Favorable alleles can improve the grain-filling rate in hybrid japonica rice by enhancing both carbon-fixing activity and the expression of a gene related to sucrose unloading.
Abstract
Hybrid rice (Oryza sativa) has been cultivated commercially for 42 years in China. However, poor grain filling still limits the development of hybrid japonica rice. We report here the map-based cloning and characterization of the GRAIN-FILLING RATE1 (GFR1) gene present at a major-effect quantitative trait locus. We elucidated and confirmed the function of GFR1 via genetic complementation experiments and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) gene editing in combination with genetic and molecular biological analyses. In addition, we conducted haplotype association analysis to mine the elite alleles of GFR1 among 117 rice accessions. We observed that GFR1 was constitutively expressed and encoded a membrane-localized protein. The allele of the rice accession Ludao (GFR1Ludao) improved the grain-filling rate of rice by increasing Rubisco initial activity in the Calvin cycle. Moreover, the increased expression of the cell wall invertase gene OsCIN1 in the near isogenic line NIL-GFR1Ludao promoted the unloading of Suc during the rice grain-filling stage. A yeast two-hybrid assay indicated that the Rubisco small subunit interacts with GFR1, possibly in the regulation of the rice grain-filling rate. Evaluation of the grain-filling rate and grain yield of F1 plants harboring GFR1Ludao and the alleles of 20 hybrids widely cultivated commercially confirmed that favorable alleles of GFR1 can be used to further improve the grain-filling rate of hybrid japonica rice.
Rice (Oryza sativa) consists of two subspecies, indica and japonica (Molina et al., 2011). The grain yield per unit area of hybrid japonica rice is higher than that of hybrid indica rice, whereas the competitive heterosis of grain yield of hybrid japonica rice is lower than that of hybrid indica rice. Because indica rice and japonica rice are two different subspecies, they are distributed in their own adaptive ecological environments, and their cultivation areas are distinctly different. Additionally, the flavor quality of japonica rice is favored by increasing numbers of consumers. Therefore, increasing the grain yield per unit area of hybrid japonica rice is needed for both farmers and consumers.
In general, the number of spikelets per panicle (as well as per unit area) of hybrid japonica rice plants is much greater than that of pure line plants, whereas the grain plumpness (GP) of hybrid japonica rice is not as high as that of pure lines. Inferior florets on the same panicle of hybrid japonica rice plants are not fully filled with starch (Hong and Leng, 2004). Poor grain filling of these inferior florets results in decreases not only in grain yield but also in commodity value (Kato and Takeda, 1996). We previously reported that the rice accession Ludao, a wild-type japonica rice grown naturally in the Liangyungang region (34°33ʹ N, 119°13ʹ E), Jiangsu Province, China, exhibited a high grain-filling rate and could be selected as a parent for improving the GP of hybrid japonica rice (Zhang et al., 2009). We were interested in using this unique accession to study the genetic basis and molecular mechanism of the grain-filling rate of rice to develop a hybrid japonica rice breeding strategy for high quality and high yields.
There are four mechanisms responsible for poor grain filling in rice. The first mechanism is source insufficiency (source-limited conditions; Bai et al., 2016; You et al., 2017). The second mechanism is regulation by plant hormones (You et al., 2016; Zhang et al., 2016). The third mechanism involves low polyamine levels and low biosynthetic activity (Yang et al., 2008). Last, the fourth mechanism involves the low transport ability of assimilates from leaves to the grain (Rennie and Turgeon, 2009; Ishibashi et al., 2014).
The enzymes related to the synthesis and transport of assimilates, including those involved in starch synthesis and Suc metabolism, have a significant effect on rice grain filling. To date, at least 15 genes related to enzyme activity and assimilate transport have been cloned. The genes Starch SynthaseI (SSI), SSIIa, SSIII, SSIV, Waxy, and Basic Leucine Zipper58 have been found to alter the structure of starch, but the effects of these types of starch synthase genes on rice grain filling have not been determined (Jiang et al., 2004; Fujita et al., 2006; Wang et al., 2013). The Suc transport-related genes Sucrose Transporter1 (OsSUT1), OsSUT2, OsSUT3, OsSUT4, and OsSUT5 have been reported to have a strong effect on grain filling (Hirose et al., 2008; Ishibashi et al., 2014). Moreover, the SWEET gene in maize (Zea mays; ZmSWEET4c) and rice (OsSWEET4) regulates the seed-filling process by mediating transepithelial hexose transport across the basal endosperm transfer layer (Sosso et al., 2015). Grain Incomplete Filling2 (GIF2), which encodes an ADP-Glc pyrophosphorylase large subunit, plays important roles in the regulation of grain filling and starch biosynthesis during caryopsis development (Wei et al., 2017).
The members of the cell wall invertase (CIN) gene family have been shown to be related to grain filling (Cho et al., 2005). OsCIN1, OsCIN2, OsCIN4, and OsCIN7 may also be related to the distribution of Suc to sinks (Cho et al., 2005). OsCIN1, OsCIN2, and OsCIN3 play important roles in regulating the unloading of assimilates during the rice grain-filling stage (Hirose et al., 2002). GIF1 is a key gene that encodes a cell wall invertase (OsCIN2) that regulates Suc transport and unloading and is required for carbon partitioning during the early grain-filling stage (Wang et al., 2008). Plant hormone-related genes have also been reported to affect grain filling. Gene-regulated brassinosteroid hormone levels can increase the assimilation of grain from Glc to starch (Wu et al., 2008). Among endosperm development-related genes, Fertilization-Independent Endosperm2 (OsFIE2) regulates rice endosperm development and grain filling via a mechanism different from that in Arabidopsis (Arabidopsis thaliana); the OsFIE2 gene product regulates H3K27me3 to alter the expression of a key helix-loop-helix transcription factor gene (Nallamilli et al., 2013). A possible functional network of microRNAs involved in rice grain filling was studied by Peng et al. (2013) and Zhao et al. (2019), who revealed important roles of 24-nucleotide small interfering RNAs and the miR1432-OsACOT (acyl-CoA thioesterase) module in the regulation of rice grain filling, respectively. Protein phosphorylation in rice has been studied as a key regulatory mechanism for regulating cell proliferation and growth, plant hormone biosynthesis, grain filling, and seed development (Qiu et al., 2016).
The objectives of this study were to (1) clone the favorable allele of GRAIN-FILLING RATE1 (GFR1) in the accession Ludao (GFR1Ludao), which is present at a major-effect quantitative trait locus (QTL) that regulates the grain-filling rate of rice; (2) elucidate the regulatory mechanism of GFR1 in terms of the grain-filling rate; (3) mine alleles favorable to the grain-filling rate in natural populations; and (4) confirm the phenotypic effects of elite alleles in terms of the grain-filling rate via four experimental F1 hybrids and 20 hybrids widely cultivated in rice production.
RESULTS AND DISCUSSION
GFR1 Is a Major Locus Controlling the Grain-Filling Rate of Rice
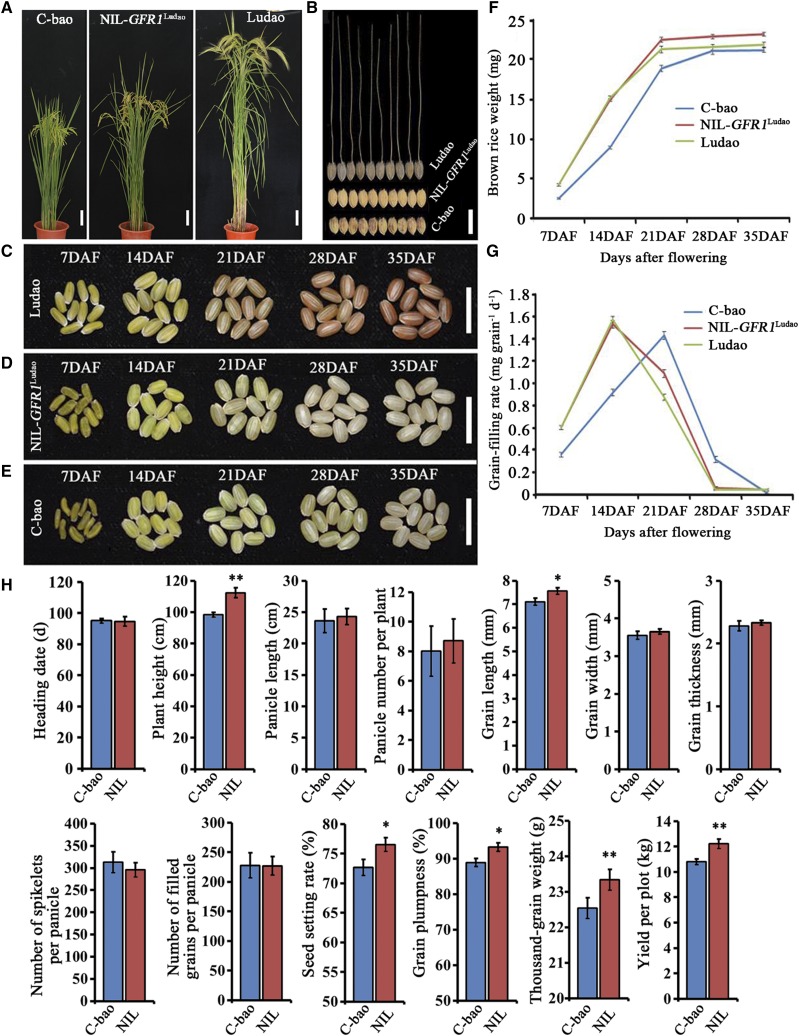
A population consisting of 102 backcross recombinant inbred lines (BILs) was constructed via two distinct accessions that exhibited natural variations in grain-filling rate (Supplemental Fig. S1A; Supplemental Table S1). Seven QTLs were detected by the conditional QTL mapping method, which involved adjusting the brown rice weight to an identical level (BRWIL), and three QTLs were detected with the time-course mapping method. Via the BRWIL method, three, two, one, and two QTLs affecting the grain-filling rate were detected at 7, 14, 28, and 35 d after flowering (DAF), respectively, and one was detected at both 7 and 14 DAF. Using the time-course QTL mapping method, we detected three and one QTLs at 7 and 14 DAF, respectively, and one was detected at both 7 and 14 DAF, all of which were among the seven QTLs detected by the BRWIL method (Supplemental Fig. S2; Supplemental Table S2). The time-course method is efficient and necessary for the study of the grain-filling rate because of the complexity of the underlying mechanism (Takai et al., 2005; Liu et al., 2015). Among the seven QTLs, we detected a major QTL, GFR1, which was detected by both the BRWIL conditional QTL method and the time-course QTL mapping method; this QTL was located at the end of the long arm of chromosome 10 between markers RM6745 and RM1146 and explained 17.18% and 33.72% of the phenotypic variation of the grain-filling rate at 7 and 14 DAF, respectively (Supplemental Fig. S2; Supplemental Table S2). The GFR1 allele from Ludao (GFR1Ludao) positively contributes to the grain-filling rate. A near-isogenic line (NIL) plant containing the QTL GFR1 from Ludao in the genomic background of C-bao was isolated (Supplemental Fig. S1). The grain-filling rates of Ludao and NIL-GFR1Ludao reached the highest values of 1.571 and 1.529 mg grain−1 d−1 at 14 DAF, respectively (Fig. 1G). The grain-filling rates of Ludao and NIL-GFR1Ludao were both higher than the grain-filling rate of C-bao at 7 and 14 DAF (Fig. 1G). With respect to the 12 agronomic traits investigated, no significant differences were detected for heading date, panicle number per plant, panicle length (PL), grain width (GW), grain thickness (GT), number of spikelets per panicle, and number of filled grains per panicle, while significant differences were detected for grain length (GL), seed setting rate (SS), GP, thousand-grain weight (TGW), and yield per plot (YPP) between C-bao and NIL-GFR1Ludao. These results showed that the relatively high grain-filling rate of NIL-GFR1Ludao also contributes to improvements in GL, SS, GP, TGW, and YPP (Fig. 1H). In addition to the significant differences in GL, SS, TGW, GP, and YPP between C-bao and NIL-GFR1Ludao, the height of the NIL-GFR1Ludao plants was much greater than that of the C-bao plants. In previous studies, the plant height QTL qPH10 was detected between markers RM228 (69 centimorgan) and c405a (83.8 centimorgan) on chromosome 10 (Xu et al., 2004) within the same region of the QTL of the GFR gene, which may explain why the NIL-GFR1Ludao plants were much taller than the C-bao plants.
Figure 1.
Phenotypic analysis of grain filling among Ludao, NIL-GFR1Ludao, and C-bao. A, Plants at the grain-filling stage. This is a composite image with three photographs. Bars = 10 cm. B, Seed shape after maturation. Bar = 1 cm. C to E, Morphological changes in brown rice grains of Ludao (C), NIL-GFR1Ludao (D), and C-bao (E) at five stages. Bars = 1 cm. F, Brown rice weight changes of Ludao, NIL-GFR1Ludao, and C-bao at five stages. G, Grain-filling rate changes of Ludao, NIL-GFR1Ludao, and C-bao at five stages. H, Comparison of yield traits between C-bao and NIL-GFR1Ludao in 2011. The data are means ± sd (n = 3). Student’s t test was used for statistical analysis (*, P ≤ 0.05 and **, P ≤ 0.01).
Map-Based Cloning of GFR1
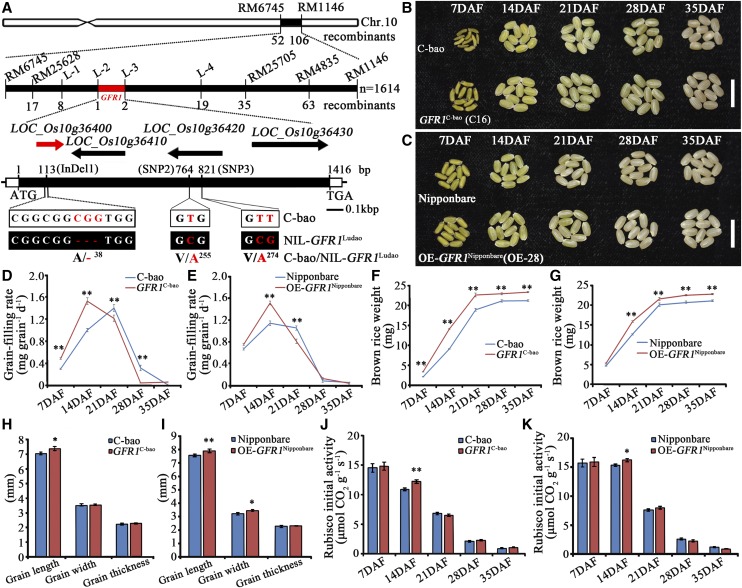
Genetic analysis of an F2 population derived from a cross between NIL-GFR1Ludao and C-bao showed a 1:2:1 segregation of the grain-filling rate trait [48 plants with the C-bao homozygous type:99 plants with the heterozygous type:53 plants with the NIL-GFR1Ludao homozygous type; χ2(1:2:1) = 0.17 < χ2(0.05,2) = 5.99] at the GFR1 locus, indicating that GFR1 behaves as a single Mendelian factor and a semidominant locus. High-resolution mapping involving 1,614 BC1F2 plants (derived from a single-locus segregating population) presenting fast grain-filling rates at 14 DAF delimited the GFR1 locus to a 34-kb chromosomal region between the insertion/deletion (InDel) markers L-2 and L-3 (Fig. 2A). This region contains four annotated open reading frames (ORFs; National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov/]): ORF1 (LOC_Os10g36400) encodes the DUF641 domain of unknown function; ORF2 (LOC_Os10g36410) encodes a protein with no putative conserved domains; ORF3 (LOC_Os10g36420) encodes a putative YABBY protein, which is a plant-specific transcription factor; and ORF4 encodes a putative retrotransposon protein (Fig. 2A; Supplemental Table S3). On the basis of a reverse transcription quantitative PCR (RT-qPCR) analysis, ORF1, ORF2, and ORF3 were expressed in young rice panicles, whereas ORF4 was not (Supplemental Fig. S3). Sequence comparison of these four ORFs between C-bao and NIL-GFR1Ludao revealed three single-nucleotide polymorphisms (SNPs) situated in the exon of ORF1 (LOC_Os10g36400; Fig. 2A), and no differences in the sequences of the other three ORFs were detected. Furthermore, the expression of only ORF1 differed between C-bao and NIL-GFR1Ludao (Supplemental Fig. S3). Therefore, ORF1 was most likely the candidate gene that affected the grain-filling rate.
Figure 2.
Map-based cloning and identification of GFR1. A, GFR1 was finely mapped onto the long arm of chromosome 10. Molecular markers and numbers of recombinants are labeled above and below the filled bars, respectively. The candidate ORF1 (LOC_Os10g36400) is marked in red. The mutation sites of ORF1 are also shown. ATG and TGA represent the start and stop codons, respectively. B, Complementation identification of GFR1: morphological changes of brown rice grains of C-bao and C16 of the GFR1C-bao line (T3 transgenic plants) at five stages. Bar = 1 cm. C, Overexpression identification of GFR1: morphological changes of brown rice grains of Nipponbare and OE-GFR1Nipponbare (T3 transgenic plants) at five stages. Bar = 1 cm. D, Grain-filling rate changes in C-bao and GFR1C-bao. E, Grain-filling rate changes in Nipponbare and OE-28 of the OE-GFR1Nipponbare line. F, Brown rice weight changes of C-bao and GFR1C-bao. G, Brown rice weight changes of Nipponbare and OE-GFR1Nipponbare. H, Grain size of C-bao and GFR1C-bao. I, Grain size of Nipponbare and OE-GFR1Nipponbare. J, Initial Rubisco activity of C-bao and GFR1C-bao. K, Initial Rubisco activity of Nipponbare and OE-GFR1Nipponbare. The data are means ± sd (n = 3). Student’s t test was used for statistical analysis (*, P ≤ 0.05 and **, P ≤ 0.01).
Confirmation of LOC_Os10g36400 as GFR1
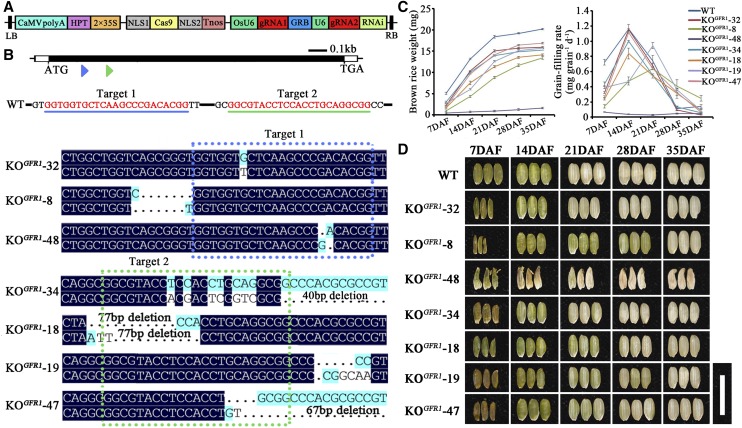
Among the 20 lines of transgenic plants (T3) carrying the full-length cDNA of LOC_Os10g36400 driven by its native promoter (1.969 kb upstream from ATG), 17 lines fully complemented the slow grain-filling rate phenotype of C-bao (GFR1C-bao; Fig. 2, B, D, and F; Supplemental Table S4). We also overexpressed OE-GFR1Ludao in a japonica variety, Nipponbare; the transgenic plants consequently exhibited a relatively fast grain-filling rate in 23 of 28 lines at 14 DAF (Supplemental Table S5). Compared with that of the control Nipponbare plants, the grain-filling rate of the transgenic plants (T3; OE-GFR1Nipponbare) increased (Fig. 2, C, E, and G; Supplemental Table S5). With respect to grain size, the GL of GFR1C-bao transgenic plants was greater than that of C-bao (Fig. 2H). In addition, the GL and GW of the transgenic plants (T3; OE-GFR1Nipponbare) were both greater than those of the Nipponbare plants (Fig. 2I). Furthermore, seven knockout (KO) mutant plants were obtained using the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-association 9 (Cas9 gene-editing technology to knock out GFR1 in the Nipponbare wild-type plants (Fig. 3, A and B). Compared with the wild-type Nipponbare plants, all seven mutants exhibited significantly lower grain-filling rates, especially during the early grain-filling stages, which resulted in significant reductions in brown rice weight at the maturity stage. Among the seven mutants, KOGFR1-48 exhibited abnormal grain filling and could not produce plump grains. KOGFR1-32 and KOGFR1-34, which had a mutation in one allele, presented higher grain-filling rates during the early stages than did the other mutants (Fig. 3, B–D). These results demonstrate that LOC_Os10g36400 of Ludao functions to improve the grain-filling rate. Therefore, ORF1 is the target gene GFR1 that regulates the grain-filling rate, and GFR1 was found to be a novel gene that controls the grain-filling rate at 7 and 14 DAF. It is better to use C-bao and NIL-GFR1Ludao along with Nipponbare for genetic modifications (overexpression and KO) comparisons. However, we failed to genetically transform NIL-GFR1Ludao. Nipponbare and Zhonghua 11 are some of the easiest japonica rice cultivars to use for genetic transformation among all rice varieties. Nipponbare has the same allele with NIL-GFR1Ludao on the GFR1 locus, so we chose the Nipponbare for genetic modification comparisons.
Figure 3.
Knocking out GFR1 in Nipponbare decreases the grain-filling rate and grain weight. A, T-DNA expression construct for Cas9 and gRNA for the GFR1 gene in rice. The Cas9 nuclease is under the control of the doubled 35S promoter, and transcription was terminated by the nos terminator. The rice U6 promoter (OsU6) was used to drive the expression of the single guide RNAs (sgRNAs). LB, Left border; RB, right border. B, Sequence comparisons of the targeting site of the KO plants generated by CRISPR-Cas9 technology. The two sgRNA target sequences are underlined in blue and green. WT, Wild type. C, Comparison of the brown rice weight and grain-filling rate changes of Nipponbare and KO plants at five stages. The data are means ± sd (n = 3). D, Morphological changes in the brown rice grain of Nipponbare and KO plants at the five grain-filling stages. Bar = 1 cm.
GFR1 Is Constitutively Expressed
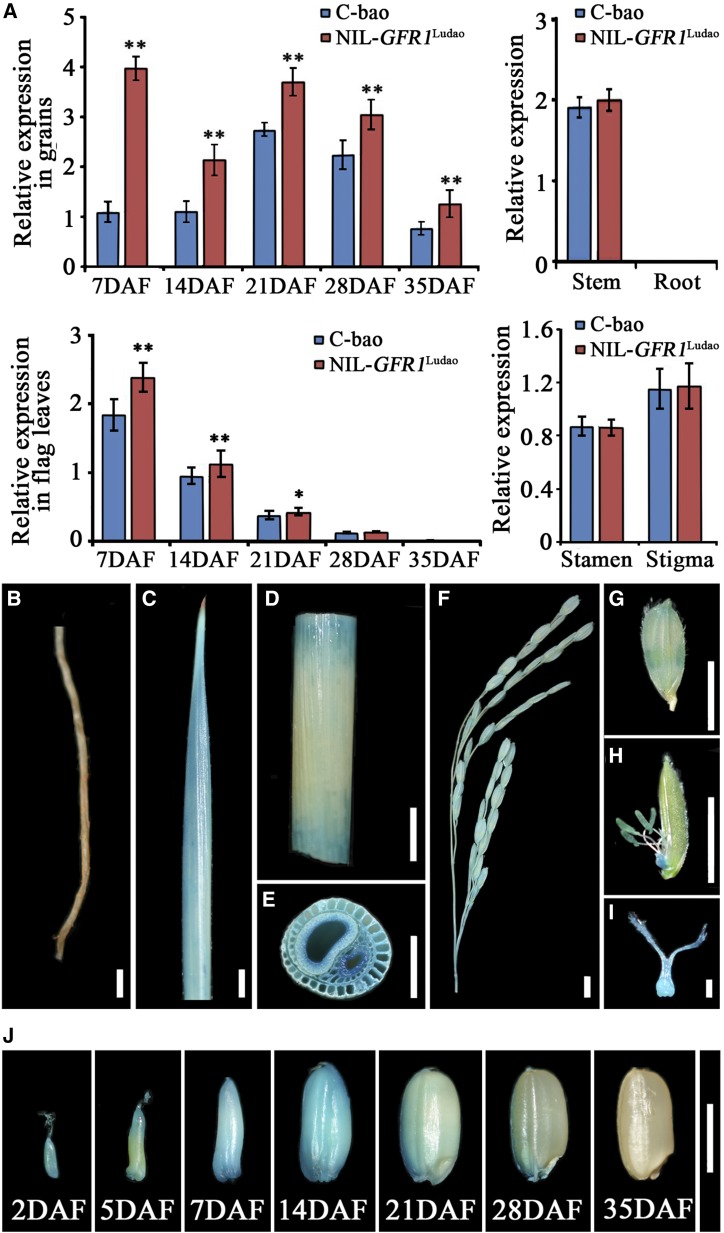
The spatial and temporal expression of GFR1 was investigated to study how it functions at the molecular level. GFR1 transcripts were detected in nearly all organs investigated except the roots via RT-qPCR (Fig. 4A). Notably, GFR1 was expressed more in the endosperm and flag leaves than in the other organs during the early grain-filling stage, which indicated that the biological function of GFR1 mainly involved the grain-filling rate (Fig. 4A). Consistent with the RT-qPCR results, GUS activity was detected predominantly in the vascular bundles of the stems, flag leaves, young panicles, spikelets, stamens, pistils, and endosperm at different DAF (Fig. 4, B–J). Together, these results indicate that GFR1 is constitutively expressed. Among the tissues in which GFR1 is expressed, the flag leaf, vascular bundles of stems, and endosperm are related to the synthesis, transport, and accumulation of carbon assimilates, which indicates that GFR1 has important biological significance in the process of carbon assimilation. To investigate the biological effects of the constitutive expression pattern of GFR1 on other agronomic traits, PH, PL, GL, GW, GT, TGW, and SS were measured in seven KO plants. The results revealed that, compared with that of wild-type Nipponbare plants, the height of all KO plants was significantly reduced (Supplemental Fig. S4, A–H). Moreover, compared with that of Nipponbare plants, the grain size of KO plants changed significantly (Supplemental Fig. S4I). The PLs of KOGFR1-19 and KOGFR1-34 were significantly smaller than those of Nipponbare (Supplemental Fig. S4J). Compared with the Nipponbare plants, three KO plants showed significantly larger GLs (Supplemental Fig. S4K), and most KO plants presented smaller GW and GT values (Supplemental Fig. S4, L and M). A relatively low grain-filling rate and small grain size led to a relatively low TGW for all KO plants (Supplemental Fig. S4N). Moreover, the SS of five KO plants was lower than that of the Nipponbare plants, especially KOGFR1-48, which reached only 65.5% (Supplemental Fig. S4O). Therefore, GFR1 may also play roles in the regulation of PH, PL, GL, GW, GT, TGW, and SS in addition to important roles in the grain-filling rate in rice. However, the underlying molecular mechanisms of these traits need to be further investigated.
Figure 4.
Analysis of the expression of GFR1. A, Relative expression levels of GFR1 are shown in the tissues of C-bao and NIL-GFR1Ludao, including grains and flag leaves at five grain-filling stages, and in stems, roots, stamens, and stigmas. The 18S rRNA gene was used as an internal control. The data are means ± sd (n = 3). Student’s t test was used for statistical analysis (*, P ≤ 0.05 and **, P ≤ 0.01). B to J, Promoter activity analysis of GFR1 in NIL-GFR1Ludao. B, Root. Bar = 1 cm. C, Center of flag leaf. Bar = 1 cm. D, Stem. Bar = 1 cm. E, Transverse section of a stem. Bar = 1 cm. F, Young panicle. Bar = 1 cm. G, Spikelet. Bar = 5 mm. H, Stamen. Bar = 5 mm. I, Pistil. Bar = 1 mm. J, Brown rice grains at different DAF. Bar = 5 mm.
GFR1 Interacts with the Rice Rubisco Small Subunit Protein
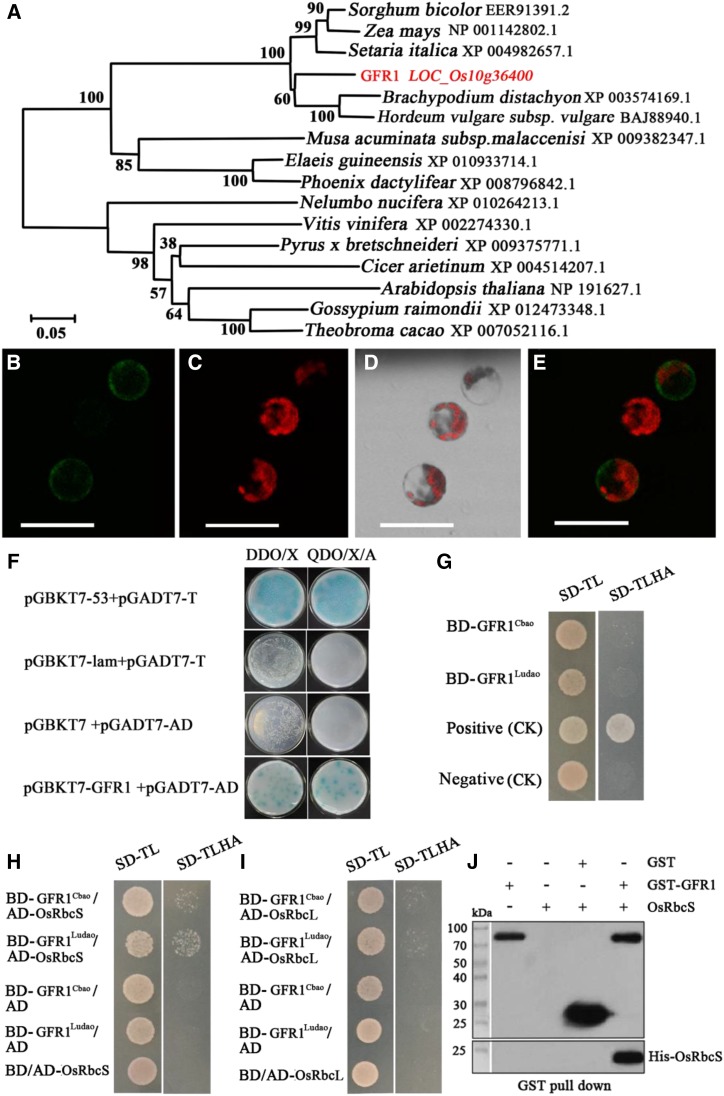
Phylogenetic analysis revealed that a homologous protein of GFR1 was detected in brome grass (Brachypodium distachyon), barley (Hordeum vulgare), sorghum (Sorghum bicolor), and maize, among other species (Fig. 5A). Subcellular localization revealed that GFR1 protein was located in the cell membrane (Fig. 5, B–E). The transcriptional activity of GFR1 was analyzed via a yeast two-hybrid (Y2H) system. No transcriptional activation was observed with the full-length GFR1 gene (Fig. 5G). Another Y2H assay was performed by selecting a cDNA library to search for the interaction protein targets of GFR1. Positive clones were sequenced after cotransformation verification (Fig. 5F). Eight possible interaction protein targets were detected (Supplemental Table S6). Among them, the proteins encoded by LOC_Os12g19381 and LOC_Os10g21268 were the most likely interaction protein targets of GFR1. LOC_Os12g19381 and LOC_Os10g21268 are related to ribulose bisphosphate carboxylase small chain precursor (OsRbcS) and ribulose bisphosphate carboxylase large chain precursor (OsRbcL), respectively. Ribulose bisphosphate carboxylase is an important carboxylase in the C3 pathway (Calvin cycle) and determines the rate of carbon assimilation during photosynthesis. The interaction between BD-GFR1Ludao and AD-OsRbcS is stronger than that between BD-GFR1C-bao and AD-OsRbcS (Fig. 5H). However, no interactions were found between BD-GFR1C-bao and AD-OsRbcL, or between BD-GFR1Ludao and AD-OsRbcL (Fig. 5I). Pull-down experiments revealed that GFR1 binds OsRbcS in vitro (Fig. 5J). These results support the hypothesis that the GFR1 gene product possibly participates in the Calvin cycle.
Figure 5.
Characterization of the GFR1 protein in rice. A, Phylogenetic tree of GFR1 in Arabidopsis, rice, and other monocotyledonous species. The phylogenetic tree was constructed using MEGA 5.0. Bootstrap values are shown as percentages at the nodes. The scale bar at the bottom represents the genetic distance. B to E, Subcellular localization of the GFR1-GFP fusion protein. Membrane localization of the GFR1-GFP fusion protein (B), the location of the chloroplast (C), a merged image of the bright field and the chloroplast (D), and a merged image (E) are shown. Bars = 50 μm. F, Y2H assay showing that GFR1 interacts with proteins of the cDNA library. DDO, Control medium (SD/-Trp-Leu); QDO, selective medium (SD/-Ade/-His/-Leu/-Trp/Aba). G, Transactivation activity assays of GFR1. H, Y2H assay showing that GFR1 of C-bao and Ludao interacts with OsRbcS. I, Y2H assay showing that GFR1 of C-bao and Ludao interacts with OsRbcL. AD, Activation domain; BD, binding domain; SD, synthetic dropout. J, Glutathione S-transferase (GST) pull-down assay of GFR1 and OsRbcS. GFR1 expressed as part of a GST fusion protein was pulled down by His-OsRbcS.
Biological Roles of GFR1
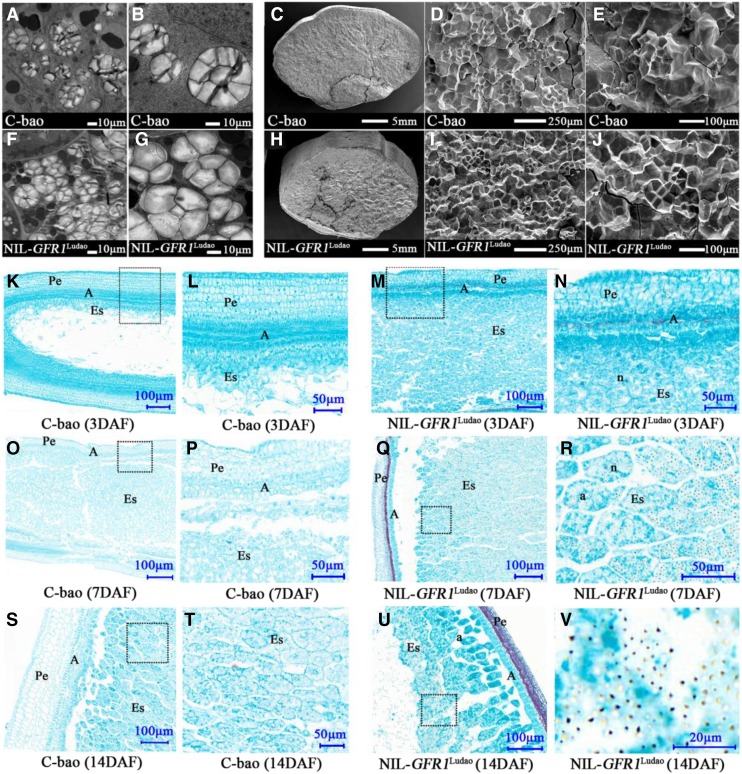
The starch granule density in the endosperm was greater in NIL-GFR1Ludao than in C-bao during the early (14 DAF) and late (35 DAF) grain-filling stages (Fig. 6, A–J). Paraffin sections showed that GFR1Ludao promotes the speed of endosperm cell division during the early grain-filling stages, including 3, 7, and 14 DAF (Fig. 6, K–V). Moreover, the net photosynthetic rate of NIL-GFR1Ludao was greater than that of C-bao at 7, 14, and 21 DAF, and there were no significant differences during other periods (Supplemental Fig. S5A). The contents of chlorophyll a and chlorophyll b in the flag leaves of C-bao were significantly greater than those of NIL-GFR1Ludao (Supplemental Fig. S5B). In addition, the initial Rubisco activity in the flag leaves of NIL-GFR1Ludao was significantly greater than that of C-bao at both 7 and 14 DAF (Supplemental Fig. S5C). With respect to the transgenic plants, the Rubisco initial activity in the flag leaves of GFR1C-bao was significantly greater than that of C-bao at both 7 and 14 DAF (Fig. 2J). In addition, the Rubisco initial activity in the flag leaves of OE-GFR1Nipponbare was significantly higher than that in Nipponbare at 14 DAF (Fig. 2K). The Suc, Glc, and Fru contents in the flag leaves and endosperm of NIL-GFR1Ludao were greater than those of C-bao during five grain-filling stages (Supplemental Fig. S5, D–F). In the flag leaves, the contents of Suc, Glc, and Fru increased during the early stages but then decreased during the late stages. The contents increased faster in NIL-GFR1Ludao than in C-bao during the early stages (Supplemental Fig. S5, D–F). In the endosperm, the contents of Suc, Glc, and Fru decreased at each stage in NIL-GFR1Ludao and C-bao. Moreover, the contents decreased faster in NIL-GFR1Ludao than in C-bao (Supplemental Fig. S5, G–I). These results indicate that GFR1Ludao can increase the Rubisco initial activity in the Calvin cycle.
Figure 6.
Microscopy assay of the endosperm between C-bao and NIL-GFR1Ludao. A and B, Transmission electron microscopy images of the central part of endosperm cells in C-bao. C to E, Scanning electron microscopy images of cross sections of seeds of C-bao. F and G, Transmission electron microscopy images of the central part of endosperm cells in NIL-GFR1Ludao. H to J, Scanning electron microscopy images of cross sections of seeds of NIL-GFR1Ludao. K to V, Paraffin sections of grains of C-bao (K, L, O, P, S, and T) and NIL-GFR1Ludao (M, N, Q, R, U, and V) at 3, 7, and 14 DAF, respectively. L, P, T, N, R, and V show magnifications of the dashed boxes in K, O, S, M, Q, and U, respectively. a, Amyloplast; A, Aleurone layer; Es, Endosperm; n, nucleus; Pe, pericarp.
Genes Regulated by GFR1
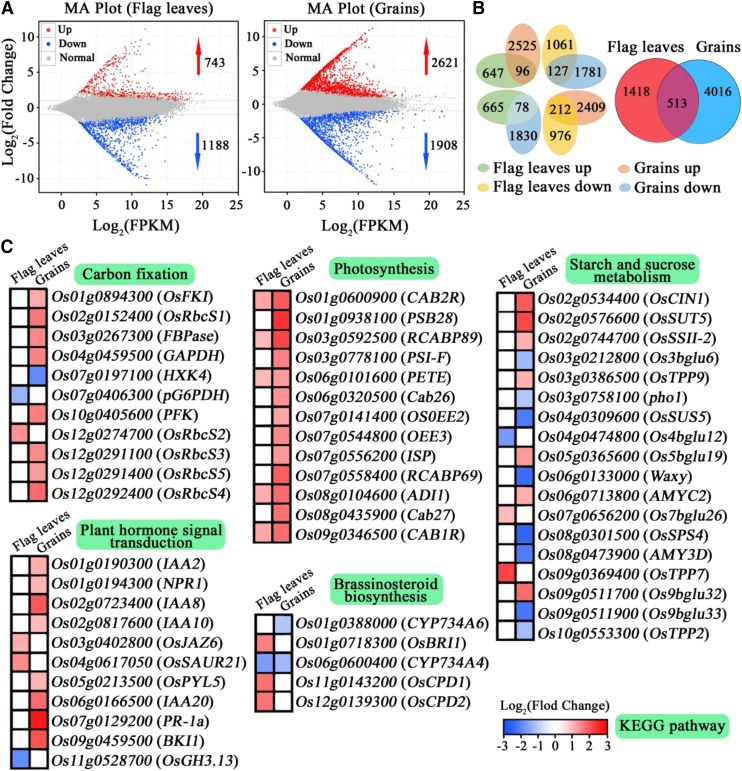
Through the RNA sequencing of flag leaves and grains at 14 DAF in C-bao and NIL-GFR1Ludao, a total of 355.6 million reads were generated. After filtering out low-quality reads, more than 353.1 million pair-end reads with Q30 > 92.64%, which indicated raw data of high quality, were obtained (Supplemental Table S7). A total of 1,931 and 4,529 differentially expressed genes (DEGs) were found in flag leaves and grains at 14 DAF between C-bao and NIL-GFR1Ludao, respectively (Fig. 7, A and B). Among these genes, 743 were up-regulated and 1,188 were down-regulated in flag leaves of NIL-GFR1Ludao. And 4,529 DEGs, including 2,621 up-regulated genes and 1,908 down-regulated genes, were found in the grains at 14 DAF (Fig. 7, A and B). These DEGs were involved in a wide spectrum of biological processes and metabolic pathways (Supplemental Figs. S6 and S7). We mainly focused on the DEGs possibly involved in the pathway of grain-filling rate according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Fig. 7C; Supplemental Table S8). These DEGs were involved in pathways including carbon fixation, photosynthesis, starch and Suc metabolism, and plant hormone signal transduction, which play important roles in grain-filling rate (Fig. 7C). Rice has five OsRbcS genes (OsRbcS1–OsRbcS5) and one OsRbcL gene (Suzuki et al., 2007). Among the 11 DEGs in the carbon fixation pathway, OsRbcS2 was up-regulated in flag leaves of NIL-GFR1Ludao, and OsRbcS1, OsRbcS3, OsRbcS4, and OsRbcS5 were up-regulated in grains of NIL-GFR1Ludao (Fig. 7C). Among the 18 DEGs in the starch and Suc metabolism pathway, the cell wall invertase gene OsCIN1, which plays important roles in regulating the unloading of assimilates, was up-regulated in grains of NIL-GFR1Ludao (Fig. 7C). Two important genes, OsSUT5 and OsSSII-2, were also up-regulated in grains of NIL-GFR1Ludao (Fig. 7C). A total of 14 DEGs involved in the photosynthesis pathway showed increased expression in flag leaves and grains of NIL-GFR1Ludao (Fig. 7C). Among the genes in plant hormone signal transduction and brassinosteroid biosynthesis pathways, Brassinosteroid Insensitive1, Constitutive Photomorphogenesis and Dwarf1 (OsCPD1), OsCPD2 and Jasmonate ZIM Domain-Containing Protein6 were up-regulated in flag leaves of NIL-GFR1Ludao (Fig. 7C). Indoleacetic Acid-Induced2 (IAA2), IAA10, and IAA20 were highly expressed in grains of NIL-GFR1Ludao (Fig. 7C).
Figure 7.
Transcriptome profiling in flag leaves and grains at 14 DAF of C-bao and NIL-GFR1Ludao. A, MA plot of DEGs in flag leaves and grains at 14 DAF of C-bao and NIL-GFR1Ludao. The number next to the arrow indicates the number of DEGs. FPKM, Fragments per kilobase of exon model per million mapped fragments. MA, M versus A. M is minus and A is add. B, Overview of DEGs in flag leaves and grains at 14 DAF of C-bao and NIL-GFR1Ludao. C, DEGs related to the GFR according to the KEGG pathway. The color in each cell indicates the value of the log2 fold change (NIL-GFR1Ludao/C-bao).
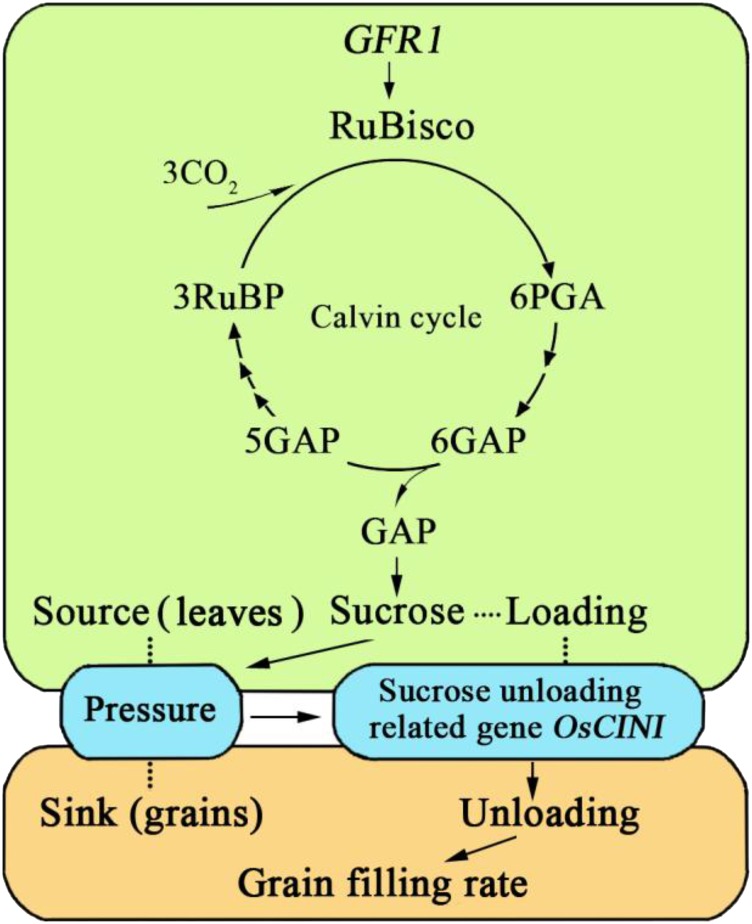
To validate the expression levels and further detect other genes regulated by GFR1, the expression levels of 19 grain-filling rate-related genes were compared between C-bao and NIL-GFR1Ludao. We found that the relative expression levels of OsCINI were greater in the young panicles of NIL-GFR1Ludao than in those of C-bao (Supplemental Fig. S8, A and B). With the exception of that of OsSSI and OsSSIIa, the expression of other related genes did not significantly differ between C-bao and NIL-GFR1Ludao (Supplemental Fig. S8, A–C). Further study indicated that the relative expression levels of OsCINI in the endosperm were greater in NIL-GFR1Ludao than in C-bao during early grain-filling stages, particularly at 7 and 14 DAF (Supplemental Fig. S8D). GST pull-down assays revealed no direct interaction between GFR1 and OsCIN1. We found that the relative expression levels of OsRbcS2, OsRbcS3, and OsRbcL were greater in the flag leaves of NIL-GFR1Ludao than in those of C-bao (Supplemental Fig. S5E). OsRbcS1, OsRbcS3, OsRbcS4, and OsRbcS5 showed high expression in the grains of NIL-GFR1Ludao, which showed consistent results with transcriptome profiling (Supplemental Fig. S5F). Considering the results of biological and expression analyses, we speculated that GFR1Ludao can increase the Rubisco initial activity and the expression of Rubisco genes in the Calvin cycle, which in turn promotes the synthesis of Suc. The greater pressure between the source and sink caused by the highly efficient and faster synthesis of Suc promotes the expression of the Suc unloading-related gene OsCINI, which in turn drives the assimilate synthesis rate at the source. GFR1Ludao promotes the expression of OsCINI, and the increased expression of OsCINI guarantees the smooth transport of Suc from the source to the sink (Fig. 8). Therefore, we speculated that GFR1 regulates the grain-filling rate by affecting Rubisco activity and the expression of OsCINI in rice.
Figure 8.
Hypothetical model of the function of GFR1 in terms of the grain-filling rate. PGA, 3-phosphoglycerate; GAP, glyceralgehyde-3-phosphate; RuBP, ribulose-1,5-bisphosphate.
Genetic Diversity Analysis of GFR1 Alleles among Rice Germplasm
By sequencing 117 accessions with an abundant diversity of grain-filling rate (Supplemental Table S9), we detected four SNPs and two InDels among the 117 rice accessions (Supplemental Table S10). The 117 accessions were divided into three subpopulations by a Bayesian model-based simulation of the population structure (Pritchard et al., 2000), and a neighbor-joining tree was constructed via MEGA 5.0 (Tamura et al., 2011) on the basis of the SNP genotype of GFR1 (Supplemental Figs. S9A and S10A). The average air temperatures at the Jiangpu Experiment Station throughout the grain-filling periods in 2014 and 2015 are shown in Supplemental Figure S9C. These data indicated that the grain filling of all rice plants proceeded under normal climatic conditions during both years, and the differences in the grain-filling rate among the accessions during the five stages were caused by genotypic differences (Supplemental Table S11). The results of a haplotype association analysis indicated that InDel1 and SNP1 were significantly associated with the grain-filling rate at 7, 14, and 21 DAF in both 2014 and 2015 (Supplemental Table S12). These results indicated that InDel1 and SNP1 are two functional mutations associated with grain-filling rate. Therefore, the effect of ORF1 on the grain-filling rate of a NIL plant is mainly due to the InDel1 (CGG deletion in the NIL plant). The SNP1 (G/A), detected only in 30 of 117 accessions, also affects grain-filling rate in addition to InDel1. Theoretically, six allele haplotypes exist for all InDel1 and SNP1 combinations; however, allele haplotypes A5 and A6 were not detected among the accessions investigated (Supplemental Fig. S9D). Among the 117 rice accessions, which included 102 japonica rice and 15 indica rice accessions, the A2 allele haplotype and the A3 allele haplotype were mainly detected in indica rice accessions growing in tropical and subtropical areas, whereas the A4 allele haplotype was detected mainly in japonica rice accessions growing in temperate areas (Supplemental Figs. S9E and S10B). Analysis of the phenotypic effects of four allele haplotypes revealed that the A2 allele haplotype had the largest positive phenotypic effect on the grain-filling rate at 7 and 14 DAF (Supplemental Table S13). Therefore, the A2 haplotype is an elite allele haplotype found mainly in indica rice. The four allele haplotypes (A1–A4) exerted different effects in terms of increasing Rubisco initial activity during the early stages, including at 7 and 14 DAF (Supplemental Fig. S9F). At 7 DAF, the A4 allele haplotype had the largest effect on improving Rubisco initial activity (Supplemental Fig. S9F). In addition, the A2 allele haplotype had the largest effect on improving Rubisco initial activity at 14 DAF (Supplemental Fig. S9F). The plant height of the 117 rice accessions was also measured (Supplemental Table S9), but no relationship was detected between plant height and grain-filling rate. Therefore, no potential linkage of GFR1 and the QTL qPH10 in the NIL plants was detected in the natural population.
Elite Alleles of GFR1 Significantly Enhance Rice Grain Yields
To explore the application effects of NIL-GFR1Ludao on enhancing the grain-filling rate of hybrid japonica rice, we crossed C-bao and NIL-GFR1Ludao (both as restorers) with two commercial cytoplasmic male-sterile lines, Xu2A (containing the A1 allele haplotype) and 9522A (containing the A4 allele haplotype), to generate F1 seeds of japonica rice. Compared with Xu2A/C-bao and 9522A/C-bao plants, the F1 plants of Xu2A/NIL-GFR1Ludao and 9522A/NIL-GFR1Ludao exhibited significantly faster grain-filling rates (Supplemental Fig. S11, A–D), resulting in grain yield increases of 15.97% and 22.34% in field trials, respectively (Supplemental Fig. S11, E–H). The functional marker B designed for GFR1 was then used to discover superior varieties that harbor elite alleles (Supplemental Fig. S10D; Supplemental Table S14). The results of 20 hybrid rice cultivars, including 10 hybrid indica rice and 10 hybrid japonica rice combinations (Supplemental Table S15), grown in the field at Tianjin Tianlong Seed Co., showed that the G1 genotype appeared only in indica hybrids, while genotypes G2 and G3 existed mainly in japonica hybrids (Supplemental Fig. S11, I and J). The results concerning grain-filling rate changes during five grain-filling stages showed that G1 (homozygous A2 allele haplotype) presents the greatest extent of grain filling at 7 and 14 DAF and that G2 and G3 exhibit poor grain filling during the early stages (Supplemental Fig. S11K). These results might explain why the grain filling of hybrid indica rice is better than that of hybrid japonica rice. Currently, the A2 and A4 elite allele haplotypes of GFR1 are not widely used in the production of hybrid japonica rice. Therefore, it is promising that these GFR1 elite allele haplotypes could be used for improving the grain-filling rate of hybrid japonica rice, and the functional marker B developed in this study could be used to discover superior varieties that harbor elite alleles (Supplemental Fig. S10D; Supplemental Table S14).
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The rice (Oryza sativa) materials used in this study included two parents and four populations. A BIL population (CLC-BIL, BC1F6) consisting of 102 lines was constructed using the parents C-bao (recurrent parent) and Ludao (donor parent) for primary mapping of GFR1. A NIL (named NIL-GFR1Ludao) was derived from a set of 56 chromosomal segment substitution lines (Supplemental Fig. S1). The F2 population, which comprised 9,280 plants, derived from a cross between NIL-GFR1Ludao and C-bao, was used for the fine-mapping and positional cloning of GFR1. Two cytoplasmic male-sterile lines, 9522A and Xu2A, were crossed with C-bao and NIL-GFR1Ludao, respectively, to produce four F1 combinations for grain yield trials. A set of 117 rice accessions was used for sequence analysis of the GFR1 gene locus. All plants were grown in a paddy field from May to October at the Jiangpu Experiment Station (32°0404′N, 118°6375′E), Nanjing Agricultural University, Nanjing, China. For the parents, BIL population, the natural population, and F1 populations, the field trials were arranged in a completely randomized block design, with three replications.
Investigation of the Grain-Filling Rate and GP
Fifty flowers on each plant bloomed on the same day, and five plants from each plot (for the F2 population, five panicles per plant) were marked with a black pen on the glume surface. Seven days after marking, the marked fresh grains of one plant were harvested and dried in an oven at 105°C to a constant weight. The dried grains were then hulled by hand, and 20 randomly selected grains of brown rice were weighed on a balance with a precision of up to 0.001 g and averaged. Similarly, the fresh grains at 7, 14, 21, 28, and 35 d after marking were harvested, dried, hulled, and weighed. The grain-filling rate at each stage was subsequently calculated as follows: GFRi = (GWi − GWi-1)/7, where GFRi is the grain-filling rate (mg grain−1 d−1) and GWi is the dried grain weight (mg grain−1) at stage i (i = 1,2,3,4,5).
The measurement of GP was based on the method described by Liu et al. (2017).
QTL Mapping of GFR1
A genetic marker map for the CLC-BIL population was constructed via 114 simple sequence repeat markers using MapMaker3.0/EXP version 3.0 (Lander et al., 1987). QTL analysis was performed by QTLMapper 2.0 in conjunction with the time-course QTL mapping method and conditional QTL method by adjusting the grain weight to identical levels (Wang et al., 1999). For fine-mapping, 1,614 plants with a fast grain-filling rate isolated from the single-locus segregating F2 population were used.
Gene Cloning, Vector Construction, and Plant Transformation
For the genetic complementation test, the 4.34-kb genomic DNA fragment containing GFR1Ludao was amplified using Q5 High-Fidelity DNA Polymerase (New England Biolabs) and then cloned into a pBWA(V)H binary vector (BioRun) using an In-Fusion Advantage PCR Cloning Kit (Clontech). The resultant plasmid was transformed into Agrobacterium tumefaciens strain EHA105, which was then introduced into C-bao via A. tumefaciens-mediated transformation. The full-length gene controlled by the Cauliflower mosaic virus 35S promoter was cloned into a pBWA(V)HS plant binary vector to generate OE-GFR1Nipponbare, which was subsequently introduced into Nipponbare by A. tumefaciens-mediated transformation as reported previously (Toki et al., 2006). The 35S promoter was also cloned into pBWA(V)HG to create pGFR1::GUS. The resultant plasmid was transformed into A. tumefaciens strain EHA105, which was then introduced into Nipponbare. At least 20 transgenic events were produced for each construct.
CRISPR/Cas9 Targeting of GFR1
For the CRISPR/Cas9 assay, 20-bp target sequences with NGG (PAM) at the 3ʹ end were selected using the CRISPR sgRNA design online tool (http://crispr.mit.edu) for GFR1. The designed sequences were BLASTed against the nucleotide collection of rice at the National Center for Biotechnology Information for putative off-targets. Then two target sequences (GGTGGTGCTCAAGCCCGACACGG and GGCGTACCTCCACCTGCAGGCGG) were selected to construct the CRISPR/Cas9 vector. The two synthesized target sgRNA sequences were inserted between the OsU6 promoter and the sgRNA scaffold by PCR. The resulting vector pCas9-GFR1 was sequence verified to confirm the presence of all elements. Then the resultant pCas9-GFR1 construct was subsequently introduced into A. tumefaciens and used to infect Nipponbare calli.
Subcellular Localization of the GFR1 Protein
The GFP coding region was fused in frame into the GFR1 C terminus under the control of the Cauliflower mosaic virus 35S promoter. The fusion construct was transformed into rice leaf protoplasts via polyethylene glycol. The cells were then examined with a confocal fluorescence microscope, and the fluorescence was observed using a Leica TCS-SP4 confocal microscope.
Microscopy and GUS Staining
Grains of C-bao and NIL-GFR1Ludao were harvested at 35 DAF and dried in an oven at 40°C to a constant weight. The samples were then fixed in 2% (w/v) OsO4 for 2 h, dehydrated via an ethanol gradient, infiltrated, and subsequently embedded in butyl methyl methacrylate. The samples were subjected to critical point drying, sputter coated with platinum, and then observed with a scanning electron microscope (Hitachi S-3000N) at an accelerating voltage of 15 kV. Grains of C-bao and NIL-GFR1Ludao harvested at 14 DAF were fixed with 2.5% (v/v) glutaraldehyde at 4°C. The samples were processed according to the method described by Xiao et al. (2015). Afterward, the samples were observed and imaged via a transmission electron microscope (Hitachi H-7650). The grains of C-bao and NIL-GFR1Ludao at 3, 7, and 14 DAF were fixed with formalin-acetic acid-alcohol fixative. Paraffin sections were created in accordance with the method described by Fang et al. (2016), and GUS staining was performed as described by Zhou et al. (2011). Various tissues or hand-cut sections of pGFR1::GUS T2 generation transgenic plants were incubated in GUS staining solution. Images were captured directly or with a stereomicroscope.
Construction of a Rice Endosperm cDNA-AD Library and Yeast Assays
A cDNA-AD library of rice endosperm was generated by Genecreate Company. A two-hybrid system was used to test transcriptional activation assays of GFR1. GFR1 was cloned into pGBKT7, yielding a pGBKT7-GFR1 construct (BD-GFR1). BD-GFR1 was used to search the cDNA-AD library. A hybridization solution of cDNA and the BD was assayed on selective medium (QDO/A). Y2H Gold (pGBKT7-53) and Y187 (pGADT7-T) were cotransfected as positive controls, and Y2H Gold (pGBKT7-lam) and Y187 (pGADT7-T) were cotransfected as negative controls. The positive single clones were sequenced. Those proteins without a shift code were selected as candidate interaction proteins. The activation ability was assayed on selective medium (QDO plus X-α-Gal) plates, and interactions were assayed on selective medium (QDO) plates.
GST Pull-Down Assays
GFR1 was cloned into a pGEX4T-1 vector to construct GST-GFR1 fusion proteins, and OsRbcS and OsCINI were cloned into a pCZNI vector to form His-OsRbcS and His-OsCINI fusion proteins. Pull-down assays were performed as described by Zheng et al. (2015). Western blots were performed with anti-His antibodies at 1:1,000 dilution, and anti-mouse secondary antibodies (1:5,000 dilution) were also used. The western blots were incubated using enhanced chemiluminescence reagent (Bio-Rad).
RNA Sequencing and Data Analysis
The flag leaves and grains of C-bao and NIL-GFR1Ludao at the 14-DAF stage were collected for RNA extraction. Construction of the cDNA library and sequencing were performed (Genepioneer) using the Illumina (HiSeq/MiSeq) system. RNA sequencing clean reads of three biological replicates were mapped to the Oryza sativa ssp. japonica reference genome (Oryza_sativa.IRGSP-1.0: ftp://ftp.ensemblgenomes.org/pub/plants/release-44/fasta/oryza_sativa/dna/) after the adaptor and low-quality nucleotides were removed by TopHat2 (Kim et al., 2013). Cufflinks was then used in conjunction with the TopHat-generated alignment to assemble a set of reference-based transcripts (Trapnell et al., 2010). The expression values were calculated in terms of fragments per kilobase of exon model per million mapped fragments, and the DEGs were further analyzed by DESeq with log2(fold change) ≥ 1 and a false discovery rate < 0.05 (Anders and Huber, 2010). In addition, both Gene Ontology functional annotation (Ashburner et al., 2000) and KEGG pathway enrichment analyses (Kanehisa et al., 2004) were conducted.
RT-qPCR Analysis
Total RNA was extracted from various plant tissues using an RNApure Plant Kit (CWBIO). RNA was reverse transcribed with the 1st Strand cDNA Synthesis Kit (Vazyme) following the manufacturer’s protocol. Then the qPCR was carried out on the Roche Applied Science LightCycler480 with SYBR Green Master Mix (Vazyme) according to the operation manual. The rice 18S rRNA gene was used as an internal control. The relative quantification of the transcript levels was performed using the comparative threshold cycle method (Livak and Schmittgen, 2001).
Photosynthesis Rate, Chlorophyll Content, Sugar Content, and Rubisco Activity
The photosynthesis rate of the flag leaves was examined by using an LI-6400XT photosynthesis analyzer (LI-COR) between 9 and 11 am on sunny days during five grain-filling stages. Leaf portions measuring 6 cm2 (3 cm in length, 2 cm in width) were placed in the instrument under white light (1,200 μmol photons m−2 s−1). The chlorophyll content was measured according to the method described by Gitelson et al. (1996), and sample preparation and analysis of sugars were performed as described by Tan et al. (2011). The Rubisco activity in the flag leaves and grains during the five grain-filling stages was determined in accordance with the protocol described by Morita et al. (2014).
Genetic Diversity and Haplotype Association Analysis
We selected 117 rice accessions that exhibited an abundant diversity of grain-filling rates for sequencing a 1.413-kb genomic DNA fragment that encompasses the coding DNA sequence (CDS) of GFR1 (Supplemental Table S9). SNPs were identified by sequence alignment via DNAMAN software (Lynnon Biosoft). The model-based program STRUCTURE 2.2 (Pritchard et al., 2000) was used to determine the population structure of the 117 rice accessions. Twenty independent runs were performed for each k (from two to 10, depending on the number of rice ecotypes used) via a burn-in length of 50,000, a run length of 100,000, and a model for the admixture and independent allele frequency. The true number of populations (K) was determined when the mean log-likelihood value after 20 runs at each K value reached the highest value for the model parameter K. A neighbor-joining tree was constructed by MEGA 5.0 on the basis of the sequence of GFR1 (Tamura et al., 2011). The general linear model in TASSEL 2.1 software was used for association mapping (Bradbury et al., 2007), and the population structure (Q) was included as a covariate in the model to test for associations between grain-filling rate and SNP.
Evaluation of the Breeding Value of GFR1Ludao
To evaluate the allelic effect of GFR1Ludao, the grain-filling rate and other agronomic traits of C-bao and NIL-GFR1Ludao were compared using two pairs of rice hybrids (9522A/C-bao and 9522A/NIL-GFR1Ludao, Xu2A/C-bao and Xu2A/NIL-GFR1Ludao). A total of 400 plants for each hybrid were grown in a plot (13.6 m2), which was replicated three times. For each plot, the grain-filling rate and other agronomic traits were measured. To understand the status of the alleles of GFR1 in actual hybrid rice production, 20 cultivars that have been widely and recently grown in China were selected to identify the genotype of GFR1. All 20 cultivars were grown in a paddy field at Tianjin Tianlong Seed Co.
Accession Numbers
Accession numbers of the major genes mentioned in this article are listed in Supplemental Table S16.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Separation process of NIL-GFR1Ludao and graphical genotype of NIL-GFR1Ludao.
Supplemental Figure S2. Linkage map of grain-filling rate QTLs on chromosomes.
Supplemental Figure S3. Expression levels of four ORFs in young panicles of NILs.
Supplemental Figure S4. Agronomic trait comparisons between wild-type Nipponbare plants and KO plants.
Supplemental Figure S5. Biological roles of GFR1.
Supplemental Figure S6. Gene Ontology classification analysis.
Supplemental Figure S7. Cluster of Orthologous Groups of proteins function classification and scatterplot of KEGG pathway enrichment statistics.
Supplemental Figure S8. qPCR analyses of the expression of rice grain-filling rate-related genes in C-bao and NIL-GFR1Ludao.
Supplemental Figure S9. Analysis of the population genetic variation in GFR1.
Supplemental Figure S10. Neighbor-joining tree and analysis of the phenotypic effects of four alleles.
Supplemental Figure S11. Evaluation of GFR1 in the actual production of hybrid rice.
Supplemental Table S1. Descriptive statistics and estimates of genetic variance components of the grain-filling rate (mg grain−1 d−1) of the BIL population at five stages.
Supplemental Table S2. Time-related QTLs mapped in the CLC-BIL population.
Supplemental Table S3. Predicted genes at the GFR1 locus according to the Gramene Web site.
Supplemental Table S4. Grain-filling rates at five filling stages and brown rice weight of complementation transgenic plants (T3).
Supplemental Table S5. Grain-filling rates at five filling stages and brown rice weight of overexpression transgenic plants (T3).
Supplemental Table S6. Interaction proteins detected via bait protein.
Supplemental Table S7. The statistics of RNA sequencing data.
Supplemental Table S8. The most promising grain-filling rate-related genes regulated by GFR1 among DEGs between C-bao and NIL-GFR1Ludao in flag leaves and grains at 14 DAF.
Supplemental Table S9. Grain-filling rates of the two parents and 117 rice accessions.
Supplemental Table S10. SNPs detected in the CDS region of GFR1 among 117 rice accessions and two parents.
Supplemental Table S11. Descriptive statistics for the grain-filling rate (mg grain−1 d−1) of two parents and 117 rice accessions at five stages.
Supplemental Table S12. Haplotype association analysis of the InDels and SNPs within GFR1 in conjunction with the grain-filling rate among 117 accessions.
Supplemental Table S13. Phenotypic effects of alleles significantly associated with grain-filling rate.
Supplemental Table S14. Primer sequences designed in this study.
Supplemental Table S15. Genotypes of GFR1 among hybrid rice varieties used commercially.
Supplemental Table S16. Accession numbers for genes referred to in this study.
ACKNOWLEDGMENTS
We thank Liu Linglong and Tan Helin (Nanjing Agriculture University) for help with the molecular biology experiments and the Life Science Experimental Center of Nanjing Agriculture University for help with the microscopy.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31571743 and 31671658), the National High-Tech R&D Program of China (863 Program; grant no. 2010AA101301), the Research Fund for the Doctoral Program of Higher Education of China (grant nos. B0201100690 and B0201300662), the China Postdoctoral Science Foundation (grant no. 2016M601830), and the Chongqing Key Project of the Foundation and Frontier Research Program (cstc2015jcyjBX0008).
Articles can be viewed without a subscription.
References
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000) Gene Ontology: Tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Bai R, Jin Y (2016) Characteristics and coordination of source-sink relationships in super hybrid rice. Open Life Sci 11: 470–475 [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 [DOI] [PubMed] [Google Scholar]
- Cho JI, Lee SK, Ko S, Kim HK, Jun SH, Lee YH, Bhoo SH, Lee KW, An G, Hahn TR, et al. (2005) Molecular cloning and expression analysis of the cell-wall invertase gene family in rice (Oryza sativa L.). Plant Cell Rep 24: 225–236 [DOI] [PubMed] [Google Scholar]
- Fang X, Fu HF, Gong ZH, Chai WG (2016) Involvement of a universal amino acid synthesis impediment in cytoplasmic male sterility in pepper. Sci Rep 6: 23357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Asakura N, Ohdan T, Miyao A, Hirochika H, Nakamura Y (2006) Function and characterization of starch synthase I using mutants in rice. Plant Physiol 140: 1070–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitelson A, Merzlyak M, Lichtenthaler H (1996) Detection of red edge position and chlorophyll content by reflectance measurements near 700 nm. J Plant Physiol 148: 501–508 [Google Scholar]
- Hirose T, Scofield GN, Terao T (2008) An expression analysis profile for the entire sucrose synthase gene family in rice. Plant Sci 174: 534–543 [Google Scholar]
- Hirose T, Takano M, Terao T (2002) Cell wall invertase in developing rice caryopsis: Molecular cloning of OsCIN1 and analysis of its expression in relation to its role in grain filling. Plant Cell Physiol 43: 452–459 [DOI] [PubMed] [Google Scholar]
- Hong D, Leng Y (2004) Genetic analysis of heterosis for number of spikelets per panicle and panicle length of F1 hybrids in japonica rice hybrids. Zhongguo Shuidao Kexue 11: 255 [Google Scholar]
- Ishibashi Y, Okamura K, Miyazaki M, Phan T, Yuasa T, Iwaya-Inoue M (2014) Expression of rice sucrose transporter gene OsSUT1 in sink and source organs shaded during grain filling may affect grain yield and quality. Environ Exp Bot 97: 49–54 [Google Scholar]
- Jiang H, Dian W, Liu F, Wu P (2004) Molecular cloning and expression analysis of three genes encoding starch synthase II in rice. Planta 218: 1062–1070 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32: D277–D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Takeda K (1996) Associations among characters related to yield sink capacity in space-planted rice. Crop Sci 36: 1135–1139 [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA (1987) MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181 [DOI] [PubMed] [Google Scholar]
- Liu E, Liu X, Zeng S, Zhao K, Zhu C, Liu Y, Breria MC, Zhang B, Hong D (2015) Time-course association mapping of the grain-filling rate in rice (Oryza sativa L.). PLoS ONE 10: e0119959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E, Zeng S, Chen X, Dang X, Liang L, Wang H, Dong Z, Liu Y, Hong D (2017) Identification of putative markers linked to grain plumpness in rice (Oryza sativa L.) via association mapping. BMC Genet 18: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Molina J, Sikora M, Garud N, Flowers JM, Rubinstein S, Reynolds A, Huang P, Jackson S, Schaal BA, Bustamante CD, et al. (2011) Molecular evidence for a single evolutionary origin of domesticated rice. Proc Natl Acad Sci USA 108: 8351–8356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K, Hatanaka T, Misoo S, Fukayama H (2014) Unusual small subunit that is not expressed in photosynthetic cells alters the catalytic properties of Rubisco in rice. Plant Physiol 164: 69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallamilli BR, Zhang J, Mujahid H, Malone BM, Bridges SM, Peng Z (2013) Polycomb group gene OsFIE2 regulates rice (Oryza sativa) seed development and grain filling via a mechanism distinct from Arabidopsis. PLoS Genet 9: e1003322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T, Du Y, Zhang J, Li J, Liu Y, Zhao Y, Sun H, Zhao Q (2013) Genome-wide analysis of 24-nt siRNAs dynamic variations during rice superior and inferior grain filling. PLoS ONE 8: e61029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Hou Y, Tong X, Wang Y, Lin H, Liu Q, Zhang W, Li Z, Nallamilli BR, Zhang J (2016) Quantitative phosphoproteomic analysis of early seed development in rice (Oryza sativa L.). Plant Mol Biol 90: 249–265 [DOI] [PubMed] [Google Scholar]
- Rennie EA, Turgeon R (2009) A comprehensive picture of phloem loading strategies. Proc Natl Acad Sci USA 106: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosso D, Luo D, Li QB, Sasse J, Yang J, Gendrot G, Suzuki M, Koch KE, McCarty DR, Chourey PS, et al. (2015) Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat Genet 47: 1489–1493 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Ohkubo M, Hatakeyama H, Ohashi K, Yoshizawa R, Kojima S, Hayakawa T, Yamaya T, Mae T, Makino A (2007) Increased Rubisco content in transgenic rice transformed with the ‘sense’ rbcS gene. Plant Cell Physiol 48: 626–637 [DOI] [PubMed] [Google Scholar]
- Takai T, Fukuta Y, Shiraiwa T, Horie T (2005) Time-related mapping of quantitative trait loci controlling grain-filling in rice (Oryza sativa L.). J Exp Bot 56: 2107–2118 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H, Yang X, Zhang F, Zheng X, Qu C, Mu J, Fu F, Li J, Guan R, Zhang H, et al. (2011) Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol 156: 1577–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47: 969–976 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DL, Zhu J, Li ZKL, Paterson AH (1999) Mapping QTLs with epistatic effects and QTL×environment interactions by mixed linear model approaches. Theor Appl Genet 99: 1255–1264 [Google Scholar]
- Wang E, Wang J, Zhu X, Hao W, Wang L, Li Q, Zhang L, He W, Lu B, Lin H, et al. (2008) Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet 40: 1370–1374 [DOI] [PubMed] [Google Scholar]
- Wang JC, Xu H, Zhu Y, Liu QQ, Cai XL (2013) OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J Exp Bot 64: 3453–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Jiao G, Lin H, Sheng Z, Shao G, Xie L, Tang S, Xu Q, Hu P (2017) GRAIN INCOMPLETE FILLING 2 regulates grain filling and starch synthesis during rice caryopsis development. J Integr Plant Biol 59: 134–153 [DOI] [PubMed] [Google Scholar]
- Wu CY, Trieu A, Radhakrishnan P, Kwok SF, Harris S, Zhang K, Wang J, Wan J, Zhai H, Takatsuto S, et al. (2008) Brassinosteroids regulate grain filling in rice. Plant Cell 20: 2130–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao GQ, Zhang HW, Lu XY, Huang RF (2015) Characterization and mapping of a novel light-dependent lesion mimic mutant lmm6 in rice (Oryza sativa L.). J Integr Agric 14: 1687–1696 [Google Scholar]
- Xu CG, Li XQ, Xue Y, Huang YW, Gao J, Xing YZ (2004) Comparison of quantitative trait loci controlling seedling characteristics at two seedling stages using rice recombinant inbred lines. Theor Appl Genet 109: 640–647 [DOI] [PubMed] [Google Scholar]
- Yang J, Yunying C, Zhang H, Liu L, Zhang J (2008) Involvement of polyamines in the post-anthesis development of inferior and superior spikelets in rice. Planta 228: 137–149 [DOI] [PubMed] [Google Scholar]
- You C, Chen L, He H, Wu L, Wang S, Ding Y, Ma C (2017) iTRAQ-based proteome profile analysis of superior and inferior spikelets at early grain filling stage in japonica rice. BMC Plant Biol 17: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You C, Zhu H, Xu B, Huang W, Wang S, Ding Y, Liu Z, Li G, Chen L, Ding C, et al. (2016) Effect of removing superior spikelets on grain filling of inferior spikelets in rice. Front Plant Sci 7: 1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Jiang J, Yao J, Hong D (2009) Characterization and genetic analysis of grain filling rate of “Ludao” and restorer line C-bao in japonica rice. Zuo Wu Xue Bao 35: 1229–1235 [Google Scholar]
- Zhang W, Cao Z, Zhou Q, Chen J, Xu G, Gu J, Liu L, Wang Z, Yang J, Zhang H (2016) Grain filling characteristics and their relations with endogenous hormones in large- and small-grain mutants of rice. PLoS ONE 11: e0165321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YF, Peng T, Sun HZ, Teotia S, Wen HL, Du YX, Zhang J, Li JZ, Tang GL, Xue HW, et al. (2019) miR1432-OsACOT (acyl-CoA thioesterase) module determines grain yield via enhancing grain filling rate in rice. Plant Biotechnol J 17: 712–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Wang Y, Wang Y, Wang C, Ren Y, Lv J, Peng C, Wu T, Liu K, Zhao S, et al. (2015) DEFORMED FLORAL ORGAN1 (DFO1) regulates floral organ identity by epigenetically repressing the expression of OsMADS58 in rice (Oryza sativa). New Phytol 206: 1476–1490 [DOI] [PubMed] [Google Scholar]
- Zhou S, Wang Y, Li W, Zhao Z, Ren Y, Wang Y, Gu S, Lin Q, Wang D, Jiang L, et al. (2011) Pollen semi-sterility1 encodes a kinesin-1-like protein important for male meiosis, anther dehiscence, and fertility in rice. Plant Cell 23: 111–129 [DOI] [PMC free article] [PubMed] [Google Scholar]