AtIQM1 positively regulates jasmonic acid content and resistance to Botrytis cinerea by interacting with and stimulating the reactive oxygen species scavenger CATALASE2.
Abstract
Calmodulin (CaM) regulates plant disease responses through its downstream calmodulin-binding proteins (CaMBPs) often by affecting the biosynthesis or signaling of phytohormones, such as jasmonic acid (JA) and salicylic acid. However, how these CaMBPs mediate plant hormones and other stress resistance-related signaling remains largely unknown. In this study, we conducted analyses in Arabidopsis (Arabidopsis thaliana) on the functions of AtIQM1 (IQ-Motif Containing Protein1), a Ca2+-independent CaMBP, in JA biosynthesis and defense against the necrotrophic pathogen Botrytis cinerea using molecular, biochemical, and genetic analyses. IQM1 directly interacted with and promoted CATALASE2 (CAT2) expression and CAT2 enzyme activity and indirectly increased the activity of the JA biosynthetic enzymes ACX2 and ACX3 through CAT2, thereby positively regulating JA content and B. cinerea resistance. In addition, in vitro assays showed that in the presence of CaM5, IQM1 further enhanced the activity of CAT2, suggesting that CaM5 may affect the activity of CAT2 by combining with IQM1 in the absence of Ca2+. Our data indicate that IQM1 is a key regulatory factor in signaling of plant disease responses mediated by JA. The study also provides new insights that CaMBP may play a critical role in the cross talk of multiple signaling pathways in the context of plant defense processes.
Plant disease resistance involves a series of signaling response pathways, including calcium (Ca2+)/calmodulin (CaM), reactive oxygen species (ROS), and phytohormones. These factors interact with each other, forming sophisticated positive or negative feedback loops to coordinate the entire plant immune system (Lecourieux et al., 2006; Ali et al., 2003; Fu and Dong., 2013). Ca2+/CaM is a common second messenger that plays crucial roles in response to diverse signaling cascades in all plant life processes. An individual CaM usually has no biochemical or enzymatic activity (Reddy et al., 2002; Kushwaha et al., 2008). However, as pervasive and versatile Ca2+ sensors, CaMs can interact with a large variety of functional proteins and modulate their structures or activities and are thus involved in almost all aspects of plant growth and development as well as in responses to environmental stresses (Zielinski, 1998; Bouché et al., 2005). Therefore, determining the target proteins of CaMs (CaM-binding proteins [CaMBPs]) and understanding their biological functions could help clarify the complex molecular mechanisms of Ca2+ signaling (Poovaiah et al., 2013; Zeng et al., 2015). To date, at least 500 CaMBPs in plants have been identified. These CaMBPs include ion channels and pumps, structural proteins, kinase/phosphatase proteins, metabolic enzymes, transcription factors, chaperonins, and other proteins of unknown functions (Reddy et al., 2002). A great deal of research has shown that CaMBPs are closely related to plant defense; for example, Calmodulin-Binding Transcription Activator3 (Galon et al., 2008; Du et al., 2009), IQ-Domain1 (Levy et al., 2005), Barley Mildew Resistance Locus O (Jørgensen, 1992; Kim et al., 2002), Cyclic Nucleotide Gated Channel2 (Chin et al., 2013; Lu et al., 2016), and Calmodulin Binding Protein-Like60g (Wang et al., 2009; Wan et al., 2012) participate in plant disease resistance by affecting ROS or phytohormone levels. However, the specific regulatory mechanism among CaMBPs, ROS, and plant hormones remains vague.
ROS are generated in various metabolic pathways of plant growth and development as well as biotic and abiotic stress responses. Appropriate control of ROS production is essential for the maintenance of growth balance when plants face external stress signals, and moderate ROS levels can help plants resist stresses, but failure to eliminate excess ROS in due time causes damage to plant cells (Apel and Hirt, 2004). Hydrogen peroxide (H2O2) is a main form of ROS that can accumulate due to rapidly rising cytosolic Ca2+ in plants triggered by leaf senescence and pathogen infection (Rentel and Knight, 2004). There are three catalases in Arabidopsis (Arabidopsis thaliana), CAT1, CAT2, and CAT3, that are located in the peroxisome lumen and function as antioxidant enzymes to scavenge excess H2O2 (Su et al., 2018). A series of reports have shown that catalases are closely involved in plant stress resistance. In Arabidopsis, the regulation of catalases occurs at both the transcriptional and protein levels. The key transcription factor of the abscisic acid (ABA) signaling pathway, ABA-INSENSITIVE5, can bind to the promoter of CAT1 and activate its expression to promote seed germination (Bi et al., 2017). The zinc finger protein LESION SIMULATING DISEASE1 interacts with all three catalases to regulate the cell death process of plants by affecting their enzyme activity (Li et al., 2013). The chaperone protein NO CATALASE ACTIVITY1 stimulates the activity of CAT2 to promote plant resistance to multiple abiotic stresses (Li et al., 2015). In addition, CAT3 activity can be modulated by CALCIUM-DEPENDENT PROTEIN KINASE in response to drought stress (Zou et al., 2015).
Both jasmonic acid (JA) and salicylic acid (SA) are essential plant hormones that regulate plant growth and developmental processes as well as disease resistance signaling. In general, JA and SA act antagonistically in the plant defense response. SA is considered to act against biotrophic pathogens, while JA is involved in resistance to wounding, insect damage, and necrotrophic pathogens (Spoel and Dong, 2008). Attack by the necrotrophic pathogen Botrytis cinerea induces the accumulation of JA and activates JA signaling molecules (Song et al., 2013, 2014). The JA biosynthesis pathway is initiated in chloroplasts, where α-linolenic acid (18:3) is converted to 12-oxo-phytodienoic acid (12-OPDA) by lipoxygenase (LOX), allene oxide synthase (AOS), and allene oxide cyclase. 12-OPDA is then exported to the peroxisome and reduced to 3-oxo-2-(2′(Z)-pentenyl)-cyclopentane-1-octanoic acid by OPDA Reductase3 (OPR3). Three cycles of β-oxidation catalyzed by acyl-CoA oxidase (ACX) complete the biosynthesis of JA (Li et al., 2017). Fluctuations in JA content lead to changes in the expression of downstream transcription factors, such as MYC2, ERF1, and PDF1.2, in the JA signaling pathway (Wasternack and Hause, 2013).
Although some reports of CaMBPs affecting plant defense have been published, the target proteins downstream of most CaMBPs and the mechanisms by which they regulate plant disease resistance are largely unclear. Previously, we identified a family of proteins in Arabidopsis that consists of six members, named IQ-Motif Containing Protein1 (IQM1) to IQM6, that each contains one copy of the IQ motif. Among them, IQM1 modulates stomatal movement by affecting the content of ROS in plants (Zhou et al., 2010, 2012). Here, we show a function of IQM1 in JA biosynthesis and defense against the necrotrophic pathogen B. cinerea. Our results reveal that IQM1 is essential in the processes of plant disease defense mediated by JA signaling.
RESULTS
IQM1 Positively Regulates JA Biosynthesis and Plant Defense against B. cinerea
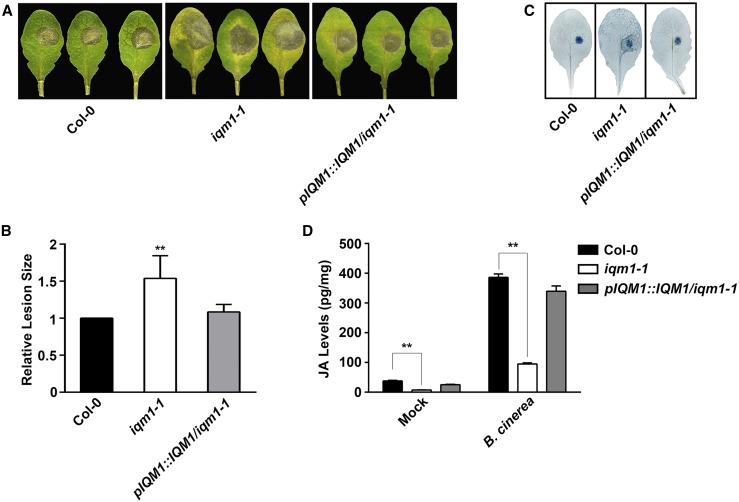
Our previous microarray data showed that numerous chitin-responsive genes and pathogen-associated genes are up-regulated in iqm1-1 (Zhou et al., 2012), suggesting that IQM1 may be involved in plant disease defense pathways. To verify this possibility, we assessed the resistance of iqm1-1 to necrotrophic pathogens by challenging wild-type and iqm1-1 plants with B. cinerea. We observed increased susceptibility to B. cinerea in iqm1-1, which showed larger lesions than the wild type; in addition, pIQM1::IQM1/iqm1-1 complement strains exhibited attenuated susceptibility compared with iqm1-1 (Fig. 1, A–C).
Figure 1.
IQM1 positively regulates plant defense against B. cinerea and the synthesis of JA. A, Photographs of rosette leaves cut from 4-week-old wild-type (Columbia-0 [Col-0]), iqm1-1 mutant, and pIQM1::IQM1/iqm1-1 complement plants at 3 d after infection with B. cinerea spores. B, Relative lesion sizes measured by ImageJ software at 3 d after inoculation with B. cinerea. The experiments were repeated at least three times, and similar results were obtained. The data are means ± sd of 10 infected leaves. C, Photographs of infected leaves with Trypan Blue staining at 3 d after inoculation with B. cinerea. D, JA levels of 4-week-old wild-type (Col-0), iqm1-1 mutant, and pIQM1::IQM1/iqm1-1 complement plants at 3 d after infection with B. cinerea. The results were obtained from at least 30 independent plants for each genotype by mixed grinding. The data are means ± sd of three replicates. Asterisks in B and D represent significant differences compared with Col-0 (Student’s t test, **, P < 0.01).
Plant resistance to the necrotrophic pathogen B. cinerea requires JA and JA signaling (Song et al., 2013). Therefore, we directly measured JA content in iqm1-1. The JA levels in iqm1-1 were only approximately one-fifth of those in wild-type plants under normal conditions without infection. Although B. cinerea-infected iqm1-1 and wild-type plants accumulated more JA than uninfected plants, the JA content in infected iqm1-1 was much lower than that in wild-type plants (Fig. 1D). This was consistent with the visual observation that the iqm1-1 mutants were more sensitive to B. cinerea. These results indicated that IQM1 is involved in the JA biosynthetic pathway and that the enhanced susceptibility of iqm1-1 to B. cinerea may have resulted from the suppression of the JA-mediated disease defense pathway due to deficiency of IQM1.
Plant resistance processes to biotrophic and necrotrophic pathogens are usually antagonistic (Thatcher et al., 2009; Nie et al., 2012), so we also examined the resistance of iqm1-1 to the biotrophic pathogen Pseudomonas syringae pv tomato (Pst) DC3000. Upon infection, iqm1-1 displayed higher resistance than wild-type plants (Supplemental Fig. S1), consistent with previous reports that coi1, myc2, and cat2 mutants are more resistant to the biotrophic pathogen Pst DC3000 and more susceptible to the necrotrophic pathogen B. cinerea than the wild type (Laurie-Berry et al., 2006; Yuan et al., 2017).
IQM1 Directly Interacts with CAT2 in Planta
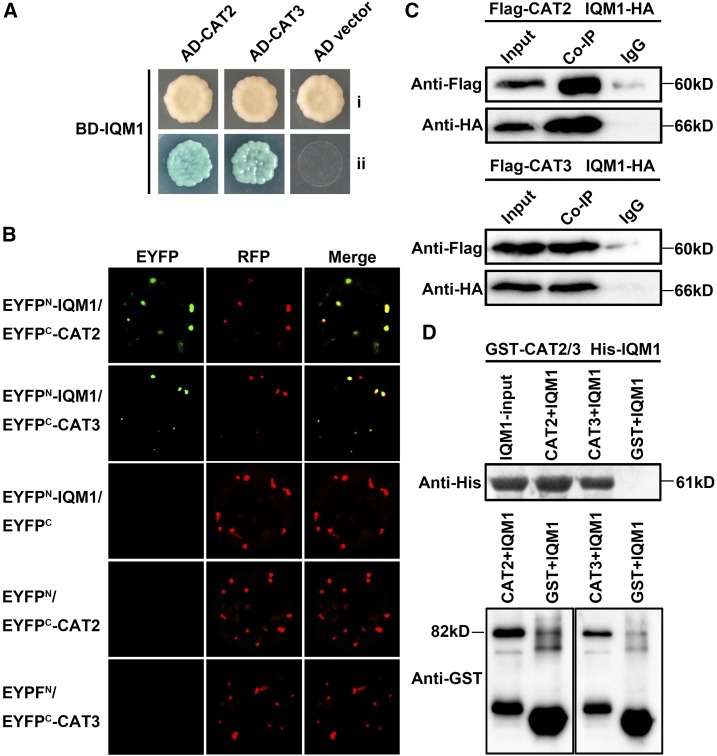
To further understand the mechanism by which IQM1 regulates JA synthesis and disease defense, we carried out yeast two-hybrid screening to search for proteins interacting with IQM1. Sixty-eight positive yeast (Saccharomyces cerevisiae) clones were screened on synthetic dropout medium (SD)/-His-Trp-Leu-Ade plates. Then, plasmids of yeast clones were extracted and transformed into an Escherichia coli strain, and 18 fragments (four genes) without frame shift were confirmed as interacting with IQM1 through sequencing analysis. We identified two catalases, CAT2 and CAT3, which are predominant peroxisomal proteins that function as H2O2 scavengers (Su et al., 2018), that may be involved in plant disease resistance response (Yuan et al., 2017). The interaction between IQM1 and CAT2/3 was confirmed upon cotransformation of AH109 yeast cells with AD-CAT2/3 and BD-IQM1. On SD-Trp-Leu-His-Ade medium with 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (x-α-gal), the clones grew well and were blue in color (Fig. 2A; Supplemental Fig. S2), indicating that IQM1 and CAT2/3 interacted in yeast.
Figure 2.
IQM1 directly interacts with CAT2 in planta. A, IQM1 interacts with CAT2/3 in yeast cells. pGADT7-CAT2/3 and pGBKT7-IQM1 were cotransformed into AH109 yeast cells, which were grown on SD/-Trp-Leu (i) and SD/-Trp-Leu-His-Ade containing 20 µg mL−1 x-α-gal (ii) for 3 d. B, BiFC assays to detect the interaction between IQM1 and CAT2/3. nEYFP-IQM1 and cEYFP-CAT2/3 plasmids were cotransformed into protoplasts isolated from the leaves of 4-week-old Col-0 plants. After 16 h of incubation, the EYFP signals were observed using a laser confocal microscope. RFP-AtKAT1 was used as a colocalization marker protein. nEYFP-IQM1 and cEYFP empty vector or nEYFP empty vector and cEYFP-CAT2/3 were set as negative controls. C, Co-IP assay to verify the interaction of IQM1 with CAT2/3 in planta. Flag-CAT2/3 and IQM1-HA plasmids were cointroduced into protoplasts isolated from the leaves of 4-week-old Col-0 plants with polyethylene glycol (PEG)/CaCl2. Protein G Plus/Protein A-Agarose and anti-Flag antibody were added to the plant protein, and the precipitated proteins were separated by 12% (w/v) SDS-PAGE for immunoblotting with anti-Flag antibody and anti-HA antibody. Proteins mixed with beads and IgG were used as negative controls. D, In vitro pull-down assay to verify the direct interaction of IQM1 with CAT2/3. His-IQM1 and GST-CAT2/3 plasmids were separately transformed into the E. coli strain Rosetta. The recombinant fusion proteins were purified with Ni-NTA agarose and Glutathione Sepharose. Agarose-conjugated His-IQM1 was then washed with imidazole. All eluates were added to agarose-conjugated GST-CAT2/3. The precipitated proteins were separated by 12% (w/v) SDS-PAGE for immunoblotting with anti-His antibody and anti-GST antibody. His-IQM1 plus GST beads and purified GST were used as negative controls. All experiments were repeated at least three times.
Furthermore, an enhanced yellow fluorescent protein (EYFP)-based bimolecular fluorescence complementation (BiFC) assay was performed. Plasmids carrying nEYFP-IQM1 and cEYFP-CAT2/3 fused expression vectors were cotransformed into protoplasts of Arabidopsis Col-0. Strong EYFP signals were yielded in peroxisomes, while the negative controls did not fluoresce. AtKAT1 (AT1G04710), a peroxisome-located gene, fused with red fluorescent protein (RFP) was used as a colocalization marker protein (Fig. 2B). Cotransforming nEYFP-IQM1 and cEYFP empty vector or nEYFP empty vector and cEYFP-CAT2/3 were set as negative controls.
We also conducted coimmunoprecipitation (Co-IP) assays to examine the interaction between IQM1 and CAT2/3 in planta. IQM1-6HA and FLAG-CAT2/3 plasmids were cointroduced into protoplasts, and the total proteins were precipitated with agarose-conjugated anti-FLAG antibody. The precipitated proteins were separated by SDS-PAGE for immunoblotting with an anti-HA antibody. Our results showed that FLAG-CAT2/3 pulled down IQM1-6HA from the plant total proteins, indicating that IQM1 indeed interacted with CAT2/3 in planta (Fig. 2C).
Additional evidence for a direct interaction between IQM1 and CAT2/3 came from in vitro pull-down assays. Purified His-IQM1 was added to agarose-conjugated glutathione S-transferase (GST)-CAT2/3. After incubation overnight, the total and immunoprecipitated proteins were analyzed by immunoblotting using anti-His and anti-GST antibodies. The results showed that GST-CAT2/3 resin could pull down His-IQM1 (Fig. 2D), demonstrating that IQM1 directly interacts with CAT2/3. Taken together, these data strongly suggest that IQM1 can directly interact with CAT2/3 in vivo and imply that this interaction has a biological purpose.
IQM1 Regulates JA Biosynthesis and Plant Disease Resistance through Interaction with CAT2
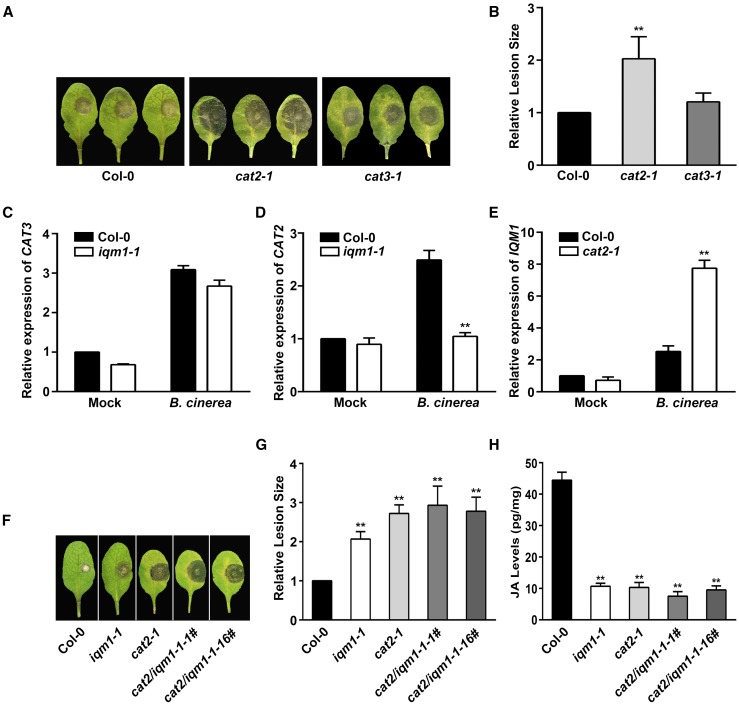
To determine whether IQM1 affects plant disease resistance through CAT2 and CAT3, we assayed the susceptibility of cat2-1 and cat3-1 mutants to B. cinerea. The cat2-1 mutant displayed greater susceptibility with larger lesions than wild-type plants. In contrast, the resistance of cat3-1 to B. cinerea was similar to that of the wild type (Fig. 3, A and B). In agreement with the results of Yuan et al. (2017), the JA levels in cat2-1 were much lower than those in the wild type without infection and under treatment with B. cinerea; however, the JA content in cat3-1 was similar to that in the wild type upon B. cinerea infection.
Figure 3.
IQM1 regulates JA biosynthesis and plant disease resistance through interaction with CAT2. A, Photographs of rosette leaves cut from 4-week-old wild-type, cat2-1, and cat3-1 plants at 3 d after infection with B. cinerea spores. B, Relative lesion sizes measured by ImageJ software at 3 d after inoculation with B. cinerea. The experiments were repeated at least three times, and similar results were obtained. The data are means ± sd of 10 infected leaves. C to E, Relative expression levels of CAT3 and CAT2 in iqm1-1 (C and D) and of IQM1 in cat2-1 (E) at 3 d after infection with B. cinerea. The differentially expressed genes were examined in leaves of 4-week-old plants harvested for RT-qPCR. ACTIN2 was used as the internal control. Independent biological assays were repeated three times, and similar results were obtained. Data from one representative replicate are shown. Values are means with sd from three technical measurements. F, Photographs of rosette leaves cut from 4-week-old wild-type, iqm1-1, cat2-1, and cat2/iqm1-1 double mutant plants at 3 d after infection with B. cinerea spores. G, Relative lesion sizes at 3 d after inoculation with B. cinerea. H, JA content of 4-week-old wild-type, iqm1-1, cat2-1, and cat2/iqm1-1 double mutant plants. The results were obtained from at least 30 independent plants for each genotype by mixed grinding. The data are means ± sd of three replicates. Asterisks represent significant differences compared with Col-0 (Student’s t test, **, P < 0.01).
We further examined the expression patterns of CAT2 and CAT3 in iqm1-1 under B. cinerea attack by reverse transcription quantitative PCR (RT-qPCR). The transcript levels of CAT3 were markedly increased with similar amplitudes in the wild type and iqm1-1 after 3 d of infection (Fig. 3C); however, CAT2 could be induced only in wild-type plants (Fig. 3D), indicating that the absence of IQM1 results in inhibition of CAT2 expression during JA-mediated defense against B. cinerea. Conversely, the expression levels of IQM1 were more strongly up-regulated in cat2-1 than in the wild type upon challenge with B. cinerea (Fig. 3E). This reciprocal cross-regulation phenomenon may be due to plant self-feedback regulation. When a gene is damaged, the expression of upstream genes at the same pathway is induced in large quantities to maintain the normal growth and development as well as to make correct responses to external stresses in mutant plants (Schilmiller et al., 2007). These results demonstrated that IQM1 could regulate JA synthesis and plant disease resistance to a necrotrophic pathogen mainly through interactions with CAT2 rather than CAT3.
Next, the CRISPR-Cas9 system was used to mutate CAT2 in the iqm1-1 mutant background to generate cat2/iqm1-1 double mutants (Supplemental Fig. S3, A and B). Compared with iqm1-1, the cat2-1 mutant was slightly more sensitive to B. cinerea and demonstrated larger lesions; in addition, the cat2/iqm1-1 double mutant also displayed decreased resistance to B. cinerea, similar to the cat2-1 mutant (Fig. 3, F and G), in accordance with the content of JA in these mutants (Fig. 3H). These data further confirmed that IQM1 and CAT2 are in the same pathway associated with JA-mediated pathogen defense.
The Expression of JA Biosynthesis and Signaling Genes Is Consistent in iqm1 and cat2 Mutants
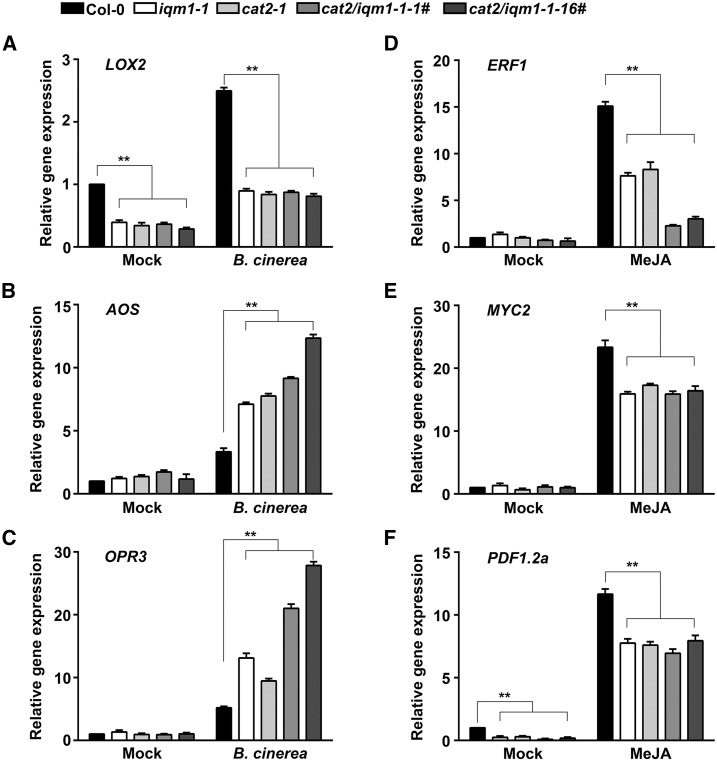
Because the JA levels in the iqm1-1 and cat2-1 mutants were much lower than those in wild-type plants, we investigated the expression of genes related to JA biosynthesis, including LOX2, AOS, and OPR3, to further identify the regulatory mechanisms of IQM1 and CAT2 in JA synthesis. The expression levels of LOX2 in iqm1-1, cat2-1, and cat2/iqm1-1 double mutants were much lower than those in the wild type regardless of whether the plants were infected with B. cinerea (Fig. 4A). The expression levels of AOS and OPR3, which are downstream of LOX2, were higher in these mutants than in the wild-type plants under infection with B. cinerea (Fig. 4, B and C). Interestingly, regardless of whether the expression of LOX2, AOS, and OPR3 was higher or lower than in wild-type plants after infection, the expression trends of each gene were always consistent in iqm1-1, cat2-1, and cat2/iqm1-1 double mutants.
Figure 4.
The expression of JA biosynthesis and signaling genes is consistent in iqm1 and cat2 mutants. A to C, Relative expression of the JA biosynthesis genes LOX2 (A), AOS (B), and OPR3 (C) after 3 d of infection with B. cinerea. Four-week-old leaves of wild-type, iqm1-1, cat2-1, and cat2/iqm1-1 double mutant plants were harvested for RT-qPCR. D to F, Relative expression of ERF1 (3-h treatment; D), MYC2 (1-h treatment; E), and PDF1.2a (3-h treatment; F) in 12-d-old seedlings after treatment with 50 µm MeJA. Total RNA was extracted at the indicated times. The transcript levels were quantified by RT-qPCR using ACTIN2 as a control. Independent biological assays were repeated three times, and similar results were obtained. Data from one representative replicate are shown. Values are means with sd from three technical measurements. Asterisks represent significant differences compared with Col-0 (Student’s t test, **, P < 0.01).
In addition, we also detected the expression of JA signaling pathway-related genes in 12-d-old seedlings of the iqm1-1, cat2-1, and cat2/iqm1-1 double mutants after treatment with methyl jasmonate (MeJA). The results showed that under normal conditions, the expression of ERF1 and MYC2 in the three mutant strains was not substantially different from that in the wild type (Fig. 4, D and E), and the expression levels of PDF1.2a in the mutants were markedly lower than those in the wild type (Fig. 4F). Under MeJA treatment, the expression levels of ERF1, MYC2, and PDF1.2a were rapidly induced, but they were significantly lower in the mutants than in the wild-type plants (Fig. 4, D–F). Overall, the findings that the expression patterns of JA biosynthesis and signaling genes were the same in the iqm1-1, cat2-1, and cat2/iqm1-1 double mutants further confirmed that IQM1 plays a key role in regulating JA biosynthesis and plant disease resistance responses triggered by necrotrophic pathogens through direct interaction with CAT2 in vivo.
Overexpression of CAT2 Can Restore the Increased Sensitivity of iqm1 to B. cinerea
To further determine the regulatory relationship between IQM1 and CAT2 in the plant disease response, we overexpressed CAT2 in iqm1-1 using the 35S promoter. The results showed that p35S::CAT2/iqm1-1 plants had almost a complete recovery of the enhanced sensitivity to B. cinerea compared with iqm1-1 plants, as the CAT2-overexpressing plants exhibited disease lesions similar to those of the wild type (Supplemental Figs. S3C and S4). This result indicated that the susceptibility of the iqm1-1 mutant to B. cinerea may have been caused by the weakening of CAT2 function due to the deletion of IQM1. In addition, the data also proved that CAT2 is not dependent only on IQM1; when CAT2 is overexpressed, other proteins that can regulate CAT2 play a greater role.
IQM1 Directly Promotes the Enzyme Activity of CAT2 through Its Interaction
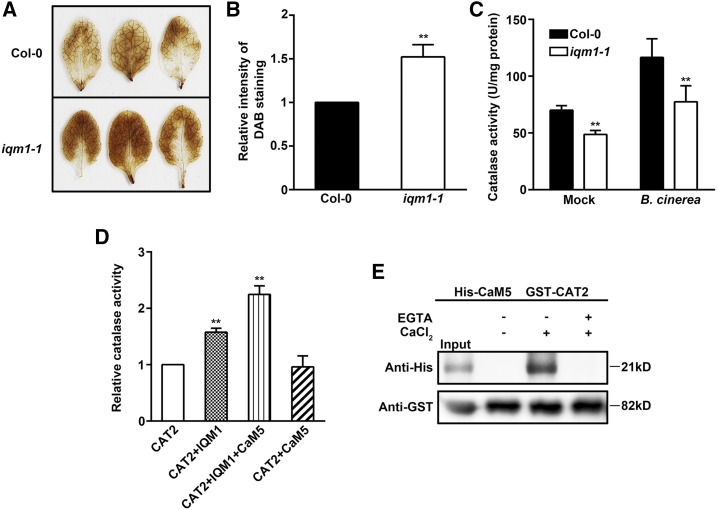
CAT2 is an important member of the catalase group. Its main function is to scavenge excess H2O2 in plants (Mhamdi et al., 2010). To study the biological significance of the interactions between IQM1 and CAT2, we first detected the content of H2O2 in plant leaves with 3,3′-diaminobenzidine (DAB) staining. The iqm1-1 mutant strain accumulated more H2O2 than the wild type, as indicated by denser staining (Fig. 5, A and B). This observation suggests that catalase activity in the iqm1-1 mutant may be reduced. Furthermore, the results of the enzyme activity test showed that the catalase activity of iqm1-1 was decreased by approximately 30% compared with that of wild-type plants under normal conditions. After B. cinerea treatment, the catalase activity in wild-type plants increased significantly, while the increase in activity in iqm1-1 was much smaller in magnitude (Fig. 5C). Previous studies have revealed that, among the three catalases, CAT2 is the key factor affecting overall catalase activity (Mhamdi et al., 2010). Therefore, these data demonstrate that the reduced activity of catalase in iqm1-1 may be caused by decreases in CAT2 activity due to the deletion of IQM1.
Figure 5.
IQM1 directly promotes the enzyme activity of CAT2. A, DAB staining images of leaves from 4-week-old wild-type and iqm1-1 mutant plants. B, Relative DAB staining intensity detected by Photoshop software. C, Catalase activity of 4-week-old wild-type and iqm1-1 mutant plants after 3 d of inoculation with B. cinerea. The catalase activity is presented as units per mg of protein. D, In vitro assays of catalase activity. Ten micrograms of purified His-IQM1 or His-CaM5 protein was added to 10 µg of His-CAT2. The mixture was incubated at 4°C overnight, and then CAT2 enzyme activity was tested. All experiments were repeated at least three times. The data are means ± sd of three biological replicates. Asterisks represent significant differences (Student’s t test, **, P < 0.01). E, Pull-down assay confirming that CAT2 interacts with CaM5 in a Ca2+-dependent manner. Immobilized GST-CAT2 was incubated with purified His-CaM5 in the presence of 1 mm Ca2+ or 5 mm EGTA (a Ca2+ chelator) and eluted for immunoblot analysis with anti-His antibody (top) and anti-GST antibody (bottom).
To provide direct evidence that IQM1 affects the activity of CAT2, we carried out an in vitro enzyme activity test. The enzyme activity of CAT2 was significantly improved after an equivalent amount of IQM1 was added to the purified CAT2 protein. Consistent with the in vivo data, the results further confirmed that the enzyme activity of CAT2 can be directly activated by IQM1 in planta (Fig. 5D). Since IQM1 is a Ca2+-independent CaM-binding protein that interacts with CaM5 in vivo (Zhou et al., 2012), the protein function of IQM1 may be affected by CaM5 in some cases. Accordingly, we investigated whether CAM5 had an effect on the function of IQM1 in activating CAT2. The enzyme activity of CAT2 did not change after the addition of CaM5 alone. When both CaM5 and IQM1 were added to CAT2, however, the activity of CAT2 increased significantly to higher levels than those achieved after the addition of IQM1 alone (Fig. 5D). This finding suggests that CaM5 may indirectly affect the enzyme activity of CAT2 through IQM1. To further prove this conclusion, we performed in vitro assays to examine interaction between CAT2 and CaM5. The interaction of CAT2 with CaM5 was Ca2+ dependent and completely suppressed by the Ca2+ chelator EGTA (Fig. 5E). In conclusion, these data suggest that IQM1 can directly activate CAT2 activity and acts as a connector between CaM5 and CAT2.
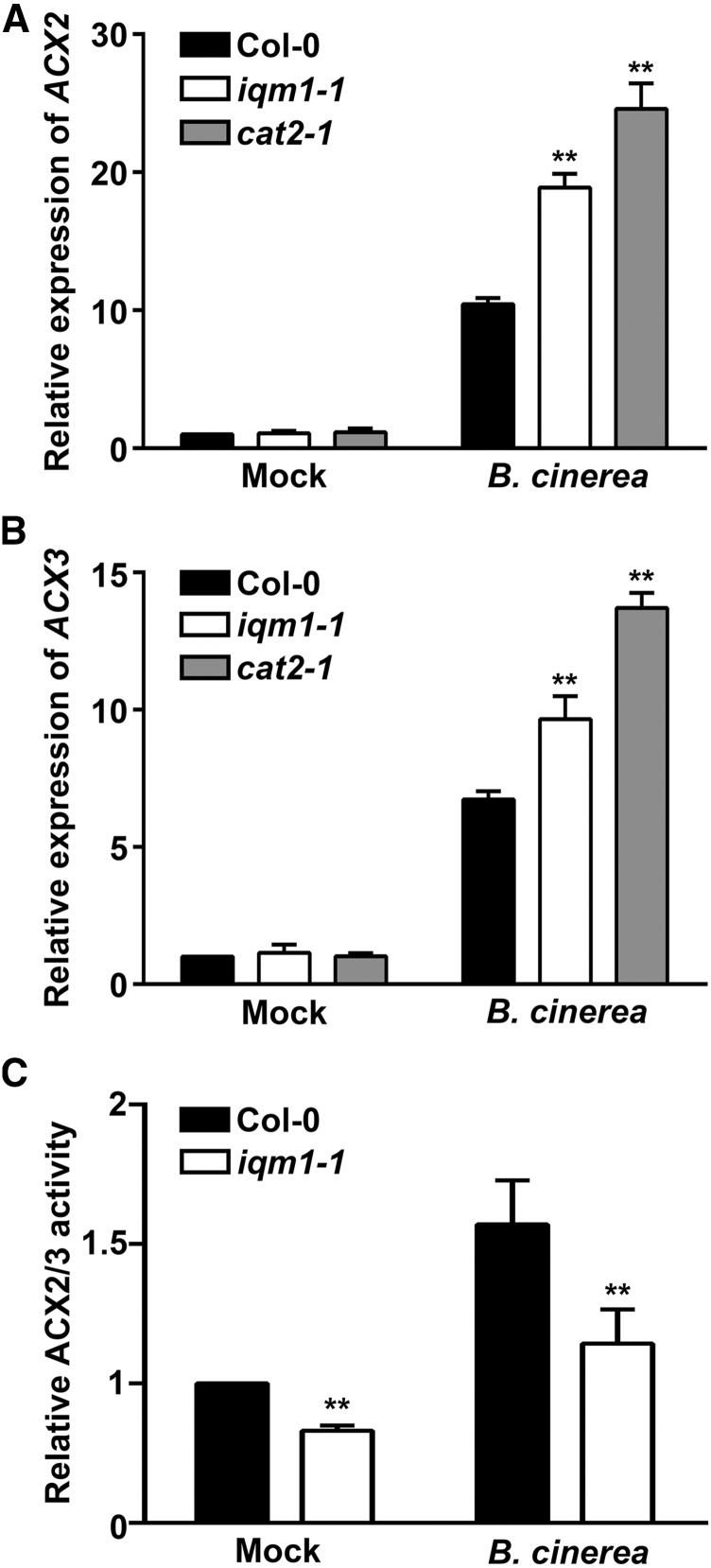
The Expression of ACX2 and ACX3 Is Increased in iqm1 and cat2 Mutants
A recent report has shown that CAT2 can interact with the JA synthetases ACX2 and ACX3 to enhance their activity and affect the synthesis of JA (Yuan et al., 2017). To clarify whether IQM1 affects JA content through ACX2/3, we first detected the expression of ACX2 and ACX3 in iqm1-1 and cat2-1 mutants using RT-qPCR. We unexpectedly found that the expression of ACX2/3 in two mutant strains was significantly higher than that in wild-type plants after infection with B. cinerea (Fig. 6, A and B). This result is in contrast to the reduced JA content of iqm1-1 and cat2-1 mutants. However, this phenomenon is very common in the process of JA synthesis regulation. For example, OPR3 is a JA biosynthetic enzyme that is upstream of ACXs in the JA synthesis pathway. The wound-induced expression of OPR3 is significantly higher in acx1 than in wild-type plants, despite the fact that acx1 mutants have notably lower JA content (Schilmiller et al., 2007). The increased expression of ACX2/3 in iqm1-1 and cat2-1 mutants was not caused by the direct regulation of IQM1 or CAT2 but resulted from self-adjustment when there were problems in some links of the JA synthesis pathway.
Figure 6.
IQM1 indirectly enhances the enzyme activity of ACX2/3 through CAT2. A and B, Expression of ACX2 and ACX3 in iqm1-1 mutants. The expression of the JA biosynthetic enzyme genes ACX2 and ACX3 was assayed in the leaves of wild-type, iqm1-1, and cat2-1 plants. Four-week-old seedlings were subjected to B. cinerea for 3 d. Total RNA was extracted at the indicated times. The transcript levels were quantified by RT-qPCR using ACTIN2 as a control. Independent biological assays were repeated three times, and similar results were obtained. Data from one representative replicate are shown. Values are means with sd from three technical measurements. C, Relative ACX2/3 activity in leaves of 4-week-old wild-type and iqm1-1 mutant plants at 3 d after infection with B. cinerea. All experiments were repeated at least three times. The data shown are means ± sd of three biological repeats. Asterisks indicate significant differences compared with Col-0 (Student’s t test, **, P < 0.01).
IQM1 Enhances the Enzyme Activity of ACX2/3 Indirectly through CAT2
Since IQM1 does not directly regulate the expression of ACX2/3, we tested ACX2/3 enzyme activity using C14-CoA as a substrate. The activity of ACX2/3 in iqm1-1 mutants was markedly lower than in the wild type. This may, at least in part, explain why the JA content of the iqm1-1 mutant was fivefold lower than that of the wild type. Under B. cinerea treatment, the ACX2/3 activity of the wild type was increased; although the ACX2/3 enzyme activity of the iqm1-1 mutant was also induced, it did not reach the same level as that in the wild type (Fig. 6C). These data suggest that even high levels of transcriptional expression cannot maintain normal levels of JA if ACX2/3 protein activity is not effectively activated.
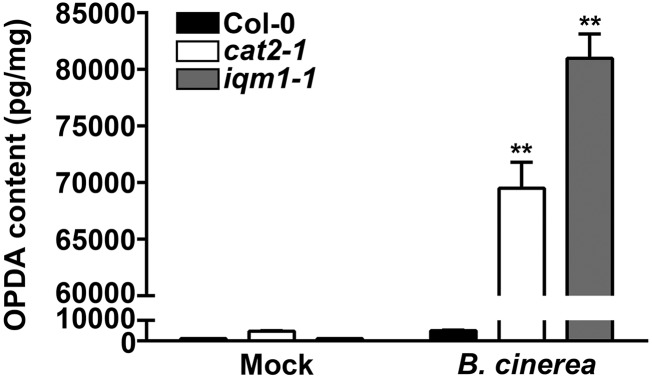
To determine whether the change in ACX2/3 enzyme activity was directly caused by IQM1, we conducted yeast two-hybrid assays upon cotransforming AH109 yeast competent cells with AD-ACX2/3 and BD-IQM1 plasmids. IQM1 and ACX2/3 could not interact with each other (Supplemental Fig. S5), indicating that IQM1 does not directly affect the activity of ACX2/3. To further prove that IQM1 and CAT2 affect JA synthesis through ACXs, we also detected the contents of OPDA in iqm1-1 and cat2-1 mutant plants. OPDA is a precursor of JA upstream of ACXs in the JA synthesis pathway. More OPDA accumulated in both iqm1-1 and cat2-1 than in the wild type under infection with B. cinerea (Fig. 7), suggesting that the decreased content of JA in iqm1-1 and cat2-1 mutants was indeed caused by the weakening of ACX activity when they challenged B. cinerea. Together, our results revealed that the main biological function of IQM1 during the plant defense response is to directly interact with and activate CAT2 and then to indirectly promote ACX activity, thus maintaining the normal JA content and the ability of plants to resist necrotrophic pathogens.
Figure 7.
Accumulation of OPDA in cat2 and iqm1 mutants. The OPDA levels of 4-week-old wild-type, cat2-1, and iqm1-1 mutant plants at 3 d after infection with B. cinerea are shown. The results were obtained from at least 30 independent plants for each genotype by mixed grinding. The data are means ± sd of three replicates. Asterisks represent significant differences compared with Col-0 (Student’s t test, **, P < 0.01).
DISCUSSION
Plant disease resistance involves a series of signaling response pathways, including Ca2+, ROS, and plant hormone pathways. These factors act coordinately, providing feedback to each other to resist external invaders, but the specific regulatory mechanism among them is still unclear (Ali et al., 2003; Lecourieux et al., 2006). A recent study revealed that CAT2 plays an important role in regulating the balance of multiple phytohormones in plants. SA inhibits the synthesis of JA and auxin by restraining the activity of CAT2 and ultimately affects plant disease resistance (Yuan et al., 2017). AtJAV1, which contains a VQ motif, interacts with CaMs in a Ca2+-dependent manner and is degraded by the RING-type E3 ubiquitin ligase JUL1 in case of injury, thus destroying the complex of JAV1-JAZ8-WRKY51 and activating JA synthesis (Yan et al., 2018; Ali et al., 2019). The Arabidopsis calmodulin-like protein AtCML37, as a potential Ca2+ sensor, affects JA synthesis and plant defense by positively regulating JAR1 expression and enzyme activity (Scholz et al., 2014). In this study, we determined that a calmodulin-binding protein, IQM1, interacts with CAT2 and CaM5 in vivo. IQM1 directly stimulates activity and increases the expression of CAT2, indirectly affecting the activity of the JA synthase ACX2/3 and thus participating in the biosynthesis of JA and in resistance to disease (Figs. 1, 2, 5, and 6). Moreover, in the presence of CaM5 and IQM1, the enzyme activity of CAT2 increased to a higher level than adding IQM1 alone; however, the addition of CaM5 alone had no effect on CAT2 activity (Fig. 5D). Previous studies have shown that AtCAT3 is a Ca2+-dependent CaM-binding protein and that Ca2+/CaM can stimulate the enzyme activity of AtCAT3 by directly binding in vitro in the presence of Ca2+ (Yang and Poovaiah., 2002). Using the binding site search and analysis function of the Calmodulin Target Database (http://calcium.uhnres.utoronto.ca), we found a potential IQ motif (LQxxxRxxxxK/R) near the 296th amino acid residue and a conserved sequence, YWSQAD(K/R)SLGQK, at the C termini of both CAT2 and CAT3, which is the interaction site between CAT3 with CaMs (Yang and Poovaiah., 2002). Therefore, it is possible that CaM5 directly binds to and activates CAT2 in the presence of Ca2+ while CaM5 stimulates CAT2 by combining with IQM1 in the absence of Ca2+, and this combination is Ca2+ independent. At this point, IQM1 may act as a bridge between CaM5 and CAT2 so the Ca2+/CaM signal can be transduced normally. We can speculate that IQM1 and other IQMs may assist CaMs in transducing signals under Ca2+-deficient or low-Ca2+ conditions. However, further research should be conducted to confirm this possibility.
There are seven CaMs in Arabidopsis, the amino acid sequences of which are highly similar (Supplemental Fig. S6), suggesting functional redundancy among CaM proteins (Toutenhoofd and Strehler, 2000). Therefore, a single mutant of CaMs may not have any distinct phenotype. We used CaM5 as the representative of CaMs to prove that CaMs or CMLs (CaM-like) can affect the activity of CAT2 enzyme through IQM1 in vitro (Fig. 5D). However, since these were in vitro experiments, does IQM1 really mediate the processes of CaM to disease resistance in planta when plants are challenged by pathogens? To this end, we generated the transgenic lines pIQM1::IQM1∆IQ/iqm1-1 with IQ motif mutation. Because the IQ motif is the binding site between IQM1 and CaMs (Zhou et al., 2012), the binding of IQM1 to all CaMs was blocked by IQ motif mutation. Our results showed that pIQM1::IQM1∆IQ cannot rescue the disease resistance defect of iqm1-1 to the wild-type level (Supplemental Fig. S1), which was consistent with the results of enzyme activity in vitro (Fig. 5D), suggesting that IQM1 is the key factor connecting CaM signal to CAT2. To sum up, our study gets through several key regulatory factors of the plant disease resistance pathway, in order CaM, CaMBP, ROS, and JA biosynthesis, and provides new insights for further understanding the mechanisms of plant defense responses.
According to previous studies, some proteins can regulate the function of CAT2 at the transcriptional and protein levels. For example, G-Box-Binding Factor1 (GBF1) inhibits the expression of CAT2 by binding to the G-box motif in the CAT2 promoter sequence in Arabidopsis, resulting in activity loss of CAT2 in leaf senescence. Overexpression of GBF1 results in the accumulation of H2O2 in plants, thus enhancing resistance to bacterial pathogens (Giri et al., 2017). In our research, IQM1 directly interacted with CAT2 in vivo. The expression of CAT2 was inhibited in the iqm1-1 mutant under B. cinerea infection (Fig. 3D), which may have been associated with direct or indirect effects of IQM1. On the other hand, IQM1 directly stimulated the enzyme activity of CAT2 through its interaction (Fig. 5, C and D). These results reveal that IQM1 can regulate the functions of CAT2 at both the transcriptional and protein levels, indicating the complexity of the JA pathway in regulating the plant disease resistance response.
Our previous study showed that IQM1 is located in the nucleus and cytoplasm in onion (Allium cepa) epidermal cells (Zhou et al., 2012). In this study, we found that IQM1 interacts with CAT2/3 in the peroxisome (Fig. 2B). It has been reported that CAT2 and CAT3 are antioxidant enzymes located in the peroxisome lumen and that catalase activity is regulated by environmental stimuli (Li et al., 2015). Whether the interaction between IQM1 and CAT2/3 was affected by abiotic stress and whether CAT2/3 recruited IQM1 to the peroxisome from the nucleus or cytoplasm remained unknown. Considering that the previous subcellular localization experiments were carried out in onion epidermal cells, due to species differences, the findings of those experiments may not reflect the true localization of the AtIQM1 gene in the native species Arabidopsis. Therefore, we again conducted a subcellular localization assay with Arabidopsis protoplast cells to confirm the localization of AtIQM1 and found that IQM1 was located in the nucleus, cytoplasm, and peroxisome under normal conditions (Supplemental Fig. S7), suggesting that the interaction between IQM1 and CAT2/3 is not induced by stress and that the role of IQM1 in promoting CAT2 is widespread and fundamental.
To adapt to multiple environmental challenges, plants have evolved a series of complex feedback regulation mechanisms to maintain the balance of growth and development (Chung et al., 2008; Spoel and Dong, 2008). With regard to posttranslational regulation of the JA biosynthetic pathway, it is hypothesized that after wounding or pathogen infection, JA is produced very rapidly in an almost instantaneous burst. However, JA levels are very low in JA synthesis-deficient plants despite the constitutive expression of JA biosynthetic genes. These situations can be explained in two possible ways. First, JA production may be limited by substrate deficiency. Second, constitutively expressed enzymes may be in an inactive state and may require posttranslational modification for activation (Schaller and Stintzi, 2009; Wasternack and Hause, 2013). In this study, the expression levels of AOS, OPR3, ACX2, and ACX3 were much higher in the iqm1-1 and cat2-1 mutants than in the wild-type plants, inconsistent with the reduced JA content (Figs. 4, B and C, and 6, A and B). This phenomenon might have been due to feedback expression and posttranslational regulation mechanisms in the plants that reinforced or amplified the capacity of the plants to synthesize JA in response to pathogen attack and thus attempted to normalize the JA content; however, such a mechanism was evidently futile and did not alter the fact that JA synthesis was blocked by the absence of IQM1 or CAT2.
Previous studies have proven that the absence of CAT2 can cause catalase activity to decrease by up to 90% (Mhamdi et al., 2010), while deletion of IQM1 only caused a 30% to 40% decrease in catalase activity in this study (Fig. 5, C and D), indicating that the activity of CAT2 is not completely dependent upon regulation by IQM1. In addition, 35S::CAT2/iqm1-1 transgenic plants had almost completely restored the increased sensitivity of the iqm1-1 mutant to B. cinerea (Supplemental Figs. S4 and S8), proving that there are some unknown factors that also affect CAT2 in response to B. cinerea. Genes of the same family may have similar or complementary functions, suggesting that other members of the IQM family might also promote or inhibit the enzyme activity of CAT2. At present, we are using the CRISPR-Cas9 gene-editing technique to construct multiple IQM family mutants to analyze the function of the IQM family in phytohormone synthesis and plant disease resistance.
Our previous microarray data showed that the expression of some pathogen-related genes is increased in iqm1-1 mutants. These results are not inconsistent with the reduced resistance of iqm1-1 to B. cinerea. First, the resistance of a plant to biotrophic and necrotrophic pathogens is usually antagonistic (Laurie-Berry et al., 2006; Thatcher et al., 2009; Nie et al., 2012; Yuan et al., 2017). In this study, the resistance of iqm1-1 mutant plants to the necrotrophic pathogen B. cinerea was weakened while that to the biotrophic pathogen Pst DC3000 was enhanced (Fig. 1; Supplemental Figs. S1 and S8). Therefore, the changes of pathogen-associated genes shown in the microarray data cannot be reflected in the real responses of iqm1-1 to a certain pathogen. Second, the microarray data were derived from direct measurement in wild-type and mutant plants without any treatment, which would not reflect the true expression of genes when plants are challenged by pathogens. Because plants have sophisticated feedback regulation mechanisms, one gene mutation may lead to the down-regulation of expression in related genes, but when plants are subjected to certain stresses, the expression of these genes in mutants may be stronger than that in the wild type (Chen et al., 2012; Piisilä et al., 2015).
In summary, we show a function of IQM1 in regulating JA biosynthesis and plant defense against the necrotrophic pathogen B. cinerea. Whether other members of the IQM family have the same function is still unknown. Our recent research found that IQM4 is involved with seed dormancy and germination in Arabidopsis by regulating ABA content (Zhou et al., 2018). Therefore, it is suggested that members of the IQM family may participate in complex hormone signaling pathways associated with plant growth and development as well as in responses to diverse biotic and abiotic stresses, thus playing a variety of important roles. Identification and characterization of more target proteins of IQMs will help in understanding the mechanisms by which IQMs regulate these diverse processes.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds of different Arabidopsis (Arabidopsis thaliana) genotypes were grown on one-half-strength Murashige and Skoog (MS) plates (Sigma-Aldrich, M5519) after sterilization with disinfectant (20% [v/v] bleach and 0.3% [w/v] SDS). After 3 d of vernalization at 4°C, the plates were transferred to growth chambers at 22°C under long-day conditions (16 h of light/8 h of dark) for 3 to 4 d. Seedlings with similar growth were transplanted into soil and grown for approximately 25 d under same growth conditions.
Trypan Blue Staining
Briefly, leaves were submerged in lactophenol-Trypan Blue staining solution (0.05% [w/v] Trypan Blue, 25% [v/v] lactic acid, 25% [v/v] water-saturated phenol, and 50% [v/v] ethanol) at 37°C for 1 h. The samples were then washed with 2.5 g mL−1 chloral hydrate solution to reduce the background and were observed with a microscope.
RNA Extraction and RT-qPCR Analyses
The RNA extraction and RT-qPCR assays were performed as previously described (Bu et al., 2014). Briefly, total RNA was isolated from 4-week-old leaves with or without infection using RNAiso plus (Takara, 9108), according to the manufacturer’s protocol. First-strand complementary DNA (cDNA) synthesis was carried out with the PrimeScript RT Reagent Kit (Takara, RR047A) following the manufacturer’s instructions. The cDNAs were used as templates in RT-qPCR with the gene-specific primers. The primer sequences are listed in Supplemental Table S1. Then, RT-qPCR was performed on 384-well plates with SYBR Premix Ex Taq II (Takara, RR820A) using a Roche Light Cycler 480 Real-Time PCR system. All experiments were repeated at least three times.
Pathogen Culture and Plant Inoculation
The Botrytis cinerea (B05.10) used in this study was maintained on 2xV8 agar plates (36 mL of V8 juice, 0.2 g of CaCO3, 2 g of agar, up to 100 mL with double distilled water) for 14 to 21 d at 22°C under dark conditions. The spores were collected with Potato Dextrose Broth (Solarbio, P9240) liquid medium. The concentration of spores was measured with a counting chamber using the microscope and adjusted to 1 × 105 mL−1. The Potato Dextrose Broth without spores was set as the control treatment. Four-week-old plants were selected for the infection assay.
For the test on detached leaves, rosette leaves at the same position and similar growth were removed. The petiole was inserted into 0.6% (w/v) agar, and each leaflet was infected with 5-μL spore suspensions on the right or left side of the main midrib. The plates were then placed under same conditions for 3 d. For the test on living plants, each leaflet was infected with 5-μL spore suspensions. The tray holding the inoculated plants was covered by a plastic cover to maintain 100% humidity. Pathogenic lesions were measured using ImageJ software.
Pseudomonas syringae pv tomato DC3000 was propagated in King’s B medium containing 50 μg mL−1 rifampicin at 28°C for 1 d. The bacterial cells were resuspended in one-tenth volumes of 10 mm MgCl2 and adjusted to a final concentration to 0.001 at OD600. In the infection experiments, 4-week-old leaves of Arabidopsis were selected and injected with bacterium suspension with a disposable sterile syringe. Plants were covered with a plastic cover to provide enough humidity for successful inoculation. To test bacterial growth, five batches of seeds were sown at the same time, cultured under the same conditions, and infected with the same concentration of bacterial suspension. Statistical analysis of biomass was sampled at 0 and 3 d post inoculation. At each time point, a 0.5-cm2 leaf disc from 10 leaves of each genotype was harvested, homogenate was spread on the tryptone Suc Glu acid plate containing 50 μg mL−1 rifampicin at 28°C for 3 d, and bacterial growth was determined.
JA and OPDA Extraction, Purification, and Quantification
The extraction, purification, and quantification of JA and OPDA were performed as described by Luo et al. (2019). Briefly, 200 mg of leaves from 4-week-old seedlings was ground into fine powder in liquid nitrogen, and the powder was homogenized and extracted for 24 h in methanol containing [2H5]JA as an internal standard. Purification was conducted with an Oasis Max solid-phase extract cartridge following centrifugation. Liquid chromatography-mass spectrometry analysis was performed on an ultra-performance liquid chromatography system (Waters) coupled to a 5500 Q-Trap system (AB Sciex). Samples were injected onto a BEH C18 column (1.7 mm, 2.1 × 150 mm; Waters) with a mobile phase consisting of 0.05% (v/v) acetic acid (A) and 0.05% (v/v) acetic acid in acetonitrile (B). The multiple reaction monitoring mode was used for quantification, and the selected multiple reaction monitoring transitions were 214 > 59 for [2H5]JA, 209 > 59 for JA, and 291 > 165 for OPDA. Each sample was obtained from at least 30 independent plants by mixed grinding and quantified three times.
Screening of Yeast cDNA Library and Yeast Two-Hybrid Assays
For screening of the yeast (Saccharomyces cerevisiae) cDNA library, the full-length coding sequence (CDS) of Arabidopsis IQM1 was cloned into pGBKT7 (Clontech) to perform a screening of the yeast cDNA library. The procedure was performed following the manufacturer’s protocols. Then, plasmids of yeast clones were extracted and transformed into the Escherichia coli DH5α strain for sequencing.
For yeast two-hybrid assays, the full-length CDSs of CAT2 and CAT3 were separately cloned into pGADT7. The plasmids of AD-CAT2 or AD-CAT3 and BD-IQM1 were cotransformed into AH109 yeast cells by the PEG/LiAC method. Interactions in yeast were tested on SD/-His-Trp-Leu-Ade plates containing 20 µg mL−1 x-α-gal. Cotransforming with the AD empty vector and BD-IQM1 was used as a control.
BiFC Assays
For the BiFC assays, the CDSs of IQM1 and CAT2/3 were cloned into the binary pSAT1-nEYFP and pSAT1-cEYFP vectors, respectively. Then, high concentrations (more than 1 µg µL−1) of pSAT1-nEYFP-IQM1, pSAT1-cEYFP-CAT2/3, and RFP-AtKAT1 plasmids were transformed into the protoplasts isolated from the leaves of 4-week-old Col-0. After 16 h of incubation at 22°C, the EYFP signal was observed using a Zeiss LSM800 confocal microscope at 488-nm absorption and 530-nm emission wavelength. Cotransforming with the nEYFP-IQM1 and cEYFP empty vector or nEYFP empty vector and cEYFP-CAT2/3 was used as negative controls.
Co-IP Assays
For Co-IP assays, the full-length CDSs of IQM1 and CAT2/3 were cloned into the 35S-pGreen-6HA and 35S-pGreenII-FLAG plant expression vectors, respectively, and then a high concentration of plasmids was cointroduced into protoplasts isolated from the leaves of 4-week-old Col-0 by PEG/CaCl2. After 16 h of incubation at 22°C, the total proteins were extracted with IP buffer (50 mm HEPES, pH 7.5, 150 mm KCl, 5 mm MgCl2, 10 µm ZnSO4, 1% [v/v] Triton X-100, 0.05% [w/v] SDS, 1 mm phenylmethylsulfonyl fluoride [PMSF], and 20 mm MG132 with Roche protease inhibitor cocktail). Protein G Plus/Protein A-Aagrose (Millipore, 2735599) beads and anti-Flag antibody (Sigma-Aldrich, F3165) were added into plant protein samples and incubated overnight at 4°C to immunoprecipitate the protein complexes. The precipitants were separated by 12% (w/v) SDS-PAGE for immunoblotting with anti-Flag antibody and anti-HA antibody (Sigma-Aldrich, H9658). The protein plus added beads and IgG was used as a negative control.
Pull-Down Assays
For the pull-down assays, the full-length CDSs of IQM1 and CAT2/3 were cloned into pDEST17 and pGEX-4T-1 to express 6×His-IQM1 and GST-CAT2/3 protein, respectively, in E. coli strain Rosetta. The total protein of E. coli was homogenized in His-lysis buffer (10 mm imidazole, 50 µm mercaptoethanol, 1% [v/v] Triton X-100, 1 mm PMSF, and 0.25 mg mL−1 lysozyme, in pH 7.5 phosphate-buffered saline ) or GST-lysis buffer (50 mm Tris-HCl, pH 8, 150 mm NaCl, 1 mm EDTA, pH 8, 1% [v/v] Triton X-100, 5 mm DTT, 1 mm PMSF, and 0.25 mg mL−1 lysozyme). The recombinant fusion protein was purified with Ni-NTA agarose (Qiagen, 30210) and Glutathione Sepharose 4B (GE Healthcare, 17-0756-01). The agarose-conjugated His-IQM1 was then successively washed with 200, 300, and 500 mm imidazole. All elutions were added into agarose-conjugated GST-CAT2/3 and incubated overnight at 4°C to immunoprecipitate the protein complexes. The precipitants were separated by 12% (w/v) SDS-PAGE for immunoblotting with anti-GST antibody (Sigma-Aldrich, SAB4200237) and anti-His antibody (Sigma-Aldrich, H1029).
Generation of the cat2/iqm1-1 Double Mutant and CAT2 Overexpression Transgenic Lines
The generation of the cat2/iqm1-1 double mutant refers to a method described previously (Su et al., 2018). The sgRNA (CCCTTCTTCACCACCAACTC and AGAAGCTTGCCAATTTCGAC) targets in the CAT2 gene were designed by CRISPR-GE (Genome Editing)-Liu YG Lab (http://skl.scau.edu.cn) and cloned into the pYLCRISPR/Cas9P35S-N vector (Ma and Liu, 2016) and then were transformed into the iqm1-1 mutant by floral dip. The transgenic T1 seeds were screened on half-strength MS medium containing 50 µg mL−1 kanamycin. The fragments covering the mutation sites were amplified from the T1 transgenic lines by PCR and sequenced to identify the successfully mutated lines. The homozygous mutants were screened from the T2 generation, and the seeds were harvested from individual lines to obtain T3 plants, of which the non-kanamycin-resistant ones were obtained.
To generate the CAT2-overexpressed transgenic lines, the full-length CDS of CAT2 was cloned into the pPZPY122-3Flag plant expression vector driven by the 35S promoter and then transformed into Col-0 and the iqm1-1 mutant by floral dip. The transgenic T1 seeds were selected and screened on half-strength MS medium containing 100 µg mL−1 gentamicin with a 3:1 separation ratio. The homozygous mutants were screened from the T2 generation.
Detection of the H2O2 Concentration
The content of H2O2 was determined by DAB staining. The specific methods are as follows. The leaves of 4-week-old wild-type and iqm1-1 mutant plants were cut and placed on a 12-well cell culture plate. Two milliliters of DAB staining solution (1 mg mL−1 DAB [Sigma-Aldrich, D8001], 10 mm Na2HPO4, and 0.05% [v/v] Tween 20, pH reduced to 3 with 0.2 m HCl) was added to each well and then gently vacuum infiltrated for 5 min to ensure that the leaves were completely immersed in liquid. The plates were then shaken 4 to 8 h at 80 to 100 rpm on a standard laboratory shaker. The DAB staining solution was then replaced with bleaching solution (ethanol:acetic acid:glycerol = 3:1:1), and the leaves were boiled for 15 min to bleach out the chlorophyll.
Measurement of Catalase Activity
The activity of catalase was measured as previously described (Su et al., 2018) using a catalase assay kit (Beyotime, S0051) following the manufacturer’s protocols. Four-week-old leaves with or without infection were collected and ground with liquid nitrogen for sample preparation. All values were normalized by the total protein concentration of the same sample. The activity of catalase is shown as units per mg of protein. All results came from three independent plant materials. For the in vitro assay, the full-length CDSs of IQM1, CAT2, and CaM5 were cloned into prokaryotic expression vector pET28a and then transformed into E. coli BL21 (DE3) competent cells. The expression and purification of proteins were performed according to a previous report (Yuan et al., 2017).
Measurement of ACX2 and ACX3 Activity
Detection of ACX2/3 activity was performed as described previously (Khan et al., 2012). Briefly, 4-week-old leaves with or without infection were ground with liquid nitrogen and then resuspended in cold extraction buffer containing 50 μm FAD, 0.025% (v/v) Triton X-100, 1× protease inhibitor cocktail (Roche, 04693132001), and 50 mm KPO4, pH 7.6. After centrifugation at 13,200g at 4°C for 15 min, the supernatant was used for assaying ACX activity. About 100 μL of the supernatant was added into 100 μL of the reaction buffer containing 100 μm C14-CoA (Myristoyl Coenzyme A; Sigma-Aldrich), 50 μm FAD, 100 μg mL−1 BSA, 0.025% (v/v) Triton X-100, 110 units of horseradish peroxidase, 50 mm p-hydroxybenzoic acid, 2 mm 4-aminoantipyrine, and 50 mm KPO4, pH 7.6. The reaction was monitored spectrophotometrically at 500 nm at 25°C for 30 min to detect H2O2 production. An H2O2 standard curve was created to calculate H2O2 concentrations (pmol H2O2 per mg of protein). All results came from three independent plant materials.
Primers Used
All primers used in this study can be found in Supplemental Table S1.
Accession Numbers
The Arabidopsis Information Resource locus identifiers for the genes mentioned in this study are as follows: AT4G33050 for IQM1, AT4G35090 for CAT2, AT1G20620 for CAT3, AT5G65110 for ACX2, AT1G06290 for ACX3, AT2G27030 for CaM5, and AT1G04710 for KAT1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypes of disease resistance in IQ motif deletion mutants.
Supplemental Figure S2. Differences in interactions between IQM1 with CAT2 and CAT3.
Supplemental Figure S3. Identification of transgenic plants.
Supplemental Figure S4. Overexpression of CAT2 can restore the increased sensitivity of iqm1 to B. cinerea.
Supplemental Figure S5. IQM1 does not interact with ACX2/3 in yeast cells.
Supplemental Figure S6. Alignment of the Arabidopsis CaM family protein.
Supplemental Figure S7. Subcellular localization of IQM1 in Arabidopsis protoplast cells.
Supplemental Figure S8. Sensitivity of all genotypes to B. cinerea in this study.
Supplemental Table S1. Primers used in this study.
ACKNOWLEDGMENTS
We thank Xingliang Hou of South China Botanical Garden, Chinese Academy of Sciences, for sharing vector and experimental methods, Yan Guo of China Agricultural University for sharing cat2 and cat3 mutants, and Chuanyou Li and Jianru Zuo of Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, for providing the B. cinerea strain and anti-CAT2 antibody.
Footnotes
This work was supported by the National Natural Science Foundation of China (311770342) and the China Postdoctoral Science Foundation (2017M622653).
References
- Ali GS, Reddy VS, Lindgren PB, Jakobek JL, Reddy ASN (2003) Differential expression of genes encoding calmodulin-binding proteins in response to bacterial pathogens and inducers of defense responses. Plant Mol Biol 51: 803–815 [DOI] [PubMed] [Google Scholar]
- Ali MRM, Uemura T, Ramadan A, Adachi K, Nemoto K, Nozawa A, Hoshino R, Abe H, Sawasaki T, Arimura GI (2019) The RING-type E3 ubiquitin ligase JUL1 targets the VQ-motif protein JAV1 to coordinate jasmonate signaling. Plant Physiol 179: 1273–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Bi C, Ma Y, Wu Z, Yu YT, Liang S, Lu K, Wang XF (2017) Arabidopsis ABI5 plays a role in regulating ROS homeostasis by activating CATALASE 1 transcription in seed germination. Plant Mol Biol 94: 197–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Yellin A, Snedden WA, Fromm H (2005) Plant-specific calmodulin-binding proteins. Annu Rev Plant Biol 56: 435–466 [DOI] [PubMed] [Google Scholar]
- Bu Q, Lv T, Shen H, Luong P, Wang J, Wang Z, Huang Z, Xiao L, Engineer C, Kim TH, et al. (2014) Regulation of drought tolerance by the F-box protein MAX2 in Arabidopsis. Plant Physiol 164: 424–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Jiang H, Li L, Zhai Q, Qi L, Zhou W, Liu X, Li H, Zheng W, Sun J, et al. (2012) The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24: 2898–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, DeFalco TA, Moeder W, Yoshioka K (2013) The Arabidopsis cyclic nucleotide-gated ion channels AtCNGC2 and AtCNGC4 work in the same signaling pathway to regulate pathogen defense and floral transition. Plant Physiol 163: 611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Koo AJK, Gao X, Jayanty S, Thines B, Jones AD, Howe GA (2008) Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol 146: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Ali GS, Simons KA, Hou J, Yang T, Reddy ASN, Poovaiah BW (2009) Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457: 1154–1158 [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Dong X (2013) Systemic acquired resistance: Turning local infection into global defense. Annu Rev Plant Biol 64: 839–863 [DOI] [PubMed] [Google Scholar]
- Galon Y, Nave R, Boyce JM, Nachmias D, Knight MR, Fromm H (2008) Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Letters 582: 943–948 [DOI] [PubMed] [Google Scholar]
- Giri MK, Singh N, Banday ZZ, Singh V, Ram H, Singh D, Chattopadhyay S, Nandi AK (2017) GBF1 differentially regulates CAT2 and PAD4 transcription to promote pathogen defense in Arabidopsis thaliana. Plant J 91: 802–815 [DOI] [PubMed] [Google Scholar]
- Jørgensen IH. (1992) Discovery, characterization and exploitation of MLO powdery mildew resistance in barley. Euphytica 63: 141–152 [Google Scholar]
- Khan BR, Adham AR, Zolman BK (2012) Peroxisomal acyl-CoA oxidase 4 activity differs between Arabidopsis accessions. Plant Mol Biol 78: 45–58 [DOI] [PubMed] [Google Scholar]
- Kim MC, Panstruga R, Elliott C, Müller J, Devoto A, Yoon HW, Park HC, Cho MJ, Schulze-Lefert P (2002) Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416: 447–451 [DOI] [PubMed] [Google Scholar]
- Kushwaha R, Singh A, Chattopadhyay S (2008) Calmodulin7 plays an important role as transcriptional regulator in Arabidopsis seedling development. Plant Cell 20: 1747–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie-Berry N, Joardar V, Street IH, Kunkel BN (2006) The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Mol Plant Microbe Interact 19: 789–800 [DOI] [PubMed] [Google Scholar]
- Lecourieux D, Ranjeva R, Pugin A (2006) Calcium in plant defence-signalling pathways. New Phytol 171: 249–269 [DOI] [PubMed] [Google Scholar]
- Levy M, Wang Q, Kaspi R, Parrella MP, Abel S (2005) Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J 43: 79–96 [DOI] [PubMed] [Google Scholar]
- Li J, Liu J, Wang G, Cha JY, Li G, Chen S, Li Z, Guo J, Zhang C, Yang Y, et al. (2015) A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 27: 908–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Li CY, Smith SM (2017) Hormone Metabolism and Signaling in Plants. Academic Press, London [Google Scholar]
- Li Y, Chen L, Mu J, Zuo J (2013) LESION SIMULATING DISEASE1 interacts with catalases to regulate hypersensitive cell death in Arabidopsis. Plant Physiol 163: 1059–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Zhang Y, Tang S, Pan J, Yu Y, Han J, Li Y, Du X, Nan Z, Sun Q (2016) AtCNGC2 is involved in jasmonic acid-induced calcium mobilization. J Exp Bot 67: 809–819 [DOI] [PubMed] [Google Scholar]
- Luo Z, Kong X, Zhang Y, Li W, Zhang D, Dai J, Fang S, Chu J, Dong H (2019) Leaf-derived jasmonate mediates water uptake from hydrated cotton roots under partial root-zone irrigation. Plant Physiol 180: 1660–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Liu YG (2016) CRISPR/Cas9-based multiplex genome editing in monocot and dicot plants. Curr Protoc Mol Biol 115: 31.6.1-31.6.21 [DOI] [PubMed] [Google Scholar]
- Mhamdi A, Queval G, Chaouch S, Vanderauwera S, Van Breusegem F, Noctor G (2010) Catalase function in plants: A focus on Arabidopsis mutants as stress-mimic models. J Exp Bot 61: 4197–4220 [DOI] [PubMed] [Google Scholar]
- Nie H, Zhao C, Wu G, Wu Y, Chen Y, Tang D (2012) SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol 158: 1847–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piisilä M, Keceli MA, Brader G, Jakobson L, Jõesaar I, Sipari N, Kollist H, Palva ET, Kariola T (2015) The F-box protein MAX2 contributes to resistance to bacterial phytopathogens in Arabidopsis thaliana. BMC Plant Biol 15: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poovaiah BW, Du L, Wang H, Yang T (2013) Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant-microbe interactions. Plant Physiol 163: 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VS, Ali GS, Reddy ASN (2002) Genes encoding calmodulin-binding proteins in the Arabidopsis genome. J Biol Chem 277: 9840–9852 [DOI] [PubMed] [Google Scholar]
- Rentel MC, Knight MR (2004) Oxidative stress-induced calcium signaling in Arabidopsis. Plant Physiol 135: 1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A, Stintzi A (2009) Enzymes in jasmonate biosynthesis: Structure, function, regulation. Phytochemistry 70: 1532–1538 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Koo AJK, Howe GA (2007) Functional diversification of acyl-coenzyme A oxidases in jasmonic acid biosynthesis and action. Plant Physiol 143: 812–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz SS, Vadassery J, Heyer M, Reichelt M, Bender KW, Snedden WA, Boland W, Mithöfer A (2014) Mutation of the Arabidopsis calmodulin-like protein CML37 deregulates the jasmonate pathway and enhances susceptibility to herbivory. Mol Plant 7: 1712–1726 [DOI] [PubMed] [Google Scholar]
- Song S, Huang H, Gao H, Wang J, Wu D, Liu X, Yang S, Zhai Q, Li C, Qi T, et al. (2014) Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26: 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Fan M, Zhang X, Gao H, Huang H, Wu D, Guo H, Xie D (2013) The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet 9: e1003653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Dong X (2008) Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3: 348–351 [DOI] [PubMed] [Google Scholar]
- Su T, Wang P, Li H, Zhao Y, Lu Y, Dai P, Ren T, Wang X, Li X, Shao Q, et al. (2018) The Arabidopsis catalase triple mutant reveals important roles of catalases and peroxisome-derived signaling in plant development. J Integr Plant Biol 60: 591–607 [DOI] [PubMed] [Google Scholar]
- Thatcher LF, Manners JM, Kazan K (2009) Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J 58: 927–939 [DOI] [PubMed] [Google Scholar]
- Toutenhoofd SL, Strehler EE (2000) The calmodulin multigene family as a unique case of genetic redundancy: Multiple levels of regulation to provide spatial and temporal control of calmodulin pools? Cell Calcium 28: 83–96 [DOI] [PubMed] [Google Scholar]
- Wan D, Li R, Zou B, Zhang X, Cong J, Wang R, Xia Y, Li G (2012) Calmodulin-binding protein CBP60g is a positive regulator of both disease resistance and drought tolerance in Arabidopsis. Plant Cell Rep 31: 1269–1281 [DOI] [PubMed] [Google Scholar]
- Wang L, Tsuda K, Sato M, Cohen JD, Katagiri F, Glazebrook J (2009) Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PLoS Pathog 5: e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B (2013) Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111: 1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Fan M, Yang M, Zhao J, Zhang W, Su Y, Xiao L, Deng H, Xie D (2018) Injury activates Ca2+/calmodulin-dependent phosphorylation of JAV1-JAZ8-WRKY51 complex for jasmonate biosynthesis. Mol Cell 70: 136–149.e7 [DOI] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW (2002) Hydrogen peroxide homeostasis: Activation of plant catalase by calcium/calmodulin. Proc Natl Acad Sci USA 99: 4097–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan HM, Liu WC, Lu YT (2017) CATALASE2 coordinates SA-mediated repression of both auxin accumulation and JA biosynthesis in plant defenses. Cell Host Microbe 21: 143–155 [DOI] [PubMed] [Google Scholar]
- Zeng H, Xu L, Singh A, Wang H, Du L, Poovaiah BW (2015) Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front Plant Sci 6: 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YP, Chen YZ, Yamamoto KT, Duan J, Tian CE (2010) Sequence and expression analysis of the Arabidopsis IQM family. Acta Physiol Plant 32: 191–198 [Google Scholar]
- Zhou YP, Duan J, Fujibe T, Yamamoto KT, Tian CE (2012) AtIQM1, a novel calmodulin-binding protein, is involved in stomatal movement in Arabidopsis. Plant Mol Biol 79: 333–346 [DOI] [PubMed] [Google Scholar]
- Zhou YP, Wu JH, Xiao WH, Chen W, Chen QH, Fan T, Xie CP, Tian CE (2018) Arabidopsis IQM4, a novel calmodulin-binding protein, is involved with seed dormancy and germination in Arabidopsis. Front Plant Sci 9: 721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski RE. (1998) Calmodulin and calmodulin-binding proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 49: 697–725 [DOI] [PubMed] [Google Scholar]
- Zou JJ, Li XD, Ratnasekera D, Wang C, Liu WX, Song LF, Zhang WZ, Wu WH (2015) Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell 27: 1445–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]