Abstract
Background
Osteoarthritis (OA) is one of the most common chronic musculoskeletal diseases, yet to date it lacks effective therapeutic strategies. Increasing evidence suggests that long noncoding RNAs (lncRNAs) serve pivotal roles in the occurrence and development of OA. However, the possible molecular mechanism involving lncRNAs, such as nuclear enriched abundant transcript 1 (NEAT1), in OA progression is still unclear.
Material/Methods
First, NEAT1 and miR-181a expression in OA synovium tissues and normal synovium tissues were detected. Then, the effect of NEAT1 on modulating growth ability, apoptosis, and inflammation in OA chondrocytes was investigated by a series of loss-function experiments. Next, the correlation between NEAT1, miR-181a, and glycerol-3-phosphate dehydrogenase 1-like (GPD1L) was fully investigated. Finally, the downregulation of miR-181a was employed as a recovery experiment to explore the functional mechanism of NEAT1 in OA.
Results
In the present study, we found that NEAT1 expression was downregulated in OA tissues, while miR-181a expression was prominently upregulated. Moreover, reduced expression of NEAT1 suppressed cell growth while elevating the apoptotic rate and increasing the abundance of inflammatory cytokines released in OA chondrocytes. Furthermore, we clarified that miR-181a was a direct sponge of NEAT1, and GPD1L was able to bind to miR-181a. Additionally, we found that downregulation of miR-181a was able to attenuate the effect of NEAT1 on apoptosis, inflammatory response, and proliferation in OA chondrocytes.
Conclusions
Our findings indicate that downregulation of NEAT1 aggravated progression of OA via modulating the miR-181a/GPD1L axis, providing a novel insight into the mechanism of OA pathogenesis.
MeSH Keywords: MicroRNAs; Osteoarthritis; RNA, Long Noncoding
Background
Osteoarthritis (OA), a degenerative joint disease characterized by progressive erosion of cartilage components, can cause pain and loss of articular function, leading to a significant reduction in quality of life of patients [1]. After the age of 65, most people will suffer from OA in at least 1 joint. It is generally believed that the prevalence of OA is rising as the global population ages and obesity rates increase, resulting in a serious public health problem in the coming decades [2]. Even worse, although numerous studies propose a variety of therapeutic options for OA – including drug treatment, electromagnetic therapy and stem cell treatment – there is still no generally satisfactory therapeutic strategy for this disease [3]. Therefore, it is now essential to explore the molecular mechanism underlying the occurrence and development of OA.
OA has long been believed to be a chronic inflammatory disease in which the abnormal release of inflammatory cytokines is triggered by different pathological stimuli. This inflammation leads to deteriorating OA pathology by changing chondrocytes to a catabolic phenotype [4]. During OA progression, inflammatory pathway dysregulation has been found in almost every pathological stage, including p38 MAPK [5], PI3K/Akt [6], Fas [7], TNF-a [8], and IL-1b [9], which is accompanied by cartilage destruction and retardation of extracellular matrix (ECM) synthesis [10,11]. Importantly, an excessive inflammatory response is always associated with apoptotic cell death in OA chondrocytes, causing a decrease in cartilage cellularity and homeostasis, which could be a potent option for a new therapeutic target [12,13]. Additionally, researchers have speculated that various genes in chondrocytes are aberrantly expressed via local activation of epigenetic mechanisms, due to the persistent inflammatory condition of the OA microenvironment [14].
MicroRNAs (miRNAs) are known as a class of small noncoding single-stranded RNA molecules of 18–25 nucleotides, and serve as pivotal regulators of gene expression by inhibiting the translation of mRNA and/or encouraging mRNA degradation by complementary base-pairing with 3′-untranslated regions (3-UTRs). miRNAs have been shown to contribute to pathological processes in many diseases, including cancers, cardiovascular disease, diabetes mellitus, and OA [14]. Ample evidence suggests that miRNAs function as key regulators for several cellular functions, including proliferation, differentiation, inflammatory response, and apoptosis [15]. Recent studies have suggested that miRNAs can serve as modulators for chondrocyte apoptosis in the occurrence and development of OA. For example, Abouheif et al. confirmed that knockdown of miR-34a can repress IL-1b-induced chondrocyte apoptosis [16]. Jing Li et al. reported that miR-146a can induce chondrocyte apoptosis by targeting Smad4 [17]. In 2015, Gabler et al. used microarray analysis to show that the miR-181 family was involved in chondrocyte maturation of successive differentiation stages, suggesting miR-181 might become a novel target for OA therapy [18]. A growing body of research has demonstrated that miR-181 functions as a key player in proliferation, apoptosis, and inflammatory response in OA chondrocytes [19,20]. Nevertheless, specific molecular mechanisms of miR-181 involved in OA progression remain unknown.
Long noncoding RNAs (lncRNAs) are a kind of noncoding transcripts, consisting of more than 200 nucleotides (nt) in length [21]. Growing evidence has shown that lncRNAs can influence various cellular processes such as cell growth, cell apoptosis, ECM synthesis, and inflammatory response in OA, which further impact the balance between the anabolic and catabolic phases of joint cartilage [22]. In addition, many studies have revealed that lncRNAs can function as competing endogenous RNA (ceRNA), thereby reducing the endogenous suppressive effect of miRNAs [23]. Nuclear enriched abundant transcript 1 (NEAT1) is a newly identified nuclear-restricted lncRNA, the dysregulation of which participates in tumorigenesis of a variety of neoplastic diseases [17]. For example, Peng et al. reported that NEAT1 was able to regulate colorectal cancer cell apoptosis via the Akt signaling pathway [23]. Wang et al. found that NEAT1 functions as a ceRNA for miR-107 to promote laryngeal squamous cell cancer proliferation [17]. However, the potential biological mechanisms of NEAT1 in OA are still poorly understand.
The aim of our present study was to determine the exact role of NEAT1 in the proliferation, apoptosis, and inflammatory response in OA chondrocytes. Moreover, the downstream targeting of NEAT1 by microRNA and the associated ceRNA mechanism were also investigated.
Material and Methods
Clinical specimens
We collected samples of 30 degenerated synovium tissues from OA patients and 30 normal synovium tissues from trauma patients who underwent arthroscopic surgery in Tianjin Hospital between Dec 2016 and Jul 2017. The enrolled 30 OA patients consisted of 14 males and 16 females, whose age ranged from 46 to 62 years, while there were 17 males and 13 females in the control group, aged 41 to 60 years. There was no notable variation in sex and age between OA patients and trauma patients. Once collected, tissue specimens were immediately frozen. All proceedures were approved by the Institute on Research Ethics Committee of Tianjin Hospital and were in accordance with the Helsinki Declaration and with institutional guidelines.
Cell culture and transfection
The chondrocytes were isolated from OA articular cartilages and were subsequently cultured. Briefly, the articular cartilages obtained from surgery were minced into small fragments and then were digested using trypsin (Beyotime, China) for 30 min, followed by stimulating with 0.3% collagenase (Beyotime, China) for 5–6 h. After that, extracted chondrocytes were cultured in RPMI Medium 1640 media with heat-inactivated 10% FBS (Gibco, USA) at 37°C with 5% CO2.
For transfection, siRNAs specifically targeting NEAT1, miR-181a mimics, miR-181a inhibitor, and corresponding special negative control (NC) were designed and obtained from GenePharma (Shanghai, China). Chondrocytes cultured in DMEM (with 10% FBS) were first cultured in 6-well plates, and then transfected with si-NEAT1 or miR-181a-5p mimics using Lipofectamine 3000 (Invitrogen, USA) at approximately 60–70% confluence. After transfection for 48 h, the cells were harvested for a series of follow-up experiments.
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA from tissues and cells was extracted using TRIzol reagent (Thermo Fisher Scientific, USA). The RNA was then reverse transcribed into cDNA with the PrimeScript™ RT Reagent kit (Takara, Japan) following the manufacturer’s specification. Subsequently, qPCR was implemented using an ABI7500 system (Applied Biosystems, USA) and a SYBR Green qPCR kit (Life Technologies, USA) in accordance with the manufacturer’s protocol. Relative gene expression levels were quantified by the 2−ΔΔCt method. GAPDH and U6 were used as controls. The primer sequences are listed in Supplementary Table 1.
MTT assay
Cell proliferation was detected using a Cell Proliferation Reagent Kit I (MTT, Sigma-Aldrich, USA). Briefly, the cells were first seeded in 96-well plates at 5×104 cells per well. After different incubation times, 20 μL of MTT solution was added into all the wells and the culture was continued at 37°C for another 4 h. Then, 150 μL of solubilisation solution (DMSO, Beyotime, China) was mixed in each well followed by shaking for 10 min. Finally, cell proliferation was tested using a Benchmark microplate spectrometer (Bio-Rad, USA).
Apoptosis assay
For the cell apoptosis assay, the transfected chondrocytes (1×104) were harvested, washed with PBS, and then added to 500 uL binding buffer containing 2 uL AnnexinV-fluorescein isothiocyanate (FITC, Beyotime, China) and 5 uL PI (Beyotime, China) for 1 h, avoiding direct light. The apoptosis rate of labeled chondrocytes was measured using BD FACS Calibur (BD Biosciences, USA).
Luciferase reporter assay
First, we predicted the downstream targets of NEAT1 and miR-181a using a bioinformatic algorithm. Next, we amplified and inserted 3′UTR of wild-type NEAT1 (NEAT1-WT), mutant-type NEAT1 (NEAT1-MUT), wild-type GPD1L (GPD1L-WT), and mutant-type GPD1L (GPD1L-MUT) into the pmirGLO vector (Invitrogen, USA). For the luciferase reporter assay, these plasmids were co-transfected with miR-181a mimic or mimic-NC into cells using Lipofectamine 3000 (Invitrogen, USA). At 48 h after transfection, the Dual-Luciferase Reporter system (Promega, USA) was used to measure luciferase activity.
Western blot analysis
Total protein was isolated from cells using RIPA buffer (Beyotime, China) and then separated by 10% SDS-PAGE. The protein was then transferred to PVDF membranes (Thermo Fisher Scientific, USA) blocking with 5% fat-free milk for 1 h. Next, the membranes were incubated overnight at 4°C with the following primary antibodies: GPD1L, tumor necrosis factor α (TNF-α), IL-1β, IL-6, IL-8, metalloprotease 13 (MMP13), cyclooxygenase 2 (COX-2), and GAPDH (1: 1,000, Abcam, UK). Samples were then hybridized with HRP-conjugated secondary antibodies (Abcam, UK) for 1 h at 37°C. Western blot bands were visualized using an enhanced chemiluminescence kit (Beyotime, China).
RNA immunoprecipitation (RIP) assay
The RNA immunoprecipitation (RIP) assay was done using the EZMagna RIP RNA-binding protein immunoprecipitation kit (Millipore, USA), following the manufacturer’s specifications. Briefly, the transfected cells were lysed and immunoprecipitated with anti-human argonaute 2 (Ago2) antibodies (Abcam, UK) and control IgG (input group). After 48-h incubation, the coprecipitated RNAs were isolated and measured using PCR.
Statistical analysis
SPSS (version 21.0, Chicago, IL) software was used to perform the statistical analysis. All data are presented as mean±standard deviation (SD), and a t test or one-way ANOVA was used to assess statistical significance. The correlations between expression level of NEAT1 and miR-181a, or miR-181a and GPD1L, were analyzed using Pearson’s correlation coefficient. P<0.05 was deemed to be statistically significant.
Results
NEAT1 and miR-181a expression level in OA tissues
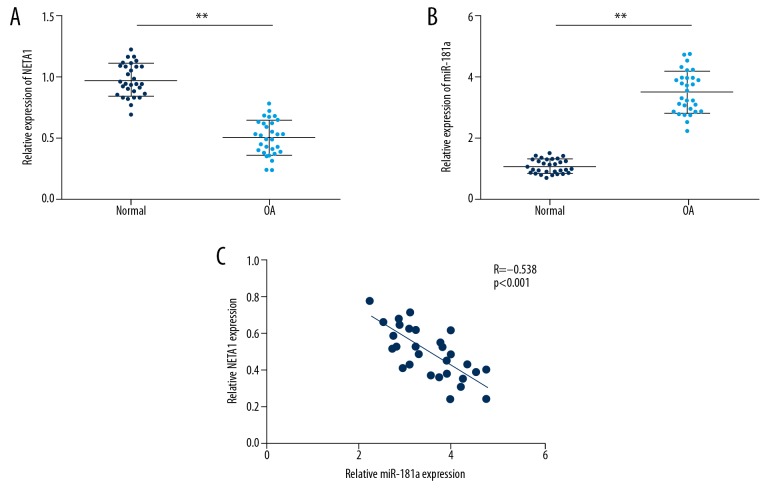
To investigate the effect of NEAT1 and miR-181a on OA, we first performed qRT-PCR to detect their expression in 30 OA patients and 30 controls. Not unexpectedly, our results showed that expression level of NETA1 conspicuously declined in OA tissues compared to normal cartilage tissue, whereas the expression level of miR-181a was significantly elevated in OA tissues (Figure 1A, 1B). As shown in Figure 1C, our findings indicated that there was a negative relationship between NETA1 and miR-181a expression level in OA patients (Figure 1C). Therefore, our results supported the idea that NETA1 and miR-181a likely participate in OA progression.
Figure 1.
The expression of NEAT1 and miR-181a in OA tissues. (A, B) The NEAT1 and miR-181a expression level in OA tissues detected by RT-PCR analysis. (C) The correlation between of NEAT1 and miR-181a expression level in OA. Data are shown as the mean±SD of 3 replicates. * P<0.05, ** P<0.01 vs. control.
Downregulation of NEAT1 regulated biological function in OA chondrocytes
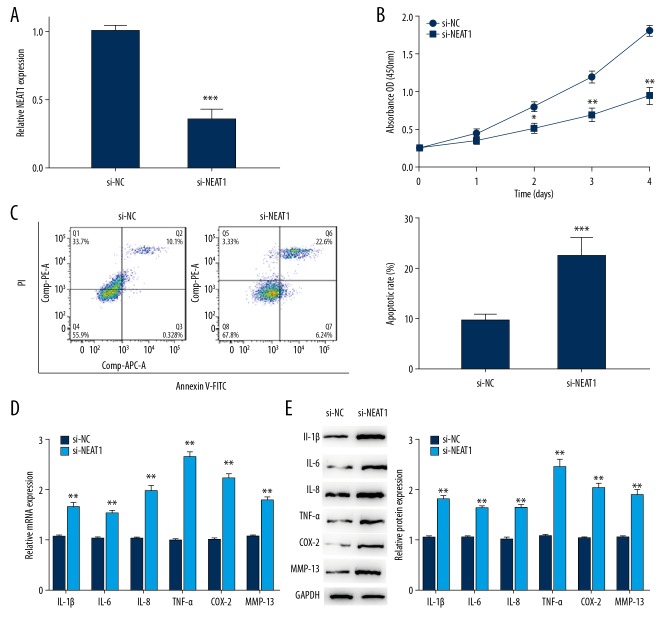
To further explore the effect of NEAT1 on OA, we examined whether NEAT1 knockdown could affect biological functions in chondrocytes. From the start, our data showed NEAT1 expression was conspicuously lower in the si-NEAT1 group when compared with the control group, making clear that si-NEAT1 could achieve a steady inhibition efficiency for subsequent experiments (Figure 2A). After that, the proliferation, apoptosis, and inflammatory responses of OA chondrocytes transfected with si-NEAT1 were assessed. Our results revealed that knockdowns of NEAT1 could inhibit cell growth and promote apoptosis in OA chondrocytes (Figure 2B, 2C). Moreover, the level of inflammatory cytokines COX-2, TNF-α, IL-1β, IL-6, IL-8, and MMP13 were measured in OA chondrocytes transfected with si-NEAT1. As illustrated in Figure 2D, 2E, the expression levels of these inflammatory factors were all prominently elevated in the si-NEAT1 group compared to the control group. Taken together, our finding suggested that NEAT1 is a key regulator of cell growth, apoptosis, and inflammatory response of OA chondrocytes.
Figure 2.
Knockdown of NEAT1 regulated proliferation, apoptosis, and inflammation in OA chondrocytes. (A) Si-NETA1 were transfected into chondrocytes to achieve NETA1 downregulation. (B, C) MTT assay and flow cytometric analysis were performed to determine the cell growth ability and apoptotic rate in chondrocytes transfected with si-NETA1. (D, E) RT-PCR and Western blot were performed to measure inflammatory cytokines in chondrocytes transfected with si-NETA1. Data are shown as the mean±SD of 3 replicates. * P<0.05, ** P<0.01 vs. NC. NC – negative control.
miR-181a level negatively modulated by NEAT1
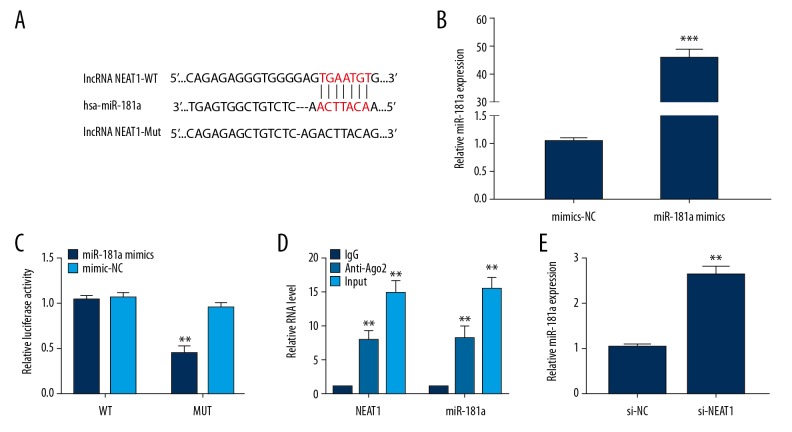
Increasing evidence has confirmed that lncRNAs can act as competitive endogenous RNAs (ceRNAs) through competitively binding miRNAs, which thereby suppress expression of the targeted miRNA [24]. With the use of online bioinformatic tools, we predicted that miR-181a had a putative binding site within NEAT1 (Figure 3A). We then verified that miR-181a mimics could successfully attain amplification of miR-181a when compared to the corresponding control (Figure 3B). Moreover, we carried out luciferase reporter assays and RIP assays to validate the correlation between NEAT1 and miR-181a in OA. As shown in Figure 3C, our results showed that miR-181a overexpression obviously restrained the luciferase activity when fused to NEAT1-WT, whereas the NEAT1-MUT group was not affected. Likewise, our results demonstrated that NEAT1 and miR-181a were preferentially enriched in the Ago2 pellet in contrast to controls, indicating that miR-181a could directly connect to the NEAT1 3′UTR (Figure 3D). In addition, the knockdown of NEAT1 was able to facilitate miR-181a expression in OA chondrocytes (Figure 3E). In summary, our findings demonstrated that NEAT1 can be regarded as a ceRNA of miR-181a.
Figure 3.
NEAT1 is targeted by miR-181a. (A) Complementary sequences of miR-181a on NETA1 mRNA 3′-UTR were obtained from publicly available algorithms. (B) miR-181a mimics were transfected into chondrocytes to achieve miR-181a overexpression. (C) Luciferase activity of a luciferase reporter plasmid containing NEAT1-WT 3′UTR or NEAT1-MUT 3′UTR co-transfected with miR-181a mimics. (D) The RIP assay was performed to confirm whether NEAT1 and miR-181a could directly bind to Ago2. (E) The endogenous miR-181a level in chondrocytes treated with si-NEAT1. Data are shown as the mean±SD of 3 replicates. * P<0.05, ** P<0.01 vs. NC. NC – negative control.
GAP1L can directly bind to miR-181a
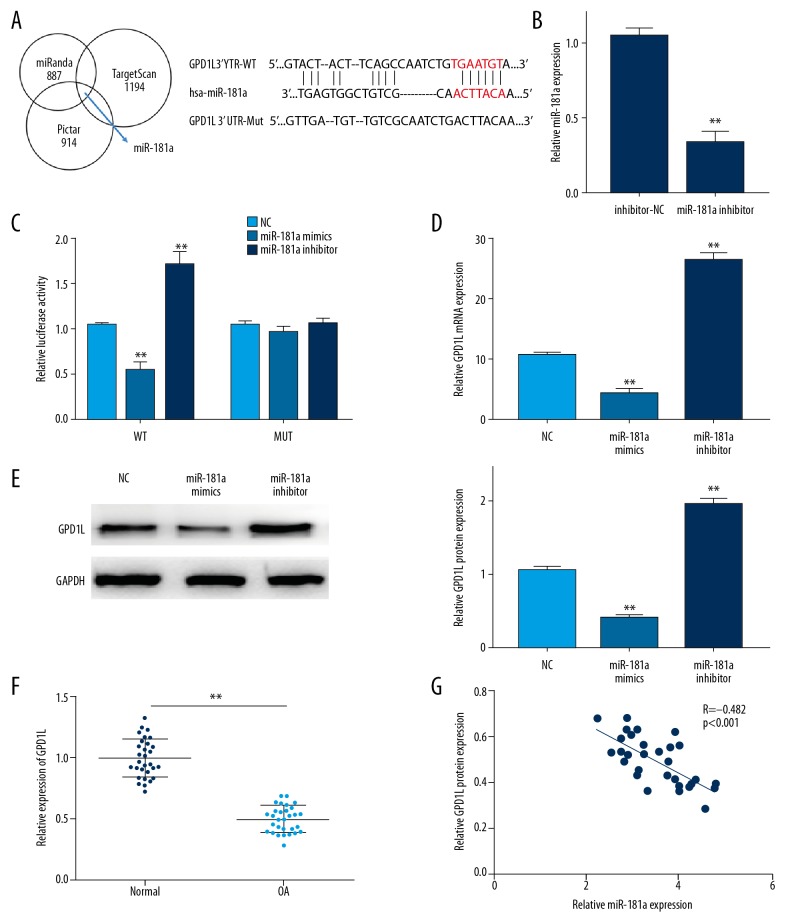
With online bioinformatic tools, we further found GAP1L was a potential downstream target of miR-181a (Figure 4A). Then, we confirmed that miR-181a inhibitor could effectively reduce miR-181a expression (Figure 4B). In order to confirm the interaction of GAP1L and miR-181a in OA, a dual-luciferase reporter assay was performed. As exhibited in Figure 4C, ectopic expression of miR-181a was able to inhibit the luciferase activity in the GAP1L-WT group, whereas downregulated miR-181a showed the opposite effect. However, neither miR-181a mimics and nor miR-181a inhibitor altered luciferase activity in the GAP1L-MUT group. Moreover, we assessed the role of miR-181a at an endogenous level of GAP1L by using RT-qPCR and Western blot. Our data showed that the expression of GAP1L was distinctly upregulated by downregulation of miR-181a. In contrast, miR-181a overexpression suppressed GAP1L expression in OA chondrocytes (Figure 4D, 4E). Additionally, we showed that expression of GAP1L was dramatically downregulated in OA patients, which was negatively related to the expression level of miR-181a (Figure 4F, 4G). Therefore, our results suggested that GAP1L could directly bind to miR-181a, and was negatively modulated by miR-181a.
Figure 4.
miR-181a directly targets GAP1L. (A) The predicted binding sites of miR-181a within the GAP1L-3′UTR (B) MiR-181a inhibitor were transfected into chondrocytes to achieve miR-181a downregulation. (C) Luciferase activity of cells co-transfected with GAP1L-WT 3′UTR or GAP1L-MUT 3′UTR and miR-181a mimics or miR-181a inhibitor. (D, E) The mRNA and protein level of GAP1L in chondrocytes in different groups. (F) The expression of GAP1L in OA tissues detected by RT-PCR analysis. (G) The correlation between miR-181a and GAP1L expression in OA tissues. Data are shown as the mean±SD of 3 replicates. * P<0.05, ** P<0.01 vs. control. NC – negative control.
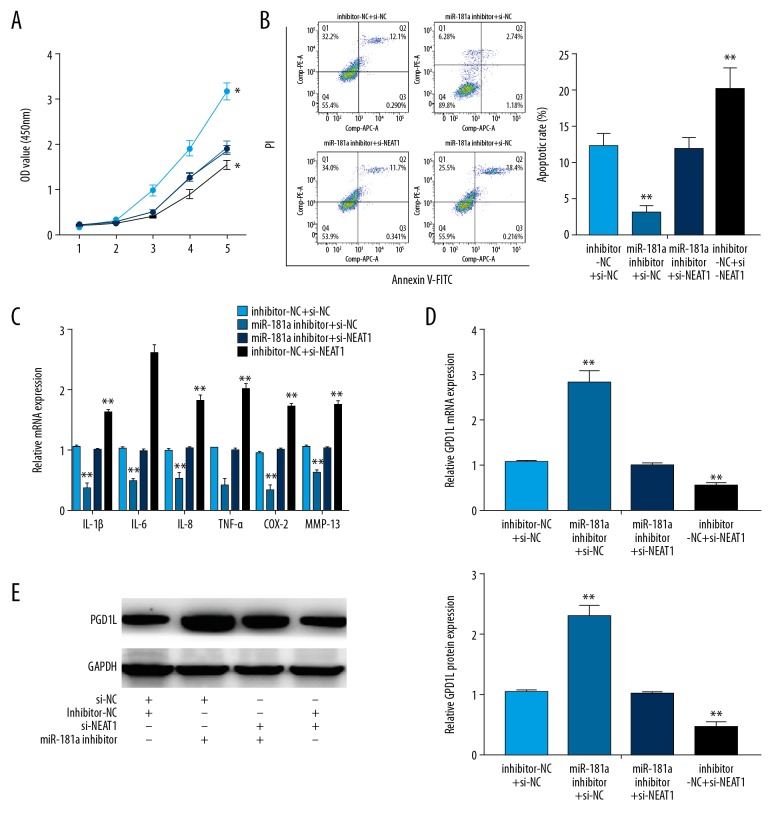
NEAT1-miR-181a-GPD1L axis regulated OA progression
To further clarify the effect of NEAT1 on OA, we established a battery of rescue experiments. First, our results found that downregulation of miR-181a could attenuate the inhibitory effect of NEAT1 knockdown on OA chondrocytes (Figure 5A). In addition, downregulated miR-181a was capable of reversing the increased apoptosis caused by the NEAT1 knockdown (Figure 5B). As shown in Figure 5C, the concentrations of inflammatory cytokines were also reversed in OA chondrocytes co-transfected with miR-181a inhibitor and si-NEAT1 when compared with the si-NEAT1 group or the miR-181a inhibitor group. Finally, our results showed that GPD1L expression could be rescued in OA chondrocytes co-transfected with si-NEAT1 and miR-181a inhibitor (Figure 5D, 5E). Therefore, these results showed that NEAT1 can modulate OA progression by modulating the miR-181a-GPD1L axis.
Figure 5.
NEAT1/miR-181a/GPD1L axis regulates OA progression. (A, B) The proliferation and the apoptosis in chondrocytes co-transfected with inhibitor-NC or miR-181a inhibitor and with either NETA1 or si-NC. (C) The expression of inflammatory cytokines in chondrocytes co-transfected with inhibitor-NC or miR-181a inhibitor and with either si-NETA1 or si-NC. (D, E) The expression of GAP1L in chondrocytes co-transfected with inhibitor-NC or miR-181a inhibitor and with either si-NETA1 or si-NC, detected by RT-PCR analysis and Western blot. Data are shown as the mean±SD of 3 replicates. * P<0.05, ** P<0.01 vs. control. NC – negative control.
Discussion
OA, one of the commonest chronic musculoskeletal diseases in elderly populations, is characterized by a progressive degeneration of articular cartilage, ultimately resulting in decreased patient quality of life [25,26]. Chondrocytes, the unique cell type making up cartilage, mediate the cartilage homeostasis of chondrocyte anabolism [27]. Abundant evidence has shown that a great diversity of factors are involved in the development and progression of OA, including composition and structural changes of the ECM, abnormal release of local inflammatory cytokines, and polytype of programmed cell death of chondrocytes [28,29]. Recently, an increasing number of studies have focused on the effects of lncRNAs and microRNAs on regulating biological activity of chondrocytes [30]. These RNAs may help to explain the underlying molecular mechanisms concerned with articular cartilage injury and maintenance, and may also lead to the development of new therapeutic interventions.
In recent years, a large body of evidence has shown that microRNAs play a pivotal role in OA due to the fact that mechanical loading was found to affect miRNAs differentiation expression [14]. In support of this view, further research proved that miRNAs could regulate multiple genes whose expression was directly correlated with cartilage development, homeostasis, and OA pathology [31]. For example, Chen et al. indicated that miR-29b-3p promoted OA chondrocyte apoptosis via negatively regulating PGRN, providing a potential target for OA treatments [31], and Etich et al. showed that dysfunction of miR-26a expression could bring about extracellular matrix changes in OA, which performed a pivotal role in OA progression [32]. In the case of miR-181, recent research has supported that it could be a promoter of OA progression through suppressing proliferation and by accelerating apoptosis and inflammatory responses in chondrocytes [19,20]. In agreement with this, our present results indicated that miR-181a expression was elevated in OA patients, the inhibition of which could promote proliferation while attenuating the inflammatory responses and apoptosis in OA chondrocytes, thus identifying miR-181a as a potential target for OA treatments.
To further understand the mechanism by which miR-181a affects biological activity regulation in chondrocytes, we used bioinformatic tools to predict the downstream targets of miR-181a. As expected, luciferase reporter assays showed that GPD1L was a potential target of miR-181a. Previous studies reported that GPD1L could regulate the hydroxylation of HIF-1a, leading to suppression of chondrocyte viability and inducing apoptosis of chondrocytes [33]. Meanwhile, Zhai et al. showed that miR-181a can play a key role in regulating chondrocyte apoptosis by negatively regulating GPD1L [34]. Similarly, our findings indicated that GPD1L expression was markedly lower in OA patients, which indicates it could be deeply involved in the functional biology of chondrocytes by counteracting miR-181a.
Coincidentally, lncRNAs can also participate in the complex biological processes of cartilage degeneration, and investigations have confirmed that numerous differentially expressed lncRNAs are present in OA tissues, including MALAT1, XIST, MIAT, and NEAT1 [30,35]. NEAT1, which was first discovered in 2007 by Hutchinson et al. [36], is located on the human chromosome 11q13 and is involved in various biological processes, including carcinogenesis, viral diseases, sepsis, and neurodegeneration [37]. Recently, Wang et al. [28] show that NEAT1/miR-181c could regulate osteopontin-mediated synoviocyte proliferation in OA tissues, indicating that NEAT1 may be a negative factor in synoviocyte proliferation [28]. Likewise, our study showed that NEAT1 was evidently downregulated in OA tissues, the inhibition of which induced the apoptosis of OA chondrocytes and facilitated expression of apoptosis-related inflammatory factors. Taken together, this evidence shows that NEAT1 serves as a vital factor in OA progression. Nevertheless, the exact molecular mechanism by which NEAT1 is involved in OA remains unclear.
Increasing evidence shows that lncRNAs can regulate miRNAs as a ceRNA by modulating the occurrence and development of OA. For example, Chen et al. demonstrated that MEG3 regulates cell growth, apoptosis, and ECM degradation in OA chondrocytes by targeting the miR-93-TGFBR2 axis [38]. Zhang et al. concluded that HOTAIRM1 facilitates cell growth and reduced apoptotic rate in OA chondrocytes through regulation of the miR-125b/BMPR2/JNK/MAPK/ERK signaling pathway, suggesting that HOTAIRM1 is a possible biomarker and treatment target for OA [39]. Using a luciferase activity reporter assay and RIP assay, our present study suggests that NEAT1 directly sponges miR-181a. Moreover, miR-181a inhibitors can reverse the acceleration of cell apoptosis and inflammatory responses by NEAT1 depletion in OA chondrocytes, as well as rescuing the anti-proliferative effect caused by the suppression of NEAT1. Therefore, our data show that NEAT1 can partly regulate OA progression by sponging the miR-181a-GPD1L axis. Nevertheless, due to our small sample size and the lack of a known underlying mechanism for this regulation, further investigation of the specific role of the NEAT1-miR-181a-GPD1L axis in OA progression is imperative.
Conclusions
Our study identified that a NEAT1 knockdown promoted apoptosis and inflammatory response and inhibited proliferation in OA chondrocytes, exerting a protective function in OA development. Furthermore, NEAT1 was able to regulate GPD1L expression by acting as a ceRNA of miR-181a. Additionally, our findings indicated that downregulation of NEAT1 aggravated progression of OA via modulating the miR-181a-GPD1L axis, providing new insight into the molecular mechanisms of OA pathogenesis.
Supplementary Data
Supplementary Table 1.
Primer sequences for RT-PCR analysis.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| miR-181a | 5′-GGAAAGCAGACATTGACCTCAC-3′ | 5′-CCATCCTTTACATCCTTCTGTCTGT-3′ |
| NETA1 | 5′-TCAAAACCTGTTCCCAGGA-3′ | 5′-TGTGCACACTCTATGTGGTAGA-3′ |
| IL-1β | 5′-ATGGCCAAAGTTCGAGACATG-3′ | 5′-TCAAGTCAGTGTTGAGATGATGCTT-3′ |
| IL-6 | 5′-AGCCACTCACCTCTTCAGAAC-3′ | 5′-GCCTCTTTGCTGCTTTCACAC-3′ |
| IL-8 | 5′-TCCAACCTGAGTGACATAGCGA-3′ | 5′-CTGACCTCCAACTCCAACGAAT-3′ |
| COX-2 | 5′-CAGCACTTCACGCATCAGTT-3′ | 5′-CGCAGTTTACGCTGTCTAGC-3′ |
| TNF-α | 5′-TGAAATATACAAGTTATATG-3′ | 5′-GTTCGAGAAGATGATTGATG-3′ |
| MMP13 | 5′-AGCCCGTTTAAAGTGCATGTGTGC-3′ | 5′-GAGTGTCCGAGGAAGATACTTGGT- 3′ |
| U6 | 5′-CTCGCTTCGGCAGCACA-3′ | 5′-AACGCTTCACGAATTTGCGT-3′ |
| GAPDH | 5′-ACAACTTTGGTATCGTGGAAGG-3′ | 5′-GCCATCACGCCACAGTTTC-3′ |
Footnotes
Source of support: Departmental sources
References
- 1.Loza E, Lopez-Gomez JM, Abasolo L, et al. Economic burden of knee and hip osteoarthritis in Spain. Arthritis Rheum. 2009;61:158–65. doi: 10.1002/art.24214. [DOI] [PubMed] [Google Scholar]
- 2.Spector TD. Epidemiology of the rheumatic diseases. Curr Opin Rheumatol. 1993;5:132–37. doi: 10.1097/00002281-199305020-00002. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Wang F, Chen G, et al. LncRNA MALAT1 promotes osteoarthritis by modulating miR-150-5p/AKT3 axis. Cell Biosci. 2019;9:54. doi: 10.1186/s13578-019-0302-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fathollahi A, Aslani S, Jamshidi A, Mahmoudi M. Epigenetics in osteoarthritis: Novel spotlight. J Cell Physiol. 2019;234:12309–24. doi: 10.1002/jcp.28020. [DOI] [PubMed] [Google Scholar]
- 5.Zhou Y, Liu SQ, Yu L, et al. Berberine prevents nitric oxide-induced rat chondrocyte apoptosis and cartilage degeneration in a rat osteoarthritis model via AMPK and p38 MAPK signaling. Apoptosis. 2015;20:1187–99. doi: 10.1007/s10495-015-1152-y. [DOI] [PubMed] [Google Scholar]
- 6.Huang Y, Wu D, Fan W. Protection of ginsenoside Rg1 on chondrocyte from IL-1beta-induced mitochondria-activated apoptosis through PI3K/Akt signaling. Mol Cell Biochem. 2014;392:249–57. doi: 10.1007/s11010-014-2035-1. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Wang J, Hu B, et al. MiR-34a promotes Fas-mediated cartilage endplate chondrocyte apoptosis by targeting Bcl-2. Mol Cell Biochem. 2015;406:21–30. doi: 10.1007/s11010-015-2420-4. [DOI] [PubMed] [Google Scholar]
- 8.Kayal RA, Siqueira M, Alblowi J, et al. TNF-alpha mediates diabetes-enhanced chondrocyte apoptosis during fracture healing and stimulates chondrocyte apoptosis through FOXO1. J Bone Miner Res. 2010;25:1604–15. doi: 10.1002/jbmr.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Gai P, Xu R, et al. Shikonin protects chondrocytes from interleukin-1beta-induced apoptosis by regulating PI3K/Akt signaling pathway. Int J Clin Exp Pathol. 2015;8:298–308. [PMC free article] [PubMed] [Google Scholar]
- 10.Troeberg L, Nagase H. Proteases involved in cartilage matrix degradation in osteoarthritis. Biochim Biophys Acta. 2012;1824:133–45. doi: 10.1016/j.bbapap.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Little CB, Barai A, Burkhardt D, et al. Matrix metalloproteinase 13-deficient mice are resistant to osteoarthritic cartilage erosion but not chondrocyte hypertrophy or osteophyte development. Arthritis Rheum. 2009;60:3723–33. doi: 10.1002/art.25002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas CM, Fuller CJ, Whittles CE, Sharif M. Chondrocyte death by apoptosis is associated with cartilage matrix degradation. Osteoarthritis Cartilage. 2007;15:27–34. doi: 10.1016/j.joca.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 13.Ryu JH, Shin Y, Huh YH, et al. Hypoxia-inducible factor-2alpha regulates Fas-mediated chondrocyte apoptosis during osteoarthritic cartilage destruction. Cell Death Differ. 2012;19:440–50. doi: 10.1038/cdd.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malemud CJ. MicroRNAs and osteoarthritis. Cells. 2018;7 doi: 10.3390/cells7080092. pii: E92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Li Y, Kong D, et al. Cross-talk between miRNA and Notch signaling pathways in tumor development and progression. Cancer Lett. 2010;292:141–48. doi: 10.1016/j.canlet.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abouheif MM, Nakasa T, Shibuya H, et al. Silencing microRNA-34a inhibits chondrocyte apoptosis in a rat osteoarthritis model in vitro. Rheumatology (Oxford) 2010;49:2054–60. doi: 10.1093/rheumatology/keq247. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Huang J, Dai L, et al. miR-146a, an IL-1beta responsive miRNA, induces vascular endothelial growth factor and chondrocyte apoptosis by targeting Smad4. Arthritis Res Ther. 2012;14:R75. doi: 10.1186/ar3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabler J, Ruetze M, Kynast KL, et al. Stage-specific miRs in chondrocyte maturation: Differentiation-dependent and hypertrophy-related miR clusters and the miR-181 family. Tissue Eng Part A. 2015;21:2840–51. doi: 10.1089/ten.TEA.2015.0352. [DOI] [PubMed] [Google Scholar]
- 19.Wu XF, Zhou ZH, Zou J. MicroRNA-181 inhibits proliferation and promotes apoptosis of chondrocytes in osteoarthritis by targeting PTEN. Biochem Cell Biol. 2017;95:437–44. doi: 10.1139/bcb-2016-0078. [DOI] [PubMed] [Google Scholar]
- 20.Zhu LM, Yang M. The suppression of miR-181 inhibits inflammatory responses of osteoarthritis through NF-kappaB signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:5567–74. doi: 10.26355/eurrev_201907_18290. [DOI] [PubMed] [Google Scholar]
- 21.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec 1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 22.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 23.Peng W, Wang Z, Fan H. LncRNA NEAT1 impacts cell proliferation and apoptosis of colorectal cancer via regulation of Akt signaling. Pathol Oncol Res. 2017;23:651–56. doi: 10.1007/s12253-016-0172-4. [DOI] [PubMed] [Google Scholar]
- 24.Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Won Y, Shin Y, Chun CH, et al. Pleiotropic roles of metallothioneins as regulators of chondrocyte apoptosis and catabolic and anabolic pathways during osteoarthritis pathogenesis. Ann Rheum Dis. 2016;75:2045–52. doi: 10.1136/annrheumdis-2015-208406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aigner T, Sachse A, Gebhard PM, Roach HI. Osteoarthritis: Pathobiology-targets and ways for therapeutic intervention. Adv Drug Deliv Rev. 2006;58:128–49. doi: 10.1016/j.addr.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 27.Kim H, Kang D, Cho Y, Kim JH. Epigenetic regulation of chondrocyte catabolism and anabolism in osteoarthritis. Mol Cells. 2015;38:677–84. doi: 10.14348/molcells.2015.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, Wang W, Zhang F, et al. NEAT1/miR-181c regulates osteopontin (OPN)-mediated synoviocyte proliferation in osteoarthritis. J Cell Biochem. 2017;118:3775–84. doi: 10.1002/jcb.26025. [DOI] [PubMed] [Google Scholar]
- 29.Song J, Ahn C, Chun CH, Jin EJ. A long non-coding RNA, GAS5, plays a critical role in the regulation of miR-21 during osteoarthritis. J Orthop Res. 2014;32:1628–35. doi: 10.1002/jor.22718. [DOI] [PubMed] [Google Scholar]
- 30.Sun H, Peng G, Ning X, et al. Emerging roles of long noncoding RNA in chondrogenesis, osteogenesis, and osteoarthritis. Am J Transl Res. 2019;11:16–30. [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L, Li Q, Wang J, et al. MiR-29b-3p promotes chondrocyte apoptosis and facilitates the occurrence and development of osteoarthritis by targeting PGRN. J Cell Mol Med. 2017;21:3347–59. doi: 10.1111/jcmm.13237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Etich J, Holzer T, Pitzler L, et al. MiR-26a modulates extracellular matrix homeostasis in cartilage. Matrix Biol. 2015;43:27–34. doi: 10.1016/j.matbio.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 33.Kelly TJ, Souza AL, Clish CB, Puigserver P. A hypoxia-induced positive feedback loop promotes hypoxia-inducible factor 1alpha stability through miR-210 suppression of glycerol-3-phosphate dehydrogenase 1-like. Mol Cell Biol. 2011;31:2696–706. doi: 10.1128/MCB.01242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhai X, Meng R, Li H, et al. miR-181a modulates chondrocyte apoptosis by targeting glycerol-3-phosphate dehydrogenase 1-like protein (GPD1L) in osteoarthritis. Med Sci Monit. 2017;23:1224–31. doi: 10.12659/MSM.899228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson MJ, Jones SW. Review: Long noncoding RNAs in the regulation of inflammatory pathways in rheumatoid arthritis and osteoarthritis. Arthritis Rheumatol. 2016;68:2575–83. doi: 10.1002/art.39759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hutchinson JN, Ensminger AW, Clemson CM, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghafouri-Fard S, Taheri M. Nuclear enriched abundant transcript 1 (NEAT1): A long non-coding RNA with diverse functions in tumorigenesis. Biomed Pharmacother. 2019;111:51–59. doi: 10.1016/j.biopha.2018.12.070. [DOI] [PubMed] [Google Scholar]
- 38.Chen K, Zhu H, Zheng MQ, Dong QR. LncRNA MEG3 inhibits the degradation of the extracellular matrix of chondrocytes in osteoarthritis via targeting miR-93/TGFBR2 axis. Cartilage. 2019 doi: 10.1177/1947603519855759. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao Y, Yan X, Yang Y, Ma X. Downregulation of long noncoding RNA HOTAIRM1 variant 1 contributes to osteoarthritis via regulating miR-125b/BMPR2 axis and activating JNK/MAPK/ERK pathway. Biomed Pharmacother. 2019;109:1569–77. doi: 10.1016/j.biopha.2018.10.181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Primer sequences for RT-PCR analysis.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| miR-181a | 5′-GGAAAGCAGACATTGACCTCAC-3′ | 5′-CCATCCTTTACATCCTTCTGTCTGT-3′ |
| NETA1 | 5′-TCAAAACCTGTTCCCAGGA-3′ | 5′-TGTGCACACTCTATGTGGTAGA-3′ |
| IL-1β | 5′-ATGGCCAAAGTTCGAGACATG-3′ | 5′-TCAAGTCAGTGTTGAGATGATGCTT-3′ |
| IL-6 | 5′-AGCCACTCACCTCTTCAGAAC-3′ | 5′-GCCTCTTTGCTGCTTTCACAC-3′ |
| IL-8 | 5′-TCCAACCTGAGTGACATAGCGA-3′ | 5′-CTGACCTCCAACTCCAACGAAT-3′ |
| COX-2 | 5′-CAGCACTTCACGCATCAGTT-3′ | 5′-CGCAGTTTACGCTGTCTAGC-3′ |
| TNF-α | 5′-TGAAATATACAAGTTATATG-3′ | 5′-GTTCGAGAAGATGATTGATG-3′ |
| MMP13 | 5′-AGCCCGTTTAAAGTGCATGTGTGC-3′ | 5′-GAGTGTCCGAGGAAGATACTTGGT- 3′ |
| U6 | 5′-CTCGCTTCGGCAGCACA-3′ | 5′-AACGCTTCACGAATTTGCGT-3′ |
| GAPDH | 5′-ACAACTTTGGTATCGTGGAAGG-3′ | 5′-GCCATCACGCCACAGTTTC-3′ |