Abstract
Background
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) has been rapidly developed and widely used as an analytical technique in clinical laboratories with high accuracy in microorganism identification.
Objective
To validate the efficacy of MALDI-TOF MS in identification of clinical pathogenic anaerobes.
Methods
Twenty-eight studies covering 6685 strains of anaerobic bacteria were included in this meta-analysis. Fixed-effects models based on the P-value and the I-squared were used for meta-analysis to consider the possibility of heterogeneity between studies. Statistical analyses were performed by using STATA 12.0.
Results
The identification accuracy of MALDI-TOF MS was 84% for species (I2 = 98.0%, P < 0.1), and 92% for genus (I2 = 96.6%, P < 0.1). Thereinto, the identification accuracy of Bacteroides was the highest at 96% with a 95% CI of 95–97%, followed by Lactobacillus spp., Parabacteroides spp., Clostridium spp., Propionibacterium spp., Prevotella spp., Veillonella spp. and Peptostreptococcus spp., and their correct identification rates were all above 90%, while the accuracy of rare anaerobic bacteria was relatively low. Meanwhile, the overall capabilities of two MALDI-TOF MS systems were different. The identification accuracy rate was 90% for VITEK MS vs. 86% for MALDI biotyper system.
Conclusions
Our research showed that MALDI-TOF-MS was satisfactory in genus identification of clinical pathogenic anaerobic bacteria. However, this method still suffers from different drawbacks in precise identification of rare anaerobe and species levels of common anaerobic bacteria.
Keywords: MALDI-TOF MS, Anaerobes, Bacteria identification
Background
Anaerobic bacteria exist as part of the normal flora in the human intestinal tract, oral cavity and urogenital tract [1], and can cause infectious diseases as a result of impairment to the microenvironment and/or immune system. Anaerobic infection can also be induced by deep wounds accompanied with facultative anaerobes and aerobic bacteria invasion. Invasive anaerobic infections are life threatening, and the mortality rate of anaerobic bacteremia is high as 40% [2]. Thus, the accurate and fast identification of anaerobic bacteria is pivotal to prompt antimicrobial treatments. Conventional anaerobe identification methods are cumbersome, time-consuming, and costly. It requires a long-term cultivation (not less than 24 h) to obtain enough inocula. In addition, the identification work is complex, including colony traits, colony morphology, and staining results. Meanwhile, it is difficult to identify rare or newly identified species by using conventional phenotyping methods and commercial kits [3]. Real-time, fast, high-throughput, high-sensitivity, high-selectivity, and low cost have been the goals pursued by analysts in modern analytical science.
The modern mass spectrometry technology enhances the understanding about the whole biological system through direct analysis of biological molecules such as proteins, lipids, carbohydrates and amino acids [4], which has been applied to the field of life science [5]. As an emerging technology, matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) has been widely used in clinical microbial diagnosis in the past decade. It is gradually replacing the traditional identification methods [6, 7]. MALDI-TOF MS is a rapid mass spectrometry technology developed in the late 1980s with relatively high sensitivity to various samples types. It is a useful, fast and accurate tool for routine laboratory analysis and has been used to identify mycobacteria [8, 9], nocardia [10], yeasts [11, 12] and anaerobes [13, 14] isolated from solid media of clinical specimens. At present, there are few studies to evaluate the efficacy of identifying anaerobic bacteria by MALDI-TOF MS. The aim of the present meta-analysis is to determine the reliability and effectiveness of mass spectrometry as a routine diagnostic method for anaerobic bacteria by searching related publications in the literature.
Methods
Search strategy
The scientific literature was extensively searched using the MeSH terms “maldi-ms” and “anaerobic bacteria” to query the electronic database of Medline and Web of science (up to 1 April 2018). Selected articles contained studies involving the identification of anaerobes by MALDI-TOF MS. The references cited in these articles were examined to determine other articles. The meta-analysis was performed by referring to (when appropriate) the PRISMA guidelines [15]. EndNote X8 (Thomson Reuters) was used for literature management. We read the titles and abstracts of each searched publication and selected only those relevant articles for full-text reading. There are no restrictions on the language, publication status and geographical distribution of publications.
Inclusion and exclusion criteria
We set up the criteria for the inclusion and exclusion of the literature. The inclusion criteria were as follows: (1) the study objective: the clinical specimens were identified as anaerobic bacteria by reference methods (16S rRNA gene sequencing); (2) the study method: the identification of anaerobes by MALDI-TOF MS; (3) the research objective: the accuracy of MALDI-TOF MS identification of anaerobes. The exclusion criteria included the following aspects: review articles, reviews, case reports, scientific abstracts and lectures; common anaerobes with fewer than 10 strains of anaerobes and less than 5 uncommon anaerobes; direct identification of bacteria in the positive blood culture bottle; The target bacteria could not be extracted, and pathogenic microorganisms or industrial environmental microbes of plants or animals were identified.
The identification criteria of MALDI-TOF MS in the included studies were as follows: a score of ≥2.0 was considered an accurate species-level identification; ≥ 1.7 but < 2.0, a probable genus-level identification; an isolate with a score < 1.7 was considered as “unidentified”; and an isolate identified as another species or genus was considered to be “misidentification”.
Quality assessment
What is important in meta-analysis is whether heterogeneity exists in the included studies and the possible reasons for the existence of heterogeneity, because heterogeneity may lead to deviations in meta-analysis results - known as “mixed apples and oranges” [16]. The sources of heterogeneity can be divided into, different research designs, different experimental conditions, different definitions of exposure and/or outcomes, different measurement methods, and the existence of other interference factors, i.e. covariates. In addition, low-quality literature will bring significant heterogeneity [17]. The following modified criteria, referring to the quality assessment for studies of diagnostic accuracy (QUADAS) [18], were used to assess the quality of original studies: study design, category and geographical distribution of strains, blinded status, reference methods, threshold, strain source, and system database.
Assessment of publication bias and influence analysis
According to statistics, the studies of positive results are more likely to publish than those of negative results, but it could not really represent the overall study population. In fact, the samples may be less representative. This result is called “publication bias” in statistics [17]. Funnel diagrams are commonly used graphical tests to assess publication bias in meta-analysis [19]. Egger’ s linear regression test of funnel plot asymmetry at the genus level and Begg’s rank correlation (with continuity correction) showed that little publication bias was detected in this review (t = − 1.54 and P = 0.123 for Egger’ s Test; z = − 0.35 and P = 0.727 for Begg’s Test).
Results
Results of the systematic literature search
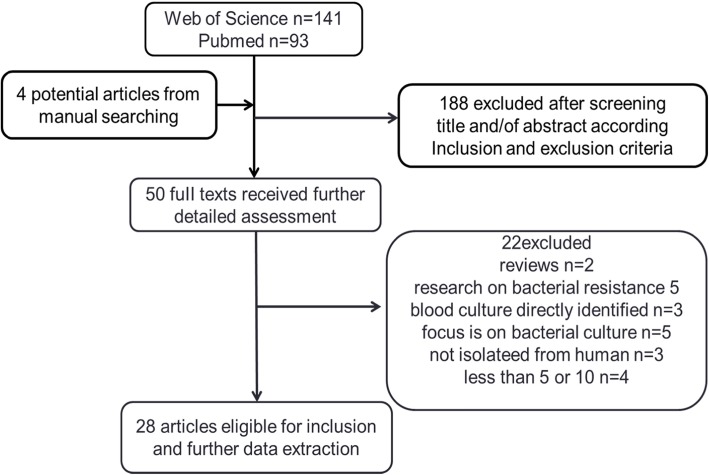
A total of 234 articles were retrieved from the electronic database. Additional four articles were identified through manual search, bibliographic search, and commentator suggestions. Finally, 28 studies were included according to the defined inclusion and exclusion criteria (Fig. 1). Countries and study periods included in all articles were shown in Table 1. The geographical distributions of the literature were Asia (5, 17.86%), Australia (1, 3.57%), South America (1, 3.57%), North America (4, 14.29%) and Europe (17, 60.71%), containing 24 cities in 14 countries.
Fig. 1.
Flow diagram for selection of studies
Table 1.
Geographical distributions and study periods of all included studies
| Author (publication year) | Country | City | Period of the study |
|---|---|---|---|
| Lucia Sanchez Ramos (2018) [20] | Germany | Leipzig | NM |
| Mervi Gürsoy (2017) [21] | Finland | Turku | NM |
| Belén Rodríguez-Sánchez (2017) [22] | Spain | Madrid | January 2010 to August 2012. |
| A.C.M. Veloo (2016) [23] | The Netherlands | Groningen | NM |
| Tomoyuki Yunoki (2016) [24] | Japan | Kyoto | June 2013 to May 2014 |
| Sung Jin Jo, M.D. (2015) [25] | Korea | Seoul | January to February 2015 |
| NINA HANDAL (2014) [26] | Norway | Lørenskog | January 2009 to December 2013 |
| Wonmok Lee, M.D. (2014) [27] | Korea | Seoul | 2011 |
| M.J. Barba (2014) [28] | Spain | A Coruña | 2007–2014 |
| Roy Chean (2014) [29] | Australia | Melbourne | 2000–2010 |
| Mariela S. Záratea (2014) [30] | Argentina | Ciudad Autónoma de Buenos Aires | NM |
| Yang Li (2014) [31] | China | Nanjing | NM |
| Yen-Michael S. Hsu (2014) [32] | USA | St. Louis | NM |
| Susanna K P Lau (2013) [33] | China | Hong Kong | NM |
| O. Garner (2013) [34] | USA | St. Louis | January 2012 to August 2012. |
| Melody Barreau(2013) [35] | France | Marseille | 2010–2013 |
| L. Coltella (2013) [7] | Italy | Rome | June 2010 to October 2011 |
| Bryan H. Schmitt (2012) [36] | USA | Minnesota | 2012 |
| N. Wüppenhorst (2012) [37] | Germany | Freiburg | NM |
| Silvia Vega-Casta˜no (2012) [38] | Spain | Salamanca | NM |
| Rémi Fournier (2012) [39] | France | Lille | NM |
| M. Knoester (2012) [40] | The Netherlands | Leiden | January 2010 to February 2011 |
| D. P. Fedorko (2012) [41] | USA | Bethesda | NM |
| Ulrik Stenz Justesen (2011) [14] | Denmark | Vejle | November 2007 to October 2010 |
| Esther Culebras (2011) [42] | Spain | Madrid | 2004–2006 |
| Bernard La Scola (2011) [13] | France | Marseille | 2009–2010 |
| A. C. M. Veloo (2011) [43] | The Netherlands | Leiden | NM |
| A.C.M. Velooa (2011) [44] | The Netherlands | Groningen | NM |
NM Not mentioned in the article
Bacterial isolates
After comprehensive and detailed data compilation, we collected 6685 (Additional file 1) strains of anaerobic bacteria. The most 4 common genera (> 500) in this article were Bacteroides spp. (1952), Clostridium spp. (1599), Propionibacterium spp. (611) and Prevotella spp. (509). A total of 5125 anaerobic bacteria were analyzed by MALDI biotyper, and VITEK MS analyzed a total of 1609 anaerobic bacteria. In addition, 49 anaerobic bacteria were analyzed by both MALDI-TOF MS systems.
Performance of the MS system
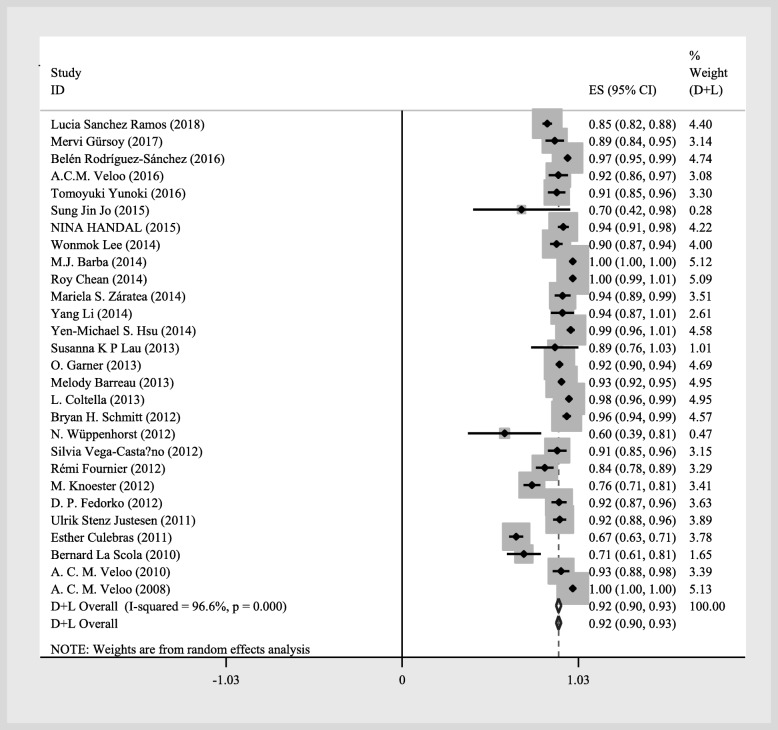
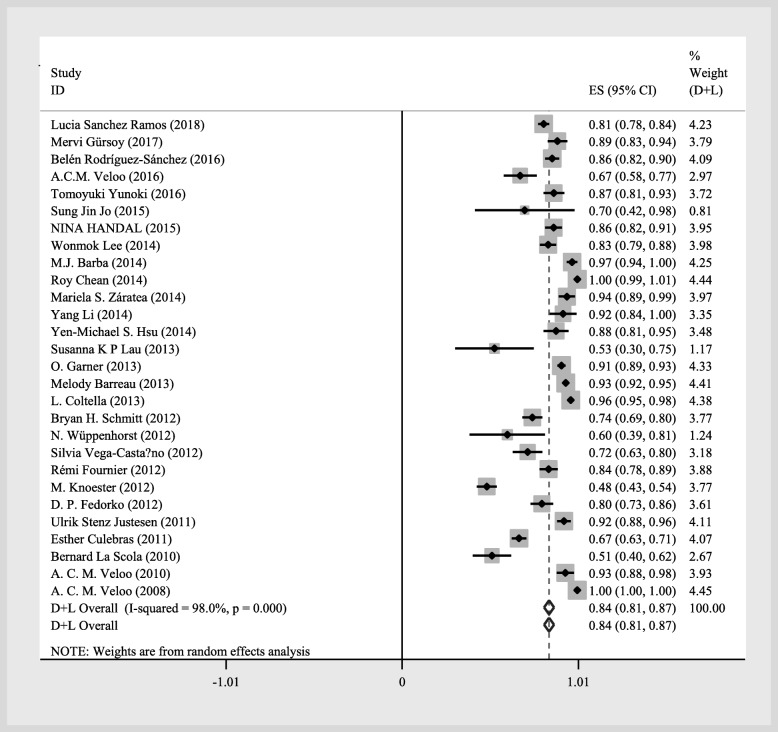
The overall statistical results of the meta-analysis at the genus and species levels identification were summarized using a forest plots of random-effects model (Figs. 2 and 3) [3, 13, 14, 20–44]. Of these, 6008 (92%; 95% CI of 90 to 93%) were correctly identified at the genus level, while 5656 (84%; 95% CI of 81 to 87%) were correctly identified at the species level by MALDI-TOF MS using a random-effects model.
Fig. 2.
Forest plot for the meta-analysis of the gross identification ratio at the genus level
Fig. 3.
Forest plot for the meta-analysis of the gross identification ratio at the species level
The pooled identification results of MALDI-TOF MS by random-effects for all anaerobic genera were shown in Table 2. The overall correct identification ratio of MALDI-TOF MS to anaerobic bacteria ranged from 60 to 100% at the genus level and ranged from 51 to 100% at the species level. Significant heterogeneity was found both at the genus level (P < 0.001; I2 = 96.6%) and the species level (P < 0.001; I2 = 98.0%). Identification accuracy of Bacteroides spp. was the highest at 96% with a 95% CI of 95 to 97%. The higher proportion of anaerobic bacteria was Lactobacillus spp., Parabacteroides spp., Clostridium spp., Propionibacterium spp., Prevotella spp., Veillonella spp. and Peptostreptococcus spp. The correct identification rate was higher than 90%. Identification accuracy of Bifidobacterium spp., Solobacterium spp., Finegoldia spp., Capnocytophaga spp., Parvimonas spp., Peptoniphilus spp., Slackia spp., Actinomyces spp., Ruminococcus spp. and Tissierella spp. was similar with an overall correct identification ratio at 80%, followed by Fusobacterium spp., Eggerthella spp. with an identification proportion above 70%. Identification accuracy of Actinobaculum spp., Atopobium spp., Anaerococcus spp. and Flavonifracter spp. was similar with an overall correct identification ratio at 60%. The lowest performance of MALDI-TOF MS was in Eubacterium spp., Bilophila spp., Butyricimonas spp. and Porphyromonas spp. (50%). Multiple factors contributed to this result, including the category of strains, the proportion of common and unusual species, or the reference library version.
Table 2.
Identification accuracy rate of all anaerobic genera
| Genus | Proportion | 95%CI | Weight% |
|---|---|---|---|
| Bacteroides | 96% | 95–97% | 6.79 |
| Lactobacillus | 95% | 89–102% | 5.16 |
| Parabacteroides | 94% | 87–101% | 4.88 |
| Clostridium | 92% | 90–93% | 6.73 |
| Propionibacterium | 91% | 89–93% | 6.55 |
| Prevotella | 91% | 88–93% | 6.48 |
| Veillonella | 91% | 85–197% | 5.25 |
| Peptostreptococcus | 90% | 85–95% | 5.60 |
| Bifidobacterium | 89% | 76–103% | 2.60 |
| Solobacterium | 88% | 73–104% | 2.27 |
| Finegoldia | 87% | 83–91% | 5.96 |
| Capnocytophaga | 86% | 60–112% | 1.01 |
| Parvimonas | 86% | 82–91% | 5.78 |
| Peptoniphilus | 86% | 81–91% | 5.59 |
| Slackia | 83% | 67–98% | 2.24 |
| Actinomyces | 81% | 74–89% | 4.66 |
| Ruminococcus | 80% | 45–115% | 0.59 |
| Tissierella | 80% | 45–115% | 0.59 |
| Fusobacterium | 79% | 74–84% | 5.61 |
| Eggerthella | 74% | 63–85% | 3.39 |
| Actinobaculum | 68% | 46–91% | 1.26 |
| Atopobium | 68% | 51–85% | 1.92 |
| Anaerococcus | 64% | 54–73% | 3.92 |
| Flavonifracter | 63% | 29–96% | 0.64 |
| Eubacterium | 57% | 20–94% | 0.55 |
| Bilophila | 56% | 23–88% | 0.68 |
| Butyricimonas | 56% | 23–88% | 0.68 |
| Porphyromonas | 50% | 36–64% | 2.59 |
Subgroup meta-analyses
We selected the genera (sample number not smaller than 5) identified by MALDI biotyper and VITEK MS to compare the identification accuracy for the same genus of the two systems (Table 3). The identification accuracy rate of MALDI biotyper was higher than VITEK MS for Parabacteroides spp., Eggerthella spp., Peptostreptococcus spp., Parvimonas spp., Bacteroides spp., Clostridium spp. and Peptoniphilus spp., and the efficacy of the two systems were similar for Prevotella spp. and Actinomyces spp. However, the heterogeneity of MALDI biotyper was more significant. In addition, the correct rate of MALDI biotyper for some strains (such as Finegoldia spp. and Fusobacterium spp.) was lower than VITEK MS, and the heterogeneity of MALDI biotype was higher than the latter. To sum up, the results of Table 3 showed that the correct rate of MALDI biotyper identification of anaerobic bacteria was higher than that of VITEK MS, while the heterogeneity of the MALDI biotyper was more significant.
Table 3.
Accuracy of MALDI-TOF MS identification
| Genus | Numbera | MALDI biotyperb | Vitekb |
|---|---|---|---|
| Parabacteroides | 42/6 | 100% (I2 = 0.0%, P > 0.01) | 72% (I2 = 83.3%, P > 0.01) |
| Peptostreptococcus | 41/91 | 100% (I2 = 0.0%, P > 0.01) | 97% (I2 = 76.6%, P > 0.01) |
| Eggerthella | 36/26 | 100% (I2 = 21.8%, P > 0.01) | 77% (I2 = 0.0%, P > 0.01) |
| Parvimonas | 205/7 | 100% (I2 = 35.5%, P > 0.01) | 98% (I2 = 19.0%, P > 0.01) |
| Clostridium | 779/820 | 98% (I2 = 63.5%, P < 0.01) | 94% (I2 = 96.7%, P < 0.01) |
| Finegoldia | 233/17 | 98% (I2 = 77.8%, P < 0.01) | 99% (I2 = 2.0%, P > 0.01) |
| Prevotella | 404/105 | 92% (I2 = 84.1%, P < 0.01) | 92% (I2 = 0.0%, P > 0.01) |
| Bacteroides | 1517/435 | 97% (I2 = 89.4%, P < 0.01) | 96% (I2 = 74.8%, P < 0.01) |
| Fusobacterium | 214/34 | 91% (I2 = 89.7%, P < 0.01) | 92% (I2 = 84.8%, P = 0.01) |
| Propionibacterium | 605/6 | 90% (I2 = 91.6%, P < 0.01) | 100% (I2 = 0.0%, P > 0.01) |
| Peptoniphilus | 41/91 | 85% (I2 = 92.4%, P < 0.01) | 62% (I2 = 96.8%, P < 0.01) |
| Actinomyces | 79/28 | 74% (I2 = 94.5%, P < 0.01) | 74% (I2 = 0.0%, P > 0.01) |
aThe left side of / is the number of samples of MALDI biotyper, and the right side of / is the number of samples of Vitek
bThe higher I-square values combined P value more than 0.1 mean the higher heterogeneity between those studies
In additional, the identification rate of anaerobic bacteria in European countries (species: 84%, genus: 88%) was lower than that in Asia (species: 84%, genus: 91%) and North America (species: 86%, genus: 94%). The protocol for the studies at different cities was the same. A total of 21 articles reported on the media, including anaerobic horse blood agar, chocolate agar, blood culture bottle, schaedler agar, bacteroides bile esculin agar, CDC anaerobic blood agar, brucella blood plates, columbia blood plates and blood plates. Among them, brucella and columbia blood plates were two most frequently used media (species 73% and genus 92%; species 73% and genus 75%).
It was worth noting that VITEK MS incorrectly identified Actinomyces georgiae as Capnocytophaga gingivalis, MALDI biotyper incorrectly identified Clostridium spp. as Enterococcus spp. (Table 4), and MALDI biotyper also incorrectly identified some rare anaerobic bacteria Mogibacterium timidum and Parvimonas micra as other bacteria, probably due to the lack of corresponding standard spectra in the database.
Table 4.
Common misidentification pattern in these studies
| Sequence identifcation | MALDI-TOF MS identifcation | System | Reference |
|---|---|---|---|
| Actinomyces georgiae | Capnocytophaga gingivalis | bioMérieux Vitek MS | [27] |
| Actinomyces graevenitzii | Actinomyces turicensis | Bruker MALDI Biotyper | [7] |
| Actinomyces meyeri | Actinomyces odontolyticus | Bruker MALDI Biotyper | [58] |
| Actinomyces viscosus | Actinomyces oris | Bruker MALDI Biotyper | [58] |
| Anaerococcus hydrogenalis | Bacteroides fragilis | Bruker MALDI Biotyper | [24] |
| Anaerococcus tetradius | Brevibacillus spp. | bioMérieux Vitek MS | [31] |
| Bacteroides cellulosilyticus | Bacteroides intestinalis | Bruker MALDI Biotyper | [3] |
| Bacteroides dorei | Bacteroides vulgatus | Bruker MALDI Biotyper | [28, 58] |
| Bacteroides faecis | Bacteroides thetaiotaomicron | Bruker MALDI Biotyper | [58] |
| Bacteroides faecis | Bacteroides thetaiotaomicron | bioMérieux Vitek MS | [27] |
| Bacteroides nordii | Bacteroides thetaiotaomicron | bioMérieux Vitek MS | [27] |
| Bacteroides vulgatus | Bacteroides eggerthii | bioMérieux Vitek MS | [31] |
| Clostridium baratii | Clostridium paraputrificum | bioMérieux Vitek MS | [20] |
| Clostridium beijerinckii | Clostridium butyricum | bioMérieux Vitek MS | [20] |
| Clostridium bifermentans | Clostridium sordellii | bioMérieux Vitek MS | [20] |
| Clostridium bolteae | Clostridium clostridioforme | bioMérieux Vitek MS | [27] |
| Clostridium butyricum | Clostridium beijerinckii | bioMérieux Vitek MS | [20] |
| Clostridium cadaveris | Clostridium sordellii | bioMérieux Vitek MS | [20] |
| Clostridium clostridioforme | Bacillus ssp. | bioMérieux Vitek MS | [20] |
| Clostridium difficile | Clostridium septicum | bioMérieux Vitek MS | [20] |
| Clostridium difficile | Enterococcus faecium | Bruker MALDI Biotyper | [24] |
| Clostridium histolyticum | C. sordellii/C. septicum | bioMérieux Vitek MS | [20] |
| Clostridium limosum | Clostridium tyrobutyricum | bioMérieux Vitek MS | [20] |
| Clostridium perfringens | Enterococcus faecalis | Bruker MALDI Biotyper | [24] |
| Clostridium sordellii | Clostridium bifermentans | bioMérieux Vitek MS | [20] |
| Clostridium sporogenes | C.difficile/ C.glycolicum | bioMérieux Vitek MS | [20] |
| Clostridium tetani | Clostridium septicum | bioMérieux Vitek MS | [20] |
| Fusobacterium nucleatum | Enterococcus faecalis | Bruker MALDI Biotyper | [24] |
| Fusobacterium nucleatum | Fusobacterium naviforme | Bruker MALDI Biotyper | [26] |
| Mogibacterium timidum | Clostridium halophilum | Bruker MALDI Biotyper | [58] |
| Parvimonas micra | Slackia exigua | Bruker MALDI Biotyper | [24] |
| Peptoniphilus Indolicus | Peptoniphilus harei | Bruker MALDI Biotyper | [28, 58] |
| Porphyromonas gulae | Porphyromonas gingivalis | Bruker MALDI Biotyper | [58] |
| Prevotella bivia | Streptococcus anginosus | Bruker MALDI Biotyper | [24] |
| Prevotella denticola | Bacteroides fragilis | Bruker MALDI Biotyper | [24] |
| Prevotella oralis | Prevotella nanciencis | Bruker MALDI Biotyper | [58] |
| Prevotella oris | Prevotella buccae | Bruker MALDI Biotyper | [24] |
| Veillonella dispar | Veillonella parvula | Bruker MALDI Biotyper | [7] |
| Veillonella dispar | Veillonella parvula | bioMérieux Vitek MS | [27] |
Discussion
MALDI-TOF MS, based on the microbial identification of characteristic protein fingerprints of bacteria, usually takes only a few minutes to rapidly identify species of different microorganisms, thus greatly shortening the detection time and improving the diagnostic efficiency of infectious diseases. It is usually difficult to isolate and culture anaerobic bacteria by conventional approaches, and MALDI-TOF MS provides a useful technology for their identification. In this study, we conducted a meta-analysis to analyze the differences in independent research results by addressing heterogeneity between studies in an attempt to shed new light on the identification of anaerobic bacteria by MALDI-TOF MS [45, 46].
According to the inclusion and exclusion criteria, 28 anaerobic genera were included and assessed critically using two currently available MALDI-TOF MS systems. It is known that anaerobes are more difficult to be identified in clinical laboratories [47]. However, using MALDI-TOF MS, the overall identification accuracy of anaerobic bacteria was 92% (95% CI of 0.90 to 0.93) at the genus level in 28 included articles with 6685 various anaerobes isolates. These results indicate that MALDI-TOF MS is a qualified method for accurate and rapid identification of pathogenic anaerobes. At the same time, we noticed that the identification property of MALDI-TOF MS against common anaerobe isolate species was variable. Among them, the correct rate was more than 80% for 18 anaerobic genera (Bacteroides spp., Lactobacillus spp., Parabacteroides spp., Clostridium spp., ect.), 60–80% for 6 anaerobic genera (Fusobacterium spp., Eggerthella spp., Actinobaculum spp., Atopobium spp., Anaerococcus spp., Flavonifracter spp.,), and lower than 60% for the other 4 anaerobic genera (Eubacterium spp., Bilophila spp., Butyricimonas spp. and Porphyromonas spp.). The different identification correct rate might be due to the difficulty of obtaining satisfactory spectra from some species, such as Mogibacterium timidum or Actinomyces georgiae, and partly due to the limit of uncommon anaerobes species spectra in commercial reference libraries. Therefore, it is increasingly important to update the library of various anaerobic species, especially those lacking or poorly represented in the current version. Fortunately, commercial databases are constantly being improved and updated at intervals of about three to 6 months [48].
In this study, we analyzed two commonly used commercial MALDI-TOFMS systemstwo identification systems: the Bruker MALDI biotyper and the bioMérieux VITEK MS. To compare the same anaerobic genus between the two systems, we focused our attention on analysis of 12 out of 28 anaerobic bacteria genera included in both systems. Among them, Bacteroides spp., Clostridium spp., Propionibacterium spp. and Prevotella spp. were the predominant anaerobes. Figures 2 and 3 showed the overall identification rates of the specimens with two identification systems, MALDI biotyper and VITEK MS. The identification capacities of the two systems in Table 3 and forest plot (Figs. 2 and 3) was different. The overall identification rate of MALDI biotyper was higher than that of VITEK MS (Table 3), though the data in forest plot was opposite. It is supposed that low equipment cost leads to a wider range of MALDI biotyper applications. The rare anaerobic specimens identified by MALDI biotyper may account for a large proportion of the reason, for most of them were not included in the relevant database as previously described, which decreased of overall identification rate. This is consistent with the data presented in the forest plot.
In addition to the instrument, the identification correct ratio of anaerobic bacteria is also related to the system paired database. As shown in Table 4, one-third of the 28 studies displayed identification errors, most of which were correct genus and wrong species, and some of which were wrong genera. These results might attribute to the similarity protein composition of the species, which made the differentiation of the quality peak difficult, and made it difficult for MALDI-TOF MS to correctly identify the strain. The similarity of the protein structures led to the incorrect identification results in both anaerobic bacteria and other genera, such as Streptococcus spp. [49], Mycobacterium spp. [50], Enterococcus spp. [51] and yeast [52]. These result-related mistakes might be attributed to the similar protein compositions of the species, making the discrepancy of differentiation of the quality peak difficult, and making it difficult for MALDI-TOF MS to correctly identify the strains. Beyond this, the lower identification scores might be related to interspecies correlation and bacterial cell wall composition [6]. Therefore, updating the existing information and perfecting the database of difficultly identified organisms (such as Fusobacterium spp. and Porphyromonas spp.) are useful to improve the identification accuracy of MALDI-TOF MS.
Another point that cannot be ignored is the impact of the geographical distribution on the identification rate of MALDI-TOF MS. The anaerobic identification rate was slightly lower in European countries than that elsewhere. In our study, we give priority to the following reasons for this situation. The geographical distribution of the collected literature was related to the level of the economic development, given the high cost of the equipment. For this reason, the strains involved in this study reflected the situation in some developed countries rather the whole world; for instance, the technology is more advanced in Europe than that in other continents, where MALDI-TOF MS has been first applied to the field of microbial identification. In addition, MALDI-TOF MS was used to identify microorganisms with imperfect databases in some of the early studies, which is also the reason for the relatively low overall identification rate. Therefore, updating and perfecting the databases are essential for improving the identification rate. On the other hand, technological improvements should be made to reduce the cost of MALDI-TOF MS equipment as much as possible so that it can be applied in more countries and cities.
There are some limitations in our research. First, Table 3 does not list all anaerobes collected because some data of the species were discarded for statistical reasons. In additional, some articles only reported the results of MALDI-TOF MS identification of the isolates at the “species” level without providing the identification results at the “genus” level. If a specimen was not identified to “species”, it would be defined as being identified neither at the species level nor at the genus level. This may lead to the negligence of the strains identified at the “genus” level, thus underestimating the accuracy of MALDI-TOF MS in identifying anaerobes at the genus level. Despite these problems, MALDI-TOF MS is still widely used in bacterial identification and other fields such as strain typing [53], detection of virulence factors [54] and evaluation of drug resistance [55–57].
Conclusions
In conclusion, the current meta-analysis showed that MALDI-TOF MS has shown a high degree of accuracy in anaerobic identification, although there is a lack of data to define its effectiveness in identifying rare anaerobic species. As a novel technology, MALDI-TOF MS has been widely used in the clinical diagnosis of pathogenic diseases. Therefore, it is necessary to analyze the comprehensive ability of this technique in clinical and microbiology diagnosis in the future.
Supplementary information
Additional file 1. The details of 6685 strains.
Acknowledgments
We are grateful for the MALDI-TOF MS provided by the Microbiology Laboratory of Xuzhou Medical University and the kind help provided by the laboratory teachers.
Abbreviations
- CI
Confidence interval
- MALDI-TOF MS
Matrix-assisted laser desorption ionization-time of flight mass spectrometry
Authors’ contributions
Conceived and designed the study: BG, HCL. Searched and screened the literature: MZS, XHM, MJY. Analyzed the data: MZS, XHM. Wrote and revised the paper: YL, MZS, ZBZ, YC, QJZ. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (No. 17KJB360014), Jiangsu Privincial Natural Science Foundation (BK20151154) and the National Natural Science Foundation of China (81871734, 81471994, 81902040). Jiangsu Privincial Medical Talent (ZDRCA2016053) and Six talent peaks project of Jiangsu Province (WSN-135) lead the design of each part of the study. The Natural Science Foundation of Jiangsu Province (Grants No BK20150209) and the Advanced health talent of six-one project of Jiangsu Province (LGY2016042) participated in revising manuscripts. The funding bodies had no role in the collection, analysis, and interpretation of data.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ying Li and Mingzhu Shan contributed equally to this work.
Contributor Information
Ying Li, Email: sdwhrsly@163.com.
Mingzhu Shan, Email: 15952183853@163.com.
Zuobin Zhu, Email: geneticszzb@163.com.
Xuhua Mao, Email: jsyxmao@163.com.
Mingju Yan, Email: 421710089@qq.com.
Ying Chen, Email: cyylotus@163.com.
Qiuju Zhu, Email: susan_9335@163.com.
Hongchun Li, Email: 56834997@qq.com.
Bing Gu, Email: gb20031129@163.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12879-019-4584-0.
References
- 1.Norin E. How normal is a “normal” flora in animal or man? Anaerobe. 2011;17(6):431–432. doi: 10.1016/j.anaerobe.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Kim J, Lee Y, Park Y, et al. Anaerobic bacteremia: impact of inappropriate therapy on mortality. Infect Chemother. 2016;48(2):91–98. doi: 10.3947/ic.2016.48.2.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau SK, Tang BS, Teng JL, et al. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for identification of clinically significant bacteria that are difficult to identify in clinical laboratories. J Clin Pathol. 2014;67(4):361–366. doi: 10.1136/jclinpath-2013-201818. [DOI] [PubMed] [Google Scholar]
- 4.Petrotchenko EV, Borchers CH. Modern mass spectrometry-based structural proteomics. Adv Protein Chem Struct Biol. 2014;95(2):193–213. doi: 10.1016/B978-0-12-800453-1.00006-3. [DOI] [PubMed] [Google Scholar]
- 5.Matsuo T, Seyama Y. Introduction to modern biological mass spectrometry. J Mass Spectrom. 2015;35(2):114–130. doi: 10.1002/(SICI)1096-9888(200002)35:2<114::AID-JMS949>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 6.Prod'Hom G, Bizzini A, Durussel C, et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for direct bacterial identification from positive blood culture pellets. J Clin Microbiol. 2010;48(4):1481–3. [DOI] [PMC free article] [PubMed]
- 7.Coltella L, Mancinelli L, Onori M, et al. Advancement in the routine identification of anaerobic bacteria by MALDI-TOF mass spectrometry. Eur J Clin Microbiol Infect Dis. 2013;32(9):1183–1192. doi: 10.1007/s10096-013-1865-1. [DOI] [PubMed] [Google Scholar]
- 8.Buckwalter SP, Olson SL, Connelly BJ, et al. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of Mycobacterium species, Nocardia species, and other aerobic Actinomycetes. J Clin Microbiol. 2016;54(2):376. doi: 10.1128/JCM.02128-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saleeb PG, Drake SK, Murray PR, et al. Identification of Mycobacteria in solid-culture media by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2011;49(5):1790–1794. doi: 10.1128/JCM.02135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segawa S, Nishimura M, Sogawa K, et al. Identification of Nocardia species using matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Clin Proteomics. 2015;12(1):6. doi: 10.1186/s12014-015-9078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aslani N, Janbabaei G, Abastabar M, et al. Identification of uncommon oral yeasts from cancer patients by MALDI-TOF mass spectrometry. BMC Infect Dis. 2018;18(1):24. doi: 10.1186/s12879-017-2916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veen SQ, Van CECJ, Kuijper EJ. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization-time of flight mass spectrometry in conventional medical microbiology laboratories. J Clin Microbiol. 2010;48(3):900–907. doi: 10.1128/JCM.02071-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scola BL, Fournier PE, Raoult D. Burden of emerging anaerobes in the MALDI-TOF and 16S rRNA gene sequencing era. Anaerobe. 2011;17(3):106–112. doi: 10.1016/j.anaerobe.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Justesen US, Holm A, Knudsen E, et al. Species identification of clinical isolates of anaerobic bacteria: a comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry systems. J Clin Microbiol. 2011;49(12):4314–4318. doi: 10.1128/JCM.05788-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. J Clin Epidemiol. 2009;6(10):e1–34. doi: 10.1016/j.jclinepi.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Georgia S, Simon S, Higgins JP. Obstacles and opportunities in meta-analysis of genetic association studies. Genet Med. 2005;7(1):13–20. doi: 10.1097/01.gim.0000151839.12032.1a. [DOI] [PubMed] [Google Scholar]
- 17.Ioannidis JP. Interpretation of tests of heterogeneity and bias in meta-analysis. J Eval Clin Pract. 2010;14(5):951–957. doi: 10.1111/j.1365-2753.2008.00986.x. [DOI] [PubMed] [Google Scholar]
- 18.Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 19.Hunter JP, Saratzis A, Sutton AJ, et al. In meta-analyses of proportion studies, funnel plots were found to be an inaccurate method of assessing publication bias. J Clin Epidemiol. 2014;67(8):897–903. doi: 10.1016/j.jclinepi.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez Ramos L, Rodloff AC. Identification of Clostridium species using the VITEK® MS. Anaerobe. 2018;54:217–223. doi: 10.1016/j.anaerobe.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Gursoy M, Harju I, Matomaki J, et al. Performance of MALDI-TOF MS for identification of oral Prevotella species. Anaerobe. 2017;47:89–93. doi: 10.1016/j.anaerobe.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 22.Rodríguez-Sánchez B, Alcalá L, Marín M, et al. Evaluation of MALDI-TOF MS (matrix-assisted laser desorption-ionization time-of-flight mass spectrometry) for routine identification of anaerobic bacteria. Anaerobe. 2016;42:101–107. doi: 10.1016/j.anaerobe.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 23.Veloo AC, de Vries ED, Jean-Pierre H, et al. The optimization and validation of the Biotyper MALDI-TOF MS database for the identification of gram-positive anaerobic cocci. Clin Microbiol Infect. 2016;22(9):793–798. doi: 10.1016/j.cmi.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 24.Yunoki T, Matsumura Y, Nakano S, et al. Genetic, phenotypic and matrix-assisted laser desorption ionization time-of-flight mass spectrometry-based identification of anaerobic bacteria and determination of their antimicrobial susceptibility at a University Hospital in Japan. J Infect Chemother. 2016;22(5):303–307. doi: 10.1016/j.jiac.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 25.Jo SJ, Park KG, Han K, et al. Direct identification and antimicrobial susceptibility testing of Bacteria from positive blood culture bottles by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and the Vitek 2 system. Ann Lab Med. 2016;36(2):117–123. doi: 10.3343/alm.2016.36.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Handal N, Bakken Jørgensen S, Smith Tunsjø H, et al. Anaerobic blood culture isolates in a Norwegian university hospital: identification by MALDI-TOF MS vs 16S rRNA sequencing and antimicrobial susceptibility profiles. APMIS. 2015;123(9):749–758. doi: 10.1111/apm.12410. [DOI] [PubMed] [Google Scholar]
- 27.Lee W, Kim M, Yong D, et al. Evaluation of VITEK mass spectrometry (MS), a matrix-assisted laser desorption ionization time-of-flight MS system for identification of anaerobic bacteria. Ann Lab Med. 2015;35(1):69–75. doi: 10.3343/alm.2015.35.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barba MJ, Fernández A, Oviaño M, et al. Evaluation of MALDI-TOF mass spectrometry for identification of anaerobic bacteria. Anaerobe. 2014;30:126–128. doi: 10.1016/j.anaerobe.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 29.Chean R, Kotsanas D, Francis MJ, et al. Comparing the identification of Clostridium spp. by two Matrix-Assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF) mass spectrometry platforms to 16S rRNA PCR sequencing as a reference standard: a detailed analysis of age of culture and sample preparation. Anaerobe. 2014;30:85–89. doi: 10.1016/j.anaerobe.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 30.Zárate MS, Romano V, Nievas J, et al. Utilidad de la espectrometría de masas MALDI-TOF en la identificación de bacterias anaerobias. Rev Argent Microbiol. 2014;46(2):98–102. doi: 10.1016/S0325-7541(14)70055-0. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Gu B, Liu G, et al. MALDI-TOF MS versus VITEK 2 ANC card for identification of anaerobic bacteria. J Thorac Dis. 2014;6(5):517–523. doi: 10.3978/j.issn.2072-1439.2014.02.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu YM, Burnham CA. MALDI-TOF MS identification of anaerobic bacteria: assessment of pre-analytical variables and specimen preparation techniques. Diagn Microbiol Infect Dis. 2014;79(2):144–148. doi: 10.1016/j.diagmicrobio.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 33.Lau SK, Tang BS, Teng JL, et al. Matrix-assisted laser desorption ionisation time-of-flight mass spectrometry for identification of clinically significant bacteria that are difficult to identify in clinical laboratories. J Clin Pathol. 2014;67(4):361–6. [DOI] [PubMed]
- 34.Garner O, Mochon A, Branda J, et al. Multi-Centre evaluation of mass spectrometric identification of anaerobic bacteria using the VITEK® MS system. Clin Microbiol Infect. 2014;20(4):335–9. [DOI] [PubMed]
- 35.Barreau M, Pagnier I, La Scola B. Improving the identification of anaerobes in the clinical microbiology laboratory through MALDI-TOF mass spectrometry. Anaerobe. 2013;22:123–5. [DOI] [PubMed]
- 36.Schmitt BH, Cunningham SA, Dailey AL, et al. Identification of Anaerobic Bacteria by Bruker Biotyper Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry with On-Plate Formic Acid Preparation. J Clin Microbiol. 2013;51(3):782–6. [DOI] [PMC free article] [PubMed]
- 37.Wüppenhorst N, Consoir C, Lörch D, et al. Direct identification of bacteria from charcoal-containing blood culture bottles using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Eur J Clin Microbiol Infect Dis. 2012;31(10):2843–50. [DOI] [PubMed]
- 38.Vega-Castaño S, Ferreira L, González-Ávila M, et al. Eficacia de la espectrometría de masas MALDI-TOF en la identificación debacterias anaerobias. Enferm Infecc Microbiol Clin. 2012;30(10):597–601. [DOI] [PubMed]
- 39.Fournier R, Wallet F, Grandbastien B, et al. Chemical extraction versus direct smear for MALDI-TOF mass spectrometry identification of anaerobic bacteria. Anaerobe. 2012;18(3):294–7. [DOI] [PubMed]
- 40.Knoester M, van Veen SQ, Claas EC, et al. Routine identification of clinical isolates of anaerobic bacteria: matrix-assisted laser desorption ionization-time of flight mass spectrometry performs better than conventional identification methods. J Clin Microbiol. 2012;50(4):1504. [DOI] [PMC free article] [PubMed]
- 41.Fedorko DP, Drake SK, Stock F, et al. Identification of clinical isolates of anaerobic bacteria using matrix-assisted laser desorption ionization-time of flight mass spectrometry. Eur J Clin Microbiol Infect Dis. 2012;31(9):2257–62. [DOI] [PubMed]
- 42.Culebras E, Rodriguez-Avial I, Betriu C, et al. Rapid identification of clinical isolates of Bacteroides species by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry. Anaerobe. 2012;18(1):163–5. [DOI] [PubMed]
- 43.Veloo AC, Erhard M, Welker M, et al. Identification of Gram-positive anaerobic cocci by MALDI-TOF mass spectrometry. Syst Appl Microbiol. 2011;34(1):58–62. [DOI] [PubMed]
- 44.Veloo AC, Knoester M, Degener JE, et al. Comparison of two matrix-assisted laser desorption ionisation-time of flight mass spectrometry methods for the identification of clinically relevant anaerobic bacteria. Clin Microbiol Infect. 2011;17(10):1501–6. [DOI] [PubMed]
- 45.Andreas W, Lukas S, Jette J, et al. MALDI-TOF MS in microbiological diagnostics-identification of microorganisms and beyond (mini review). Appl Microbiol Biotechnol. 2012;93(3):965–74. [DOI] [PubMed]
- 46.Neelja S, Manish K, Kanaujia PK, et al. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol. 2015;6:791. [DOI] [PMC free article] [PubMed]
- 47.Marinach-Patrice C, Lethuillier A, Marly A, et al. Use of mass spectrometry to identify clinical Fusarium isolates. Clin Microbiol Infect. 2010;15(7):634–42. [DOI] [PubMed]
- 48.Lavigne JP, Espinal P, Dunyach-Remy C, et al. Mass spectrometry: a revolution in clinical microbiology? Clin Chem Lab Med. 2013;51(2):257–70. [DOI] [PubMed]
- 49.Fan WT, Qin TT, Bi RR, et al. Performance of the matrix-assisted laser desorption ionization time-of-flight mass spectrometry system for rapid identification of streptococci: a review. Eur J Clin Microbiol Infect Dis. 2017;36(6):1005–12. [DOI] [PubMed]
- 50.Zingue D, Flaudrops C, Drancourt M. Direct matrix-assisted laser desorption ionisation time-of-flight mass spectrometry identification of mycobacteria from colonies. Eur J Clin Microbiol Infect Dis. 2016;35(12):1983–7. [DOI] [PubMed]
- 51.Lallemand E, Arvieux C, Coiffier G, et al. Use of MALDI-TOF mass spectrometry after liquid enrichment (BD Bactec™) for rapid diagnosis of bone and joint infections. Res Microbiol. 2017;168(2):122–29. [DOI] [PubMed]
- 52.Ling H, Yuan Z, Shen J, et al. Accuracy of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of clinical pathogenic fungi: a meta-analysis. J Clin Microbiol. 2014;52(7):2573–82. [DOI] [PMC free article] [PubMed]
- 53.Sauer S, Freiwald A, Maier T, et al. Classification and identification of bacteria by mass spectrometry and computational analysis. PLoS One. 2008;3(7):e2843. [DOI] [PMC free article] [PubMed]
- 54.Bittar F, Ouchenane Z, Smati F, et al. MALDI-TOF-MS for rapid detection of staphylococcal Panton-Valentine leukocidin. Int J Antimicrob Agents. 2009;34(5):467–70. [DOI] [PubMed]
- 55.De Carolis E, Vella A, Florio AR, et al. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry for caspofungin susceptibility testing of Candida and Aspergillus species. J Clin Microbiol. 2012;50(7):2479–83. [DOI] [PMC free article] [PubMed]
- 56.Lata M, Sharma D, Deo N, et al. Proteomic analysis of ofloxacin-mono resistant Mycobacterium tuberculosis isolates. J Proteomics. 2015;127:114–21. [DOI] [PubMed]
- 57.Sharma D, Lata M, Singh R, et al. Cytosolic Proteome Profiling of Aminoglycosides Resistant Mycobacterium tuberculosis Clinical Isolates Using MALDI-TOF/MS. Front Microbiol. 2016;7:1816. [DOI] [PMC free article] [PubMed]
- 58.Huang YL, Sun QL, Li JP, et al. Evaluation of an in-house MALDI-TOF MS rapid diagnostic method for direct identification of micro-organisms from blood cultures. J Med Microbiol. 2019;68(1):41–7. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. The details of 6685 strains.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.