A sequence of physiological, morphological, and molecular events in aging plants suggests that roots undergo a genetically determined developmental program typical of organ senescence.
Abstract
Aging-related processes in plant tissues are associated with changes in developmental and physiological processes relevant for stress tolerance and plant performance. While senescence-regulated processes have been extensively characterized in leaves, they remain poorly described in roots. Here, we investigated the physiological processes and molecular determinants underlying the senescence of seminal roots in hydroponically grown barley (Hordeum vulgare). Transcriptome profiling in apical and basal root tissues revealed that several NAC-, WRKY-, and APETALA2 (AP2)-type transcription factors were upregulated just before the arrest of root elongation, when root cortical cell lysis and nitrate uptake, as well as cytokinin concentrations ceased. At this time point, root abscisic acid levels peaked, suggesting that abscisic acid is involved in root aging-related processes characterized by expression changes of genes involved in oxidative stress responses. This temporal sequence of aging-related processes in roots is highly reminiscent of typical organ senescence, with the exception of evidence for the retranslocation of nutrients from roots. Supported by the identification of senescence-related transcription factors, some of which are not expressed in leaves, our study indicates that roots undergo an intrinsic genetically determined senescence program, predominantly influenced by plant age.
In plants, age-dependent degenerative processes have been widely studied in readily accessible plant organs, particularly in leaves, but also in flowers, fruits, or specialized organs such as nodules (Oeller et al., 1991; Cam et al., 2012; Wu et al., 2017; Grosskinsky et al., 2018). These studies have built the concept of organ senescence, which defines a genetically determined developmental program following the completion of growth, with the major purpose to recycle nutrients within cells, organs, or whole plants (Lim et al., 2007). Surprisingly, roots have rarely been considered for senescence-related studies, although root functions are critical to overcome age- or stress-related disorders but cease with plant age (Schippers et al., 2015). Also, in agricultural plant production, aging-related processes determine the lifespan of a physiologically active root and thus postflowering nutrient and water uptake, which is a highly relevant but poorly characterized agronomic trait.
Even though the progression of senescence varies among organs or species and strongly depends on environmental factors (Navas et al., 2003), several morphological and physiological processes appear typical. In leaves, senescence starts with chlorosis, resulting from breakdown of chlorophyll and dismantling of chloroplasts, allowing proteins like Rubisco to be engulfed in vesicles and transported to the vacuole for proteolysis (Ishida et al., 2008). Protein catabolism is generally associated with a decrease in total amino acids, as many of these are translocated to sink organs. However, some amino acids, especially Trp and Phe, accumulate and partly serve as precursors, e.g. for the synthesis of melatonin and serotonin or of salicylic acid (SA), which act as plant growth regulators in leaf senescence (Morris et al., 2000; Kang et al., 2009; Erland et al., 2015). Senescence-induced decomposition of macromolecules is further accompanied by enhanced mobilization of mineral elements, in particular of nitrogen (N), phosphorus (P), potassium (K), or iron (Fe; Himelblau and Amasino, 2001). Remobilization of these nutrients requires metabolic activity to bring these minerals into phloem-mobile forms, e.g. via the induction of phosphatases to release phosphate from nucleic acids or via the synthesis of nicotianamine and phytosiderophores to chelate metals for phloem loading (Shi et al., 2012; Veneklaas et al., 2012). At the cellular level, senescence induces autophagy, an intracellular trafficking and degradation pathway guiding organelles or proteins to the vacuole for proteolysis and nutrient recycling (Avila-Ospina et al., 2014). With progressing leaf age, plasma membranes eventually disintegrate and allow uncontrolled efflux of cellular metabolites and constituents (Kim et al., 2009).
Morphological and physiological events occurring during leaf senescence are under control of a complex regulatory network that coordinates responses to environmental factors with the plant developmental program and involves transcriptional regulators as well as phytohormones (Schippers et al., 2015). In Arabidopsis (Arabidopsis thaliana), high-resolution temporal profiling of gene expression during leaf senescence and clustering of coexpressed genes revealed a chronological sequence of distinct senescence-associated processes (Breeze et al., 2011). Transcriptional regulation of leaf senescence is dominated by NAC- and WRKY-type transcription factors and marked by an early increase in genes involved in reactive oxygen species (ROS) formation or in abscisic acid (ABA), ethylene, and SA synthesis and signaling. Further analyses showed that leaf senescence is governed by individual leaf age as well as by the developmental stage of the whole plant (Zentgraf et al., 2004; Miao et al., 2013; Bohner et al., 2015). This concept has been expanded by the hypothesis that initiation and progression of leaf senescence depend on previous experiences of a leaf and thus likely differ from leaf to leaf, while reflecting the developmental stage of the entire plant (Kim et al., 2018). Unlike aboveground organs, such type of age-related studies in roots is scarce. Microscopic analysis of basal root segments from graminaceous species showed progressing lysis of the cortical cell layer with age (Schneider et al., 2017). This so-called root cortical senescence appears associated with a decrease of P and N concentrations as well as of root respiration. How these morphological changes temporally integrate into a sequence of root aging-related processes remains to be elucidated.
So far, aging-related studies in plant roots have described changes of individual root properties over time, such as root browning, the decrease in nutrient uptake rate, or the degeneration of individual cell layers (Eissenstat et al., 2000; Bingham, 2007; Schneider and Lynch, 2018). It has thus remained unclear whether these age-related processes in roots are biologically linked and coordinated by a developmental program or just the consequence of apoptosis in individual cell types (Kosslak et al., 1997; Schneider and Lynch, 2018). Here, we tested the hypothesis whether aging-related processes in roots reflect typical senescence-related processes, such as those described in other plant organs. Our root zone-specific phytohormone and transcriptome studies indicate that the majority of aging-related processes are governed by plant age rather than by tissue age. Dynamic changes in metabolites, phytohormones, and gene expression identify candidates triggering or regulating root senescence. Thereby, this study allows composing a root-specific chronological sequence of physiological and developmental processes that govern root senescence in barley (Hordeum vulgare).
RESULTS
Morphological Changes during Seminal Root Aging
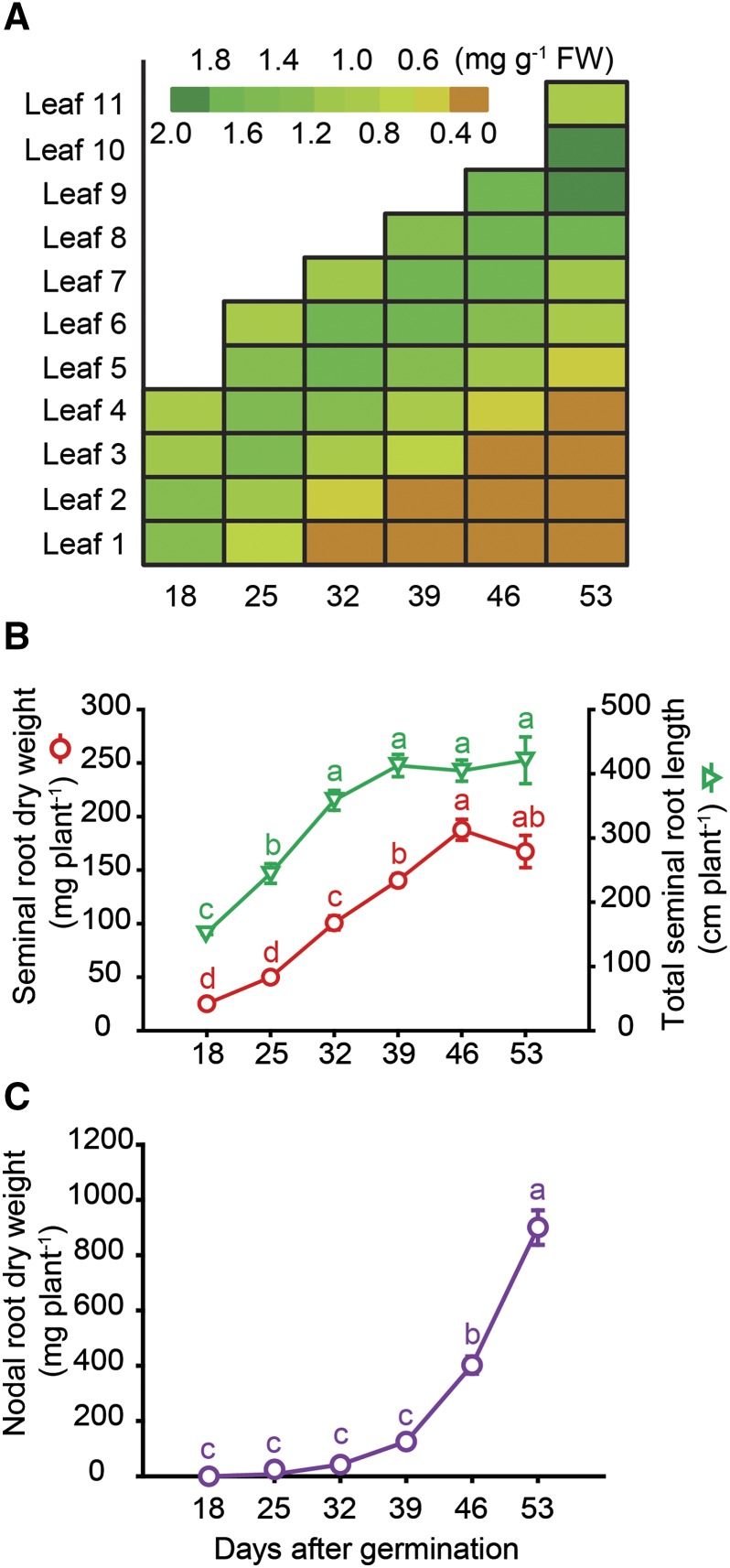
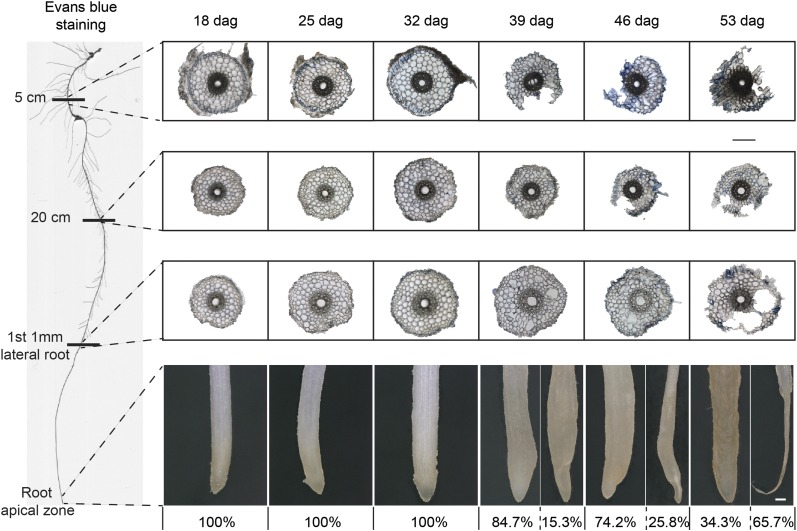
The root system of barley plants consists of embryonic and postembryonic roots. Embryonic roots, referred to as seminal roots in graminaceous species, are formed during embryogenesis. Postembryonic roots are formed after germination and include lateral roots and nodal roots, which develop from the basal part of the shoot (Naz et al., 2014). Respecting that root types differ in development and nodal roots proliferate progressively over time, we focused here on aging-related processes only in seminal, i.e. embryonic roots. We cultivated barley plants in hydroponics over a period of almost 8 weeks and harvested seminal roots on a weekly basis between days 18 and 53. At the same time points, individual leaves of the main stem were subjected to chlorophyll analysis, showing that in the first leaf, chlorophyll levels decreased from 25 d after germination (dag), while in each of the subsequent leaves, loss of chlorophyll set in approximately 1 week later (Fig. 1A). Nevertheless, shoots appeared visually green until 53 dag, as the main stem was covered by newly formed tillers (Supplemental Fig. S1). Seminal root biomass increased almost linearly between 18 and 46 dag and then leveled off, while seminal root elongation began to stagnate at 39 dag (Fig. 1B). This was also the time point when root browning set in (Supplemental Fig. S1). As root browning has been found to correlate with the reduction of nutrient uptake and root respiration (Baldi et al., 2010), root browning has been widely used as a phenotypical trait to describe the transition of a physiologically active to an inactive or dead root (Comas et al., 2000). The first nodal roots emerged approximately 25 dag, while strongest proliferation was observed around 46 dag, followed by nodal root browning at 53 dag (Supplemental Fig. S1). Meanwhile, nodal root dry weight increased almost exponentially (Fig. 1C), justifying our approach to dismiss this root type from characterizing aging-related processes. A closer look at the root tips of the seminal roots revealed that at 39 dag, 15.3% of the seminal root tips were degraded (Fig. 2). This proportion of degraded seminal root tips increased up to 65.7% at final harvest.
Figure 1.
Development of leaf chlorosis and growth of seminal and nodal roots with progressing plant age. A, Color-coded heat map of chlorophyll concentrations in individual leaves (numbers 1–11) of the main tiller. B, Dry mass and total length of the seminal root system. C, Dry mass of the nodal root system. Hydroponically grown barley plants were harvested weekly from 18 to 53 d after germination (dag). Symbols represent means ± se. Different letters indicate significant differences among means according to Tukey’s test at P < 0.05 (n = 5 independent biological replicates).
Figure 2.
Structural changes in the seminal root tissue and phenotype of root tips with progressing plant age. Light microscopy of root cross sections at 5 cm below the hypocotyl, 20 cm below the hypocotyl, or at the position where the first lateral root had emerged to a maximum length of 1 mm. Roots of hydroponically grown barley plants were examined weekly from 18 to 53 d after germination (dag). To test membrane integrity, seminal roots were stained with Evans blue. Percentage (%) indicates the abundance of degraded apical root tips. Representative images of root tips with percent values of intact and degraded phenotypes at a given plant age (n ≥ 70 independent biological replicates). Bars = 200 µm.
To investigate whether structural changes were responsible for arrested elongation and root tip decay, we inspected cross sections of different root zones by light microscopy. Initially, seminal root tips were intact and whitish, but Evans blue staining, indicating plasma membrane damage, showed that already 18 dag, some epidermal cells in the most basal root section were leaky (Fig. 2). From 39 dag on, root tips turned brown and outer cell layers of the basal root zone degraded. With plant age, this degradation process progressed acropetally with largest lesions observed in the cortex, most likely reflecting root cortical senescence (Schneider et al., 2017). At 39 dag, epidermal cell leakage had partially reached the apical root zone, coinciding with the onset of tip degradation in individual roots (Fig. 2). As a reference for the developmental stage of the main stem, we also inspected the spike meristem. Between 18 and 25 dag, the spike meristem had passed the double-ridge stage and progressed until the awn primordium stage at 53 dag (Supplemental Fig. S2), indicating that the apical shoot meristem of the main tiller had not yet formed a major sink organ. Taken together, seminal roots underwent degenerative processes including root browning, arrested elongation, and partial cell decay, which set in between 32 and 39 dag, i.e. when nodal roots began their exponential growth.
Physiological and Metabolic Rearrangements during Seminal Root Aging
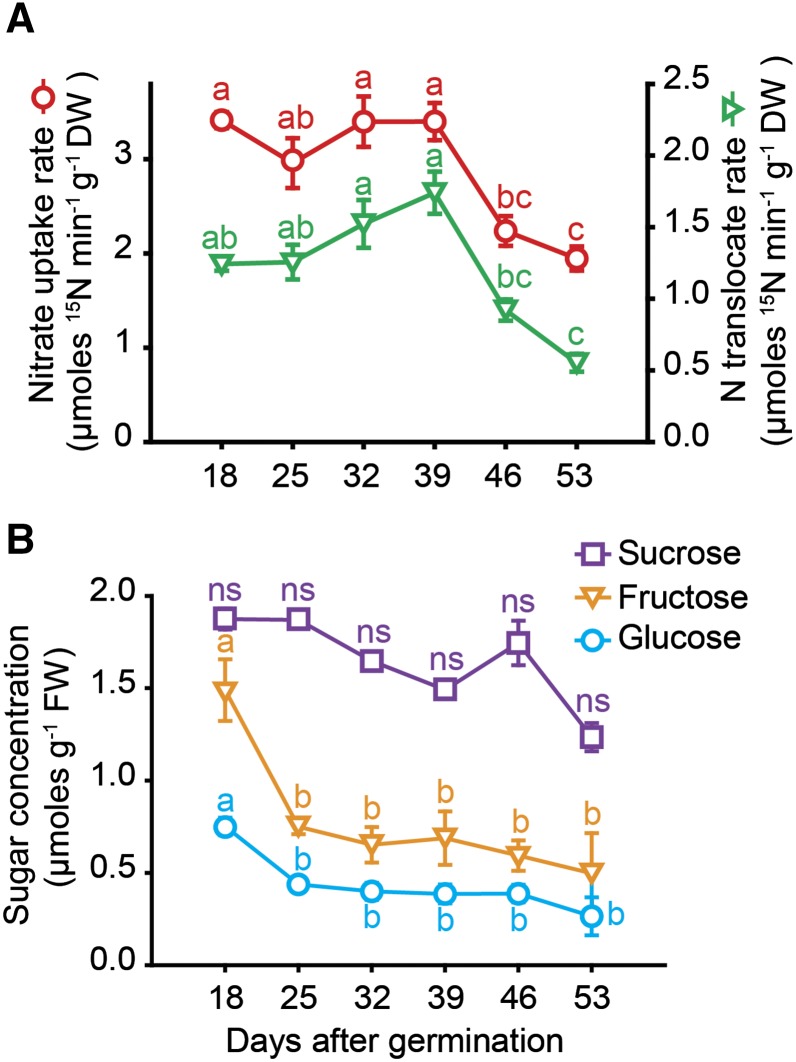
As a fundamental physiological root function, nutrient uptake was assessed in seminal roots of progressing age. Between 18 and 39 dag, nitrate uptake rate was constant at approximately 3 μmol min−1 g−1 dry weight, before a significant drop set in (Fig. 3A). In parallel, root-to-shoot translocation of 15N slightly increased until 39 dag but sharply declined afterward. In part, this decline in 15N translocation was a direct consequence of decreasing nitrate uptake rates. We hypothesized that this decline in nitrate uptake and translocation resulted from sugar depletion, since this type of root activity strongly relies on assimilate supply (Lejay et al., 2003). Although carbohydrate analysis in seminal roots revealed an initial decline of Fru and Glc between 18 and 25 dag, followed by a phase of constant levels, Suc concentrations did not decrease significantly (Fig. 3B). As these changes did not temporally coincide with the decline in nitrate uptake rate, it was assumed that assimilate depletion in roots was unlikely the cause for the decreasing root activity.
Figure 3.
Uptake and translocation of nitrate and sugar levels in seminal roots with progressing plant age. A, Nitrate uptake rate and root-to-shoot translocation rate of labeled nitrogen. Seminal roots of different plant age were exposed to 1 mm 15N-labeled KNO3 for 20 min. B, Concentrations of Suc, Fru, and Glc in seminal roots of hydroponically grown barley plants that were examined weekly from 18 to 53 d after germination (dag). Symbols represent means ± se. ns, not significant. Different letters indicate significant differences among means according to Tukey’s test at P < 0.05 (n = 6 independent biological replicates).
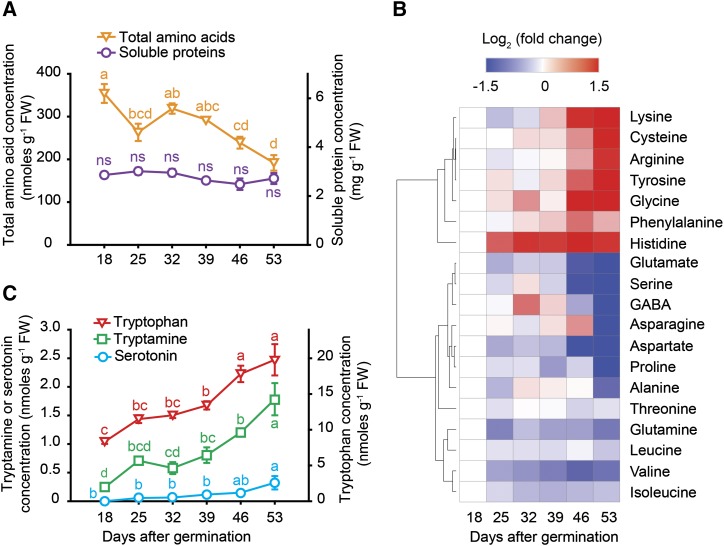
Leaf senescence is typically associated with protein degradation (Hörtensteiner, 2009). In seminal roots, however, the concentration of soluble proteins remained rather stable despite progressing plant age (Fig. 4A). In contrast, the concentration of total amino acids decreased, which was mainly attributed by a gradual decrease in Glu, Asp, and Ser but less in Asn and Gln (Fig. 4, A and B). At the same time, levels of Cys, Gly, Lys, Arg, and of the aromatic amino acids Tyr, Phe, and Trp increased (Fig. 4, B and C), indicating profound changes in amino acid metabolism and suggesting a possible requirement of aromatic amino acids as precursors for the synthesis of secondary metabolites (Watanabe et al., 2013). In a downstream pathway, the Trp-derived metabolites serotonin and its precursor tryptamine gradually accumulated during aging of seminal roots (Fig. 4C), which was highly reminiscent of age-dependent changes during leaf senescence, as serotonin has been shown to increase in senescing rice (Oryza sativa) leaves (Kang et al., 2009).
Figure 4.
Plant age-dependent profiles of proteins, amino acids, and amines in seminal roots with progressing plant age. A, Soluble protein and total amino acid concentrations. B, Log2-fold change of individual amino acid concentrations represented in a heat map. C, Concentrations of Trp, tryptamine, and serotonin. GABA, Gamma-aminobutyric acid; FW, fresh weight. Symbols represent means ± se. ns, not significant. Different letters indicate significant differences among means according to Tukey’s test at P < 0.05 (n = 5 independent biological replicates). For the heat map, amino acids concentrations are referred to their levels at 18 d after germination (dag) and resulting fold changes were log2 transformed (log2FC) before being clustered according to the k-means clustering method.
Changes in Mineral Element Contents during Root Aging
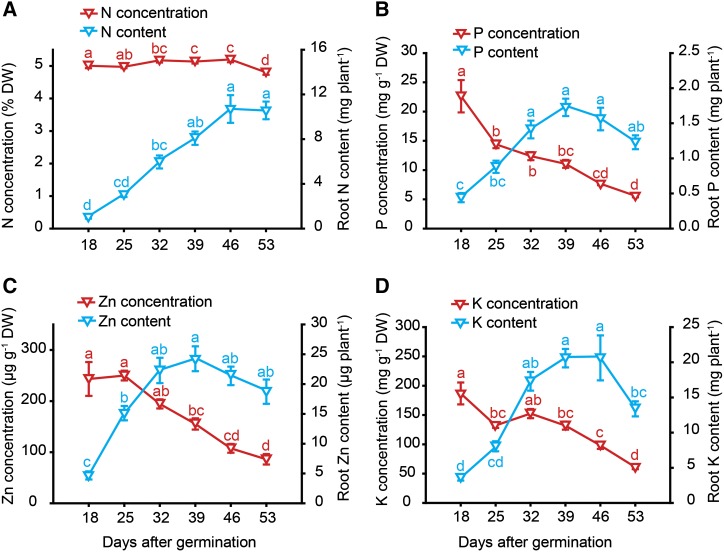
To address the question whether nutrient remobilization, the central biological function of leaf senescence, also occurs during root aging, seminal roots were subjected to elemental analysis. From 18 dag on, concentrations of most mineral elements decreased, except for N that remained constantly at 5% and decreased just slightly at 53 dag (Fig. 5A; Supplemental Fig. S3). Decreasing nutrient concentrations were mostly related to a dilution effect, because root biomass increased steadily until 46 dag (Fig. 1B). Nutrient content, which is defined as the total element amount of a given tissue and reflected here the whole nutrient pool in seminal roots, increased at least until 39 dag for every element. From then on, P and zinc (Zn) contents tended to decrease, and with a delay also those of K (Fig. 5, B–D), suggesting that seminal roots may have remobilized a part of these elements during aging. Most other essential elements, including sulfur (S), Fe, copper (Cu), or molybdenum (Mo), which were reported to decrease by up to 40% in senescing leaves (Himelblau and Amasino, 2001), did not show any substantial decrease of their contents, while the nutritional status of these elements in seminal roots—as reflected by their concentrations—remained adequate and largely unaffected by plant age (Supplemental Fig. S3). Thus, clear evidence for nutrient remobilization out of roots was not obtained.
Figure 5.
Plant age-dependent profiles of nutrient concentrations and contents in seminal roots with progressing plant age. Concentrations and of contents of nitrogen (A), phosphorus (B), zinc (C), and potassium (D) in seminal roots of hydroponically grown barley plants that were examined weekly from 18 to 53 d after germination (dag). Root element contents represent the total amount in the seminal root biomass. Symbols represent means ± se. Different letters indicate significant differences among means according to Tukey’s test at P < 0.05 (n = 6 independent biological replicates).
Plant-Age- and Tissue-Age-Dependent Phytohormone Dynamics during Root Aging
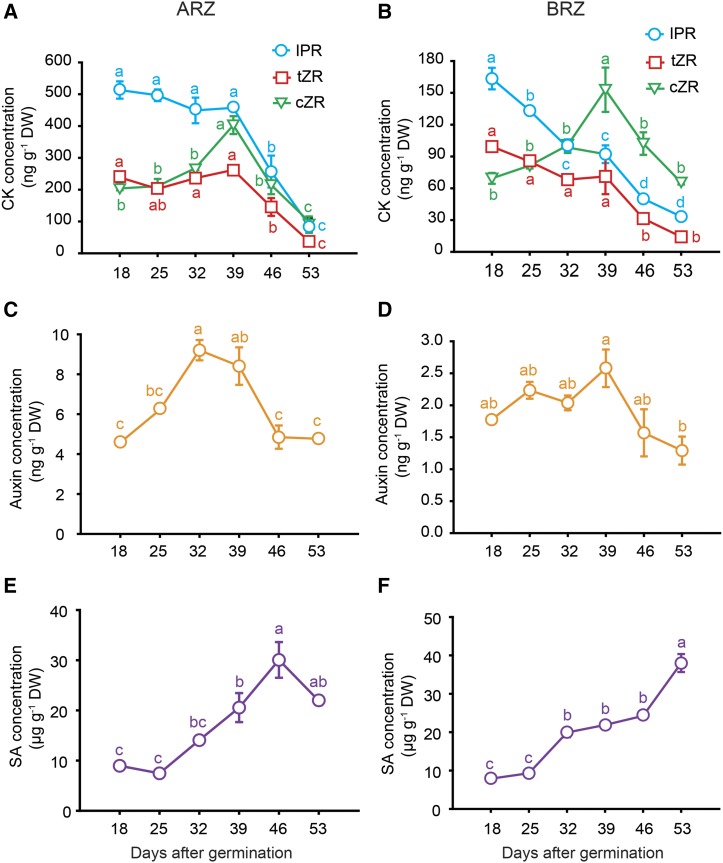
As the degradation of epidermal and cortical root cells set in earlier than that of inner root cells (Fig. 2), one may speculate that seminal roots degrade in a tissue-age-dependent manner. On the other hand, incipient root tip decay from 39 dag (Fig. 2) suggested that this degradation process is not controlled by tissue age but rather by plant age. We therefore harvested root tissue separately from the basal root zone (BRZ) and apical root zone (ARZ). In view of the fact that the majority of cells in these root samples represent differentiated, mitotically inactive cells, we refer here to postmitotic senescence processes (Gan and Amasino, 1997; Woo et al., 2013). Whereas BRZ samples reflected progressing tissue age and plant age, continuous proliferation of new cells in the growing ARZ resulted in constant tissue age over time despite progressing plant age. Both fractions were first subjected to phytohormone analysis, since phytohormones take over critical roles in regulating leaf senescence. In general, auxin and cytokinin (CK) concentrations were much higher in the ARZ than in the BRZ (Fig. 6, A–D), which likely related to the fact that the root apical meristem is a major sink for rootward transported auxin and a major site of CK biosynthesis. Among the CKs, nonribosylated CKs were apparently below the detection limits, whereas three ribosylated transport forms were detected, which all showed a marked decline at 46 and 53 dag in either tissue. In the BRZ, transzeatin-riboside (tZR) and isopentenyladenosine decreased continuously from 18 dag on, while cis-zeatin-riboside (cZR) peaked at 39 dag (Fig. 6, A and B). Thus, synthesis of cZR, which represents a form of weaker physiological activity (Heyl et al., 2006), or its conversion from tZR apparently increased until this time point before degradation set in or synthesis of all ribosylated CK forms slowed down. Concentrations of auxin, which has been described as a negative regulator of leaf senescence (Kim et al., 2011), increased between 18 and 32 dag in the ARZ, while it remained constant in the BRZ, before dropping sharply at 46 dag (Fig. 6, C and D). Concentrations of SA, which regulates a subset of senescence-promoting genes in leaves (Morris et al., 2000), showed no such transition point but accumulated steadily during root aging in the BRZ and until 46 dag also in the ARZ (Fig. 6, E and F).
Figure 6.
Hormone profiles in apical and basal root zones with progressing plant age. Concentrations of cytokinin ribosides (CK; A and B), auxin (C and D), and SA (E and F) in the apical (ARZ) or basal root zone (BRZ) of hydroponically grown barley plants sampled weekly from 18 to 53 d after germination (dag). DW, Dry weight. Symbols represent means ± se. Different letters indicate significant differences among means according to Tukey’s test at P < 0.05 (n = 5 independent biological replicates). IPR, Isopentenyladenosin; tZR, transzeatin riboside; cZR, cis-zeatin riboside.
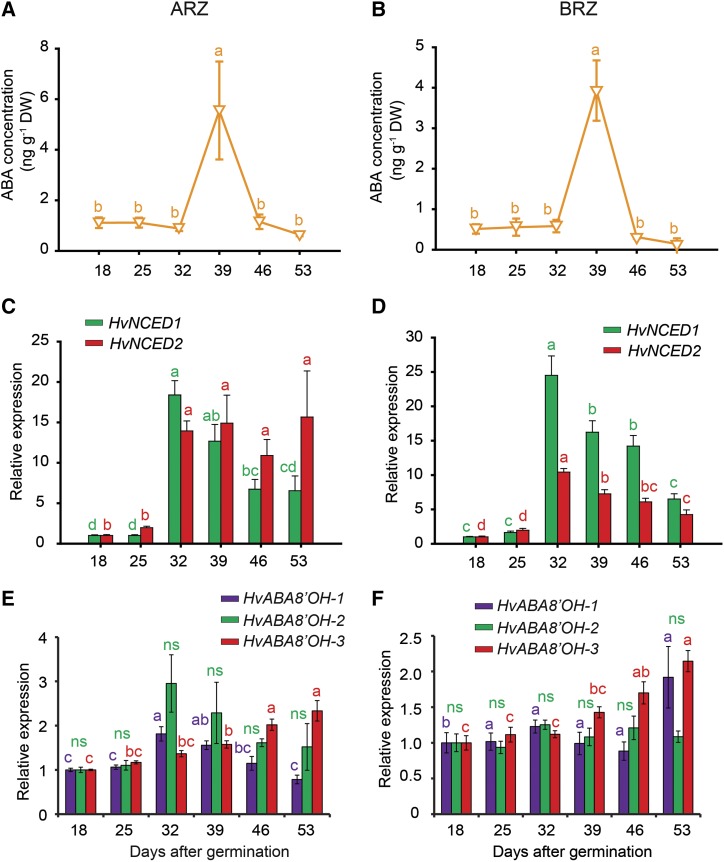
Remarkably, ABA levels exhibited a completely different temporal pattern during root aging. ABA concentrations in the ARZ and BRZ were in general low but showed a pronounced peak at 39 dag with approximately 4-fold higher levels than at any other day (Fig. 7, A and B). This striking ABA peak was associated with a massive accumulation of transcript levels of the 9-cis-EPOXYCAROTENOID DIOXYGENASE (NCED) genes HvNCED1 and HvNCED2 from 32 dag on (Fig. 7, C and D). These genes encode the rate-limiting enzymes for ABA biosynthesis in barley (Seiler et al., 2011). In the ARZ, 32 dag was also the starting point for an increase in transcript levels of ABA8′-HYDROXYLASE1 (ABA8′-OH-1) and ABA8′-OH-2, while ABA8′-OH-3 increased later in both root fractions (Fig. 7, E and F). These genes are involved in ABA degradation, and their transcript levels are known to be induced by elevated levels of ABA (Kushiro et al., 2004). Thus, enhanced transcript levels of ABA biosynthesis and degradation genes at day 32 were preceding or concomitant with the ABA peak at 39 dag when auxin and CK levels dropped, suggesting a marked transition in the phytohormonal regulation of developmental and physiological processes in apical as well as BRZs.
Figure 7.
Hormone profile and relative expression of genes involved in ABA metabolism with progressing plant age. Concentrations of ABA (A and B); transcript levels of barley 9-CIS-EPOXYCAROTENOID DIOXYGENASE (HvNCED1 and HvNCED2; C and D), and of ABSCISIC ACID 8′-HYDROXYLASE (HvABA8′OH-1, HvABA8′OH-2, and HvABA8′OH-3; E and F) in the apical (ARZ) and basal root zone (BRZ) of plants sampled weekly from 18 to 53 d after germination (dag). Relative expression values of target genes were normalized to transcript levels of UBIQUITIN C (AY220735.1), and values at 18 dag were set to 1. DW, Dry weight. Triangles and bars represent means ± se. ns, not significant. Different letters indicate significant differences among means according to Tukey’s test at P < 0.05 (n = 5 independent biological replicates).
Plant-Age- and Tissue-Age-Dependent Transcriptome Analysis of Roots
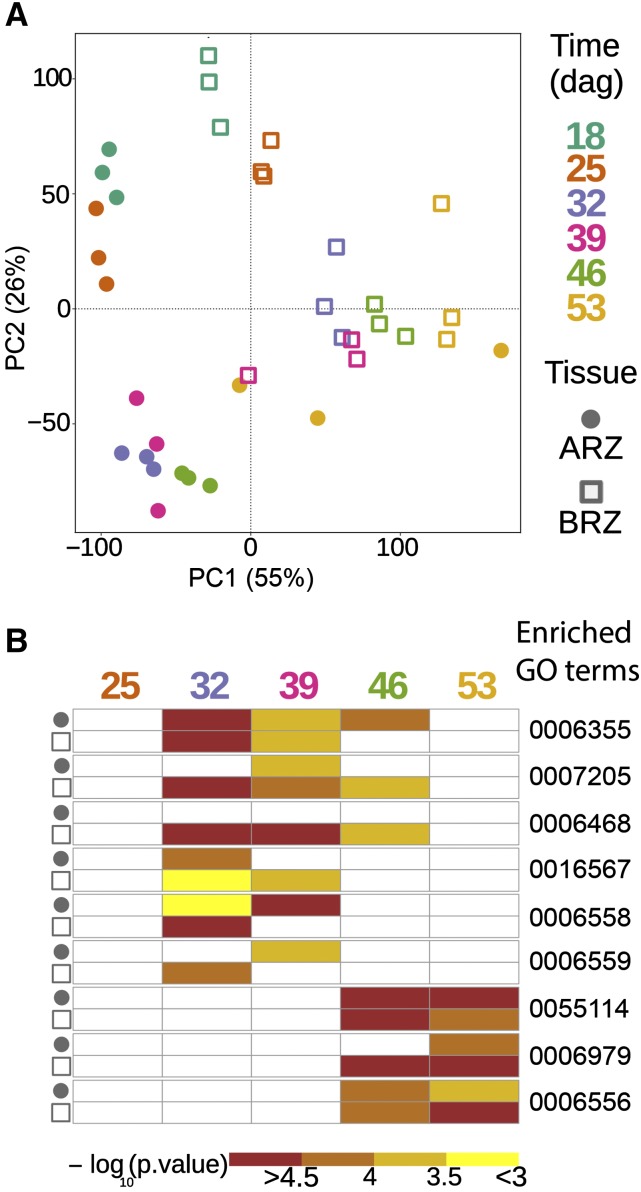
The two root tissues, ARZ and BRZ, were then subjected separately to microarray analysis. Raw feature intensities were background corrected to normalize between arrays. Before calculating differential gene expression, the normalized data were subjected to principal component analysis, which showed gradual changes in transcriptome profiles over time (Fig. 8A). In both tissues, age-dependent transcriptome changes separated along PC2 between 18 and 32 dag but along PC1 from 32 to 54 dag, suggesting a qualitative shift in transcriptome responses at 32 dag. We then took expression values of individual transcripts at 18 dag as reference for calculating relative changes at later time points and conducted a nonsupervised cluster analysis. This approach revealed that the top three gene clusters of the BRZ and ARZ showed their most prominent changes at 32 dag (Supplemental Fig. S4), supporting day 32 as most prominent transition point for gene regulation in the BRZ and ARZ.
Figure 8.
Variation in transcriptomes and enrichment of gene ontology (GO) terms in genes expressed in the apical and basal root zone during seminal root aging. A, Principal component analysis (PCA) plot of 36 barley transcriptome datasets. B, Heat map of enriched GO terms in the apical and basal root zones (ARZ and BRZ, respectively) at each time point. Each data point in the PCA plot represents the transcriptome of either ARZ or BRZ samples at one of the six time points. ARZ tissues are marked with a circle and BRZ tissues with a square. Color corresponds to six different time points. In the GO term heat map, colors indicate −log10 transformed P values of each term. The complete list of enriched GO terms (Supplemental Tables S1–S5) was further filtered by two criteria to display the GO term heat map: (1) GO terms that are terminal nodes in the GO hierarchy in at least one time point of either ARZ or BRZ, and (2) the enriched GO term must be significant (P value < 0.05) in at least two out of 10 tissue-specific time points; dag, days after germination. GO:0006355, regulation of transcription, DNA-templated; GO:0007205, protein kinase C-activating G-protein coupled receptor signaling pathway; GO:0006468, protein phosphorylation; GO:0016567, protein ubiquitination. GO:0006558, l-Phe metabolic process; GO:0006559, l-Phe catabolic process; GO:0055114, oxidation-reduction process; GO:0006979, response to oxidative stress; GO:0006556, S-adenosyl-Met biosynthetic process. ARZ, Apical root zone; BRZ, basal root zone.
Employing GO enrichment analysis, we subsequently explored changes in the abundance of transcripts that are involved in known biological processes at different stages of root development. Based on all significantly enriched GO terms in the ARZ and BRZ (Supplemental Tables S1–S5), we focused on those that represented terminal nodes in the GO term hierarchy and appeared at least twice across all time points and tissues. At 25 dag, a relatively small number of genes showed altered expression, and only a few GO terms, including “response to acid chemical” and “response to water,” were significantly enriched in the BRZ, indicating that both tissues were still in a similar developmental or physiological state as at 18 dag (Fig. 8B; Supplemental Tables S1–S5). At 32 dag, several GO terms became significantly enriched in the two root tissues, in particular “regulation of transcription,” which remained enriched at 39 dag and in the ARZ even until 46 dag. In addition, both tissues were characterized by changes in the expression of genes involved in “protein ubiquitination” and “l-Phe metabolic process,” the latter coinciding with an increase in root Phe concentrations (Fig. 4B). The terms “l-protein phosphorylation” and “protein kinase C-activating G-protein coupled receptor signaling pathway” appeared preferentially in the BRZ. At 46 and 53 dag, both tissues were marked by a sudden enrichment of genes belonging to “oxidation-reduction process” and “response to oxidative stress,” together with the term “S-adenosyl-Met biosynthetic process” that was also significant in the BRZ one time point before (Fig. 8B; Supplemental Tables S1–S5). Taken together, principal component analysis, gene clustering, and GO term analysis revealed major quantitative and qualitative shifts in gene expression in both root tissues at 32 dag and another qualitative shift at 46 dag. Since these predominant changes in the transcriptome were common to the ARZ and BRZ, plant age rather than tissue age appeared responsible for this transition in global gene expression patterns.
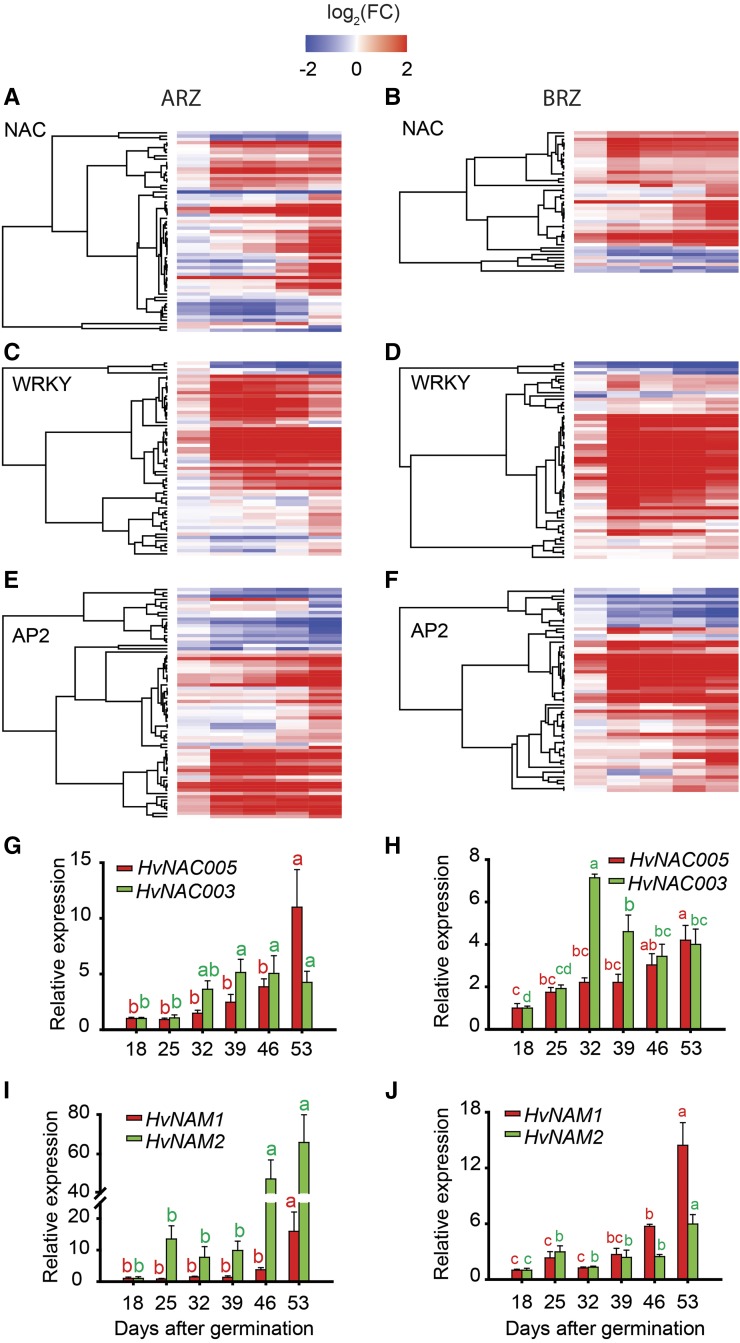
In leaves, the Cys protease gene SAG12 has been widely used as senescence marker (Guo et al., 2004). Transcript levels in roots of the putative barley ortholog HvSAG12/HvPAP17 strongly increased in the ARZ at 32 dag, whereas in the BRZ the closely related homolog HvPAP15 increased at 39 dag (Supplemental Fig. S5). Based on the highly significant enrichment of the term “regulation of transcription” at 32 dag in the ARZ and BRZ, we queried the transcriptome data for transcription factors constituting this GO term. In Arabidopsis and barley, onset of leaf senescence is characterized by up-regulation especially of NAC- and WRKY-type transcription factors (Balazadeh et al., 2008; Christiansen and Gregersen, 2014). Indeed, more than 50% of significantly altered NAC-, WRKY-, and AP2-type transcription factors were upregulated from 32 dag onwards (Fig. 9, A–F; Supplemental Tables S6–S11). We further verified transcript levels of a few of these transcription factors by quantitative PCR, including HvNAC005, HvNAC003, HvNAM1, and HvNAM2, which all were upregulated during root aging, albeit with slightly distinct temporal patterns (Fig. 9, G–J). Since these transcription factors or their close orthologs have been identified as leaf senescence regulators (Distelfeld et al., 2014; Christiansen et al., 2016; Mao et al., 2017), these observations emphasize that several transcriptional regulators, previously associated with leaf senescence, showed an age-dependent increase in expression also in roots. Moreover, according to the barley transcriptome atlas (https://webblast.ipk-gatersleben.de/barley_ibsc/), HvWRKY53, HvWRKY70-like, and two AP2-type transcription factors were upregulated in old versus young roots but not expressed in leaves (Supplemental Table S12). Based on changes in the GO term “response to oxidative stress” at 46 dag (Fig. 8B), we inspected genes belonging to this GO term and verified their deregulation in the BRZ and to a weaker extent also in the ARZ (Supplemental Fig. S6, A and B; Supplemental Tables S13 and S14). Representative for this group, the catalase-encoding genes HvCAT2 and HvCAT3 were found to be upregulated with progressing root age (Supplemental Fig. S6, C and D), which is reminiscent to the induction of catalase genes in senescing Arabidopsis leaves (Zimmermann et al., 2006).
Figure 9.
Relative expression of NAC-, WRKY-, and AP2-type transcription factors with progressing plant age. Heat maps of NAC-type (A and B), WRKY-type (C and D), and AP2-type (E and F) transcription factors in apical and basal root zones (ARZ, BRZ, respectively) of plants sampled weekly from 18 to 53 d after germination (dag). G to J, Relative transcript levels of HvNAC003 (G and H) and HvNAC005, and HvNAM1 and HvNAM2 (I and J), as determined by quantitative PCR. For heat maps, transcript levels at 18 dag were taken as references for later time points, and relative differences were log2-transformed (log2FC) and displayed along a color scale. Relative expression values of target genes were normalized to transcript levels of UBIQUITIN C (AY220735.1), and values at 18 dag were set to 1. Bars represent means ± se. Different letters indicate significant differences among means according to Tukey’s test at P < 0.05 (n = 5 independent biological replicates).
To further explore regulatory hubs for the transcriptional changes observed at 32 dag, we performed weighted gene coexpression network analysis (Zhang and Horvath, 2005). Using a threshold value of 0.8 for Pearson’s correlation coefficient between two genes yielded a total of 315 coexpressed genes (Supplemental Table S15). Out of the top five hubs, i.e. genes with most edges, three genes showed no clear homology to genes in other plant species and encoded proteins with unknown function (Supplemental Fig. S7). One hub was represented by a barley PREPHENATE DEHYDRATASE1 gene, which encodes the enzyme catalyzing prephenate to phenylpyruvate, the precursor of Phe (Warpeha et al., 2006). This agrees with accumulating Phe levels in aging roots (Fig. 4B). Another major hub encoded a putative ortholog of ZAT6 (ZINC FINGER OF ARABIDOPSIS6), which functions there as repressor of primary root growth (Devaiah et al., 2007) and promotes melatonin-mediated cold tolerance (Shi and Chan, 2014). These two examples may highlight the importance of metabolites in the regulation of aging-related processes in barley roots.
DISCUSSION
To date, aging-related studies in plant roots have remained rare and restricted to describe changes in individual root properties over time, such as root browning, the decrease in nutrient uptake rate, or the degeneration of individual cell layers (Eissenstat et al., 2000; Bingham, 2007; Schneider and Lynch, 2018). It has thus remained unclear whether and to what extent these processes are biologically linked. We examined metabolite, nutrient, and phytohormone levels simultaneously with structural changes of the root tissue and employed transcriptome profiling to uncover genes underlying the regulation of aging in seminal roots of barley. This study shows that age-dependent changes in several morphological, physiological, and molecular traits constitute a temporal sequence in the regulation of transcriptional and metabolic processes and are part of a concerted developmental program.
The Intrinsic Regulation of Seminal Root Senescence
We first tested the hypothesis of whether roots undergo organ senescence by examining in roots processes that are typical for leaf senescence: Leaf senescence is marked by chlorosis, disintegration of mesophyll cells, and declining integrity of plasma membranes (Dhindsa et al., 1982; Lim et al., 2007). Related processes were observed in roots. From 39 dag on, epidermal cells in the BRZ became leaky and subsequently epidermal and cortical cells began to disintegrate (Fig. 2). This process, which partly refers to root cortical senescence (Liljeroth, 1995; Bingham, 2007), expanded with progressing plant age along the seminal root axis toward the tip and went along with root browning and declining nitrate uptake activity (Figs. 2 and 3A; Supplemental Fig. S1). Although the expression of nitrate transporters strongly depends on photoassimilate supply from shoots (Lejay et al., 2003), sugar concentrations in roots remained constant (Fig. 3B) and unrelated to the progression of leaf chlorosis (Fig. 1A), indicating that, at least under our growth conditions, the onset of degeneration processes in roots was not under direct control of the shoot. Such a view agrees with leaf pruning experiments, which reduced sugar delivery to roots but did not affect the progression of root cortical senescence in wheat or of root browning in grape (Lascaris and Deacon, 1991; Comas et al., 2000). So far, root browning has been widely used as a phenotypical trait to describe the transition of a physiologically active to a senescing root, because, e.g. root browning correlates with the reduction of nutrient uptake and root respiration (Baldi et al., 2010). To date, there is no solid experimental data to demonstrate which metabolites are responsible for the brown color. Moreover, the study by Kosslak et al. (1997) has shown that root browning is linked with cell death and suppressed in a soybean mutant, while the underlying gene has not yet been identified. Moreover, at 39 dag, root tips degraded and overall seminal root elongation arrested (Figs. 1B and 2). Such early degeneration of the apical root tissue was unexpected, because growth conditions were still favorable for root development, as demonstrated by the continued increase in nodal root biomass (Fig. 1C). Hence, we conclude that these age-dependent degeneration processes are subject to intrinsic regulation by the seminal roots themselves and reflect organ-specific senescence.
Senescence processes in leaves proceed with plant age and tissue age (Zentgraf et al., 2004; Bohner et al., 2015). To dissect the impact of plant or tissue age on the regulation of root senescence, we subjected apical and basal root segments separately to phytohormone analysis. In contrast to basal root segments that integrate over plant and tissue age, all sampled apical segments were newly formed and of similar tissue age. In both root segments, changes in phytohormone concentrations went in parallel, in particular at 39 dag when ABA peaked and auxin and CK levels decreased sharply (Figs. 6 and 7). Since this time point of phytohormone transitions coincided with the decline in root activity and tissue integrity (Figs. 2 and 3A), we assume that peaking ABA and the drop in auxin and CK levels triggered the progression of morphological and physiological degeneration. In contrast to its regulatory role in leaf senescence (Morris et al., 2000), SA appeared unrelated to this hormonal transition point in roots as it increased gradually with plant age (Fig. 6, E and F). Considering further that apical root segments reflected the same symptoms of tissue degeneration despite their younger and relatively constant tissue age (Fig. 2), we concluded that plant age rather than tissue age had a dominant impact on the regulation of senescence-related processes in seminal roots.
Distinct and Common Features of Root versus Leaf Senescence
The major physiological purpose of leaf senescence is the recycling of nutrients. In leaves, nutrient remobilization is most pronounced for N and starts with the degradation of proteins, going along with decreasing concentrations of soluble proteins, amino acids, and total N, while less abundant amino acids and amines accumulate (Diaz et al., 2008; Kang et al., 2009; Bohner et al., 2015). Preceding is a marked increase in autophagy and in the expression of vacuolar Cys proteases (Martínez et al., 2007; Parrott et al., 2010), both critical factors for N remobilization (Avila-Ospina et al., 2014; Li et al., 2015). In roots, however, neither soluble protein nor total N concentrations showed a marked decrease (Figs. 4A and 5A). This may be related to the fact that roots lack highly abundant proteins like Rubisco that represent a major reservoir of protein-bound N undergoing massive degradation during senescence (Yoshida, 2003). In the barley plants examined here, roots remained continuously supplied with all mineral elements through continuous exchange of the nutrient solution, which may have suppressed nutrient retranslocation processes. Lacking up-regulation of autophagy-related genes in the root transcriptome supported this notion. Considering further that root contents of nutrients, except K, did not decline significantly toward final harvest (Fig. 5; Supplemental Fig. S3), it is obvious that nutrient retranslocation out of roots was quantitatively negligible or did not take place here.
With decreasing amino acid levels, barley roots also underwent profound changes in amino acid composition with progressing root age, most remarkably at 46 and 53 dag (Fig. 4B). A pronounced increase of Trp went along with enhanced concentrations of tryptamine and serotonin (Fig. 4C). In rice, serotonin synthesis increases in senescing rice leaves and, if further increased by overexpression of Trp decarboxylase, serotonin is able to attenuate leaf senescence, probably because of its antioxidant activity (Kang et al., 2009). Further evidence for raising oxidative stress arose from a massive deregulation of genes involved in oxidation-reduction reactions or in responses to oxidative stress (Fig. 8B). Among the latter were two members of the catalase family (Supplemental Fig. S6), which are induced typically by their substrate H2O2 (Xing et al., 2007). With the decay of H2O2, the corresponding enzymes contribute not only to the decrease of an early intracellular senescence signal (Zimmermann et al., 2006) but also to the detoxification of ROS. ROS accumulate during late stages of leaf senescence and induce apoptosis, which concludes the final phase of leaf senescence (Zimmermann and Zentgraf, 2005). Despite a weaker potential for ROS production in roots than in leaves, most likely because of the absence of light-induced ROS generation, this response of the root transcriptome to progressing plant age is reminiscent of ROS-mediated apoptosis in developmental leaf senescence (Lim et al., 2007). Thus, an oxidative burst may trigger also the decay of outer root cells and seminal root tips (Fig. 2), thereby marking the final phase of seminal root senescence in barley.
Global changes in the transcriptomes of the two root segments were highly similar, as four major gene clusters displayed the same pattern of age-dependent changes in gene expression (Supplemental Fig. S4). Besides clusters with continuously up- and down-regulated genes, a remarkably big cluster harbored genes being upregulated at 32 dag in both segments. Among these genes were prominent regulators of leaf senescence, such as NAC-, WRKY-, and AP2-type transcription factors. Many of these reached highest expression levels at 53 dag (Fig. 9; Supplemental Tables S6–S11), indicating that despite advanced degeneration of the cortex and epidermis, roots were still under active transcriptional control. Enhanced expression of genes like HvNAC005, HvNAC003, HvNAM1, and HvNAM2 (Fig. 9), themselves or their close orthologs being proven transcriptional regulators of leaf senescence (Uauy et al., 2006; Distelfeld et al., 2014; Christiansen et al., 2016), implies their involvement in transcriptional control of senescence processes also in roots. In addition, roots expressed a number of NAC-, WRKY-, and AP2-type transcription factors, such as HvWRKY53 that show no presence in young or senescing leaves (Supplemental Table S12). Hence, we expect that several of these are involved in the onset of senescence-initiating processes intrinsic to seminal roots.
A Chronological Sequence of Senescence Processes in Barley Seminal Roots
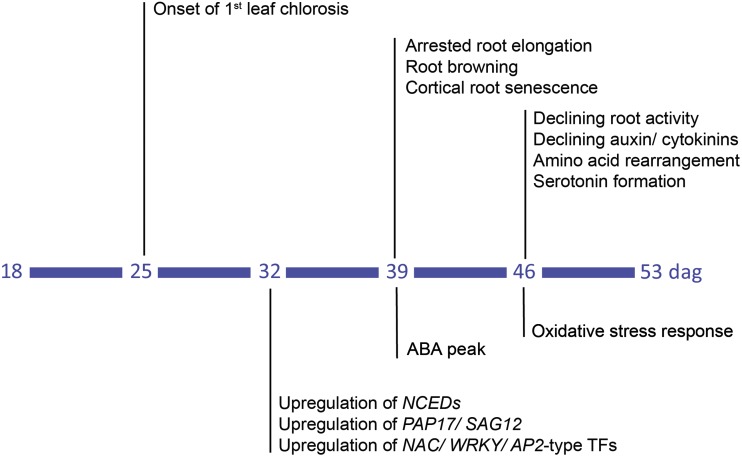
Based on the present results, we propose as an early regulatory event in the initiation of root senescence the up-regulation NAC-, WRKY-, and AP2-type transcription factors, such as HvNAC003 (Figs. 9 and 10). Their up-regulation goes along with the expression of HvSAG12/HvPAP17 and of HvNCED1 and HvNCED2 that, with a delay in time, lead to an ABA peak (Fig. 7; Supplemental Fig. S5). Such a temporal cascade is highly reminiscent of the NAC-ABA-SAG regulatory module that is induced by OsNAC2 in senescing rice leaves (Mao et al., 2017). Overexpression of the transcription factor OsNAC2, a homolog of HvNAC003, accelerates the onset of leaf senescence by binding to promoters of genes involved in chlorophyll degradation and in ABA biosynthesis (Christiansen et al., 2016; Mao et al., 2017). Hormonal signaling by ABA may additionally be involved in ceasing root elongation and root activity, and subsequently, in the decrease of auxin and CK levels (Figs. 1B, 3, and 6; Rowe et al., 2016). As the ABA peak is simultaneously generated in the ARZ and BRZ (Fig. 7), it is most likely under control of plant age and may act as a physiological signal reinforcing further senescence-related processes in roots (Fig. 10). Such second-stage processes likely involve an oxidative burst and related oxidative stress responses that predominate metabolic activities in both root zones (Fig. 8B; Supplemental Fig. S6). Oxidative stress may then accelerate the degradation of outer root cells and contribute to the decay of root apices (Fig. 2). At current stage, the above-described scenario provides only a crude frame for the chronological sequence of senescence-related processes in roots, as more frequent root sampling is required to define senescence-related processes at higher resolution. In contrast to previous studies assigning age-dependent degenerative processes in roots to sudden cell death (Kosslak et al., 1997; Schneider and Lynch, 2018), the present work now allows adding roots to those plant organs that undergo a senescence program.
Figure 10.
Temporal sequence of morphological, physiological, and molecular changes with progressing plant age. Following onset of chlorosis in the first leaf at 25 d after germination (dag), 32 dag is marked by enhanced expression of transcription factors and genes involved in protein degradation and ABA biosynthesis. At 39 dag, ABA concentrations increase and roots arrest elongation and undergo browning and lesion of the cortex. At 46 dag, decreasing auxin and cytokinin concentrations are accompanied by declining root activity, rearrangements in amino acid metabolism and serotonin formation, as well as altered expression in oxidative stress-related genes.
CONCLUSION
Despite the importance of aging-related processes in roots for root activity and longevity, previous studies have described changes of individual root traits over time, leaving it open whether roots are senescing. Here, we have conducted in parallel morphological, physiological, and molecular analyses in seminal roots of barley, showing that with progressing plant age, roots undergo a concerted sequence of metabolic and hormonal rearrangements that finally associate with tissue deterioration. These changes are preceded by transcriptional up- or down-regulation of specific transcription factors, some of which are confirmed regulators of leaf senescence while others appear root specific, indicating that root aging follows an intrinsically regulated developmental program resembling organ senescence. The identified associations between phytohormones, transcription factors, and genes representing major hubs in gene networks now pave the way for hypothesis testing using gene editing, ideally by employing root-specific promoters. Exploring the role of key metabolites and regulatory factors governing root senescence and longevity promises to improve crop performance, especially under premature senescence as induced by nutrient deficiency or drought stress.
MATERIALS AND METHODS
Plant Culture and Sampling
Barley (Hordeum vulgare) seeds, cv Golden Promise, were germinated on wet filter paper for 5 d at 4°C in the dark. Germinated seedlings were transferred to half-strength nutrient solution without Fe supply for 7 d in climate chamber under short-day conditions at a 20°C/18°C and 10h/14h light-dark regime and a light intensity of 250 μmol m−2 sec−1 at 70% humidity. Then, seedlings were transferred to full nutrient solution, containing 2 mm Ca(NO3)2, 0.5 mm K2SO4, 0.5 mm MgSO4, 0.1 mm KH2PO4, 0.1 mm KCl, 1 μm H3BO3, 0.5 μm MnSO4, 0.5 μm ZnSO4, 0.2 μm CuSO4, 0.01 μm (NH4)6Mo7O24, and 0.1 mm Fe-EDTA. With plant transfer to hydroponics, conditions were switched to long days following a 20°C/18°C and 16h/8h light-dark regime with a light intensity of 250 μmol m−2 sec−1 at 70% humidity. The nutrient solution was replaced every 3 to 4 d.
From 18 dag on, plants were sampled weekly, i.e. at 18, 25, 32, 39, 46, and 53 dag. Individual leaves of the main tiller were frozen in liquid nitrogen for chlorophyll determination. The whole root system was first washed by 1 mm CaSO4 for 1 min and briefly blotted by paper tissue before the whole seminal root fraction and the nodal root fraction were harvested separately. In addition, segments of the ARZ and the BRZ of seminal roots were sampled separately in five biological replicates at each time point. Each biological replicate consisted of root segments from 15 to 30 independent plants, depending on plant age. The ARZ was defined as the root zone from root cap up to the lateral root branching zone, i.e. when the first lateral root reached 1 mm length, while the BRZ consisted of the remaining seminal root tissue after ARZ removal. Root samples from the ARZ and BRZ were frozen in liquid nitrogen for transcriptome and phytohormone analysis. For all other analyses, nonfractionated seminal root samples were used. The experiment was conducted twice with similar results, the results shown here represents the first experiment.
Analysis of Morphological Root Traits and Microscopy
Before weighing, seminal and nodal roots were dried at 65°C. Seminal root length was measured using a ruler. Freshly sampled seminal root tips were photographed under a VHX-5000 digital microscope (Keyence Neuisenburg). Before cross sectioning, seminal roots were incubated in 0.25% (w/v) Evans blue aqueous solution for 15 min at room temperature under vacuum. Stained seminal roots were washed three times for 10 min with distilled water before root segments of 0.5 cm length were collected 5 cm below the hypocotyl, 20 cm below the hypocotyl, or at the position where the first lateral root had emerged to a maximum length of 1 mm. All segments were embedded in 4% (w/v) agar, sectioned by a vibratome (VT1000S, Zeiss) and photographed under a light microscope (Axio Imager 2, Zeiss).
Physiological Analyses
Chlorophyll concentration was quantified as described (Porra et al., 1989). Protein concentrations were determined by the 2-d Quant Kit (GE Healthcare). Nitrate uptake rates were determined by incubating seminal roots for 20 min in full nutrient solution containing 1 mm 15N-labeled KNO3, while nodal roots remained in a separate pot containing full nutrient solution with nonlabeled nitrate. Total nitrogen and 15N were quantified by an elemental analyzer (Euro-EA, HEKAtech) coupled to isotope ratio mass spectrometry (Horizon, NU Instruments). Other elements were analyzed as described in Lešková et al. (2017).
Soluble sugars and amino acids were determined according to Hajirezaei et al. (2000) and Hilo et al. (2017). Extraction and analysis of Trp, tryptamine, and serotonin was performed as described previously (Cao et al., 2006; Kang et al., 2009). Phytohormones were extracted and quantified as described in Eggert and von Wirén (2017).
Transcriptome Profiling and Quantitative PCR
Three biological replicates of basal and apical root segments at each time point were subjected to RNA extraction using an RNeasy plant mini kit (Qiagen) with on-column DNase treatment according to the manufacturer’s instructions. cDNA was prepared using random hexamer primers and SuperScript II reverse transcriptase (Life Technologies) according to the manufacturer’s protocol. RNA amplification, labeling, and hybridization to 60K Agilent custom-made gene expression microarrays (design available at EMBL-EBI ArrayExpress, accession no. A-MTAB-530) were conducted as described (Koppolu et al., 2013). Analysis of microarray data were performed with the R package limma (Ritchie et al., 2015). Raw feature intensities were background corrected using “normexp,” and the “quantile” normalization method was used to normalize between arrays. Differential expression was determined by fitting a linear model to log2-transformed data by an empirical Bayes method (Smyth, 2004). The Benjamini-Hochberg method was used to correct for multiple testing. By mapping the microarray probe IDs to the functional description of the barley reference sequence v 1.0 (Beier et al., 2017; Mascher et al., 2017), the microarray gene annotation was adjusted to include GO terms. We computed enrichment analysis applying Fisher’s exact test and reduced the results to GO terms in the category “biological process” when these passed Benjamini-Hochberg multiple hypothesis correction (P ≤ 0.05). Corresponding results were visualized using the R package GOplot (Walter et al., 2015). Tissue-specific expression of genes was verified using the barley transcriptome atlas (https://webblast.ipk-gatersleben.de/barley_ibsc/) with a false discovery rate < 0.05 at 32 dag and a log2 fold-change > |2|.
The cDNA samples for real-time PCR were prepared in the same way as for microarray analysis. Real-time PCR was conducted by using a Mastercycler ep realplex (Eppendorf) and QuantiTect SYBR Green qPCR mix (QIAGEN). Gene-specific primers are listed in Supplemental Table S16. Primer specificity was confirmed by analysis of melting curves. Relative expression levels were calculated according to Pfaffl (2001).
Statistical Analysis
All statistics were performed using SigmaPlot 11.0. Mean values were compared by one-way ANOVA (P < 0.05) followed by Tukey’s post-hoc multiple comparisons test, as indicated in the legends of each figure.
Accession Numbers
The transcriptome dataset is stored in the PGP-Repository (Arend et al., 2016) and can be accessed at e!DAL (http://dx.doi.org/10.5447/IPK/2019/15).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotype of shoots and seminal and nodal roots with progressing plant age.
Supplemental Figure S2. Developmental stages of the apical shoot meristem with progressing plant age.
Supplemental Figure S3. Plant-age-dependent profiles of macro- and micronutrient concentrations and contents in seminal roots.
Supplemental Figure S4. Clustering of significantly altered genes in the BRZ and ARZ of seminal roots with progressing plant age.
Supplemental Figure S5. Relative transcript levels of HvPAP15 and HvPAP17 with progressing plant age.
Supplemental Figure S6. Relative expression of oxidative stress-related genes with progressing plant age.
Supplemental Figure S7. Coexpression network of differentially expressed genes in the ARZ at 32 dag.
Supplemental Table S1. Overrepresented GO terms in the ARZ and BRZ at 25 dag.
Supplemental Table S2. Overrepresented GO terms in the ARZ and BRZ at 32 dag.
Supplemental Table S3. Overrepresented GO terms in the ARZ and BRZ at 39 dag.
Supplemental Table S4. Overrepresented GO terms in the ARZ and BRZ at 46 dag.
Supplemental Table S5. Overrepresented GO terms in the ARZ and BRZ at 53 dag.
Supplemental Table S6. Relative expression of significantly altered NAC transcription factors in the ARZ during seminal root aging.
Supplemental Table S7. Relative expression of significantly altered NAC transcription factors in the BRZ during seminal root aging.
Supplemental Table S8. Relative expression of significantly altered WRKY transcription factors in the ARZ during seminal root aging.
Supplemental Table S9. Relative expression of significantly altered WRKY transcription factors in the BRZ during seminal root aging.
Supplemental Table S10. Relative expression of significantly altered AP2 transcription factors in the ARZ during seminal root aging.
Supplemental Table S11. Relative expression of significantly altered AP2 transcription factors in the BRZ during seminal root aging.
Supplemental Table S12. Root- or leaf-specific expression of transcription factors.
Supplemental Table S13. Relative transcript levels of oxidative stress-related genes significantly altered in the ARZ during seminal root aging.
Supplemental Table S14. Relative transcript levels of oxidative stress-related genes significantly altered in the BRZ during seminal root aging.
Supplemental Table S15. List of coexpressed genes at 32 dag in the ARZ.
Supplemental Table S16. Primers used in this study.
Acknowledgments
We thank Dr. Kai Eggert, Barbara Kettig, Yudelsy Tandron-Moya, Elis Fraust, Nicole Schäfer, Guozheng Liu, and Dagmar Böhmert (Leibniz Institute of Plant Genetics and Crop Plant Research) for excellent technical assistance and Dr. Ricardo Giehl and Dr. Ying Liu (Leibniz-Institute of Plant Genetics and Crop Plant Research, Gatersleben) for critically reading the manuscript.
Footnotes
This work was supported by the Leibniz Graduate School ‘Yield formation in cereals’ program, with a grant from the Leibniz Association, Berlin, to N.v.W.
Articles can be viewed without a subscription.
References
- Arend D, Junker A, Scholz U, Schüler D, Wylie J, Lange M (2016) PGP repository: A plant phenomics and genomics data publication infrastructure. Database (Oxford) 2016: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-Ospina L, Moison M, Yoshimoto K, Masclaux-Daubresse C (2014) Autophagy, plant senescence, and nutrient recycling. J Exp Bot 65: 3799–3811 [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Riaño-Pachón DM, Mueller-Roeber B (2008) Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol (Stuttg) 10(Suppl 1): 63–75 [DOI] [PubMed] [Google Scholar]
- Baldi E, Wells CE, Marangoni B (2010) Nitrogen absorption and respiration in white and brown peach roots. J Plant Nutr 33: 461–469 [Google Scholar]
- Beier S, Himmelbach A, Colmsee C, Zhang X-Q, Barrero RA, Zhang Q, Li L, Bayer M, Bolser D, Taudien S, et al. (2017) Construction of a map-based reference genome sequence for barley, Hordeum vulgare L. Sci Data 4: 170044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham IJ. (2007) Quantifying the presence and absence of turgor for the spatial characterization of cortical senescence in roots of Triticum aestivum (Poaceae). Am J Bot 94: 2054–2058 [DOI] [PubMed] [Google Scholar]
- Bohner A, Kojima S, Hajirezaei M, Melzer M, von Wirén N (2015) Urea retranslocation from senescing Arabidopsis leaves is promoted by DUR3-mediated urea retrieval from leaf apoplast. Plant J 81: 377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim Y-S, Penfold CA, Jenkins D, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam Y, Pierre O, Boncompagni E, Hérouart D, Meilhoc E, Bruand C (2012) Nitric oxide (NO): A key player in the senescence of Medicago truncatula root nodules. New Phytol 196: 548–560 [DOI] [PubMed] [Google Scholar]
- Cao J, Murch SJ, O’Brien R, Saxena PK (2006) Rapid method for accurate analysis of melatonin, serotonin and auxin in plant samples using liquid chromatography-tandem mass spectrometry. J Chromatogr A 1134: 333–337 [DOI] [PubMed] [Google Scholar]
- Christiansen MW, Gregersen PL (2014) Members of the barley NAC transcription factor gene family show differential co-regulation with senescence-associated genes during senescence of flag leaves. J Exp Bot 65: 4009–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen MW, Matthewman C, Podzimska-Sroka D, O’Shea C, Lindemose S, Møllegaard NE, Holme IB, Hebelstrup K, Skriver K, Gregersen PL (2016) Barley plants over-expressing the NAC transcription factor gene HvNAC005 show stunting and delay in development combined with early senescence. J Exp Bot 67: 5259–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas LH, Eissenstat DM, Lakso AN (2000) Assessing root death and root system dynamics in a study of grape canopy pruning. New Phytol 147: 171–178 [Google Scholar]
- Devaiah BN, Nagarajan VK, Raghothama KG (2007) Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiol 145: 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhindsa RS, Plumb-Dhindsa PL, Reid DM (1982) Leaf senescence and lipid peroxidation: Effects of some phytohormones, and scavengers of free radicals and singlet oxygen. Physiol Plant 56: 453–457 [Google Scholar]
- Diaz C, Lemaître T, Christ A, Azzopardi M, Kato Y, Sato F, Morot-Gaudry J-F, Le Dily F, Masclaux-Daubresse C (2008) Nitrogen recycling and remobilization are differentially controlled by leaf senescence and development stage in Arabidopsis under low nitrogen nutrition. Plant Physiol 147: 1437–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A, Avni R, Fischer AM (2014) Senescence, nutrient remobilization, and yield in wheat and barley. J Exp Bot 65: 3783–3798 [DOI] [PubMed] [Google Scholar]
- Eggert K, von Wirén N (2017) Response of the plant hormone network to boron deficiency. New Phytol 216: 868–881 [DOI] [PubMed] [Google Scholar]
- Eissenstat D, Wells C, Yanai R, Whitbeck J (2000) Building roots in a changing environment: implications for root longevity. New Phytol 147: 33–42 [Google Scholar]
- Erland LAE, Murch SJ, Reiter RJ, Saxena PK (2015) A new balancing act: The many roles of melatonin and serotonin in plant growth and development. Plant Signal Behav 10: e1096469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S, Amasino RM (1997) Making sense of senescence (molecular genetic regulation and manipulation of leaf senescence). Plant Physiol 113: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskinsky DK, Syaifullah SJ, Roitsch T (2018) Integration of multi-omics techniques and physiological phenotyping within a holistic phenomics approach to study senescence in model and crop plants. J Exp Bot 69: 825–844 [DOI] [PubMed] [Google Scholar]
- Guo Y, Cai Z, Gan S (2004) Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ 27: 521–549 [Google Scholar]
- Hajirezaei MR, Takahata Y, Trethewey RN, Willmitzer L, Sonnewald U (2000) Impact of elevated cytosolic and apoplastic invertase activity on carbon metabolism during potato tuber development. J Exp Bot 51: 439–445 [DOI] [PubMed] [Google Scholar]
- Heyl A, Werner T, Schmülling T (2006) Cytokinin metabolism and signal transduction In Hedden P, and Thomas SG, eds, Plant Hormone Signaling. Blackwell, Oxford, pp 93–123 [Google Scholar]
- Hilo A, Shahinnia F, Druege U, Franken P, Melzer M, Rutten T, von Wirén N, Hajirezaei M-R (2017) A specific role of iron in promoting meristematic cell division during adventitious root formation. J Exp Bot 68: 4233–4247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himelblau E, Amasino RM (2001) Nutrients mobilized from leaves of Arabidopsis thaliana during leaf senescence. J Plant Physiol 158: 1317–1323 [Google Scholar]
- Hörtensteiner S. (2009) Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci 14: 155–162 [DOI] [PubMed] [Google Scholar]
- Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, Ohsumi Y, Hanson MR, Mae T (2008) Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol 148: 142–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K, Kim Y-S, Park S, Back K (2009) Senescence-induced serotonin biosynthesis and its role in delaying senescence in rice leaves. Plant Physiol 150: 1380–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim JH, Lyu JI, Woo HR, Lim PO (2018) New insights into the regulation of leaf senescence in Arabidopsis. J Exp Bot 69: 787–799 [DOI] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG (2009) Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Kim JI, Murphy AS, Baek D, Lee S-W, Yun D-J, Bressan RA, Narasimhan ML (2011) YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J Exp Bot 62: 3981–3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppolu R, Anwar N, Sakuma S, Tagiri A, Lundqvist U, Pourkheirandish M, Rutten T, Seiler C, Himmelbach A, Ariyadasa R, et al. (2013) Six-rowed spike4 (Vrs4) controls spikelet determinacy and row-type in barley. Proc Natl Acad Sci USA 110: 13198–13203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosslak RM, Chamberlin MA, Palmer RG, Bowen BA (1997) Programmed cell death in the root cortex of soybean root necrosis mutants. Plant J 11: 729–745 [DOI] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J 23: 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascaris D, Deacon J (1991) Relationship between root cortical senescence and growth of wheat as influenced by mineral nutrition, Idriella bolleyi (Sprague) von Arx and pruning of leaves. New Phytol 118: 391–396 [Google Scholar]
- Lejay L, Gansel X, Cerezo M, Tillard P, Müller C, Krapp A, von Wirén N, Daniel-Vedele F, Gojon A (2003) Regulation of root ion transporters by photosynthesis: Functional importance and relation with hexokinase. Plant Cell 15: 2218–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lešková A, Giehl RFH, Hartmann A, Fargašová A, von Wirén N (2017) Heavy metals induce iron deficiency responses at different hierarchic and regulatory levels. Plant Physiol 174: 1648–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Chung T, Pennington JG, Federico ML, Kaeppler HF, Kaeppler SM, Otegui MS, Vierstra RD (2015) Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 27: 1389–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljeroth E. (1995) Comparisons of early root cortical senescence between barley cultivars, Triticum species and other cereals. New Phytol 130: 495–501 [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Mao C, Lu S, Lv B, Zhang B, Shen J, He J, Luo L, Xi D, Chen X, Ming F (2017) A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol 174: 1747–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez DE, Bartoli CG, Grbic V, Guiamet JJ (2007) Vacuolar cysteine proteases of wheat (Triticum aestivum L.) are common to leaf senescence induced by different factors. J Exp Bot 58: 1099–1107 [DOI] [PubMed] [Google Scholar]
- Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J, et al. (2017) A chromosome conformation capture ordered sequence of the barley genome. Nature 544: 427–433 [DOI] [PubMed] [Google Scholar]
- Miao Y, Jiang J, Ren Y, Zhao Z (2013) The single-stranded DNA-binding protein WHIRLY1 represses WRKY53 expression and delays leaf senescence in a developmental stage-dependent manner in Arabidopsis. Plant Physiol 163: 746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K, MacKerness SAH, Page T, John CF, Murphy AM, Carr JP, Buchanan-Wollaston V (2000) Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J 23: 677–685 [DOI] [PubMed] [Google Scholar]
- Navas ML, Ducout B, Roumet C, Richarte J, Garnier J, Garnier E (2003) Leaf life span, dynamics and construction cost of species from Mediterranean old-fields differing in successional status. New Phytol 159: 213–228 [DOI] [PubMed] [Google Scholar]
- Naz AA, Arifuzzaman M, Muzammil S, Pillen K, Léon J (2014) Wild barley introgression lines revealed novel QTL alleles for root and related shoot traits in the cultivated barley (Hordeum vulgare L.). BMC Genet 15: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeller PW, Lu MW, Taylor LP, Pike DA, Theologis A (1991) Reversible inhibition of tomato fruit senescence by antisense RNA. Science 254: 437–439 [DOI] [PubMed] [Google Scholar]
- Parrott DL, Martin JM, Fischer AM (2010) Analysis of barley (Hordeum vulgare) leaf senescence and protease gene expression: A family C1A cysteine protease is specifically induced under conditions characterized by high carbohydrate, but low to moderate nitrogen levels. New Phytol 187: 313–331 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra R, Thompson W, Kriedemann P (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe JH, Topping JF, Liu J, Lindsey K (2016) Abscisic acid regulates root growth under osmotic stress conditions via an interacting hormonal network with cytokinin, ethylene and auxin. New Phytol 211: 225–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schippers JH, Schmidt R, Wagstaff C, Jing H-C (2015) Living to die and dying to live: The survival strategy behind leaf senescence. Plant Physiol 169: 914–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider HM, Lynch JP (2018) Functional implications of root cortical senescence for soil resource capture. Plant Soil 423: 13–26 [Google Scholar]
- Schneider HM, Wojciechowski T, Postma JA, Brown KM, Lücke A, Zeisler V, Schreiber L, Lynch JP (2017) Root cortical senescence decreases root respiration, nutrient content and radial water and nutrient transport in barley. Plant Cell Environ 40: 1392–1408 [DOI] [PubMed] [Google Scholar]
- Seiler C, Harshavardhan VT, Rajesh K, Reddy PS, Strickert M, Rolletschek H, Scholz U, Wobus U, Sreenivasulu N (2011) ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J Exp Bot 62: 2615–2632 [DOI] [PubMed] [Google Scholar]
- Shi H, Chan Z (2014) The cysteine2/histidine2-type transcription factor ZINC FINGER OF ARABIDOPSIS THALIANA 6-activated C-REPEAT-BINDING FACTOR pathway is essential for melatonin-mediated freezing stress resistance in Arabidopsis. J Pineal Res 57: 185–191 [DOI] [PubMed] [Google Scholar]
- Shi R, Weber G, Köster J, Reza-Hajirezaei M, Zou C, Zhang F, von Wirén N (2012) Senescence-induced iron mobilization in source leaves of barley (Hordeum vulgare) plants. New Phytol 195: 372–383 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: e3. [DOI] [PubMed] [Google Scholar]
- Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC Gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314: 1298–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veneklaas EJ, Lambers H, Bragg J, Finnegan PM, Lovelock CE, Plaxton WC, Price CA, Scheible WR, Shane MW, White PJ, et al. (2012) Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol 195: 306–320 [DOI] [PubMed] [Google Scholar]
- Walter W, Sánchez-Cabo F, Ricote M (2015) GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics 31: 2912–2914 [DOI] [PubMed] [Google Scholar]
- Warpeha KM, Lateef SS, Lapik Y, Anderson M, Lee B-S, Kaufman LS (2006) G-protein-coupled receptor 1, G-protein Galpha-subunit 1, and prephenate dehydratase 1 are required for blue light-induced production of phenylalanine in etiolated Arabidopsis. Plant Physiol 140: 844–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Balazadeh S, Tohge T, Erban A, Giavalisco P, Kopka J, Mueller-Roeber B, Fernie AR, Hoefgen R (2013) Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis. Plant Physiol 162: 1290–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Kim HJ, Nam HG, Lim PO (2013) Plant leaf senescence and death - regulation by multiple layers of control and implications for aging in general. J Cell Sci 126: 4823–4833 [DOI] [PubMed] [Google Scholar]
- Wu L, Ma N, Jia Y, Zhang Y, Feng M, Jiang C-Z, Ma C, Gao J (2017) An ethylene-induced regulatory module delays flower senescence by regulating cytokinin content. Plant Physiol 173: 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Jia W, Zhang J (2007) AtMEK1 mediates stress-induced gene expression of CAT1 catalase by triggering H2O2 production in Arabidopsis. J Exp Bot 58: 2969–2981 [DOI] [PubMed] [Google Scholar]
- Yoshida S. (2003) Molecular regulation of leaf senescence. Curr Opin Plant Biol 6: 79–84 [DOI] [PubMed] [Google Scholar]
- Zentgraf U, Jobst J, Kolb D, Rentsch D (2004) Senescence-related gene expression profiles of rosette leaves of Arabidopsis thaliana: leaf age versus plant age. Plant Biol (Stuttg) 6: 178–183 [DOI] [PubMed] [Google Scholar]
- Zhang B, Horvath S (2005) A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4: e17. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Heinlein C, Orendi G, Zentgraf U (2006) Senescence-specific regulation of catalases in Arabidopsis thaliana (L.) Heynh. Plant Cell Environ 29: 1049–1060 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Zentgraf U (2005) The correlation between oxidative stress and leaf senescence during plant development. Cell Mol Biol Lett 10: 515–534 [PubMed] [Google Scholar]