number of DMRs were identified between Lakes 1 and 2, as well as between Lakes 2 and 3. The comparison of Lakes 1 and 3 identified a lower number of DMRs at P < 1e-05 with higher number of P < 0.001, Figs 3 and 4. Therefore, DMRs for all were identified but the Lakes 1 versus 3 had reduced numbers of DMRs at the same statistical threshold for both cDNA DMRs and de novo DMRs. In all comparisons, epigenetic alterations were observed between the different lakes. The epigenetic alterations (i.e. DMRs) identified had negligible overlap at the higher stringency threshold of P < 1e-05. However, at a lower stringency comparison, additional overlap was observed. Because the threshold of P < 1e-05 is relatively stringent, lower stringency comparisons were included to illustrate how the threshold chosen affects the results of the genome-wide analysis.
The presence of epigenetic differences among populations inhabiting various sites has several potential explanations. If epigenetic marks are stably inherited and act like genetic mutations, then observed differences might be the consequence of either random genetic drift or natural selection in the absence of sufficient gene flow to prevent divergence. If so, the number of epigenetic differences may be explained by geographic distance (in the case of random genetic drift) or divergent selection between sites, particularly in an expanding invasive population. However, there was no consistent pattern attributed to geographic distance between sites. The most distant sites (Lakes 1 and 3) did not necessarily have the most DMRs. The comparisons between closer sites showed higher DMR numbers (774 from Lakes 1 to 2, 1004 from Lakes 2 to 3).
However, if epigenetic marks are not stably inherited but instead environmentally sensitive, then any observed epigenetic differences may result from a response to environmental factors, such as the proximity of a site to urbanization and industrialization. For example, environmental toxicants unique to each sample site may explain the presence of epigenetic alterations (as in [11–13]). Lake 2 appears to be the most distinct in terms of methylation alterations. This is shown in both the number of DMRs identified and the overlap of cDNA DMRs in comparing Lake 1 or 3 with Lake 2. This potentially indicates a difference due to ecological toxicants, where Lake 2 has significant nutrient enrichment and fecal coliform contamination and Lake 3 has heavy metals, volatile organic compounds and dichloro-diphenyl-trichloroethane (DDT) contamination (Fig. 1). The environmental and ecological conditions at each of the three lakes are so different, each providing a unique combination of selection pressures, that our results implicate a complex epigenetic response unique to each population. A further analysis might seek to replicate multiple combinations of urban and rural environmental conditions and examine the epigenetic response of the replicate populations.
The epigenetic analysis focuses on DNA methylation while other epigenetic processes such as non-coding RNA or histone modification could be considered in future studies. Many physiological and phenotypic aspects of P. antipodarum were not examined, such as reproductive fitness or longevity. It is anticipated that a more thorough analysis and comparison of physiological and phenotypic differences would reveal additional differences between lake habitats. Future phenotypic and physiological analyses will need to more thoroughly assess a variety of different parameters to more clearly understand the adaptative impacts of the environmental epigenetics observed.
The importance of epigenetic variation as an additional mechanism for the regulation of gene expression or developmental variation that can ultimately serve to facilitate natural selection has been proposed [25], but not yet well supported in natural populations. Examples in Darwin’s finches [12], alligators [11] and dandelion [26] have suggested a role for epigenetics in adaptation and evolution [25]. Such comparisons could provide strong support for epigenetic response to environmental stressors as an alternative path to adaptation, independent of genetic sequence changes, in natural populations.
This study was designed to determine if a comparison of the rural and urban lakes would identify epigenetic alterations that correlate with the different populations sampled. Epigenetic alterations were identified between the different lake environments, as well as some phenotypic variation between the lakes. Therefore, the DMRs and epigenetic alterations suggest the ecology of a rural versus urban environment has the ability to promote epigenetic variation in isolated populations. Similar observations in the American alligator and two species of Darwin’s Finch support this epigenetic mechanism [11, 12, 15].
Habitat-specific patterns of methylation could be particularly important in populations lacking genetic variation. A potential explanation for the persistence of genetically limited populations in heterogeneous environments is the generation of a response to the environment through epigenetic alterations and changes in gene expression [5]. Associated shifts in the methylation pattern between sites and pollution levels suggest a potential crucial role for epigenetic mechanisms in the persistence and spread of natural populations in heterogeneous environments.
A more complete genome would be useful in future analyses of this species. Since P. antipodarum does not have a complete genome assembly to allow effective comparison, a de novo assembly and cDNA library sequence were used. The incomplete and fragments of a de novo reference is anticipated to result in an under representation of the potential DMR numbers. However, a combination of de novo assembly and the cDNA analysis provides a more complete genome analysis. This is recommended if only a cDNA library is available and allows intergenic regions to be investigated. Clearly a complete genome sequence would improve the genome-wide analysis. Although a limited genome is currently available, the analysis performed clearly identified epigenetic alterations between the different lake environments. A number of gene associations were identified, but a more complete genome and annotation will be useful for further analysis. The causal link between the epigenetic shift and response to distinct ecology and environmental toxicants is yet to be determined in P. antipodarum. Without this link, it is difficult to determine whether the epigenetic response is adaptive. In addition, many environmental toxicants and exposures have unique epigenetic alterations [27]. A link between the effects of the altered methylation and the specific environmental stressors would provide further support for epigenetic response as an important aspect of adaptation of a population.
Methods
Sample Collection
Potamopyrgus antipodarum individuals were sampled from three lakes representing a similar habitat for water current speeds. This is an important aspect of the habitat because water flow rates influence gastropod shell morphology and adaptation.
The snails were collected from Lake Lytle (Lake 1) in Rockaway Beach, OR (45.6272°N, 123.9392°W), which serves as a relatively pristine habitat as the lake is located in a rural, coastal town and is subjected to little contamination. Lake Lytle water quality analyses show the lake to have a high level of invasive weeds, but this problem is not attributed to nutrient enrichment [16]. There is little evidence of environmental stressors due to nutrient enrichment or ecotoxicants in this lake.
Snails were also collected from two urban lake sites which represent more contaminated conditions. The first of these contaminated sample sites is Marathon Park on Capitol Lake (Lake 2) in Olympia, WA (47.0384°N, 122.9119°W). Capitol Lake is a man-made reservoir that is filling with sediment from its tributaries resulting in a shallow lake that heats up more than other lakes in summer which can cause stress for aquatic organisms [17]. The same analysis found Capitol Lake also has enhanced weed growth due to the high water temperatures in summer, high levels of fecal coliform bacteria from storm water runoff, and high levels of phosphorous. There is little evidence that the water is contaminated with industrial pollutants or toxic chemicals [17].
The second contaminated site is a beach area of Lake Washington (Lake 3) in Seattle, WA (47.6971°N, 122.2711°W). Lake Washington has a decades-long history of contamination of heavy metals, polycyclic aromatic hydrocarbons, volatile organic compounds (VOHs) and a more recent contamination of DDT [18].
For simplicity, we refer to Lake Lytle as Lake 1, Capitol Lake as Lake 2 and Lake Washington as Lake 3. Lake 1 represents a relatively pristine habitat while Lakes 2 and 3 are urban, polluted lakes.
Samples were obtained by searching the substrate and scraping snails off the underside of rocks, woody debris and vegetation. The samples were maintained on wet paper towels and kept cool until they reached the laboratory at Washington State University, Pullman, WA. In the laboratory, the samples were kept in water in an environmental chamber held at 14°C with a 12:12 day/night cycle for several days until they were processed for phenotypic and molecular analysis. The time spent in the laboratory for processing was not sufficient for the accumulation of epigenetic changes, particularly in the single, specific cell type used for epigenetic analysis. Major epigenetic shifts generally require months as mitosis is required to dramatically shift the epigenetics. Phenotypic analysis included measurements of the aperture width and total shell spire height on ImageJ software (described in [10]). To examine allometric difference in the relationship between aperture width and spire height, the r package smatr was used [19].
Tissue
Thirty individual samples were collected from each site. To harvest tissue, the shell was cracked open and the soft tissue was separated from the shell. A thin slice of the foot pad tissue was separated from the remainder of the soft tissue and this foot tissue was stored in Nanopure™ water and immediately frozen. The foot muscle tissue was sampled because foot muscle is a homogeneous marker tissue in the adult [28]. This homogeneous tissue records epigenetic shifts due to environmental inductions that are passed onto adult somatic cells in the developing embryo since formation of the egg cell.
Genomic DNA Preparation
Genomic DNA from snail foot tissue was utilized in this study. DNA was extracted from a thin slice of foot muscle tissue. Samples were pooled due to the small volume of tissue, to ensure sufficient DNA yields. For the individuals collected from Lake Lytle and Lake Washington, foot pad tissue samples were pooled in three pools of 10 individuals for each sample site (a total of six pools for these two sites) and the genomic DNA was isolated following a DNeasy blood and tissue kit (Qiagen, Valencia, CA). The pooled tissue from 10 individuals was cut, suspended in 1× Phosphate Buffered Saline (PBS) solution and 20 µl of Proteinase K (20 mg/ml) was added followed by incubation at 56°C for 12 h. Following this incubation, the manufacturer’s protocol was followed, using water to resuspend the DNA in the last step. Genomic DNA was isolated from the samples collected from Capitol Lake starting with foot pad tissue from 10 individuals in each of three pools, which was sonicated and prepared for analysis. Snail tissue was suspended in 100 μl of 1× PBS, then 820 μl DNA extraction buffer (50 mM Tris pH8, 10 mM EDTA pH8, 0.5% SDS) and 80 μl Proteinase K (20 mg/ml) was added and the sample incubated on a rotator at 55 °C for 2–3 h. After incubation, 300 μl of protein precipitation solution (Promega, A795A) was added, the sample mixed and incubated on ice for 15 min, then spun at 4 °C at 13 000 rpm for 20 min. The supernatant was transferred to a fresh tube, then precipitated overnight with the same volume 100% isopropanol and 2 μl glycoblue at −20 °C. The sample was then centrifuged and the pellet washed with 75% ethanol, then air-dried and resuspended in 100 μl H2O. DNA concentration was measured using the Nanodrop (Thermo Fisher).
Methylated DNA Immunoprecipitation
MeDIP from genomic DNA was performed to quantify levels of methylation. For the snail samples, ∼6 μg of each genomic DNA pool was diluted to 130 μl with TE buffer into the appropriate Covaris tube. Covaris was set to 300 bp program. Ten microlitres of each sonicated DNA were run on 1.5% agarose gel to verify fragment size. The sonicated DNA was transferred from the Covaris tube to a 1.7 ml microfuge tube and the volume measured. The sonicated DNA was then diluted with TE buffer (10 mM Tris HCl, pH7.5; 1 mM EDTA) to 400 μl, heat-denatured for 10 min at 95°C, then immediately cooled on ice for 5 min. Then 100 μl of 5× IP buffer and 5 μg of antibody (monoclonal mouse anti 5-methylcytidine; Diagenode #C15200006) were added to the denatured sonicated DNA. The DNA-antibody mixture was incubated overnight on a rotator at 4°C.
The following day magnetic beads (Dynabeads M-280 Sheep anti-Mouse IgG; 11201 D) were pre-washed as follows: The beads were resuspended in the vial, then the appropriate volume (50 μl per sample) was transferred to a microfuge tube. The same volume of Washing Buffer (at least 1 ml) was added and the bead sample was resuspended. The tube was then placed into a magnetic rack for 1–2 min and the supernatant discarded. The tube was removed from the magnetic rack and the washed beads were resuspended in the same volume of 1× IP buffer as the initial volume of beads. Fifty microlitres of beads were added to the 500 μl of DNA-antibody mixture from the overnight incubation, then incubated for 2 h on a rotator at 4°C.
After the incubation, the beads were washed three times with 1× IP buffer as follows: The tube was placed into magnetic rack for 1–2 min and the supernatant discarded, then washed with 1× IP buffer three times. The washed beads were then resuspended in 250 μl digestion buffer (50 mM Tris-HCI pH8, 10 mM EDTA pH8, 0.5% SDS) with 3.5 μl Proteinase K (20 mg/ml). The sample was then incubated for 2–3 h on a rotator at 55°C. After incubation the tube was put again into the magnetic rack for 3 min and the supernatant removed to a new microfuge tube. The beads were discarded. Two hundred and fifty microlitres of buffered Phenol–Chloroform–Isoamylalcohol solution were added to the supernatant and the tube vortexed for 30 s then centrifuged at 14 000 rpm for 5 min at room temperature. The aqueous supernatant was carefully removed and transferred to a fresh microfuge tube. Then 250 μl chloroform were added to the supernatant from the previous step, vortexed for 30 s and centrifuged at 14 000 rpm for 5 min at room temperature. The aqueous supernatant was removed and transferred to a fresh microfuge tube. To the supernatant 2 μl of glycoblue (20 mg/ml), 20 μl of 5 M NaCl and 500 μl ethanol were added and mixed well, then precipitated in a −20°C freezer for 1 h to overnight.
The precipitate was centrifuged at 14 000 rpm for 20 min at 4°C and the supernatant removed, while not disturbing the pellet. The pellet was washed with 500 μl cold 70% ethanol in -20°C freezer for 15 min. then centrifuged again at 14 000 rpm for 5 min. at 4°C and the supernatant discarded. The tube was spun again briefly to collect residual ethanol at bottom of tube and then as much liquid as possible was removed with gel loading tip. Pellet was air-dried at RT until it looked dry (∼5 min) then resuspended in 25 μl H2O or TE. DNA concentration was measured in Qubit with ssDNA kit.
MeDIP-Seq
Snail foot pad MeDIP pools were used to create libraries for next-generation sequencing (NGS) at Washington State University, Genomics Core Laboratory with an Illumina® platform. All the pools consisted of 10 individuals (see above), with 3 pools each for the three lake populations. For library preparation, the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® was used, starting at step 1.4 of the manufacturer’s protocol to generate double stranded DNA. After this step the manufacturer’s protocol was followed. Each pool received a separate index primer. NGS was performed at the WSU Spokane Genomic Core laboratory using the Illumina® HiSeq 2500 with a PE50 application, with a read size of ∼50 bp and 60 million reads per pool. The cDNA analysis had a 25% mapping, and de novo a 40% mapping efficiency. The use of 50 bp reads is confirmed to function well with MeDIP protocols without any variation or alignment issues. Three libraries each were run in one lane comparing each of these three lake populations. All sequencing data have been deposited to NCBI GEO (GEO # GSE133502).
De Novo Assembly and Genome Sequence Analysis
For the de novo analysis, samples were cleaned and filtered to remove low quality base pairs using Trimmomatic [29]. Reads that lost one of the paired ends due to the Trimmomatic cleaning were not included in the de novo reference assembly. All samples were then concatenated into a single pair of files. The combined sample reads were then assembled into contigs using ABySS [30, 31]. ABySS was run with default parameters and a 25 bp k-mer size.
To generate the reference genome, all ABySS generated contigs <500 bp were concatenated into a series of larger scaffolds. Contigs were separated on the scaffolds by ambiguous bases (Ns). Contigs that were at least 500 bp in length were kept as individual sequences. The scaffolds and contigs were then used as the reference genome for all remaining analysis steps.
The Bowtie 2 mapping and DMR identification analyses were performed using the same methods and parameters as the transcriptome reference analysis except for the use of the ABySS generated contigs as the reference genome. The overall mapping percentage of reads to the de novo reference was around 40% for each sample. DMRs identified using the de novo reference were then examined to ensure that they did not span multiple contigs.
To identify DMRs present in both the de novo and transcriptome analyses, the sequence for each DMR in the de novo analysis was aligned with each sequence for the DMRs in the transcriptome analysis. This was done using the Smith–Waterman local alignment algorithm implemented in the pairwise Alignment function in the Biostrings R package [32]. Sequence alignments with a score >50 were examined further to determine whether there was a substantial overlap.
Bioinformatics and Statistics
Basic read quality was verified using summaries produced by the FastQC program [33]. Reads were trimmed and filtered to remove low quality base pairs using Trimmomatic [29]. The reads for each sample for DMR analyses were mapped to a P. antipodarum transcriptome reference [21] using Bowtie2 [34] with default parameter options. Due to using this incomplete transcriptome reference, mapping efficiency was low. Only 20–25% of reads mapped successfully to the reference. The mapped read files were then converted to sorted BAM files using SAMtools [35]. To identify DMR, the reference genome was broken into 100 bp windows. The MEDIPS R package [36] was used to calculate differential coverage between sample groups. The edgeR P-value [37] was used to determine the relative difference between the two groups for each genomic window. Windows with an edgeR P-value <10−5 were considered DMRs for subsequent data analysis, but DMRs were also present at less stringent thresholds. The DMR edges were extended until no genomic window with an edgeR P-value <0.1 remained within 1000 bp of the DMR. CpG density and other information were then calculated for the DMR based on the reference genome.
Supplementary Material
Acknowledgements
We thank Ms Jayleana Barton for technical assistance, Ms Amanda Quilty for assistance in editing the manuscript and Ms Heather Johnson for assistance in preparation of the manuscript.
Funding
This study was supported by a John Templeton Foundation grant (61174 to M.K.S.) and NIH grant (ES012974 to M.K.S.).
Accession Number
All sequencing data have been deposited to NCBI GEO (GEO # GSE93836 and # GSE133502).
Conflict of interest statement. None declared.
References
- 1. Jirtle RL, Skinner MK.. Environmental epigenomics and disease susceptibility. Nat Rev Genet 2007;8:253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Skinner MK. Environment, epigenetics and reproduction. Mol Cell Endocrinol 2014;398:1–3. [DOI] [PubMed] [Google Scholar]
- 3. Skinner MK. Environmental epigenetic transgenerational inheritance and somatic epigenetic mitotic stability. Epigenetics 2011;6:838–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Skinner MK, Ben Maamar M, Sadler-Riggleman I, Beck D, Nilsson E, McBirney M, Klukovich R, Xie Y, Tang C, Yan W.. Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT-induced epigenetic transgenerational inheritance of disease. Epigenet Chromatin 2018;11:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Verhoeven KJ, Preite V.. Epigenetic variation in asexually reproducing organisms. Evolution 2014;68:644–55. [DOI] [PubMed] [Google Scholar]
- 6. Drown DM, Levri EP, Dybdahl MF.. Invasive genotypes are opportunistic specialists not general purpose genotypes. Evol Appl 2011;4:132–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kistner EJ, Dybdahl MF.. Adaptive responses and invasion: the role of plasticity and evolution in snail shell morphology. Ecol Evol 2013;3:424–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paczesniak D, Jokela J, Larkin K, Neiman M.. Discordance between nuclear and mitochondrial genomes in sexual and asexual lineages of the freshwater snail Potamopyrgus antipodarum. Mol Ecol 2013;22:4695–710. [DOI] [PubMed] [Google Scholar]
- 9. Kronholm I, Collins S.. Epigenetic mutations can both help and hinder adaptive evolution. Mol Ecol 2016;25:1856–68. [DOI] [PubMed] [Google Scholar]
- 10. Thorson JLM, Smithson M, Beck D, Sadler-Riggleman I, Nilsson E, Dybdahl M, Skinner MK.. Epigenetics and adaptive phenotypic variation between habitats in an asexual snail. Sci Rep 2017;7:14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guillette LJ Jr, Parrott BB, Nilsson E, Haque MM, Skinner MK.. Epigenetic programming alterations in alligators from environmentally contaminated lakes. Gen Comp Endocrinol 2016;238:4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McNew SM, Beck D, Sadler-Riggleman I, Knutie SA, Koop JAH, Clayton DH, Skinner MK.. Epigenetic variation between urban and rural populations of Darwin's finches. BMC Evol Biol 2017;17:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Skinner MK, Guerrero-Bosagna C, Haque MM, Nilsson EE, Koop JAH, Knutie SA, Clayton DH.. Epigenetics and the evolution of Darwin's Finches Genome Biology &. Evolution 2014;6:1972–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmidt A, Schmid MW, Klostermeier UC, Qi W, Guthorl D, Sailer C, Waller M, Rosenstiel P, Grossniklaus U.. Apomictic and sexual germline development differ with respect to cell cycle, transcriptional, hormonal and epigenetic regulation. PLoS Genet 2014;10:e1004476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Benson AJ, Kipp RM, Larson J, Fusaro A.. Potamopyrgus antipodarum (J.E. Gray, 1853): U.S. Geological Survey, Nonindigenous Aquatic Species Database, Gainesville, FL, https://nas.er.usgs.gov/queries/factsheet.aspx?SpeciesID=1008, 2019 (15 October 2019, date last accessed).
- 16. Johnson Y, Drake D.. Water Quality Status and Action Plan: North Coast Basin Oregon: State of Oregon Department of Environmental Quality, 2011.
- 17. FloydSnider I. Phase 1 Report on the Capitol Lake/Lower Deschutes Watershed Long-term Management Planning. Washington State Department of Enterprise Services. http://des.wa.gov/sites/default/files/public/documents/About/CapitolLake/2016MeetingDocs/ProvisoReport-Phase1-2016-12-30.pdf, 2016. (15 October 2019, date last accessed).
- 18. Moshenberg KL. Sediment Triad Analysis of Lakes Sammamish, Washington, and Union. King County Department of Natural Resources and Parks. https://your.kingcounty.gov/dnrp/library/2004/kcr1880.pdf, 2004. (15 October 2019, date last accessed).
- 19. Warton DI, Duursma RA, Falster DS, Taskinen S.. smatr 3—an R package for estimation and inference about allometric lines. Methods Ecol Evol 2012;3:257–9. [Google Scholar]
- 20. Ben Maamar M, Nilsson E, Sadler-Riggleman I, Beck D, McCarrey JR, Skinner MK.. Developmental origins of transgenerational sperm DNA methylation epimutations following ancestral DDT exposure. Dev Biol 2019;445:280–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wilton PR, Sloan DB, Logsdon JM Jr, Doddapaneni H, Neiman M.. Characterization of transcriptomes from sexual and asexual lineages of a New Zealand snail (Potamopyrgus antipodarum). Mol Ecol Resour 2013;13:289–94. [DOI] [PubMed] [Google Scholar]
- 22. Kaspi A, Ziemann M, Keating ST, Khurana I, Connor T, Spolding B, Cooper A, Lazarus R, Walder K, Zimmet P, El-Osta A.. Non-referenced genome assembly from epigenomic short-read data. Epigenetics 2014;9:1329–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Skinner MK, Guerrero-Bosagna C.. Role of CpG deserts in the epigenetic transgenerational inheritance of differential DNA methylation regions. BMC Genomics 2014;15:692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmitz RJ, Schultz MD, Lewsey MG, O'Malley RC, Urich MA, Libiger O, Schork NJ, Ecker JR.. Transgenerational epigenetic instability is a source of novel methylation variants. Science 2011;334:369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Skinner MK. Environmental epigenetics and a unified theory of the molecular aspects of evolution: a neo-Lamarckian concept that facilitates neo-Darwinian evolution. Genome Biol Evol 2015;7:1296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wilschut RA, Oplaat C, Snoek LB, Kirschner J, Verhoeven KJ.. Natural epigenetic variation contributes to heritable flowering divergence in a widespread asexual dandelion lineage. Mol Ecol 2016;25:1759–68. [DOI] [PubMed] [Google Scholar]
- 27. Nilsson E, Sadler-Riggleman I, Skinner MK.. Environmentally induced epigenetic transgenerational inheritance of disease. Environ Epigenet 2018;4:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Skinner M, Guerrero-Bosagna C, Haque MM, Nilsson E, Bhandari R, McCarrey J.. Environmentally induced transgenerational epigenetic reprogramming of primordial germ cells and subsequent germline. PLoS One 2013;8:1–15, e66318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jackman SD, Vandervalk BP, Mohamadi H, Chu J, Yeo S, Hammond SA, Jahesh G, Khan H, Coombe L, Warren RL, Birol I.. ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res 2017;27:768–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I.. ABySS: a parallel assembler for short read sequence data. Genome Res 2009;19:1117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pagès HH, Aboyoun P, Gentleman R, DebRoy S.. Biostrings: Efficient Manipulation of Biological Strings. R Package Version 1.2-1, 2019.
- 33. Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/, 2010 (October 2019, date last accessed).
- 34. Langmead B, Salzberg SL.. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R.. Genome project data processing S. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lienhard M, Grimm C, Morkel M, Herwig R, Chavez L.. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 2014;30:284–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robinson MD, McCarthy DJ, Smyth GK.. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.