Abstract
Adenosine-to-inosine (A-to-I) RNA editing is a very common co-/posttranscriptional modification that can lead to A-to-G changes at the RNA level and compensate for G-to-A genomic changes to a certain extent. It has been shown that each healthy individual can carry dozens of missense variants predicted to be severely deleterious. Why strongly detrimental variants are preserved in a population and not eliminated by negative natural selection remains mostly unclear. Here, we ask if RNA editing correlates with the burden of deleterious A/G polymorphisms in a population. Integrating genome and transcriptome sequencing data from 447 human lymphoblastoid cell lines, we show that nonsynonymous editing activities (prevalence/level) are negatively correlated with the deleteriousness of A-to-G genomic changes and positively correlated with that of G-to-A genomic changes within the population. We find a significantly negative correlation between nonsynonymous editing activities and allele frequency of A within the population. This negative editing-allele frequency correlation is particularly strong when editing sites are located in highly important genes/loci. Examinations of deleterious missense variants from the 1000 Genomes Project further show a significantly higher proportion of rare missense mutations for G-to-A changes than for other types of changes. The proportion for G-to-A changes increases with increasing deleterious effects of the changes. Moreover, the deleteriousness of G-to-A changes is significantly positively correlated with the percentage of editing enzyme binding motifs at the variants. Overall, we show that nonsynonymous editing is associated with the increased burden of G-to-A missense mutations in healthy individuals, expanding RNA editing in pathogenomics studies.
Generally, most mutations at functionally important loci, especially those that cause nonsynonymous changes (missense substitutions), are destined for selective elimination because of their deleteriousness. However, it was observed that each healthy individual could carry hundreds of missense substitutions, some of which were homozygous and predicted to be severely deleterious or disease-causing (Lohmueller et al. 2008; Chun and Fay 2009; MacArthur et al. 2012; Tennessen et al. 2012; Xue et al. 2012; Lohmueller 2014). Recent analysis of the Human Gene Mutation Database (HGMD) (Stenson et al. 2009) also revealed that 5132 out of 92,331 missense variants were classified as disease-causing mutations (Stenson et al. 2017). For pathogenomics studies, deleterious variants are often observed in well-established disease-associated genes in population controls, making it difficult to extract pathogenic variants (MacArthur et al. 2012). Many studies have investigated the burden of deleterious variants, or the so-called mutation load, carried by a population and indicated that the persistence of deleterious variants in a population is primarily affected by the strength of genetic drift and negative selection (Kimura et al. 1963; King and Jukes 1969; Lohmueller 2014; Henn et al. 2015). It is understandable that mildly deleterious variants contribute more to mutation load than severely deleterious ones because the former are subject to relatively weaker selective constraints than the latter (Lohmueller 2014; Henn et al. 2015). However, the reason that strongly detrimental variants (particularly those in the homozygous state) are preserved in a population and not eliminated by negative natural selection remains mostly unclear.
Adenosine-to-inosine (A-to-I) RNA editing is a very common co-/posttranscriptional modification mechanism in metazoans (Porath et al. 2017; Hung et al. 2018). It is catalyzed by the protein families of adenosine deaminases acting on RNA (ADAR), which convert adenosine (A) to inosine (I), leading to differences between the RNA products and the corresponding genomic sequences. A-to-I editing is also known as A-to-G editing because inosine is subsequently recognized as guanosine (G) by the cellular translation machinery. Since nonsynonymous A-to-G RNA editing events can result in amino acid changes at the RNA level (even though most observed coding editing sites are edited at a very low level [Li et al. 2009; Tan et al. 2017]), they may compensate for G-to-A genomic mutations to a certain extent and thus partially reduce the deleteriousness of such mutations. Hence, we are curious about whether nonsynonymous A-to-G RNA editing is associated with the persistence of deleterious A/G genomic variants in a population.
To address the above-mentioned issue, we conducted the first population-based analysis to examine the association between A-to-G RNA editing activities and the allele frequency of A/G genomic variants in a human population. Since minor alleles tend to be risk alleles (Park et al. 2011; Kido et al. 2018), an ancestral or relatively nondeleterious allele should reach a higher allele frequency within a population. If nonsynonymous RNA editing contributes to the prevalence of deleterious A/G genomic changes in a population, there should be a correlation between the editing activity at As and the allele frequency of A. To neutralize the deleterious effect of G-to-A genomic changes at the RNA level, we speculate that As with a lower allele frequency (Gs tend to be relatively nondeleterious alleles) should be edited more frequently than As with a higher allele frequency (As tend to be relatively nondeleterious alleles). If this is true, then a negative correlation should be observed between A-to-G RNA editing activities (prevalence and level) and the allele frequency of As within a population; meanwhile, this negative correlation should be particularly strong in highly important, evolutionarily conserved loci/genes. However, identification of RNA editing is often hampered by difficulties in distinguishing between true editing sites and A/G genomic variants because of the lack of genomic and transcriptomic data from the same samples. To minimize false positives arising from single nucleotide polymorphisms (SNPs), the editing detection pipelines generally discarded sites overlapping with known SNPs (e.g., dbSNP) (Eisenberg 2012; Ramaswami et al. 2012; Kurmangaliyev et al. 2016; Brümmer et al. 2017; Walkley and Li 2017). Thus, such a relationship between A-to-G RNA editing and the allele frequency of A/G genomic variants within a population remains uninvestigated. Recently, the human Geuvadis lymphoblastoid cell line (LCL) data (Lappalainen et al. 2013), which encompass both genotype data and transcriptome sequencing data from each LCL sample of 447 individuals, have provided a unique opportunity to determine RNA editing activities at A/G polymorphic sites within a human population. In other words, this data set provides an ideal resource for us to explore A-to-I RNA editing sites in some LCL individuals while genomically encoded as G in other LCL ones (or the so-called “polymorphic editing sites” [An et al. 2019]). Moreover, we examined the relationship between the distribution of different SNP types from the human population of apparently healthy individuals (The 1000 Genomes Project Consortium 2015) and the deleterious effects of the corresponding missense changes. We thus tested the contribution of nonsynonymous A-to-G RNA editing to the persistence of deleterious variants in the healthy human population.
Results
Nonsynonymous A-to-G RNA editing activity is associated with both the deleteriousness and direction of A/G missense changes
To assess the correlation between coding A-to-G RNA editing activity and the deleteriousness of A/G genomic variants in a population, we first extracted sites with A/G SNPs in coding regions from the Geuvadis LCLs of 447 individuals (derived from the 1000 Genomes Project [The 1000 Genomes Project Consortium 2015]) and the RNA-seq data of the corresponding LCL samples from the Geuvadis project (Lappalainen et al. 2013). For each A/G SNP site, there are three possible genotypes across the LCL population: AA, AG, and GG. We selected individuals with the homozygous genotype AA and used the RNA-seq data from the corresponding LCL samples to determine RNA editing (Fig. 1A). Of note, throughout this study we only calculated editing levels at sites with the homozygous genotype AA to eliminate the expression effect of allele G. An A/G SNP site was defined as a SNP editing site if it was found to be edited at a level >5% in at least two LCL samples from individuals with homozygous genotype AA. In this study, we only considered SNP editing sites within coding regions (1712 sites) for the following analyses, in which editing causes a nonsynonymous change at 889 sites and causes a synonymous change at 823 sites (Supplemental File S1). We also detected A-to-G RNA editing at the sites without the corresponding genotype information (see Methods and Supplemental File S1) and showed that the vast majority of all the detected RNA-DNA variants were A-to-G variants for each LCL individual (Supplemental Fig. S1A), supporting the effectiveness of our detection procedures. Of all the detected A-to-G RNA editing sites (85,565 sites), only 1% (889 sites) were nonsynonymous SNP editing sites (Supplemental File S1). On average, a sample (individual) carried 22.1 nonsynonymous SNP editing sites. More than 97% of the examined samples (437 out of 447 samples) carried <40 nonsynonymous SNP editing sites (see Supplemental File S1). To test the reliability of the identified SNP editing sites, we first examined the allelic ratio (the ratio of number of G reads to the sum of numbers of A and G reads) for the SNP editing sites, non-SNP editing sites (coding editing at non-SNP sites), and known SNPs with heterozygous genotype AG within coding regions (“known SNPs”). We found that the median allelic ratio of known SNPs was indeed centered in 0.5 and significantly higher than those of SNP and non-SNP editing sites (both P-values < 10−15 by two-tailed Wilcoxon rank-sum test) (Supplemental Fig. S1B). Next, we examined the primary ADAR sequence motif (Lehmann and Bass 2000; Eggington et al. 2011) for G depletion and G enrichment at the 5′ and 3′ neighboring nucleotides of the editing sites. Using known SNPs as the control, we calculated the observed-to-expected (O/E) ratio of the presence of “non-G” immediately upstream of and “G” immediately downstream from the editing sites and showed that the O/E ratios for both SNP and non-SNP editing sites were indeed significantly greater than 1 (both P-values < 10−10 by χ2 test) (Supplemental Fig. S1C), indicating a significantly higher ADAR preference for SNP and non-SNP editing sites than for known SNPs. These results suggest that the identified SNP editing sites are less likely to be derived from heterozygous polymorphisms.
Figure 1.
A-to-G RNA editing activities at A/G genomic variant sites. (A) Schematic diagram of an editing event at a variant site with homozygous genotype AA in a population. A and G represent the minor and major alleles in this population, respectively. (B,C) Comparisons of (B) nonsynonymous and (C) synonymous editing activities (prevalence and median level) at A/G genomic variant sites with harmless (CADD score ≤10) and deleterious (CADD score >10) A-to-G/G-to-A ancestral-to-derived allele changes in the LCL population. The number of editing sites examined in each group was provided in parentheses. P-values were determined using a two-tailed Wilcoxon rank-sum test. (**) P-value < 0.01, (***) P-value < 0.001, (NS) not significant.
We proceeded to examine the correlation between RNA editing activity and the deleterious effect of genomic changes in the population. We divided the A/G SNP sites where editing events would occur in the individuals with homozygous genotype AA into A-to-G and G-to-A ancestral-to-derived allele changes based on the human-chimpanzee-rhesus macaque orthologs. We used the Combined Annotation Dependent Depletion (CADD) tool (Kircher et al. 2014; Rentzsch et al. 2019), a well-developed tool for measuring the molecular functionality and pathogenicity of genomic changes, to assess the deleteriousness of A/G genomic changes. Two phenomena were observed for the nonsynonymous editing sites (Fig. 1B). First, both the prevalence and level of editing were higher for G-to-A genomic changes than for A-to-G ones. Second, both the prevalence and level of editing were reduced for deleterious (CADD score >10) A-to-G genomic changes and elevated for deleterious G-to-A changes. Here, the prevalence of editing at each site was defined as the percentage of individuals with editing done at the site over all individuals with homozygous genotype AA and a read coverage ≥10 (i.e., all individuals that were testable for this editing site). Of note, the median levels of nonsynonymous editing at sites with neutral/harmless (CADD score ≤10) A-to-G and G-to-A genomic changes were not statistically different (P-value = 0.73), whereas the median editing level was significantly higher at sites with G-to-A genomic changes than at those with A-to-G changes when the changes were deleterious (P-value < 0.001) (Fig. 1B, right). In contrast, these phenomena were not observed for synonymous editing sites, although the phenomenon of a higher prevalence/level of editing for G-to-A genomic changes than for A-to-G ones held (Fig. 1C). Meanwhile, we found that the host gene expression of the examined editing events at sites with neutral and deleterious genomic changes was not significantly different, regardless of direction (i.e., A-to-G or G-to-A) of the corresponding genomic changes and whether the changes were nonsynonymous or synonymous (Supplemental Fig. S2). This suggested that the host gene expression of the editing sites was not the major cause for the trends observed above. These results thus suggest that A-to-G RNA editing activities are associated with both the deleterious effect and direction of the corresponding genomic changes in a population.
Nonsynonymous A-to-G RNA editing activity is negatively correlated with the allele frequency of A in a population
We then examined the correlation between nonsynonymous A-to-G RNA editing activity and the allele frequency of A in the LCL population. We first showed that the proportion of deleterious nonsynonymous genomic changes (CADD score >10) markedly decreased with an increasing minor allele frequency (Supplemental Fig. S3), reflecting the observation that minor alleles tend to be risk alleles (Park et al. 2011; Kido et al. 2018). Next, the above results (Fig. 1B) showed that both the prevalence and level of nonsynonymous editing were negatively correlated with the deleteriousness of A-to-G genomic changes and positively correlated with the deleteriousness of G-to-A genomic changes within a population. Accordingly, for an A/G genomic variant, if G is a minor allele within a population (which implies that A is an ancestral or relatively nondeleterious allele), then the A allele should be edited less to prevent the conversion of A into I (which is then recognized as G) at the RNA level. In contrast, if A is a minor allele (which implies that G is an ancestral or relatively nondeleterious allele), then editing of the A allele should be promoted to compensate for the deleterious G-to-A change to a certain extent. In other words, we speculated that both the prevalence and level of nonsynonymous editing should negatively correlate with the allele frequency of A within the LCL population. Indeed, we observed that both the prevalence (Fig. 2A) and level (Fig. 2B) of nonsynonymous RNA editing markedly decreased with an increasing allele frequency of A (or with a decreasing allele frequency of G) within the LCL population. We further performed partial correlation analysis (Kim and Yi 2007) to control for the host gene expression of the examined editing sites and the GC content of the host genes. We found that the negative correlations remained significant after the control (Supplemental Fig. S4), suggesting that the host gene expression of the editing sites was not the major cause for the correlations observed in Figure 2. Again, these results support the negative correlation between nonsynonymous RNA editing activities and the deleteriousness of A/G missense changes in the human population.
Figure 2.
Correlation between nonsynonymous editing activities and allele frequency of A. Box (left) and scatter (right) plots represent the correlations between nonsynonymous editing activities ([A] prevalence, and [B] median editing level) and allele frequency of A within the LCL population. The number of nonsynonymous editing sites examined in each group is provided in parentheses. Statistical significance was estimated using a two-tailed Wilcoxon rank-sum test (left) and Spearman's rank coefficient of correlation (ρ) (right). (**) P-value < 0.01, (***) P-value < 0.001.
The negative RNA editing-allele frequency correlation is stronger in functionally more important loci/genes than in less important ones
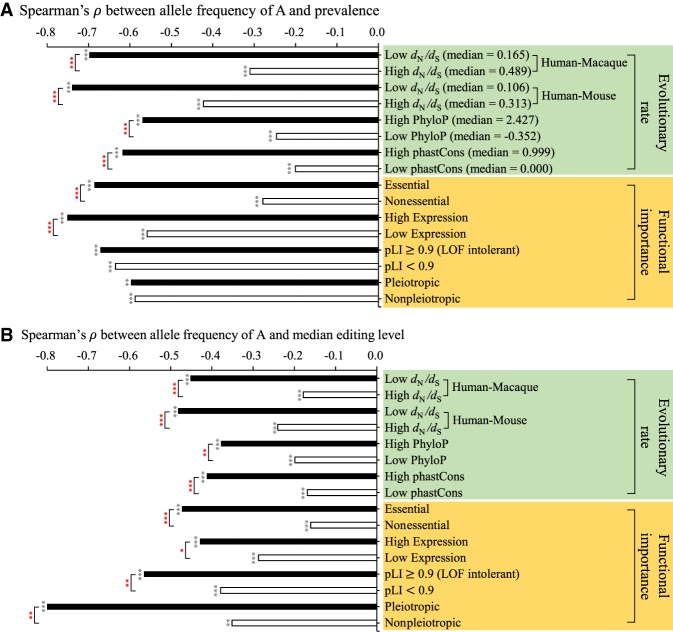
We have observed a significant correlation between nonsynonymous RNA editing activities and the deleteriousness of A/G missense changes in a population (Fig. 2). We then asked whether such a correlation was stronger in functionally more important loci/genes than in less important ones. If it was, we should observe a stronger correlation between nonsynonymous RNA editing activities and allele frequency of A within a population in functionally more important loci/genes than in less important ones. To this end, we performed the following evolutionary and functional analyses (see Fig. 3A,B and Supplemental Fig. S5). First, we considered the selective constraints of the genes that contained the nonsynonymous editing sites examined. We divided nonsynonymous editing sites into two equally sized groups (i.e., editing sites located within genes under strong and weak selection pressure) according to the ratios of nonsynonymous to synonymous substitution rates (dN/dS) calculated using one-to-one human-rhesus macaque and human-mouse orthologs, respectively. Indeed, the negative RNA editing-allele frequency correlations were significantly stronger for editing sites located within genes under strong selective constraints than for those located within genes under weak selective constraints (all P-values < 0.001 by two-tailed Z-score test). Second, similar to the above analysis, we divided the nonsynonymous editing sites into two equally sized groups: highly and lowly conserved sites (as determined by phyloP [Pertea et al. 2011] or phastCons [Siepel et al. 2005] scores). We observed that the correlations were significantly stronger for highly conserved sites than for lowly conserved ones. Third, on the basis of the Online GEne Essentiality (OGEE) database (Chen et al. 2017), we examined the effect of gene essentiality on the correlation because essential genes are those that are functionally indispensable for the survival of an organism. We also found a significantly stronger negative RNA editing-allele frequency correlation for editing sites located within essential genes than for those located within nonessential ones. Fourth, we examined the effect of the host gene expression of the examined editing sites on the RNA editing-allele frequency correlation. We divided nonsynonymous editing sites into two groups: sites located within highly expressed genes and those located within lowly expressed genes (see Methods). We observed a significantly stronger negative correlation for editing sites located within highly expressed genes than for those located within lowly expressed ones. Fifth, we further predicted that such a negative correlation should be stronger for editing sites located within genes intolerant of a loss-of-function (LOF) mutation than for those located within genes with LOF tolerance. We considered gene variant intolerance (pLI) scores estimated by the Exome Aggregation Consortium (ExAC) (Lek et al. 2016), which range from 0 to 1, with higher pLI scores indicating greater LOF intolerance levels. Human genes were divided into two groups using the 90th percentile as a cut-off. The resulting data also supported our prediction. Finally, we examined the effect of gene pleiotropy on the correlation. Generally, we found more strongly negative correlations between nonsynonymous editing activities (editing level, particularly) and the allele frequency of A for editing sites located within pleiotropic genes than for those located within nonpleiotropic ones. Taken together, if A is a minor allele (e.g., allele frequency of A ≤33%), the median editing levels are generally higher in functionally more important loci/genes than in less important ones, whereas the reversed trend is observed if A is a major allele (e.g., allele frequency of A >66%) (Supplemental Table S1). These results support that the negative RNA editing-allele frequency correlation is stronger in functionally more important loci/genes than in less important ones in the human population; of note, the above-mentioned trends were observed for nonsynonymous editing (Fig. 3) but not for synonymous editing (Supplemental Fig. S6).
Figure 3.
Functional and evolutionary analysis of the RNA editing-allele frequency correlations. The histograms represent the correlations between nonsynonymous editing activities ([A] prevalence and [B] median editing level) and allele A frequency within the LCL population in four categories of evolutionary rates and four categories of functional importance (see the text) of the target genes/loci where the editing sites were located. For the four categories of evolutionary rates, editing sites were divided into two equal groups according to the high and low scores of the target genes/loci. The statistical significance of Spearman's rank correlation coefficients (ρ) are represented by black stars. Significant differences between two independent correlations (represented by red stars) were estimated using a two-tailed Z-score test with the paired.r function within the psych R library. (*) P-value < 0.05, (**) P value < 0.01, (***) P-value < 0.001.
Phylogenetic variation in the RNA editing-allele frequency correlation
The pattern of phylogenetic variation of a site is often used to assess its evolutionary/functional importance. Regarding the patterns of phylogenetic variations for the nonsynonymous editing sites, we can assess the RNA editing-allele frequency correlation in different phylogenetic types of nonsynonymous A-to-G editing sites. To this end, we retrieved human, chimpanzee, rhesus macaque, and mouse orthologous nucleotides and defined five types of human nonsynonymous editing sites according to their phylogenetic variations (Fig. 4A). First, “G-conserved” sites were those for which a G allele was observed in all three nonhuman orthologs. Second, “A-conserved” sites were those for which an A allele was observed in all three nonhuman orthologs. Third, “hardwired” sites were those for which either an A or G was observed in all three nonhuman orthologs. Fourth, “G-unfound” sites were those for which G was not observed in any of the three nonhuman orthologs, excluding sites already designated as A-conserved. Fifth, “diversified” sites were those for which either a G or non-G was observed in all nonhuman orthologs, excluding sites already designated as hardwired. This categorization resulted in 100 G-conserved sites, 299 A-conserved sites, 114 hardwired sites, 35 G-unfound sites, and 26 diversified sites. Of note, A/G coincident SNPs (Hodgkinson et al. 2009; Chen et al. 2016), which were orthologous sites observed to be A/G polymorphic in any two examined species, were considered as the hardwired sites. Regarding the patterns of phylogenetic variations, diversified sites may be subject to the weakest selective constraints compared to other types of sites because these positions allow various amino acid changes (Xu and Zhang 2014). Thus, we predicted that the RNA editing-allele frequency correlation should be the weakest at diversified sites because editing would cause the smallest impact on these sites, given their tolerance for variation in amino acids. Indeed, we found that the correlation between the median editing level and the allele frequency of A at diversified sites was insignificant; in contrast, the negative RNA editing-allele frequency correlations held significantly for all the other types of sites (Fig. 4B; Supplemental Fig. S7).
Figure 4.

Phylogenetic analysis of nonsynonymous A-to-G editing sites. (A) Definition of five types of human nonsynonymous editing sites (A-conserved, G-unfound, hardwired, G-conserved, and diversified) based on their evolutionary variations among human, chimpanzee, rhesus macaque, and mouse. (B) Correlations between median editing levels and allele frequency of A within the LCL population for the five groups of editing sites. The number of nonsynonymous editing sites examined in each group is provided in parentheses. P-values were determined using a two-tailed Wilcoxon rank-sum test. (*) P-value < 0.05, (**) P-value < 0.01, (***) P-value < 0.001.
Deleterious G-to-A missense changes are more tolerable than other types of changes in a population
The above results that nonsynonymous editing activities are correlated with the deleteriousness of A/G genomic changes raise the question of whether nonsynonymous editing is associated with the distribution of missense variants (or nonsynonymous SNPs) in a population, especially when the missense changes are damaging. To this end, we extracted rare missense mutations (see Methods) from the 1000 Genomes Project (The 1000 Genomes Project Consortium 2015) and examined the correlation between the distribution of six possible SNP types and the deleterious effects of the corresponding missense changes (Supplemental Table S2). The SNPs were classified into six groups rather than 12 because genomic variants could potentially cause variants in both the sense and antisense transcripts (e.g., A-to-G changes on one strand and T-to-C changes on the opposite strand). It was not surprising that there was a higher proportion of rare missense mutations in the two transition groups (A-to-G/T-to-C and G-to-A/C-to-T changes) than in the four transversion groups (Fig. 5A). Of note, the two transition groups exhibited quite different trends in terms of the proportions of rare missense mutations. Proportions of rare missense mutations were markedly negatively correlated with the corresponding CADD scores for A-to-G/T-to-C changes, but the reversed trend was observed for G-to-A/C-to-T changes (Fig. 5A). Particularly, regarding the very deleterious missense changes (e.g., CADD scores >30), the vast majority (83%) of rare missense variants were G-to-A/C-to-T changes while only 2% were A-to-G/T-to-C changes (Fig. 5A). The result suggests that G-to-A/C-to-T missense changes are more acceptable than other types of changes in the human population when the changes are deleterious. Considering all human coding regions, we further compared the frequencies of two types of transitions (i.e., ƒ(G-to-A/C-to-T) vs. ƒ(A-to-G/T-to-C)) in different CADD scores (see Methods and Supplemental Table S2). We found that the ƒ(G-to-A/C-to-T)/ƒ(A-to-G/T-to-C) ratio was relatively close to 1 when the rare missense changes were neutral/harmless (CADD score ≤10); however, such ratios were significantly higher than 1 when the changes were deleterious (CADD score >10) (Fig. 5B). The ƒ(G-to-A/C-to-T)/ƒ(A-to-G/T-to-C) ratios markedly increased with increasing CADD scores (Fig. 5B). Moreover, we calculated the proportions of rare missense mutations for A-to-G/T-to-C and G-to-A/C-to-T changes per individual from the 1000 Genomes Project and examined the individual mutational burden of these two types of rare missense changes. We found that the median proportion of G-to-A/C-to-T missense changes was significantly higher than that of A-to-G/T-to-C changes, particularly when the changes were deleterious (Fig. 5C, top). The differences in mutational burden between these two types of rare missense changes markedly increased with increasing deleterious effects of the changes (Fig. 5C, bottom). These results further support that damaging G-to-A/C-to-T missense mutations are especially tolerable in a healthy population. We then examined whether the preference for damaging G-to-A/C-to-T missense mutations in a population was associated with the primary ADAR sequence motif (Lehmann and Bass 2000; Eggington et al. 2011) at the variants. Indeed, sites of G-to-A/C-to-T missense mutations have a significantly higher percentage of the ADAR motif than those of non-G-to-A/C-to-T ones; such differences markedly increased with increasing CADD scores (Fig. 5D). These observations also reflect a recent comment that G-to-A mutation sites tend to be favorable locations for the origination of robust A-to-I RNA editing (An et al. 2019).
Figure 5.
Relationship between the distributions of different types of rare missense mutations and A-to-G RNA editing. (A) Correlation between the distributions of different types of rare missense mutations from the 1000 Genomes Project and deleteriousness (measured by CADD scores) of the corresponding genomic changes. (B) Fraction of the frequency of nonsynonymous G-to-A/C-to-T changes to the frequency of nonsynonymous A-to-G/T-to-C changes (i.e., the ƒ(G-to-A/C-to-T)/ƒ(A-to-G/T-to-C) ratio) for different deleterious effects of the genomic changes. (C) Comparisons of the individual mutational burden of the two types of rare transition missense mutations (A-to-G/T-to-C and G-to-A/C-to-T) in the 1000 Genomes Project. The top panel represents the proportions of rare A-to-G/T-to-C and G-to-A/C-to-T missense mutations per individual for different CADD scores. The bottom panel represents the differences (effect sizes) in mutational burden between these two types of rare missense variants. Effect sizes were measured by Cohen's d, which was defined as the difference between both mean numbers of these two types of rare missense mutations divided by the standard deviation of the paired differences. The estimated 95% confidence intervals of effect sizes were plotted (see also Supplemental Table S3). (D) Comparisons of the proportions of SNP sites with the ADAR motif for different deleterious effects of G-to-A/C-to-T and non-G-to-A/C-to-T rare missense mutations. P-values were determined using a two-tailed Fisher's exact test (B and D) or a two-tailed Wilcoxon signed-rank test (C). (*) P-value < 0.05, (***) P-value < 0.001.
To test the robustness of our results, we performed similar analyses using another tool (SIFT [Ng and Henikoff 2003]) to assess the deleteriousness of the rare missense mutations and found similar trends (Supplemental Fig. S8). In addition, since the risk of passing on deleterious mutations to future generations is changing and evolving rapidly, different human populations may carry substantially distinctive mutational spectra (Harris and Pritchard 2017). To ask whether the differences influence our results, we performed similar analyses in five human subpopulations and observed similar results in different subpopulations (Supplemental Fig. S9). These results revealed the trends observed above to be independent of tools for measuring the deleteriousness of amino acid substitutions and human subpopulations. Moreover, on the basis of genome and transcriptome sequencing data from the LCL samples, we asked whether non-A-to-G RNA-DNA variants would also play a role in reducing the deleterious effect of predicted damaging mutations. We calculated the ratio of the number of RNA-DNA variant events at SNP sites with deleterious (CADD score >10) genomic changes to those at SNP sites with neutral/harmless (CADD score ≤10) ones. Since G-to-A RNA-DNA mismatches often reflected sequencing errors (Bahn et al. 2012; Liscovitch-Brauer et al. 2017) and the corresponding RNA-DNA variant events detected in the four transversion SNP groups were few (<50 events for each transversion SNP group), we only compared these two transition SNP groups for the ratios. Our results revealed that the ratio was close to 1 for G-to-A/C-to-T RNA-DNA variant events at A-to-G/T-to-C SNP sites (P-value = 0.7 by χ2 test) but significantly larger than one (P-value <0.001) for A-to-G/T-to-C RNA-DNA variant events at G-to-A/C-to-T SNP sites (Supplemental Fig. S10). We thus suggest that nonsynonymous A-to-G RNA editing is highly associated with the distribution of existing nonsynonymous polymorphisms at functionally important loci, contributing to the persistence of predicted deleterious missense variants in the human population.
Discussion
This study conducted the first population-based analysis to examine the relationship between A-to-G RNA editing activities and the allele frequency of A/G genomic variants. Our results suggested that editing activities were associated with both the functional importance and direction (i.e., A-to-G or G-to-A) of the genomic changes in a population. We found that RNA editing activities were negatively correlated with the allele frequency of A in the LCL population and such a negative editing-allele frequency correlation was significantly stronger in functionally more important loci/genes than in less important ones. We further observed that G-to-A/C-to-T missense mutations were much more prevalent than A-to-G/T-to-C ones (and other types of changes) at functionally important sites, in which the differences in the mutational burden of missense changes was significantly positively correlated with the deleteriousness of the changes. For an existing A/G missense variant, if the G-to-A genomic change has severely deleterious effects on protein function, RNA editing at this site with a higher editing level is more likely to neutralize the deleterious effect of the genomic change at the RNA level, making the deleterious effect weaker than expected. RNA editing may facilitate the escape of this existing deleterious A/G variant from negative natural selection and therefore aid the tolerance for this deleterious variant (i.e., the deleterious allele A) in a population. We thus suggest that nonsynonymous A-to-G RNA editing is associated with the missense variant distribution and contributes to damaging mutations in a population.
As it is known that DNA methylation at CpG dinucleotides can significantly increase the rate of spontaneous C-to-T transitions (Coulondre et al. 1978; Bird 1980), we were curious about whether such a high mutation rate at CpG dinucleotides might affect the preference for G-to-A/C-to-T missense mutations at functionally important loci in a population. We first used the Mr. Eel tool (Carlson et al. 2018) to measure the mutation rates at the sites with rare missense mutations. The Mr. Eel mutation rates were estimated using extremely rare variants and local sequence context, which were demonstrated to be more accurate than the estimates based on ancestrally older variants (Carlson et al. 2018). Indeed, we observed that sites with G-to-A/C-to-T rare missense mutations had the highest mutation rate as compared with sites with other types of mutations (Supplemental Fig. S11A). We also found a marginal correlation between mutation rates and CADD scores at the sites with rare missense mutations (Supplemental Fig. S11B). To control for the effect of mutation rates, we divided the examined SNP sites into two equally sized groups (sites with high and low mutation rates) according to the Mr. Eel mutation rates. We found that the trends of the preference for G-to-A/C-to-T missense mutations and the elevated ƒ(G-to-A/C-to-T)/ƒ(A-to-G/T-to-C) ratios at functionally important loci still held in both groups (Supplemental Fig. S11C). We further reevaluated ƒ(G-to-A/C-to-T) with excluding sites at CpG dinucleotides and observed a similar pattern of ƒ(G-to-A/C-to-T)/ƒ(A-to-G/T-to-C) ratios illustrated in Figure 5B (Supplemental Fig. S11D). These results thus suggest that mutation rate is not the major cause of the preference for damaging G-to-A/C-to-T missense variants in the human population. We proceeded to probe the effect of DNA methylation on the trend observed in Figure 5. In human, it was observed that the majority of gene bodies were heavily methylated (Keller et al. 2016). Position-dependent correlations have also been shown between CpG methylation level and dN of the target genes (Chuang and Chiang 2014) or exons (Chuang et al. 2012); such methylation-dN correlations were observed to be negative for gene bodies (or internal/last exons) and positive for promoter regions (or first exons). Accordingly, we asked whether our results shown in Figure 5 were biased toward first exons. We divided nonsynonymous SNP sites into two groups: sites located within the first and nonfirst (internal/last) exons. We performed a similar analysis and found that all the trends observed in Figure 5 held well in both first and nonfirst exons (Supplemental Fig. S12), suggesting that the effect of CpG methylation is less likely to be responsible for the preference for damaging G-to-A/C-to-T missense mutations in a population.
That nonsynonymous editing may help tolerate existing deleterious G-to-A/C-to-T missense variants in a population raises the question of how common advantageous RNA editing is. Actually, this issue remains controversial. Since most A-to-G RNA editing events are considered to originate through promiscuous targeting by ADAR proteins (Xu and Zhang 2014), several studies support that editing events are generally nonadaptive. For example, human coding RNA editing events are rare and tend to be synonymous (Kleinman et al. 2012; Chen 2013). Only a small number of conserved mammalian editing sites have been observed (Pinto et al. 2014; Liscovitch-Brauer et al. 2017). The level of coding editing was generally <10% (Tan et al. 2017) except for a very few events of high editing levels (e.g., the editing site at position 607 of GRIA2 [Sommer et al. 1991], or see the summary table in the Hung et al. study [Hung et al. 2018]). In addition, it was observed that edited As were more likely to be replaced with Gs than unedited As in evolution (Xu and Zhang 2014). However, other studies speculated that nonsynonymous editing is more beneficial for enhancing transcriptome (and therefore proteome) diversity and fitness than the direct replacement of As with Gs at the DNA level, and thus it is maintained by natural selection (Gommans et al. 2009; Li et al. 2009; Nishikura 2010; Porath et al. 2017; Eisenberg and Levanon 2018). Sequences around some editing sites were demonstrated to be under strong selective constraints in human and rhesus macaque, suggesting the functional regulation of these sites during primate evolution (Chen et al. 2014). Recent analyses also suggested that newly originated A-to-I RNA editing events are generally selectively constrained (An et al. 2019) and edited As have a relatively high fitness (Popitsch et al. 2017). Accumulating evidence indicates that RNA editing is crucial in the neuronal dynamic in the mammalian central nervous system (Gal-Mark et al. 2017). A handful of editing events have been demonstrated to be highly regulated during brain development or neural differentiation (Barbon et al. 2003; Kawahara et al. 2004; Wahlstedt et al. 2009; Osenberg et al. 2010) and involved in neuronal diseases, such as inflammation, epilepsy (Brusa et al. 1995; Srivastava et al. 2017), depression (Gurevich et al. 2002), and amyotrophic lateral sclerosis (ALS) (Hideyama et al. 2012). These above-mentioned observations reveal the obscurity of the RNA editing effect on genome evolution. The effect of RNA editing on the efficacy of natural selection awaits further investigation in the future.
In addition, according to phylogenetic analysis of some specific cases, RNA editing was suggested to be advantageous by extending sequence divergence at the DNA level, acting as a safeguard by correcting G-to-A mutations at the RNA level and thus mediating the RNA memory of evolution (Tian et al. 2008; Chen 2013). However, as mentioned above, most coding A-to-G RNA editing events are edited at a level <10% (Tan et al. 2017). With such a low level of coding editing, most nonsynonymous editing events cannot fully posttranscriptionally compensate for the deleterious effects of G-to-A genomic mutations. In addition, regarding the five phylogenetic types of nonsynonymous A-to-G editing (Fig. 4A), we observed that the median editing level at diversified sites is similar to that at G-conserved or hardwired sites (both P-values > 0.05 by two-tailed Wilcoxon rank-sum test) (Supplemental Fig. S13A). This result was consistent with a previous study (Xu and Zhang 2014), which performed a similar analysis by categorizing human nonsynonymous editing sites into four phylogenetic types (i.e., A-conserved, hardwired, G-unfound, and diversified sites) based on phylogenetic variations among 44 nonhuman vertebrates (Supplemental Fig. S13B). Diversified sites are tolerant of many different amino acids; in contrast, Gs at both G-conserved and hardwired sites are under relatively stronger selective constraints. If A-to-G editing acts as a safeguard of G-to-A substitutions, then the editing level should be significantly higher at G-conserved and hardwired sites than at diversified sites. Therefore, although A-to-G RNA editing events would reduce the damaging effect of G-to-A mutations to a certain extent, human RNA editing does not seem to exhibit the signals of a safeguard overall.
Of note, as shown in Figure 4B, we found that A-conserved and G-unfound sites had a greater negative correlation between the allele frequency of A and the median editing level than the other types of sites. The reason is that the vast majority of the A-conserved (95%) and G-unfound (100%) sites have a high allele frequency (≥50%) of A in the LCL population and these sites with a high allele frequency of A have a relatively low editing level (see Supplemental Fig. S7). Such a greater negative RNA editing-allele frequency correlation for A-conserved and G-unfound sites is not due to a higher editing level at these sites. In contrast, the median editing level at A-conserved and G-unfound sites was significantly lower than the level at the other types of sites (P-value < 0.0001) (see Supplemental Fig. S13A). Since Gs at both A-conserved and G-unfound sites are not selectively permitted during mammalian protein evolution, most editing events at genomic As should be constrained. This result is consistent with a previous study (Xu and Zhang 2014) (see also Supplemental Fig. S13B). As mentioned above, we emphasize that the strength of the negative RNA editing-allele frequency correlation is dependent on the following two factors. First, the A allele tends to be edited less to prevent the conversion of A into G at the RNA level if G is a minor allele within a population (which implies the A-conserved sites). Second, editing of the A allele tends to be promoted to compensate for the deleterious G-to-A change to a certain extent if A is a minor allele (which implies the G-conserved sites). Since the median editing level at G-conserved sites is higher than that at A-conserved sites (Supplemental Fig. S13A), a possible explanation for a relatively weaker correlation for G-conserved sites than for A-conserved sites (Fig. 4B) may be due to the fact that the strength of the first factor is stronger than that of the second one.
Moreover, previous studies have observed that nonsynonymous editing prevalence and levels were lower in functionally more important genes than in less important one (Solomon et al. 2014; Xu and Zhang 2014). However, this study showed a different result. We found that nonsynonymous RNA editing at SNP sites of the two types of genomic changes (i.e., A-to-G or G-to-A changes) exhibited quite different trends: The prevalence and level of editing were reduced for deleterious A-to-G genomic changes but elevated for deleterious G-to-A changes (Fig. 1B). The previous analyses did not consider the direction of the corresponding genomic changes because of the lack of genome and transcriptome sequencing data from the same samples at a population scale. Therefore, their results cannot reflect the difference between nonsynonymous RNA editing activities at the two types of genomic changes. Our result thus suggests that nonsynonymous RNA editing activities at As are associated not only with the importance of the genes/loci where the sites were located but also with the direction (A-to-G or G-to-A) of the corresponding genomic changes.
In conclusion, this study highlights the association between nonsynonymous RNA editing and existing missense variants in the human population. Although the cause and distribution of damaging variants within a population are more complicated than expected (Henn et al. 2015), our findings reveal that nonsynonymous A-to-G RNA editing is associated with the increased burden of deleterious G-to-A missense variants in the healthy human population. Particularly for pathogenomics studies, deleterious variants are often observed in well-established disease-associated genes in population controls. Our results thus call for paying special attention to nonsynonymous A-to-G RNA editing in pathogenomics studies for extracting pathogenic variants.
Methods
Identification of RNA editing sites in the LCL population
The human gene annotation and the strands of sites were downloaded from the Ensembl genome browser at http://www.ensembl.org/ (version 87). The genotype data were extracted from the Geuvadis LCLs of 447 individuals (derived from the 1000 Genomes Project [The 1000 Genomes Project Consortium 2015]). The RNA-seq data of the corresponding LCL samples were extracted from the Geuvadis project at https://www.ebi.ac.uk/Tools/geuvadis-das/ (Lappalainen et al. 2013). To prevent potential allelic mapping bias of allelic-specific expression quantification, the maskOutFa tool (https://github.com/ENCODE-DCC/kentUtils/tree/master/src/hg/maskOutFa) was used to mask the SNP sites of the corresponding LCL samples at the human reference genome (GRCh38.p7) and generate the pseudogenome for each LCL individual. The corresponding RNA-seq reads were then aligned against the pseudogenome using STAR aligner (version 2.5.1b) (Dobin et al. 2013). We then genome-wide called RNA-DNA variants using a stringent computational pipeline (Hung et al. 2018). For the variant calls at the sites without the corresponding genotype information, the clustering strategy (with the parameter setting of Ncluster = 3) (Hung et al. 2018) was applied to minimize the effect of SNPs or mutations on the identification of RNA editing. For the variant calls at the sites with the corresponding genotype data, we took the identification of A-to-G RNA editing at A/G polymorphisms as an example to describe our processes. For A/G polymorphisms, there are three possible genotypes for an A/G SNP site: AA, AG, and GG. We only considered the sites with ≥10 matched reads and called variants at the A/G SNP sites with the homozygous genotype AA for each individual. To eliminate the expression effect of allele G, we only calculated editing levels at sites with the homozygous genotype AA. The editing level of a site was determined by the ratio of number of G reads to the sum of numbers of A and G reads. A site was defined as an editing site if it satisfied two rules: (1) The base quality score must be ≥25 with the STAR-mapping quality score (Dobin et al. 2013) ≥255; and (2) the site was found to be edited at a level >5% in at least two LCL samples from individuals with homozygous genotype AA. To minimize potential false positives caused by sequencing errors, an editing event was not considered if its editing level was ≥90%. We also redivided the A/G SNP sites that would have editing events occurring in the individuals with homozygous genotype AA into A-to-G and G-to-A reference-to-alternative allele changes based on the human reference genome. We reexamined the similar analysis illustrated in Figure 1C and found that editing levels in synonymous A-to-G and G-to-A SNPs are not significantly different (Supplemental Fig. S14), suggesting that the potential allelic mapping bias was not the cause for our results.
Deleterious effects of genomic changes
For the genomic changes from the 1000 Genomes Project, the CADD scores of the changes were downloaded directly from the 1000 Genomes Project. For all possible nonsynonymous genomic changes in all human coding regions, the CADD scores of the changes were downloaded from the CADD browser (release v1.4) at http://cadd.gs.washington.edu/. From the 1000 Genomes Project, we extracted rare missense mutations from derived alleles (based on the human-chimpanzee-rhesus macaque orthologs) with minor allele frequency <0.01. We took G-to-A/C-to-T rare missense mutations as an example to describe the calculation of frequencies of the genomic changes with different deleterious effects in the 1000 Genomes Project. As shown in Supplemental Table S2, there are 30,112 G-to-A/C-to-T rare mutations that cause nonsynonymous changes with a CADD score ≤10 in the 1000 Genomes Project. Of all human coding regions, 366,956 G (or C) sites would have nonsynonymous changes with a CADD score ≤10 if changed to A (or T). The frequency of G-to-A/C-to-T rare missense mutations with a CADD score ≤10 is ƒ(G-to-A/C-to-T) = 30,112/366,956 = 8.2 × 10−2. The Mr. Eel mutation rates (Carlson et al. 2018) at the examined SNP sites were downloaded from the browser at http://mutation.sph.umich.edu/.
Evolutionary and functional analysis
The dN/dS ratios between human-rhesus macaque orthologs and between human and mouse orthologs were downloaded from the Ensembl genome browser (version 87). Both phyloP (Pertea et al. 2011) and phastCons (Siepel et al. 2005) scores were downloaded from the UCSC Genome Browser at https://genome.ucsc.edu. The expression levels of the host genes were measured by TPM (transcripts per kilobase million) (Wagner et al. 2012). The expression level of each Ensembl-annotated protein-coding gene was represented by the mean TPM value of the 447 LCL individuals. These expressed genes (TPM ≥ 0.01) were then divided into two equally sized groups (highly and lowly expressed genes) according to the TPM values. The essentiality of human genes was obtained from the Online GEne Essentiality (OGEE) (Chen et al. 2017) database. OGEE essential and conditionally essential genes were both considered as essential genes for this study. The pLI scores (Lek et al. 2016) of human genes were retrieved from the ExAC browser at http://exac.broadinstitute.org/. The information on 412 pleiotropic and 1345 nonpleiotropic genes was downloaded from PleiotropyDB (Ittisoponpisan et al. 2017) at http://www.sbg.bio.ic.ac.uk/pleiotropydb/home/.
Phylogenetic variation of nonsynonymous editing sites
We retrieved the pattern of phylogenetic variation for each analyzed nonsynonymous editing site from the human, chimpanzee, rhesus macaque, and mouse orthologous nucleotides. The corresponding genome coordinates of orthologous sites among these species were determined using the UCSC liftOver tool (https://genome.ucsc.edu/cgi-bin/hgLiftOver). An orthologous site was determined as an A/G coincident SNP if it was observed to be A/G polymorphic in any two examined species. The SNP information on these nonhuman species were downloaded from the Ensembl database (version 87).
Statistical analysis
Statistical analyses were performed using R software version 3.4.2 (http://www.R-project.org/) (R Core Team 2017). Statistically significant differences were determined using two-tailed Wilcoxon rank-sum tests, two-tailed Wilcoxon signed-rank tests, two-tailed Fisher's exact tests, χ2 tests, Spearman's rank correlation, and partial correlation, as appropriate. Statistically significant differences between two independent correlations were estimated from a two-tailed Z-score test using the paired.r function within the psych R library.
Supplementary Material
Acknowledgments
We thank Drs. Ben-Yang Liao, Hurng-Yi Wang, and Chia-Ying Chen for discussions and Yu-Shiang Zeng for computational assistance. This work was supported by the Genomics Research Center (GRC), Academia Sinica, Taiwan; and the Ministry of Science and Technology, Taiwan, under the contracts MOST 103-2628-B-001-001-MY4, MOST 107-2311-B-001-046, and MOST 108-2311-B-001-020-MY3 (all to T.-J.C.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.246033.118.
Freely available online through the Genome Research Open Access option.
References
- The 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An NA, Ding W, Yang XZ, Peng J, He BZ, Shen QS, Lu F, He A, Zhang YE, Tan BC, et al. 2019. Evolutionarily significant A-to-I RNA editing events originated through G-to-A mutations in primates. Genome Biol 20: 24 10.1186/s13059-019-1638-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn JH, Lee JH, Li G, Greer C, Peng G, Xiao X. 2012. Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res 22: 142–150. 10.1101/gr.124107.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbon A, Vallini I, La Via L, Marchina E, Barlati S. 2003. Glutamate receptor RNA editing: a molecular analysis of GluR2, GluR5 and GluR6 in human brain tissues and in NT2 cells following in vitro neural differentiation. Brain Res Mol Brain Res 117: 168–178. 10.1016/S0169-328X(03)00317-6 [DOI] [PubMed] [Google Scholar]
- Bird AP. 1980. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res 8: 1499–1504. 10.1093/nar/8.7.1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brümmer A, Yang Y, Chan TW, Xiao X. 2017. Structure-mediated modulation of mRNA abundance by A-to-I editing. Nat Commun 8: 1255 10.1038/s41467-017-01459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusa R, Zimmermann F, Koh DS, Feldmeyer D, Gass P, Seeburg PH, Sprengel R. 1995. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science 270: 1677–1680. 10.1126/science.270.5242.1677 [DOI] [PubMed] [Google Scholar]
- Carlson J, Locke AE, Flickinger M, Zawistowski M, Levy S, Myers RM, Boehnke M, Kang HM, Scott LJ, Li JZ, et al. 2018. Extremely rare variants reveal patterns of germline mutation rate heterogeneity in humans. Nat Commun 9: 3753 10.1038/s41467-018-05936-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. 2013. Characterization and comparison of human nuclear and cytosolic editomes. Proc Natl Acad Sci 110: E2741–E2747. 10.1073/pnas.1218884110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Peng Z, Zhang R, Yang XZ, Tan BC, Fang H, Liu CJ, Shi M, Ye ZQ, Zhang YE, et al. 2014. RNA editome in rhesus macaque shaped by purifying selection. PLoS Genet 10: e1004274 10.1371/journal.pgen.1004274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Hung LY, Wu CS, Chuang TJ. 2016. Purifying selection shapes the coincident SNP distribution of primate coding sequences. Sci Rep 6: 27272 10.1038/srep27272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WH, Lu G, Chen X, Zhao XM, Bork P. 2017. OGEE v2: an update of the online gene essentiality database with special focus on differentially essential genes in human cancer cell lines. Nucleic Acids Res 45: D940–D944. 10.1093/nar/gkw1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang TJ, Chiang TW. 2014. Impacts of pretranscriptional DNA methylation, transcriptional transcription factor, and posttranscriptional microRNA regulations on protein evolutionary rate. Genome Biol Evol 6: 1530–1541. 10.1093/gbe/evu124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang TJ, Chen FC, Chen YZ. 2012. Position-dependent correlations between DNA methylation and the evolutionary rates of mammalian coding exons. Proc Natl Acad Sci 109: 15841–15846. 10.1073/pnas.1208214109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S, Fay JC. 2009. Identification of deleterious mutations within three human genomes. Genome Res 19: 1553–1561. 10.1101/gr.092619.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. 1978. Molecular basis of base substitution hotspots in Escherichia coli. Nature 274: 775–780. 10.1038/274775a0 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggington JM, Greene T, Bass BL. 2011. Predicting sites of ADAR editing in double-stranded RNA. Nat Commun 2: 319 10.1038/ncomms1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E. 2012. Bioinformatic approaches for identification of A-to-I editing sites. Curr Top Microbiol Immunol 353: 145–162. 10.1007/82_2011_147 [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. 2018. A-to-I RNA editing—immune protector and transcriptome diversifier. Nat Rev Genet 19: 473–490. 10.1038/s41576-018-0006-1 [DOI] [PubMed] [Google Scholar]
- Gal-Mark N, Shallev L, Sweetat S, Barak M, Li JB, Levanon EY, Eisenberg E, Behar O. 2017. Abnormalities in A-to-I RNA editing patterns in CNS injuries correlate with dynamic changes in cell type composition. Sci Rep 7: 43421 10.1038/srep43421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommans WM, Mullen SP, Maas S. 2009. RNA editing: a driving force for adaptive evolution? Bioessays 31: 1137–1145. 10.1002/bies.200900045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C. 2002. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron 34: 349–356. 10.1016/S0896-6273(02)00660-8 [DOI] [PubMed] [Google Scholar]
- Harris K, Pritchard JK. 2017. Rapid evolution of the human mutation spectrum. eLife 6: e24284 10.7554/eLife.24284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, Botigué LR, Bustamante CD, Clark AG, Gravel S. 2015. Estimating the mutation load in human genomes. Nat Rev Genet 16: 333–343. 10.1038/nrg3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideyama T, Yamashita T, Aizawa H, Tsuji S, Kakita A, Takahashi H, Kwak S. 2012. Profound downregulation of the RNA editing enzyme ADAR2 in ALS spinal motor neurons. Neurobiol Dis 45: 1121–1128. 10.1016/j.nbd.2011.12.033 [DOI] [PubMed] [Google Scholar]
- Hodgkinson A, Ladoukakis E, Eyre-Walker A. 2009. Cryptic variation in the human mutation rate. PLoS Biol 7: e1000027 10.1371/journal.pbio.1000027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung LY, Chen YJ, Mai TL, Chen CY, Yang MY, Chiang TW, Wang YD, Chuang TJ. 2018. An evolutionary landscape of A-to-I RNA editome across metazoan species. Genome Biol Evol 10: 521–537. 10.1093/gbe/evx277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittisoponpisan S, Alhuzimi E, Sternberg MJ, David A. 2017. Landscape of pleiotropic proteins causing human disease: structural and system biology insights. Hum Mutat 38: 289–296. 10.1002/humu.23155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Ito K, Sun H, Ito M, Kanazawa I, Kwak S. 2004. Regulation of glutamate receptor RNA editing and ADAR mRNA expression in developing human normal and Down's syndrome brains. Brain Res Dev Brain Res 148: 151–155. 10.1016/j.devbrainres.2003.11.008 [DOI] [PubMed] [Google Scholar]
- Keller TE, Han P, Yi SV. 2016. Evolutionary transition of promoter and gene body DNA methylation across invertebrate–vertebrate boundary. Mol Biol Evol 33: 1019–1028. 10.1093/molbev/msv345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido T, Sikora-Wohlfeld W, Kawashima M, Kikuchi S, Kamatani N, Patwardhan A, Chen R, Sirota M, Kodama K, Hadley D, et al. 2018. Are minor alleles more likely to be risk alleles? BMC Med Genomics 11: 3 10.1186/s12920-018-0322-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Yi SV. 2007. Understanding relationship between sequence and functional evolution in yeast proteins. Genetica 131: 151–156. 10.1007/s10709-006-9125-2 [DOI] [PubMed] [Google Scholar]
- Kimura M, Maruyama T, Crow JF. 1963. The mutation load in small populations. Genetics 48: 1303–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JL, Jukes TH. 1969. Non-Darwinian evolution. Science 164: 788–798. 10.1126/science.164.3881.788 [DOI] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46: 310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman CL, Adoue V, Majewski J. 2012. RNA editing of protein sequences: a rare event in human transcriptomes. RNA 18: 1586–1596. 10.1261/rna.033233.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurmangaliyev YZ, Ali S, Nuzhdin SV. 2016. Genetic determinants of RNA editing levels of ADAR targets in Drosophila melanogaster. G3 (Bethesda) 6: 391–396. 10.1534/g3.115.024471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Sammeth M, Friedländer MR, ‘t Hoen PA, Monlong J, Rivas MA, Gonzàlez-Porta M, Kurbatova N, Griebel T, Ferreira PG, et al. 2013. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501: 506–511. 10.1038/nature12531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann KA, Bass BL. 2000. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry 39: 12875–12884. 10.1021/bi001383g [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Levanon EY, Yoon J-K, Aach J, Xie B, LeProust E, Zhang K, Gao Y, Church GM. 2009. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 324: 1210–1213. 10.1126/science.1170995 [DOI] [PubMed] [Google Scholar]
- Liscovitch-Brauer N, Alon S, Porath HT, Elstein B, Unger R, Ziv T, Admon A, Levanon EY, Rosenthal JJ, Eisenberg E. 2017. Trade-off between transcriptome plasticity and genome evolution in cephalopods. Cell 169: 191–202.e11. 10.1016/j.cell.2017.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE. 2014. The distribution of deleterious genetic variation in human populations. Curr Opin Genet Dev 29: 139–146. 10.1016/j.gde.2014.09.005 [DOI] [PubMed] [Google Scholar]
- Lohmueller KE, Indap AR, Schmidt S, Boyko AR, Hernandez RD, Hubisz MJ, Sninsky JJ, White TJ, Sunyaev SR, Nielsen R, et al. 2008. Proportionally more deleterious genetic variation in European than in African populations. Nature 451: 994–997. 10.1038/nature06611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB, et al. 2012. A systematic survey of loss-of-function variants in human protein-coding genes. Science 335: 823–828. 10.1126/science.1215040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. 2003. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 31: 3812–3814. 10.1093/nar/gkg509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. 2010. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem 79: 321–349. 10.1146/annurev-biochem-060208-105251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osenberg S, Yaacov NP, Safran M, Moshkovitz S, Shtrichman R, Sherf O, Jacob-Hirsch J, Keshet G, Amariglio N, Itskovitz-Eldor J. 2010. Alu sequences in undifferentiated human embryonic stem cells display high levels of A-to-I RNA editing. PLoS One 5: e11173 10.1371/journal.pone.0011173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Gail MH, Weinberg CR, Carroll RJ, Chung CC, Wang Z, Chanock SJ, Fraumeni JF Jr, Chatterjee N. 2011. Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proc Natl Acad Sci 108: 18026–18031. 10.1073/pnas.1114759108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Salzberg SL. 2011. Detection of lineage-specific evolutionary changes among primate species. BMC Bioinformatics 12: 274 10.1186/1471-2105-12-274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto Y, Cohen HY, Levanon EY. 2014. Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol 15: R5 10.1186/gb-2014-15-1-r5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popitsch N, Huber CD, Buchumenski I, Eisenberg E, Jantsch M, von Haeseler A, Gallach M. 2017. A-to-I RNA editing uncovers hidden signals of adaptive genome evolution in animals. bioRxiv 10.1101/228734 [DOI] [PMC free article] [PubMed]
- Porath HT, Schaffer AA, Kaniewska P, Alon S, Eisenberg E, Rosenthal J, Levanon EY, Levy O. 2017. A-to-I RNA editing in the earliest-diverging eumetazoan phyla. Mol Biol Evol 34: 1890–1901. 10.1093/molbev/msx125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Ramaswami G, Lin W, Piskol R, Tan MH, Davis C, Li JB. 2012. Accurate identification of human Alu and non-Alu RNA editing sites. Nat Methods 9: 579–581. 10.1038/nmeth.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. 2019. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 47: D886–D894. 10.1093/nar/gky1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050. 10.1101/gr.3715005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon O, Bazak L, Levanon EY, Amariglio N, Unger R, Rechavi G, Eyal E. 2014. Characterizing of functional human coding RNA editing from evolutionary, structural, and dynamic perspectives. Proteins 82: 3117–3131. 10.1002/prot.24672 [DOI] [PubMed] [Google Scholar]
- Sommer B, Köhler M, Sprengel R, Seeburg PH. 1991. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67: 11–19. 10.1016/0092-8674(91)90568-J [DOI] [PubMed] [Google Scholar]
- Srivastava PK, Bagnati M, Delahaye-Duriez A, Ko JH, Rotival M, Langley SR, Shkura K, Mazzuferi M, Danis B, van Eyll J, et al. 2017. Genome-wide analysis of differential RNA editing in epilepsy. Genome Res 27: 440–450. 10.1101/gr.210740.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN. 2009. The Human Gene Mutation Database: 2008 update. Genome Med 1: 13 10.1186/gm13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, Hussain M, Phillips AD, Cooper DN. 2017. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet 136: 665–677. 10.1007/s00439-017-1779-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MH, Li Q, Shanmugam R, Piskol R, Kohler J, Young AN, Liu KI, Zhang R, Ramaswami G, Ariyoshi K, et al. 2017. Dynamic landscape and regulation of RNA editing in mammals. Nature 550: 249–254. 10.1038/nature24041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, et al. 2012. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337: 64–69. 10.1126/science.1219240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian N, Wu X, Zhang Y, Jin Y. 2008. A-to-I editing sites are a genomically encoded G: implications for the evolutionary significance and identification of novel editing sites. RNA 14: 211–216. 10.1261/rna.797108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Kin K, Lynch VJ. 2012. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci 131: 281–285. 10.1007/s12064-012-0162-3 [DOI] [PubMed] [Google Scholar]
- Wahlstedt H, Daniel C, Ensterö M, Öhman M. 2009. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res 19: 978–986. 10.1101/gr.089409.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkley CR, Li JB. 2017. Rewriting the transcriptome: adenosine-to-inosine RNA editing by ADARs. Genome Biol 18: 205 10.1186/s13059-017-1347-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Zhang J. 2014. Human coding RNA editing is generally nonadaptive. Proc Natl Acad Sci 111: 3769–3774. 10.1073/pnas.1321745111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Chen Y, Ayub Q, Huang N, Ball EV, Mort M, Phillips AD, Shaw K, Stenson PD, Cooper DN, et al. 2012. Deleterious- and disease-allele prevalence in healthy individuals: insights from current predictions, mutation databases, and population-scale resequencing. Am J Hum Genet 91: 1022–1032. 10.1016/j.ajhg.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.