Genetic mapping in Arabidopsis and analysis of sequences in the 1001 Genomes database reveal allelic variation in a chloroplast division gene is associated with natural variation in chloroplast size.
Abstract
Chloroplast size varies considerably in nature, but the underlying mechanisms are unknown. By exploiting a near-isogenic line population derived from a cross between the Arabidopsis (Arabidopsis thaliana) accessions Cape Verde Islands (Cvi-1), which has larger chloroplasts, and Landsberg erecta (Ler-0), with smaller chloroplasts, we determined that the large-chloroplast phenotype in Cvi-1 is associated with allelic variation in the gene encoding the chloroplast-division protein FtsZ2-2, a tubulin-related cytoskeletal component of the contractile FtsZ ring inside chloroplasts. Sequencing revealed that the Cvi-1 FtsZ2-2 allele encodes a C-terminally truncated protein lacking a region required for FtsZ2-2 interaction with inner-envelope proteins, and functional complementation experiments in a Columbia-0 ftsZ2-2 null mutant confirmed this allele as causal for the increased chloroplast size in Cvi-1. Comparison of FtsZ2-2 coding sequences in the 1001 Genomes database showed that the Cvi-1 allele is rare and identified additional rare loss-of-function alleles, including a natural null allele, in three other accessions, all of which had enlarged-chloroplast phenotypes. The ratio of nonsynonymous to synonymous substitutions was higher among the FtsZ2-2 genes than among the two other FtsZ family members in Arabidopsis, FtsZ2-1, a close paralog of FtsZ2-2, and the functionally distinct FtsZ1-1, indicating more relaxed constraint on the FtsZ2-2 coding sequence than on those of FtsZ2-1 or FtsZ1-1. Our results establish that allelic variation in FtsZ2-2 contributes to natural variation in chloroplast size in Arabidopsis, and they also demonstrate that natural variation in Arabidopsis can be used to decipher the genetic basis of differences in fundamental cell biological traits, such as organelle size.
During leaf growth and development, chloroplast numbers increase to maximize photosynthetic capacity (Leech and Baker, 1983). In mesophyll cells, chloroplast division takes place primarily during cell expansion and increases plastid numbers from ∼10–20 in leaf primordia to ∼100 or more in mature mesophyll cells (Possingham and Saurer, 1969; Leech and Baker, 1983; Pyke, 1997, 1999). Chloroplasts divide in the middle, producing populations of organelles that are generally similar in size and shape. However, chloroplast size in mesophyll cells can vary considerably between and within species (Honda et al., 1971; Jellings et al., 1983). While various environmental and genetic factors are known to influence numerous aspects of chloroplast morphology and function (Jarvis and Lopez-Juez, 2013; Pogson et al., 2015), the molecular determinants of natural variation in chloroplast size are unknown. However, studies in Arabidopsis (Arabidopsis thaliana) and other plant species have shown that altered expression or mutations in numerous genes important for chloroplast division often result in significant changes in chloroplast size and number (Osteryoung et al., 1998; Strepp et al., 1998; Colletti et al., 2000; Itoh et al., 2001; Vitha et al., 2003; Maple et al., 2007; Glynn et al., 2009; Schmitz et al., 2009; Zhang et al., 2009; Miyagishima et al., 2011; Osteryoung and Pyke, 2014).
The cytoskeletal filamenting temperature-sensitive Z (FtsZ) proteins are tubulin homologs that play a critical role in the division of bacteria and chloroplasts by assembling into a medial ring, called the Z ring, that helps constrict the cell or organelle (Bi and Lutkenhaus, 1991; Ma et al., 1996; Löwe and Amos, 1998; Miyagishima et al., 2001; Vitha et al., 2001; Erickson et al., 2010; Du and Lutkenhaus, 2017). While most bacteria have only a single FtsZ gene, plants possess two highly conserved nuclear FtsZ families, FtsZ1 and FtsZ2, that presumably arose by duplication of a single ancestral FtsZ gene of cyanobacterial origin (Osteryoung and Vierling, 1995; Osteryoung et al., 1998; TerBush et al., 2013; Grosche and Rensing, 2017; Chen et al., 2018). FtsZ1 and FtsZ2, which localize primarily to the stroma, copolymerize in the chloroplast Z ring, where they have nonoverlapping but complementary functions in chloroplast division (Schmitz et al., 2009; Olson et al., 2010; Yoshida et al., 2016). FtsZ2 plays a more structural role by imparting stability to the Z ring, while FtsZ1 promotes the exchange of FtsZ subunits from the ring, thereby making the Z ring more dynamic, which is critical for Z-ring constriction (Yoder et al., 2007; TerBush and Osteryoung, 2012; Yoshida et al., 2016). Like many plants, Arabidopsis has one FtsZ1 gene, called FtsZ1-1, and two FtsZ2 genes, called FtsZ2-1 and FtsZ2-2, all encoded in the nucleus and targeted to the chloroplast by cleavable transit peptides (Osteryoung and Vierling, 1995; Osteryoung et al., 1998; Fujiwara and Yoshida, 2001; McAndrew et al., 2001). Mutants null for FtsZ1-1 or FtsZ2-1 have heterogeneous but severe defects in chloroplast division, as indicated by the increased size and decreased numbers of chloroplasts in leaf mesophyll cells of mutants compared to those in the wild type. The phenotype of the ftsZ2-2 null mutant is milder, and chloroplast size and number are less variable than in the other two mutants (Yoder et al., 2007; McAndrew et al., 2008; Schmitz et al., 2009). FtsZ2-1 and FtsZ2-2 comprise about two-thirds and one-third, respectively, of the total FtsZ2 pool, and mutant complementation experiments have shown that the FtsZ2-1 and FtsZ2-2 proteins, which share 85% amino acid identity downstream of their transit peptides, can substitute for one another as long as the total FtsZ2 protein level is close to that in the wild type. Consequently, the difference in the severity of the mesophyll cell phenotypes in the ftsZ2-1 and ftsZ2-2 mutants was proposed to be attributable to differences in total FtsZ2 level in the two mutants (McAndrew et al., 2008; Schmitz et al., 2009). The three FtsZ proteins in Arabidopsis have also been shown to associate partly with thylakoid membranes (El-Kafafi et al., 2008; Karamoko et al., 2011) and to make distinct contributions to plastid division in the shoot apex (Swid et al., 2018).
In this study, we exploited a near-isogenic line (NIL) population (Keurentjes et al., 2007) derived from a cross between the Arabidopsis accessions Cape Verde Islands (Cvi-1), which has larger chloroplasts, and Landsberg erecta (Ler-0), with smaller chloroplasts, to identify loci contributing to natural variation in chloroplast size and number in leaf mesophyll cells. We found that the large-chloroplast phenotype in Cvi-1 is associated specifically with a rare allelic variant of FtsZ2-2 that encodes a truncated gene product. Comparison of FtsZ2-2 coding sequences among all accessions in the 1001 Genomes database (1001 Genomes Consortium, 2016) combined with assessment of chloroplast-size phenotypes revealed other rare loss-of-function alleles of FtsZ2-2 in other accessions that also had enlarged chloroplasts. Further analysis showed that allelic variation was higher for the FtsZ2-2 than for the FtsZ2-1 or FtsZ1-1 coding sequences, indicating more relaxed functional constraint on FtsZ2-2. Our results demonstrate that allelic variation in FtsZ2-2 contributes to natural variation in chloroplast size in Arabidopsis.
RESULTS
Natural Variation in Chloroplast Size in Arabidopsis
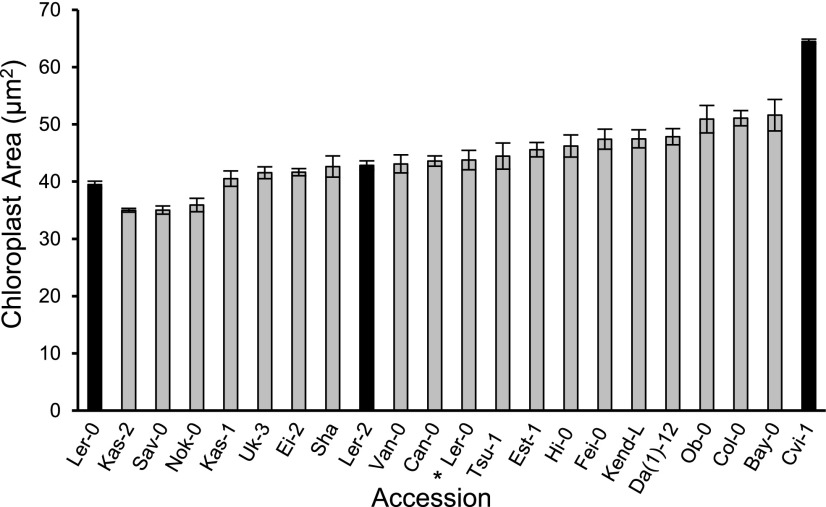
The extent of natural variation in chloroplast size in wild strains of Arabidopsis has not been previously studied. Therefore, as a prelude to our analysis, we first measured chloroplast sizes (areas) in 22 Arabidopsis accessions representing the parents of 12 available recombinant inbred line (RIL) populations and one NIL population (Supplemental Table S1; Keurentjes et al., 2007) to identify parents with the greatest differences in this trait. Further, to assess whether chloroplast size differences between any two parent accessions might be more pronounced under different environmental conditions or at different stages of development, we initially grew plants under four light regimes and harvested rosette leaves (leaf 6) from 26- and 40-d-old plants. Chloroplast sizes varied among accessions, ranging from ∼35 μm−2 to 70 μm−2, depending on growth conditions and time of harvest, but they were always biggest in Cvi-1 (Fig. 1; Supplemental Fig. S1). Further, the Cvi-1 and Ler-2 RIL parents (Alonso-Blanco et al., 1998; Juenger et al., 2006) and related Cvi-1 and Ler-0 NIL parents (Keurentjes et al., 2007) consistently exhibited greater differences in chloroplast size than any other combination of parental accessions (Fig. 1 [black bars]; Fig. 2A [left images]). Although the difference in chloroplast size between Cvi-1 and Ler-2 was greatest in 40-d-old plants grown at a light intensity of 50 µmol m−2 s−1 with a 16-h photoperiod (Supplemental Fig. S1), leaves from 40-d-old plants grown at 100 µmol m−2 s−1 with a 12-h photoperiod displayed a wider range of cell sizes, which facilitates quantitative analysis of chloroplast size phenotypes (Pyke and Leech, 1991). Therefore, these latter conditions were used for all further analyses of chloroplast size.
Figure 1.
Chloroplast areas in the 22 Arabidopsis accessions used as parents in the construction of 12 RIL populations and the Ler-0 × Cvi-1 NIL population. Accession names shown are those used in the ABRC stock information (see Supplemental Tables S1 and S2). Black bars highlight the comparison between Cvi-1 and the Ler-2 RIL parent or Ler-0 NIL parent. The asterisk (*) denotes a different Ler-0 genetic background (CS9994) from that of the other Ler-0 shown on the graph (CS-20). Plants were grown at a light intensity of 100 µmol m−2 s−1 with a 12-h light period. Samples for imaging were taken from leaf 6 harvested from 40-d-old plants. Error bars indicate the SE. Sample sizes are described in the “Experimental Design” and “Sampling and Microscopy” sections of “Materials and Methods.”
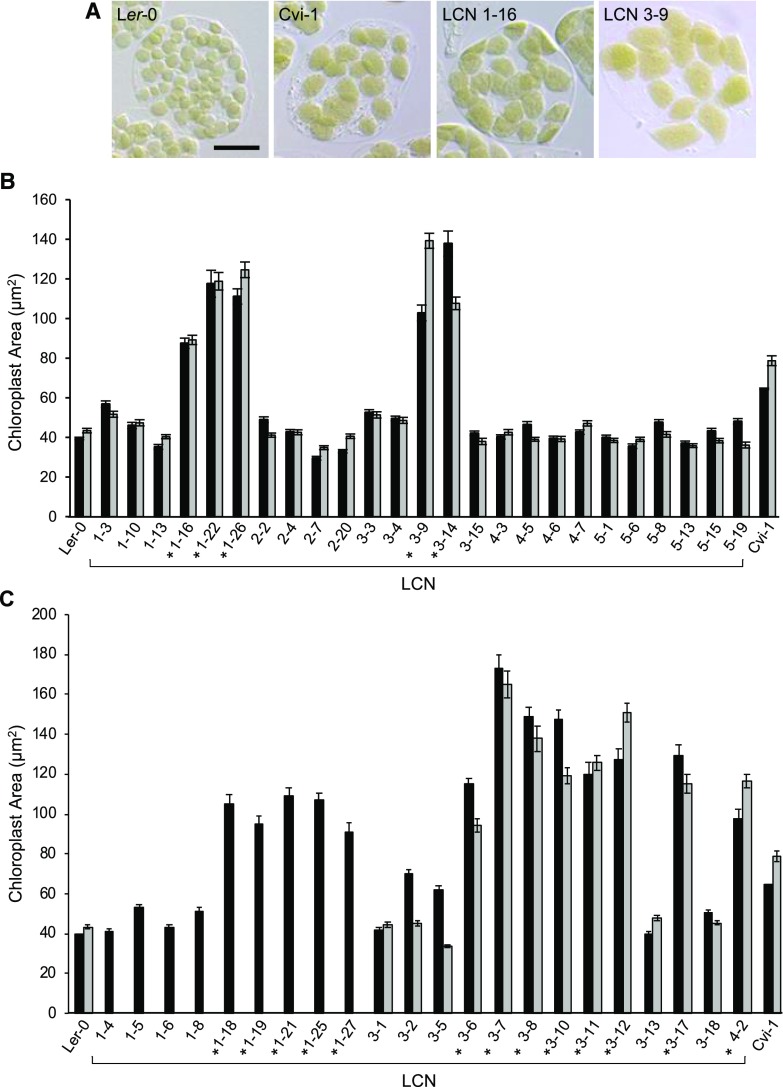
Figure 2.
Chloroplast sizes in the Ler-0 × Cvi-1 NIL population. Data are shown for the Ler-0 and Cvi-1 parent accessions and individual Ler-0 × Cvi-1 NIL (LCN) lines within the NIL population (Keurentjes et al., 2007). A, Images showing chloroplasts in mesophyll cells of Ler-0 and Cvi-1, and in LCN 1-16 and LCN 3-9 as examples of NILs with large-chloroplast phenotypes. Scale bar = 20 µm. B, Core set of 25 NILs containing Cvi-1 introgressions into the Ler-0 genome across all five chromosomes. C, An additional set of NILs containing Cvi-1 introgressions in chromosomes 1, 3, and/or 4 (see Supplemental Fig. S2). Asterisks in B and C indicate NILs with introgressions near the bottom of chromosome 3. Black and gray bars represent measurements from two independently grown sets of plants. Some NILs shown in C were only grown and scored once. Error bars indicate the SE. Sample sizes are described in the “Experimental Design” and “Sampling and Microscopy” sections of “Materials and Methods.” The same data for the Ler-0 and Cvi-1 parent accessions are repeated in B and C.
Larger Chloroplasts in Cvi-1 Are Linked to FtsZ2-2
To identify genomic regions involved in conferring the increased chloroplast size in Cvi-1, we took advantage of a set of 92 NILs in which Cvi-1 genomic DNA is introgressed across all five chromosomes in the Ler-0 background (Supplemental Fig. S2; Supplemental Table S2; Keurentjes et al., 2007). We first scored a core set of 25 NILs bearing Cvi-1 introgressions covering >90% of the genome and identified five lines with chloroplasts considerably larger than in the Ler-0 parent, similar to or larger than those in Cvi-1 (Fig. 2, A and B). Among these five lines, three (LCN 1-16, LCN 1-22, and LCN 1-26) had Cvi-1 introgressions in chromosomes 1 and 3, while two (LCN 3-9 and LCN 3-14) only had introgressions in chromosome 3 (Supplemental Fig. S2). Upon screening the remaining 67 NILs from the full set of 92 lines, we identified an additional 13 lines with enlarged Cvi-1-like chloroplasts (Figs. 2C and 3C; Supplemental Figs. S2 and S3A). Most of the lines were rescored in a second planting to confirm their phenotypes. All 18 NILs with Cvi-1-like phenotypes (Fig. 2, B and C, asterisks; Supplemental Fig. S2, green) contained introgressions near the bottom of chromosome 3, indicating that this region of the genome is responsible for the larger chloroplasts in Cvi-1.
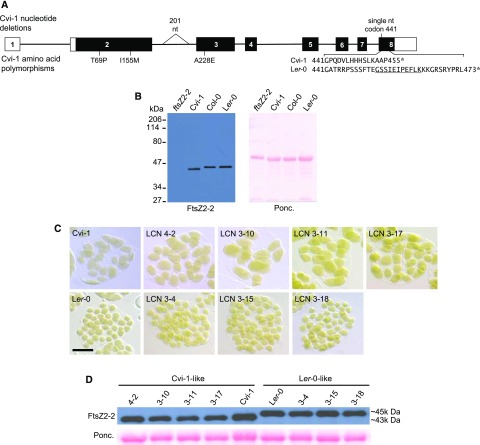
Figure 3.
Major differences between Ler-0 and Cvi-1 FtsZ2-2. A, Key polymorphisms in Cvi-1 FtsZ2-2 relative to the Ler-0 sequence, which is identical to that in Col-0. The gene model for Col-0 FtsZ2-2 annotated in The Arabidopsis Information Resource (AT3G57250.1) is diagrammed. Boxes represent the exons, numbered in order; white boxes represent UTRs and black boxes represent coding regions. Thin lines indicate introns. Significant nucleotide (nt) deletions and all AAPs in Cvi-1 are shown above and below the diagram, respectively. The triangle indicates a 201-nt deletion in intron 2. The amino acid sequence encoded by the Ler-0 allele from codon 441 to the stop codon (*) are shown below the Cvi-1 sequence for comparison. The C-terminal peptide is underlined. The full set of polymorphisms in the Cvi-1 allele is shown in Supplemental Fig. S5. B, Immunoblot detection of FtsZ2-2 (left) in leaf extracts from the indicated genotypes. Ponceau S staining of the blot (Ponc., right) served as a loading control. C, Images of mesophyll cells showing chloroplasts in Ler-0, Cvi-1, and NILs (LCN) with Cvi-1-like chloroplasts (top row) or Ler-0-like chloroplasts (bottom row). Scale bar = 20 µm. D, Immunoblot detection of FtsZ2-2 in leaf extracts from the lines shown in C. Ponceau S staining of Rubisco served as a loading control.
Based on the linkage map of the Ler-0 x Cvi-1 NIL population (Keurentjes et al., 2007), we determined that the shared Cvi-1 introgression in the 18 NILs with the Cvi-1 phenotype was between 62.1 and 68.2 centiMorgans on the genetic map of chromosome 3, which corresponds to a physical interval from 18.85 to 19.81 Mb. Since the recombinant breakpoints in this region had not been fine-mapped, we developed molecular markers and fine-mapped the region of chromosome 3 conferring the Cvi-1-like phenotype to an interval spanning only two loci: At3g52750, encoding the chloroplast division protein FtsZ2-2 (Osteryoung et al., 1998; McAndrew et al., 2008; Schmitz et al., 2009), and At3g52760, encoding a protein of unknown function (Supplemental Fig. S4). Because a knockout allele of FtsZ2-2 in ecotype Columbia-0 (Col-0) has previously been shown to result in enlarged chloroplasts resembling those in Cvi-1 (Fig. 4A; McAndrew et al., 2008; Schmitz et al., 2009), we considered FtsZ2-2 to be a strong candidate for the gene responsible for the large-chloroplast phenotype in Cvi-1.
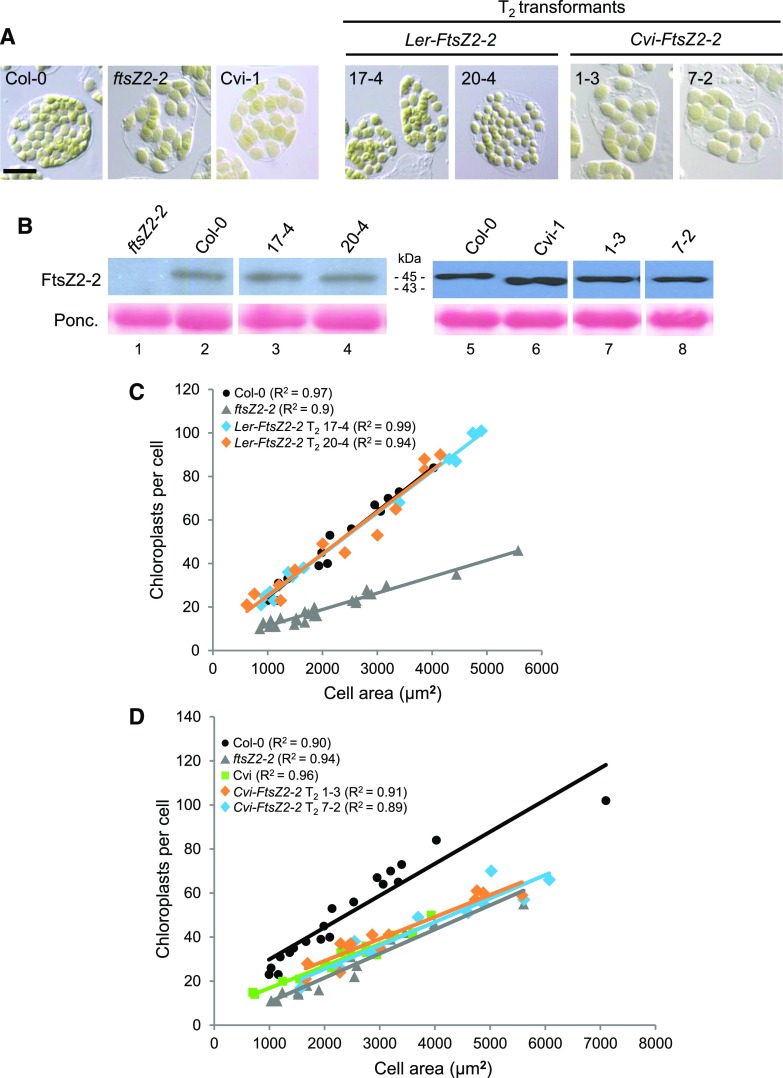
Figure 4.
Test for complementation of the Col-0 ftsZ2-2 chloroplast-size phenotype by the Ler-FtsZ2-2 and Cvi-FtsZ2-2 transgenes. Analysis was performed on transgenic plants that accumulated FtsZ2-2 protein at levels similar to those in wild-type Col-0 or Cvi-1. A, Images of mesophyll cells showing chloroplasts in Col-0, the Col-0 ftsZ2-2 null mutant, and Cvi-1 (left images), and representative independent T2 ftsZ2-2 mutants transformed with the Ler-FtsZ2-2 or Cvi-FtsZ2-2 transgene (right images). Scale bar = 20 µm. B, Immunoblot analysis of FtsZ2-2 in leaf extracts from the genotypes shown in A. Lanes 1–4 and 5–8 were from two separate blots. Ponceau S staining of Rubisco (Ponc.) served as a loading control. C and D, Graphs of chloroplast number versus mesophyll cell size for the genotypes shown in A. Best-fit lines are drawn. R2 values for each line are indicated in parentheses. Overlapping lines indicate similar chloroplast-size phenotypes (Pyke and Leech, 1992). Data for the indicated T2 transgenic plants expressing the Ler-FtsZ2-2 and Cvi-FtsZ2-2 transgenes are shown in C and D, respectively.
Since the sequence of the Cvi-1 accession was not available in the 1001 Genomes database (1001 Genomes Consortium, 2016), we isolated and sequenced an FtsZ2-2 genomic fragment from Cvi-1, including ∼1 kb flanking the start and stop codons, and the equivalent genomic fragment from Ler-0. The Ler-0 sequence was identical to the Ler-0 and Col-0 sequences in the 1001 Genomes database. In contrast, when compared to the sequence of Ler-0/Col-0 (hereafter Ler-0) FtsZ2-2, the Cvi-1 gene had a total of 73 polymorphisms, including several small insertion-deletions of one to six nucleotides, a larger deletion of 201 nucleotides in intron 2, and many single-nucleotide polymorphisms (SNPs), the majority of which were in introns (Supplemental Fig. S5). Within the coding region, three nonsynonymous SNPs were identified, two in exon 2 and one in exon 3 (Fig. 3A). In addition, a single-nucleotide deletion in the codon for Gly 441 occurred in the last exon, exon 8, creating a frameshift that altered the downstream 14 amino acids before producing a premature stop codon (Supplemental Fig. S5). This deletion yielded a predicted gene product of 455 amino acids, 18 amino acids shorter than the Ler-0 FtsZ2-2 protein (Fig. 3A).
To further narrow down the recombinant breakpoints, we sequenced FtsZ2-2 in four potentially informative large-chloroplast NILs (Supplemental Fig. S4, top four rows). All four bore the Cvi-1 deletion polymorphism in exon 8 but carried the Ler-0 SNP at the next downstream polymorphic site in the 3′ untranslated region (UTR; Supplemental Figs. S4 and S5). These results indicated recombinant breakpoints between these two sites in these NILs and suggested an association between the presence of the Cvi-1 exon 8 deletion polymorphism and the large-chloroplast phenotype. To further explore this association, we carried out immunoblotting of FtsZ2-2 in leaf extracts from Cvi-1, Ler-0, and Col-0 using an FtsZ2-2-specific antibody (McAndrew et al., 2008). Consistent with the predicted C-terminal truncation produced by this polymorphism, FtsZ2-2 migrated about 2 kD smaller in Cvi-1 than in Ler-0 and Col-0 (Fig. 3B). Further, all NILs with the larger Cvi-1-like chloroplasts also exhibited this smaller form of FtsZ2-2, whereas NILs with smaller Ler-0-like chloroplasts did not (Fig. 3, C and D; Supplemental Fig. S3), indicating a tight association between the FtsZ2-2 truncation and the chloroplast size phenotype in Cvi-1.
Because the Cvi-1 FtsZ2-2 allele bore the large deletion in intron 2 (Fig. 3A; Supplemental Fig. S5), we also considered the possibility that this might result in alternative splicing and contribute to the large-chloroplast phenotype. To assess this, we isolated FtsZ2-2 complementary DNAs (cDNAs) from Cvi-1 and Ler-0 by reverse-transcription PCR (RT-PCR). A single RT-PCR product that migrated at ∼1.7 kb was obtained in both cases. Sequencing of the cDNAs confirmed that the intron in Cvi-1 was faithfully spliced at the same sites as in Ler-0.
To address the possibility that the adjacent At3g52760 locus might instead be responsible for the Cvi-1 phenotype, we characterized a transfer DNA (T-DNA) insertion mutant in the Col-0 background (SAIL_61_A02). Sequencing confirmed that the T-DNA was inserted after the second nucleotide of the start codon, as annotated in The Arabidopsis Information Resource (http://www.arabidopsis.org), implying that this mutant allele is null. Chloroplasts in homozygous mutants were indistinguishable in size from those in wild-type Col-0 plants (Supplemental Fig. S6), indicating that the Cvi-1 chloroplast phenotype is not associated with At3g52760. Taken together, the combination of results was consistent with the exon 8 deletion polymorphism in Cvi-1 FtsZ2-2 being the likely cause of the large-chloroplast phenotype in Cvi-1.
Confirmation that the FtsZ2-2 Protein in Cvi-1 Causes Bigger Chloroplasts
To confirm that FtsZ2-2 is causal for the chloroplast-size phenotype in Cvi-1, we carried out complementation experiments in an ftsZ2-2 Col-0 knockout mutant in which the FtsZ2-2 protein is undetectable (Figs. 3B and 4B; McAndrew et al., 2008). For these experiments, we quantified chloroplast numbers in mesophyll cells of different sizes. In such analyses similar regression lines indicate similar chloroplast-size phenotypes because of the reciprocal relationship between chloroplast size and number and the linear relationship between cell size and total chloroplast compartment size (Honda et al., 1971; Ellis and Leech, 1985; Pyke and Leech, 1992; Pyke, 1999). Initial measurements showed that the phenotype of the ftsZ2-2 null mutant is similar to that of Cvi-1 (Fig. 4D, gray triangles and green squares).
A genomic copy of FtsZ2-2 from either Cvi-1 (Cvi-FtsZ2-2) or Ler-0 (Ler-FtsZ2-2), each bearing its native promoter, was introduced into the ftsZ2-2 mutant. In 22 independent T2 transformants expressing Ler-FtsZ2-2, the FtsZ2-2 protein migrated at the same mass as in wild-type Col-0 (Fig. 4B, lanes 2–4; Supplemental Fig. S7D) and chloroplast sizes were visibly smaller than those in ftsZ2-2, resembling those in wild-type Col-0 and Ler-0 (Fig. 4A; Supplemental Fig. S7, A and B). In contrast, in 28 independent T2 plants expressing Cvi-FtsZ2-2, FtsZ2-2 migrated at the same mass as in Cvi-1 (Fig. 4B, lanes 6–8; Supplemental Fig. S7E) and chloroplast sizes appeared similar to or larger than those in ftsZ2-2 and Cvi-1 (Fig. 4A; Supplemental Fig. S7, A and C). Because changes in FtsZ levels result in dose-dependent increases in chloroplast size and decreases in chloroplast number (Stokes et al., 2000; Schmitz et al., 2009), to determine whether the Ler-0 and Cvi-1 transgenes could complement the ftsZ2-2 phenotype we quantified chloroplast numbers in cells of different sizes in transgenic plants in which FtsZ2-2 protein levels were close to those in wild-type Col-0 and Cvi-1, as determined by immunoblotting (Fig. 4B). In these individuals, the ftsZ2-2 chloroplast-size defect was complemented by the Ler-0 allele, as indicated by the overlapping regression lines between wild-type Col-0 and the Ler-FtsZ2-2 transgenic plants (Fig. 4C), but not by the Cvi-1 allele, as the regression lines for the Cvi-FtsZ2-2 transgenics were similar to that of ftsZ2-2 (Fig. 4D). These results confirmed that the Cvi-1 FtsZ2-2 gene is causal for the large-chloroplast phenotype in Cvi-1.
Z-Ring Formation Is Altered in Cvi-1
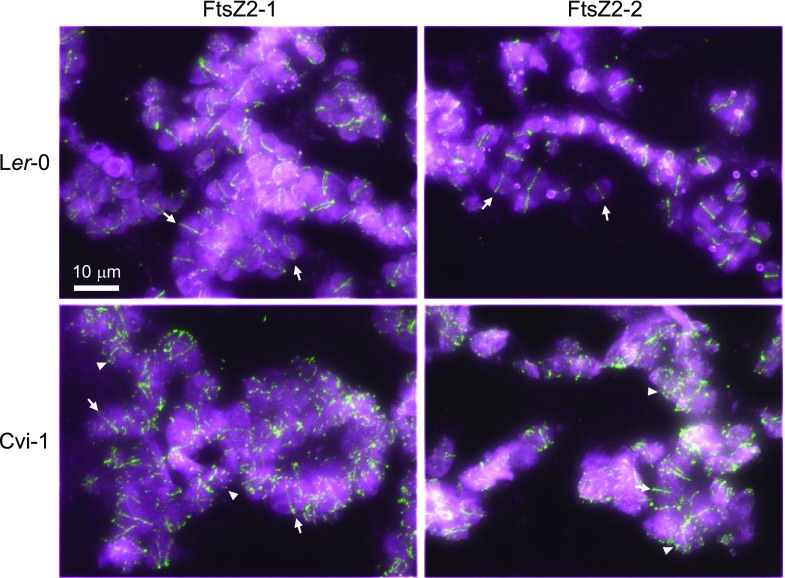
To further explore the functional basis for the increased chloroplast-size phenotype in Cvi-1, we investigated FtsZ2-2 and FtsZ2-1 localization in both Cvi-1 and Ler-0 by immunofluorescence labeling. We have shown previously that all three FtsZs colocalize to Z rings in Arabidopsis and that the anti-FtsZ2-1 and anti-FtsZ2-2 antibodies are specific for detection of their respective antigens (Stokes et al., 2000; Vitha et al., 2001; Yoder et al., 2007; McAndrew et al., 2008). In Ler-0, FtsZ2-1 and FtsZ2-2 localized primarily to the midplastid Z ring (Fig. 5, top; Supplemental Fig. S8A). In contrast, in Cvi-1, while the FtsZ2 proteins were still often detected in midplastid rings, shorter filaments and punctate structures that were not confined to the midplastid were observed much more frequently than in Ler-0 (Fig. 5, bottom; Supplemental Fig. S8B). These results suggest that alterations in Z-ring organization contribute to chloroplast enlargement in Cvi-1.
Figure 5.
Immunofluorescence localization of FtsZ2 proteins in mesophyll cells of Ler-0 and Cvi-1. Merged images of fluorescent signals from FtsZ2-1 or FtsZ2-2 (left and right, respectively; green) and chlorophyll autofluorescence (magenta) are shown. Arrows, FtsZ rings; arrowheads, FtsZ punctate structures or very short filaments in Cvi-1. Scale bar = 10 µm. Additional images are shown in Supplemental Figure S8.
Natural Variation in Arabidopsis FtsZ2-2 Protein Sequences
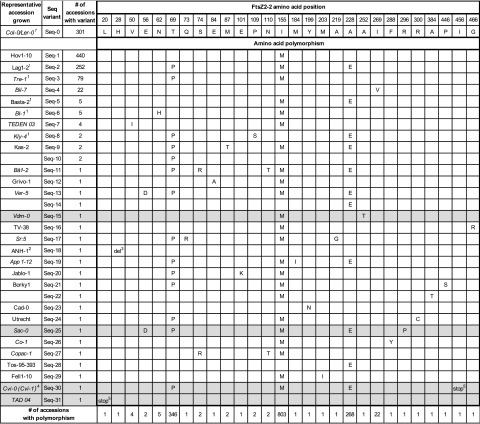
To probe the extent of natural variation in the Arabidopsis FtsZ2-2 protein in greater detail, we extracted and translated the FtsZ2-2 coding sequences from the 1,135 accessions available in the 1001 Genomes database (1001 Genomes Consortium, 2016) and compared the resulting amino acid sequences to that of the Col-0 FtsZ2-2 reference sequence. In total there were 32 unique FtsZ2-2 protein haplotypes, most with one or more amino acid polymorphisms (AAPs; Fig. 6; Supplemental Datasets S1 and S2). Three accessions had deletions in the FtsZ2-2 protein. One was Cvi-0, whose FtsZ2-2 protein sequence was identical to that in Cvi-1 (Cvi-1 is not in the 1001 Genomes database; Fig. 6, Seq-30). As in Cvi-1, chloroplasts in Cvi-0 were enlarged compared to those in Ler-0, and the FtsZ2-2 protein migrated at the same mass as in Cvi-1 (Fig. 6, gray shading; Supplemental Fig. S9, A and B). PCR analysis indicated that Cvi-0 FtsZ2-2 likely has the same deletion in intron 2 as Cvi-1 (Fig. 3A; Supplemental Fig. S9C) and that the Cvi-0 and Cvi-1 alleles may be identical. The accession TAD 04 had a two-nucleotide deletion at the first two positions of codon 5, producing a frameshift that resulted in a premature stop codon and predicted FtsZ2-2 gene product of only 19 amino acids (Fig. 6, Seq-31; Supplemental Datasets S1 and S2), indicating that the TAD 04 FtsZ2-2 allele is null. Consistent with this finding, TAD 04 had enlarged chloroplasts approximately the same size as those in the Col-0 ftsZ2-2 null mutant (Fig. 7, A and B; Supplemental Fig. S10B) and FtsZ2-2 protein was undetectable in TAD 04 by immunoblotting (Fig. 7C). The accession ANH-1 (Fig. 6, Seq-18) lacked the codon for amino acid 28, which is in the middle of the predicted chloroplast transit peptide (estimated to be 50 amino acids by ChloroP (http://www.cbs.dtu.dk/services/ChloroP/; Emanuelsson et al., 1999). Such a deletion is unlikely to affect FtsZ2-2 import into the chloroplast, and chloroplasts in ANH-1 were similar in size to those in Col-0 (Supplemental Fig. S10B).
Figure 6.
FtsZ2-2 (At3g52750) AAPs among the 1,135 Arabidopsis accessions in the 1001 Genomes database (1001 Genomes Consortium, 2016). Accessions in the first column were those in which chloroplast-size phenotypes were imaged for each sequence variant (Seq variant). Italics denote accessions that were also subjected to quantitative analysis of chloroplast size and immunoblot detection of FtsZ proteins. Gray shading highlights accessions with large-chloroplast phenotypes. Blank cells indicate sequence variants for which no accessions were grown due to lack of seed availability or germination. All accessions with each FtsZ2-2 sequence variant are listed in Supplemental Dataset S1. Superscripts indicate the following: 1Additional accessions with the same sequence variant were grown and imaged as listed in Supplemental Dataset S2. 2Accession possibly misidentified (Pisupati et al., 2017). 3Codon 28 deleted in this accession. 4The Cvi-0 and Cvi-1 FtsZ2-2 coding sequences are identical, but only the Cvi-0 genome sequence is in the 1001 Genomes database. Therefore, the number of accessions in the database with Seq-30 is shown as 1. 5Premature stop codons at the indicated positions were preceded by nucleotide deletions that altered multiple upstream amino acids. See Supplemental Dataset S1 for complete sequences.
Figure 7.
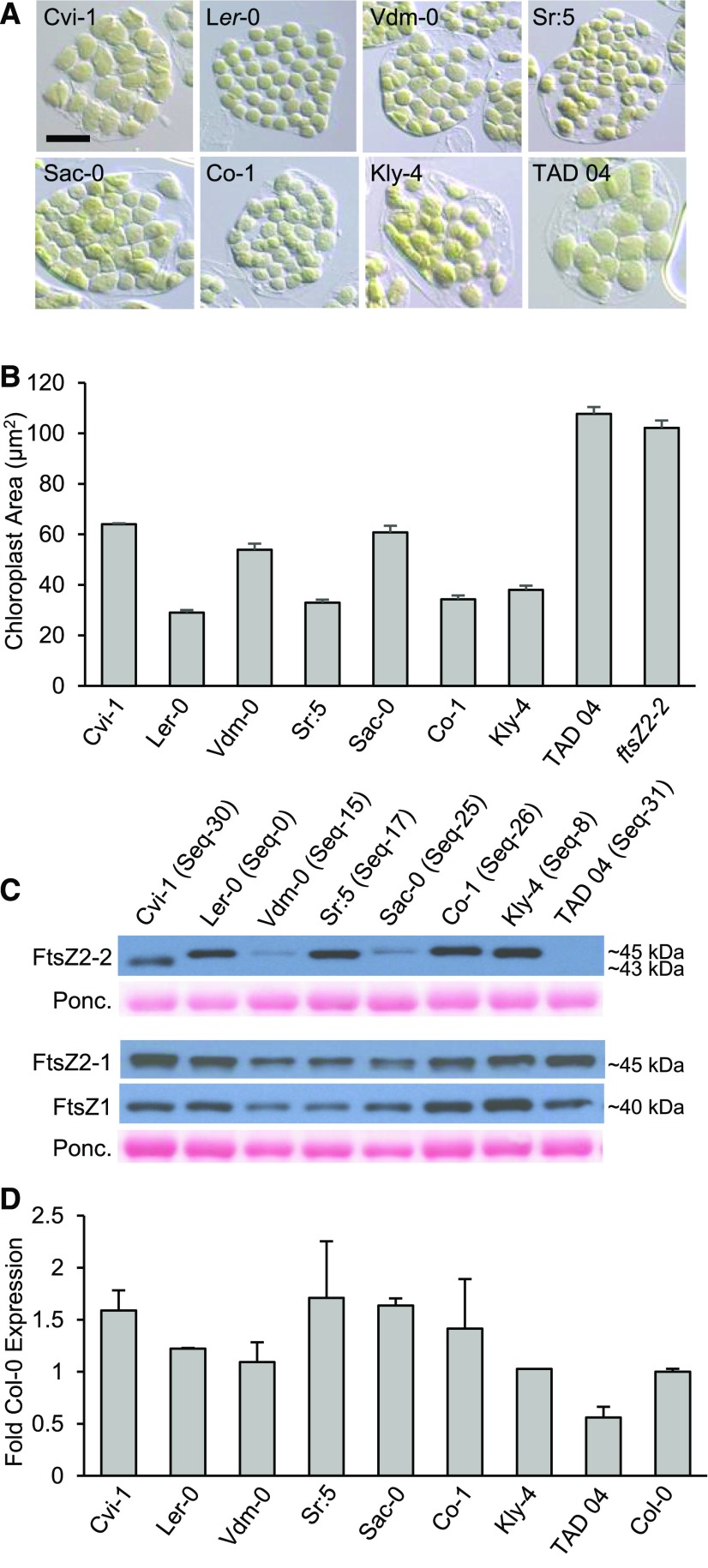
Chloroplast sizes, FtsZ protein levels, and FtsZ2-2 transcript levels in representative accessions from different polymorphic groups. Protein sequence variant (Seq) numbers are specified in C and the sequences are shown in Fig. 6 and Supplemental Dataset S1. A, Images of mesophyll cells showing chloroplasts in Cvi-1, Ler-0, and accessions from different polymorphic groups. Scale bar = 20 µm. B, Chloroplast areas in the indicated accessions and the Col-0 ftsZ2-2 null mutant. Error bars represent the se. Sample sizes are described in the “Experimental Design” and “Sampling and Microscopy” sections of “Materials and Methods.” C, Immunoblot analysis of FtsZ proteins in leaf extracts from the accessions shown in A. FtsZ2-1 and FtsZ1-1 were probed together on the same blot. Ponceau S staining of Rubisco (Ponc.) served as a loading control. Figs. 3B and 4B show that FtsZ2-2 protein is undetectable in ftsZ2-2. D, FtsZ2-2 transcript levels in the accessions shown in C relative to those in Col-0 determined by RT-qPCR. Error bars represent the se except for the accession Kly-4. Sample sizes are described in the “RT-qPCR” section of “Materials and Methods.”
The other 1,131 accessions in the 1001 Genomes database encoded FtsZ2-2 proteins that were identical in length to the Col-0 reference sequence (473 amino acids) with a total of 301 lines having a sequence identical to that in Col-0 (and Ler-0; Fig. 6, Seq-0). The other protein variants contained one or more AAPs. The most common AAP, which occurred in ∼70% of the accessions, was replacement of Ile 155 by Met (155:I→M). This AAP was the only one present in 440 accessions (Seq-1), but also occurred along with other AAPs in an additional 363 accessions (Fig. 6; Supplemental Dataset S2). Two other AAPs were also common: 69:T→P occurred in 346 accessions and 228:A→E in 268 accessions. The AAP 269:I→V occurred in a single sequence variant (Seq-4) present in 22 accessions. The other 21 AAPs were found in only five or fewer accessions, all in combination with one or more of the three most common AAPs except for 199:Y→N (Seq-23), which occurred in a single accession (Fig. 6; Supplemental Datasets S1 and S2).
FtsZ2-2 Polymorphisms and Natural Variation in Chloroplast Size
To evaluate whether chloroplast sizes might differ in accessions with FtsZ2-2 AAPs other than the deletions described above, we imaged chloroplasts in at least one representative accession from 25 of the 28 polymorphic groups that did not have amino acid deletions (Figs. 6 and 7A; Supplemental Fig. S10C), and in additional accessions from the nonpolymorphic group bearing the same sequence (Seq-0) as in Col-0 (Supplemental Dataset S2). Among these, quantitative measurements of chloroplast size and examination of FtsZ levels by immunoblotting were carried out in one accession from a subset of 13 polymorphic groups (Figs. 6 [italicized accessions] and 7, B and C; Supplemental Fig. S11) selected based on the extent of conservation of the polymorphic amino acids in FtsZ2 proteins from diverse plant species (TerBush et al., 2018). Among this subset, two accessions, Vdm-0 (Seq-15) and Sac-0 (Seq-25), had enlarged chloroplasts (Fig. 7, A and B; Supplemental Fig. S10C). Each of these accessions carries a unique AAP not found in other accessions: 252:A→T in Vdm-0 and 296:R→P in Sac-0 (Fig. 6; Supplemental Dataset S2). Immunoblotting revealed substantial reductions in the levels of FtsZ2-2 protein in these two accessions, but not in the levels of FtsZ2-1 or FtsZ1-1 (Fig. 7C). RT-quantitative PCR (RT-qPCR) showed that the lower FtsZ2-2 protein levels in Vdm-0 and Sac-0 were not due to reduced FtsZ2-2 transcript levels relative to those in Col-0, Ler-0, and other accessions (Fig. 7D). Except in Cvi-1 and TAD 04, as described above, no visibly obvious increases in chloroplast size or major differences in FtsZ levels were observed in the other accessions analyzed (Fig. 7, A–C; Supplemental Figs. S10C and S11). These results suggest that reduced accumulation of FtsZ2-2 protein is a likely cause of the increased chloroplast sizes in Vdm-0 and Sac-0, possibly due to decreased protein stability.
Comparison of AAPs between FtsZ Proteins
We also analyzed the degree of AAP in the two other FtsZ proteins in Arabidopsis, FtsZ2-1 and FtsZ1-1, by extracting and translating the FtsZ2-1 and FtsZ1-1 coding sequences from the 1001 Genomes database as described above for FtsZ2-2. In Col-0, full-length FtsZ2-2 shares 82% identity with FtsZ2-1 and 61% identity with FtsZ1-1. The lengths of all FtsZ2-1 and FtsZ1-1 protein variants were identical to those of the Col-0 reference sequences, 478 and 433 amino acids, respectively. FtsZ2-1 encoded 18 unique protein haplotypes (Supplemental Dataset S3), with 927 accessions having a sequence identical to that in Col-0 (Seq-0; Supplemental Dataset S4). FtsZ1-1 encoded only 11 unique protein haplotypes (Supplemental Datasets S5 and S6). These results revealed that AAPs are more common in FtsZ2-2 than in FtsZ2-1 and FtsZ1-1.
Comparison of Nucleotide Polymorphisms between FtsZ Genes
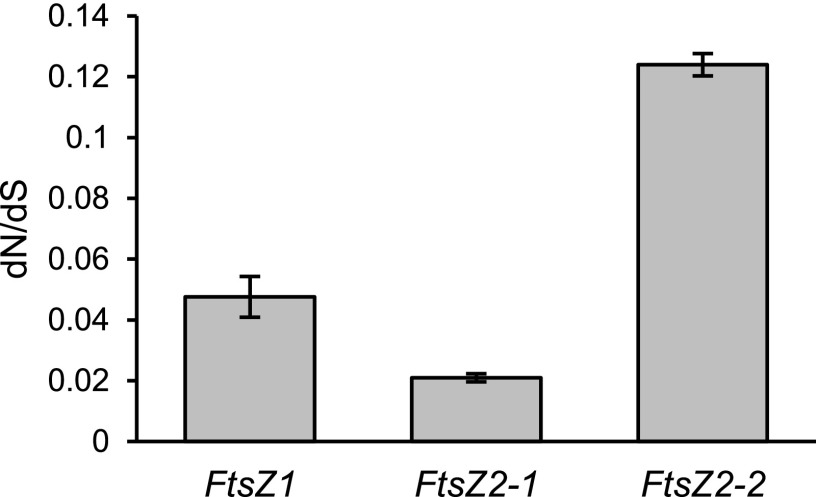
To compare the extent of nucleotide polymorphism in the coding sequences of the three FtsZ genes, we calculated the number of substitutions per synonymous site (dS) and nonsynonymous site (dN), and their ratio, dN/dS, by pairwise comparisons of the Col-0 FtsZ1-1, FtsZ2-1, and FtsZ2-2 coding sequences with those in the other accessions in the 1001 Genomes database. FtsZ1-1 had the smallest values of both dS and dN. FtsZ2-1 and FtsZ2-2 had similar dS values, but dN was larger for FtsZ2-2. As a result, dN/dS for FtsZ2-2 was 2.6-fold higher than for FtsZ1-1 and 5.9-fold higher than for FtsZ2-1 (Fig. 8). While the dN/dS ratios were <1 for all three genes, indicating negative selection that generally selects against polymorphisms impacting protein function, the higher dN/dS ratio for FtsZ2-2 suggests more relaxed functional constraint on the FtsZ2-2 than on the FtsZ2-1 or FtsZ1-1 coding sequence (Nielsen, 2005; Buschiazzo et al., 2012).
Figure 8.
dN/dS values for the three FtsZ genes in the 1,135 accessions in the 1001 Genomes database (1001 Genomes Consortium, 2016). The Col-0 genes were used as references to calculate dN and dS. dS values were 0.00332 ± 0.00007 (FtsZ1-1), 0.0199 ± 0.0001 (FtsZ2-1), and 0.0168 ± 0.0007 (FtsZ2-2). dN values were 0.000298 ± 0.000037 (FtsZ1-1), 0.000403 ± 0.000026 (FtsZ2-1), and 0.00187 ± 0.00003 (FtsZ2-2). dN/dS is 0.0476 ± 0.0067 for FtsZ1-1, 0.021 ± 0.001 for FtsZ2-1, and 0.124 ± 0.004 for FtsZ2-2. dN/dS values were all significantly different from each other (P < 0.0001) as determined by the Tukey adjustment for multiple comparisons.
DISCUSSION
Large-effect rare alleles have facilitated identification of causal genes for quantitative trait loci (QTLs) controlling flowering time in Arabidopsis (Johanson et al., 2000; El-Din El-Assal et al., 2001; Maloof et al., 2001; Balasubramanian et al., 2006; Filiault et al., 2008), grain size and grain yield in rice (Oryza sativa; Zhang et al., 2012; Hu et al., 2015), sulfate content (Loudet et al., 2007), and numerous other life-history, whole-plant and metabolic traits (Alonso-Blanco et al., 2009). In this study, a rare FtsZ2-2 allele in Cvi-1 led to our identification of FtsZ2-2 as a key locus contributing to natural variation in chloroplast size in Arabidopsis. This analysis was facilitated by the large difference in chloroplast size between Ler-0 and Cvi-1 (Figs. 1 and 2A), which enabled us to exploit the Ler-0 x Cvi-1 NIL population (Keurentjes et al., 2007) to fine-map the genomic region conferring the large-chloroplast phenotype in Cvi-1. The four other accessions with similarly enlarged chloroplasts discovered through analysis of FtsZ2-2 coding sequences in the 1001 Genomes database also carried rare alleles of FtsZ2-2 that influence its functionality (Fig. 6). To our knowledge, this is the first instance in which natural variation has been exploited to uncover a molecular determinant of phenotypic differences in a fundamental cell biological trait in plants.
FtsZ2 and FtsZ1 are conserved throughout the green lineage, and in land plants, both are components of the chloroplast Z ring (McAndrew et al., 2001; Mori et al., 2001; Vitha et al., 2001; Kuroiwa et al., 2002; Osteryoung and Pyke, 2014). Like all FtsZ proteins, both FtsZ2 and FtsZ1 are composed of a highly conserved globular core, comprising GTP-binding and GTPase-activating domains, that alone mediates FtsZ polymerization; the conserved core is flanked by N- and C-terminal extensions (Löwe and Amos, 1998; Oliva et al., 2004; TerBush et al., 2013; Yoshida et al., 2016). A key feature that only FtsZ2 shares with bacterial FtsZs is the presence of a short peptide near the C terminus called the C-terminal peptide (Ma and Margolin, 1999). The C-terminal peptides of FtsZ2-2 and its close paralog FtsZ2-1 interact with the inner envelope chloroplast division protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6 (ARC6), and this is presumed to be the primary interaction mediating membrane-tethering of the chloroplast Z ring (Maple et al., 2005; Schmitz et al., 2009; Johnson et al., 2013). The truncated protein encoded by the Cvi-1 FtsZ2-2 allele lacks the C-terminal peptide (in addition to downstream residues; Fig. 3A), presumably preventing its interaction with ARC6. Nevertheless, the presence of FtsZ2-2 in rings and filaments in Cvi-1 (Fig. 5) implies that the truncated protein coassembles with FtsZ2-1 and FtsZ-1, as expected since it retains the globular core responsible for polymerization, and also accumulates at levels similar to that of the nontruncated FtsZ2-2 protein in Col-0 and Ler-0 (Fig. 3B). However, the more disorganized morphology of FtsZ rings in Cvi-1 (Fig. 5) indicates that C-terminal truncation of FtsZ2-2 compromises its function, probably by reducing the total number of FtsZ2 subunits available for ARC6 interaction and membrane tethering, in turn resulting in reduced division capacity and hence bigger chloroplasts. These results suggest that the ability of both FtsZ2 proteins to interact with ARC6 (Maple et al., 2005; Schmitz et al., 2009) influences the efficiency of the division process.
The FtsZ2-2 alleles in TAD 04, Vdm-0, and Sac-0 were also associated with increased chloroplast size. Consistent with previous studies showing dose-dependent effects of FtsZ protein levels on chloroplast size (Stokes et al., 2000; Schmitz et al., 2009), chloroplasts in TAD 04 bearing the FtsZ2-2 null allele were similar in size to those in the Col-0 null mutant and larger than those in Vdm-0 and Sac-0, which accumulated FtsZ2-2 at low levels (Fig. 7, A–C). In Col-0, FtsZ2-2 accounts for about a third of the total FtsZ2 pool, and studies in various mutants and transgenic plants have consistently shown that deficiency or absence of any one or two FtsZ proteins does not alter accumulation of the others (Yoder et al., 2007; McAndrew et al., 2008; Schmitz et al., 2009). Likewise, TAD 04, Vdm-0, and Sac-0 all had FtsZ2-1 and FtsZ1-1 levels similar to those in other accessions (Fig. 7C). Therefore, the most likely explanation for the chloroplast-size phenotypes in these three accessions (Fig. 7, A and B) is their reduced dosage of FtsZ2-2 protein. However, although variations in protein abundance are correlated with variations in gene expression for many quantitative traits (Ghazalpour et al., 2011; Battle et al., 2015), this was not the case in Vdm-0 and Sac-0, because their FtsZ2-2 transcript levels were similar to those in other accessions with greater FtsZ2-2 protein levels and smaller chloroplasts (Fig. 7D). We suggest that reduced protein stability may account for the low FtsZ2-2 levels in Vdm-0 and Sac-0. Sac-0 has multiple AAPs, but one, 296:R→P, occurs only in this accession (Fig. 6, Seq-25). Alignment of the Arabidopsis FtsZ2 proteins with 63 FtsZ2 proteins in other angiosperms revealed conservation of this Arg in all sequences analyzed. Modeling of FtsZ2 onto the crystal structure of a bacterial FtsZ showed that this Arg resides in an interior α-helix, H7, that connects the GTP-binding and GTPase-activating domains within the globular core (Oliva et al., 2004). Similarly, in Vdm-0, the Ala substituted in the unique AAP 252:A→T (Fig. 6, Seq-15) is conserved in all other plant, as well as bacterial, FtsZs analyzed and is located in another α-helix, H5, that forms part of the interface joining polymerized FtsZ subunits (Li et al., 2013a). The 296:R→P and 252:A→T polymorphisms in Sac-0 and Vdm-0 would likely disrupt these helices (Richardson, 1981), potentially making these protein variants more susceptible to degradation. However, we cannot rule out that other mechanisms, such as altered posttranslational modification, contribute to the reduced FtsZ2-2 accumulation in these two accessions (Hansen et al., 2008; Gargano et al., 2012; Bartlett and Whipple, 2013).
While large-effect QTLs are often associated with allelic variation in regulatory regions that impact gene expression (Maloof et al., 2001; Bartlett and Whipple, 2013), our data show that variation in the FtsZ2-2 coding sequence influences chloroplast size through multiple protein-based mechanisms. Similarly, polymorphisms producing various truncated and other predicted loss-of-function proteins or altering single amino acids have been shown to be important determinants of several life-history and whole-plant traits, including flowering time, flower color and pollinator preference, drought tolerance, trichome patterning, and photomorphogenesis (Le Corre et al., 2002; Shindo et al., 2005; Balasubramanian et al., 2006; Hoballah et al., 2007; Monroe et al., 2018). In these cases, the relatively high frequency of such mutations suggests that they may confer a selective advantage. In contrast, rare allelic variants that introduce premature stop codons or alter highly conserved amino acids, such as those in FtsZ2-2 associated with increased chloroplast size, are generally assumed to be deleterious (Clark et al., 2007; Cao et al., 2011). Cvi has been noted to have a particularly high incidence of such mutations, perhaps because of its origination in an isolated island population (Günther and Schmid, 2010). Whether the rare FtsZ2-2 alleles associated with the large-chloroplast phenotype are mildly deleterious mutations that have not been eliminated from the natural populations in which they occur or may provide some advantage in local adaptation remains unclear.
Data from well-annotated plant and algal genomes indicate that many diploid land plants have two FtsZ2 genes and one FtsZ1 gene, whereas green algae have only one of each, suggesting that duplication of FtsZ2 and retention of the second paralog conferred a selective advantage during land-plant evolution. Complementation experiments in an ftsZ2-1 ftsZ2-2 double-mutant background established that Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally interchangeable in vivo as long as total FtsZ2 protein levels are close to those in the wild type, indicating that the two FtsZ2 paralogs have biochemically equivalent functions (Schmitz et al., 2009). These findings suggest that retention of both FtsZ2 paralogs may be important for maintaining stoichiometric balance within the Z ring and chloroplast-division complex, consistent with models proposed for retention of paralogs that function in macromolecular complexes (Birchler and Veitia, 2010; Panchy et al., 2016). However, the higher dN/dS ratio for FtsZ2-2 compared to FtsZ2-1 and FtsZ1-1 (Fig. 8) indicates more relaxed functional constraint on the FtsZ2-2 coding sequence. Consistent with this result, no sequence variants producing truncated gene products were identified for FtsZ2-1 or FtsZ1-1 in the 1001 Genomes database (Supplemental Datasets S3 and S5), suggesting that such mutations are not well tolerated in these two genes. While it is clear that the C-terminal truncation of FtsZ2-2 in Cvi-1 and null FtsZ2-2 allele in TAD 04 are responsible for the increased chloroplast size in these accessions, it is possible that other, less dramatic polymorphisms in the FtsZ2-2 coding sequence contribute in more subtle ways to natural variation in chloroplast size in Arabidopsis (Fig. 1) and could represent a difference between FtsZ2-2 and FtsZ2-1 that is functionally important in nature.
Natural variation in chloroplast size or other aspects of chloroplast morphology has rarely been systematically investigated within a single species (Jellings et al., 1983). However, such differences have been reported between species (Honda et al., 1971; Jellings and Leech, 1984; Pyke and Leech, 1987), in different cell and tissue types within a species (Dengler et al., 1996; Pyke, 1997; Ahmadabadi and Bock, 2012; Stata et al., 2014; Swid et al., 2018), and under different environmental conditions (Boardman, 1977; Tsuji et al., 1979; Possingham et al., 1988; Filek et al., 2010; Li et al., 2013b; Takemura et al., 2017). Chloroplast size differences have been particularly well documented in response to light conditions. Plants adapted to low light typically develop larger chloroplasts with grana that are more highly stacked, wider, and more irregularly orientated than in plants adapted to high light (Björkman et al., 1971; Anderson et al., 1973, 1988; Boardman 1977; Lichtenthaler et al., 1981). The increased chloroplast size in low-light plants may be necessary to accommodate this enhanced grana stacking and altered thylakoid architecture. The chloroplast size differences in plants grown under different light intensities results, at least in part, from differences in the frequency of chloroplast division; chloroplasts divide less often under low light but continue to expand (Possingham, 1973). Because there is a tightly regulated relationship between total chloroplast compartment size and cell size (which varies between species), and because chloroplast division and expansion are mutually compensatory but independent processes (Honda et al., 1971; Ellis and Leech, 1985; Pyke and Leech, 1992; Pyke, 1999), a reduced frequency of division will be accompanied by a reduced number of chloroplasts that are larger in size. In the case of Cvi-1 and the other Arabidopsis accessions with enlarged chloroplasts identified in this study, altered FtsZ2-2 function or accumulation explains their reduced division frequency and enlarged chloroplasts. Whether changes in the levels of FtsZ2 or other chloroplast-division proteins also contribute to chloroplast size changes in response to light or other environmental factors remains to be explored.
While the adaptive advantage of chloroplast size variation has not been investigated, experiments with Arabidopsis ftsZ and other chloroplast-division mutants have shown that plants with greatly enlarged chloroplasts exhibit defects in chloroplast movement and impairment of some photosynthetic responses as well as reduced mesophyll conductance, and that somewhat altered responses could also be observed in division mutants with less dramatic phenotypes, including ftsZ2-2 (Jeong et al., 2002; Austin and Webber, 2005; Königer et al., 2008; Dutta et al., 2015, 2017; Weise et al., 2015; Xiong et al., 2017). These results, along with the influence of environmental factors on chloroplast size and number discussed above, suggest that fine-tuning of chloroplast size is important for physiological fitness. Studies of chloroplast movement and photosynthetic performance in different Arabidopsis accessions could shed more light on the physiological significance of natural variation and phenotypic plasticity in chloroplast size.
MATERIALS AND METHODS
Plant Materials
The following Arabidopsis (Arabidopsis thaliana) seed stocks were obtained from the Arabidopsis Biological Resource Center (ABRC; Ohio State University; http://abrc.osu.edu/): the Ler-0 and Cvi-1 accessions used as parents in the construction of the NIL population (Keurentjes et al., 2007) and other accessions shown in Figure 1 (Supplemental Table S1); the 92 NILs scored for chloroplast size (Supplemental Table S2); the ftsZ2-2 null mutant (SALK 050397); the At3g52760 null mutant (SAIL_61_A02); Vdm-0 (CS78837); and TV-38 (CS78770). TAD04 was a gift from Joy Bergelson (University of Chicago). All other accessions analyzed in this study were propagated from the set of 1,135 accessions obtained from 1001genomes.org (stock ID CS78942; 1001 Genomes Consortium, 2016), and progeny were used for analysis.
Plant Growth Conditions
Seeds were imbibed in distilled water in plastic tubes and stratified at 4°C for 2 d in the dark. Next, 2–4 seeds/pot were sown in the corners of square pots (70 mm) filled with a sterile mixture of equal parts potting soil (Sure-mix), perlite, and medium vermiculite. Pots were randomized across flats and transferred into controlled-environment chambers. After testing four different growth regimes (Supplemental Fig. S1), plants were grown under white fluorescent light (100 µmol m−2 s−1, 12 h/12 h light/dark photoperiod; bulb types PHILIPS, F17T8/TL841 ALTO, 17 Watts, with two tubes; SYLVANIA FO96/841/ECO, 59 Watts, with five tubes) at 21°C in relative humidity of 60%. Plants were fertilized with one-half strength Hoagland’s solution once per week and watered additionally twice per week. To avoid positional light and temperature effects on plant growth, trays were moved three times per week randomly within the chamber.
For selection of transgenic plants, seeds were surface-sterilized and sown on plates in 0.7% (w/v) Phytagar (Gibco BRL) containing one-half strength Linsmaier and Skoog (LS) medium (Caisson Laboratories) supplemented with 1% (w/v) Suc and the relevant selection agent. Following cold treatment at 4°C for 2 d in the dark, plants on plates were grown in growth chambers under the conditions described above. After 8–10 d, resistant plants were transferred to soil and grown as above.
Experimental Design
To determine whether chloroplast size differences between any two parent accessions might be more pronounced under a particular set of conditions, the 22 Arabidopsis accessions shown in Figure 1 (Supplemental Table S1) were grown in two replicates under four different light conditions (50 or 100 µmol m−2 s−1 light intensity and 12 or 16 h light duration) and rosette leaves (leaf 6) were harvested from 26- and 40-d-old plants. The set of 92 NILs (Supplemental Table S2; Keurentjes et al., 2007) was grown in 4 replicates and their Cvi-1 and Ler-0 parents were grown in 16 replicates. The core set of 25 NILs was grown twice. Some NILs from the full set of 92 lines were grown once.
Sampling and Microscopy
A day before sampling, a toothpick was placed next to leaf 6. The following morning, plants were removed from the growth chamber before the lights came on and leaves were harvested immediately. Tissue from the leaf tip was fixed (Pyke and Leech, 1991) and chloroplasts in mesophyll cells were visualized (Osteryoung et al., 1998) under a DMI3000B inverted microscope (Leica Microsystems) using differential interference contrast optics under a 40× objective. Images were acquired with a Leica DFC320 camera mounted on the microscope. A minimum of 10 images and a maximum of 50 images per sample were taken from the fixed tissue and saved for quantitative analysis. Chloroplast plan area, number, and mesophyll cell plan area were measured using ImageJ, version 1.42 (National Institutes of Health). Average chloroplast area ± se in micrometers squared within a confidence interval of 95% with a margin of error of 5 µm2 was calculated for a range of cell sizes between ∼3,000 and 6,000 µm2. For each individual plant sampled, a minimum of 36 chloroplasts were measured in three randomly chosen chloroplasts in 12 randomly chosen mesophyll cells.
Immunofluorescence Labeling
Immunofluorescence labeling of FtsZ (Vitha et al., 2001; Yoder et al., 2007) was carried out on mature leaves of 28-d-old plants using antibodies against Arabidopsis FtsZ2-1 and FtsZ2-2 (Stokes et al., 2000; McAndrew et al., 2008). Dilution of the FtsZ2-1 antibody was 1:3,500 and the FtsZ2-2 antibody was 1:1,000. Sectioning of the wax-embedded samples was conducted on an 820 Spencer rotatory microtome (American Optical) with the thickness set at 5 µm. Alexa Fluor 488-conjugated goat antirabbit secondary antibody (1:500 dilution; lot no. 514957, Invitrogen) was used in this study. The fluorescence signals were detected under a DMRA2 microscope (Leica) equipped with a 100× oil immersion objective (HCX PL FLUOTAR; NA 1.30–0.60) and a charge-coupled device camera (Retiga Exi, QImaging), as described in a previous study (Chen et al., 2019). Filters L5 (480 nm excitation/505 nm emission) and TX2 (560 nm excitation/595 nm emission) were used to observe Alexa Fluor 488 and chlorophyll fluorescence, respectively. Exposure times of 400 ms and z stacks with a 0.5-µm interval were used for both channels. All images were captured with Image-Pro Plus 7.0 software (Media Cybernetics). Nearest-neighbor deconvolution with 70% haze removal was conducted for all obtained images. Projection of the z stacks was carried out using Fiji (ImageJ) software (http://fiji.sc/Fiji) with the maximum intensity algorithm. False colors are indicated in the figure legends.
Protein Extraction, Quantification, and Immunoblot Analysis
About 40 mg of expanding leaf tissue from 28-d-old plants was collected in 2-mL SealRite microcentrifuge tubes (USA Scientific) containing two stainless steel 3.2-mm beads (BioSpecProducts) and frozen in liquid nitrogen. The tissue was ground to powder using an automated Retsch TissueLyser II bead mill (Qiagen). The samples were suspended in 2× sample buffer (0.25 m Tris-HCl, pH 6.8, 4.2% [w/v] SDS, 5% [v/v] glycerol, 200 mm dithiothreitol, and 0.0006% [w/v] bromophenol blue), boiled for 5 min, and centrifuged at 10,000g for 10 min. The pellet was discarded, and soluble protein extract was placed on ice or stored at −80°C. Protein concentration was quantified using Pierce 660 nm Protein Assay Reagent (Pierce Biotechnology). Ten micrograms of protein were separated by SDS-PAGE on 10% (w/v) polyacrylamide gels and transferred to 0.45 μm NitroBind membrane (GE Water and Process Technologies) using a Genie Apparatus (Idea Scientific Company). The membrane was dried for 30 min at room temperature before incubation with anti-FtsZ2-2 antibody (McAndrew et al., 2008) diluted 1:12,000 or anti-FtsZ2-1 antibody (Stokes et al., 2000) diluted 1:16,000 and anti-FtsZ1-1 antibody (Stokes et al., 2000) diluted 1:20,000 in Tris-buffered saline plus Tween 20 (TBST; 0.2 m Tris-HCl, pH 7.4, 0.8 m NaCl, and 0.1% [v/v] Tween 20) containing 2% (w/v) nonfat dry milk. After washing with TBST, blots were incubated for 2 h with horseradish peroxidase-conjugated goat antirabbit secondary antibody (Pierce Biotechnology) diluted 1:10,000 in TBST containing 2% (w/v) nonfat dry milk. Following washes in TBST, the signal was detected using Super Signal West Pico Chemiluminescent Substrate or Super Signal West Dura Extended Duration Substrate (Pierce Biotechnology) and recorded on blue-sensitive autoradiography film (Denville Scientific). Following signal detection, membranes were stained with Ponceau S as a loading control.
DNA Constructs and Plant Transformation
The DNA constructs for complementation were made using the Gateway system (Invitrogen). Genomic DNA from rosette leaves of 28-d-old plants of Cvi-1 and Ler-0 was isolated using the Wizard Genomic DNA Purification Kit (Promega). A 4.4 kb genomic fragment containing ∼2.4 kb of the FtsZ2-2 coding region and ∼1 kb flanking the start and stop codons was amplified from genomic DNA using ExTaq polymerase (Clontech) and the primers GWAt3g32750F and GWAt3g32750R (Supplemental Table S3). PCR was performed under conditions described in Supplemental Methods. The PCR products were cloned into pDONOR207, verified by sequencing, and subsequently cloned into pMDC123 to create Cvi-FtsZ2-2 or Ler-FtsZ2-2 for complementation. The vectors were introduced into Agrobacterium tumefaciens strain C58CIRifR containing pGV3101 via electroporation. The ftsZ2-2 (SALK 050397) mutant plants (McAndrew et al., 2008) were transformed by floral dipping (Clough and Bent, 1998). Transformants were selected on plates containing 10 mg/L of the herbicide BASTA (glufosinate ammonium; Crescent Chemical Company).
Isolation of RNA and cDNA Cloning
Total RNA from Cvi-1 and Ler-0 was isolated from rosette leaves of 15-d-old plants using the RNeasy Plant Mini Kit (Qiagen). Reverse transcription (RT) was performed using the GoScript Reverse Transcription System (Promega) with oligo(dT)15 annealed to 1 µg total RNA. Following first-strand cDNA synthesis, PCR amplification was done using FtsZ2-2 gene-specific primers KS 24F and DK 9R (Supplemental Table S3) and ExTaq DNA Polymerase (Clontech) to generate a partial (without the 3′ UTR) FtsZ2-2 cDNA from Cvi-1 and Ler-0. PCR was performed under conditions described in Supplemental Methods. RT-PCR products were separated on agarose gels containing ethidium bromide and visualized using an Azure Biosystems C600 Imager (Azure Biosystems) at subsaturation settings. Both partial FtsZ2-2 cDNAs were cloned into pGEM-T Easy vector (Promega) and sequenced.
RT-qPCR
Total RNA from rosette leaves of 15-d-old plants was isolated as described above. First-strand cDNA was synthesized from 1 μg total RNA using Thermo Scientific Maxima H Minus First Strand cDNA Synthesis kit (K1651) with the oligo dT primer provided. RT-qPCR was performed in 96-well plates (Applied Biosystems, MicroAmp Fast Optical 96-well reaction plate with barcode, 0.1 mL, 4346906) using the Applied Biosystems 7500 Fast PCR System, with the Hot Start-IT SYBR Green qPCR Master mix (2×; USB Affymetrix, 75762). In brief, 0.5 μL of a 1:5 dilution of the cDNA, 0.5 μL 10 μm forward and reverse primer, 5 μL Hot Start-IT SYBR Green qPCR Master mix, and 3.5 μL water containing 0.7% of the passive reference dye ROX (Affymetrix) were combined in a total of 10 μL reaction volume. The gene-specific primers qPCR Z2-2 F and qPCR Z2-2 R (Supplemental Table S3) were used to determine FtsZ2-2 transcript levels. Primers for the gene encoding Protein Phosphatase 2A Subunit A3 (PP2AA3 [At1g13320]), qPCR PPAA3F, and qPCR PPAA3R (Supplemental Table S3; Huot et al., 2017), were used to determine the transcript level of the constitutively expressed reference gene. Each reaction was performed with two technical replicates and two biological replicates, except for one accession (Kly-4), and the averages were reported. Melt curve analysis was performed on each reaction after the initial amplification to confirm specificity of the PCR reaction. Average FtsZ2-2 transcript levels were presented relative to the average in Col-0.
Molecular Markers and Fine Mapping
For fine mapping, molecular markers detecting polymorphisms between Ler-0 and Cvi_0 (CVI_0.SALK) were developed based on the genomic sequences available at the 1001 Genomes database available at the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/atg1001/3.0/gebrowser.php).We generated one insertion-deletion and 11 cleaved amplified polymorphic sequence (Neff et al., 2002) molecular markers (Supplemental Table S3). Genomic DNA from rosette leaves of 28-d-old plants of the 92 NILs and their Ler-0 and Cvi-1 parents was isolated using the Wizard Genomic DNA Purification Kit (Promega) and genotyped with molecular markers. PCR was performed under conditions described in Supplemental Methods.
Analysis of FtsZ Sequences of Arabidopsis Accessions in the 1001 Genomes Database
The FtsZ2-2, FtsZ2-1, and FtsZ1-1 full-genomic DNA sequences were generated with the VCF (variant-calling format) records archived in the 1001 Genomes project at 1001genomes.org (1001 Genomes Consortium, 2016) using the FastaAlternateReferenceMaker function in GATK (McKenna et al., 2010). Multiple sequence alignments for each gene were performed with MUSCLE (Edgar, 2004). Coding sequence alignments were extracted and translated into protein sequences using customized Python scripts with the BioPython package (Cock et al., 2009). AAPs were annotated using a customized Python script. dN/dS ratio values were calculated by parsing the SnpEff (Cingolani et al., 2012) output using a customized Python script. The barplot was generated using the R package ggplot2 (Wickham, 2016). dN/dS values were all significantly different from each other (P < 0.0001), as determined by the Tukey adjustment for multiple comparisons at α = 0.05.
Accession Numbers
Sequences for these genes can be found in The Arabidopsis Information Resource database (https://www.arabidopsis.org/) under the following names and accession numbers: Col-0 FtsZ2-2, AT3G52750; Col-0 FtsZ2-1, AT2G36250; and Col-0 FtsZ1-1, AT5G55280. The sequence for the Cvi-1 FtsZ2-2 gene can be found in GenBank under accession number MN401146.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Chloroplast areas in Cvi-1 and Ler-2 grown under different conditions and harvested at different times.
Supplemental Figure S2. Diagram of Cvi-1 introgressions in Ler-0 in 92 lines of the Ler-0 x Cvi-1 NIL population.
Supplemental Figure S3. Characterization of NILs with Cvi-1 introgressions in chromosome 1.
Supplemental Figure S4. Fine-mapping of the region of chromosome 3 conferring the Cvi-1-like large-chloroplast phenotype in the Ler-0 x Cvi-1 NIL population.
Supplemental Figure S5. Nucleotide polymorphisms between the Cvi-1 and Ler-0/Col-0 alleles of FtsZ2-2.
Supplemental Figure S6. Chloroplast morphology phenotype of At3g52760 mutant SAIL_61_A02.
Supplemental Figure S7. Additional transgenic Col-0 ftsZ2-2 mutants expressing the Ler-FtsZ2-2 or Cvi-FtsZ2-2 transgenes.
Supplemental Figure S8. Additional images showing immunofluorescence staining of FtsZ2 proteins in mesophyll cells of Ler-0 and Cvi-1.
Supplemental Figure S9. Comparison of Cvi-0 and Cvi-1 phenotypes.
Supplemental Figure S10. Chloroplast phenotypes in accessions representing different FtsZ2-2 polymorphic groups.
Supplemental Figure S11. Chloroplast size and FtsZ protein phenotypes in representative accessions from different polymorphic groups.
Supplemental Table S1. List of RIL and NIL population parent accessions and ABRC seed stocks used for analysis.
Supplemental Table S2. Ler-0 x Cvi-1 NIL population seed stocks used for analysis.
Supplemental Table S3. Primers used in this study.
Supplemental Methods. PCR reaction conditions.
Supplemental Dataset S1. FtsZ2-2 amino acid sequence variants in Arabidopsis accessions in the 1001 Genomes database.
Supplemental Dataset S2. FtsZ2-2 AAPs in Arabidopsis accessions in the 1001 Genomes database.
Supplemental Dataset S3. FtsZ2-1 amino acid sequence variants in Arabidopsis accessions in the 1001 Genomes database.
Supplemental Dataset S4. FtsZ2-1 AAPs in Arabidopsis accessions in the 1001 Genomes database.
Supplemental Dataset S5. FtsZ1-1 amino acid sequence variants in Arabidopsis accessions in the 1001 Genomes database.
Supplemental Dataset S6. FtsZ1-1 AAPs in Arabidopsis accessions in the 1001 Genomes database.
ACKNOWLEDGMENTS
We thank Joy Bergelson for providing the TAD 04 seeds, Maria Magallanes-Lundback for providing seeds from the 1001 Genomes collection, and Joost Keurentjes for genotype information on the NIL population. We thank Miyah Williams, Amber Bedore, Matthew Mills, and Emily Graham for help with chloroplast phenotyping, Emily Jennings for assistance with alignments, and Shin-han Shiu, Christopher Oakley, and members of the Osteryoung laboratory for helpful discussions.
Footnotes
This work was supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences (grant no. DE-FG02-06ER15808 to K.W.O.) and by the National Science Foundation (grant no. 1719376 to K.W.O.).
Articles can be viewed without a subscription.
References
- 1001 Genomes Consortium (2016) 1,135 Genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadabadi M, Bock R (2012) Plastid division and morphology in the genus Peperomia. Biol Plant 56: 301–306 [Google Scholar]
- Alonso-Blanco C, Aarts MG, Bentsink L, Keurentjes JJ, Reymond M, Vreugdenhil D, Koornneef M (2009) What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21: 1877–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Peeters AJ, Koornneef M, Lister C, Dean C, van den Bosch N, Pot J, Kuiper MT (1998) Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J 14: 259–271 [DOI] [PubMed] [Google Scholar]
- Anderson JM, Chow WS, Goodchild DJ (1988) Thylakoid membrane organisation in sun/shade acclimation. Aust J Plant Physiol 15: 11–26 [Google Scholar]
- Anderson JM, Goodchild DJ, Boardman NK (1973) Composition of the photosystems and chloroplast structure in extreme shade plants. Biochim Biophys Acta 325: 573–585 [DOI] [PubMed] [Google Scholar]
- Austin J II, Webber AN (2005) Photosynthesis in Arabidopsis thaliana mutants with reduced chloroplast number. Photosynth Res 85: 373–384 [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Agrawal M, Michael TP, Wessinger C, Maloof JN, Clark R, Warthmann N, Chory J, Weigel D (2006) The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat Genet 38: 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett ME, Whipple CJ (2013) Protein change in plant evolution: Tracing one thread connecting molecular and phenotypic diversity. Front Plant Sci 4: 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle A, Khan Z, Wang SH, Mitrano A, Ford MJ, Pritchard JK, Gilad Y (2015) Genomic variation. Impact of regulatory variation from RNA to protein. Science 347: 664–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi EF, Lutkenhaus J (1991) FtsZ ring structure associated with division in Escherichia coli. Nature 354: 161–164 [DOI] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA (2010) The gene balance hypothesis: Implications for gene regulation, quantitative traits and evolution. New Phytol 186: 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkman O, Boardman NK, Anderson JM, Thorne SW, Goodchild DJ, Pyliotis NA (1971) Effect of light intensity during growth of Atriplex patula on the capacity of photosynthetic reactions, chloroplast components and structure. Year B Carnegie Inst Wash 71: 115–135 [Google Scholar]
- Boardman NK. (1977) Comparative photosynthesis of sun and shade plants. Annu Rev Plant Physiol Plant Mol Biol 28: 355–377 [Google Scholar]
- Buschiazzo E, Ritland C, Bohlmann J, Ritland K (2012) Slow but not low: Genomic comparisons reveal slower evolutionary rate and higher dN/dS in conifers compared to angiosperms. BMC Evol Biol 128: 1471–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Gunther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Chen C, Cao L, Yang Y, Porter KJ, Osteryoung KW (2019) ARC3 activation by PARC6 promotes FtsZ-Ring remodeling at the chloroplast division site. Plant Cell 31: 862–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, MacCready JS, Ducat DC, Osteryoung KW (2018) The molecular machinery of chloroplast division. Plant Physiol 176: 138–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al. (2007) Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317: 338–342 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, et al. (2009) Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25: 1422–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colletti KS, Tattersall EA, Pyke KA, Froelich JE, Stokes KD, Osteryoung KW (2000) A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr Biol 10: 507–516 [DOI] [PubMed] [Google Scholar]
- Dengler NG, Donnelly PM, Dengler RE (1996) Differentiation of bundle sheath, mesophyll, and distinctive cells in the C-4 grass Arundinella hirta (Poaceae). Am J Bot 83: 1391–1405 [Google Scholar]
- Du S, Lutkenhaus J (2017) Assembly and activation of the Escherichia coli divisome. Mol Microbiol 105: 177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Cruz JA, Imran SM, Chen J, Kramer DM, Osteryoung KW (2017) Variations in chloroplast movement and chlorophyll fluorescence among chloroplast division mutants under light stress. J Exp Bot 68: 3541–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Cruz JA, Jiao Y, Chen J, Kramer DM, Osteryoung KW (2015) Non-invasive, whole-plant imaging of chloroplast movement and chlorophyll fluorescence reveals photosynthetic phenotypes independent of chloroplast photorelocation defects in chloroplast division mutants. Plant J 84: 428–442 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Din El-Assal S, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M (2001) A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet 29: 435–440 [DOI] [PubMed] [Google Scholar]
- Ellis JR, Leech RM (1985) Cell size and chloroplast size in relation to chloroplast replication in light-grown wheat leaves. Planta 165: 120–125 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, Anderson DE, Osawa M (2010) FtsZ in bacterial cytokinesis: Cytoskeleton and force generator all in one. Microbiol Mol Biol Rev 74: 504–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kafafi E-S, Karamoko M, Pignot-Paintrand I, Grunwald D, Mandaron P, Lerbs-Mache S, Falconet D (2008) Developmentally regulated association of plastid division protein FtsZ1 with thylakoid membranes in Arabidopsis thaliana. Biochem J 409: 87–94 [DOI] [PubMed] [Google Scholar]
- Filek M, Gzyl-Malcher B, Zembala M, Bednarska E, Laggner P, Kriechbaum M (2010) Effect of selenium on characteristics of rape chloroplasts modified by cadmium. J Plant Physiol 167: 28–33 [DOI] [PubMed] [Google Scholar]
- Filiault DL, Wessinger CA, Dinneny JR, Lutes J, Borevitz JO, Weigel D, Chory J, Maloof JN (2008) Amino acid polymorphisms in Arabidopsis phytochrome B cause differential responses to light. Proc Natl Acad Sci USA 105: 3157–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Yoshida S (2001) Chloroplast targeting of chloroplast division FtsZ2 proteins in Arabidopsis. Biochem Biophys Res Commun 287: 462–467 [DOI] [PubMed] [Google Scholar]
- Gargano D, Maple-Grødem J, Møller SG (2012) In vivo phosphorylation of FtsZ2 in Arabidopsis thaliana. Biochem J 446: 517–521 [DOI] [PubMed] [Google Scholar]
- Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, Farber CR, Sinsheimer J, Kang HM, Furlotte N, et al. (2011) Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet 7: e1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn JM, Yang Y, Vitha S, Schmitz AJ, Hemmes M, Miyagishima SY, Osteryoung KW (2009) PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant J 59: 700–711 [DOI] [PubMed] [Google Scholar]
- Grosche C, Rensing SA (2017) Three rings for the evolution of plastid shape: A tale of land plant FtsZ. Protoplasma 254: 1879–1885 [DOI] [PubMed] [Google Scholar]
- Günther T, Schmid K (2010) Deleterious amino acid polymorphisms in Arabidopsis thaliana and rice. Theor Appl Genet 121: 157–168 [DOI] [PubMed] [Google Scholar]
- Hansen BG, Halkier BA, Kliebenstein DJ (2008) Identifying the molecular basis of QTLs: eQTLs add a new dimension. Trends Plant Sci 13: 72–77 [DOI] [PubMed] [Google Scholar]
- Hoballah ME, Gubitz T, Stuurman J, Broger L, Barone M, Mandel T, Dell’Olivo A, Arnold M, Kuhlemeier C (2007) Single gene-mediated shift in pollinator attraction in Petunia. Plant Cell 19: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda SI, Hongladarom-Honda T, Kwanyuen P, Wildman SG (1971) Interpretations on chloroplast reproduction derived from correlations between cells and chloroplasts. Planta 97: 1–15 [DOI] [PubMed] [Google Scholar]
- Hu J, Wang Y, Fang Y, Zeng L, Xu J, Yu H, Shi Z, Pan J, Zhang D, Kang S, et al. (2015) A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant 8: 1455–1465 [DOI] [PubMed] [Google Scholar]
- Huot B, Castroverde CDM, Velasquez AC, Hubbard E, Pulman JA, Yao J, Childs KL, Tsuda K, Montgomery BL, He SY (2017) Dual impact of elevated temperature on plant defence and bacterial virulence in Arabidopsis. Nat Commun 8: 1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh R, Fujiwara M, Nagata N, Yoshida S (2001) A chloroplast protein homologous to the eubacterial topological specificity factor minE plays a role in chloroplast division. Plant Physiol 127: 1644–1655 [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, Lopez-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14: 787–802 [DOI] [PubMed] [Google Scholar]
- Jellings AJ, Leech RM (1984) Anatomical variation in first leaves of nine Tritucum genotypes, and its relationship to photosynthetic capacity. New Phytol 96: 371–382 [Google Scholar]
- Jellings AJ, Usher MB, Leech RM (1983) Variation in the chloroplast to cell area index in Deschampsia antarctica along a 16° latitudinal gradient. Brit Antarct Surv Bull 61: 13–20 [Google Scholar]
- Jeong WJ, Park Y-I, Suh K, Raven JA, Yoo OJ, Liu JR (2002) A large population of small chloroplasts in tobacco leaf cells allows more effective chloroplast movement than a few enlarged chloroplasts. Plant Physiol 129: 112–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Johnson CB, Tang LK, Smith AG, Ravichandran A, Luo Z, Vitha S, Holzenburg A (2013) Single particle tracking analysis of the chloroplast division protein FtsZ anchoring to the inner envelope membrane. Microsc Microanal 19: 507–512 [DOI] [PubMed] [Google Scholar]
- Juenger TE, Wayne T, Boles S, Symonds VV, McKay J, Coughlan SJ (2006) Natural genetic variation in whole-genome expression in Arabidopsis thaliana: The impact of physiological QTL introgression. Mol Ecol 15: 1351–1365 [DOI] [PubMed] [Google Scholar]
- Karamoko M, El-Kafafi E-S, Mandaron P, Lerbs-Mache S, Falconet D (2011) Multiple FtsZ2 isoforms involved in chloroplast division and biogenesis are developmentally associated with thylakoid membranes in Arabidopsis. FEBS Lett 585: 1203–1208 [DOI] [PubMed] [Google Scholar]
- Keurentjes JJ, Bentsink L, Alonso-Blanco C, Hanhart CJ, Blankestijn-De Vries H, Effgen S, Vreugdenhil D, Koornneef M (2007) Development of a near-isogenic line population of Arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics 175: 891–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Königer M, Delamaide JA, Marlow ED, Harris GC (2008) Arabidopsis thaliana leaves with altered chloroplast numbers and chloroplast movement exhibit impaired adjustments to both low and high light. J Exp Bot 59: 2285–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa H, Mori T, Takahara M, Miyagishima S, Kuroiwa T (2002) Chloroplast division machinery as revealed by immunofluorescence and electron microscopy. Planta 215: 185–190 [DOI] [PubMed] [Google Scholar]
- Le Corre V, Roux F, Reboud X (2002) DNA polymorphism at the FRIGIDA gene in Arabidopsis thaliana: Extensive nonsynonymous variation is consistent with local selection for flowering time. Mol Biol Evol 19: 1261–1271 [DOI] [PubMed] [Google Scholar]
- Leech RM, Baker NR (1983) The development of photosynthetic capacity in leaves In Dale JE, and Milthorpe FL, eds, The Growth and Functioning of Leaves. Cambridge University Press, Cambridge, United Kingdom, pp 271–307 [Google Scholar]
- Li Y, Hsin J, Zhao LY, Cheng YW, Shang WN, Huang KC, Wang HW, Ye S (2013a) FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation. Science 341: 392–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ren B, Ding L, Shen Q, Peng S, Guo S (2013b) Does chloroplast size influence photosynthetic nitrogen use efficiency? PLoS One 8: e62036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK, Buschmann C, Döll M, Fietz H-J, Bach T, Kozel U, Meier D, Rahmsdorf U (1981) Photosynthetic activity, chloroplast ultrastructure, and leaf characteristics of high-light and low-light plants and of sun and shade leaves. Photosynth Res 2: 115–141 [DOI] [PubMed] [Google Scholar]
- Loudet O, Saliba-Colombani V, Camilleri C, Calenge F, Gaudon V, Koprivova A, North KA, Kopriva S, Daniel-Vedele F (2007) Natural variation for sulfate content in Arabidopsis thaliana is highly controlled by APR2. Nat Genet 39: 896–900 [DOI] [PubMed] [Google Scholar]
- Löwe J, Amos LA (1998) Crystal structure of the bacterial cell-division protein FtsZ. Nature 391: 203–206 [DOI] [PubMed] [Google Scholar]
- Ma X, Ehrhardt DW, Margolin W (1996) Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA 93: 12998–13003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Margolin W (1999) Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J Bacteriol 181: 7531–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloof JN, Borevitz JO, Dabi T, Lutes J, Nehring RB, Redfern JL, Trainer GT, Wilson JM, Asami T, Berry CC, et al. (2001) Natural variation in light sensitivity of Arabidopsis. Nat Genet 29: 441–446 [DOI] [PubMed] [Google Scholar]
- Maple J, Aldridge C, Møller SG (2005) Plastid division is mediated by combinatorial assembly of plastid division proteins. Plant J 43: 811–823 [DOI] [PubMed] [Google Scholar]
- Maple J, Vojta L, Soll J, Møller SG (2007) ARC3 is a stromal Z-ring accessory protein essential for plastid division. EMBO Rep 8: 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAndrew RS, Froehlich JE, Vitha S, Stokes KD, Osteryoung KW (2001) Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol 127: 1656–1666 [PMC free article] [PubMed] [Google Scholar]
- McAndrew RS, Olson BJ, Kadirjan-Kalbach DK, Chi-Ham CL, Vitha S, Froehlich JE, Osteryoung KW (2008) In vivo quantitative relationship between plastid division proteins FtsZ1 and FtsZ2 and identification of ARC6 and ARC3 in a native FtsZ complex. Biochem J 412: 367–378 [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010) The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima S, Takahara M, Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T (2001) Plastid division is driven by a complex mechanism that involves differential transition of the bacterial and eukaryotic division rings. Plant Cell 13: 2257–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima SY, Nakanishi H, Kabeya Y (2011) Structure, regulation, and evolution of the plastid division machinery. Int Rev Cell Mol Biol 291: 115–153 [DOI] [PubMed] [Google Scholar]
- Monroe JG, Powell T, Price N, Mullen JL, Howard A, Evans K, Lovell JT, McKay JK (2018) Drought adaptation in Arabidopsis thaliana by extensive genetic loss-of-function. eLife 7: 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Kuroiwa H, Takahara M, Miyagishima S, Kuroiwa T (2001) Visualization of an FtsZ ring in chloroplasts of Lilium longiflorum leaves. Plant Cell Physiol 42: 555–559 [DOI] [PubMed] [Google Scholar]
- Neff MM, Turk E, Kalishman M (2002) Web-based primer design for single nucleotide polymorphism analysis. Trends Genet 18: 613–615 [DOI] [PubMed] [Google Scholar]
- Nielsen R. (2005) Molecular signatures of natural selection. Annu Rev Genet 39: 197–218 [DOI] [PubMed] [Google Scholar]
- Oliva MA, Cordell SC, Lowe J (2004) Structural insights into FtsZ protofilament formation. Nat Struct Mol Biol 11: 1243–1250 [DOI] [PubMed] [Google Scholar]
- Olson BJ, Wang Q, Osteryoung KW (2010) GTP-dependent heteropolymer formation and bundling of chloroplast FtsZ1 and FtsZ2. J Biol Chem 285: 20634–20643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung KW, Pyke KA (2014) Division and dynamic morphology of plastids. Annu Rev Plant Biol 65: 443–472 [DOI] [PubMed] [Google Scholar]
- Osteryoung KW, Stokes KD, Rutherford SM, Percival AL, Lee WY (1998) Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung KW, Vierling E (1995) Conserved cell and organelle division. Nature 376: 473–474 [DOI] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu SH (2016) Evolution of gene duplication in plants. Plant Physiol 171: 2294–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisupati R, Reichardt I, Seren U, Korte P, Nizhynska V, Kerdaffrec E, Uzunova K, Rabanal FA, Filiault DL, Nordborg M (2017) Verification of Arabidopsis stock collections using SNPmatch, a tool for genotyping high-plexed samples. Sci Data 4: 170184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Ganguly D, Albrecht-Borth V (2015) Insights into chloroplast biogenesis and development. Biochim Biophys Acta 1847: 1017–1024 [DOI] [PubMed] [Google Scholar]
- Possingham J. (1973) Effect of light quality on chloroplast replication in spinach. J Exp Bot 24: 1247–1260 [Google Scholar]
- Possingham JV, Hashimoto H, Oross J (1988) Factors that influence plastid division in higher plants In Boffey SA, and Lloyd D, eds, Division and Segregation of Organelles. Cambridge University Press, Cambridge, United Kingdom, pp 1–20 [Google Scholar]
- Possingham JV, Saurer W (1969) Changes in chloroplast number per cell during leaf development in spinach. Planta 86: 186–194 [DOI] [PubMed] [Google Scholar]
- Pyke K. (1997) The genetic control of plastid division in higher plants. Am J Bot 84: 1017–1027 [PubMed] [Google Scholar]
- Pyke KA. (1999) Plastid division and development. Plant Cell 11: 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA, Leech RM (1987) The control of chloroplast number in wheat mesophyll cells. Planta 170: 416–420 [DOI] [PubMed] [Google Scholar]
- Pyke KA, Leech RM (1991) Rapid image analysis screening procedure for identifying chloroplast number mutants in mesophyll cells of Arabidopsis thaliana (L.) Heynh. Plant Physiol 96: 1193–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA, Leech RM (1992) Chloroplast division and expansion is radically altered by nuclear mutations in Arabidopsis thaliana. Plant Physiol 99: 1005–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JS. (1981) The anatomy and taxonomy of protein structure. Adv Protein Chem 34: 167–339 [DOI] [PubMed] [Google Scholar]
- Schmitz AJ, Glynn JM, Olson BJ, Stokes KD, Osteryoung KW (2009) Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development. Mol Plant 2: 1211–1222 [DOI] [PubMed] [Google Scholar]
- Shindo C, Aranzana MJ, Lister C, Baxter C, Nicholls C, Nordborg M, Dean C (2005) Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol 138: 1163–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stata M, Sage TL, Rennie TD, Khoshravesh R, Sultmanis S, Khaikin Y, Ludwig M, Sage RF (2014) Mesophyll cells of C4 plants have fewer chloroplasts than those of closely related C3 plants. Plant Cell Environ 37: 2587–2600 [DOI] [PubMed] [Google Scholar]
- Stokes KD, McAndrew RS, Figueroa R, Vitha S, Osteryoung KW (2000) Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2 genes in Arabidopsis. Plant Physiol 124: 1668–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strepp R, Scholz S, Kruse S, Speth V, Reski R (1998) Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc Natl Acad Sci USA 95: 4368–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swid N, Nevo R, Kiss V, Kapon R, Dagan S, Snir O, Adam Z, Falconet D, Reich Z, Charuvi D (2018) Differential impacts of FtsZ proteins on plastid division in the shoot apex of Arabidopsis. Dev Biol 441: 83–94 [DOI] [PubMed] [Google Scholar]
- Takemura K, Kamachi H, Kume A, Fujita T, Karahara I, Hanba YT (2017) A hypergravity environment increases chloroplast size, photosynthesis, and plant growth in the moss Physcomitrella patens. J Plant Res 130: 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush AD, MacCready JS, Chen C, Ducat DC, Osteryoung KW (2018) Conserved dynamics of chloroplast cytoskeletal FtsZ proteins across photosynthetic lineages. Plant Physiol 176: 295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush AD, Osteryoung KW (2012) Distinct functions of chloroplast FtsZ1 and FtsZ2 in Z-ring structure and remodeling. J Cell Biol 199: 623–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush AD, Yoshida Y, Osteryoung KW (2013) FtsZ in chloroplast division: Structure, function and evolution. Curr Opin Cell Biol 25: 461–470 [DOI] [PubMed] [Google Scholar]
- Tsuji H, Naito K, Hatakeyama I, Ueda K (1979) Benzyladenine-induced increase in DNA content per cell, chloroplast size, and chloroplast number per cell in intact bean leaves. J Exp Bot 30: 1145–1151 [Google Scholar]
- Vitha S, Froehlich JE, Koksharova O, Pyke KA, van Erp H, Osteryoung KW (2003) ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15: 1918–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S, McAndrew RS, Osteryoung KW (2001) FtsZ ring formation at the chloroplast division site in plants. J Cell Biol 153: 111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise SE, Carr DJ, Bourke AM, Hanson DT, Swarthout D, Sharkey TD (2015) The arc mutants of Arabidopsis with fewer large chloroplasts have a lower mesophyll conductance. Photosynth Res 124: 117–126 [DOI] [PubMed] [Google Scholar]
- Wickham H. (2016) Ggplot2: Elegant graphics for data analysis, Ed 2 Springer, Houston, Texas [Google Scholar]
- Xiong D, Huang J, Peng S, Li Y (2017) A few enlarged chloroplasts are less efficient in photosynthesis than a large population of small chloroplasts in Arabidopsis thaliana. Sci Rep 7: 5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder DW, Kadirjan-Kalbach D, Olson BJSC, Miyagishima SY, DeBlasio SL, Hangarter RP, Osteryoung KW (2007) Effects of mutations in arabidopsis FtsZ1 on plastid division, FtsZ ring formation and positioning, and FtsZ filament morphology in vivo. Plant Cell Physiol 48: 775–791 [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Mogi Y, TerBush AD, Osteryoung KW (2016) Chloroplast FtsZ assembles into a contractible ring via tubulin-like heteropolymerization. Nat Plants 2: 16095. [DOI] [PubMed] [Google Scholar]
- Zhang M, Hu Y, Jia J, Li D, Zhang R, Gao H, He Y (2009) CDP1, a novel component of chloroplast division site positioning system in Arabidopsis. Cell Res 19: 877–886 [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang J, Huang J, Lan H, Wang C, Yin C, Wu Y, Tang H, Qian Q, Li J, et al. (2012) Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc Natl Acad Sci USA 109: 21534–21539 [DOI] [PMC free article] [PubMed] [Google Scholar]