Abstract
MicroRNAs (miRNAs) play a critical role as posttranscriptional regulators of gene expression. The ENCODE Project profiled the expression of miRNAs in an extensive set of organs during a time-course of mouse embryonic development and captured the expression dynamics of 785 miRNAs. We found distinct organ-specific and developmental stage–specific miRNA expression clusters, with an overall pattern of increasing organ-specific expression as embryonic development proceeds. Comparative analysis of conserved miRNAs in mouse and human revealed stronger clustering of expression patterns by organ type rather than by species. An analysis of messenger RNA expression clusters compared with miRNA expression clusters identifies the potential role of specific miRNA expression clusters in suppressing the expression of mRNAs specific to other developmental programs in the organ in which these miRNAs are expressed during embryonic development. Our results provide the most comprehensive time-course of miRNA expression as part of an integrated ENCODE reference data set for mouse embryonic development.
Development is a well-orchestrated process primarily controlled by transcriptional regulators with posttranscriptional regulators such as microRNAs (miRNAs) playing an essential role in fine tuning gene expression dynamics. miRNAs are small ∼22 nucleotide (nt) endogenous noncoding RNAs that regulate gene expression by mediating the posttranscriptional degradation of messenger RNA (mRNA) or by hindering the translation of proteins (Bartel 2004; He and Hannon 2004). miRNA biogenesis occurs in several steps, starting with transcription of typically polyadenylated primary miRNA (pri-miRNA) transcripts (>200 nt), sometimes referred to as the “host genes.” These pri-miRNAs have a characteristic hairpin structure that is cleaved in the nucleus by the enzyme Drosha into pre-miRNA (∼60 nt), which are exported to the cytoplasm before finally being processed into 21- to 24-nt mature miRNA by the enzyme Dicer (Han et al. 2006). The first miRNA was discovered in the nematode Caenorhabditis elegans as perturbing its cell developmental lineage (Lee et al. 1993), and since then, thousands of miRNAs have been discovered in diverse plants, metazoans, and some viruses (Kozomara and Griffiths-Jones 2011).
Many studies have shown that the deletion of key players in the biogenesis of miRNA such as Ago2, Dicer1, and Dgcr8 leads to embryonic lethality and arrest (Bernstein et al. 2003; Alisch et al. 2007; Morita et al. 2007; Wang et al. 2007). However, the loss of single miRNAs does not have as dramatic an effect as knocking out all the miRNAs in the organism (Park et al. 2010). This could be because of the redundancy of miRNA–mRNA interactions as each mRNA could be targeted by multiple miRNAs, and thus, the lack of one miRNA would be compensated by others. Hence, there is a strong rationale for studying the role of miRNAs as a functional group or unit. Studies have shown that most genes are potential targets of miRNAs (Friedman et al. 2009) and that miRNAs are involved in regulating diverse cellular processes during development and homeostasis (Vidigal and Ventura 2015). Dysregulation of miRNA expression is known to underlie numerous diseases and developmental defects such as cancer (Lin and Gregory 2015), cardiovascular diseases (Romaine et al. 2015; Zhao et al. 2015), and neurological diseases (Cao et al. 2016).
miRNAs have been profiled in various tissues and primary cells in diverse metazoans and plants (Lagos-Quintana et al. 2002; Wienholds et al. 2005; Ehrenreich and Purugganan 2008). Mineno et al. (2006) used massively parallel signature sequencing (MPSS) technology to profile miRNAs in mouse whole embryos during three embryonic stages (E9.5, E10.5, and E11.5) and were able to detect 390 distinct miRNAs. Chiang et al. (2010) extended this work by sequencing small RNAs from mouse brain, ovary, testes, embryonic stem cells, embryonic stages of complete embryos from three developmental stages, and whole newborns to profile the expression of 398 annotated and 108 novel miRNAs. Landgraf et al. (2007) cloned and sequenced more than 250 small RNA libraries from 26 different organs and cell types from humans and rodents to profile miRNA expression and describe various other miRNA characteristics. More recently, the FANTOM5 project has created a miRNA expression atlas using deep-sequencing data from 396 human and 47 mouse RNA samples (De Rie et al. 2017); however, many of these mouse samples were simply replicates of a handful of mouse cell lines. Previous efforts by the ENCODE Consortium affiliates focused on a meta-analysis of previously published 501 human and 236 mouse small RNA sequencing data sets from a multitude of sources to characterize splicing-derived miRNAs (mirtrons) in the human and mouse genomes (Ladewig et al. 2012). However, the diversity of the source tissues and the different underlying experimental protocols from the disparate primary sources complicated any sort of systematic quantitative analysis. Last but not least, many individual studies have focused on the expression of particular miRNAs in certain tissues in a handful of (typically two to three) mouse developmental time points. Therefore, a complete and systematic atlas of miRNA expression during development of the major organ systems and broad number of mouse embryonic stages is still missing. This is helpful not only for understanding mouse development but also for studying the potential role of miRNAs in human development, in which access to the same time points is either very difficult or outright impossible.
With the growing evidence of the critical role of miRNAs in homeostasis and disease, multiple techniques have been developed for profiling the expression of mature miRNAs, each with their own strengths (Mestdagh et al. 2014). RNA-seq typically refers to the profiling of expressed transcripts ≥200 nt, including the mRNAs and long noncoding RNAs (lncRNAs) (Mortazavi et al. 2008), which in this work we will refer to as mRNA-seq, whereas there are also multiple miRNA-specific sequencing protocols such as miRNA-seq (Alon et al. 2011; Roberts et al. 2015) and short RNA-seq (Fejes-Toth et al. 2009). There are also hybridization-based assays, such as microarrays, as well as molecule counting such as NanoString, which involves hybridization and counting of color-coded molecular barcodes (Geiss et al. 2008; Wyman et al. 2011). As mature miRNAs are processed from longer host pri-miRNAs and the annotated pri-miRNAs are predominantly protein-coding or lncRNA transcripts (Cai et al. 2004), we expect that mRNA-seq should be able to profile the expression of pri-miRNAs. However, there is a significant number of miRNAs whose host genes have not been characterized yet. Furthermore, an important question is whether the expression of pri-miRNAs can reliably predict the expression of their corresponding mature miRNAs. As previously reported (Zeng et al. 2016; De Rie et al. 2017), this would allow the simultaneous profiling of mature miRNA expression along with mRNAs using mRNA-seq. The availability of matching mRNA-seq and miRNA-seq data sets for the same samples in our study provides a unique opportunity to answer this question using a broader set of organs and developmental time points. Furthermore, the corresponding mRNA data can shed light into the targeting of these miRNAs and their functional role during embryonic development.
Each miRNA targets a set of mRNAs through Watson–Crick pairing between miRNA seed region (positions 2–7 from 5′ end) and the binding sites on their targets (Bartel 2009). Such complementary base-pairing has been used to computationally predict miRNA targets (Bartel 2009). The expression of miRNAs and mRNAs in matching samples has been used to identify miRNA–mRNA interactions, for example, in cancer (The Cancer Genome Atlas Research Network 2008). Several methods such as biclustering (Jin and Lee 2015) have been used to infer miRNA–mRNA interactions from gene expression data. However, the expression levels of mRNAs are often affected by multiple factors, and comparison of mRNA and miRNA expressions cannot establish a functional relationship by itself. Therefore, an approach that integrates miRNA and mRNA expression data and their predicted interactions should provide better inference of their functional interaction networks.
In this study, we used miRNA-seq and NanoString to characterize the expression patterns of known miRNAs using a set of 16 different mouse organs (we use the term interchangeably with tissues in this study) at eight embryonic (E10.5–P0) stages that were specifically selected by the ENCODE Consortium for a wide variety of functional sequencing assays such as mRNA-seq, chromatin immunoprecipitation followed by sequencing (ChIP-seq), and DNase-seq. The value of this data set is that the samples and stages are all matched. We show one example of integrative analysis of the mRNA-seq data with matching ENCODE mRNA-seq and ChIP-seq data (Gorkin et al. 2017) to compare the characteristics and dynamics of miRNA expression to characterize the changes in overall tissue specificity of particular miRNAs during mouse development. In particular, we compute the enrichment of computationally predicted miRNA targets in specific mouse organs along with the negative partial correlation analysis of miRNA and mRNA expression clusters during mouse development to identify developmental processes targeted by miRNAs. We find that groups of miRNAs expressed in one or more organs target groups of developmentally important mRNAs highly expressed in other organs.
Results
A reference miRNA catalog across mouse development
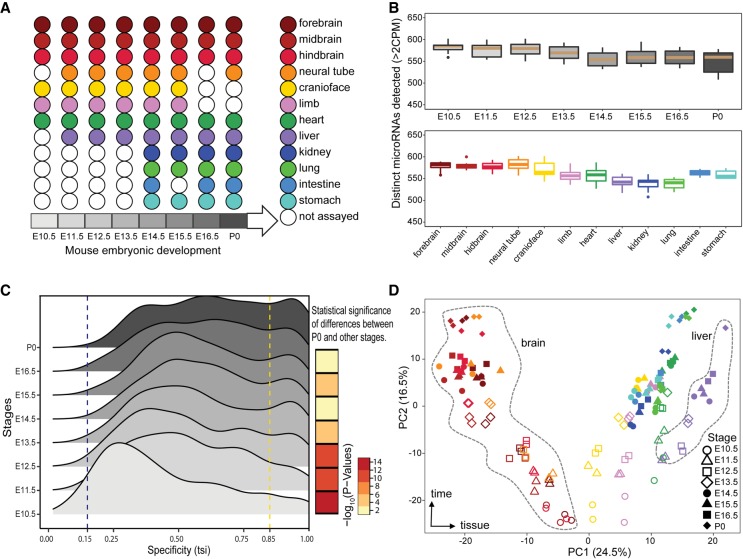
As part of the ENCODE Project, we used miRNA-seq and NanoString to profile mature miRNAs during mouse embryonic development and matched them to mRNA-seq in order to profile the expression of pri-miRNAs (Supplemental Fig. S1A). This study encompasses 156 miRNA-seq and 154 NanoString data sets in matching mouse organs with two biological replicates each (Fig. 1A). We found a high correlation between miRNA-seq and NanoString data in the same organs at the same time points (median Spearman's correlation = 0.68) (Supplemental Fig. S1B), which matches the reproducibility between platforms reported previously (Mestdagh et al. 2014). We subsampled one of our deeply sequenced data sets (heart E11.5 with 60 million reads) to evaluate the impact of sequencing depth on the robust detection of miRNAs. Although we detect more miRNAs as we sequence deeper, we detect a constant number using a counts per million (CPM)–based cutoff and we selected 1 million mapped reads to be the sufficient depth of sequencing required for detecting most of miRNAs expressed in a sample (Supplemental Fig. S2). All of our samples in this study were sequenced to a minimum of 2 million mapped reads per replicate.
Figure 1.
Overview of mouse ENCODE miRNA data sets. (A) Representative major organ systems were profiled in a time-course of mouse embryonic development. (B) Number of distinct miRNAs detected in different organs and developmental stages (minimum two CPM). There are no significant differences between the number of miRNAs detected at different stages or within different organs. Developmental stage and organ colors correspond to A. (C) The distribution of tissue specificity of miRNAs expressed at each developmental stage measured as tissue-specific index (TSI). The miRNAs are significantly more tissue-specific at stage of P0 compared with E10.5 (Kolmogorov–Smirnov [KS] test P-value < 2.2 × 10−16). (D) Principal component analysis (PCA) of 12 mouse organs across eight developmental stages. Organs are represented by various colors corresponding to A, whereas shapes denote the different developmental stages.
We used a set of three spike-ins of different sequence lengths (22, 25, and 30 bp) in decreasing concentrations (5000, 500, and 50 pM, respectively) in our miRNA-seq samples to assess replicate concordance for different library normalization strategies (Supplemental Fig. S1C). Although the spike-in counts were highly concordant for biological replicates for each sample, they differ for different stages of mouse embryonic development using CPM normalization only. We found that TMM normalization of miRNA CPMs ameliorates such differences in spike-in expression across developmental stages. Therefore, we normalized our data using TMM normalization for downstream analysis.
We used miRNA-seq reads to quantify miRNA expression levels using miRBase version 22 annotations, which include 1981 mature miRNAs. We detected 785 of these mature miRNAs expressed in at least one of the samples; About 80% of these mature miRNAs correspond to the pre-miRNAs identified as high confidence by miRBase (Kozomara and Griffiths-Jones 2014). This set of miRNAs encompasses 72% of high-confidence miRNAs in miRBase compared with the 65% recovered by the FANTOM data set (De Rie et al. 2017) and 71% of miRNAs annotated in MirGeneDB (Supplemental Fig. S3; Fromm et al. 2018). We detect additional miRNAs if we use a more lenient cutoff of one read as opposed to two CPM (miRBase: 72%, miRBase high confidence: 92%, and MirGeneDB: 77%). There are no significant differences in the number of distinct miRNAs expressed in mouse organs and developmental stages, although stage P0 has the highest number of organs profiled as well as the least number of distinct miRNAs detected (Fig. 1B). This result is in contrast to the finding that the absolute numbers of expressed miRNAs increase over the developmental time in other model organisms such as Drosophila melanogaster (Ninova et al. 2014). At the organ level, we find that the nervous system samples show the highest number of distinct miRNAs expressed (Fig. 1B).
Dynamics of miRNA tissue specificity during development
As previous studies have shown, a few highly expressed miRNAs are responsible for most of the detected expression (Lagos-Quintana et al. 2002; Landgraf et al. 2007), with ∼50% of the expression corresponding to the top 10 expressed miRNAs (Supplemental Fig. S4; Supplemental Table S2). Only 42 miRNAs fall within the top 10 expressed miRNAs across our 72 distinct tissue-stage samples. Six of these miRNAs are in the top 10 expressed list for more than half of the samples, with miR-16-5p and miR-26a-5p being one of the top expressed miRNAs in every single experiment. To study the specificity of the miRNAs at each stage, we used the tissue specificity index (TSI) as defined previously (Ludwig et al. 2016); by using this metric, we found that 40% of the top expressed miRNAs are tissue specific in at least one of the stages that they are highly expressed in. These miRNAs include miR-1a-3p, miR-208b-3p, and miR-351-5p in the heart (the last one is only specific in the earlier stages); miR-9-3p, miR-9-5p, miR-124-3p, miR-125b-5p, and miR-92b-3p in the brain; miR-122-5p and miR-142a-3p in the liver; miR-10a-5p in the kidney; miR-194-5p in the intestine; and miR-196b-5p in the limbs (Supplemental Fig. S5).
Although there are few miRNAs that are expressed ubiquitously (TSI < 0.15) at the earlier stages of embryonic development, most miRNAs become more tissue-specific as the embryo develops further and the expression of miRNAs shifts from ubiquitous to being organ specific (Fig. 1C; Supplemental Fig. S6). This shift is partly owing to changes in the specificity of the miRNAs throughout development with the following miRNAs showing the most change: miR-128-3p, miR-181a-1-3p, miR-138-5p, and miR-3099-3p in the brain; miR-101a-3p and miR-496a-3p in the liver; and miR-140-5p in the limbs (Supplemental Fig. S7). All of these miRNAs increase in their specificity from being almost ubiquitous to become tissue-specific. However, there is a group of more than 20 miRNAs that stay mostly tissue-specific throughout the developmental time points captured in our study. This group includes some of the well-studied tissue-specific miRNAs such as miR-9 and miR-92b-3p in the brain; miR-1a-3p, miR-208a-3p, and miR-133a-3p in the heart; and miR-122-5p in the liver (Supplemental Fig. S8). Finally, there is a group of miRNAs that are present in almost all the tissues at every stage of development, including miR-421-3p, miR-361-5p, and miR-744-5p. (Supplemental Fig. S9). In summary, our high-resolution time-course captures the distinct patterns of miRNA expression during mouse embryonic development.
Clustering of miRNAs recovers distinct tissue-specific clusters
Global analysis of mouse tissues and developmental stages shows distinct miRNA expression patterns as revealed by principal component analysis (PCA) (Fig. 1D). Principal component (PC) 1 accounts for 24.5% of the variation and clearly separates the various tissues with the nervous system and liver tissues at the extremes, whereas PC2 (16.5% variation) represents the time component of mouse development with a temporal gradient between early development at E10.5 and postnatal samples right after birth (P0) (Fig. 1D), PC3 (10.8% variation) separates kidney samples from liver, PC4 (6.1% variation) separates heart samples from other tissues, and PC5 (4.5% variation) separates kidney samples from limb and craniofacial samples. Overall the first five PCs explained >60% of the variation in the data set with most of that variation corresponding to specific tissues.
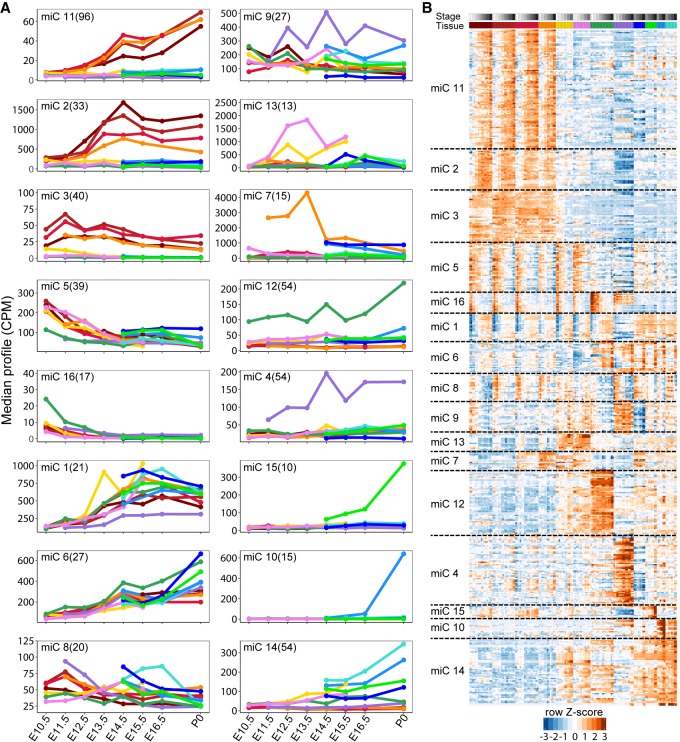
We used maSigPro (Nueda et al. 2014) to cluster the 785 expressed miRNAs based on the tissue-specific changes in their expression during the development. maSigPro identified 535 of these miRNAs as being differentially expressed (Supplemental Table S3) during embryonic development into 16 clusters based on regression of their expression levels in each of the tissues (Fig. 2; Supplemental Fig. S10). Clustering of the matching NanoString miRNA data recovers 14 clusters, the majority of which match the miRNA-seq clusters (Supplemental Fig. S11). We focus on the miRNA-seq clusters going forward. Cluster 11 has the highest number of miRNAs (96 miRNAs) that are highly expressed in the brain. Additionally, the expression of these miRNAs increases during embryonic development, whereas in cluster 2, another brain-specific cluster, the expression of miRNAs goes up initially and comes down after E14. miRNA clusters 4, 12, and 14 are the second largest clusters, with 54 miRNAs each. Clusters 4 and 12 are composed of miRNAs mostly expressed in the liver and heart, respectively, whereas miRNAs in cluster 14 are highly expressed in all the tissues except in the liver and brain. Analysis of tissue-specific miRNAs in each cluster reveals that more than half of the miRNA clusters are enriched for specificity to only one organ with the rest of clusters enriched for specificity in two or three different organs (Supplemental Fig. S12). Thus, miRNAs during development show distinct clustered expression in select tissues.
Figure 2.
Clustering of mouse miRNAs during embryonic development time-course. (A) Clustering of miRNAs using maSigPro into 16 nonredundant groups based on median expression level of the miRNAs in each cluster. Organ colors correspond to Figure 1A. (B) Heatmap of the normalized expression levels (z-scores) of miRNAs in each cluster from A. Organ and stage colors correspond to Figure 1A.
Comparative dynamics of conserved miRNAs during development
The ENCODE Consortium also collected a limited set of human developmental samples that were characterized with short RNA-seq to profile pre-miRNAs and mature miRNAs for a total of 32 data sets in various tissues during weeks 19–40 of human fetal development that correspond primarily to P0 of mouse development (Supplemental Fig. S13A). We compared the consistency of miRNA-seq and short RNA-seq in human K562 and GM12878 cells and compared them to data sets from a previous phase of ENCODE. We show that there is high correlation between miRNA-seq and short RNA-seq (Pearson's correlation = 0.99) (Supplemental Fig. S13B). This level of reproducibility clearly allows us to differentiate between different cell types as well as across different methods and batches (Supplemental Fig. S13C), which allows us to compare human and mouse miRNA expression levels across the two sequencing techniques.
We quantified human known and novel miRNAs using short RNA-seq and GENCODE v. 25 annotations consisting of 1569 known miRNAs supplemented with the novel miRNAs. A global PCA analysis of miRNA expression shows the brain samples clustering as previously seen in mouse (Supplemental Fig. S14A). Although the availability of human samples was more limited compared with mouse, we identified 279 tissue-specific miRNAs (Supplemental Table S4), most of which (83%) are preferentially expressed in neuronal and muscular tissues (Supplemental Fig. S14B).
To compare miRNA expression across development, we first searched for orthologous miRNAs between mouse and human (Supplemental Fig. S15A). We found that a subset of miRNAs is conserved between mouse and human, with 304 miRNAs having a one-to-one orthologous relationship (Supplemental Table S5). Our analysis also revealed that 838 and 516 miRNAs in human and mouse, respectively, lack a clear ortholog in the other species (Supplemental Fig. S15A). We used the set of one-to-one miRNA orthologs to perform PCA on matching organs in mouse and human, which revealed clustering of samples based on tissue type (Supplemental Fig. S15B). Furthermore, comparative clustering of organs in mouse and human reveals distinct miRNA expression patterns similar to the clustering of organs in mouse only. As in the mouse developmental time-course, the nervous system and liver tissues cluster separately from the rest of the organs. We compared the sets of organ-specific orthologous miRNAs across all the available organs in mouse and human and represented each comparison as pie charts, where the sizes of the pie charts are in proportion to the number of tissue-specific miRNAs (Supplemental Fig. S15C). We found that muscle organs in mouse and human show the highest conservation (>50%) of expression and that the conservation of expression among the corresponding brain, neural tube, and lung organs is significant (∼50%), whereas the conservation of expression between the liver samples is low (Supplemental Fig. S15C; Supplemental Table S6). Therefore, the conservation of organ expression of homologous miRNAs between human and mouse is dependent on the specific organ.
Correlation of expression among pri-miRNAs and their corresponding mature miRNAs
The availability of matching miRNA-seq and mRNA-seq data allowed us to evaluate whether the expression of pri-miRNAs is predictive of the expression of their corresponding mature miRNAs. Fewer than 50% of miRNAs in mouse have annotated primary transcripts in GENCODE version M10 (Supplemental Fig. S16). We used mRNA-seq data to assemble additional transcript models and supplement the GENCODE annotations, which increased the number of pri-miRNAs in mouse and human by 7% and 17%, respectively. A representative novel model transcript in mouse, assembled using all mouse mRNA-seq data sets, overlaps miR-let7a and miR-let7f that were lacking annotated pri-miRNAs and is supported by stage-matched ChIP-seq for both H3K4me3 marking the putative promoter (Supplemental Figs. S16A,B, S17) and H3K36me3, as correlate of transcription. Global correlation analysis of the expression levels of the pri-miRNAs (measured by mRNA-seq) and their corresponding miRNAs (measured by miRNA-seq) shows that 143 (41% of the miRNAs expressed at a minimum of 10 CPM) are well correlated (Spearman's correlation ≥ 0.6) with their corresponding pri-miRNAs across the developmental time-course. The median Spearman's correlation for all the miRNAs and their corresponding pri-miRNAs is 0.51 (Supplemental Fig. S16C,D; Supplemental Table S7). Thus, miRNA expression can be imputed from the expression of its primary transcript as measured by mRNA-seq and confirmed using the matching ENCODE ChIP-seq data resources.
Integrative analysis of miRNA and mRNA expression profiles during mouse development identifies significant anticorrelations of developmentally important genes with miRNAs predicted to target them
To understand the connection between miRNAs and the expression of their targets, we developed an integrative analysis pipeline to connect miRNAs to their mRNA targets (Fig. 3A). As a first step, we quantified the tissue specificity of miRNA clusters by computing a tissue specificity matrix. The tissue specificity of each miRNA cluster was determined based on the expression changes of miRNAs in each tissue during development. The tissues that had the highest standard deviation of a given miRNA cluster's expression in different stages were identified as the tissue specificity of that cluster. The tissue specificity of the miRNA clusters calculated in this manner are highly concordant with the tissue specificities obtained by the specificity analysis of the individual miRNAs in each cluster. There is at least one miRNA cluster identified for each tissue and at least one tissue identified as tissue-specificity of each miRNA cluster (Supplemental Fig. S12; Supplemental Table S8).
Figure 3.
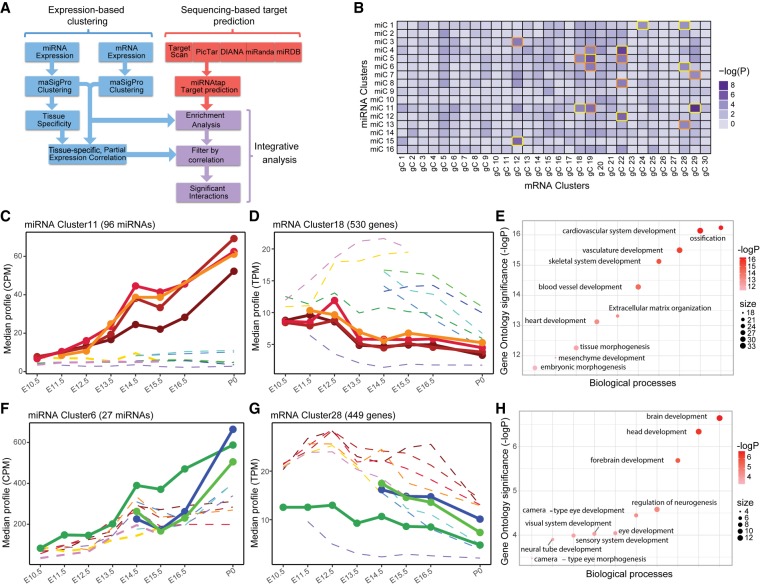
Identification of miRNA–mRNA cluster interactions. (A) Potential targets of each miRNA cluster were obtained by applying an ensemble approach. Interactions were called as significant if they had a negative tissue-specific partial correlation and were enriched beyond the Bonferroni-corrected P-value of 10−4. (B) Heatmap of miRNA cluster target enrichment calculated using χ2 statistics. The 18 interactions identified as enriched are boxed in orange and gold. The interactions boxed in gold have negative partial correlation and are identified as significant interactions. (C) miRNA cluster 11 corresponds to brain-specific miRNAs up-regulated during development. (D) mRNA cluster 18 genes are highly expressed in other organs such as limbs, cranioface, and heart. (E) Gene Ontology of miRNA cluster 11 targets in mRNA cluster 18 shows enrichments in developmentally important genes with roles outside the brain. (F) miRNA cluster 6 increases significantly during heart development. (G) mRNA cluster 28 genes are overexpressed in the brain. (H) Gene Ontology analysis of miRNA cluster 6 targets in mRNA cluster 28 revealed terms such as brain, head, and forebrain development.
We clustered mouse developmental mRNAs from ENCODE using maSigPro into 30 clusters incorporating 14,827 differentially expressed genes out of the 20,686 genes that were expressed at least once during the development with a replicate average expression of at least two TPM (Supplemental Fig. S18; Supplemental Table S9). About one third of these clusters are specific to a single tissue, with the rest being expressed in multiple tissues. The largest three clusters are clusters 9, 3, and 12 with 1773, 1214, and 884 genes in them, respectively, and all three of these clusters correspond to genes expressed in brain. Most of the tissue-specific clusters correspond to the liver, heart, and lung after the brain.
After identifying the clusters of miRNA and mRNA that are dynamically expressed during development, we calculated the partial correlation between each of the miRNA clusters and each of the mRNA clusters. The partial correlation matrix was built using Pearson's correlation between each pair of clusters (miRNA–mRNA) within the context of tissues that the miRNA cluster was active in (using only the miRNA tissue specificity as the context) (Supplemental Fig. S19; Supplemental Table S10). Using this partial correlation approach, 60% of the miRNA–mRNA cluster interactions are anticorrelated with a mean correlation coefficient value of −0.47. This anticorrelation was used to filter out the positive interactions after target enrichment analysis.
We collected the predicted targets of each of the miRNAs from five different resources and prediction algorithms using miRNAtap (https://bioconductor.org/packages/release/bioc/html/miRNAtap.html). We used the unique set of all the predicted targets for miRNAs in each of the miRNA clusters to build a contingency table that contains the distribution of each of these unique target sets among the mRNA clusters. We then performed a χ2 test on the contingency table to study the enrichment of targets in different mRNA clusters (Fig. 3B; Supplemental Table S11) and applied a P-value cutoff of 10−4 (Bonferroni corrected P-value: 0.05/[16 × 30]) to determine the mRNA clusters that were significantly enriched for miRNA cluster targets. Eighteen interactions between 11 unique miRNA clusters and seven unique mRNA cluster were identified as significant; however, only nine of these interactions passed the filter for negative partial correlation (Supplemental Fig. S20; Supplemental Table S12). We also evaluated the conservation of the 3′ UTR 8mer target sites for different miRNA seeds and gene cluster interactions (Supplemental Fig. S21). We found that eight out of our nine significant interactions fall in the top 30 percentile of conserved 8mer target sites. In particular, miR clusters 11, 14, and 6 have the highest number of conserved targets across the gene clusters. From these nine significant interactions, we chose to further analyze two pairs. The interaction between miRNA cluster 11 and mRNA cluster 18 (Fig. 3C–E; Supplemental Table S13) had a P-value of 10−5 for the target enrichment and a correlation coefficient of −0.73. The miRNAs involved in this interaction are highly expressed and increase during time in brain, whereas the target genes are expressed more highly in other tissues such as limbs, cranioface, and heart at the same stages of development. Gene Ontology of the targets revealed that this miRNA cluster targets genes involved in the development of skeletal system, cardiac development, and vasculature development (P-values <10−15), by presumably down-regulating them in the brain. Another interaction between miRNA cluster 6 and mRNA cluster 28 has a P-value of 5.2 × 10−5and a negative correlation coefficient of −0.68. This miRNA cluster increases expression mainly in the heart, lung, and kidney (Fig. 3F), whereas the mRNA targets are highly expressed in the brain organ and their expression is very limited in the heart (Fig. 3G). Gene Ontology analysis of this interaction enriches for terms involved with head and brain development (P-values <10−5) (Fig. 3H; Supplemental Table S14). In both of these cases, as well as several of the others, the miRNA cluster is enriched for targets that are developmentally important genes for tissues other than the tissue in which the miRNAs are highly expressed.
Discussion
In this study, we provide a comprehensive resource of miRNA expression dynamics across mouse developmental stages in multiple organs. Our catalog of organ-specific and developmental stage–specific miRNAs provides a valuable resource for elucidating the role of miRNAs and highlighting certain key properties of miRNAs during mouse development. We detected that only 42% of the annotated miRNAs in mouse expressed a minimum of two CPM (72% of miRNAs annotated as highly confident) in the 16 different organs that are representative of major organ systems during mouse embryogenesis. This result suggests that only a subset of miRNAs might be involved in regulating gene expression during mouse development, with the remaining either expressed in other tissues or more likely expressed later in postnatal development and adult tissues (Ludwig et al. 2016). There is also little variability in the number of miRNAs detected per tissues, with the heart and nervous system tissues showing the highest number of detected miRNAs. The miRNA output of most embryonic samples is dominated by the expression of a few highly expressed miRNAs that usually consist of nontissue-specific and ubiquitously but highly expressed miRNAs, which matches reports from human and mouse cell types (De Rie et al. 2017).
Although tissue specificity of miRNAs has been well studied and well reported in multiple model organisms (Lagos-Quintana et al. 2002; Gao et al. 2011; Ludwig et al. 2016), a comprehensive study of the dynamics of such tissue-specific miRNAs across mouse development was lacking. Our analysis fills this knowledge gap. We show that most of the tissue-specific miRNAs are dynamically regulated across development, with different subsets of miRNAs in the brain and heart expressed at different levels during embryonic development.
We provide evidence that the tissue-specific expression of a subset of miRNAs is conserved in human and mouse, although the overall transcriptional programs are known to have considerably diverged in the two species (Yue et al. 2014). Although the number of one-to-one miRNA orthologs in human and mouse is low as a fraction of the known miRNAs in each species (∼20% of annotated miRNAs in human have one-to-one orthologs in mouse), we show that the tissue-specific expression patterns of the miRNA orthologs closely resemble the overall patterns observed in each individual species. The conservation of miRNA expression in human and mouse tissues is driven by core sets of miRNAs. We show that the expression of tissue-specific miRNAs is well conserved in some tissues (brain, muscle, and lung), although less conserved in other tissues (liver). The fraction of conserved miRNAs is significantly lower than the number of conserved genes between human and mouse (Herrero et al. 2016), which suggests that miRNAs are evolving more frequently.
Finally, the clustering of the miRNAs based on the dynamics of their expression in different tissues allowed us for a unique opportunity to study the functionality and role of these miRNAs in a cooperative way. This approach revealed that some of these tissue-specific clusters of miRNAs likely act as suppressors of genes involved in the development of other tissues than those in which the cluster of miRNAs are expressed. Although the coregulation of the mRNAs could be simply owing to the sharing of cis-regulatory elements, we note that many of the target genes of our miRNAs are transcription factors that are themselves important for mouse development, which strongly suggests that posttranscriptional regulation needs to be incorporated into models of transcriptional regulation being built from ChIP-seq, open chromatin, and mRNA expression data. The availability of miRNA expression levels in matching tissues and time points of the Mouse ENCODE data set of embryonic development provides a unique opportunity to integrate the analysis of miRNAs with other functional genomic data used to build the Mouse Encyclopedia of DNA Elements.
Methods
Mouse tissue collection and total RNA isolation
For each of the embryonic stages assayed (Fig. 1A), a single pregnant female was sacrificed and dissected for embryo removal. Tissues from embryos and a newborn mouse at day 0 were collected. The detailed protocol of tissue collection can be accessed at https://www.encodeproject.org/documents/631aa21c-8e48-467e-8cac-d40c875b3913/@@download/attachment/StandardTissueExcisionProtocol_02132017.pdf.
Total RNA was extracted from each sample using the mirVana miRNA isolation kit (Thermo Fisher Scientific, AM1561), followed by genomic DNA removal using TURBO DNA-free kit (Thermo Fisher Scientific, AM1907).
miRNA profiling of mouse embryonic and postnatal samples
Mouse miRNA-seq libraries were constructed by following the miRNA-seq protocol described previously (Alon et al. 2011; Roberts et al. 2015) without the blocking of highly abundant miRNAs and with some minor modifications (for details, see Supplemental Methods). The library concentrations were measured by a library quantification kit (KAPA Biosystems, KK4824). The library loading concentration for the sequencing was determined using the concentration obtained by KAPA and the estimated fragment size of 140 bp (because we could not use Bioanalyzer to determine the fragment sizes of these libraries). The miRNA-seq libraries were sequenced as 50-bp single-end reads on an Illumina HiSeq 2000 sequencer.
Mouse miRNA-seq read adapter trimming and mapping
We used Cutadapt v. 1.7.1 (Martin 2014) with Python 2.7.10 to sequentially trim 5′ and 3′ adapters from raw reads. Trimmed reads were mapped to mouse miRBase v. 22 (Kozomara and Griffiths-Jones 2011) mature miRNA sequences with STAR v. 2.4.2a (for details, see Supplemental Methods; Dobin et al. 2013). Counts of reads mapping to each miRBase mature miRNA were obtained from STAR output. The counts were normalized for sequencing depth and further TMM normalized using edgeR (Robinson et al. 2010) to obtain CPM. Furthermore, for cross-referencing with the high-confidence subset of miRNAs in miRBase (Kozomara and Griffiths-Jones 2014), the miRNAs in MirGeneDB (Fromm et al. 2018), and FANTOM project novel miRNAs (De Rie et al. 2017), the trimmed reads were mapped to their corresponding mature miRNA reference files and quantified using STAR v. 2.4.2a.
Tissue specificity analysis of individual miRNAs
miRNA tissue specificity was determined using TSI as previously described (Ludwig et al. 2016):
To prevent any biases introduced by multiple tissues of neural origin, we excluded the samples from the hindbrain, midbrain, and neural tube and used only the forebrain samples for tissue specificity study of individual miRNAs. Also because some of the organs were not assayed at the earlier stages of embryonic development (because they start development later), we decided to restrict the number of organs considered for TSI calculations to cranioface, forebrain, heart, limb, and liver for the first four stages (E10.5–E13.5) and to cranioface, forebrain, heart, limb, liver, stomach, kidney, lung, and intestine for the last four stages (E14.5–P0). Alternative methods for this calculation were used as described in Supplemental Figure S6.
Clustering of mouse miRNA-seq data
Time-series analysis of the mouse miRNA-seq time-course was performed using maSigPro v. 1.48.0 (Nueda et al. 2014) in R 3.4.4 (R Core Team 2018). Briefly, each organ (12 in total) that was assayed in at least two developmental time points was analyzed using a degree 3 and maSigPro functions “p.vector(data, design = design.matrix, counts = TRUE),” “T.fit(p.vector_output, alfa = 0.01),” and “get.siggenes(T.fir_output, rsq = 0.7, vars=‘all’).” Different numbers of clusters (k) were tested to obtain a robust clustering of miRNAs by comparing the clusters at each step of k with the previous ones using the command “see.genes(get$sig.genes, k = …).” The best clustering of miRNAs was obtained with k = 16. The median profiles of the genes were plotted using ggplot2 package (Wickham 2016) in R. The R code used to generate these clusters and plots can be found at https://github.com/sorenar/mouse_embryonic_miRNAs/blob/master/miRNA_maSigPro.R.
Analysis of the mouse mRNA-seq data
mRNA-seq reads were mapped to the mouse genome (assembly mm10) using STAR v. 2.4.2a. The alignments to the genome were assembled into ab initio transcripts using StringTie v. 1.2.4 (Pertea et al. 2015). The expression levels of the GENCODE v. M10 and the StringTie model transcripts were obtained using STAR and RSEM v. 1.2.25 (for details, see Supplemental Methods; Li and Dewey 2011).
Time-series analysis of the mouse mRNA-seq time-course was performed similar to the clustering of miRNAs. In this case, higher numbers of clusters (k = 20–35) were tested, and k = 30 was selected as the number that gave the best results. Similarly, the median profiles of these clusters were plotted using ggplot2 in R.
Target enrichment analysis
The R package miRNAtap v. 1.10.0 (https://bioconductor.org/packages/release/bioc/html/miRNAtap.html) was used as an ensemble method to compile the predicted targets for each miRNA in our data. miRNAtap uses five different sources to generate a list of predicted targets: TargetScan (Friedman et al. 2009), DIANA (Maragkakis et al. 2011), miRanda (Enright et al. 2003), PicTar (Lall et al. 2006), and miRDB (Wong and Wang 2015). We used getPredictedTargets(miRNA, species = “mmu”, method = “geom”, min_src = 3) to obtain the list of predicted targets for miRNA. The parameter “min_src” indicates that if the miRNA has targets that are present in more than “min_src” value, the reported list would be only limited to those targets; otherwise, the method will reduce the “min_src” until it gets a list of targets or no target at all.
For each significant interaction with a negative partial correlation (as described in Supplemental Methods), the list of the target genes in the interaction was compiled. The Gene Ontology analysis of each of these target lists was performed via Metascape (Zhou et al. 2019), and the top 10 most enriched terms were plotted for these analyses.
Data access
All the miRNA-seq, short RNA-seq, and NanoString data from this study can be accessed through the ENCODE data portal (https://www.encodeproject.org) under the accession numbers listed in Supplemental Table S1.
Supplementary Material
Acknowledgments
This work was supported by the NIH, National Human Genome Research Institute under grants U54 HG006998 and UM1 HG009443.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.248997.119.
Freely available online through the Genome Research Open Access option.
References
- Alisch RS, Jin P, Epstein M, Caspary T, Warren ST. 2007. Argonaute2 is essential for mammalian gastrulation and proper mesoderm formation. PLoS Genet 3: e227 10.1371/journal.pgen.0030227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon S, Vigneault F, Eminaga S, Christodoulou DC, Seidman JG, Church GM, Eisenberg E. 2011. Barcoding bias in high-throughput multiplex sequencing of miRNA. Genome Res 21: 1506–1511. 10.1101/gr.121715.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. 2003. Dicer is essential for mouse development. Nat Genet 35: 215–217. 10.1038/ng1253 [DOI] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10: 1957–1966. 10.1261/rna.7135204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068. 10.1038/nature07385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao DD, Li L, Chan WY. 2016. MicroRNAs: key regulators in the central nervous system and their implication in neurological diseases. Int J Mol Sci 17: 842 10.3390/ijms17060842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009. 10.1101/gad.1884710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rie D, Abugessaisa I, Alam T, Arner E, Arner P, Ashoor H, Åström G, Babina M, Bertin N, Burroughs AM, et al. 2017. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat Biotechnol 35: 872–878. 10.1038/nbt.3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich IM, Purugganan M. 2008. MicroRNAs in plants: possible contributions to phenotypic diversity. Plant Signal Behav 3: 829–830. 10.4161/psb.3.10.5914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. 2003. MicroRNA targets in Drosophila. Genome Biol 5: R1 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejes-Toth K, Sotirova V, Sachidanandam R, Assaf G, Hannon GJ, Kapranov P, Foissac S, Willingham AT, Duttagupta R, Dumais E, et al. 2009. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457: 1028–1032. 10.1038/nature07759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105. 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm B, Domanska D, Hackenberg M, Mathelier A, Høye E, Johansen M, Hovig E, Flatmark K, Peterson K. 2018. MirGeneDB2.0: the curated microRNA Gene Database. bioRxiv doi.org/10.1101/258749
- Gao Y, Schug J, McKenna LB, Le Lay J, Kaestner KH, Greenbaum LE. 2011. Tissue-specific regulation of mouse MicroRNA genes in endoderm-derived tissues. Nucleic Acids Res 39: 454–463. 10.1093/nar/gkq782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. 2008. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26: 317–325. 10.1038/nbt1385 [DOI] [PubMed] [Google Scholar]
- Gorkin DU, Barozzi I, Zhang Y, Lee AY, Li B, Zhao Y, Wildberg A, Ding B, Zhang B, Wang M, et al. 2017. Systematic mapping of chromatin state landscapes during mouse development. bioRxiv 10.1101/166652v2 [DOI]
- Han J, Lee Y, Yeom K-H, Nam J-W, Heo I, Rhee J-K, Sohn SY, Cho Y, Zhang B-T, Kim VN. 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125: 887–901. 10.1016/j.cell.2006.03.043 [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531. 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- Herrero J, Muffato M, Beal K, Fitzgerald S, Gordon L, Pignatelli M, Vilella AJ, Searle SMJ, Amode R, Brent S, et al. 2016. Ensembl comparative genomics resources. Database 2016: baw053 10.1093/database/baw053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D, Lee H. 2015. A computational approach to identifying gene-microRNA modules in cancer. PLoS Comput Biol 11: e1004042 10.1371/journal.pcbi.1004042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2011. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39(Database issue): D152–D157. 10.1093/nar/gkq1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42(Database issue): D68–D73. 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladewig E, Okamura K, Flynt AS, Westholm JO, Lai EC. 2012. Discovery of hundreds of mirtrons in mouse and human small RNA data. Genome Res 22: 1634–1645. 10.1101/gr.133553.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. 2002. Identification of tissue-specific microRNAs from mouse. Curr Biol 12: 735–739. 10.1016/S0960-9822(02)00809-6 [DOI] [PubMed] [Google Scholar]
- Lall S, Grün D, Krek A, Chen K, Wang Y-L, Dewey CN, Sood P, Colombo T, Bray N, MacMenamin P, et al. 2006. A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol 16: 460–471. 10.1016/j.cub.2006.01.050 [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414. 10.1016/j.cell.2007.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Gregory RI. 2015. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 15: 321–333. 10.1038/nrc3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stähler C, Meese E, et al. 2016. Distribution of miRNA expression across human tissues. Nucleic Acids Res 44: 3865–3877. 10.1093/nar/gkw116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M, Vergoulis T, Alexiou P, Reczko M, Plomaritou K, Gousis M, Kourtis K, Koziris N, Dalamagas T, Hatzigeorgiou AG. 2011. DIANA-microT web server upgrade supports Fly and Worm miRNA target prediction and bibliographic miRNA to disease association. Nucleic Acids Res 39: W145–W148. 10.1093/nar/gkr294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2014. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, Cheo D, D'Andrade P, DeMayo M, Dennis L, et al. 2014. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods 11: 809–815. 10.1038/nmeth.3014 [DOI] [PubMed] [Google Scholar]
- Mineno J, Okamoto S, Ando T, Sato M, Chono H, Izu H, Takayama M, Asada K, Mirochnitchenko O, Inouye M, et al. 2006. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res 34: 1765–1771. 10.1093/nar/gkl096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Horii T, Kimura M, Goto Y, Ochiya T, Hatada I. 2007. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics 89: 687–696. 10.1016/j.ygeno.2007.01.004 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods 5: 621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Ninova M, Ronshaugen M, Griffiths-Jones S. 2014. Fast-evolving microRNAs are highly expressed in the early embryo of Drosophila virilis. RNA 20: 360–372. 10.1261/rna.041657.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nueda MJ, Tarazona S, Conesa A. 2014. Next maSigPro: updating maSigPro bioconductor package for RNA-seq time series. Bioinformatics 30: 2598–2602. 10.1093/bioinformatics/btu333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Choi YS, McManus MT. 2010. Analysis of microRNA knockouts in mice. Hum Mol Genet 19: R169–R175. 10.1093/hmg/ddq367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnology 33: 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Roberts BS, Hardigan AA, Kirby MK, Fitz-Gerald MB, Wilcox CM, Kimberly RP, Myers RM. 2015. Blocking of targeted microRNAs from next-generation sequencing libraries. Nucleic Acids Res 43: e145 10.1093/nar/gkv724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaine SPR, Tomaszewski M, Condorelli G, Samani NJ. 2015. MicroRNAs in cardiovascular disease: an introduction for clinicians. Heart 101: 921–928. 10.1136/heartjnl-2013-305402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidigal JA, Ventura A. 2015. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol 25: 137–147. 10.1016/j.tcb.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. 2007. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39: 380–385. 10.1038/ng1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York: http://ggplot2.org. [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. 2005. MicroRNA expression in zebrafish embryonic development. Science 309: 310–311. 10.1126/science.1114519 [DOI] [PubMed] [Google Scholar]
- Wong N, Wang X. 2015. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 43: D146–D152. 10.1093/nar/gku1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M. 2011. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res 21: 1450–1461. 10.1101/gr.118059.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515: 355–364. 10.1038/nature13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Jiang S, Kong X, El-Ali N, Ball AR Jr, Ma CI, Hashimoto N, Yokomori K, Mortazavi A. 2016. Single-nucleus RNA-seq of differentiating human myoblasts reveals the extent of fate heterogeneity. Nucleic Acids Res 44: e158 10.1093/nar/gkw739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Zhao S-P, Zhao Y-H. 2015. MicroRNA-143/-145 in cardiovascular diseases. BioMed Res Int 2015: 531740 10.1155/2015/531740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. 2019. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10: 1523 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.