Abstract
Alzheimer’s disease (AD) is highly heritable and recent studies have identified over 20 disease-associated genomic loci. Yet these only explain a small proportion of the genetic variance, indicating that undiscovered loci remain. Here, we performed a large genome-wide association study of clinically diagnosed AD and AD-by-proxy (71,880 cases, 383,378 controls). AD-by-proxy, based on parental diagnoses, showed strong genetic correlation with AD (rg=0.81). Meta-analysis identified 29 risk loci, implicating 215 potential causative genes. Associated genes are strongly expressed in immune-related tissues and cell types (spleen, liver and microglia). Gene-set analyses indicate biological mechanisms involved in lipid-related processes and degradation of amyloid precursor proteins. We show strong genetic correlations with multiple health-related outcomes, and Mendelian randomisation results suggest a protective effect of cognitive ability on AD risk. These results are a step forward in identifying the genetic factors that contribute to AD risk and add novel insights into the neurobiology of AD.
Alzheimer’s disease (AD) is the most frequent neurodegenerative disease with roughly 35 million people affected.1 AD is highly heritable, with estimates ranging between 60 and 80%.2 Genetically, AD can be roughly divided into 2 subgroups: 1) familial early-onset cases that are often explained by rare variants with a strong effect,3 and 2) late-onset cases that are influenced by multiple common variants with low effect sizes.4 Segregation analyses have linked several genes to the first subgroup, including APP5, PSEN16 and PSEN27. The identification of these genes has resulted in valuable insights into a molecular mechanism with an important role in AD pathogenesis, the amyloidogenic pathway,8 exemplifying how gene discovery can add to biological understanding of disease etiology.
Besides the identification of a few rare genetic factors (e.g. TREM29 and ABCA710), genome-wide association studies (GWASs) have mostly discovered common risk variants for the more complex late-onset type of AD. APOE is the strongest genetic risk locus for late-onset AD, responsible for a 3- to 15-fold increase in risk.11 A total of 19 additional GWAS loci have been described using a discovery sample of 17,008 AD cases and 37,154 controls, followed by replication of the implicated loci with 8,572 AD patients and 11,312 controls.4 The currently confirmed AD risk loci explain only a fraction of the heritability of AD and increasing the sample size is likely to boost the power for detection of more common risk variants, which will aid in understanding biological mechanisms involved in the risk for AD.
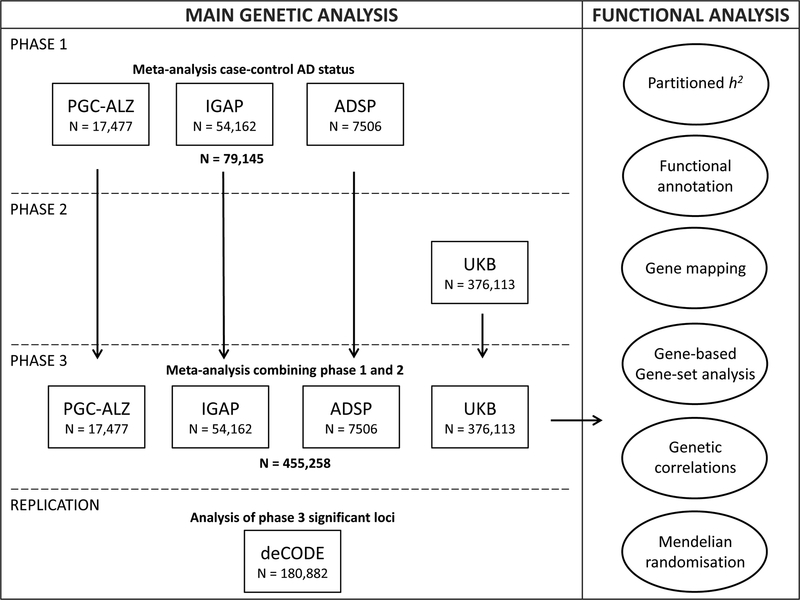
In the current study, we included 455,258 individuals (Nsum) of European ancestry, meta-analysed in 3 phases (Figure 1). Phase 1 consisted of 24,087 clinically diagnosed late-onset AD cases, paired with 55,058 controls. In phase 2, we analysed an AD-by-proxy phenotype, based on individuals in the UK Biobank (UKB) for whom parental AD status was available (N proxy cases=47,793; N proxy controls=328,320). The value of by-proxy phenotypes for GWAS was recently demonstrated by Liu et al.12 for 12 common diseases, including substantial gains in statistical power for AD. The high heritability of AD implies that case status for offspring can be partially inferred from parental case status and that offspring of AD parents are likely to have a higher genetic AD risk load. We thus defined individuals with one or two parents with AD as proxy cases, while upweighting cases with 2 parents. Similarly, the proxy controls include subjects with 2 parents without AD, where older cognitively normal parents were upweighted to account for the higher likelihood that younger parents may still develop AD (see Methods). As the proxy phenotype is not a pure measure of an individual’s AD status and may include individuals that never develop AD, genetic effect sizes will be somewhat underestimated. However, the proxy case-control sample is very large, and therefore substantially increases power to detect genetic effects for AD12, as was also demonstrated in a more recent study using UKB13. Finally, in phase 3, we meta-analysed all individuals of phase 1 and phase 2 together and tested for replication in an independent sample.
Figure 1. Overview of analysis steps.
The main genetic analysis encompasses the procedures to detect GWAS risk loci for AD. The functional analysis includes the in silico functional follow-up procedures with the aim to put the genetic findings in biological context. N=total of individuals within specified dataset.
Results
Genome-wide meta-analysis for AD status
Phase 1 involved a genome-wide meta-analysis for clinically-diagnosed AD case-control status using cohorts collected by 3 independent consortia (Alzheimer’s disease working group of the Psychiatric Genomics Consortium (PGC-ALZ), the International Genomics of Alzheimer’s Project (IGAP), and the Alzheimer’s Disease Sequencing Project (ADSP)), totalling 79,145 individuals (Nsum; effective sample size Neff=72,500) of European ancestry and 9,862,738 genetic variants passing quality control (Figure 1, Supplementary Table 1). The ADSP subset encompassed whole exome sequencing data from 4,343 cases and 3,163 controls, while the remaining datasets consisted of genotype single nucleotide polymorphism (SNP) arrays. For PGC-ALZ and ADSP, raw genotypic data were subjected to a standardized quality control pipeline. GWA analyses were run per cohort and then included in a meta-analysis alongside IGAP, for which only summary statistics were available (see Methods). As described in detail in the Supplementary Note, the phase 1 analysis identified 18 independent loci meeting genome-wide significance (GWS; P<5×10−8), all of which have been identified by previous GWASs (Table 1, Supplementary Figure 1, Supplementary Table 2).
Table 1.
Summary statistics of significantly associated regions identified in the genome-wide association analysis of Alzheimer’s disease (AD) case-control status, AD-by-proxy phenotype and meta-analysis.
| Region | Case-control status (Phase 1) | AD-by-proxy (Phase 2) | Overall (Phase 3) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Chr | Gene | SNP | p | SNP | p | SNP | bp | A1 | A2 | MAF | Z | p | direction |
| 1 | 1 | ADAMTS4 | rs4575098 | 1.57E-04 | rs4575098 | 6.88E-08 | rs4575098 | 161155392 | A | G | 0.240 | 6.36 | 2.05E-10 | ?+++ |
| 2 | 1 | CR1 | rs6656401 | 1.39E-17 | rs679515 | 8.85E-10 | rs2093760 | 207786828 | A | G | 0.205 | 8.82 | 1.10E-18 | ++++ |
| 3 | 2 | BIN1 | rs4663105 | 3.58E-29 | rs4663105 | 5.46E-26 | rs4663105 | 127891427 | C | A | 0.415 | 13.94 | 3.38E-44 | ?+++ |
| 4 | 2 | INPPD5 | rs10933431 | 1.67E-06 | rs10933431 | 2.51E-06 | rs10933431 | 233981912 | G | C | 0.235 | −6.13 | 8.92E-10 | ?--- |
| 5 | 3 | HESX1 | NA | rs184384746 | 1.24E-08 | rs184384746 | 57226150 | T | C | 0.002 | 5.69 | 1.24E-08 | ???+ | |
| 6 | 4 | CLNK | rs6448453 | 0.024 | rs6448451 | 1.19E-08 | rs6448453 | 11026028 | A | G | 0.252 | 6.00 | 1.93E-09 | ?+-+ |
| -- | 4 | HS3ST1 | rs7657553 | 2.16E-08 | rs7657553 | 0.790 | rs7657553 | 11723235 | A | G | 0.291 | 1.95 | 0.051 | ?++- |
| 7 | 6 | HLA-DRB1 | rs9269853 | 2.66E-08 | rs6931277 | 1.78E-07 | rs6931277 | 32583357 | T | A | 0.153 | −6.49 | 8.41E-11 | ?--- |
| 8 | 6 | TREM2 | NA | rs187370608 | 1.45E-16 | rs187370608 | 40942196 | A | G | 0.002 | 8.26 | 1.45E-16 | ???+ | |
| 9 | 6 | CD2AP | rs9381563 | 5.35E-09 | rs9381563 | 8.10E-06 | rs9381563 | 47432637 | C | T | 0.355 | 6.33 | 2.52E-10 | ?+++ |
| 10 | 7 | ZCWPW1 | rs1859788 | 6.05E-09 | rs7384878 | 2.38E-10 | rs1859788 | 99971834 | A | G | 0.310 | −7.93 | 2.22E-15 | ---- |
| 11 | 7 | EPHA1 | rs11763230 | 2.58E-11 | rs7810606 | 1.01E-06 | rs7810606 | 143108158 | T | C | 0.500 | −6.62 | 3.59E-11 | ?--- |
| 12 | 7 | CNTNAP2 | NA | rs114360492 | 2.10E-09 | rs114360492 | 145950029 | T | C | 0.000 | 5.99 | 2.10E-09 | ???+ | |
| 13 | 8 | CLU/PTK2B | rs4236673 | 6.36E-20 | rs1532278 | 7.45E-09 | rs4236673 | 27464929 | A | G | 0.391 | −8.98 | 2.61E-19 | ---- |
| 14 | 10 | ECHDC3 | rs11257242 | 2.38E-08 | rs11257238 | 5.84E-05 | rs11257238 | 11717397 | C | T | 0.375 | 5.69 | 1.26E-08 | ?+++ |
| 15 | 11 | MS4A6A | rs7935829 | 8.21E-13 | rs1582763 | 4.72E-09 | rs2081545 | 59958380 | A | C | 0.381 | −7.97 | 1.55E-15 | ---- |
| 16 | 11 | PICALM | rs10792832 | 1.12E-17 | rs3844143 | 5.31E-11 | rs867611 | 85776544 | G | A | 0.314 | −8.75 | 2.19E-18 | ?--- |
| 17 | 11 | SORL1 | rs11218343 | 5.57E-11 | rs11218343 | 2.81E-06 | rs11218343 | 121435587 | C | T | 0.040 | −6.79 | 1.09E-11 | ?--- |
| 18 | 14 | SLC24A4 | rs12590654 | 1.98E-08 | rs12590654 | 3.70E-06 | rs12590654 | 92938855 | A | G | 0.344 | −6.39 | 1.65E-10 | ?--- |
| 19 | 15 | ADAM10 | rs442495 | 3.09E-04 | rs442495 | 2.65E-07 | rs442495 | 59022615 | C | T | 0.320 | −6.07 | 1.31E-09 | ?--- |
| 20 | 15 | APH1B | rs117618017 | 0.022 | rs117618017 | 2.64E-07 | rs117618017 | 63569902 | T | C | 0.132 | 5.52 | 3.35E-08 | ++++ |
| 21 | 16 | KAT8 | rs59735493 | 8.25E-04 | rs59735493 | 3.72E-06 | rs59735493 | 31133100 | A | G | 0.300 | −5.49 | 3.98E-08 | ?--- |
| 22 | 17 | SCIMP | rs113260531 | 3.21E-06 | rs9916042 | 4.73E-08 | rs113260531 | 5138980 | A | G | 0.120 | 6.12 | 9.16E-10 | ?+++ |
| 23 | 17 | ABI3 | rs28394864 | 7.29E-05 | rs28394864 | 6.80E-06 | rs28394864 | 47450775 | A | G | 0.473 | 5.62 | 1.87E-08 | ?+++ |
| -- | 17 | BZRAP1-AS1 | rs2632516 | 1.42E-09 | rs2632516 | 0.005 | rs2632516 | 56409089 | C | G | 0.455 | −4.90 | 9.66E-07 | ?--- |
| -- | 18 | SUZ12P1 | rs8093731 | 4.63E-08 | rs8093731 | 0.766 | rs8093731 | 29088958 | T | C | 0.010 | −2.17 | 0.030 | ?-?- |
| 24 | 18 | ALPK2 | rs76726049 | 0.039 | rs76726049 | 1.83E-07 | rs76726049 | 56189459 | C | T | 0.014 | 5.52 | 3.30E-08 | ?+++ |
| 25 | 19 | ABCA7 | rs4147929 | 8.64E-09 | rs3752241 | 2.87E-08 | rs111278892 | 1039323 | G | C | 0.161 | 6.50 | 7.93E-11 | ?+++ |
| 26 | 19 | APOE | rs41289512 | 2.70E-194 | rs75627662 | 9.51E-296 | rs41289512 | 45351516 | G | C | 0.039 | 35.50 | 5.79E-276 | ?+++ |
| 27 | 19 | AC074212.3 | rs76320948 | 1.54E-05 | rs76320948 | 1.80E-05 | rs76320948 | 46241841 | T | C | 0.046 | 5.46 | 4.64E-08 | ?+?+ |
| 28 | 19 | CD33 | rs3865444 | 4.25E-08 | rs3865444 | 4.97E-05 | rs3865444 | 51727962 | A | C | 0.320 | −5.81 | 6.34E-09 | ?--- |
| 29 | 20 | CASS4 | rs6014724 | 8.72E-08 | rs6014724 | 6.32E-06 | rs6014724 | 54998544 | G | A | 0.089 | −6.18 | 6.56E-10 | ?--- |
Note: Independent lead SNPs are defined by r2 < 0.1; distinct genomic loci are >250kb apart. The locus column indicates the loci number based on phase 3 (-- indicates that this locus is non-significant). The gene symbols are included to conveniently compare the significant loci with previously discovered loci. The bolded genes correspond to the novel loci indicating the genes in closest proximity to the most significant SNP, while emphasizing this is not necessarily the causal gene. Allele1 is the effect allele for the meta association statistic. The directions of effect of the distinct cohorts are in the following order: ADSP, IGAP, PGC-ALZ, and UKB; note that the first cohort is often missing as this concerns exome sequencing data. Corrected P value for significance=5E-08 (marked as bold and underlined values). Note that the lead SNP can differ between the distinct analyses, while it tags the same locus.
We next (phase 2) performed a GWAS using 376,113 individuals of European ancestry from UKB with parental AD status weighted by age to construct an AD-by-proxy status (Figure 1). Here, we identified 13 independent GWS loci, 8 of which overlapped with phase 1 (Table 1, Supplementary Note). We observed a strong genetic correlation of 0.81 (s.e.m=0.185) between AD status and AD-by-proxy, as well as substantial concordance in the individual SNP effects, as described in the Supplementary Note.
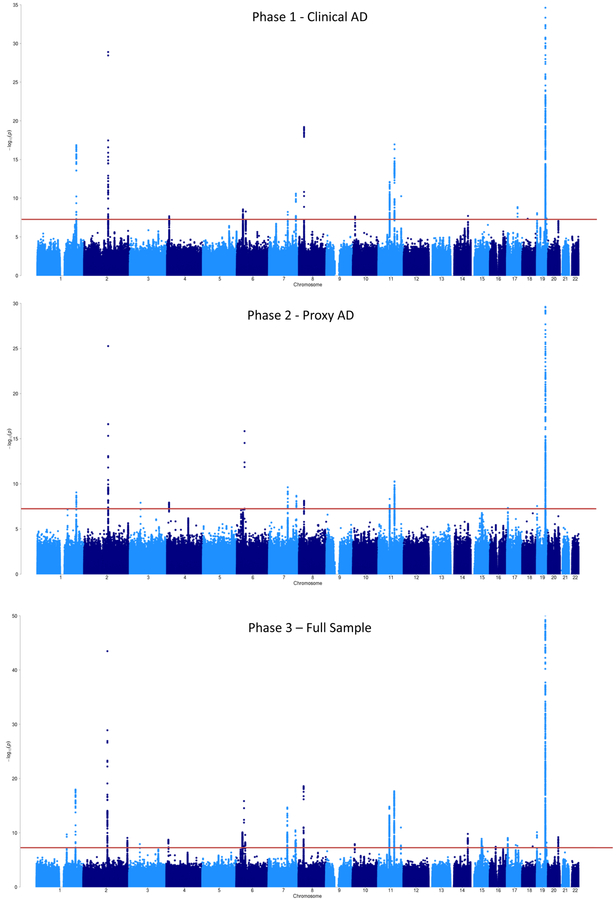
Given the high genetic overlap, in phase 3 we conducted a meta-analysis of the clinical AD GWAS and the AD-by-proxy GWAS (Figure 1), comprising a total sample size of 455,258 (Neff=450,734), including 71,880 (proxy) cases and 383,378 (proxy) controls. The linkage disequilibrium (LD) score intercept14 was 1.0018 (s.e.m=0.0109) and the sample size-adjusted15 λ1000 was 1.044, indicating that most of the inflation in genetic signal (λGC=1.0833) could be explained by polygenicity (Supplementary Figure 1B). There were 2,357 GWS variants, which were represented by 94 lead SNPs, located in 29 distinct loci (Table 1, Figure 2, Supplementary Figure 2). These included 15 of the 18 loci detected in Phase 1, all of the 13 detected in Phase 2, as well as 9 loci that were sub-threshold in both individual analyses but reached significance in the meta-analysis. A large proportion of the lead SNPs (60 of 94) was concentrated in the established APOE risk locus on chromosome 19. This region is known to have a complex LD structure and a very strong effect on AD risk; thus, we consider these SNPs likely to represent a single association signal. Conditional analysis indicated that most loci represented a single fully independent signal, while the TREM2, PTK2B/CLU, and APOE loci contained multiple possible causal signals (Supplementary Note; Supplementary Tables 3–4).
Figure 2. GWAS meta-analysis for AD risk (N=455,258).
Manhattan plot displays all associations per variant ordered according to their genomic position on the x-axis and showing the strength of the association with the −log10 transformed P-values on the y-axis. The y-axis is limited to enable visualization of non-APOE loci. For the Phase 3 meta-analysis, the original −log10 P-value for the APOE locus is 276.
Of the 29 associated loci, 16 overlapped 1 of the 20 genomic regions previously identified by the GWAS of Lambert et al.4, replicating their findings, while 13 were novel. The association signals of five loci (CR1, ZCWPW1, CLU/PTK2B, MS4A6a and APH1B) are partly based on the ADSP exome-sequencing data. Re-analysis of these loci excluding ADSP resulted in similar association signals (Supplementary Table 5), implying that we have correctly adjusted for partial sample overlap between IGAP and ADSP. The lead SNPs in three loci (with nearest genes HESX1, TREM2 and CNTNAP2) were only available in the UKB cohort (Table 1), but were of good quality (INFO>0.91, HWE P>0.19, missingness<0.003). These SNPs were all rare (minor allele frequency (MAF) <0.003), meaning that they will require future confirmation in another similarly large sample. However, variants in TREM2 have been robustly linked to AD in prior research.9
Verifying the 13 novel loci against other recent genetic studies on AD9,16,12,17,18, 4 loci (TREM2, ECHDC3, SCIMP and ABI3) have been previously discovered in addition to the 16 identified by Lambert et al., leaving 9 novel loci at the time of this writing (ADAMTS4, HESX1, CLNK, CNTNAP2, ADAM10, APH1B, KAT8, ALPK2, AC074212.3). The ADAMTS4 and KAT8 loci have also since been identified in a recent analysis in a partially overlapping sample.13 Comparing our meta-analysis results with all loci of Lambert et al.4 to determine differences in associated loci, we were unable to observe 4 loci (MEF2C, NME8, CELF1 and FERMT2) at a GWS level (observed P-values were 1.6×10−5 to 0.0011), which was mostly caused by a lower association signal in the UKB dataset (Supplementary Table 6). By contrast, Lambert et al.4 were unable to replicate the DSG2 and CD33 loci in the second stage of their study. In our study, DSG2 was also not supported (meta-analysis P=0.030; UKB analysis P=0.766), implying invalidation of this locus, while the CD33 locus (rs3865444 in Table 1) was significantly associated with AD (meta-analysis P=6.34×10−9; UKB analysis P=4.97×10−5), implying a genuine genetic association with AD risk.
Next, we aimed to find further support for the novel findings by using an independent Icelandic cohort (deCODE19,20), including 6,593 AD cases and 174,289 controls (Figure 1; Supplementary Table 7) to test replication of the lead SNP or an LD-proxy of the lead SNP (r2>0.9) in each locus. We were unable to test two loci as the lead SNPs (and SNPs in high LD) either were not present in the Icelandic reference panel or were not imputed with sufficient quality. For 6 of the 7 novel loci tested for replication, we observed the same direction of effect in the deCODE cohort. Furthermore, 4 loci (CLNK, ADAM10, APH1B, AC074212.3) showed nominally significant association results (P<0.05) for the same SNP or a SNP in high LD (r2>0.9) within the same locus (two-tailed binomial test P=1.9×10−4). The locus on chromosome 1 (ADAMTS4) was very close to significance (P=0.053), implying stronger evidence for replication than for non-replication. Apart from the novel loci, we also observed sign concordance for 96.3% of the top (per-locus) lead SNPs in all loci from the meta-analysis (two-tailed binomial test P=4.17×10−7) that were available in deCODE (26 of 27).
As an additional method of testing for replication, we used genome-wide polygenic score prediction in two independent samples.21 The current results explain 7.1% of the variance in clinical AD at a low best fitting P-threshold of 1.69×10−5 in 761 individuals with case-control diagnoses (P=1.80×10−10). When excluding the APOE locus (chr19: 45020859–45844508), the results explain 3.9% of the variance with a best fitting P-threshold of 3.5×10−5 (P=1.90×10−6). We also predict AD status in a sample of 1,459 pathologically confirmed cases and controls22 with an R2=0.41 and an area under the curve (AUC) of 0.827 (95% confidence interval (95% CI): 0.805–0.849, P=9.71×10−70) using the best-fitting model of SNPs with a GWAS P<0.50, as well as R2=0.23 and AUC=0.733 (95% CI: 0.706–0.758, P=1.16×10−45) using only APOE SNPs. This validation sample contains a small number of individuals overlapping with IGAP; previous simulations with this sample have indicated that this overfitting increases the margin of error of the estimate approximately 2–3%.22 This sample, however, represented severe, late-stage AD cases contrasted with supernormal controls, so the polygenic prediction may be higher than expected for typical case-control or population samples.
Functional interpretation of genetic variants
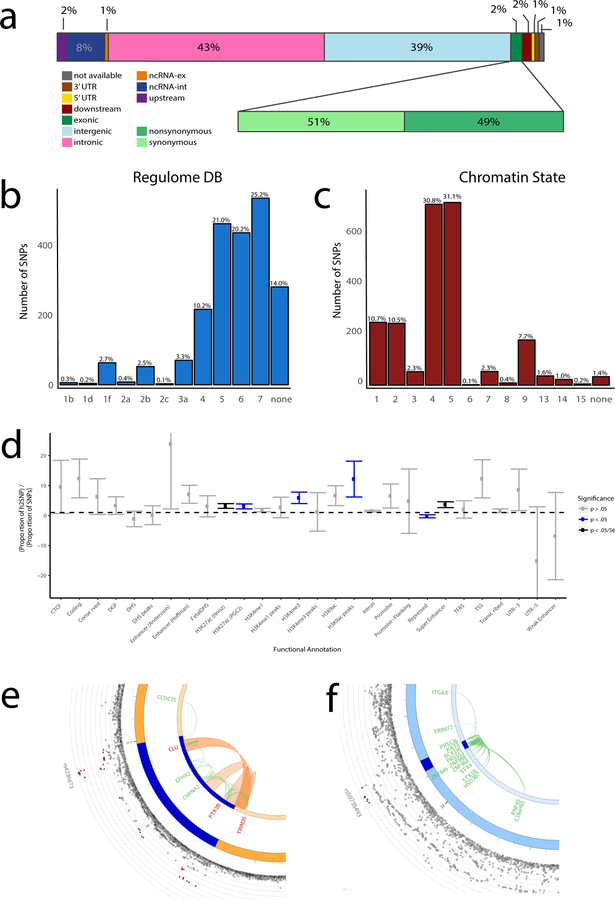
Functional annotation of all GWS SNPs (n=2,357) in the associated loci showed that SNPs were mostly located in intronic/intergenic areas, but also in regions that were enriched for chromatin states 4 and 5, implying effects on active transcription (Figure 3; Supplementary Table 8). Twenty-five GWS SNPs were exonic non-synonymous (Figure 3A; Supplementary Table 9) with likely deleterious impacts on gene function. Converging evidence of strong association (Z>|7|) and a high observed probability of a deleterious variant effect (CADD23 score≥30) was found for rs75932628 (TREM2), rs142412517 (TOMM40) and rs7412 (APOE). The first two missense mutations are rare (MAF=0.002 and 0.001, respectively) and the alternative alleles were associated with higher risk for AD. The latter APOE missense mutation is the well-established protective allele Apoε2. Supplementary Tables 8 and 9 present a detailed annotation catalogue of variants in the associated genomic loci. We also applied a fine-mapping model24 to identify credible sets of causal SNPs from the identified GWS variants (Supplementary Table 8). The proportion of plausible causal SNPs varied drastically between loci; for example, 30 out of 854 SNPs were selected in the APOE locus (no. 26), while 345 out of 434 SNPs were nominated in the HLA-DRB1 locus (no. 7). Credible causal SNPs were not limited to known functional categories such as ExNS, indicating more complicated causal pathways that merit investigation with the set of variants prioritized by these statistical and functional annotations.
Figure 3. Functional annotation of GWAS results.
a) Functional effects of variants in genomic risk loci of the meta-analysis (the colours of the legend are ordered from right to left in the figure) – the second bar shows distribution for exonic variants only; b) Distribution of RegulomeDB score for variants in genomic risk loci, with a low score indicating a higher probability of having a regulatory function (see Methods); c) Distribution of minimum chromatin state across 127 tissue and cell types for variants in genomic risk loci, with lower states indicating higher accessibility (see Methods); d) Heritability enrichment of 28 functional variant annotations calculated with stratified LD score regression (bars represent standard errors). UTR=untranslated region; CTCF=CCCTC-binding factor; DHS=DNaseI Hypersensitive Site; TFBS=transcription factor binding site; DGF=DNAaseI digital genomic footprint; e) Zoomed-in circos plot of chromosome 8; f) Zoomed-in circos plot of chromosome 16. Circos plots show implicated genes by significant loci, where dark blue areas indicate genomic risk loci, green lines indicate eQTL associations and orange lines indicate chromatin interactions. Genes mapped by both eQTL and chromatin interactions are in red. The outer layer shows a Manhattan plot containing the negative log10-transformed P-value of each SNP in the GWAS meta-analysis of AD. Full circos plots of all autosomal chromosomes are provided in Supplementary Figure 4.
Partitioned heritability analysis,25 excluding SNPs with extremely large effect sizes (that is, APOE variants) showed enrichment for the SNP-heritability (h2SNP) for variants located in H3K27ac marks (enrichment=3.18, P=9.63×10−5), which are associated with activation of transcription, and in super enhancers (enrichment=3.62, P=2.28×10−4), which are genomic regions where multiple epigenetic marks of active transcription are clustered (Figure 3D; Supplementary Table 10). Heritability was also enriched in variants on chromosome 17 (enrichment=3.61, P=1.63×10−4) and we observed a trend of enrichment for heritability in common rather than rarer variants (Supplementary Figure 3; Supplementary Tables 11 and 12). Although a large proportion (23.9%) of the heritability can be explained by SNPs on chromosome 19, this enrichment is not significant, due to the large standard errors around this estimate (Supplementary Table 11). Overall these results suggest that, despite some nonsynonymous variants contributing to AD risk, most of the GWS SNPs are located in non-coding regions and are enriched for regions that have an activating effect on transcription.
Implicated genes
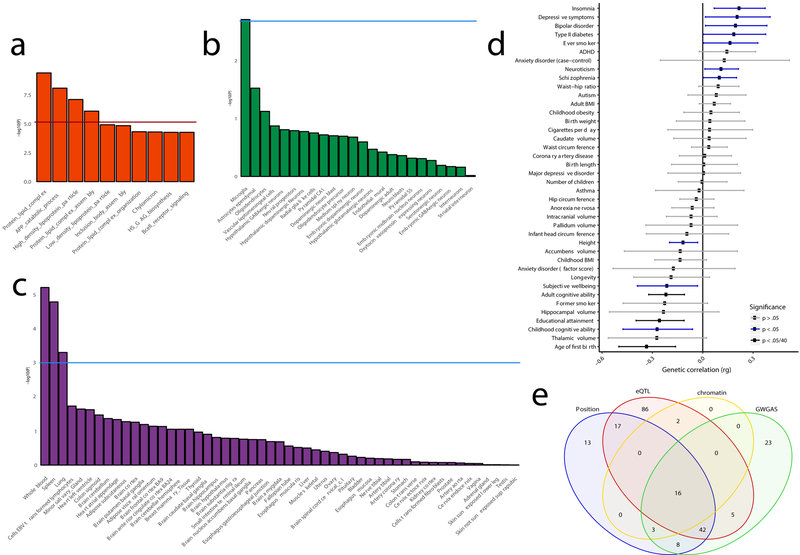
To link the associated variants to genes, we applied three gene-mapping strategies implemented in Functional Mapping and Annotation (FUMA)26 (see Methods). We used all SNPs with a P-value<5×10−8 for gene-mapping. Positional gene-mapping aligned SNPs to 99 genes by their location within or immediately up/downstream (±10 kilobases (kb)) of known gene boundaries, eQTL (expression quantitative trait loci) gene-mapping matched cis-eQTL SNPs to 168 genes whose expression levels they influence in one or more tissues, and chromatin interaction mapping linked SNPs to 21 genes based on three-dimensional DNA-DNA interactions between each SNP’s genomic region and nearby or distant genes, which we limited to include only interactions between annotated enhancer and promoter regions (Supplementary Figure 4; Supplementary Tables 13 and 14). This resulted in 192 uniquely mapped genes, 80 of which were implicated by at least two mapping strategies and 16 by all 3 (Figure 4E).
Figure 4. Functional implications based on gene-set analysis, genetic correlations and functional annotations.
The gene-set results are displayed per category of biological mechanisms (a), brain cell-types (b) and tissue types (c). The red horizontal line indicates the significance threshold corrected for all gene-set tests of all categories, while the blue horizontal lines display the significance threshold corrected only for the number of tests within the three categories (that is, gene-ontology, tissue expression or single cell expression); d) Genetic correlations between AD and other heritable traits (bars represent 95% confidence intervals); e) Venn diagram showing the number of genes mapped by four distinct strategies. ADHD=attention deficit hyperactivity disorder; BMI=body mass index; EBV=Epstein-Barr virus.
Of special interest is the locus on chromosome 8 (CLU/PTK2B). In the GWAS by Lambert et al.4, this locus was defined as 2 distinct loci (CLU and PTK2B). Although our conditional analysis based on genetic data also specified this locus as having at least 2 independent association signals (Supplementary Table 4), the chromatin interaction data in two immune-related tissues – the spleen and liver (Supplementary Table 14) – suggests that the genomic regions indexed by PTK2B and CLU loci might physically interact (Figure 3E), therefore putatively affecting AD pathogenesis via the same biological mechanism. The patterns of tissue-specific gene expression are largely dissimilar between CLU and PTK2B, although both are expressed relatively highly in the brain and lymph nodes.27 Future studies should thus consider the joint effects of how these two genes simultaneously impact AD risk.
Eight genes (HLA-DRB5, HLA-DRB1, HLA-DQA, HLA-DQB1, KAT8, PRSS36, ZNF232 and CEACAM19) are particularly notable as they are implicated via eQTL association in the hippocampus, a brain region highly affected early in AD pathogenesis (Supplementary Table 13). Chromosome 16 contains a locus implicated by long-range eQTL association (Figure 3F), clearly illustrating how the more distant genes C16orf93, RNF40 and ITGAX can be affected by a genetic factor (rs59735493) in various body tissues (for example, blood and skin), including a change in expression for RNF40 observed in the dorsolateral prefrontal cortex. These observations emphasize the relevance of considering putative causal genes or regulatory elements not solely on the physical location but also on epigenetic influences. As detailed in the Supplementary Note, eQTLs were overrepresented in the risk loci and a number of quantitative trait locus (QTL) associations (including eQTLs, methylation quantitative trait loci (mQTLs), and histone acetylation quantitative trait loci (haQTLs)) were identified in relevant brain regions, providing interesting targets for future functional follow-up and biological interpretation (Supplementary Tables 15–17).
Although these gene-mapping strategies imply multiple putative causal genes per GWAS locus, several genes are of particular interest, as they have functional or previous genetic association with AD. For locus 1 in Supplementary Table 13, ADAMTS4 encodes a protein of the ADAMTS family which has a function in neuroplasticity and has been extensively studied for its role in AD pathogenesis.28 For locus 19, the obvious most likely causal gene is ADAM10, as this gene has been associated with AD by research focusing on rare coding variants in ADAM10.29 However, this is the first time that this gene is implicated as a common risk factor for AD, and is supported by the putative causal molecular mechanism observed in dorsolateral prefrontal cortex eQTL and mQTL data (Supplementary Tables 15 and 16) for multiple common SNPs in LD. The lead SNP for locus 20 is a nonsynonymous variant in exon 1 of APH1B, which encodes for a protein subunit of the γ-secretase complex cleaving APP.30 A highly promising candidate gene for locus 21 is KAT8, as the lead SNP of this locus is located within the third intron of KAT8, and multiple significant variants within this locus influence the expression or methylation levels of KAT8 in multiple brain regions (Supplementary Tables 13 and 16) including hippocampus. The chromatin modifier KAT8 is regulated by KANSL1, a gene associated with AD in absence of Apoɛ4. A study on Parkinson’s disease reported KAT8 as potential causal gene based on GWAS and differential gene expression results, implying a putative shared role in neurodegeneration of KAT8 in AD and Parkinson’s disease.31 Although previously reported functional information on genes can be of great value, it is preferable to consider all implicated genes as putative causal factors to guide potential functional follow-up experiments.
We next performed genome-wide gene-based association analysis (GWGAS) using Multi-marker Analysis of GenoMic Annotation (MAGMA).32 This method annotates SNPs to known protein-coding genes to estimate aggregate associations based on all SNPs in a gene. It differs from FUMA as it provides a statistical gene-based test, whereas FUMA maps individually significant SNPs to genes. With GWGAS, we identified 97 genes that were significantly associated with AD (Supplementary Figure 5; Supplementary Table 18), of which 74 were also mapped by FUMA (Figure 4E). In total, 16 genes were implicated by all four strategies (Supplementary Table 19), of which 7 genes (HLA-DRA, HLA-DRB1, PTK2B, CLU, MS4A3, SCIMP and RABEP1) are not located in the APOE-locus, and therefore of high interest for further investigation.
Gene-sets implicated in AD and AD-by-proxy
Using the gene-based P-values, we performed gene-set analysis for curated biological pathways and tissue/single-cell expression. Four Gene Ontology (GO)33 gene-sets were significantly associated with AD risk: Protein lipid complex (P=3.93×10−10), Regulation of amyloid precursor protein catabolic process (P=8.16×10−9), High density lipoprotein particle (P=7.81×10−8), and Protein lipid complex assembly (P=7.96×10−7) (Figure 4A; Supplementary Tables 20 and 21). Conditional analysis on the APOE locus showed associations with AD for these four gene-sets to be independent of the effect of APOE, though part of the association signal was also attributable to APOE. All 25 genes of the High density lipoprotein particle pathway are also part of the Protein lipid complex; conditional analysis showed that these gene-sets are not interpretable as independent associations (P=0.18), but the other three sets are independently significant (Supplementary Table 20).
Linking gene-based P-values to tissue- and cell-type-specific gene-sets, no association survived the stringent Bonferroni correction, which corrected for all tested gene-sets (that is, 6,994 GO categories, 53 tissues and 39 cell types). However, we did observe suggestive associations across immune-related tissues when correcting only for the number of tests within all tissue types or cell-types (Figure 4C; Supplementary Table 22), particularly whole blood (P=5.61×10−6), spleen (P=1.50×10−5) and lung (P=4.67×10−4), which were independent from the APOE locus. In brain single-cell expression gene-set analyses, we found association for microglia in the mouse-based expression dataset (P=1.96×10−3), though not surviving the stringent Bonferroni correction (Figure 4B; Supplementary Table 23). However, we observed a similar association signal for microglia in a second independent single-cell expression dataset in humans (P=2.56×10−3) (Supplementary Figure 6; Supplementary Table 24). As anticipated, both microglia signals are partly depending on APOE, though a large part is independent (Supplementary Tables 23 and 24).
Cross-trait genetic influences
As described in the Supplementary Note and Supplementary Tables 25–26, we observed that the genetic influences on AD overlapped with a number of other diseases and psychological traits including cognitive ability and educational attainment, replicating previous studies.34,35 To extend these findings, we used Generalised Summary-statistic-based Mendelian Randomisation36 (GSMR) to test for potential credible causal associations of genetically correlated outcomes which may directly influence the risk for AD. Due to the nature of AD being a late-onset disorder and summary statistics for most other traits being obtained from younger samples, we do not report tests for the opposite direction of potential causality (that is, we did not test for a causal effect of a late-onset disease on an early-onset disease). In this set of analyses, SNPs from the summary statistics of genetically correlated phenotypes were used as instrumental variables to estimate the putative causal effect of these “exposure” phenotypes on AD risk by comparing the ratio of SNPs’ associations with each exposure to their associations with AD outcome (see Methods). Association statistics were standardized, such that the reported effects reflect the expected difference in odds ratio (OR) for AD as a function of every SD increase in the exposure phenotype. We observed a protective effect of cognitive ability (OR=0.89, 95% CI: 0.85–0.92, P=5.07×10−9), educational attainment (OR=0.88, 95%CI: 0.81–0.94, P=3.94×10−4), and height (OR=0.96, 95%CI: 0.94–0.97, P=1.84×10−8) on risk for AD (Supplementary Table 27; Supplementary Figure 7). No substantial evidence of pleiotropy was observed between AD and these phenotypes, with <1% of overlapping SNPs being filtered as outliers (Supplementary Table 27).
Discussion
By using an unconventional approach of including a proxy phenotype for AD to increase sample size, we have identified nine novel loci and gained novel biological knowledge on AD etiology. We were able to test seven of the nine novel loci for replication, of which four loci showed clear replication, one locus showed marginal replication and two loci were not replicated at this moment. Both the high genetic correlation between the standard case-control status and the UKB by proxy phenotype (rg=0.81) and the high rate of novel loci replication in the independent deCODE cohort suggest that this strategy is robust. Through in silico functional follow-up analysis, and in line with previous research,18,37 we emphasise the crucial causal role of the immune system - rather than immune response as a consequence of disease pathology - by establishing variant enrichments for immune-related body tissues (whole blood, spleen, liver) and for the main immune cells of the brain (microglia). Of note, the enrichment observed for liver could alternatively indicate the genetic involvement of the lipid system in AD pathogenesis.38 Furthermore, we observe informative eQTL associations and chromatin interactions within immune-related tissues for the identified genomic risk loci. Together with the AD-associated genetic effects on lipid metabolism in our study, these biological implications (which are based on genetic signals and unbiased by prior biological beliefs) strengthen the hypothesis that AD pathogenesis involves an interplay between inflammation and lipids, as lipid changes might harm immune responses of microglia and astrocytes, and vascular health of the brain.39
In accordance with previous clinical research, our study suggests an important role for protective effects of several human traits on AD. Cognitive reserve has been proposed as a protective mechanism in which the brain aims to control brain damage with prior existing cognitive processing strategies.40 Our findings imply that some component of the genetic factors for AD might affect cognitive reserve, rather than being involved in AD-pathology-related damaging processes, influencing AD pathogenesis in an indirect way through cognitive reserve. Furthermore, a large-scale community-based study observed that AD incidence rates declined over decades, which was specific for individuals with at minimum a high school diploma.41 Combined with our Mendelian randomisation results for educational attainment, this suggests that the protective effect of educational attainment on AD is influenced by genetics. Similarly, the observed positive effects of height could be a result of the genetic overlap between height and intracranial volume42,43, a measure associated to decreased risk of AD.44 This indirect association is furthermore supported by the observed increase in cognitive reserve for taller individuals.45 Alternatively, genetic variants influencing height might also affect biological mechanisms involved in AD aetiology, such as IGF1 that codes for the insulin-like growth factor and is associated with cerebral amyloid.46
The results of this study could furthermore serve as a valuable resource for selection of promising genes for functional follow-up experiments and identify targets for drug development and stratification approaches. We anticipate that functional interpretation strategies and follow-up experiments will result in a comprehensive understanding of late-onset AD aetiology, which will serve as a solid foundation for improvement of AD therapy.
URLs
UK Biobank: http://ukbiobank.ac.uk
Database of Genotypes and Phenotypes (dbGaP): https://www.ncbi.nlm.nih.gov/gap
Functional Mapping and Annotation (FUMA) software: http://fuma.ctglab.nl
Multi-marker Analysis of GenoMic Annotation (MAGMA) software: http://ctg.cncr.nl/software/magma
mvGWAMA and effective sample size calculation: https://github.com/Kyoko-wtnb/mvGWAMA
LD Score Regression software: https://github.com/bulik/ldsc
LD Hub (GWAS summary statistics): http://ldsc.broadinstitute.org/
LD scores: https://data.broadinstitute.org/alkesgroup/LDSCORE/
Psychiatric Genomics Consortium (GWAS summary statistics): http://www.med.unc.edu/pgc/results-and-downloads
MSigDB curated gene-set database: http://software.broadinstitute.org/gsea/msigdb/collections.jsp
NHGRI GWAS catalog: https://www.ebi.ac.uk/gwas/
Generalised Summary-data-based Mendelian Randomisation software: http://cnsgenomics.com/software/gsmr/
Credible SNP set analysis software: https://github.com/hailianghuang/FM-summary
Online Methods
Participants
Participants in this study were obtained from multiple sources, including raw data from case-control samples collected by the Psychiatric Genomics Consortium (PGC-ALZ) and the Alzheimer’s Disease Sequencing Project (ADSP; made publicly available through dbGaP [see URLs]), summary data from the case-control samples in the International Genomics of Alzheimer’s Project (IGAP), and raw data from the population-based UK Biobank (UKB) sample which was used to create a weighted AD-proxy phenotype. An additional independent case-control sample (deCODE) was used for replication. Full descriptions of the samples and their respective phenotyping and genotyping procedures are provided in the Supplementary Note and the Life Sciences Reporting Summary.
Data Analysis
Single-marker association analysis
Genome-wide association analysis (GWAS) for each of the ADSP, PGC-ALZ and UKB datasets was performed in PLINK47, using logistic regression for dichotomous phenotypes (cases versus controls for ADSP and PGC-ALZ cohorts), and linear regression for phenotypes analysed as continuous outcomes (proxy phenotype constructed as the number of parents with AD for UKB cohort). For the ADSP and PGC-ALZ cohorts, association tests were adjusted for gender, batch (if applicable), and the first 4 ancestry principal components. Twenty principal components were calculated, and depending on the dataset being tested, additional principal components (on top of the standard of 4) were added if significantly associated to the phenotype. Furthermore, for the PGC-ALZ cohorts age was included as a covariate. For 4,537 controls of the DemGene cohort (subset of PGC-ALZ), no detailed age information was available, besides the age range the subjects were in (20–45 years). We therefore set the age of these individuals conservatively to 20 years. For the ADSP dataset, age was not included as a covariate due to the enrichment for older controls (mean age cases = 73.1 years (s.e.m.=7.8); mean age controls = 86.1 years (s.e.m.=4.5)) in their collection procedures. Correcting for age in ADSP would remove a substantial part of genuine association signals (e.g. well-established APOE locus rs11556505 is strongly associated to AD (P=1.08×10−99), which is lost when correcting for age (P=0.0054). For the UKB dataset, 12 ancestry principal components were included as covariates, as well as age, sex, genotyping array, and assessment centre. We used the genome-wide threshold for significance of P<5×10−8).
Multivariate genome-wide meta-analysis
Two meta-analyses were performed, including: phase 1) cohorts with case-control phenotypes (IGAP, ADSP and PGC-ALZ datasets), and phase 3) all cohorts, also including the UKB proxy phenotype.
Because of partial overlap between cohorts, the per SNP test statistics was defined by
where wi and Zi are the squared root of the sample size and the test statistics of SNP k in cohort i, respectively. CTI is the cross-trait LD score intercept estimated by LDSC14,48 using genome-wide summary statistics. This is equal to48
where Ni and Nj are the sample sizes of cohorts i and j and Nsij the number of samples overlapping between them, and ρij the phenotypic correlation between the measures used in the two cohorts for the overlapping samples. Under the null hypothesis of no association any correlation between Zi and Zj is determined only by that phenotypic correlation, scaled by the relative degree of overlap. As such, this correlation can be estimated by the CTI.
The test statistics per SNP per GWAS were converted from the P-value, incorporating the sign of either beta or odds ratio. When direction is aligned the conversion is two-sided. To avoid infinite values, we replaced P-value 1 with 0.999999 and P-value < 1e-323 to 1e-323 (the minimum >0 value in Python). The script for the multivariate GWAS is available online (see URLs).
Effective sample size
The effective sample size (Neff) is computed for each SNP k from the matrix M, containing the sample size Ni of each cohort i on the diagonal and the estimated number of shared data points for each pair of cohorts i and j as the off-diagonal values. A recursive approach is used to compute Neff. Going from the first cohort to the last the (remaining) size of the current cohort is added to the total Neff. Then for each remaining other cohort it overlaps with, the size of those other cohorts is reduced by the expected number of samples shared by the current cohort; overlap between the remaining cohorts is similarly adjusted. This process ensures that each overlapping data point is counted only once in Neff.
The computation proceeds as follows. Starting with the first cohort in M, Neff is first increased by M1,1, corresponding to the sample size of that cohort. The proportion of samples shared between cohort 1 and each other cohort j is then computed as p1,j = M1,j/Mj,j, and M is adjusted to remove this overlap, multiplying all values in each column j by 1-p1,j. This amounts to reducing the sample size of each other cohort j by the number of samples it shares with cohort 1 and reducing the shared samples between cohort j and subsequent cohorts by the same proportion. After this, the first row and column of M are discarded, and the same process is applied to the new M matrix. This is repeated until M is empty.
The effective sample size is used as a parameter in the MAGMA analysis (see Methods “Gene based-analysis”) and reported in the main text as the combined sample sizes for the meta-analysis. We use the term Nsum to indicate the total number of individuals when simply summing them over the distinct cohorts. The script for the Neff computation is available online (see URLs).
Genomic risk loci definition
We used FUMA26 v1.2.8, an online platform for functional mapping and annotation of genetic variants, to define genomic risk loci and obtain functional information of relevant SNPs in these loci. We first identified independent significant SNPs that have a genome-wide significant P-value (<5×10−8) and are independent from each other at r2<0.6. These SNPs were further represented by lead SNPs, which are a subset of the independent significant SNPs that are in approximate linkage equilibrium with each other at r2>0.6. We then defined associated genomic risk loci by merging any physically overlapping lead SNPs (LD blocks <250kb apart). LD information was calculated using the UK Biobank genotype data as a reference.
For GWS SNPs in the defined risk loci, we applied a summary statistic-based fine-mapping model to identify credible causal SNPs within each locus, as previously described24. This Bayesian model estimates a per-SNP posterior probability of a true disease association using maximum likelihood estimation and the steepest descent approach, creating a set of SNPs in each locus that contains the causal SNP in 99% of cases, given that the causal variants are among the genotyped/imputed SNPs. The software used, FM-summary, is available online (see URLs).
Independent sample replication
For novel SNPs identified in the phase 3 meta-analysis, replication was tested in the independent deCODE sample using logistic regression with Alzheimer’s disease status as the response and genotype counts and a set of nuisance variables including sex, county of birth, and current age as predictors.20 Correction for inflation of test statistics due to relatedness and population stratification in this Icelandic cohort was performed using the intercept estimate (1.29) from LD score regression14.
Conditional analysis
We performed conditional analysis with GCTA-COJO49 to assess the independence of association signals, either within or between GWAS risk loci. COJO enables conditional analysis of GWAS summary statistics without individual-level genotype data. We therefore performed conditional analysis on the phase 3 summary statistics, using 10,000 randomly selected unrelated samples from the UKB dataset as a reference dataset to determine LD-patterns. Conditional analysis was run per chromosome or per locus with the default settings of the software.
Heritability and genetic correlation
LD score regression14 was used to estimate clinical AD heritability and to calculate genetic correlations48 between the case-control and proxy phenotypes using summary statistics. Pre-calculated LD scores from the 1000 Genomes European reference population were obtained online (see URLs). Liability heritability was calculated with a population prevalence of 0.0431 (the population prevalence of age group 70–75 in the Western European population, resembling the average age of onset of 74.5 for the clinical case group) and a sample prevalence of 0.304. The genetic correlation was calculated on HapMap3 SNPs only to ensure high quality LD score calculation.
Stratified heritability
To test whether specific categories of SNP annotations were enriched for heritability, we partitioned the SNP heritability for binary annotations using stratified LD score regression14. Heritability enrichment was calculated as the proportion of heritability explained by a SNP category divided by the proportion of SNPs that are in that category. Partitioned heritability was computed by 28 functional annotation categories, by minor allele frequency (MAF) in six percentile bins, and by 22 chromosomes. Annotations for binary categories of functional genomic characteristics (for example, coding or regulatory regions) were obtained online (see URLs). The Bonferroni-corrected significance threshold for 56 annotations was set at: P<0.05/56=8.93×10−4.
Polygenic risk scoring
We calculated polygenic scores (PGS) using two independent genotype datasets. First, 761 individuals (379 cases and 382 controls) from the ADDNeuroMed study50 were included, using the same QC and imputation approach as for the other datasets with genotype-level data (see Supplementary Note). Second, 1459 individuals (912 severe, late-stage cases and 547 age-matched controls with little to no cognitive dysfunction) from the TGEN study22 were assessed and their diagnostic status was confirmed via post-mortem neuropathology. Imputed SNPs in this sample were filtered based on INFO score>0.9 and MAF>0.01. PGS were created using PLINK47 for the TGEN dataset and PRSice21 for the ADDNeuroMed dataset. In both samples, PGS were calculated on hard-called imputed genotypes using P-value thresholds from 0.0 to 0.5 and using PLINK’s clumping procedure to prune for LD. Clumping was based on the effect size estimates of SNPs originating from the phase 3 meta-analysis for the ADDNeuroMed sample. For TGEN, clumping was previously performed using the IGAP summary statistics; these clumped SNPs were filtered for overlap with the phase 3 SNPs. PGS were calculated in both samples using the SNP effect size estimates from the phase 3 meta-analysis. The explained variance (ΔR2) was derived from a linear model in which the AD phenotype was regressed on each PGS while controlling for GWAS covariates, compared to a linear model with covariates only. In the TGEN dataset, sensitivity, specificity, and area under the curve (AUC) of predicting confirmed case/control status were calculated, using the R package pROC51 and bootstrapped confidence intervals. Of note, approximately 3% of the TGEN sample overlapped with the IGAP cohort included in the meta-analysis; previous simulation work using PGS in this sample has shown that this overfitting leads to only a modest increase (2–3%) in the margin of error around the AUC estimate.22
Functional annotation
Functional annotation of GWS SNPs implicated in the meta-analysis was performed using FUMA26 v1.2.8. Functional consequences for these SNPs were obtained by matching SNPs to databases containing known functional annotations, including ANNOVAR52 categories, Combined Annotation Dependent Depletion (CADD) scores23, RegulomeDB53 (RDB) scores, and chromatin states54,55. ANNOVAR annotates the functional consequence of SNPs on genes (for example, intron, exon, intergenic). CADD scores predict how deleterious the effect of a SNP with higher scores referring to higher deleteriousness. A CADD score above 12.37 is the threshold to be potentially pathogenic56. The RegulomeDB score is a categorical score based on information from expression quantitative trait loci (eQTLs) and chromatin marks, ranging from 1a to 7 with lower scores indicating an increased likelihood of having a regulatory function. The chromatin state represents the accessibility of genomic regions (every 200bp) with 15 categorical states predicted by a hidden Markov model based on 5 chromatin marks in the Roadmap Epigenomics Project.55 A lower state indicates higher accessibility, with states 1–7 referring to open chromatin states. We annotated the minimum chromatin state across tissues to SNPs. A legend describing the RegulomeDB and chromatin state scores can be found in the Supplementary Note.
Gene-mapping
Genome-wide significant loci obtained by GWAS were mapped to genes in FUMA26 using three strategies:
Positional mapping maps SNPs to genes based on physical distance (within a 10kb window) from known protein coding genes in the human reference assembly (GRCh37/hg19).
eQTL mapping maps SNPs to genes with which they show a significant eQTL association (i.e. allelic variation at the SNP is associated with the expression level of that gene). eQTL mapping uses information from 45 tissue types in 3 data repositories (GTEx57 v6, Blood eQTL browser58, BIOS QTL browser59), and is based on cis-eQTLs which can map SNPs to genes up to 1Mb apart. We used a false discovery rate (FDR) of 0.05 to define significant eQTL associations.
Chromatin interaction mapping was performed to map SNPs to genes when there is a three-dimensional DNA-DNA interaction between the SNP region and another gene region. Chromatin interaction mapping can involve long-range interactions as it does not have a distance boundary. FUMA currently contains Hi-C data of 14 tissue types from the study of Schmitt et al60. Since chromatin interactions are often defined in a certain resolution, such as 40kb, an interacting region can span multiple genes. If a SNP is located in a region that interacts with a region containing multiple genes, it will be mapped to each of those genes. To further prioritize candidate genes, we selected only genes mapped by chromatin interaction in which one region involved in the interaction overlaps with a predicted enhancer region in any of the 111 tissue/cell types from the Roadmap Epigenomics Project55 and the other region is located in a gene promoter region (250bp up- and 500bp downstream of the transcription start site and also predicted by Roadmap to be a promoter region). This method reduces the number of genes mapped but increases the likelihood that those identified will have a plausible biological function. We used an false discovery rate of 1×10−5 to define significant interactions, based on previous recommendations60 modified to account for the differences in cell lines used here.
Brain-specific QTL annotation
As AD is characterized by neurodegeneration, we annotated the significant genomic loci with publicly available databases of expression, methylation, and histone acetylation QTLs, as catalogued in BRAINEAC61, CommonMind Consortium Portal62 and xQTL Serve63, as an extension of the GTEx tissue eQTL mapping performed in FUMA. Descriptions of these brain eQTL databases and settings we used are in the Supplementary Note.
Gene-based analysis
To account for the distinct types of genetic data in this study, genotype array (PGC-ALZ, IGAP, UKB) and whole-exome sequencing data (ADSP), we first performed two gene-based genome-wide association analysis (GWGASs) using MAGMA32, followed by a meta-analysis. SNP-based P-values from the meta-analysis of the 3 genotype-array-based datasets were used as input for the first GWGAS, while the unimputed individual-level sequence data of ADSP was used as input for the second GWGAS. A total of 18,233 protein-coding genes (each containing at least one SNP in the GWAS) from the National Center for Biotechnology Information (NCBI) 37.3 gene definitions were used as basis for GWGAS in MAGMA. Bonferroni correction was applied to correct for multiple testing (P<2.74×10−6).
Gene-set analysis
Results from the GWGAS analyses were used to test for association in 7,086 predefined gene-sets of four categories:
1. 6,994 curated gene-sets representing known biological and metabolic pathways derived from Gene Ontology (5917 gene-sets), Biocarta (217 gene-sets), KEGG (186 gene-sets), Reactome (674 gene-sets) catalogued by and obtained from the MsigDB version 6.164 (see URLs)
Gene expression values from 53 tissues obtained from GTEx57, log2 transformed with pseudocount 1 after winsorization at 50 and averaged per tissue.
Cell-type specific expression in 24 broad categories of brain cell types, which were calculated following the method described in ref.37. Briefly, brain cell-type expression data was drawn from single-cell RNA sequencing data from mouse brains. For each gene, the value for each cell-type was calculated by dividing the mean Unique Molecular Identifier (UMI) counts for the given cell type by the summed mean UMI counts across all cell types. Single-cell gene-sets were derived by grouping genes into 40 equal bins based on specificity of expression.
Nucleus specific gene expression of 15 distinct human brain cell-types from the study described in65. The value for each cell-type was calculated as in point 3.
These gene-sets were tested using MAGMA. We computed competitive P-values, which represent the test of association for a specific gene-set compared with genes not in the gene-set to correct for baseline level of genetic association in the data. The Bonferroni-corrected significance threshold was 0.05/7,087 gene-sets=7.06×10−6. The suggestive significance threshold was defined by the number of tests within the category. Conditional analyses were performed as a follow-up using MAGMA to test whether each significant association observed was independent of APOE (a gene-set including all genes within region chr19:45,020,859–45,844,508). Furthermore, the association between each of the significant gene-sets was tested conditional on each of the other significantly associated gene-sets. Gene-sets that retained their association after correcting for other sets were considered to represent independent signals. We note that this is not a test of association per se, but rather a strategy to identify, among gene-sets with known significant associations and overlap in genes, which set(s) are responsible for driving the observed association.
Cross-trait genetic correlation
Genetic correlations (rg) between AD and 41 phenotypes were computed using LD score regression14, based on GWAS summary statistics obtained from publicly available databases (see URLs and Supplementary Table 26). The Bonferroni-corrected significance threshold was 0.05/41 traits=1.22×10−3.
Mendelian randomization
To infer credible causal associations between AD and traits that are genetically correlated with AD, we performed Generalised Summary-data based Mendelian Randomisation36 (GSMR; see URLs). This method utilizes summary-level data to test for putative causal associations between a risk factor (exposure) and an outcome by using independent genome-wide significant SNPs as instrumental variables as an index of the exposure. HEIDI-outlier detection was used to filter genetic instruments that showed clear pleiotropic effects on the exposure phenotype and the outcome phenotype. We used a threshold p-value of 0.01 for the outlier detection analysis in HEIDI, which removes 1% of SNPs by chance if there is no pleiotropic effect. To test for a potential causal effect of various outcomes on risk for AD, we selected phenotypes in non-overlapping samples that showed (suggestive) significant (P<0.05) genetic correlations (rg) with AD. With this method it is typical to test for bi-directional causation by repeating the analyses while switching the role of the exposure and the outcome; however, because AD is a late-onset disease, it makes little sense to estimate its causal effect on outcomes that develop earlier in life, particularly when the summary statistics for these outcomes were derived mostly from younger samples than those of AD cases. Therefore, we conducted these analyses only in one direction. For genetically correlated phenotypes, we selected independent (r2=<0.1), GWS lead SNPs as instrumental variables in the analyses. The method estimates a putative causal effect of the exposure on the outcome (bxy) as a function of the relationship between the SNPs’ effects on the exposure (bzx) and the SNPs’ effects on the outcome (bzy), given the assumption that the effect of non-pleiotropic SNPs on an exposure (x) should be related to their effect on the outcome (y) in an independent sample only via mediation through the phenotypic causal pathway (bxy). The estimated causal effect coefficients (bxy) are approximately equal to the natural log odds ratio (OR)36 for a case-control trait. An OR of 2 can be interpreted as a doubled risk compared to the population prevalence of a binary trait for every SD increase in the exposure trait. This method can help differentiate the causal direction of association between two traits, but cannot make any statement about the intermediate mechanisms involved in any potential causal process.
Data Availability
Summary statistics will be made available for download upon publication (https://ctg.cncr.nl).
Code Availability
The analyses were produced with standard code for software programs utilized, which can be made available on request from the first author. All software used is freely available online. Custom code for the meta-analysis correcting for overlapping samples is available at https://github.com/Kyoko-wtnb/mvGWAMA
Supplementary Material
Acknowledgments
This work was funded by The Netherlands Organization for Scientific Research (NWO VICI 453-14-005). The analyses were carried out on the Genetic Cluster Computer, which is financed by the Netherlands Scientific Organization (NWO: 480-05-003), by the VU University, Amsterdam, The Netherlands, and by the Dutch Brain Foundation, and is hosted by the Dutch National Computing and Networking Services SurfSARA. The work was also funded by The Research Council of Norway (#251134, #248778, #223273, #213837, #225989), KG Jebsen Stiftelsen, The Norwegian Health Association, European Community’s JPND Program, ApGeM RCN #237250, and the European Community’s grant # PIAPP-GA-2011–286213 PsychDPC. This research has been conducted using the UK Biobank resource under application number 16406 and the public ADSP dataset, obtained through the Database of Genotypes and Phenotypes (dbGaP) under accession number phs000572. Full acknowledgments for the studies that contributed data can be found in the Supplementary Note. We thank the numerous participants, researchers, and staff from many studies who collected and contributed to the data.
Footnotes
Competing Interests Statement
The authors report the following potentially competing financial interests. P.F.S.: Lundbeck (advisory committee), Pfizer (Scientific Advisory Board member), and Roche (grant recipient, speaker reimbursement). J.H.L.: Cartana (Scientific Advisor) and Roche (grant recipient). O.A.A.: Lundbeck (speaker’s honorarium). St.St., H.S., and K.S. are employees of deCODE Genetics/Amgen. J.H. is a cograntee of Cytox from Innovate UK (UK Department of Business). D.A. has received research support and/or honoraria from Astra-Zeneca, Lundbeck, Novartis Pharmaceuticals, and GE Health, and serves as a paid consultant for Lundbeck, Eisai, Heptares, and Axovant. All other authors declare no financial interests or potential conflicts of interest.
References
- 1.Prince M, Bryce R, Albanese E, Wimo A, Ribeiro W, Ferri CP. The global prevalence of dementia: a systematic review and metaanalysis. Alzheimer’s & dementia : the journal of the Alzheimer’s Association 2013; 9(1): 63–75.e2. [DOI] [PubMed] [Google Scholar]
- 2.Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry 2006; 63(2): 168–74. [DOI] [PubMed] [Google Scholar]
- 3.Cacace R, Sleegers K, Van Broeckhoven C. Molecular genetics of early-onset Alzheimer’s disease revisited. Alzheimer’s & dementia : the journal of the Alzheimer’s Association 2016; 12(6): 733–48. [DOI] [PubMed] [Google Scholar]
- 4.Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 2013; 45(12): 1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goate A, Chartier-Harlin MC, Mullan M, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature 1991; 349(6311): 704–6. [DOI] [PubMed] [Google Scholar]
- 6.Sherrington R, Rogaev EI, Liang Y, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature 1995; 375(6534): 754–60. [DOI] [PubMed] [Google Scholar]
- 7.Sherrington R, Froelich S, Sorbi S, et al. Alzheimer’s disease associated with mutations in presenilin 2 is rare and variably penetrant. Human molecular genetics 1996; 5(7): 985–8. [DOI] [PubMed] [Google Scholar]
- 8.Karran E, Mercken M, De Strooper B. The amyloid cascade hypothesis for Alzheimer’s disease: an appraisal for the development of therapeutics. Nature reviews Drug discovery 2011; 10(9): 698–712. [DOI] [PubMed] [Google Scholar]
- 9.Jonsson T, Stefansson H, Steinberg S, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. The New England journal of medicine 2013; 368(2): 107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steinberg S, Stefansson H, Jonsson T, et al. Loss-of-function variants in ABCA7 confer risk of Alzheimer’s disease. Nature genetics 2015; 47(5): 445–7. [DOI] [PubMed] [Google Scholar]
- 11.Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nature reviews Neurology 2013; 9(2): 106–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu JZ, Erlich Y, Pickrell JK. Case-control association mapping by proxy using family history of disease. Nat Genet 2017; 49(3): 325–31. [DOI] [PubMed] [Google Scholar]
- 13.Marioni RE, Harris SE, Zhang Q, et al. GWAS on family history of Alzheimer’s disease. Transl Psychiatry 2018; 8: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulik-Sullivan BK, Loh PR, Finucane HK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015; 47(3): 291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Bakker PIW, Ferreira MAR, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet 2008; 17(R2): R122–R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerreiro R, Wojtas A, Bras J, et al. TREM2 variants in Alzheimer’s disease. N Engl J Med 2013; 368(2): 117–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desikan RS, Schork AJ, Wang Y, et al. Polygenic Overlap Between C-Reactive Protein, Plasma Lipids, and Alzheimer Disease. Circulation 2015; 131(23): 2061–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sims R, van der Lee SJ, Naj AC, et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nature genetics 2017; 49(9): 1373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gudbjartsson DF, Helgason H, Gudjonsson SA, et al. Large-scale whole-genome sequencing of the Icelandic population. Nature genetics 2015; 47(5): 435–44. [DOI] [PubMed] [Google Scholar]
- 20.Steinthorsdottir V, Thorleifsson G, Sulem P, et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nature genetics 2014; 46(3): 294–8. [DOI] [PubMed] [Google Scholar]
- 21.Euesden J, Lewis CM, O’Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics 2015; 31(9): 1466–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Escott-Price V, Myers AJ, Huentelman M, Hardy J. Polygenic risk score analysis of pathologically confirmed Alzheimer disease. Annals of Neurology 2017; 82(2): 311–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nature genetics 2014; 46(3): 310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511(7510): 421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finucane HK, Bulik-Sullivan B, Gusev A, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nature genetics 2015; 47(11): 1228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nature communications 2017; 8(1): 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fagerberg L, Hallstrom BM, Oksvold P, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Molecular & Cellular Proteomics : MCP 2014; 13(2): 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gurses MS, Ural MN, Gulec MA, Akyol O, Akyol S. Pathophysiological Function of ADAMTS Enzymes on Molecular Mechanism of Alzheimer’s Disease. Aging and disease 2016; 7(4): 479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suh J, Choi SH, Romano DM, et al. ADAM10 missense mutations potentiate beta-amyloid accumulation by impairing prodomain chaperone function. Neuron 2013; 80(2): 385–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dries DR, Yu G. Assembly, maturation, and trafficking of the gamma-secretase complex in Alzheimer’s disease. Current Alzheimer research 2008; 5(2): 132–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dumitriu A, Golji J, Labadorf AT, et al. Integrative analyses of proteomics and RNA transcriptomics implicate mitochondrial processes, protein folding pathways and GWAS loci in Parkinson disease. BMC medical genomics 2016; 9: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015; 11(4): e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Expansion of the Gene Ontology knowledgebase and resources. Nucleic acids research 2017; 45(D1): D331–d8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anttila V, Bulik-Sullivan B, Finucane HK, et al. Analysis of shared heritability in common disorders of the brain. Science 2018; 360(6395). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savage JE, Jansen PR, Stringer S, et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet 2018; 50(7): 9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Z, Zheng Z, Zhang F, et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nature communications 2018; 9(1): 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skene NG, Grant SG. Identification of Vulnerable Cell Types in Major Brain Disorders Using Single Cell Transcriptomes and Expression Weighted Cell Type Enrichment. Frontiers in neuroscience 2016; 10: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang J, Rivest S. Lipid metabolism and neuroinflammation in Alzheimer’s disease: a role for liver X receptors. Endocrine reviews 2012; 33(5): 715–46. [DOI] [PubMed] [Google Scholar]
- 39.Loewendorf A, Fonteh A, Mg H, Me C. Inflammation in Alzheimer’s Disease: Cross-talk between Lipids and Innate Immune Cells of the Brain; 2015.
- 40.Stern Y Cognitive reserve in ageing and Alzheimer’s disease. The Lancet Neurology 2012; 11(11): 1006–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satizabal C, Beiser AS, Seshadri S. Incidence of Dementia over Three Decades in the Framingham Heart Study. The New England journal of medicine 2016; 375(1): 93–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams HH, Hibar DP, Chouraki V, et al. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nature neuroscience 2016; 19(12): 1569–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ikram MA, Fornage M, Smith AV, et al. Common variants at 6q22 and 17q21 are associated with intracranial volume. Nature genetics 2012; 44(5): 539–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graves AB, Mortimer JA, Larson EB, Wenzlow A, Bowen JD, McCormick WC. Head circumference as a measure of cognitive reserve. Association with severity of impairment in Alzheimer’s disease. The British journal of psychiatry : the journal of mental science 1996; 169(1): 86–92. [DOI] [PubMed] [Google Scholar]
- 45.Abbott RD, White LR, Ross GW, et al. Height as a marker of childhood development and late-life cognitive function: the Honolulu-Asia Aging Study. Pediatrics 1998; 102(3 Pt 1): 602–9. [DOI] [PubMed] [Google Scholar]
- 46.Giuffrida ML, Tomasello F, Caraci F, Chiechio S, Nicoletti F, Copani A. Beta-amyloid monomer and insulin/IGF-1 signaling in Alzheimer’s disease. Molecular neurobiology 2012; 46(3): 605–13. [DOI] [PubMed] [Google Scholar]
- 47.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 2015; 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bulik-Sullivan B, Finucane HK, Anttila V, et al. An atlas of genetic correlations across human diseases and traits. Nature genetics 2015; 47(11): 1236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J, Ferreira T, Morris AP, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet 2012; 44(4): 369–75, s1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lovestone S, Francis P, Kloszewska I, et al. AddNeuroMed--the European collaboration for the discovery of novel biomarkers for Alzheimer’s disease. Annals of the New York Academy of Sciences 2009; 1180: 36–46. [DOI] [PubMed] [Google Scholar]
- 51.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC bioinformatics 2011; 12: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research 2010; 38(16): e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome research 2012; 22(9): 1790–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nature methods 2012; 9(3): 215–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature 2015; 518(7539): 317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amendola LM, Dorschner MO, Robertson PD, et al. Actionable exomic incidental findings in 6503 participants: challenges of variant classification. Genome research 2015; 25(3): 305–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science (New York, NY) 2015; 348(6235): 648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Westra HJ, Peters MJ, Esko T, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nature genetics 2013; 45(10): 1238–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhernakova DV, Deelen P, Vermaat M, et al. Identification of context-dependent expression quantitative trait loci in whole blood. Nature genetics 2017; 49(1): 139–45. [DOI] [PubMed] [Google Scholar]
- 60.Schmitt AD, Hu M, Jung I, et al. A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell reports 2016; 17(8): 2042–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramasamy A, Trabzuni D, Guelfi S, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nature neuroscience 2014; 17(10): 1418–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fromer M, Roussos P, Sieberts SK, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nature neuroscience 2016; 19(11): 1442–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ng B, White CC, Klein HU, et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nature neuroscience 2017; 20(10): 1418–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledgebased approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 2005; 102(43): 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Habib N, Avraham-Davidi I, Basu A, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nature methods 2017; 14(10): 955–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary statistics will be made available for download upon publication (https://ctg.cncr.nl).