Abstract
Background
MicroRNAs (miRNAs) are a class of 21–24 nucleotide endogenous non-coding small RNAs that play important roles in plant development and defense responses to biotic and abiotic stresses. Tobacco curly shoot virus (TbCSV) is a monopartite begomovirus, cause leaf curling and plant stunting symptoms in many Solanaceae plants. The betasatellite of TbCSV (TbCSB) induces more severe symptoms and enhances virus accumulation when co-infect the plants with TbCSV.
Methods
In this study, miRNAs regulated by TbCSV and TbCSB co-infection in Nicotiana benthamiana were characterized using high-throughput sequencing technology.
Results
Small RNA sequencing analysis revealed that a total of 13 known miRNAs and 42 novel miRNAs were differentially expressed in TbCSV and TbCSB co-infected N. benthamiana plants. Several potential miRNA-targeted genes were identified through data mining and were involved in both catalytic and metabolic processes, in addition to plant defense mechanisms against virus infections according to Gene Ontology (GO) analyses. In addition, the expressions of several differentially expressed miRNAs and their miRNA-targeted gene were validated through quantitative real time polymerase chain reaction (qRT-PCR) approach.
Conclusions
A large number of miRNAs are identified, and their target genes, functional annotations also have been explored. Our results provide the information on N. benthamiana miRNAs and would be useful to further understand miRNA regulatory mechanisms after TbCSV and TbCSB co-infection.
Keywords: MicroRNA, Tobacco curly shoot virus, Betasatellite, Nicotiana benthamiana, Small RNA-sequencing
Background
MicroRNAs (miRNAs) are a large group of small, non-coding RNAs that are 21–24 nucleotides (nt) long and regulate gene expression at the post-transcriptional level [1, 2]. In plants, miRNA is initially produced by RNA polymerase II (Pol II) or III (Pol III) and is known as the primary miRNA (pri-miRNA). Then the pri-miRNA is processed by a Dicer-like (DCL) enzyme, which produces an miRNA precursor (pre-miRNA), which contains both a miRNA:miRNA* intermediate duplex and a stem-loop-like structure [3]. After excision, the mature miRNA is bound to an argonaute (AGO) protein to form a RNA-induced silencing complex (RISC) [4]. Finally, the RISC complex recognizes and binds with the target RNA transcript, in a sequence-specific manner, and then cleaves the transcript or represses the translation process [2].
Since the first report of plant miRNA in Arabidopsis thaliana [5, 6], numerous studies have shown that plant miRNAs play critical roles in regulating plant development [7, 8], gene translational repression [9], flowering and sex determination [10], phytohormone signaling [11], and plant responses to abiotic and/or biotic stresses [12] such as drought stress [13] and infections with viruses or other pathogens [14–16].
In addition, many studies have indicated that miRNAs are highly conserved in eukaryotes, and virus infections in plants often alter miRNA expressions. For example, tobacco mosaic virus (TMV) infection in Nicotiana tabacum changes the expressions of several specific miRNAs [17]. Infection of cotton plants with cotton leaf roll dwarf virus (CLRDV) affects miRNA expression, downregulating the expression of specific miRNAs and causing disease symptoms in cotton leaves [18]. In A. thaliana and N. benthamiana plants, the expressions of miR168 and the Argonaute 1 gene are upregulated after infection with several plant viruses [19]. The expression of miR159 is upregulated by cucumber green mottle mosaic virus (CGMMV) infection cucumber at 10 post-inoculation [20]. In rice, downregulation of osa-miR171b expression in plants infected with rice stripe virus (RSV) enhances RSV disease symptoms [21]. The expression of miR319 in rice is increased after infection with rice ragged stunt virus (RRSV), and upregulation of miR319 promotes RRSV infection and disease symptoms by inhibiting JA-mediated host defense [22]. Amin et al. (2011) reported that begomovirus infection upregulates the accumulation of miRNAs controlling plant development [23].
Geminiviridae has nine genera: Mastrevirus, Curtovirus, Begomovirus, Topocuvirus, Becurtovirus, Turncurtovirus, Eragrovirus, Capulavir, and Euphorbia [24]. The family Geminiviridae currently consists of 468 species, in which Begomovirus is the largest genus (https://talk.ictvonline.org/taxonomy/). Begomovirus has about 409 species or members that cause severe damage to economically important food crops [25–27]. TbCSV was initially identified in tobacco plants and is a monopartite begomovirus [28]. Monopartite begomoviruses often co-infect their host plants with specific betasatellite DNA [29]. Betasatellite DNA genomes are typically about half the size of begomovirus DNA genomes, and play essential roles in the symptoms induction by monopartite begomoviruses [30, 31]. Host plants infected with TbCSV often display clear disease symptoms [32, 33]. When host plants are co-infected with TbCSV and TbCSB (TbCSV/TbCSB), they often show enhanced disease symptoms [33, 34].
In this study, a miRNA-sequencing approach was used to identify N. benthamiana miRNAs regulated by TbCSV/TbCSB co-infection. Then we predicted the target genes of the differentially expressed miRNAs. To investigate the interactions between the differentially expressed miRNAs and their target genes during TbCSV/TbCSB co-infection in N. benthamiana, the expression levels of several identified miRNAs together with their target genes were analyzed using qRT-PCR. The results not only shed light on the possible roles of miRNAs in N. benthamiana development and physiology, but also their possible roles in N. benthamiana resistance to TbCSV/TbCSB co-infection.
Methods
Plant growth and virus inoculation
N. benthamiana plants were grown inside a greenhouse at 24 °C and a 16/8 h (light/dark) photoperiod. Infectious clones of TbCSV isolate Y35 and its betasatellite (TbCSB) were individually introduced to Agrobacterium tumefaciens strain EHA105 [33]. After overnight culturing and centrifugation, agrobacterium pellets were resuspended in infiltration buffer (10 mM MES, 10 mM MgCl2 and 200 μM acetosyringone in sterile water) until they reached an OD600 value of 2, followed by 3 h incubation at room temperature. The two agrobacterium cultures were mixed 1:1 (v/v) and then co-infiltrated into the leaves of N. benthamiana plants at the six to eight leaf stage as described previously [35]. Plants infiltrated with the infiltration buffer only were used as non-infected controls.
Sample preparation and total RNA isolation
At 20 days-post co-infiltration (dpi), systemic leaves of three TbCSV/TbCSB-infected and three non-infected control plants were harvested. Total RNA was isolated from leaf samples using the TRIzol reagent, following the manufacturer’s instructions (Invitrogen, Waltham, MA, USA).
Small RNA library construction and Illumina sequencing
The integrity and concentration of RNA samples were checked using a NanoDrop spectrophotometer and an Agilent 2100 Bioanalyzer, following the manufacturer’s instructions (Agilent, Santa Clara, CA, USA). High-quality samples were identified and used to construct small RNA libraries followed by Illumina sequencing using an IlluminaHiseq™2500 instrument. Small RNA libraries and Illumina sequencing were performed by the Novogene Bioinformatics Technology Company, Beijing, China.
Analyses of Illumina sequencing date
Raw reads were processed to remove both adaptors and low-quality reads. Both the commercial GeneChip® Tomato Genome Array and the miRBase (http://www.mirbase.org/) were searched to identify known miRNAs in the two small RNA libraries (i.e., TbCSV/TbCSB and mock). The reads matched to the N. benthamiana genome shotgun-sequence assemblies were kept for further identifications.
Prediction of miRNA-targeted genes and gene function analyses
Potential miRNA-targeted genes were predicted using the psRNATarget program (http://plantgrn.noble.org/psRNATarget). The rules used for the predictions were as described by Schwab et al. [36]. To explore the possible functions of the predicted target genes, Gene Ontology (GO) analyses were performed as described previously [37]. The results were split into three different categories: Biological process, Molecular functions and Cellular components. Through this process, all potential target genes were mapped to GO terms described in the database (http://www.geneontology.org).
Validations of miRNA and target gene expression
qRT-PCR was performed to validate the expressions of miRNAs together with their target genes. Total RNA was isolated at 20 dpi from N. benthamiana leaves either infected with TbCSV/TbCSB or not, using TRIzol reagent. For miRNAs, the cDNA was synthesized by RT using the PrimeScript RT reagent Kit with gDNA Eraser with a special stem-loop RT primer according to the manufacturer’s instructions (TaKaRa, Dalian, China). The cDNA products were used as templates for real-time PCR analyses. The primers used for qRT-PCR assays are listed in (Additional file 1 Table S1). In addition, we also used qRT-PCR to analyze the expression of target genes, and the primers are also listed in Table S1. Purified total RNA was used as a template and reverse-transcribed using the PrimeScript RT reagent Kit (TaKaRa, Dalian, China) to obtain cDNA. The reactions consisted of incubation in 96-well plates at 98 °C for 2 min, followed by 40 cycles of 98 °C for 10 s, 60 °C for 10 s, and 68 °C for 30 s. The expression level of the N. benthamiana Ubiquitin C gene (UBC) was used as the reference [38, 39]. Three biological samples were used for each treatment and each biological sample had a further three technical replicates during qPCR. The qPCR results were calculated using the 2-ΔΔCT method [38].
Results
Small RNA library construction
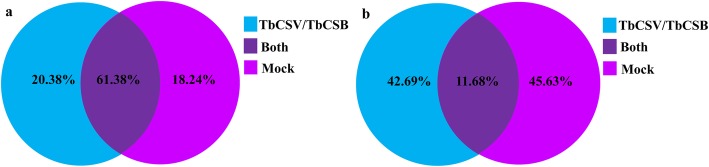
Small RNA libraries were constructed using total RNA from N. benthamiana leaves either infected with TbCSV/TbCSB or not. A total of 16,753,586 and 17,822,708 raw reads were generated via Illumina sequencing to represent these samples. After removing raw reads without 3′ adapters, with 5′ added adapters, with more than 10% unidentified nucleotides (nt), with poly (A/T/G/C) stretches, and that were less than 18 nt or more than 30 nt, a total of 13,896,237 and 15,227,477 clean reads were obtained from infected and non-infected libraries, respectively (Table 1). Of these, 5,936,641 (20.38%) were only in the infected library and 5,311,979 (18.24%) were only in the non-infected library (Fig. 1a). As shown in Fig. 1b, 10,438,652 unique reads were revealed, including 1,219,644 common reads and 44,562,19 and 47,627,89 specific reads for TbCSV/TbCSB and mock, respectively.
Table 1.
Summary of read data produced by small RNA sequencing
| sRNA | TbCSV/TbCSB | Mock |
|---|---|---|
| Total reads | 16,753,586 | 17,822,708 |
| Low quality reads | 14,843 | 16,495 |
| 3′ adapter null or insert null reads | 252,259 | 312,162 |
| 5′ adapter contaminants reads | 3772 | 9622 |
| N% > 10% | 1274 | 1347 |
| With ployA/T/G/C reads | 58,177 | 63,927 |
| < 18 nt, > 30 nt | 2,527,024 | 2,191,678 |
| Clean reads | 13,896,237 | 15,227,477 |
| sRNAs mapping to genome | 10,080,639 | 12,671,385 |
| Unique reads mapping to genome | 3,745,058 | 4,241,949 |
Fig. 1.
Percentages of the common and specific sequences of the clean reads (a) and unique sRNAs (b) from the TbCSV/TbCSB-infected and non-infected (Mock) control libraries
The resulting clean small RNA reads belonged to different categories, including exon sense and antisense, intron sense and antisense, rRNA, tRNA, snRNA, snoRNA, RNA repeats, natural antisense transcripts (NAT), trans-acting siRNA (TAS), and other unannotated reads. Among these, there were 404,032 (4.01%) miRNA tags from the infected library and 408,746 (3.22%) from the non-infected library (Table 2).
Table 2.
Distribution of small RNA sequences among the two constructed libraries
| Mock | TbCSV/TbCSB | |||
|---|---|---|---|---|
| Total sRNA reads (percent %) | Unique sRNA reads (percent %) | Total sRNA reads (percent %) | Unique sRNA reads (percent %) | |
| Total | 12,671,385 (100%) | 4,241,949 (100%) | 10,080,639 (100%) | 3,745,058 (100%) |
| Exon sense | 470,589 (3.71%) | 170,880 (4.03%) | 369,364 (3.66%) | 119,349 (3.19%) |
| Exon antisense | 567,077 (4.48%) | 44,166 (1.04%) | 387,710 (3.85%) | 44,418 (1.19%) |
| Intron sense | 1,057,103 (8.34%) | 363,908 (8.58%) | 975,273 (9.67%) | 349,541 (9.33%) |
| Intron antisense | 675,481 (5.33%) | 198,623 (4.68%) | 505,824 (5.02%) | 179,369 (4.79%) |
| miRNA | 408,746 (3.22%) | 1501 (0.04%) | 404,032 (4.01%) | 1488 (0.04%) |
| rRNA | 638,241 (5.04%) | 31,900 (0.75%) | 443,250 (4.4%) | 27,223 (0.73%) |
| tRNA | 1 (0%) | 1 (0%) | 1 (0%) | 1 (0%) |
| snRNA | 3576 (0.03%) | 841 (0.02%) | 3584 (0.04%) | 1054 (0.03%) |
| snoRNA | 26,219 (0.21%) | 1688 (0.04%) | 17,338 (0.17%) | 1678 (0.04%) |
| Repeat | 3,991,675 (31.5%) | 801,555 (18.9%) | 2,985,768 (29.62%) | 741,474 (19.8%) |
| NAT | 297,399 (2.35%) | 9280 (0.22%) | 186,878 (1.85%) | 7343 (0.2%) |
| TAS | 15,340 (0.12%) | 688 (0.02%) | 15,305 (0.15%) | 704 (0.02%) |
| Unannotated | 4,519,938 (35.67%) | 2,616,918 (61.69%) | 3,786,312 (37.56%) | 2,271,416 (60.65%) |
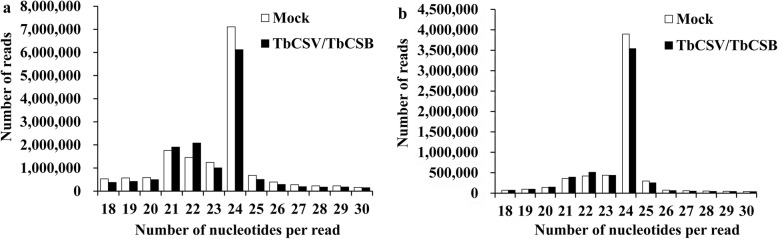
When all of the reads (18 to 30 nt) were analyzed, those with 24 nt were the most abundant. Of these, 6,119,284 (44.04%) were found in the infected library and 7,109,221 (46.69%) were found in the non-infected library. The second most abundant reads (1,760,916 reads or 11.56%) in the non-infected library had 21 nt, and that (2,079,345 reads or 14.96%) in the infected library had 22 nt (Fig. 2 a). When only unique reads were considered, 24 nt reads were the most abundant class with a total of 3,538,301 reads (62.34%) in the infected library and 3,896,777 reads (65.14%) in the non-infected library. The second most abundant unique read (510,867 reads or 9.0%) in the infected library had 22 nt while that (438,923 reads or 7.34%) in the non-infected library had 23 nt (Fig. 2b).
Fig. 2.
Numbers of total miRNA reads (a) and unique reads (b) with specific numbers of nucleotides from the TbCSV/TbCSB-infected and non-infected (Mock) N. benthamiana libraries
Identification of known miRNAs
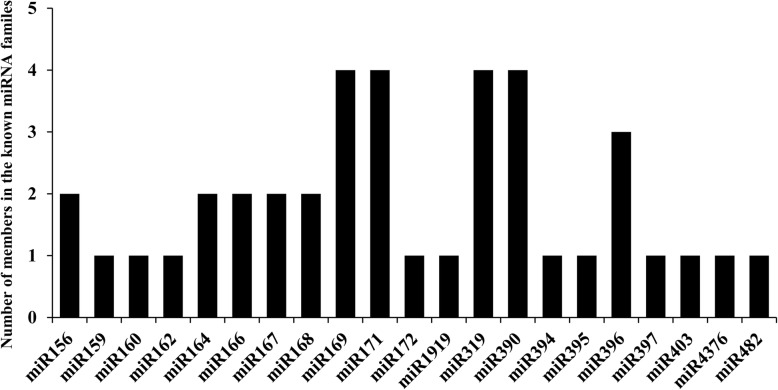
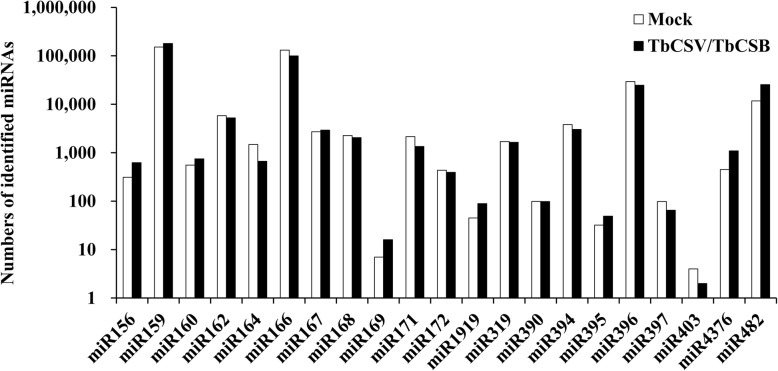
To identify known miRNAs in the two libraries, all small RNA reads were used to blast search the miRBase site with known mature plant miRNAs and then the N. benthamiana genome database. Through this approach, a total of 349,937 small RNA reads from the infected library and 346,265 reads from the non-infected library were mapped to the Solanum lycopersicum genome. Excluding miRNAs expressed at extremely low levels, 40 miRNAs in 21 known miRNA families were identified. The number of small RNA reads matched to known miRNA families are summarized in Figs. 3 and 4. The expression levels of miRNAs changed slightly after infection, as shown in Table 3. There were 13 differentially expressed miRNAs within the two libraries, with 2 downregulated miRNAs and 11 upregulated miRNAs after TbCSV/TbCSB co-infection.
Fig. 3.
Numbers of miRNA members identified in each conserved miRNA family in the small RNA libraries
Fig. 4.
Numbers of identified miRNAs in each conserved miRNA family from the TbCSV/TbCSB-infected and non-infected (Mock) libraries
Table 3.
miRNAs expressed differentially between the TbCSV/TbCSB and Mock
| miRNA | TbCSV/TbCSB | Mock | Fold-change | P-value | Mode | Sig-lable | ||
|---|---|---|---|---|---|---|---|---|
| name | Counts | Normalized | Counts | Normalized | log2(TbCSV/TbCSB/ Mock) | |||
| novel-10 | 887 | 2212.16 | 633 | 1561.89 | 0.61 | 2.29E-26 | up | ** |
| novel-103 | 75 | 187.05 | 55 | 135.71 | 0.57 | 0.0042 | up | ** |
| novel-104 | 27 | 67.34 | 44 | 108.57 | −0.59 | 0.0018 | down | ** |
| novel-105 | 17 | 42.40 | 64 | 157.92 | −1.79 | 3.96E-17 | down | ** |
| novel-112 | 2161 | 5389.50 | 1656 | 4086.07 | 0.50 | 3.20E-41 | up | ** |
| novel-113 | 9 | 22.45 | 24 | 59.22 | −1.30 | 3.42E-05 | down | ** |
| novel-115 | 51 | 127.19 | 118 | 291.16 | −1.09 | 4.40E-16 | down | ** |
| novel-121 | 56 | 139.66 | 21 | 51.82 | 1.53 | 9.87E-11 | up | ** |
| novel-127 | 118 | 294.29 | 249 | 614.39 | −0.96 | 6.72E-27 | down | ** |
| novel-128 | 8 | 19.95 | 15 | 37.01 | − 0.79 | 0.0227 | down | * |
| novel-129 | 214 | 533.71 | 124 | 305.96 | 0.91 | 2.51E-15 | up | ** |
| novel-131 | 32 | 79.81 | 0 | 0.00 | 7.37 | 1.64E-19 | up | ** |
| novel-133 | 43 | 107.24 | 90 | 222.07 | −0.95 | 1.59E-10 | down | ** |
| novel-137 | 23 | 57.36 | 46 | 113.50 | −0.88 | 1.45E-05 | down | ** |
| novel-138 | 39 | 97.27 | 27 | 66.62 | 0.65 | 0.0163 | up | * |
| novel-140 | 8 | 19.95 | 2 | 4.93 | 2.12 | 0.0019 | up | ** |
| novel-142 | 4 | 9.98 | 9 | 22.21 | −1.05 | 0.0290 | down | * |
| novel-15 | 723 | 1803.15 | 553 | 1364.49 | 0.51 | 5.49E-15 | up | ** |
| novel-16 | 348 | 867.91 | 745 | 1838.24 | −0.98 | 1.73E-79 | down | ** |
| novel-18 | 320 | 798.07 | 218 | 537.90 | 0.67 | 8.97E-13 | up | ** |
| novel-21 | 365 | 910.30 | 265 | 653.87 | 0.58 | 7.87E-11 | up | ** |
| novel-31 | 244 | 608.53 | 176 | 434.27 | 0.59 | 6.23E-08 | up | ** |
| novel-32 | 113 | 281.82 | 239 | 589.72 | −0.96 | 5.22E-26 | down | ** |
| novel-42 | 135 | 336.69 | 103 | 254.15 | 0.51 | 0.00067 | up | ** |
| novel-5 | 3355 | 8367.31 | 6145 | 15,162.39 | −0.75 | 0 | down | ** |
| novel-50 | 104 | 259.37 | 67 | 165.32 | 0.75 | 4.54E-06 | up | ** |
| novel-52 | 64 | 159.61 | 115 | 283.76 | −0.73 | 2.89E-09 | down | ** |
| novel-6 | 3704 | 9237.71 | 1040 | 2566.13 | 1.95 | 0 | up | ** |
| novel-60 | 19 | 47.39 | 34 | 83.89 | −0.72 | 0.0013 | down | ** |
| novel-61 | 56 | 139.66 | 40 | 98.70 | 0.60 | 0.0078 | up | ** |
| novel-66 | 55 | 137.17 | 87 | 214.67 | −0.54 | 3.32E-05 | down | ** |
| novel-69 | 6 | 14.96 | 39 | 96.23 | −2.58 | 5.08E-16 | down | ** |
| novel-70 | 11 | 27.43 | 45 | 111.03 | −1.91 | 2.03E-13 | down | ** |
| novel-71 | 45 | 112.23 | 16 | 39.48 | 1.61 | 1.64E-09 | up | ** |
| novel-75 | 21 | 52.37 | 38 | 93.76 | −0.74 | 0.0006 | down | ** |
| novel-78 | 11 | 27.43 | 23 | 56.75 | −0.95 | 0.0012 | down | ** |
| novel-82 | 14 | 34.92 | 8 | 19.74 | 0.93 | 0.0387 | up | * |
| novel-9 | 687 | 1713.37 | 1732 | 4273.60 | −1.22 | 1.30E-248 | down | ** |
| novel-91 | 29 | 72.33 | 14 | 34.54 | 1.17 | 0.0002 | up | ** |
| novel-92 | 45 | 112.23 | 82 | 202.33 | −0.75 | 3.09E-07 | down | ** |
| novel-94 | 7 | 17.46 | 29 | 71.56 | −1.93 | 2.99E-09 | down | ** |
| novel-96 | 1 | 2.49 | 4 | 9.87 | −1.88 | 0.0302 | down | * |
| miR156a | 224 | 558.65 | 101 | 249.21 | 1.27 | 2.70E-28 | up | ** |
| miR156d-5p | 401 | 1000.09 | 210 | 518.16 | 1.05 | 9.17E-36 | up | ** |
| miR160a | 747 | 1863.01 | 554 | 1366.96 | 0.55 | 2.04E-18 | up | ** |
| miR164a-5p | 655 | 1633.56 | 1469 | 3624.66 | −1.05 | 1.32E-170 | down | ** |
| miR168b-3p | 670 | 1670.97 | 477 | 1176.97 | 0.61 | 1.55E-20 | up | ** |
| miR169a | 2 | 4.99 | 0 | 0.00 | 3.37 | 0.0423 | up | * |
| miR169c | 13 | 32.42 | 6 | 14.80 | 1.23 | 0.0094 | up | ** |
| miR171b | 443 | 1104.83 | 1070 | 2640.16 | −1.15 | 2.75E-143 | down | ** |
| miR1919c-5p | 89 | 221.96 | 45 | 111.03 | 1.10 | 8.28E-10 | up | ** |
| miR390a-3p | 2 | 4.99 | 0 | 0.00 | 3.37 | 0.0423 | up | * |
| miR395a | 49 | 122.21 | 32 | 78.96 | 0.73 | 0.0022 | up | ** |
| miR4376 | 1091 | 2720.94 | 451 | 1112.81 | 1.39 | 1.24E-153 | up | ** |
| miR482a | 25,539 | 63,693.84 | 11,753 | 28,999.78 | 1.24 | 0 | up | ** |
* represnt Fold-change (log2 TbCSV/TbCSB/Mock) > 1.0 or Fold-change (log2 TbCSV/TbCSB/Mock) < −1.0, and 0.01 < =P -values < 0.05; ** represnt Fold-change (log2 TbCSV/TbCSB/Mock) > 1.0 or Fold-change (log2 TbCSV/TbCSB/Mock) < − 1.0, and P -values < 0.01
Identification of novel candidate miRNAs
To predict hairpin-like structures in the identified miRNA precursors and identify the corresponding miRNAs that could be used to further identify novel miRNAs, we utilized the miREvo and miRDeep2 software as described [40, 41]. We also used the mfold web server (http://unafold.rna.albany.edu) to predict the secondary structures and the minimum free energy of the annotated small RNA tags that were mapped to the N. benthamiana genome, as described previously [42]. After removing miRNAs with extremely low expressions, a total of 42 miRNAs were found to show differential expression between the two libraries. The novel-6 miRNA showed the highest abundance and had 9238 transcripts per million (TPM) followed by the novel-5 miRNA (8367 TPM) in the infected library. In the non-infected control library, the novel-5 miRNA showed the highest abundance (15,162 TPM) followed by the novel-9 miRNA (4273 TPM) (Table 3, Additional file 2 Figure S1). Among the predicted novel miRNA candidates, the base bias in the first position showed that the majority of these novel miRNA candidates started with a 5′ uridine (U) as shown in Fig. S2a and Fig. S2b. Furthermore, novel miRNA candidate nucleotide bias at each position were also analyzed (Additional file 3 Figure S2c and Figure S2d). In addition, we also found among the 42 differentially-expressed novel miRNAs, twenty-five had complementary miRNA*s, with precursor lengths ranging from 43 to 295 nt and predicted minimal folding energy (MFE) ranging from − 14.2 to − 130.7 kcal/mol. Also, 17 differentially-expressed novel miRNAs without miRNA*s detected were identified as candidate miRNAs (Additional file 4 Table S2). Fifteen out of 42 new miRNAs were 21 nt in length, while three, one, ten and thirteen miRNA had lengths of 19, 20, 22, and 24 nt, respectively (Additional file 4 Table S2). All of the novel miRNAs’ loci, pre-miRNA sequences and structures, and reads from deep sequencing were shown in Additional file 4. This is in agreement with published criteria for novel miRNA [43, 44], and suggests that these candidate miRNAs are most likely to be new miRNA family members in N. benthamiana.
Prediction of potential miRNA-targeted genes
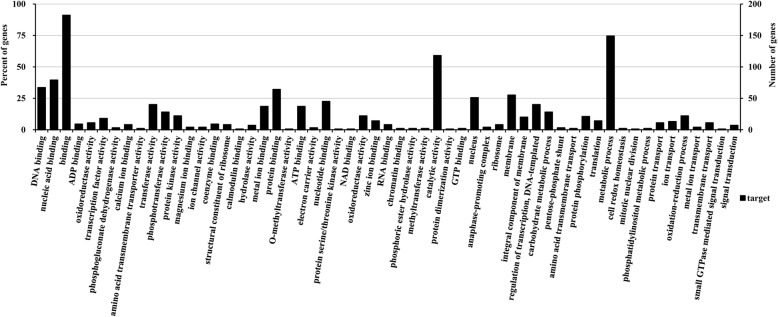
Numerous genes are responsive to virus infections and to differentially expressed miRNAs. Our results indicate that many potential miRNA-targeted genes encode transcription or non-transcription factor proteins, which are important for physiological processes. To explore the regulatory functions of the identified miRNAs in the infected library, potential target genes of nine conserved and four novel miRNAs were predicted by GO analyses. The GO annotated terms Biological process, Cellular components, and Molecular function were further analyzed to determine genes that could potentially be targeted by the identified miRNAs (Fig. 5). For the Biological process category, genes involved in metabolic processes (GO: 0008152) were the most represented GO terms. Membrane (GO: 0016020) and nucleus (GO: 0005634) were the major GO terms within the Cellular component category. For the Molecular function category, the major GO terms were binding (GO: 0005488) and catalytic activity (GO: 0003824). Many of the identified miRNA-targeted genes have previously been reported to play a role in defense against pathogens.
Fig. 5.
Gene ontology (GO) analysis using the predicted target genes regulated by the differentially expressed miRNAs
Validations of miRNA and target gene expressions by qRT-PCR
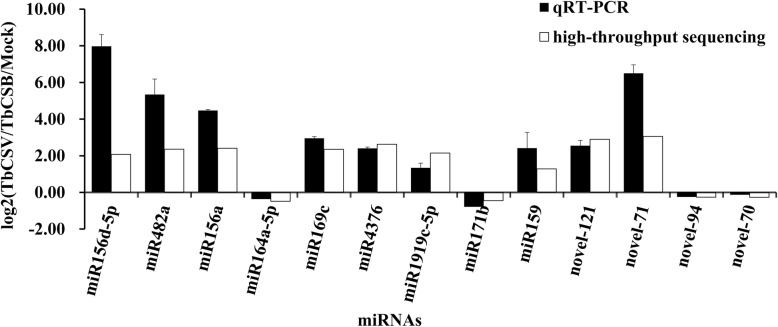
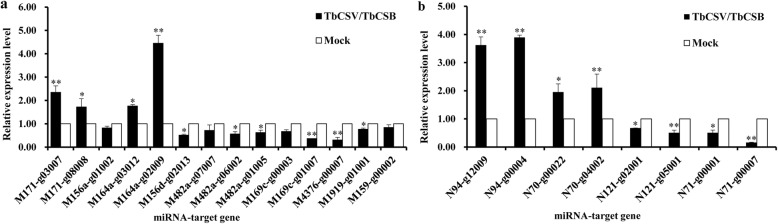
To validate the high-throughput sequencing results, nine known and four novel miRNAs that showed differential expression between the two libraries were selected and analyzed for expression by stem-loop qRT-PCR (Table 4 and Fig. 6). The PCR primers are listed in Table S1. The expressions of miR156d-5p, miR169c, miR4376, miR156a, miR1919c-5p, miR159, novel-121, novel-71, and miR482a in the infected library were all upregulated, whereas the expressions of miR171b, miR164a-5p, novel-94, and novel-70 were all downregulated. To examine if the expressions of TbCSV/TbCSB infection-regulated miRNAs could influence the expressions of their target genes, we analyzed the predicted target genes through qRT-PCR. The expressions of squamosa promoter-binding-like protein (targeted by miR156a), disease resistance protein (targeted by miR482a), nuclear transcription factor Y subunit (targeted by miR169c), calcium-transporting ATPase (targeted by miR4376), conserved hypothetical protein (targeted by miR4376), MYB-like transcription factor (targeted by miR159), heavy metal transport (targeted by novel-121), and N-acetyltransferase (targeted by novel-71) were all downregulated after TbCSV/TbCSB infection. By contrast, GRAS family transcription factor (targeted by miR171), NAC domain-containing protein (targeted by miR164a), transcription factor (targeted by novel-70), and 1-aminocyclopropane-1-carboxylate oxidase (targeted by novel-94) were all upregulated after TbCSV/TbCSB co-infection (Table 4 and Fig. 7).
Table 4.
Expression of differentially expressed miRNAs and targeted mRNA genes analyzed
| ID miRNA | log2(TbCSV/TbCSB / Mock) | qRT-PCR | ID target gene | qRT-PCR | Functional annotation |
|---|---|---|---|---|---|
| miR156a | 1.27(up) | 4.47(up) | Niben101Scf19266g01002 | 0.82(down) | Squamosa promoter-binding protein |
| miR156d-5p | 1.05(up) | 7.97(up) | Niben101Scf10743g02013 | 0.51(down) | Squamosa promoter-binding-like protein |
| miR164a-5p | −1.05(down) | −0.36(down) | Niben101Scf02318g03012 | 1.76(up) | NAC domain-containing protein |
| Niben101Scf04745g02009 | 4.45(up) | NAC domain-containing protein | |||
| miR169c | 1.23(up) | 2.95(up) | Niben101Scf15723g00003 | 0.67(down) | YA2 |
| Niben101Scf10191g01007 | 0.36(down) | Nuclear transcription factor Y subunit A | |||
| miR171b | −1.15 (down) | −0.79(down) | Niben101Scf03072g03007 | 2.35(up) | GRAS family transcription factor |
| Niben101Scf03693g08008 | 1.72(up) | GRAS family transcription factor | |||
| miR1919c-5p | 1.10(up) | 1.34(up) | Niben101Scf02655g01001 | 0.76(down) | conserved hypothetical protein |
| miR4376 | 1.39(up) | 2.41(up) | Niben101Scf04808g00007 | 0.30(down) | calcium-transporting ATPase |
| miR482a | 1.24 (up) | 5.34(up) | Niben101Scf01683g07007 | 0.71(down) | Cyclin-dependent kinase |
| Niben101Scf01052g06002 | 0.56(down) | Disease resistance protein (NB-ARC) | |||
| Niben101Scf01941g01005 | 0.62(down) | Disease resistance protein (NB-ARC) | |||
| miR159 | 0.36(up) | 2.42(up) | Niben101Scf11569g00002 | 0.84(down) | MYB-like transcription factor |
| novel 121 | 1.53(up) | 2.55(up) | Niben101Scf00504g02001 | 0.68(down) | Heavy metal transport |
| Niben101Scf02139g05001 | 0.51(down) | Heavy metal transport | |||
| novel 70 | −1.91(down) | −0.12(down) | Niben101Scf02825g00022 | 1.96(up) | fiber protein Fb11 |
| Niben101Scf00578g04002 | 2.11(up) | Transcription factor | |||
| novel 71 | 1.61 (up) | 6.50(up) | Niben101Scf12157g00001 | 0.51(down) | N-acetyltransferase 10 homolog |
| Niben101Scf03820g00007 | 0.16(down) | N-acetyltransferase 10 homolog | |||
| novel 94 | −1.93(down) | −0.25(down) | Niben101Scf19336g00004 | 3.90(up) | 1-aminocyclopropane-1-carboxylate oxidase |
| Niben101Scf04528g12009 | 3.62(up) | GRAS family transcription factor |
Fig. 6.
qRT-PCR and high-throughput sequencing analyses of the relative expressions of nine known and four new miRNAs. Total RNA was isolated from the TbCSV/TbCSB-infected or non-infected (Mock) N. benthamiana leaves. The x-axis shows the names of the miRNAs analyzed in this study. The y-axis shows the Log2ratio between the expression values from a TbCSV/TbCSB-infected sample versus its Mock sample. Three biological replicates were analyzed for each miRNA through qRT-PCR. Expression level of N. benthamiana UBC gene was used as the reference gene during qRT-PCR assays
Fig. 7.
Relative expressions of miRNA-targeted genes. Relative expression of each gene was determined through qRT-PCR. a Relative expressions of the known miRNA-targeted genes. b Relative expressions of the novel miRNA-targeted genes. The x-axis shows the names of the miRNA-targeted genes analyzed in this study. The y-axis shows the Log2ratio between the expression values from a TbCSV/TbCSB-infected sample versus its Mock sample. Three biological replicates were analyzed for each miRNA-targeted gene through qRT-PCR. Expression level of N. benthamiana UBC gene was used as the reference gene during qRT-PCR assays. Asterisks indicate statistically significant differences compared with Mock, “*” indicate significant difference (0.01 ≤ p < 0.05), “**” indicate extremely significant difference (P < 0.01)
Discussion
High-throughput sequencing technology has been used extensively in small RNA research [45]. More and more studies have illustrated that various virus infections in plants often alter miRNA expressions [17–20, 46]. A large number of miRNAs have been identified in plants and the functions of many miRNAs have also been investigated [47, 48]. To better understand the roles of the N. benthamiana miRNAs in host resistance to TbCSV/TbCSB co-infection, in this study, two libraries were constructed, using total RNA from N. benthamiana plants either infected with TbCSV/TbCSB or not.
The most abundant small RNA reads in the two libraries were those with 24 nt. In addition, the 24 nt small RNA reads were the predominant unique small RNA reads. This finding agrees with earlier reports which have shown that 24 nt small RNAs are more abundant in several other diseased plants, such as tomato plants infected with Phytophthora infestans, tomato infected with cucumber mosaic virus (CMV) and wheat plants infected with powdery mildew pathogen [13, 49, 50]. The length distribution of small RNA reads may reflect their compositions [51]. We found more 24 nt small RNAs in the non-infected library than in the infected library. By contrast, 21 and 22 nt miRNAs were more abundant in the infected library. Our results also indicate that the expression profiles of miRNAs were significantly altered after TbCSV/TbCSB co-infection in N. benthamiana, and the differentially regulated expressions of miRNAs suggested that miRNAs play important roles during TbCSV/TbCSB co-infection.
In all, 13 known and 42 potentially novel miRNAs were differentially regulated by TbCSV/TbCSB co-infection. To better understand the relative abundances of miRNAs in the two libraries, we analyzed sequence frequencies and used them as indexes. When the two libraries were compared, the normalized reads varied from about 2 (novel-96) to 63,693 (miR482a) in the infected library and from 0 (novel-131) to 28,999 (miR482a) in the non-infected control library, indicating significant variation in the relative abundances of different miRNA sequences. This finding was later confirmed through qRT-PCR analyses using several selected differentially expressed miRNAs.
miRNAs may regulate host defenses against pathogens, including viruses, by suppressing pathogen multiplication at the post-transcriptional level [13, 52]. Several stress-responsive miRNAs (e.g., miR168, miR169, and miR482) have been reported to target transcription factors controlling host resistance to virus infection [53–55]. In N. benthamiana, virus infection may regulate the expression of miR168 to alleviate the anti-viral function of AGO1 protein [53]. In our study, N. benthamiana miR168 was found to be responsive to TbCSV/TbCSB co-infection. In addition, the expression of miR169 was upregulated after co-infection. In a previous study, rice miR169 was found to negatively regulate rice immunity against Magnaporthe oryzae infection by differentially repressing its target genes [54]. Studies also showed that rice miR164 plays an important role in rice resistance to southern rice black-streaked dwarf virus (SRBSDV) infection as well as rice resistance to drought stresses by differentially regulating its target genes [56, 57]. In addition, miR482 can regulate the expression of NBS-LRR defense genes during fungal pathogen infection in cotton [55]. In our study, when the expressions of miR164a and miR482 in the two libraries were compared, the normalized miRNA164a and miR482 reads were 3624 and 28,999 in the non-infected control library, and 1633 and 63,693 in the infected library, respectively, suggesting that these two miRNAs may play roles in N. benthamiana resistance to TbCSV/TbCSB co-infection. The further studies will be continued to unravel the functions of these miRNAs.
Conclusion
In this study, miRNAs regulated by TbCSV and TbCSB co-infection in N. benthamiana were characterized using high-throughput sequencing technology, and some miRNAs involved in plant defense system were found to be significantly regulated after TbCSV and TbCSB infection. The molecular functions of these miRNAs in N. benthamiana resistance to TbCSV/TbCSB co-infection may require further investigation. Nonetheless, our results improve knowledge of the infection of TbCSV/TbCSB in host plants while also providing additional information for the development of management strategies for TbCSV/TbCSB infection in the future.
Supplementary information
Additional file 1: Table S1. Primers used for qRT-PCR.
Additional file 2: Figure S1. The precursors of 42 novel microRNAs and their hairpin structures in N. benthamiana.
Additional file 3: Figure S2. First nucleotide bias and nucleotide bias analysis.
Additional file 4: Table S2. 42differentially-expressed novel miRNAs and 25 equally-expressed novel miRNAs with miRNAs in N. benthamiana.
Acknowledgements
We are grateful to Professor Xueping Zhou in the Biotechnology Institute of Zhejiang University in China for providing the infectious clones of TbCSV isolate Y35 and its betsatellite (TbCSB), and to professor Xinshun Ding in the Samuel Roberts Noble Foundation for helping to revise this manuscript.
Abbreviations
- miRNA
MicroRNA
- nt
Nucleotide
- qRT-PCR
Quantitative real time polymerase chain reaction
- TbCSB
Tobacco curly shoot betasatellite
- TbCSV
Tobacco curly shoot virus
- TPM
Transcripts per million reads
Authors’ contribution
LQ conceived and designed the study and revised the paper. JD, GW, ML, ZZ and MS performed the experiments and made analysis of the data. KJ and JZ participated preparation including discussion and editing. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Founding of China (Grant No. 31772127), the Fundamental Research Funds for the Central Universities (XDJK2017A006 and XDJK2017D164), the China Postdoctoral Science Foundation (2015 M572431), and the Chongqing Postdoctoral Science Special Foundation (Xm2015120).
Availability of data and materials
All data and materials described in the manuscript are available in the Additional files 2,3 and 4.
Ethics approval and consent to participate
Not applicable.
Consent for publication
All the authors consent to publish.
Competing interests
The authors declare no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jiang Du, Email: dujiang20080803@163.com.
Gentu Wu, Email: wugtu@163.com.
Zhongpiao Zhou, Email: 997498645@qq.com.
Jiayuan Zhang, Email: 1095340861@qq.com.
Mingjun Li, Email: lmj603@163.com.
Miao Sun, Email: 445879215@qq.com.
Kairong Jiang, Email: 1938446949@qq.com.
Ling Qing, Phone: 0086-023-68251541, Email: qling@swu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12985-019-1234-5.
References
- 1.Obernosterer G, Leuschner PJF, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. Rna-a Publication of the Rna Society. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through dicer-like 1 protein functions. Proc Natl Acad Sci U S A. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci U S A. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park W, Li JJ, Song RT, Messing J, Chen XM. CARPEL FACTORY, a dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- 7.Yang TW, Xue LG, An LZ. Functional diversity of miRNA in plants. Plant Sci. 2007;172:423–432. [Google Scholar]
- 8.Lelandais-Briere C, Sorin C, Declerck M, Benslimane A, Crespi M, Hartmann C. Small RNA Diversity in plants and its impact in development. Current Genomics 2010;11:14–23. [DOI] [PMC free article] [PubMed]
- 9.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 10.Chuck G, Candela H, Hake S. Big impacts by small RNAs in plant development. Curr Opin Plant Biol. 2009;12:81–86. doi: 10.1016/j.pbi.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Liu Q, Chen YQ. Insights into the mechanism of plant development: interactions of miRNAs pathway with phytohormone response. Biochem Biophys Res Commun. 2009;384:1–5. doi: 10.1016/j.bbrc.2009.04.028. [DOI] [PubMed] [Google Scholar]
- 12.Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D. A collection of target mimics for comprehensive analysis of MicroRNA function in Arabidopsis thaliana. PLoS Genet. 2010;6. [DOI] [PMC free article] [PubMed]
- 13.Feng J, Liu S, Wang M, Lang Q, Jin C. Identification of microRNAs and their targets in tomato infected with Cucumber mosaic virus based on deep sequencing. Planta. 2014;240:1335–1352. doi: 10.1007/s00425-014-2158-3. [DOI] [PubMed] [Google Scholar]
- 14.Katiyar-Agarwal S, Jin HL. Role of Small RNAs in Host-Microbe Interactions. Annual Review of Phytopathology, Vol 48 2010;48:225–246. [DOI] [PMC free article] [PubMed]
- 15.Chen L, Ren Y, Zhang Y, Xu J, Zhang Z, Wang Y. Genome-wide profiling of novel and conserved Populus microRNAs involved in pathogen stress response by deep sequencing. Planta. 2012;235:873–883. doi: 10.1007/s00425-011-1548-z. [DOI] [PubMed] [Google Scholar]
- 16.Li HY, Dong YY, Yin HL, Wang N, Yang J, Liu XM, Wang YF, Wu JY, Li XK. Characterization of the stress associated microRNAs in Glycine max by deep sequencing. BMC Plant Biol. 2011;11. [DOI] [PMC free article] [PubMed]
- 17.Bazzini AA, Hopp HE, Beachy RN, Asurmendi S. Infection and coaccumulation of tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development. Proc Natl Acad Sci U S A. 2007;104:12157–12162. doi: 10.1073/pnas.0705114104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romanel E, Silva TF, Correa RL, Farinelli L, Hawkins JS, Schrago CE, Vaslin MF. Global alteration of microRNAs and transposon-derived small RNAs in cotton (Gossypium hirsutum) during Cotton leafroll dwarf polerovirus (CLRDV) infection. Plant Mol Biol. 2012;80:443–460. doi: 10.1007/s11103-012-9959-1. [DOI] [PubMed] [Google Scholar]
- 19.Varallyay E, Havelda Z. Unrelated viral suppressors of RNA silencing mediate the control of ARGONAUTE1 level. Mol Plant Pathol. 2013;14:567–575. doi: 10.1111/mpp.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu HW, Luo LX, Liang CQ, Jiang N, Liu PF, Li JQ. High-Throughput Sequencing Identifies Novel and Conserved Cucumber (Cucumis sativus L.) microRNAs in Response to Cucumber Green Mottle Mosaic Virus Infection. PLoS One 2015;10:e0129002. [DOI] [PMC free article] [PubMed]
- 21.Tong A, Yuan Q, Wang S, Peng J, Lu Y, Zheng H, Lin L, Chen H, Gong Y, Chen J, Yan F. Altered accumulation of Osa-miR171b contributes to rice stripe virus infection by regulating disease symptoms. J Exp Bot. 2017;68:4357–4367. doi: 10.1093/jxb/erx230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang C, Ding Z, Wu K, Yang L, Li Y, Yang Z, Shi S, Liu X, Zhao S, Yang Z, et al. Suppression of Jasmonic acid-mediated defense by viral-inducible MicroRNA319 facilitates virus infection in Rice. Mol Plant. 2016;9:1302–1314. doi: 10.1016/j.molp.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Amin I, Patil BL, Briddon RW, Mansoor S, Fauquet CM. A common set of developmental miRNAs are upregulated in Nicotiana benthamiana by diverse begomoviruses. Virol J. 2011;8. [DOI] [PMC free article] [PubMed]
- 24.Varsani A, Roumagnac P, Fuchs M, Navas-Castillo J, Moriones E, Idris A, Briddon RW, Rivera-Bustamante R, Zerbini FM, Martin DP. Capulavirus and Grablovirus: two new genera in the family Geminiviridae. Arch Virol. 2017;162:1819–1831. doi: 10.1007/s00705-017-3268-6. [DOI] [PubMed] [Google Scholar]
- 25.Fauquet CM, Bisaro DM, Briddon RW, Brown JK, Harrison BD, Rybicki EP, Stenger DC, Stanley J. Revision of taxonomic criteria for species demarcation in the family Geminiviridae, and an updated list of begomovirus species. Arch Virol. 2003;148:405–421. doi: 10.1007/s00705-002-0957-5. [DOI] [PubMed] [Google Scholar]
- 26.Fauquet CM, Briddon RW, Brown JK, Moriones E, Stanley J, Zerbini M, Zhou X. Geminivirus strain demarcation and nomenclature. Arch Virol. 2008;153:783–821. doi: 10.1007/s00705-008-0037-6. [DOI] [PubMed] [Google Scholar]
- 27.Yang X, Guo W, Li F, Sunter G, Zhou X. Geminivirus-associated Betasatellites: exploiting chinks in the antiviral arsenal of plants. Trends Plant Sci. 2019;24:519–529. doi: 10.1016/j.tplants.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Xie Y, Zhou XP, Zhang ZK, Qi YJ. Tobacco curly shoot virus isolated in Yunnan is a distinct species of Begomovirus. Chin Sci Bull. 2002;47:197–200. [Google Scholar]
- 29.Zhou X. Advances in understanding Begomovirus satellites. Annu Rev Phytopathol. 2013;51:357–381. doi: 10.1146/annurev-phyto-082712-102234. [DOI] [PubMed] [Google Scholar]
- 30.Cui X, Tao X, Xie Y, Fauquet CM, Zhou X. A DNAbeta associated with Tomato yellow leaf curl China virus is required for symptom induction. J Virol. 2004;78:13966–13974. doi: 10.1128/JVI.78.24.13966-13974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Briddon RW, Brown JK, Moriones E, Stanley J, Zerbini M, Zhou X, Fauquet CM. Recommendations for the classification and nomenclature of the DNA-beta satellites of begomoviruses. Arch Virol. 2008;153:763–781. doi: 10.1007/s00705-007-0013-6. [DOI] [PubMed] [Google Scholar]
- 32.Jiang T, Liu P, Liao BL, Wu JX, Huang CJ. Malvastrum yellow vein Yunnan virus is a monopartite begomovirus. Acta Virol. 2010;54:21–26. doi: 10.4149/av_2010_01_21. [DOI] [PubMed] [Google Scholar]
- 33.Li Z, Xie Y, Zhou X. Tobacco curly shoot virusDNAβ is not necessary for infection but intensifies symptoms in a host-dependent manner. Phytopathology. 2005;95:902–908. doi: 10.1094/PHYTO-95-0902. [DOI] [PubMed] [Google Scholar]
- 34.Li K, Wu G, Li M, Ma M, Du J, Sun M, Sun X, Qing L. Transcriptome analysis of Nicotiana benthamiana infected by Tobacco curly shoot virus. Virol J. 2018;15:138. doi: 10.1186/s12985-018-1044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou X, Xie Y, Tao X, Zhang Z, Li Z, Fauquet CM. Characterization of DNAbeta associated with begomoviruses in China and evidence for co-evolution with their cognate viral DNA-A. J Gen Virol. 2003;84:237–247. doi: 10.1099/vir.0.18608-0. [DOI] [PubMed] [Google Scholar]
- 36.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 37.Liu J, Zhang X, Zhang F, Hong N, Wang G, Wang A, Wang L. Identification and characterization of microRNAs from in vitro-grown pear shoots infected with Apple stem grooving virus in response to high temperature using small RNA sequencing. BMC Genomics. 2015;16:945. doi: 10.1186/s12864-015-2126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen CF, Ridzon DA, Broomer AJ, Zhou ZH, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33. [DOI] [PMC free article] [PubMed]
- 39.He QJ, Peng JJ, Yan F, Lin L, Lu YW, Zheng HY, Chen HR, Chen JP. Intron retention and 3 '-UTR analysis of Arabidopsis dicer-like 2 transcripts. Mol Biol Rep. 2012;39:3271–3280. doi: 10.1007/s11033-011-1095-5. [DOI] [PubMed] [Google Scholar]
- 40.Wen M, Shen Y, Shi SH, Tang T. miREvo: an integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. Bmc Bioinformatics. 2012:13. [DOI] [PMC free article] [PubMed]
- 41.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonnet E, Wuyts J, Rouze P, Van De Peer Y. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics. 2004;20:2911–2917. doi: 10.1093/bioinformatics/bth374. [DOI] [PubMed] [Google Scholar]
- 44.Krol J, Sobczak K, Wilczynska U, Drath M, Jasinska A, Kaczynska D, Krzyzosiak WJ. Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. J Biol Chem. 2004;279:42230–42239. doi: 10.1074/jbc.M404931200. [DOI] [PubMed] [Google Scholar]
- 45.Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu JK. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 2008;8:25. doi: 10.1186/1471-2229-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niu QW, Lin SS, Reyes JL, Chen KC, Wu HW, Yeh SD, Chua NH. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol. 2006;24:1420–1428. doi: 10.1038/nbt1255. [DOI] [PubMed] [Google Scholar]
- 47.Perez-Quintero AL, Neme R, Zapata A, Lopez C. Plant microRNAs and their role in defense against viruses: a bioinformatics approach. BMC Plant Biol. 2010;10:138. doi: 10.1186/1471-2229-10-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tenoever BR. RNA viruses and the host microRNA machinery. Nat Rev Microbiol. 2013;11:169–180. doi: 10.1038/nrmicro2971. [DOI] [PubMed] [Google Scholar]
- 49.Luan Y, Cui J, Zhai J, Li J, Han L, Meng J. High-throughput sequencing reveals differential expression of miRNAs in tomato inoculated with Phytophthora infestans. Planta. 2015;241:1405–1416. doi: 10.1007/s00425-015-2267-7. [DOI] [PubMed] [Google Scholar]
- 50.Xin MM, Wang Y, Yao YY, Xie CJ, Peng HR, Ni ZF, Sun QX. Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 2010;10. [DOI] [PMC free article] [PubMed]
- 51.Ding SW, Lu R. Virus-derived siRNAs and piRNAs in immunity and pathogenesis. Curr Opin Virol. 2011;1:533–544. doi: 10.1016/j.coviro.2011.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Al-Abdallat AM, Ali-Sheikh-Omar MA, Alnemer LM. Overexpression of two ATNAC3-related genes improves drought and salt tolerance in tomato (Solanum lycopersicum L.) Plant Cell, Tissue and Organ Culture (PCTOC) 2014;120:989–1001. [Google Scholar]
- 53.Varallyay E, Valoczi A, Agyi A, Burgyan J, Havelda Z. Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J. 2010;29:3507–3519. doi: 10.1038/emboj.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, Zhao SL, Li JL, Hu XH, Wang H, Cao XL, Xu YJ, Zhao ZX, Xiao ZY, Yang N, et al. Osa-miR169 negatively regulates Rice immunity against the blast fungus Magnaporthe oryzae. Front Plant Sci. 2017;8. [DOI] [PMC free article] [PubMed]
- 55.Zhu QH, Fan LJ, Liu Y, Xu H, Llewellyn D, Wilson I. miR482 Regulation of NBS-LRR Defense Genes during Fungal Pathogen Infection in Cotton. PLoS One. 2013:8. [DOI] [PMC free article] [PubMed]
- 56.Xu D, Mou G, Wang K, Zhou G. MicroRNAs responding to southern rice black-streaked dwarf virus infection and their target genes associated with symptom development in rice. Virus Res. 2014;190:60–68. doi: 10.1016/j.virusres.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 57.Fang Y, Xie K, Xiong L. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J Exp Bot. 2014;65:2119–2135. doi: 10.1093/jxb/eru072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Primers used for qRT-PCR.
Additional file 2: Figure S1. The precursors of 42 novel microRNAs and their hairpin structures in N. benthamiana.
Additional file 3: Figure S2. First nucleotide bias and nucleotide bias analysis.
Additional file 4: Table S2. 42differentially-expressed novel miRNAs and 25 equally-expressed novel miRNAs with miRNAs in N. benthamiana.
Data Availability Statement
All data and materials described in the manuscript are available in the Additional files 2,3 and 4.