An established regulator of photosynthetic processes in leaves influences tomato fruit quality by regulating starch accumulation, redox balance, and fruit growth.
Abstract
NADPH-thioredoxin reductase C (NTRC) forms a separate thiol-reduction cascade in plastids, combining both NADPH-thioredoxin reductase and thioredoxin activities on a single polypeptide. While NTRC is an important regulator of photosynthetic processes in leaves, its function in heterotrophic tissues remains unclear. Here, we focus on the role of NTRC in developing tomato (Solanum lycopersicum) fruits representing heterotrophic storage organs important for agriculture and human diet. We used a fruit-specific promoter to decrease NTRC expression by RNA interference in developing tomato fruits by 60% to 80% compared to the wild type. This led to a decrease in fruit growth, resulting in smaller and lighter fully ripe fruits containing less dry matter and more water. In immature fruits, NTRC downregulation decreased transient starch accumulation, which led to a subsequent decrease in soluble sugars in ripe fruits. The inhibition of starch synthesis was associated with a decrease in the redox-activation state of ADP-Glc pyrophosphorylase and soluble starch synthase, which catalyze the first committed and final polymerizing steps, respectively, of starch biosynthesis. This was accompanied by a decrease in the level of ADP-Glc. NTRC downregulation also led to a strong increase in the reductive states of NAD(H) and NADP(H) redox systems. Metabolite profiling of NTRC-RNA interference lines revealed increased organic and amino acid levels, but reduced sugar levels, implying that NTRC regulates the osmotic balance of developing fruits. These results indicate that NTRC acts as a central hub in regulating carbon metabolism and redox balance in heterotrophic tomato fruits, affecting fruit development as well as final fruit size and quality.
Reduction-oxidation (redox) regulation appears to be a fundamental integrator of metabolic pathways in different subcellular compartments (Geigenberger and Fernie, 2014). In plant chloroplasts there are two different thiol redox systems, the ferredoxin (Fdx)-thioredoxin (Trx) system, which depends on the reduction of Fdx by photosynthetic electron transport in response to light, and the NADPH-dependent Trx reductase C (NTRC) system, which relies on NADPH and thus may be linked to Fdx-NADPH reductase in the light or sugar metabolism in the dark (Buchanan and Balmer, 2005; Zaffagnini et al., 2018). NTRC is an unusual protein, since it harbors both NADPH-Trx reductase and Trx domains on the same polypeptide (Serrato et al., 2004). This feature allows NTRC to use NADPH as a source of electrons to regulate different chloroplast target proteins via thiol-disulfide modulation (Spínola et al., 2008; Geigenberger et al., 2017).
On the analysis of Arabidopsis (Arabidopsis thaliana) transfer DNA insertion mutants, NTRC is determined to be involved in various chloroplast functions. There is in planta evidence that NTRC is important for reducing 2-Cys peroxiredoxins (2-Cys Prxs) for the removal of hydrogen peroxide (Pérez-Ruiz et al., 2006; Kirchsteiger et al., 2009; Pérez-Ruiz and Cejudo, 2009) and for facilitating starch synthesis via redox regulation of ADP-Glc pyrophosphorylase (AGPase; Michalska et al., 2009; Lepistö et al., 2013). Furthermore, NTRC has overlapping functions with the Fdx-dependent Trx systems to optimize photosynthetic processes such as nonphotochemical quenching of light energy (Thormählen et al., 2015, 2017; Naranjo et al., 2016; Yoshida and Hisabori, 2016), Calvin-Benson cycle activity (Thormählen et al., 2015; Nikkanen et al., 2016; Yoshida and Hisabori, 2016), chlorophyll biosynthesis (Richter et al., 2013; Pérez-Ruiz et al., 2014; Da et al., 2017), and ATP synthesis (Carrillo et al., 2016; Nikkanen et al., 2016). In this context, NTRC indirectly maintains the reducing capacity of the pool of Fdx-TRXs by restricting their reoxidation via an oxidation loop involving hydrogen peroxide and oxidized 2-Cys Prxs (Pérez-Ruiz et al., 2017). NTRC is, therefore, of crucial importance in balancing oxidation and reduction pathways in the chloroplast to optimize photosynthesis, overall plant growth, and development in response to light (Thormählen et al., 2015, 2017; Carrillo et al., 2016; Yoshida and Hisabori, 2016).
In addition to its role in chloroplasts, NTRC is also present in nonphotosynthetic plastids together with Fdx-dependent Trxs (Kirchsteiger et al., 2012). However, as yet little is known on the role of NTRC in heterotrophic plant tissues that lack photosynthetic functions. There is evidence that NTRC affects starch synthesis in Arabidopsis roots (Michalska et al., 2009), but this does not contribute to the growth phenotype of the Arabidopsis ntrc mutant (Kirchsteiger et al., 2012). Complementation of the ntrc mutant by NTRC overexpression under the control of a leaf-specific promoter led to wild-type phenotypes, but the mutant phenotype remained unaltered when a root-specific promoter was used (Kirchsteiger et al., 2012). These data thus indicate a role of NTRC in photosynthetic leaves, rather than in nonphotosynthetic roots.
Tomato (Solanum lycopersicum) is an important horticultural crop and major component of the human diet, serving as the primary model system to study fruit physiology and development (Kanayama, 2017). Throughout the fruit developing process, a myriad of changes in color, firmness, taste, and flavor occur that are associated with developmentally regulated metabolic shifts (e.g. changes in the levels of carbohydrates, acids, and volatile compounds; Carrari et al., 2006). Notably, the abundance of carbohydrates is a key determinant for fruit yield and quality (Kanayama, 2017). During development, immature tomato fruits act as strong sink tissues, into which massive amounts of carbon are imported as Suc from leaf tissues (Osorio and Fernie, 2014). Most of the Suc is mobilized to Glc and Fru and subsequently converted to starch as an intermediate carbon store to promote the sink activity of the immature fruits (Osorio and Fernie, 2014). In later ripening stages, starch is degraded to soluble sugars, which are key determinants of fruit taste and quality (Davies et al., 1981; Gierson and Kader, 1986).
Alterations of the transient starch pool of immature fruits have been proposed to be important for the accumulation of soluble sugar in ripening fruits, which determines the final fruit size and quality (Schaffer et al., 2000; Petreikov et al., 2009). In this context, AGPase serves as a rate-limiting enzyme for starch synthesis in developing tomato fruits (Petreikov et al., 2009). AGPase is a heterotetrameric enzyme located in the plastid and comprised of two large and two small subunits, which are subject to allosteric activation by glycerate-3-phosphate (3-PGA) and inhibition by inorganic phosphate (Pi; Preiss, 1973). AGPase is also subject to redox regulation involving reversible reduction of a regulatory disulfide linking the small subunits, which affects the allosteric properties of the enzyme (Fu et al., 1998; Tiessen et al., 2002). Manipulation of the cellular redox poise during tomato fruit development has previously been demonstrated to lead to changes in the NADPH-to-NADP+ ratio, which were accompanied by changes in the redox-activation states of NADP-malate dehydrogenase and AGPase of the developing fruits (Centeno et al., 2011). The changes in AGPase redox state were additionally associated with alterations in starch synthesis in immature fruits and subsequent alterations in soluble sugar content in mature fruits (Centeno et al., 2011). Although these studies provide evidence that redox regulation via thiol-disulfide modulation is involved in tomato fruit development, the underlying fruit-specific thiol-redox systems remain unresolved.
In this study, we downregulated NTRC gene expression specifically in fruit tissues by generating a RNA interference (RNAi) construct under the control of the fruit-specific patatin B33 promoter. The NTRC-RNAi lines were characterized by a 60% to 80% decrease in NTRC transcripts and protein levels in developing fruits. In immature fruits, NTRC downregulation decreased transient starch accumulation by decreasing the redox-activation state of AGPase and the activity of soluble starch synthase, which subsequently led to a decreased accumulation of soluble sugars during ripening and to decreased fruit yield and quality in fully ripe fruits. This was accompanied by an increased reduction state of the NAD(H) and NADP(H) redox couples. These results provide evidence for a previously unknown function of NTRC as a central hub in regulating carbon metabolism and redox balance in developing fruits.
RESULTS
Generation of Transgenic Tomato Plants with Decreased Expression of NTRC under Control of a Fruit-Specific Promoter
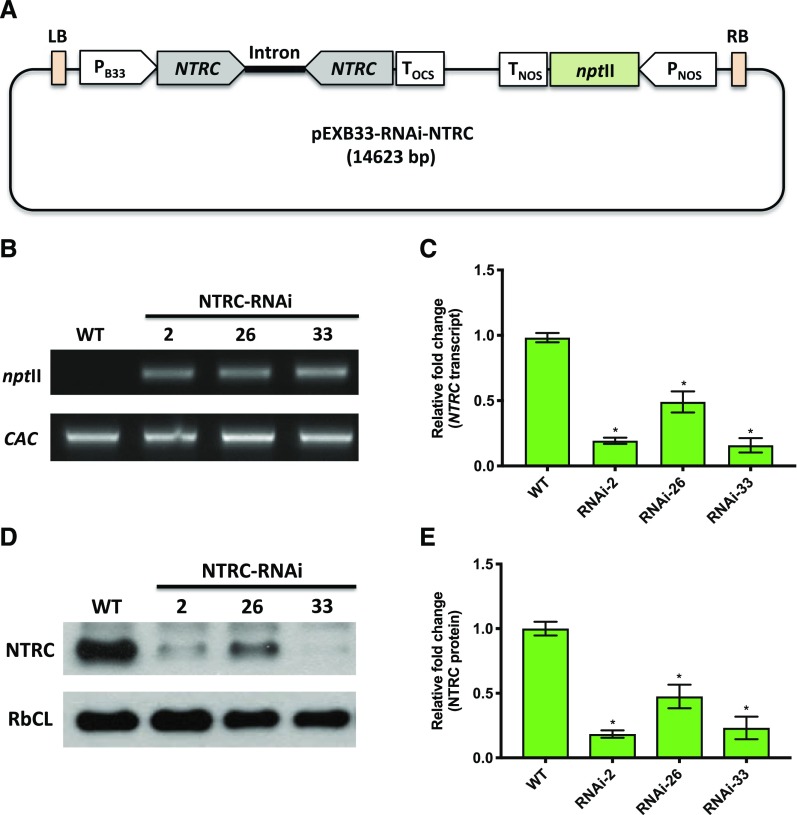
To silence NTRC gene expression specifically in fruit tissues, we generated a NTRC-RNAi construct under the control of the patatin B33 promoter, which has previously been identified to confer fruit specific expression in tomato plants (Rocha-Sosa et al., 1989; Frommer et al., 1994; Obiadalla-Ali et al., 2004), with kanamycin serving as a marker for selection (Fig. 1A). The resulting construct was transformed via an Agrobacterium tumefaciens-mediated process into tomato plants. To identify functional NTRC-RNAi lines, samples from fruits harvested 35 and 65 d after flowering (DAF) were selected for genotyping. As shown in Figure 1B, NTRC-RNAi lines 2, 26, and 33 all harbored the neomycin phosphotransferase II (nptII) gene, indicating the presence of the NTRC-RNAi cassette within their genome. The transcript level of the NTRC gene (LOC101254347) decreased by 60% to 80% in comparison to the wild type in the three RNAi lines in both 35- (Fig. 1C) and 65-DAF fruit samples (Supplemental Fig. S1A). Furthermore, the NTRC protein levels of RNAi-2, RNAi-26, and RNAi-33 decreased by 50% to 80% compared to the wild type, both in 35- (Fig. 1, D and E) and 65-DAF fruit samples (Supplemental Fig. S1, B and C). The expression of the second NTRC gene (LOC101266017; Nájera et al., 2017) was also analyzed, but its expression was too low to be detectable in fruit tissues. Thus, we concluded that NTRC-RNAi lines 2, 26, and 33 were appropriate to study the function of NTRC in tomato fruit.
Figure 1.
Molecular characterization of NTRC-RNAi lines 2, 26, and 33 compared to the wild type (WT) in 35 DAF tomato fruits. A, The RNAi construct targeting the NTRC gene. B, Genotyping by PCR analysis for detection of the nptII gene in NTRC-RNAi lines. C, Detection of the NTRC transcript level using RT-qPCR. D, Detection of NTRC proteins by immunoblot analysis. E, Quantification of NTRC protein abundance using Image-J software. The level of clathrin adaptor complexes (CAC) serves as a reference gene for genotyping and RT-qPCR. The protein level of ribulose 1,5-bisphosphate carboxylase/oxygenase large subunit (RbcL) served as a loading control for immunoblotting. Results are the mean ± se from six biological replicates. Statistical analysis used ANOVA with Dunnett’s test (*P < 0.05, compared to the wild type).
Fruit-Specific Silencing of NTRC Has No Substantial Effects on the Process of Fruit Development
The different stages of tomato fruit development are characterized by changes in pigmentation and ripening. To analyze whether decreased expression of NTRC affects the process of tomato fruit development, we analyzed chlorophyll and light-harvesting complex II (Lhcb2) protein levels as well as the expression of genes involved in ethylene synthesis and ripening in 35-DAF and 65-DAF fruits. In 35-DAF fruits, chlorophyll content and protein level of Lhcb2 were not significantly changed in NTRC-RNAi lines compared to the wild type, while neither parameter was detectable in 65-DAF fruits in all genotypes (Supplemental Fig. S2, A and B). There were no visible changes in fruit color at 35 DAF (Fig. 2A) or 65 DAF (Supplemental Fig. S3A) when NTRC-RNAi lines were compared to the wild type. To further analyze whether NTRC downregulation affected the ripening processes, we assayed the expression of genes involved in ethylene production (S-adenosylhomo-Cys hydrolase [SAHH2]; Yang et al., 2017) and ripening (NON-RIPENING [NOR]; Ma et al., 2019) in both 35-DAF and 65-DAF fruits. Both genes were upregulated when fruits became mature, but their expression levels were not significantly changed between wild-type and NTRC-RNAi lines (Supplemental Fig. S2, C and D). Taken together, NTRC downregulation did not affect the process of fruit development. While 35-DAF fruits represent the mature green stage and 65-DAF fruits represent the fully ripened stage, there were no stage-dependent differences between NTRC-RNAi lines and the wild type.
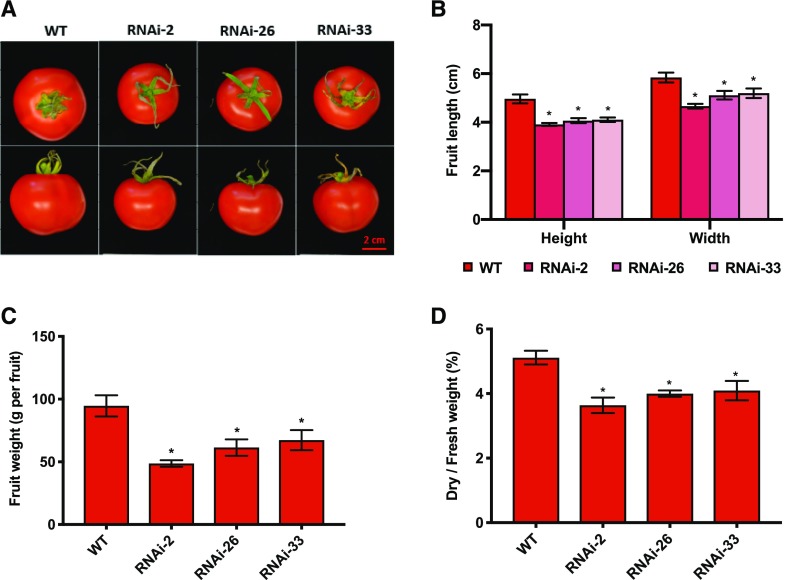
Figure 2.
Phenotypic analysis of fully ripened tomato fruits (65 DAF) of NTRC-RNAi lines 2, 26, and 33 compared to the wild type (WT). A, Visible phenotype of ripe fruits. Scale bar = 2 cm. B, Fruit length and width. C, Fruit fresh weight. D, Ratio of fruit dry weight to fresh weight. Results are the mean ± se from six biological replicates (each replicate contains data from one to three fruits). Statistical analysis used ANOVA with Dunnett’s test (*P < 0.05, compared to the wild type).
Fruit-Specific Silencing of NTRC Leads to Smaller and Lighter Fruits with Decreased Dry Weight-to-Fresh Weight Ratios
To investigate whether silencing of NTRC affects overall fruit growth, fully mature fruits (65 DAF) were harvested for the measurement of final fruit size and weight. As shown in Figure 2A, although both the wild type and RNAi lines were fully ripe, the final fruit size of NTRC-RNAi lines was clearly smaller than that of the wild type. Furthermore, the NTRC-RNAi lines exhibited a 20% reduction in fruit height and a 15% reduction in fruit width compared to the wild type (Fig. 2B). The final tomato fruit fresh weight in NTRC-RNAi lines was 40% lower than in the wild type (Fig. 2C), and the NTRC-RNAi tomato fruits also contained less dry matter and more water than wild-type fruits (Fig. 2D). The effects of NTRC silencing on fruit growth and dry-matter content were also observed at 35 DAF, indicating that they were not caused by developmental effects (Supplemental Fig. S3). Taken together, fruit-specific silencing of NTRC led to the generation of smaller and lighter fruits consisting of less dry matter in tomato plants.
Fruit-Specific Silencing of NTRC Leads to Lower Accumulation of Starch and Soluble Sugars during Fruit Development
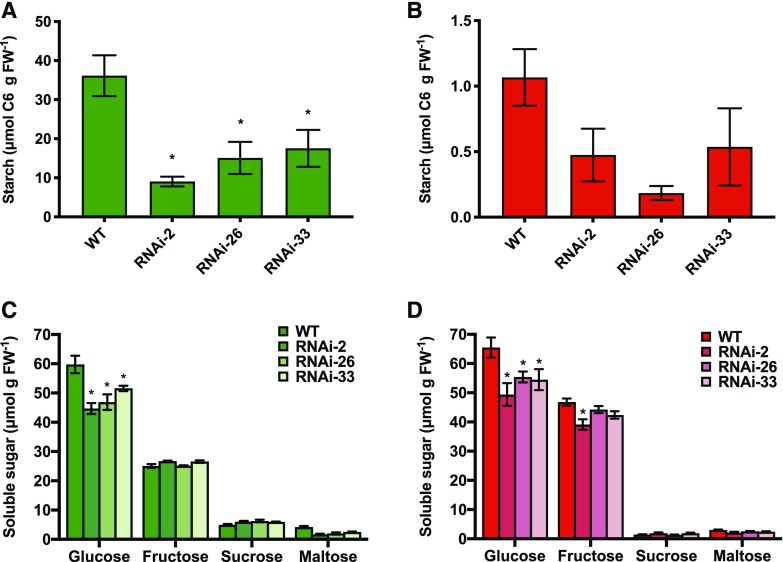
Tomato fruit development is characterized by a transient accumulation of starch in immature fruits, which is subsequently mobilized to contribute to the accumulation of soluble sugars, specifically Glc, in mature fruits (Schaffer et al., 2000). To investigate whether the inhibition of fruit growth in NTRC-RNAi lines is due to a decrease in the transient starch pool, we analyzed the levels of starch and soluble sugars during fruit development. In immature fruits (35 DAF), NTRC downregulation led to a strong decrease in starch accumulation to 25% to 50% of the wild-type level (Fig. 3A). In mature fruits (65 DAF), where starch has been almost completely mobilized in all genotypes, the NTRC-RNAi lines still exhibited a trend of decreased residual starch levels compared to the wild type (Fig. 3B). In contrast to this, NTRC silencing did not affect the protein content of the fruits at either developmental stage (Supplemental Fig. S4).
Figure 3.
The accumulation of starch and soluble sugars in tomato fruits of the wild type (WT) and NTRC-RNAi lines 2, 26, and 33. The levels of starch (A and B) and soluble sugars (C and D) were measured in fruits harvested at 35 (A and C) and 65 DAF (B and D). Results are the mean ± se from six biological replicates. Statistical analysis used ANOVA with Dunnett’s test (*P < 0.05, compared to the wild type). FW, fresh weight
To investigate whether the inhibition of starch accumulation was accompanied by an increase in soluble sugars, we analyzed the levels of Glc, Fru, Suc, and maltose in NTRC-RNAi lines compared to the wild type. At both developmental stages, the level of Glc, the major soluble sugar in tomato fruits, was strongly decreased in NTRC-RNAi lines compared to the wild type, while Fru, Suc, and maltose levels showed only slight decreases (Figs. 3, C and D, and 4A; Supplemental Table S1). Thus, interfering with NTRC expression in tomato fruits greatly compromised transient starch accumulation and final soluble sugar accumulation during their development, while there were no effects on the protein content of the fruits.
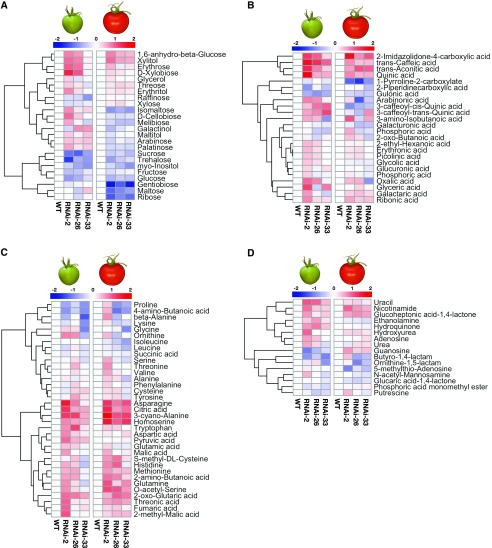
Figure 4.
Metabolite profile of NTRC-RNAi tomato fruits of NTRC-RNAi lines 2, 26, and 33 compared to the wild type (WT). The metabolite profile was measured in fruits harvested at 35 (left) and 65 DAF (right) by using GC-TOF-MS. A, Sugars and sugar alcohols. B, Organic acids. C, Amino acids and tricarboxylic acid cycle intermediates. D, Miscellaneous. Results are normalized to the wild type with log2 transformation and visualized as a heatmap with hierarchical clustering done by R software. Data are taken from Supplemental Table S1.
Interfering with NTRC Expression in Tomato Fruits Has Minor Effects on the Levels of Phosphorylated Intermediates and Adenine Nucleotides
To investigate whether the decrease in starch and sugar accumulation is related to changes in sugar and energy metabolism, we further assayed the level of phosphorylated metabolites and adenine nucleotides in developing tomato fruits. In NTRC-RNAi lines, the levels of Glc-6-phosphate (G6P), Fru-6-phosphate, and Glc-1-phosphate (G1P) showed no consistent changes at 35 or 65 DAF when compared to the wild type (Supplemental Tables S1 and S2). Also, no substantial changes were detected in the level of the glycolytic intermediate 3-PGA between the wild type and NTRC-RNAi lines at either developmental stage. Triose phosphates were also assayed, but the level was under the detection limit, which is characteristic for the glycolytic pathway operating in heterotrophic tissues (Hatzfeld and Stitt, 1990). Furthermore, the levels of ATP and ADP and the ATP-to-ADP ratio in NTRC-RNAi lines were not significantly changed during fruit development when compared to the wild type (Supplemental Table S2). The data also show that the levels of the substrates (G1P and ATP) and the allosteric regulators (3-PGA and Pi) of AGPase were not substantially changed in the transgenic fruits. Taken together, these results demonstrate that interfering with NTRC expression does not perturb glycolytic metabolism or adenylate energy state in developing fruits.
Interfering with NTRC Expression in Tomato Fruits Leads to Global Changes in Their Metabolite Profile
To obtain in-depth insight into more global metabolite changes in NTRC-RNAi tomato lines, we next performed metabolite profiling via gas chromatography time-of-flight mass spectrometry (GC-TOF-MS). We first confirmed that the NTRC-RNAi lines showed generally lower sugar levels compared to the wild type (Fig. 4A, left), which was the case for Glc (55% to 80% of the wild-type level), Fru (65% to 85% of the wild-type level), and Suc (40% to 55% of the wild-type level), as well as other sugars. Moreover, upon NTRC silencing, the levels of several sugars decreased more strongly at 65 DAF than at 35 DAF (Fig. 4A, right), including gentiobiose (3.6-fold), maltose (2-fold), and ribose (2.3-fold). With the exception of myo-inositol, there was no great change in sugar alcohols in NTRC-RNAi lines (Fig. 4A).
By contrast, levels of several organic acids showed an increasing pattern in NTRC-RNAi lines compared to wild-type levels, including those of 2-imindazolidone-4-carboxylic acid, transaconitic acid, transcaffeic acid, and quinic acid, which are involved in the synthesis of phenolic compounds. Levels of others, such as 1-pyrroline-2-carboxylate and gulonic acid, decreased (Fig. 4B). The decrease in gulonic acid level might be attributable to lower Glc content, since this acid is formed by oxidation of Glc. The intermediates of the tricarboxylic acid cycle also showed an increasing pattern compared to the wild type. Levels of citric acid and 2-oxo-glutaric acid were 2 to 4 times increased in NTRC-RNAi lines at both stages of fruit development, while malic acid, fumaric acid, and succinic acid only showed slight changes compared to the wild type (Fig. 4C). Interestingly, the tricarboxylic acid cycle precursor, pyruvic acid, increased only in immature fruits of NTRC-RNAi lines compared to the wild type (Fig. 4C, left).
In addition to organic acids, levels of amino acids also showed an increasing pattern, while NTRC expression was reduced. Asn, 3-cyano-Ala, and homo-Ser were 1.5- to 2-fold higher in NTRC-RNAi lines in immature fruits (Fig. 4C, left), and this change became more severe as fruits ripened (Fig. 4C, right). Other amino acids, such as Trp, His, Gln, and Met, showed mild increases in NTRC-RNAi lines compared to the wild type. Metabolites, such as urea, amines, nucleosides, and their derivates, were also analyzed, but the results revealed only minor differences between the NTRC-RNAi lines and the wild type at both stages (Fig. 4D). Overall, these results indicate that interfering with NTRC expression affects the balance between sugars, organic acids, and amino acids during fruit development.
Interfering with NTRC Expression in Tomato Fruits Leads to an Increase in the Cellular Redox State of Pyridine Nucleotides
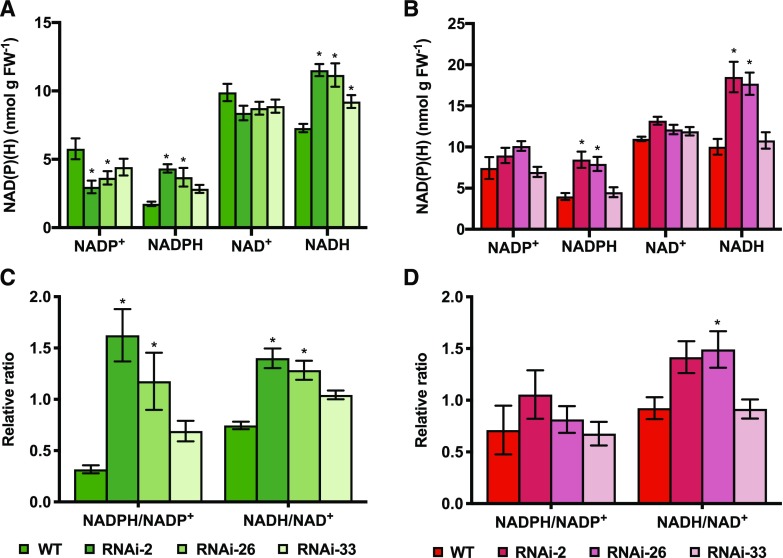
NAD(H) and NADP(H) redox couples are involved in various metabolic processes in different subcellular compartments (Berger et al., 2004; Geigenberger and Fernie, 2014) and also serve as cofactors for NTRC to reduce and regulate downstream plastidial proteins (Pérez-Ruiz and Cejudo, 2009). To investigate whether NTRC downregulation affects the levels of these pyridine nucleotides, we next analyzed the levels of NAD(P)H and NAD(P)+ in developing tomato fruits. In immature fruits (35 DAF), perturbing NTRC expression led to a strong increase in the levels of the reduced forms, NADPH and NADH, while the levels of the oxidized forms, NADP+ and NAD+, decreased (Fig. 5A). This resulted in a 2- to 5-fold increase in NADPH-to-NADP+ ratios and a 1.5- to 2-fold increase in NADH-to-NAD+ ratios in fruits of NTRC-RNAi lines compared to those of the wild type (Fig. 5C).
Figure 5.
Changes in pyridine nucleotide level in tomato fruits of NTRC-RNAi lines in comparison to the wild type (WT). The levels of NADP+, NADPH, NAD+, and NADH (A and B) and the ratios of NADPH to NADP+ and NADH to NAD+ (C and D) were measured in fruits harvested at 35 (A and C) and 65 DAF (B and D). Results are the mean ± se from six biological replicates. Statistical analysis used ANOVA with Dunnett’s test (*P < 0.05, compared to the wild type). FW, fresh weight
In fully ripe fruits (65 DAF), NTRC downregulation also led to an increase in NADPH and NADH, but the levels of the oxidized forms NADP+ and NAD+ were not substantially altered (Fig. 5B), which led to a minor increase in the ratios of NADPH to NADP+ and NADH to NAD+ in NTRC-RNAi lines (Fig. 5D). Together, these results indicate that reducing NTRC expression caused an increased reduction state of the NAD(H) and NADP(H) redox couples in developing tomato fruits, with the effect being mitigated when fruits matured.
Interfering with NTRC Expression in Tomato Fruits Leads to a Decrease in the Redox-Activation State of AGPase and Decreased Levels of ADP-Glc
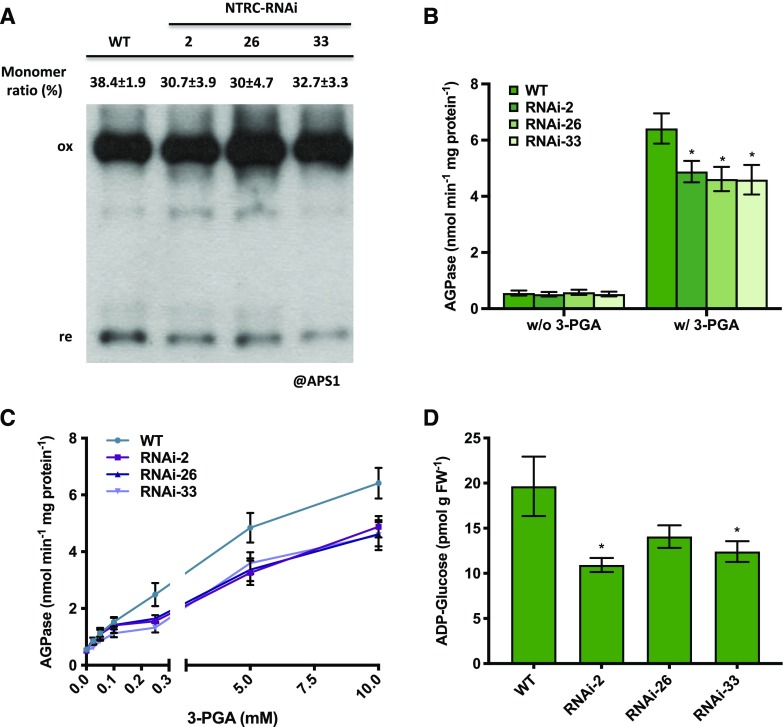
To investigate possible mechanisms involved in the inhibition of fruit starch accumulation in response to decreased NTRC expression, we measured activities of enzymes involved in starch metabolism. We first analyzed AGPase, which catalyzes the first committed step of starch biosynthesis, since its activity is regulated via thiol-disulfide modulation involving Trx and NTRC systems in the plastid (Geigenberger and Fernie, 2014; Geigenberger et al., 2017). Since AGPase is activated by reduction of an intermolecular disulfide bond between the two small subunits (AGPase small-subunit 1 [APS1]) of the heterotetrameric holoenzyme (Ballicora et al., 2000; Tiessen et al., 2002; Hendriks et al., 2003), we assayed the changes in APS1 redox state by analyzing its monomerization pattern when NTRC expression was reduced. In developing fruits (35 DAF), perturbing NTRC expression slightly decreased the monomerization ratios of APS1 (Fig. 6A) by ∼20% to 30%, indicating increased oxidation of the protein. Samples from ripe fruits (65 DAF) were also tested, but there were no consistent differences between wild-type and RNAi lines in the monomerization of APS1 (Supplemental Fig. S5A).
Figure 6.
Redox and allosteric properties of AGPase and endogenous ADP-Glc content in 35 DAF tomato fruits of NTRC-RNAi lines 2, 26, and 33 compared to the wild type (WT). A, Redox state of the intermolecular disulfide bond in APS1. B, AGPase activity assayed with or without the activation of 10 mm 3-PGA. C, AGPase activity kinetics in the presence of varied concentration of 3-PGA as indicated in the figure. D, Content of endogenous ADP-Glc. The monomer-to-dimer ratios of the respective proteins were quantified by Image-J software. Results are the mean ± se (n = 5–6). Statistical analysis used ANOVA with Dunnett’s test (*P < 0.05, compared to the wild type). FW, fresh weight
AGPase is also subject to allosteric regulation, being activated by 3-PGA and inhibited by Pi (Kleczkowski, 1999; Hwang et al., 2005). Redox regulation of AGPase alters the kinetic properties of the holoenzyme, with oxidation of APS1 leading to an enzyme less sensitive for allosteric activation (Tiessen et al., 2002; Hendriks et al., 2003). To investigate whether perturbing NTRC expression in tomato fruits leads to a change in the kinetic properties of AGPase, we assayed AGPase activity in the absence or presence of its allosteric activator, 3-PGA, in 35-DAF fruits. As shown in Figure 6B, the NTRC-RNAi lines showed AGPase activity comparable to that of the wild type when 3-PGA was omitted from the enzyme assay, but there was a 30% decrease in AGPase activity compared to the wild type in the presence of 10 mm 3-PGA. To investigate 3-PGA activation kinetics in more detail, we performed AGPase enzyme assays in the presence of varied concentrations of 3-PGA. Figure 6C shows that, in the wild type, increasing concentrations of 3-PGA from 0.025 to 10 mm led to a progressive increase in AGPase activity from 0.2 to 6.0 nmol min−1 mg−1 protein. AGPase from NTRC-RNAi lines was less sensitive to 3-PGA activation, leading to 30% to 50% lower activities when incubated in the presence of 0.25 to 10 mm 3-PGA. This concentration range corresponds to the in vivo concentration of 3-PGA in the plastid, estimated to be ∼0.3 to 0.5 mm using the nonaqueous fraction of growing potato (Solanum tuberosum) tubers (Farré et al., 2001; Tiessen et al., 2002) and developing barley (Hordeum vulgare) seeds (Tiessen et al., 2012), and 0.7 to 1 mm using the 3-PGA levels from Supplemental Table S1 (70–100 nmol g−1 fresh weight), given that the vacuole contains no 3-PGA and comprises 90% of the cell volume. The overall activities of AGPase in fully ripe fruits (65 DAF), which are characterized by an overall suppression of starch synthesis (Fig. 3B), were 10 times lower than in immature fruits (35 DAF) in both the presence and absence of 3-PGA in the enzyme assay, and there was no significant difference between wild-type and NTRC-RNAi lines (Supplemental Fig. S5B).
ADP-Glc is the direct product of the reaction catalyzed by AGPase and represents the ultimate precursor of the final polymerizing reactions of starch synthesis. In 35-DAF fruits, NTRC downregulation decreased ADP-Glc concentration to 50% to 75% of the wild-type level (Fig. 6D). This provides independent confirmation that NTRC silencing leads to an inhibition of AGPase activity in vivo.
NTRC Downregulation Leads to Decreased Activity of Soluble Starch Synthase
In addition to AGPase, we also assayed other enzymes involved in starch metabolism in 35-DAF fruits. Soluble and granule-bound starch synthases catalyze the final polymerizing reactions of starch biosynthesis. As shown in Table 1, perturbing NTRC expression decreased the activity of soluble starch synthase to 25% to 40% of the wild-type level, while the activity of granule-bound starch synthase only slightly decreased. Soluble starch synthase is subject to posttranslational redox regulation and a potential target of NTRC in vitro (Skryhan et al., 2015, 2018). In contrast, NTRC downregulation had no significant effect on the activities of starch phosphorylases and starch hydrolases, which are involved in starch degradation (Table 1). Together these results imply that downregulation of NTRC expression leads to decreased starch accumulation via a joint inhibition of both AGPase and soluble starch synthase.
Table 1. Changes in the activity of starch metabolism enzymes in 35-DAF tomato fruits of the wild type and NTRC-RNAi lines.
Results are the mean with se from six biological replicates.Statistical analysis used ANOVA with Dunnett’s test (*P < 0.05, compared to the wild type). FW, fresh weight.
| Enzymes | Wild Type | RNAi-2 | RNAi-26 | RNAi-33 |
|---|---|---|---|---|
| Soluble starch synthase (nmol min−1 mg−1 protein) | 44.8 ± 4.2 | 16.8 ± 2.6* | 11.4 ± 3.3* | 18.6 ± 6.2* |
| Granule-bound starch synthase (pmol min−1 mg−1 FW) | 2.9 ± 0.2 | 2.0 ± 0.1* | 2.0 ± 0.1* | 2.4 ± 0.2 |
| Starch phosphorylase (nmol min−1 mg−1 protein) | 49.6 ± 10.9 | 76.5 ± 18.3 | 30.7 ± 4.2 | 40.0 ± 11.3 |
| Starch hydrolases (μmol min−1 mg−1 protein) | 2.4 ± 0.4 | 2.3 ± 0.7 | 1.1 ± 0.2 | 1.0 ± 0.4 |
NTRC Downregulation Has Minor Effects on the Redox State of 2-Cys Prxs in Tomato Fruits
2-Cys Prxs are well-known targets of NTRC in Arabidopsis leaves, where they are involved in hydrogen peroxide removal during photosynthesis (Cejudo et al., 2012). Activation of 2-Cys Prxs involves reduction of an intermolecular disulfide bond linking the two subunits of the homodimer (Pérez-Ruiz et al., 2006, 2017). To investigate the role of NTRC in reduction of 2-Cys Prxs in tomato fruits, we assayed the monomer-to-dimer ratio of 2-Cys Prxs in tomato fruit tissues of NTRC-RNAi lines compared to the wild type using nonreducing SDS gels. NTRC downregulation slightly decreased the monomerization ratio of 2-Cys Prxs in both 35- and 65-DAF tomato fruits (Supplemental Fig. S6), even though the decrease was not consistent in all transgenic lines. The role of NTRC in regulating the redox state of 2-Cys Prxs seems to be less important in tomato fruit than in Arabidopsis leaves, where NTRC deficiency leads to a strong degree of 2-Cys-Prx oxidation (Cejudo et al., 2012). However, other Trxs might have compensated for the deficiency of NTRC with respect to 2-Cys-Prx reduction in tomato fruits.
DISCUSSION
NTRC is a NADPH-dependent thiol redox enzyme that has been found in chloroplasts as well as in nonphotosynthetic plastids (Kirchsteiger et al., 2012). In photosynthetic tissues, the role of NTRC in carbon metabolism and redox balance of chloroplasts in response to fluctuating light environments has been well characterized (Geigenberger et al., 2017; Zaffagnini et al., 2019). Previous studies led to the perspective that NTRC might function differently in heterotrophic tissues; however, our understanding as to how remains fragmentary (Kirchsteiger et al., 2012). In this work, we used genetic and biochemical approaches to directly test the role of NTRC in developing tomato fruits, which represent heterotrophic tissues dependent on Suc import with no substantial photosynthetic carbon assimilation (Lytovchenko et al., 2011). Our results show that NTRC does not affect developmental and ripening programs of the fruit but promotes transient starch accumulation by activating AGPase and soluble starch synthase to increase overall sink activity and hence final fruit size, dry matter, and sugar content. In contrast to this, NTRC leads to a decreasing pattern of organic and amino acids and a decrease in the NAD(P)(H) reduction state. This indicates that NTRC serves as an important hub to orchestrate carbon metabolism and redox balance in heterotrophic fruit tissues to optimize fruit growth and quality in response to Suc supply.
NTRC Is Important to Optimize Fruit Size and Quality
Tomato is an important horticultural crop and serves as a major component of our diet (Carrari and Fernie, 2006). There has been considerable interest in optimizing tomato fruit yield and quality by using genetic approaches to alter sink metabolism and ripening (Giovannoni, 2004; Carrari and Fernie, 2006), but there is a shortage of studies investigating the role of redox regulation in this context. In the present work, NTRC expression was specifically decreased by 50% to 75% in developing tomato fruits using RNAi under the control of a fruit-specific promoter (Fig. 1). Perturbing NTRC expression did not affect developmental and ripening programs of the fruit (Supplemental Fig. S2) but led to a decrease in final fruit size and weight, with fully mature fruits containing less dry matter and more water than wild-type fruits (Fig. 2). The phenotype was accompanied by a decrease in the levels of soluble sugars (Figs. 3D and 4A; Supplemental Table S1) and an opposing increase in many organic acids and amino acids, as well as phenolic compounds (Fig. 4, B, C, and D; Supplemental Table S1). Domesticated tomatoes tend to accumulate high amounts of Glc and Fru. Thus, the total abundance of Glc, Fru, and other minor sugars determines the sweetness of tomato fruits. The acid-to-sugar ratio determines the sourness of fruits, and it is also an important quality parameter (Davies et al., 1981; Gierson and Kader, 1986). Thus, it is very likely that the changes we observed in response to NTRC downregulation would eventually influence the taste and flavor of tomato fruits.
NTRC affects growth and development of Arabidopsis plants (Pérez-Ruiz et al., 2006; Lepistö et al., 2009; Toivola et al., 2013; Thormählen et al., 2015), but this is mainly ascribed to its role in photosynthesis and leaf metabolism, rather than in nonphotosynthetic tissues (Kirchsteiger et al., 2012). The current study provides direct evidence for a previously unrecognized role of NTRC in regulating the development of heterotrophic storage organs to optimize yield quantity and quality parameters. This shows that NTRC has important functions beyond photosynthesis that affect growth and development. Although immature fruits are green and contain chlorophyll, it is unlikely that the changes in fruit growth reported above are due to a role of NTRC in regulating photosynthetic processes within the fruit (Supplemental Fig. S2). Indeed, previous studies have shown that knockout of chlorophyll synthesis in pale tomato fruits does not have significant effects on fruit physiology, growth, and development (Lytovchenko et al., 2011).
There are conflicting studies concerning the effect of NTRC overexpression on the growth of Arabidopsis plants. While Toivola et al. (2013) provide evidence that mild overexpression of NTRC results in bigger plants, there is evidence from Ojeda et al. (2018) that strong NTRC overexpression may have the opposite effects. It is tempting to speculate as to whether increased expression of NTRC may lead to larger tomato fruits. However, given the results in Arabidopsis, this may depend on the level of NTRC overexpression. More studies will be necessary to clarify this question.
NTRC Regulates Transient Starch Accumulation in Developing Fruits
Tomato fruit development is characterized by a transient accumulation of starch in immature fruits, which serves as a carbon reserve for soluble sugar accumulation during ripening and finally affects fruit size (Schaffer et al., 2000). In this study, interfering with NTRC expression resulted in a 50% to 75% decrease in the level of the transient starch pool in immature fruits (Fig. 3A), which was accompanied by a decrease in soluble sugars during maturation and decreased fruit size (Figs. 3, C and D, and 4A). Our results show that the decrease in transient starch accumulation is associated with a decrease in the redox activation of AGPase (Fig. 6), which is the rate-limiting enzyme in the pathway of starch synthesis in developing tomato fruits (Schaffer et al., 2000) and other plant tissues (Geigenberger et al., 2004; Vigeolas et al., 2004; Neuhaus and Stitt, 1990; Faix et al., 2012) and a known target of NTRC (Geigenberger et al., 2017). NTRC downregulation led to a decrease in the reduction state of the small subunit of AGPase (Fig. 6A), which was accompanied by a 30% to 50% decrease in the sensitivity of the AGPase holoenzyme for its allosteric activator 3-PGA (Fig. 6, B and C). While the subcellular concentration of 3-PGA in the plastid has been estimated to yield 0.3 to 0.5 mm (Farré et al., 2001; Tiessen et al., 2002, 2012), the results of Figure 6B indicate that NTRC downregulation will lead to a 30% to 50% lower AGPase activity under in vivo conditions in developing tomato fruits. In confirmation of this, NTRC silencing led to a 40% decrease in the in vivo level of ADP-Glc (Fig. 6D), which is the immediate product of the reaction catalyzed by AGPase. This provides independent in vivo evidence that NTRC downregulation restricts AGPase activity in developing fruits.
This is consistent with previous studies documenting the primary role of AGPase in regulating starch accumulation, soluble sugar content, and growth of tomato fruits. First, increased AGPase activity in near-isogenic tomato lines led to elevated starch accumulation in immature fruits, followed by increased soluble sugar content and larger size of fully ripe fruits (Petreikov et al., 2009). Second, increased AGPase redox activation by manipulation of NAD(P)(H) redox states led to elevated rates of starch synthesis in immature fruits and a subsequent increase in the content of soluble sugars in fully ripe fruits (Centeno et al., 2011).
Interestingly, NTRC downregulation also led to a strong decrease in the activity of soluble starch synthase catalyzing the subsequent step in the pathway of starch synthesis, while enzyme activities involved in starch degradation were not significantly affected in developing fruits (Table 1). Soluble starch synthase has been identified as a redox-regulated enzyme and potential target of NTRC in vitro, with reduction via NTRC or Trx f leading to an increase in its activity in vitro (Skryhan et al., 2015, 2018). Our results provide evidence that the relationship between NTRC and soluble starch synthase, which has been shown in vitro, also extends to the in vivo situation of developing fruits. Intriguingly, our data show a combined effect of NTRC on both AGPase and soluble starch synthase, catalyzing the first committed and final polymerizing steps of starch biosynthesis, respectively. This indicates a role of NTRC in stimulating starch accumulation and ultimately growth and quality of tomato fruits by combined redox activation of both enzymes.
NTRC is also involved in transient starch accumulation in leaves of Arabidopsis plants during day-night cycles (Lepistö et al., 2013; Thormählen et al., 2015). NTRC deficiency in Arabidopsis transfer DNA insertion mutants leads to a 30% to 40% restriction in redox activation of AGPase and starch synthesis upon illumination, both parameters being further attenuated in double mutants with combined deficiencies in NTRC and Trx f1 (Thormählen et al., 2015). In leaves, NTRC is dependent on NADPH produced by the photosynthetic light reactions. Furthermore, it is involved in the regulation of the Calvin-Benson cycle, and therefore in the provision of the substrates and the allosteric activator of AGPase, which will indirectly contribute to the regulation of starch synthesis (Thormählen et al., 2015). This phenomenon contrasts with the situation in heterotrophic tissues, where AGPase redox state and starch synthesis are both regulated by the transient supply of sugars via the phloem (Tiessen et al., 2002; Michalska et al., 2009). Sugar translocation from leaves to heterotrophic organs has been shown to vary in response to development (Walker and Ho, 1977) and diurnal alterations (Bénard et al., 2015). Changes in sugar availability may affect the activity of the oxidative pentose phosphate pathway (OPPP; Geigenberger and Fernie, 2014), leading to alterations in the NADP(H) redox state and, thus, in NTRC activity. Little is known about changes in NADP(H) levels in plant tissues. There is evidence for diurnal alterations in NADP(H) levels and redox states in Arabidopsis leaves (Thormählen et al., 2015). Moreover, Glc—but not Suc—feeding leads to an increase in the NADPH-to-NADP+ ratio in heterotrophic potato tuber tissue (Kolbe et al., 2005). This implies that in developing fruits, NTRC modulates starch synthesis via redox regulation of AGPase and soluble starch synthase in response to changes in sugar import, which lead to alterations of NADPH by affecting the OPPP. This is important to adjust the sink activity of the fruit to the supply of Suc from the leaves to optimize fruit growth.
NTRC Regulates the Redox Balance of NAD(P)H in Developing Fruits
In heterotrophic plastids lacking photochemical reactions, redox homeostasis depends mainly on NADPH, so NTRC, which depends on NADPH, may play an essential role in maintaining the plastidial redox poise. Our results show that interfering with NTRC expression strongly affects the cellular redox states of NAD(H) and NADP(H) in developing tomato fruits (Fig. 5), providing evidence that NTRC is a major regulator of cellular redox homeostasis in this heterotrophic tissue. While at the moment the underlying mechanisms are unresolved, NTRC uses NADPH as a substrate and thus may influence the NADPH-to-NADP+ ratio directly or via regulation of plastidial pathways consuming or producing NADPH, such as the OPPP. However, direct effects of NTRC on enzymes of the OPPP have not been studied yet. A further possibility is that NTRC affects the redox shuttles between plastids and mitochondria, leading to changes in interorganellar redox regulation. Previous studies in tomato fruits provide evidence that plastidial and mitochondrial NAD(P)H systems are interconnected via the malate valve (Centeno et al., 2011). An increase in the NADPH redox state in response to NTRC downregulation in the plastid may therefore be transmitted to mitochondria, leading to an increase in NADH redox state, and vice versa. Since adenylate energy states were not significantly changed in NTRC-RNAi lines (Supplemental Table S2), it is quite unlikely that the changes in redox states are due to changes in respiration rates.
NTRC deficiency also leads to an increase in NADP(H) and NAD(H) reduction states in Arabidopsis plants (Thormählen et al., 2015, 2017). However, this is most probably due to NTRC regulating the balance between photosynthetic light reactions and the Calvin-Benson cycle, as well as NADPH export via NADP-dependent malate dehydrogenase of the malate valve. Taken together, these results identify NTRC as a central hub controlling the cellular redox poise in nonphotosynthetic as well as photosynthetic tissues.
Based on the work in Arabidopsis plants, NTRC has been found to play a major role in detoxification of hydrogen peroxide by providing reductants to 2-Cys Prxs, with NTRC deficiency leading to a strong decrease in the 2-Cys Prxs reduction state in photosynthesizing leaves (Pérez-Ruiz et al., 2006, 2017). Compared to this, our results show that NTRC downregulation leads to a relatively minor decrease in 2-Cys Prxs reduction in developing and ripe tomato fruits (Supplemental Fig. S6). However, it must be noted that photosynthetic activity leads to increased rates of hydrogen peroxide production, which requires increased antioxidant activity via the NTRC-2-Cys Prxs system (Pérez-Ruiz et al., 2006, 2017). These results suggest that the antioxidant function of NTRC is less important in nonphotosynthetic than in photosynthetic tissues.
NTRC Affects the Carbon-Nitrogen Balance in Developing Fruits
While our results show that NTRC downregulation leads to a decrease in soluble sugar levels, there is an opposing increase in the levels of several amino acids in developing fruits (Fig. 4; Supplemental Table S1). Since the total protein level remains unchanged (Supplemental Fig. S4), it is more likely that NTRC downregulation leads to stimulation of amino acid synthesis, rather than protein degradation. Because amino acid biosynthesis requires reducing equivalents as substrates, production of amino acids may have been promoted by the increased NAD(P)H-to-NAD(P)+ ratios in developing fruits of the NTRC-RNAi lines (Fig. 5). Increased levels of amino acids in the face of decreased sugar levels are also observed in leaves of Arabidopsis mutants lacking NTRC (Lepistö et al., 2009; Thormählen et al., 2015), indicating that there are similar effects of NTRC on amino acid synthesis in photosynthetic and nonphotosynthetic tissues. Inverse relationships between sugars and amino acids have also been found in other plant tissues, such as barley seeds (Faix et al., 2012) or potato tubers (Trethewey et al., 1998; Fernie et al., 2003; Roessner-Tunali et al., 2003) across different genetic and physiological manipulations. Overall, these results suggest that NTRC is involved in the regulation of the sugar-amino acid balance to prevent osmotic imbalances in the developing fruit.
CONCLUSIONS
NTRC is a NADPH-dependent thiol redox system located in photosynthetic as well as in nonphotosynthetic plastids. While previous approaches have been mainly performed in Arabidopsis and photosynthetic leaves to date, there has been a lack of knowledge on the role of NTRC in heterotrophic tissues and crop plants. By establishing NTRC-RNAi specifically in tomato fruit tissues, our results uncover the importance of NTRC in developing tomato fruits. In immature fruits, NTRC promotes transient starch accumulation by activating AGPase and soluble starch synthase to increase the carbohydrate stock for soluble sugar accumulation during ripening to optimize final fruit yield and quality. NTRC also plays a pivotal role in regulating the redox poise of NAD(P)(H) during fruit development, which represents the primary redox system in nonphotosynthetic tissues. In summary, NTRC acts as a central hub in regulating carbon metabolism and redox balance in heterotrophic tomato fruits, similar to its established role in photosynthetic tissues. Our results indicate that the activity of NTRC is indispensable to optimization of fruit growth as well as tomato yield quantity and quality.
MATERIALS AND METHODS
Plant Cultivation and Harvest
Tomato (Solanum lycopersicum ‘Moneymaker’) were grown as detailed in the literature (Carrari et al., 2003; Nunes-Nesi et al., 2005). Fruits were harvested at the mature-green (35 DAF) and ripe (65 DAF) stages with the intact pericarp being immediately frozen in liquid nitrogen for further analysis.
NTRC-RNAi Construct and Plant Transformation
The NTRC gene fragment was amplified from potato (Solanum tuberosum) cDNA with attB1-RNAi-NTRC (5′-ggggacaagtttgtacaaaaaagcaggctcagatgtcaggaattttgatca-3′) and attB2-RNAi-NTRC (5′-ggggaccactttgtacaagaaagctgggtcgcttgtcagatatctctccactgata-3′), and cloned into modified pBINAR-RNAi vector (pBIN19 backbone containing a patatin B33 promoter and a Gateway cassette) using a commercial Gateway cloning kit (Invitrogen). Tomato plants were transformed as described before by Tauberger et al. (2000) and the transformants were selected by kanamycin. The patatin B33 promoter acts as a fruit-specific promoter when expressed in tomato plants (Rocha-Sosa et al., 1989; Frommer et al., 1994; Obiadalla-Ali et al., 2004).
Molecular Characterization of NTRC-RNAi Lines
Genomic DNA extraction followed the cetyl-trimethyl-ammonium bromide-based protocol (Doyle et al., 1987). The kanamycin resistance gene (nptII) was amplified with primer pair Kana-F (5′-aactgttcgccaggctcaag-3′) and Kana-R (5′-tcagaagaactcgtcaagaagg-3′), and the internal control (CAC, SGN-U314153) was amplified with CAC-F (5′-cctccgttgtgatgtaactgg-3′) and CAC-R (5′-attggtggaaagtaacatcatcg-3′). The RNAzol (Sigma-Aldrich) was used for RNA extraction, and 500 μg of total RNA was used for preparing cDNA with Maxima Reverse Transcriptase (Thermo). The SYBG reagent (Biorad) was used for real-time PCR, and the signal was recorded via iQ5 Multicolor Real-Time PCR Detection System (Biorad). The primer pairs NTRC-F (5′-aggtgtatttgcagctggagatg-3′) and NTRC-R (5′-ttcttcagtagggggctggtggaa-3′), and NTRC-2F (5′-ggaccaactaagggcatcagcaa-3′) and NTRC-2R (5′-cgagcacagactcttctcctg-3′), were used to amplify NTRC genes LOC101254347 and LOC101266017, respectively. For detecting the expression of ethylene production and ripening genes, the primer pairs, SAHH2-F (5′-gacgctcacagtggaacaacga-3′) and SAHH2-R (5′-tcaactttccagcaaaatatcccc-3′), and NOR-F (5′-agagaacgatgcatggaggtttgt-3′) and NOR-R (5′-actggctcaggaaattggcaatgg-3′), were used for amplification of SAHH2 and NOR genes, respectively (Yang et al., 2017; Ma et al., 2019). The CAC gene described above served as an internal control (Expósito-Rodríguez et al., 2008). Total protein was extracted from 35 mg of pulverized sample with 150 μL of double-strength Laemmli buffer (Laemmli, 1970) containing 10 mm dithiothreitol, and 10 μL of protein extract was used for SDS-PAGE and immunoblotting. The membrane was blocked with 5% (w/v) nonfat milk and incubated with the first antibody of NTRC (1:1,000 dilution), Lhcb2 (Agrisera, 1:5,000), or ribulose 1,5-bisphosphate carboxylase/oxygenase large subunit (Agrisera, 1:10,000 dilution) at 4°C overnight for detecting different target proteins. The secondary antibody conjugating horseradish peroxidase (Agrisera, 1:10,000 dilution) was applied onto the membrane, and the membrane was incubated at 25°C for 1 h. The membrane reacted with Prime ECL solution (GE Healthcare), and the signal was visualized with x-ray film (Fuji).
Spectrophotometric Metabolite Measurements
Starch and Soluble Sugar Quantification
The extraction and detection of starch and soluble sugar followed the method described previously (Hendriks et al., 2003). In brief, 20 mg of pulverized sample was extracted with 250 μL of 80% (v/v) ethanol twice followed by another 250 μL of 50% (v/v) ethanol at 90°C for 30 min. The supernatant was used for the detection of soluble sugar, and the pellet was dried out for the detection of starch. For measuring starch level, the pellet was resuspended with 400 μL of 0.1 m sodium hydroxide and incubated at 95°C for 1 h. The solution was then neutralized to pH 7 with HCl-acetate buffer (0.5 m HCl prepared in 0.1 m acetate-NaOH buffer, pH 4.9), and 40 μL of aliquot was incubated with 110 μL of starch degradation buffer (2.8 U mL−1 amyloglucosidase, 4 U mL−1 α-amylase, and 50 mm acetate-NaOH buffer) at 37°C overnight. Fifty microliters of digested sample was first mixed with 160 μL of Glc determination buffer (3.4 U mL−1 G6PDH, 3 mm ATP, 1.3 mm NADP, 3 mm MgCl2, and 100 mm HEPES-KOH, pH 7.0). When the reaction saturated, 0.45 U hexokinase was applied to detect the formation of NADPH, which was proportional to the starch amount, at 340 nm via a spectrophotometer (FilterMax F5, Molecular Device).
For measuring the soluble sugar level, 30 μL of supernatant was first mixed with 200 μL of detection buffer (0.7 U mL−1 G6PDH, 1.7 mm ATP, 0.8 mm NADP, 5 mm MgCl2, and 50 mm HEPES-KOH, pH 7). When the reaction was stable, 0.5 U hexokinase, 1.2 U phosphoglucoisomerase, and 133 U invertase were applied to detect Glc, Fru, and Suc, respectively, via the formation of NADPH at 340 nm.
For the detection of maltose, the method described previously was followed with several modifications (Shirokane et al., 2000). Briefly, the above ethanol extract was dried out by heating at 95°C and substituted with an equal volume of double distilled water. Then, 30 μL of extract was mixed with 200 μL of detection buffer described above with addition of 0.5 U hexokinase. When the reaction was stable, 0.2 U maltose phosphorylase was applied to detect maltose via NADPH formation in the florescence mode of spectrometer (excitation [Ex] 340 nm, emission [Em] 465 nm). Varied concentration of maltose (Sigma-Aldrich) served as the standard for calculation.
Hexose Phosphates and 3-PGA
The assay of hexose phosphates and 3-PGA was performed as described before (Häusler et al., 2000). Briefly, 50 mg of pulverized sample was suspended in 500 μL of ice-cold 0.5 m HClO4 and incubated at 4°C for 10 min. Then, leaf residue was removed by centrifuging for 10 min at 20,000g. The pH of the supernatant was adjusted to the range from 5.0 to 6.0 using 2.5 m K2CO3 solution, and the supernatant was then centrifuged for 10 min at 20,000g to remove formed precipitant. Around 5 mg of active charcoal was applied into the solution, which was incubated on ice for 20 min. Charcoal was further removed via centrifuging for 10 min at 20,000g in 4°C. For measuring hexose phosphates, 50 μL of supernatant was mixed with 50 μL of water and reacted with 100 μL of detection buffer (200 mm Tris-HCl, pH 8.1, 1.6 mm NADP, 10 mm MgCl2, and 2 μm Glc-1,6-bisphosphate). When the reaction was stable, 0.1 U Glc-6-phosphate dehydrogenase, 0.35 U phosphoglucoisomerase, and 0.04 U phosphoglucomutase were applied to detect G6P, Fru-6-phosphate, and G1P, respectively, via NADPH formation in the fluorescence mode of the spectrometer (Ex 340 nm, Em 465 nm). For detecting 3-PGA, 50 μL of supernatant was mixed with 50 μ L of water and reacted with 100 μL of detection buffer (200 mm HEPES-NaOH, pH 7.0, 2 mm MgCl2, 2 mm ATP, 8 μm NADH, and 0.8 U mL −1 glyceraldehyde 3-phosphate dehydrogenase). When the reaction was stable, 0.45 U phosphoglycerate kinase was applied to detect 3-PGA via NADH consumption in the florescence mode of the spectrometer described above. Varied concentration of NADPH and NADH (Carl Roth) served as the standard for calculation.
Pyridine Nucleotides
The assay of pyridine nucleotides (NAD+, NADP+, NADH, and NADPH) was performed as described previously (Lintala et al., 2014). In short, 25 mg of pulverized sample was suspended in 250 μL of 0.1 m HClO4 for detecting NAD+ and NADP+. Another 25 mg of sample was suspended in 250 μL of 0.1 m KOH for detecting NADH and NADPH. Samples were further incubated on ice for 10 min and then centrifuged at 20,000g for 10 min at 4°C to remove the pellet. The supernatant was incubated at 95°C for 2 min, and the pH value was adjusted to the range from 8.0 to 8.5 with 0.1 m KOH prepared in 0.2 m Tris-HCl (pH 8.4) or HClO4 prepared in 0.2 m Tris-HCl (pH 8.4), respectively. For measuring NAD(H), 50 μL of extract was mixed with 50 μL of water and reacted with 50 μL of detection buffer containing 300 mm Tricine-KOH (pH 9.0), 12 mm EDTA, 1.5 m ethanol, 0.3 mm phenazine ethosulfate, 1.8 mm methylthiazolydiphenyl-tetrazolium bromide, and 18 U mL−1 alcohol dehydrogenase. For measuring NADP(H), another 50 μL of extract was mixed with 50 μL of water and reacted with 50 μL of detection buffer containing 300 mm Tricine-KOH (pH 9), 12 mm EDTA, 9 mm Glc-6-P, 0.3 mm phenazine ethosulfate, 1.8 mm methylthiazolydiphenyl-tetrazolium bromide, and 9 U mL−1 Glc-6-P dehydrogenase. The absorbance at wavelength 570 nm was recorded in a spectrometer (FilterMax F5, Molecular Device). Pyridine nucleotide (Carl Roth) solutions prepared in water were used to generate the standard curve for calculation.
ADP-Glc
The extraction of ADP-Glc was performed as described previously with minor modification (Lunn et al., 2006). Briefly, 50 mg of pulverized sample was suspended in 375 μL of ice-cold chloroform/methanol mixture (3:7) and vortexed vigorously at 4°C for 10 min followed by the addition of 300 μL of double distilled water for phase partitioning. After centrifugation at 18,000g at 4°C for 10 min, the supernatant was transferred to a new tube and dehydrated in a vacuum at ambient temperature. The dried pellet was resuspended in 70 μL of water, and 10 μL of the suspension was used for detecting ADP-Glc.
ADP-Glc was analyzed by liquid chromatography-mass spectrometry (LC-MS) using a Dionex Ultimate 3000 UHPLC (Thermo Fisher Scientific) and a timsTOF QTOF (Bruker Daltonik) in combination. The assay supernatant was injected into a C18 column (Ultra AQ C18, 150 × 2.1 mm, 3 µm, Restek) with 300 µL min−1 flow at 22°C. The solvents used were (A) water and (B) acetonitrile, both including 0.1% (v/v) formic acid. The gradient started at 1% B for 1 min, followed by a ramp to 25% B within 10 min. After a 4 min washing step at 95% B, the gradient turned back to 1% B and kept constant for 3 min equilibration. After the column compartment, the analytes passed a diode array detector at 254 nm before entering the mass spectrometer. An electrospray ionization source in negative mode was used at 5,000 V capillary voltage and nitrogen as dry gas, 8 L min−1 at 200°C. The timsTOF mass spectrometer (Bruker Daltonik) was run in MS mode from 50 to 1300 m/z mass range. Compounds were identified using the specific mass (m/z), isotope pattern, and retention time after standard injection. Data were acquired by otofControl 4.0. The evaluation was performed using DataAnalysis 5.1. All software tools were provided by Bruker (Bruker Daltonik). All solvents were supplied in LC-MS grade by Biosolve. The ADP-Glc standard was bought from Sigma-Aldrich.
Total Chlorophyll
The quantification of total chlorophyll was performed according to a previous approach (Porra et al., 1989). In short, 35 mg of pulverized sample was extracted with 1 mL of ice-cold 80% (v/v) acetone and vortexed at 4°C for 2 min followed by centrifugation to remove insoluble residues. Then, 250 μL of supernatant was mixed with 750 μL of 80% (v/v) acetone and applied into a quartz cuvette. The optical density of extract was determined by measuring the absorbance at wavelength 664 and 647 nm.
Measurement of Adenylate Nucleotide
The extraction of adenylate nucleotides followed a previously published method (Jelitto et al., 1992). Briefly, 50 mg of pulverized sample was extracted with 600 μL of extraction buffer (16% [w/v] TCA and 5 mm EGTA), and rigorously vortexed at 4°C for 1 h. The supernatant was then washed twice with 4 mL of cold water-saturated diethyl ether to remove TCA. The lower water phase was transferred to a new microtube, and the pH was adjusted to between 6.0 and 7.0 with neutralization buffer (5 m KOH and 1 m triethanoamine). The sample was placed under a hood for 1 h to let the remaining diethylether evaporate. The detection procedure was described before (Haink and Deussen, 2003) with several modifications: 85 μL of extract reacted with 20 μL of citrate buffer (62 mm citric acid and 76 mm KH2PO4, pH 5.2) and 20 μL of 50% (v/v) chloroacetaldehyde at 80°C for 15 min. Then, 20 μL of sample was applied into a NUCLEODUR 100-5 C18 ec column (MACHEREY-NAGEL) and eluted with the mixture of mobile buffer A (10 mm KH2PO4 and 5.7 mm tetrabutylammoniumhydrogensulfate, pH 5.4) and mobile phase B (90% [v/v] acetonitrile) under a flow rate of 0.8 mL min−1. The gradient was as follows: 0 min (100% A/0% B), 18 min (86% A/14% B), 36 min (56% A/44% B), 38.4 min (17.5% A/82.5% B), 39.6 min (100% A/0% B), and 42 min (100% A/0% B). The signal was analyzed by a fluorescence detector (FINNIGAN SURVEYOR PDA Plus Detector, Thermo Fisher Scientific), which was set to wavelengths Ex 280 nm, Em 410 nm. Standards were provided by Sigma-Aldrich.
Metabolite Profiling: GC-TOF-MS
Metabolic profiling was performed using a method previously described, with modification (Roessner et al., 2001; Lisec et al., 2006; Erban et al., 2007). In brief, for the extraction, 50 mg of tissue (fresh weight) was ground in 360 µL of cold (−20°C) methanol containing internal standards (10 μL of ribitol, 0.2 mg mL−1 in water and 10 μL of 13C-sorbitol, 0.2 mg mL−1 in water) for the relative quantification. First, the samples were incubated at 4°C for 30 min, and then the extract was carefully mixed with 200 μL of chloroform and 400 µL of water. To force the separation of the polar and nonpolar phase, a 15 min centrifugation step at 25,000g was performed. For derivatization, 50 μL of the upper (polar) phase was dried in vacuum. The pellet was resuspended in 10 μL of methoxyaminhydrochloride (20 mg mL−1 in pyridine) and agitated for 90 min at 40°C. After the addition of 20 µL of BSTFA (N,O-Bis[trimethylsilyl]trifluoroacetamide) containing 2.5 μL of retention time standard mixture of linear alkanes (n-decane, n-dodecane, n-pentadecane, n-nonadecane, n-docosane, n-octacosane, n-dotriacontane), the whole mixture was incubated at 40°C for 45 min. Finally, 1 μL of sample was injected into a GC-TOF-MS system (Pegasus HT, Leco). Samples were prepared by an auto-sampler system (Combi PAL, CTC Analytics AG). Helium acted as carrier gas at a constant flow rate of 1 mL min−1. Gas chromatography was performed on an Agilent GC (7890A, Agilent) using a 30 m VF-5ms column with 10 m EZ-Guard column. The injection temperature of the split/splitless injector, as well as the transfer line and the ion source, was set to 250°C. The initial oven temperature (70°C) was permanently increased to a final temperature of 350°C by 9°C min−1. To avoid solvent contaminations, the solvent delay time was set to 340 s. Compounds were ionized and fractionated at 70 eV. Mass spectra were recorded at 20 scans s−1 with an m/z of 35 to 800 scanning range. Chromatograms and mass spectra were evaluated using ChromaTOF 4.5 and TagFinder 4.1 software (Luedemann et al., 2008). All solvents were supplied in LC-MS grade by Biosolve. Standards were provided by Sigma-Aldrich.
Redox Shift Assay of APS1 and 2-Cys Prxs
To preserve the redox state, the total protein was extracted using a TCA-based method (Hendriks et al., 2003). In short, 35 mg of frozen fruit tissue was extracted with 1 mL of ice-cold 16% (w/v) TCA prepared in diethylether and incubated at −20°C overnight. After centrifugation, the supernatant was discarded, and the pellet was washed with ice-cold acetone three times. The pellet was then suspended with double-strength Laemmli buffer (Laemmli, 1970) without reducing reagent and incubated at 90°C for 5 min with 1,400 rpm shaking speed. Then, 10 μL of protein extract was applied into 10% or 12% SDS-acrylamide gel to perform immunoblot analyses detecting APS1 or 2-Cys Prxs, respectively. The immunoblotting procedure was performed as described before (Hendriks et al., 2003; Pérez-Ruiz et al., 2006), and Image-J was used for quantification of the protein signals.
Enzyme Activity Assay of Starch Metabolism
AGPase
The AGPase activity assay followed a previously described method (Tiessen et al., 2002) with several modifications. Briefly, 25 mg of pulverized frozen fruit tissue was extracted with 150 μL of 50 mm HEPES-KOH buffer (pH 7.5), incubated on ice for 10 min, then centrifuged to remove leaf residue. The crude protein extract was applied into a Zeba desalting column (Thermo Fisher Scientific) as described in the manual. Then, 20 μL of protein extract was incubated with 180 μL of AGPase reaction buffer (50 mm HEPES-KOH, pH 7.5, 7 mm MgCl2, 1.5 mm ATP, 0.5 mm G1P, and varied concentration of 3-PGA) at 25°C for 30 min with 900 rpm shaking speed, and the reaction was terminated by mixing rigorously with 100 μL of chloroform. Two microliters of the supernatant was used for quantification of ADP-Glc produced during the enzyme assay. ADP-Glc was detected as described in the section titled “ADP-Glc,” with the modification that an electrospray ionization source in negative mode was used at 4,000 V capillary voltage with nitrogen as dry gas, 8 L min−1 at 250°C.
Soluble Starch Synthase and Granule-Bound Starch Synthase
The assay was performed as previously described with modification (Kulichikhin et al., 2016). Briefly, 25 mg of pulverized sample was suspended in 150 μL of ice-cold buffer (100 mm HEPES-NaOH, pH 7.5, 8 mm MgCl2, 2 mm EDTA, 12.5% [v/v] glycerol, 5% [w/v] PVP-10, protease inhibitor cocktail), and incubated on ice for 10 min followed by centrifugation to separate the supernatant and insoluble pellet. For detecting soluble starch synthase, 50 μL of the supernatant was applied into a self-packed Sephadex G25 column equilibrated with 50 mm HEPES-NaOH (pH 7.5) for desalting and mixed with 50 μL of double-strength reaction buffer (100 mm HEPES-NaOH, pH 7.5, 2 mm ADP-Glc, and 0.2% [w/v] amylopectin) followed by incubation at 30°C for 30 min. The reaction was terminated by the addition of one-third volume chloroform with vigorous vortex, and the upper phase was used for detecting the production of ADP during the reaction. For the granule-bound starch synthase, the insoluble pellet was washed twice with 1 mL of 50 mm HEPES-NaOH (pH 7.5) and resuspended in 100 μL of reaction buffer (50 mm HEPES-NaOH, pH 7.5, and 1 mm ADP-Glc) followed by incubation at 30°C for 30 min with mild shaking. The reaction was terminated by the addition of chloroform, and 10 μL of the upper phase was used for detecting ADP. ADP was analyzed as described in the section titled “AGPase.” ADP standards were provided by Sigma-Aldrich.
Starch Phosphorylase
The activity of starch phosphorylase was assayed by detecting the number of G1Ps produced in the reaction (Zeeman et al., 1998; Häusler et al., 2000). In brief, 25 mg of pulverized sample was extracted with 150 μL of extraction buffer (40 mm HEPES-NaOH, pH 7.5, and 1 mm EDTA) and incubated on ice for 10 min followed by centrifugation to remove fruit residues. Then, 25 μL of protein extract was mixed with 75 μL of water and 100 μL of double-strength reaction buffer (200 mm Tris-HCl, pH 8.1, 1.6 mm NADP+, 10 mm MgCl2, 2 μm Glc-1,6-bisphosphate, 20 mm sodium phosphate buffer, pH 6.9, 0.1% [w/v] amylopectin, 0.11 U Glc-6-phosphate dehydrogenase, and 0.07 U phosphoglucomutase). The NADPH formation was detected in the fluorescence mode of the spectrophotometer (Ex 340 nm, Em 465 nm).
Starch Hydrolase
The activity of starch hydrolase was assayed by detecting the number of reducing groups released from starch with 3,5-dinitrosalicylic acid (DNS) reagent (Miller, 1959). In short, 25 mg of pulverized sample was extracted with 150 μL of extraction buffer (20 mm sodium phosphate buffer, pH 6.9, and 6 mm NaCl) and incubated on ice for 10 min followed by centrifugation to remove fruit residues. The supernatant was applied to a self-packed Sephadex G-25 column for desalting. Then, 50 μL of protein extract was mixed with an equal volume of 1% (w/v) soluble starch followed by incubation at 60°C for 30 min, and protein extract without the input of starch served as the blank reaction. The reaction was terminated by the addition of 100 μL of DNS reagent (1% [w/v] DNS and 30% [w/v] sodium potassium tartrate tetrahydrate), and the whole mixture was incubated at 95°C for 5 min. The amount of reducing sugar released from starch was determined in the absorbance mode of the spectrophotometer (wavelength 540 nm). Maltose served as the standard for quantification.
The protein concentration was measured by Bradford reagent (Carl Roth) and Pierce 660 nm protein reagent (Thermo) according to the manual.
Statistical Analysis
All figures and statistical analyses were made via Graphpad Prism 8. ANOVA and Dunnett’s test were used to evaluate the difference between the wild type and NTRC-RNAi lines unless otherwise stated.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers LOC101254347 and LOC101266017 (SlNTRC), Solyc09g092380 (SlSAHH2), Solyc10g006880 (SlNOR), and SGN-U314153 (SlCAC).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The molecular characterization in 65-DAF tomato fruits of NTRC-RNAi lines compared to the wild type.
Supplemental Figure S2. Characterization of developmental stage in tomato fruits.
Supplemental Figure S3. Phenotypic analysis of mature green tomato fruits (35 DAF) of NTRC-RNAi lines 2, 26, and 33 compared to the wild type.
Supplemental Figure S4. Total protein content of fruits of NTRC-RNAi lines compared to the wild type.
Supplemental Figure S5. Redox and allosteric properties of AGPase in 65-DAF tomato fruits of NTRC-RNAi lines 2, 26, and 33 compared to the wild type.
Supplemental Figure S6. Redox state of and 2-Cys Prxs in 35- and 65-DAF tomato fruits of wild type and NTRC-RNAi lines.
Supplemental Table S1. Fold changes in metabolite profiles in tomato fruits of wild type and NTRC-RNAi lines.
Supplemental Table S2. Changes in the level of hexose phosphates, 3-PGA, and adenine nucleotides in wild-type and NTRC-RNAi tomato fruits.
ACKNOWLEDGMENTS
We thank Anne Bierling (Ludwig-Maximilians University, Munich) for excellent technical assistance. We are also grateful to all greenhouse staff in the Max-Planck-Institute of Molecular Plant Physiology for taking care of tomato plants.
Footnotes
This work was supported by the Deutscher Akademischer Austauschdienst (Graduate School Scholarship Programme [57243780], grant to L.-Y.H. and P.G.) and the Deutsche Forschungsgemeinschaft (TRR 175, The Green Hub).
Articles can be viewed without a subscription.
References
- Ballicora MA, Frueauf JB, Fu Y, Schürmann P, Preiss J (2000) Activation of the potato tuber ADP-glucose pyrophosphorylase by thioredoxin. J Biol Chem 275: 1315–1320 [DOI] [PubMed] [Google Scholar]
- Bénard C, Bernillon S, Biais B, Osorio S, Maucourt M, Ballias P, Deborde C, Colombié S, Cabasson C, Jacob D, et al. (2015) Metabolomic profiling in tomato reveals diel compositional changes in fruit affected by source-sink relationships. J Exp Bot 66: 3391–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Ramírez-Hernández MH, Ziegler M (2004) The new life of a centenarian: Signalling functions of NAD(P). Trends Biochem Sci 29: 111–118 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y (2005) Redox regulation: A broadening horizon. Annu Rev Plant Biol 56: 187–220 [DOI] [PubMed] [Google Scholar]
- Carrari F, Baxter C, Usadel B, Urbanczyk-Wochniak E, Zanor M-I, Nunes-Nesi A, Nikiforova V, Centero D, Ratzka A, Pauly M, et al. (2006) Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol 142: 1380–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Fernie AR (2006) Metabolic regulation underlying tomato fruit development. J Exp Bot 57: 1883–1897 [DOI] [PubMed] [Google Scholar]
- Carrari F, Nunes-Nesi A, Gibon Y, Lytovchenko A, Loureiro ME, Fernie AR (2003) Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol 133: 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo LR, Froehlich JE, Cruz JA, Savage LJ, Kramer DM (2016) Multi-level regulation of the chloroplast ATP synthase: The chloroplast NADPH thioredoxin reductase C (NTRC) is required for redox modulation specifically under low irradiance. Plant J 87: 654–663 [DOI] [PubMed] [Google Scholar]
- Cejudo FJ, Ferrández J, Cano B, Puerto-Galán L, Guinea M (2012) The function of the NADPH thioredoxin reductase C-2-Cys peroxiredoxin system in plastid redox regulation and signalling. FEBS Lett 586: 2974–2980 [DOI] [PubMed] [Google Scholar]
- Centeno DC, Osorio S, Nunes-Nesi A, Bertolo ALF, Carneiro RT, Araújo WL, Steinhauser M-C, Michalska J, Rohrmann J, Geigenberger P, et al. (2011) Malate plays a crucial role in starch metabolism, ripening, and soluble solid content of tomato fruit and affects postharvest softening. Plant Cell 23: 162–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Q, Wang P, Wang M, Sun T, Jin H, Liu B, Wang J, Grimm B, Wang H-B (2017) Thioredoxin and NADPH-dependent thioredoxin reductase C regulation of tetrapyrrole biosynthesis. Plant Physiol 175: 652–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JN, Hobson GE, McGlasson WB (1981) The constituents of tomato fruit—the influence of environment, nutrition, and genotype. Crit Rev Food Sci Nutr 15: 205–280 [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL, Doyle JA, Doyle FJ (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19: 11–15 [Google Scholar]
- Erban A, Schauer N, Fernie AR, Kopka J (2007) Nonsupervised construction and application of mass spectral and retention time index libraries from time-of-flight gas chromatography-mass spectrometry metabolite profiles. In Weckwerth W, ed, Metabolomics: Methods and Protocols, Methods in Molecular Biology, Vol 358, Humana Press, Totowa, NJ, pp 19–38 [DOI] [PubMed] [Google Scholar]
- Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix B, Radchuk V, Nerlich A, Hümmer C, Radchuk R, Emery RJN, Keller H, Götz KP, Weschke W, Geigenberger P, et al. (2012) Barley grains, deficient in cytosolic small subunit of ADP-glucose pyrophosphorylase, reveal coordinate adjustment of C:N metabolism mediated by an overlapping metabolic-hormonal control. Plant J 69: 1077–1093 [DOI] [PubMed] [Google Scholar]
- Farré EM, Tiessen A, Roessner U, Geigenberger P, Trethewey RN, Willmitzer L (2001) Analysis of the compartmentation of glycolytic intermediates, nucleotides, sugars, organic acids, amino acids, and sugar alcohols in potato tubers using a nonaqueous fractionation method. Plant Physiol 127: 685–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Tiessen A, Stitt M, Willmitzer L, Geigenberger P (2003) Altered metabolic fluxes result from shifts in metabolite levels in sucrose phosphorylase-expressing potato tubers. Plant Cell Environ 25: 1219–1232 [Google Scholar]
- Frommer WB, Mielchen C, Martin T (1994) Metabolic control of patatin promoters from potato in transgenic tobacco and tomato plants. Plant Physiol (Life Sci Adv) 13: 329–334 [Google Scholar]
- Fu Y, Ballicora MA, Leykam JF, Preiss J (1998) Mechanism of reductive activation of potato tuber ADP-glucose pyrophosphorylase. J Biol Chem 273: 25045–25052 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Fernie AR (2014) Metabolic control of redox and redox control of metabolism in plants. Antioxid Redox Signal 21: 1389–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P, Stitt M, Fernie AR (2004) Metabolic control analysis and regulation of the conversion of sucrose to starch in growing potato tubers. Plant Cell Environ 27: 655–673 [Google Scholar]
- Geigenberger P, Thormählen I, Daloso DM, Fernie AR (2017) The unprecedented versatility of the plant thioredoxin system. Trends Plant Sci 22: 249–262 [DOI] [PubMed] [Google Scholar]
- Gierson D, Kader AA (1986) Fruit ripening and quality In Atherton JG, and Rudich J, eds, The Tomato Crop: A Scientific Basis for Improvement. Springer, New York, pp 241–280 [Google Scholar]
- Giovannoni JJ. (2004) Genetic regulation of fruit development and ripening. Plant Cell 16(Suppl): S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haink G, Deussen A (2003) Liquid chromatography method for the analysis of adenosine compounds. J Chromatogr B Analyt Technol Biomed Life Sci 784: 189–193 [DOI] [PubMed] [Google Scholar]
- Hatzfeld WD, Stitt M (1990) A study of the rate of recycling of triose phosphates in heterotrophic Chenopodium rubrum cells, potato tubers, and maize endosperm. Planta 180: 198–204 [DOI] [PubMed] [Google Scholar]
- Häusler RE, Fischer KL, Flügge UI (2000) Determination of low-abundant metabolites in plant extracts by NAD(P)H fluorescence with a microtiter plate reader. Anal Biochem 281: 1–8 [DOI] [PubMed] [Google Scholar]
- Hendriks JHM, Kolbe A, Gibon Y, Stitt M, Geigenberger P (2003) ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SK, Salamone PR, Okita TW (2005) Allosteric regulation of the higher plant ADP-glucose pyrophosphorylase is a product of synergy between the two subunits. FEBS Lett 579: 983–990 [DOI] [PubMed] [Google Scholar]
- Jelitto T, Sonnewald U, Willmitzer L, Hajirezeai M, Stitt M (1992) Inorganic pyrophosphate content and metabolites in potato and tobacco plants expressing E. coli pyrophosphatase in their cytosol. Planta 188: 238–244 [DOI] [PubMed] [Google Scholar]
- Kanayama Y. (2017) Sugar metabolism and fruit development in the tomato. Hortic J 86: 417–425 [Google Scholar]
- Kirchsteiger K, Ferrández J, Pascual MB, González M, Cejudo FJ (2012) NADPH thioredoxin reductase C is localized in plastids of photosynthetic and nonphotosynthetic tissues and is involved in lateral root formation in Arabidopsis. Plant Cell 24: 1534–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchsteiger K, Pulido P, González M, Cejudo FJ (2009) NADPH thioredoxin reductase C controls the redox status of chloroplast 2-Cys peroxiredoxins in Arabidopsis thaliana. Mol Plant 2: 298–307 [DOI] [PubMed] [Google Scholar]
- Kleczkowski LA. (1999) A phosphoglycerate to inorganic phosphate ratio is the major factor in controlling starch levels in chloroplasts via ADP-glucose pyrophosphorylase regulation. FEBS Lett 448: 153–156 [DOI] [PubMed] [Google Scholar]
- Kolbe A, Tiessen A, Schluepmann H, Paul M, Ulrich S, Geigenberger P (2005) Trehalose 6-phosphate regulates starch synthesis via posttranslational redox activation of ADP-glucose pyrophosphorylase. Proc Natl Acad Sci USA 102: 11118–11123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulichikhin K, Mukherjee S, Ayele BT (2016) Extraction and assays of ADP-glucose pyrophosphorylase, soluble starch synthase and granule bound starch synthase from wheat (Triticum aestivum L.) grains. Bio Protoc 6: e1929 [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lepistö A, Kangasjärvi S, Luomala E-M, Brader G, Sipari N, Keränen M, Keinänen M, Rintamäki E (2009) Chloroplast NADPH-thioredoxin reductase interacts with photoperiodic development in Arabidopsis. Plant Physiol 149: 1261–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepistö A, Pakula E, Toivola J, Krieger-Liszkay A, Vignols F, Rintamäki E (2013) Deletion of chloroplast NADPH-dependent thioredoxin reductase results in inability to regulate starch synthesis and causes stunted growth under short-day photoperiods. J Exp Bot 64: 3843–3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintala M, Schuck N, Thormählen I, Jungfer A, Weber KL, Weber APM, Geigenberger P, Soll J, Bölter B, Mulo P (2014) Arabidopsis tic62 trol mutant lacking thylakoid-bound ferredoxin-NADP+ oxidoreductase shows distinct metabolic phenotype. Mol Plant 7: 45–57 [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Luedemann A, Strassburg K, Erban A, Kopka J (2008) TagFinder for the quantitative analysis of gas chromatography-mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 24: 732–737 [DOI] [PubMed] [Google Scholar]
- Lunn JE, Feil R, Hendriks JHM, Gibon Y, Morcuende R, Osuna D, Scheible W-R, Carillo P, Hajirezaei M-R, Stitt M (2006) Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J 397: 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytovchenko A, Eickmeier I, Pons C, Osorio S, Szecowka M, Lehmberg K, Arrivault S, Tohge T, Pineda B, Anton MT, et al. (2011) Tomato fruit photosynthesis is seemingly unimportant in primary metabolism and ripening but plays a considerable role in seed development. Plant Physiol 157: 1650–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Balazadeh S, Mueller-Roeber B (2019) Tomato fruit ripening factor NOR controls leaf senescence. J Exp Bot 70: 2727–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska J, Zauber H, Buchanan BB, Cejudo FJ, Geigenberger P (2009) NTRC links built-in thioredoxin to light and sucrose in regulating starch synthesis in chloroplasts and amyloplasts. Proc Natl Acad Sci USA 106: 9908–9913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GL. (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31: 426–428 [Google Scholar]
- Nájera VA, González MC, Pérez-Ruiz JM, Cejudo FJ (2017) An event of alternative splicing affects the expression of the NTRC gene, encoding NADPH-thioredoxin reductase C, in seed plants. Plant Sci 258: 21–28 [DOI] [PubMed] [Google Scholar]
- Naranjo B, Mignée C, Krieger-Liszkay A, Hornero-Méndez D, Gallardo-Guerrero L, Cejudo FJ, Lindahl M (2016) The chloroplast NADPH thioredoxin reductase C, NTRC, controls non-photochemical quenching of light energy and photosynthetic electron transport in Arabidopsis. Plant Cell Environ 39: 804–822 [DOI] [PubMed] [Google Scholar]
- Neuhaus HE, Stitt M (1990) Control analysis of photosynthate partitioning: Impact of reduced activity of ADP-glucose pyrophosphorylase or plastid phosphoglucomutase on the fluxes to starch and sucrose in Arabidopsis thaliana (L.) Heynh. Planta 182: 445–454 [DOI] [PubMed] [Google Scholar]
- Nikkanen L, Toivola J, Rintamäki E (2016) Crosstalk between chloroplast thioredoxin systems in regulation of photosynthesis. Plant Cell Environ 39: 1691–1705 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Lytovchenko A, Smith AM, Loureiro ME, Ratcliffe RG, Sweetlove LJ, Fernie AR (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137: 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obiadalla-Ali H, Fernie AR, Kossmann J, Lloyd JR (2004) Developmental analysis of carbohydrate metabolism in tomato (Lycopersicon esculentum cv. Micro-Tom) fruits. Physiol Plant 120: 196–204 [DOI] [PubMed] [Google Scholar]
- Ojeda V, Pérez-Ruiz JM, Cejudo FJ (2018) 2-Cys peroxiredoxins participate in the oxidation of chloroplast enzymes in the dark. Mol Plant 11: 1377–1388 [DOI] [PubMed] [Google Scholar]
- Osorio S, Fernie AR (2014) Fruit ripening: Primary metabolism In Nath P, Bouzayen M, Mattoo AK, and Pech JC, eds, Fruit Ripening Physiology, Signalling and Genomics. CABI, Boston, pp 15–27 [Google Scholar]
- Pérez-Ruiz JM, Cejudo FJ (2009) A proposed reaction mechanism for rice NADPH thioredoxin reductase C, an enzyme with protein disulfide reductase activity. FEBS Lett 583: 1399–1402 [DOI] [PubMed] [Google Scholar]
- Pérez-Ruiz JM, Guinea M, Puerto-Galán L, Cejudo FJ (2014) NADPH thioredoxin reductase C is involved in redox regulation of the Mg-chelatase I subunit in Arabidopsis thaliana chloroplasts. Mol Plant 7: 1252–1255 [DOI] [PubMed] [Google Scholar]
- Pérez-Ruiz JM, Naranjo B, Ojeda V, Guinea M, Cejudo FJ (2017) NTRC-dependent redox balance of 2-Cys peroxiredoxins is needed for optimal function of the photosynthetic apparatus. Proc Natl Acad Sci USA 114: 12069–12074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Ruiz JM, Spínola MC, Kirchsteiger K, Moreno J, Sahrawy M, Cejudo FJ (2006) Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 18: 2356–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreikov M, Yeselson L, Shen S, Levin I, Schaffer AA, Efrati A, Bar M (2009) Carbohydrate balance and accumulation during development of near-isogenic tomato lines differing in the AGPase-L1 allele. J Am Soc Hortic Sci 134: 134–140 [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Preiss J. (1973) Adenosine diphosphoryl glucose pyrophosphorylase In Boyer PD, ed, The Enzymes, Vol 8 Academic Press, New York, pp 73–119 [Google Scholar]
- Richter AS, Peter E, Rothbart M, Schlicke H, Toivola J, Rintamäki E, Grimm B (2013) Posttranslational influence of NADPH-dependent thioredoxin reductase C on enzymes in tetrapyrrole synthesis. Plant Physiol 162: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha-Sosa M, Sonnewald U, Frommer W, Stratmann M, Schell J, Willmitzer L (1989) Both developmental and metabolic signals activate the promoter of a class I patatin gene. EMBO J 8: 23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie A (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133: 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AA, Levin I, Oguz I, Petreikov M, Cincarevsky F, Yeselson Y, Shen S, Gilboa N, Bar M (2000) ADPglucose pyrophosphorylase activity and starch accumulation in immature tomato fruit: The effect of a Lycopersicon hirsutum-derived introgression encoding for the large subunit. Plant Sci 152: 135–144 [Google Scholar]
- Serrato AJ, Pérez-Ruiz JM, Spínola MC, Cejudo FJ (2004) A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J Biol Chem 279: 43821–43827 [DOI] [PubMed] [Google Scholar]
- Shirokane Y, Ichikawa K, Suzuki M (2000) A novel enzymic determination of maltose. Carbohydr Res 329: 699–702 [DOI] [PubMed] [Google Scholar]
- Skryhan K, Cuesta-Seijo JA, Nielsen MM, Marri L, Mellor SB, Glaring MA, Jensen PE, Palcic MM, Blennow A (2015) The role of cysteine residues in redox regulation and protein stability of Arabidopsis thaliana starch synthase 1. PLoS One 10: e0136997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skryhan K, Gurrieri L, Sparla F, Trost P, Blennow A (2018) Redox regulation of starch metabolism. Front Plant Sci 9: 1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spínola MC, Pérez-Ruiz JM, Pulido P, Kirchsteiger K, Guinea M, González M, Cejudo FJ (2008) NTRC new ways of using NADPH in the chloroplast. Physiol Plant 133: 516–524 [DOI] [PubMed] [Google Scholar]
- Tauberger E, Fernie AR, Emmermann M, Renz A, Kossmann J, Willmitzer L, Trethewey RN (2000) Antisense inhibition of plastidial phosphoglucomutase provides compelling evidence that potato tuber amyloplasts import carbon from the cytosol in the form of glucose-6-phosphate. Plant J 23: 43–53 [DOI] [PubMed] [Google Scholar]
- Thormählen I, Meitzel T, Groysman J, Öchsner AB, von Roepenack-Lahaye E, Naranjo B, Cejudo FJ, Geigenberger P (2015) Thioredoxin f1 and NADPH-dependent thioredoxin reductase C have overlapping functions in regulating photosynthetic metabolism and plant growth in response to varying light conditions. Plant Physiol 169: 1766–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormählen I, Zupok A, Rescher J, Leger J, Weissenberger S, Groysman J, Orwat A, Chatel-Innocenti G, Issakidis-Bourguet E, Armbruster U, et al. (2017) Thioredoxins play a crucial role in dynamic acclimation of photosynthesis in fluctuating light. Mol Plant 10: 168–182 [DOI] [PubMed] [Google Scholar]
- Tiessen A, Hendriks JHM, Stitt M, Branscheid A, Gibon Y, Farré EM, Geigenberger P (2002) Starch synthesis in potato tubers is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase: A novel regulatory mechanism linking starch synthesis to the sucrose supply. Plant Cell 14: 2191–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiessen A, Nerlich A, Faix B, Hümmer C, Fox S, Trafford K, Weber H, Weschke W, Geigenberger P (2012) Subcellular analysis of starch metabolism in developing barley seeds using a non-aqueous fractionation method. J Exp Bot 63: 2071–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toivola J, Nikkanen L, Dahlström KM, Salminen TA, Lepistö A, Vignols HF, Rintamäki E (2013) Overexpression of chloroplast NADPH-dependent thioredoxin reductase in Arabidopsis enhances leaf growth and elucidates in vivo function of reductase and thioredoxin domains. Front Plant Sci 4: 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trethewey RN, Geigenberger P, Riedel K, Hajirezaei MR, Sonnewald U, Stitt M, Riesmeier JW, Willmitzer L (1998) Combined expression of glucokinase and invertase in potato tubers leads to a dramatic reduction in starch accumulation and a stimulation of glycolysis. Plant J 15: 109–118 [DOI] [PubMed] [Google Scholar]
- Vigeolas H, Möhlmann T, Martini N, Neuhaus HE, Geigenberger P (2004) Embryo-specific reduction of ADP-Glc pyrophosphorylase leads to an inhibition of starch synthesis and a delay in oil accumulation in developing seeds of oilseed rape. Plant Physiol 136: 2676–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AJ, Ho LC (1977) Carbon translocation in the tomato: Carbon import and fruit growth. Ann Bot 41: 813–823 [Google Scholar]
- Yang L, Hu G, Li N, Habib S, Huang W, Li Z (2017) Functional characterization of SlSAHH2 in tomato fruit ripening. Front Plant Sci 8: 1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Hisabori T (2016) Two distinct redox cascades cooperatively regulate chloroplast functions and sustain plant viability. Proc Natl Acad Sci USA 113: E3967–E3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini M, Fermani S, Marchand CH, Costa A, Sparla F, Rouhier N, Geigenberger P, Lemaire SD, Trost P (2019) Redox homeostasis in photosynthetic organisms: Novel and established thiol-based molecular mechanisms. Antioxid Redox Signal 31: 155–210 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Northrop F, Smith AM, Rees T (1998) A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolysing enzyme. Plant J 15: 357–365 [DOI] [PubMed] [Google Scholar]