Tocopherol biosynthesis is required for effective basal resistance of Arabidopsis to Pseudomonas syringae infection.
Abstract
Tocopherols are lipid-soluble antioxidants synthesized in plastids of plants and other photosynthetic organisms. The four known tocopherols, α-, β-, γ-, and δ-tocopherol, differ in number and position of methyl groups on their chromanol head group. In unstressed Arabidopsis (Arabidopsis thaliana) leaves, α-tocopherol constitutes the main tocopherol form, whereas seeds predominantly contain γ-tocopherol. Here, we show that inoculation of Arabidopsis leaves with the bacterial pathogen Pseudomonas syringae induces the expression of genes involved in early steps of tocopherol biosynthesis and triggers strong accumulation of γ-tocopherol, moderate production of δ-tocopherol, and generation of the benzoquinol precursors of tocopherols. The pathogen-inducible biosynthesis of tocopherols is promoted by the immune regulators ENHANCED DISEASE SUSCEPTIBILITY1 and PHYTOALEXIN-DEFICIENT4. In addition, tocopherols accumulate in response to bacterial flagellin and reactive oxygen species. By quantifying tocopherol forms in inoculated wild-type plants and biosynthetic pathway mutants, we provide biochemical insights into the pathogen-inducible tocopherol pathway. Notably, vitamin E deficient2 (vte2) mutant plants, which are compromised in both tocopherol and benzoquinol precursor accumulation, exhibit increased susceptibility toward compatible P. syringae and possess heightened levels of markers of lipid peroxidation after bacterial infection. The deficiency of triunsaturated fatty acids in vte2-1 fatty acid desaturase3-2 (fad3-2) fad7-2 fad8 quadruple mutants prevents increased lipid peroxidation in the vte2 background and restores pathogen resistance to wild-type levels. Therefore, the tocopherol biosynthetic pathway positively influences salicylic acid accumulation and guarantees effective basal resistance of Arabidopsis against compatible P. syringae, possibly by protecting leaves from the pathogen-induced oxidation of trienoic fatty acid-containing lipids.
Upon inoculation with microbial pathogens, plants activate a diverse array of metabolic pathways. Thereby biosynthesized plant metabolites can positively or negatively influence plant pathogen resistance (Zeier, 2013). For instance, the shikimate pathway product salicylic acid (SA) accumulates in the leaves of pathogen-inoculated plants and orchestrates the induction of local resistance responses against biotrophic and hemibiotrophic pathogen attack, such as pattern-triggered immunity (PTI; also referred to as basal immunity) and effector-triggered immunity (ETI; Wildermuth et al., 2001; Spoel and Dong, 2012; Cui et al., 2015; Klessig et al., 2018). Plants can also extend their defense efforts from inoculated to distal leaf tissue, a phenomenon known as systemic acquired resistance (SAR; Sticher et al., 1997). SAR confers broad-spectrum immunity and is triggered by a pathogen-inducible l-Lys catabolic pathway that produces the nonprotein amino acid pipecolic acid (Pip) and its oxidized derivative N-hydroxypipecolic acid (NHP). NHP accumulation is necessary for SAR and the associated defense-priming phenomenon, and a positive interplay between NHP and SA ensures the strong resistance elevation of SAR-induced plants (Návarová et al., 2012; Bernsdorff et al., 2016; Hartmann et al., 2017, 2018). The defensive proteins ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) and PHYTOALEXIN-DEFICIENT4 (PAD4) are important components of PTI, ETI, and SAR, and they positively regulate the induction of both SA and NHP biosynthesis (Jirage et al., 1999; Feys et al., 2001; Wiermer et al., 2005; Hartmann et al., 2018; Hartmann and Zeier, 2019).
Constitutively synthesized phytoanticipins and/or inducible phytoalexins represent another category of plant defensive metabolites. They contribute to pathogen resistance by directly exerting antimicrobial activity on attacking microbes (Bednarek and Osbourn, 2009; Ahuja et al., 2012). Arabidopsis (Arabidopsis thaliana) possesses a comprehensive Trp-derived metabolism that leads to the pathogen-inducible accumulation of several indolic defense compounds with potential antimicrobial activity, such as camalexin, indole-3-carboxylic acid (ICA), 4-hydroxyindole-3-carbonyl nitrile, and the indole glucosinolate-hydrolysis product indole-3-ylmethylamine (Glawischnig, 2007; Bednarek et al., 2009; Rajniak et al., 2015; Stahl et al., 2016).
Leaves of Arabidopsis inoculated with the hemibiotrophic bacterial pathogen Pseudomonas syringae and other necrosis-inducing plant pathogens enzymatically form oxylipins, such as jasmonic acid (JA) and 12-oxophytodienoic acid (OPDA; Block et al., 2005; Grun et al., 2007; Mishina and Zeier, 2007). In concert with ethylene (ET), JA activates a defense signaling network that provides plant protection against pathogens with necrotrophic feeding mode (Thomma et al., 2001). The trienoic fatty acids α-linolenic acid (18:3) and hexadecatrienoic acid (16:3) constitute the biosynthetic precursors of JA and related oxylipins (Vick and Zimmerman, 1984, Weber et al., 1997). In Arabidopsis, they are generated by the desaturation of fatty acids via the combined action of FATTY ACID DESATURASE3 (FAD3), FAD7, and FAD8 (McConn et al., 1997).
Terpenoids are considered to be the largest and most versatile class of plant natural products (Bohlmann and Keeling, 2008). Upon P. syringae attack, Arabidopsis increases the generation of the volatile terpenoid (E,E)-4,8,12-trimethyl-1,3,7,11-tridecatetraene (Attaran et al., 2008). In addition, Arabidopsis leaves synthesize the unsaturated sterol stigmasterol from β-sitosterol upon P. syringae inoculation. Stigmasterol integrates into cell membranes and favors susceptibility to bacterial infection (Griebel and Zeier, 2010). Furthermore, the levels of the carotenoid-derived plant stress hormone abscisic acid (ABA) rise in leaves infected with virulent P. syringae (de Torres Zabala et al., 2009; Gruner et al., 2013). ABA thereby favors postinvasive multiplication of bacteria in leaves by interfering with the SA defense pathway (de Torres Zabala et al., 2009; Ding et al., 2016).
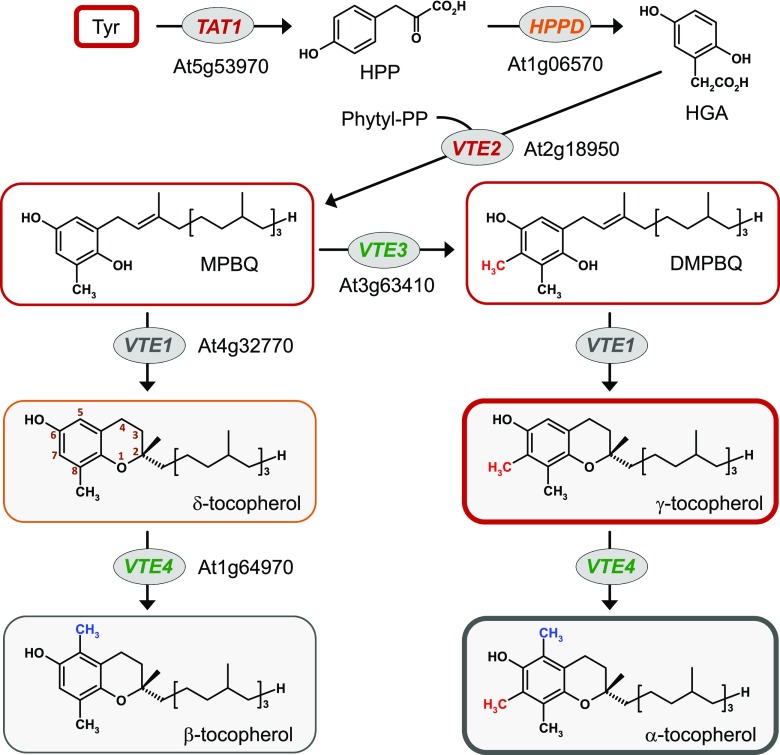
Specific branches of the shikimate and terpenoid pathways converge for the biosynthesis of tocopherols. Together with tocotrienols, tocomonoenols, and plastochromanol-8 (PC-8), tocopherols belong to a group of lipophilic hydroxychroman derivatives termed tocochromanols (DellaPenna and Pogson, 2006; Dörmann, 2007; Mène-Saffrané, 2017). Tocochromanols are almost exclusively synthesized by photosynthetic organisms, possess antioxidant capacity, and are essential for the human diet; hence, they are paraphrased by the term vitamin E (Schneider, 2005; Mène-Saffrané, 2017). They are composed of a methylated chroman-6-ol head group and a lipophilic polyprenyl side chain that varies in different tocochromanol types. In tocopherols, the most abundant tocochromanols in vascular plants (Esteban et al., 2009; Falk and Munné-Bosch, 2010), the lipophilic chain is derived from phytyl-diphosphate (phytyl-PP; Vom Dorp et al., 2015; Mène-Saffrané, 2017). The four naturally occurring tocopherol forms are α-, β-, γ-, and δ-tocopherol, which differ in number and position of methyl groups on their chromanol group (DellaPenna and Pogson, 2006).
The endogenous levels of the different tocopherol forms in plants vary between different tissues. Seeds mainly produce γ-tocopherol, whereas leaf chloroplast membranes predominantly contain α-tocopherol. Tocopherols possess antioxidant properties, can chemically scavenge various types of reactive oxygen species (ROS), and prevent the oxidation of polyunsaturated lipids. By this means, they exert critical protective functions in seeds and in the chloroplast membrane (Dörmann, 2007; Maeda and DellaPenna, 2007; Falk and Munné-Bosch, 2010). Studies on tocopherol-deficient Arabidopsis mutants show that tocopherols are vital for seed longevity, early seedling development, and overall plant fitness (Sattler et al., 2004; Mène-Saffrané et al., 2010). Moreover, tocopherol levels increase in plants in response to various abiotic stressors, and mutant analyses suggest that they are involved in plant tolerance to high light, cold, drought, heavy metal, and salinity stress (Bergmüller et al., 2003; Collakova and DellaPenna, 2003b; Havaux et al., 2005; Maeda et al., 2006; Collin et al., 2008; Ellouzi et al., 2013).
In this study, we show that inoculation of Arabidopsis leaves with P. syringae significantly activates tocopherol biosynthesis, which results in a strong accumulation of γ-tocopherol in infected leaves. We provide biochemical and regulatory information on this pathogen-inducible tocopherol response and show that vitamin E deficient2 (vte2) mutant plants impaired in the generation of both tocopherols and tocopherol precursors exhibit increased pathogen-induced generation of lipid peroxidation markers, attenuated SA biosynthesis, and increased susceptibility to compatible P. syringae. Our data indicate that tocopherol biosynthesis is subject to EDS1/PAD4 regulation and required for proper basal immunity of Arabidopsis to compatible bacterial pathogens.
RESULTS
Inoculation with P. syringae Induces Tocopherol Biosynthesis in Arabidopsis Leaves and Results in a Strong Accumulation of γ-Tocopherol
Genome-wide expression analyses show that the inoculation of Arabidopsis leaves with P. syringae triggers a large transcriptional response in both the locally inoculated and systemic leaf tissue (Gruner et al., 2013; de Torres Zabala et al., 2015; Lewis et al., 2015; Bernsdorff et al., 2016). Evaluation of two different sets of microarray data from leaf samples of Columbia-0 (Col-0) inoculated with compatible P. syringae pv maculicola ES4326 (Psm), compatible P. syringae pv tomato DC3000 (Pst), or incompatible Pst avrRpm1 suggested that the expression levels of several tocopherol biosynthetic genes are changed following bacterial attack (Supplemental Table S1).
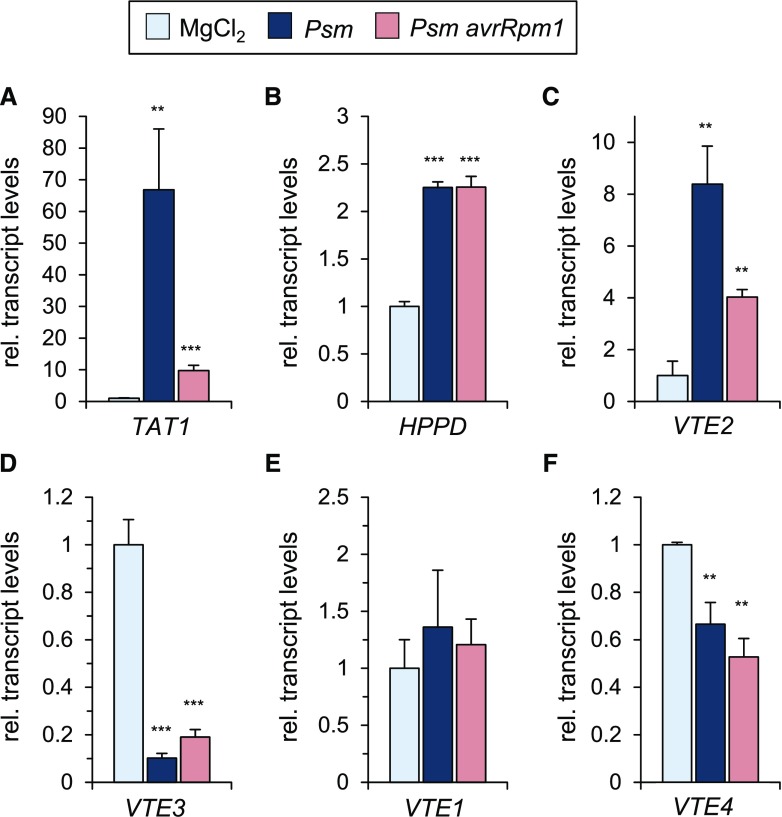
The biosynthesis of tocopherols in plants involves a two-step conversion of l-Tyr to homogentisic acid (HGA), the precursor for the chromanol ring of tocopherols (Fig. 1). l-Tyr is first deaminated to p-hydroxyphenylpyruvate (HPP) by Tyr aminotransferase. In Arabidopsis, TYROSINE AMINOTRANSFERASE1 (TAT1) takes over a significant portion of this transamination reaction (Riewe et al., 2012; Wang et al., 2019). In a second step, HPP is oxygenated to HGA by p-hydroxyphenylpyruvate dioxygenase (HPPD; DellaPenna and Pogson, 2006). The lipophilic side chain of tocopherols originates from phytyl-PP that is either synthesized de novo out of geranylgeranyl diphosphate or arises from the degradation of chlorophyll (Vom Dorp et al., 2015). The HGA phytyltransferase VTE2 condenses phytyl diphosphate and HGA to generate the benzoquinol derivative 2-methyl-6-phytyl-1,4-benzoquinol (MPBQ), which can subsequently be methylated to 2,3-dimethyl-6-phytyl-1,4-benzoquinol (DMPBQ) by the methyltransferase VTE3 (Fig. 1). The tocopherol cyclase VTE1 then catalyzes the intramolecular cyclization of MPBQ and DMPBQ, respectively, to δ- and γ-tocopherol. Finally, VTE4, another methyltransferase, methylates the chromanol head groups of δ- and γ-tocopherol to generate β- and α-tocopherol, respectively (Fig. 1). We examined the transcript levels of the tocopherol biosynthetic genes TAT1, HPPD, VTE1, VTE2, VTE3, and VTE4 by reverse transcription quantitative PCR (RT-qPCR) analysis in leaves of plants inoculated with virulent Psm and avirulent, hypersensitive response (HR)-inducing Psm avrRpm1 at 48 h post inoculation (hpi). Notably, the leaf expression levels of the early tocopherol biosynthetic genes TAT1, HPPD, and VTE2 significantly increased in response to inoculation with Psm and Psm avrRpm1 (Fig. 2). By contrast, VTE3 and VTE4 transcript levels decreased after bacterial inoculation, and expression levels of VTE1 remained unchanged in response to Psm or Psm avrRpm1 inoculation (Fig. 2). These observations were consistent with the tendencies displayed by the public microarray data (Supplemental Table S1).
Figure 1.
The tocopherol biosynthetic pathway in Arabidopsis and its P. syringae-triggered induction in leaves. Tyr, l-Tyr. In Arabidopsis, TAT1 and HPPD are localized in the cytosol, while other tocopherol pathway enzymes are in the chloroplast (Mène-Saffrané, 2017). The coloring of the tocopherol biosynthetic genes represents their degree of regulation in response to P. syringae inoculation (red, strongly up-regulated; orange, moderately up-regulated; black, no significant change; green, down-regulated). The coloring of the frames surrounding the metabolites indicates the degree of pathogen-induced metabolite accumulation (red, strong; orange, moderate; gray, no significant change), and the frame line widths illustrate the absolute levels of the metabolites in inoculated leaves of wild-type plants.
Figure 2.
Inoculation of Arabidopsis leaves with compatible Psm or HR-inducing Psm avrRpm1 trigger transcriptional changes of genes involved in tocopherol biosynthesis. Relative expression levels are shown for tocopherol biosynthetic genes upon Psm and Psm avrRpm1 inoculation (OD600 = 0.005) at 48 hpi. A, TAT1. B, HPPD. C, VTE2. D, VTE3. E, VTE1. F, VTE4. Transcript levels represent means ± sd of three biological replicate leaf samples from different plants and are expressed relative to the levels of mock-control (MgCl2) samples. Asterisks above the bars denote statistically significant differences between Psm or Psm avrRpm1 inoculation and mock inoculation: ***, P < 0.001 and **, P < 0.01 (two-tailed Student’s t test).
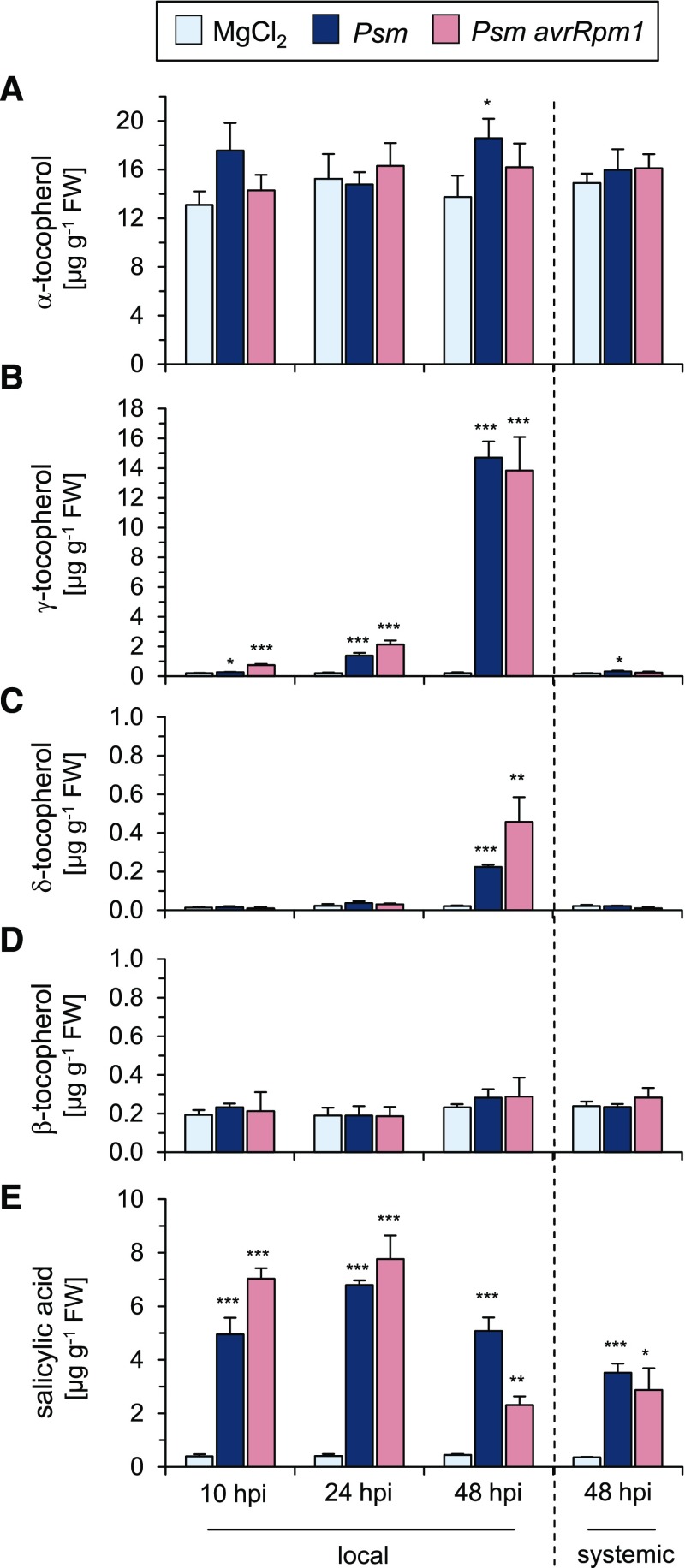
To examine if the altered leaf expression levels of tocopherol biosynthetic genes after bacterial attack were associated with changes in tocopherol levels, we conducted gas chromatography-mass spectrometry (GC-MS)-based metabolite analysis of extracts from Psm-, Psm avrRpm1-, and mock-infiltrated Col-0 leaves (Supplemental Fig. S1). We thereby determined the levels of the four different tocopherols in inoculated leaves at 10, 24, and 48 hpi and compared them with the accumulation of SA in the same time period (Fig. 3). This analysis confirmed that α-tocopherol, which had basal levels of about 15 µg g−1 fresh weight in the leaves of mock-control plants, constitutes the main leaf tocopherol form in unstressed Arabidopsis Col-0 plants (DellaPenna, 2005). The levels of α-tocopherol did not substantially alter in response to bacterial attack in the majority of performed experiments (Fig. 3A; Supplemental Figs. S2B and S3). With values around 0.2 µg g−1 fresh weight, the basal levels of γ-tocopherol were markedly lower than those of α-tocopherol. Notably, γ-tocopherol started to accumulate from 10 h after inoculation upon attack by both the compatible and the incompatible P. syringae strain and reached high values of about 15 µg g−1 fresh weight at 48 hpi (Fig. 3B; Supplemental Fig. S2A). Moreover, the contents of δ-tocopherol rose from barely detectable basal levels to pathogen-induced levels between 0.2 and 0.5 µg g−1 fresh weight (Fig. 3C; Supplemental Fig. S2C), and the levels of β-tocopherol remained unchanged at basal levels between 0.2 and 0.3 µg g−1 fresh weight after bacterial inoculation (Fig. 3D; Supplemental Fig. S2D). Comparatively, the phenolic defense hormone SA accumulated strongly at 10 h after both Psm and Psm avrRpm1 treatment (Fig. 3E), indicating that SA production starts earlier than tocopherol biosynthesis in response to bacterial inoculation. In accordance with previous results (Attaran et al., 2009), SA also markedly accumulated in systemic leaves distant from P. syringae inoculation, while the levels of tocopherols hardly changed in the systemic tissue (Fig. 3). Together, our data indicate that early genes of the tocopherol biosynthetic pathway are transcriptionally activated following leaf attack by virulent and avirulent P. syringae and that this culminates in a strong accumulation of γ-tocopherol and more moderate increases in δ-tocopherol in the inoculated tissue.
Figure 3.
Temporal accumulation of tocopherols and SA upon Psm and Psm avrRpm1 inoculation. Endogenous levels of α-tocopherol (A), γ-tocopherol (B), δ-tocopherol (C), β-tocopherol (D), and SA (E) are shown in treated (local) leaves of Arabidopsis Col-0 plants at 10, 24, and 48 h following MgCl2 treatment, Psm inoculation, or Psm avrRpm1 inoculation (OD600 = 0.005 each) and in distant (systemic) leaves at 48 h post treatment. Values are given in µg g−1 fresh weight (FW) and represent means ± sd of at least three biological replicates. Each replicate sample consisted of tissue from different plants. Six leaves from two plants were pooled for one replicate. Asterisks denote statistically significant differences between control (MgCl2) values and values of Psm- or Psm avrRpm1-treated leaf samples: ***, P < 0.001; **, P < 0.01; and *, P < 0.05 (two-tailed Student’s t test).
γ-Tocopherol Accumulates in Response to Exogenous Flagellin or ROS Treatments, and Its Pathogen-Inducible Biosynthesis Is Boosted by EDS1/PAD4 Signaling
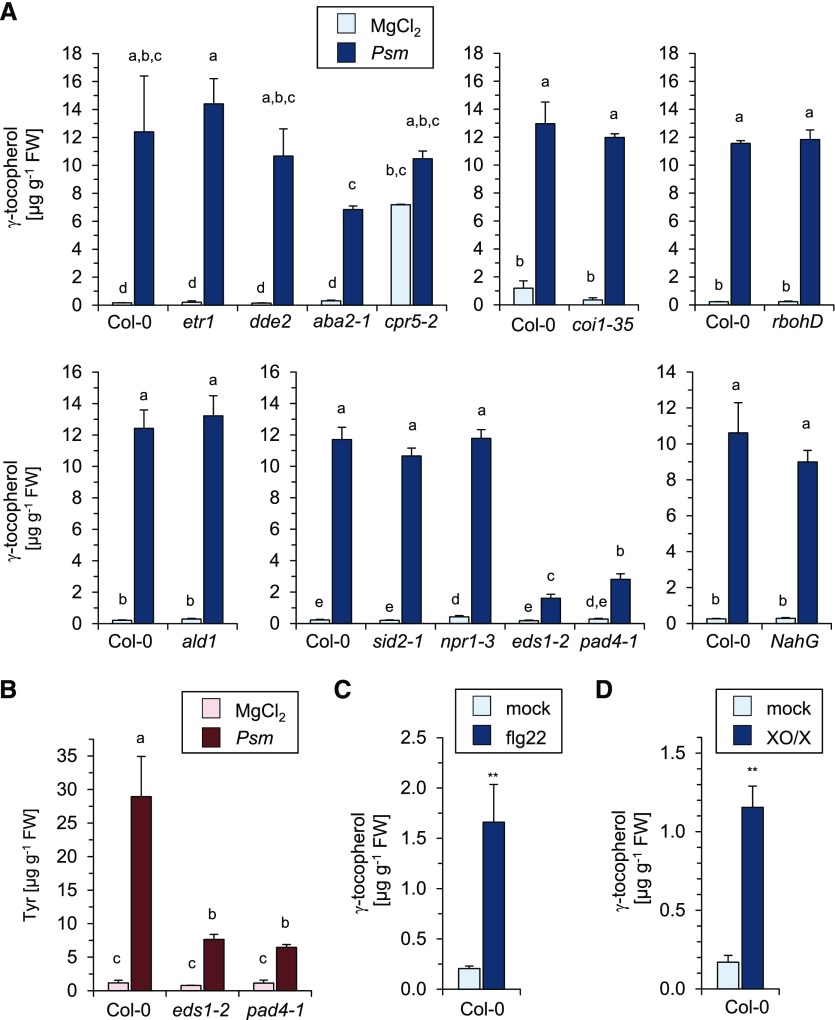
To examine the regulatory principles of the P. syringae-inducible activation of tocopherol biosynthesis, we determined the levels of the most strongly accumulating tocopherol variant, γ-tocopherol, in Arabidopsis mutant plants that exhibit defects in defense- or stress hormone-related signaling pathways (Fig. 4). At 48 hpi, the accumulation of γ-tocopherol upon Psm inoculation was wild type like in the JA biosynthetic mutant delayed dehiscence2 (dde2; von Malek et al., 2002), the JA perception-defective mutant coronatine insensitive1 (coi1-35; Xie et al., 1998), the ET signaling mutant ethylene response1 (etr1; Bleecker et al., 1988), the ABA biosynthetic mutant ABA deficient2 (aba2-1; Léon-Kloosterziel et al., 1996), the Pip and NHP biosynthetic mutant agd2-like defense response protein1 (ald1; Návarová et al., 2012; Hartmann et al., 2018), the SA biosynthetic mutant salicylic acid induction deficient2 (sid2-1; Wildermuth et al., 2001), the NahG transgenic line deficient in SA accumulation (Lawton et al., 1995), and the SA perception-defective mutant nonexpressor of pathogen-related gene1 (npr1-3; Fig. 4A; Cao et al., 1997). By contrast, γ-tocopherol accumulation at 48 hpi was greatly reduced in the eds1-2 and pad4-1 defense signaling mutants (Fig. 4A; Jirage et al., 1999; Bartsch et al., 2006). This compromised induction of tocopherol biosynthesis in eds1-2 and pad4-1 was associated with an attenuated pathogen-induced accumulation of Tyr, the tocopherol precursor amino acid, in these mutants (Fig. 4B). In addition, Arabidopsis constitutive expression of PR genes5 (cpr5-2; Bowling et al., 1997; Mateo et al., 2006), which exhibits constitutively activated immune signaling and ROS production, contained elevated γ-tocopherol levels in the absence of pathogen challenge (Fig. 4A). However, respiratory burst oxidase homolog D (rbohD), which is compromised in the oxidative burst that follows attack by avirulent P. syringae strains such as Psm avrRpm1 or Pst avrRpm1 (Fig. 4A; Torres et al., 2002; Griebel and Zeier, 2010), showed increases in γ-tocopherol levels similar to the wild type upon Psm or Psm avrRpm1 inoculation (Fig. 4A).
Figure 4.
Regulatory aspects of inducible tocopherol biosynthesis. A, The Psm-triggered induction of γ-tocopherol biosynthesis in Arabidopsis leaves is dependent on EDS1 and PAD4 but independent of SA signaling. Accumulation of γ-tocopherol is shown in the leaves of wild-type Col-0 and different mutant plants impaired in defense- or stress hormone-related signaling at 48 h after Psm inoculation or mock treatment: etr1 (not responsive to ET), dde2 (deficient in JA biosynthesis), aba2-1 (deficient in ABA biosynthesis), cpr5-2 (constitutive defense responses and ROS production), coi1-35 (JA insensitive), rbohD (deficient in oxidative burst-associated O2− production), ald1 (deficient in Pip and NHP biosynthesis), sid2-1 (deficient in SA biosynthesis), npr1-3 (deficient in SA signal transduction), eds1-2 (defective in EDS1), pad4-1 (defective in PAD4), and NahG (deficient in SA accumulation). Different letters above the bars denote statistically significant differences (P < 0.05, ANOVA and posthoc Tukey’s honestly significant difference [HSD] test). Other details are as described in Figure 3. B, Psm-inducible accumulation of the tocopherol precursor amino acid Tyr is promoted by EDS1 and PAD4 signaling. Levels of Tyr are shown in control (MgCl2-infiltrated) leaves and Psm-inoculated leaves of Col-0, eds1-2, and pad4-1 plants at 24 h post infiltration. Details are as described in (A). C and D, Exogenous treatment with flagellin and generation of O2− both trigger the induction of γ-tocopherol biosynthesis. C, Leaves of Col-0 plants were infiltrated with 100 nm flg22 peptide or water (mock). D, A mixture of 0.5 units mL−1 XO and 0.5 mm X or 0.5 mm X without enzyme (mock treatment) was infiltrated into leaves. Leaves were harvested 48 h post treatment for analyses. Other details are as described in Figure 3. FW, Fresh weight.
Treatment with bacterial flagellin and exogenously applied ROS can induce defense responses and metabolic alterations, such as stigmasterol accumulation in Arabidopsis leaves (Mishina and Zeier, 2007; Griebel and Zeier, 2010). We therefore examined if flagellin acts as a trigger for inducible γ-tocopherol generation by leaf-infiltrating Col-0 plants with a solution of 100 nm flg22, the elicitor-active 22-mer peptide corresponding to the consensus sequence of bacterial flagellin (Gómez-Gómez et al., 1999). γ-Tocopherol significantly accumulated in the flg22-treated leaves, albeit to lower absolute levels than in P. syringae-inoculated leaves (Fig. 4C). One of the earliest responses of plants after flagellin perception is O2− production (Qi et al., 2017). We thus tested whether an enzyme/substrate mix of 0.5 units mL−1 xanthine oxidase (XO) and 0.5 mm xanthine (X), which generates O2− in Arabidopsis leaves to stress-physiological levels (Delledonne et al., 1998), would induce γ-tocopherol accumulation. Indeed, leaf infiltration with an XO/X mixture caused a similar increase in γ-tocopherol levels compared with treatment with flg22 (Fig. 4D).
Together, these results indicate that the pathogen-inducible biosynthesis of γ-tocopherol and its precursor amino acid Tyr in Arabidopsis is strongly promoted by the EDS1/PAD4 defense signaling node. Moreover, activation of tocopherol biosynthesis proved to be independent of SA signaling and of the NHP-, JA-, ET-, and ABA-associated defense pathways, and stimulation with bacterial flagellin as well as exogenous treatment with O2− was sufficient to induce tocopherol biosynthesis.
Pathogen-Inducible Tocopherol Biosynthesis Requires Intact VTE1 and VTE2 Genes, Largely Depends on TAT1, and Is Qualitatively Altered in Plants with Mutated VTE4 and VTE3 Alleles
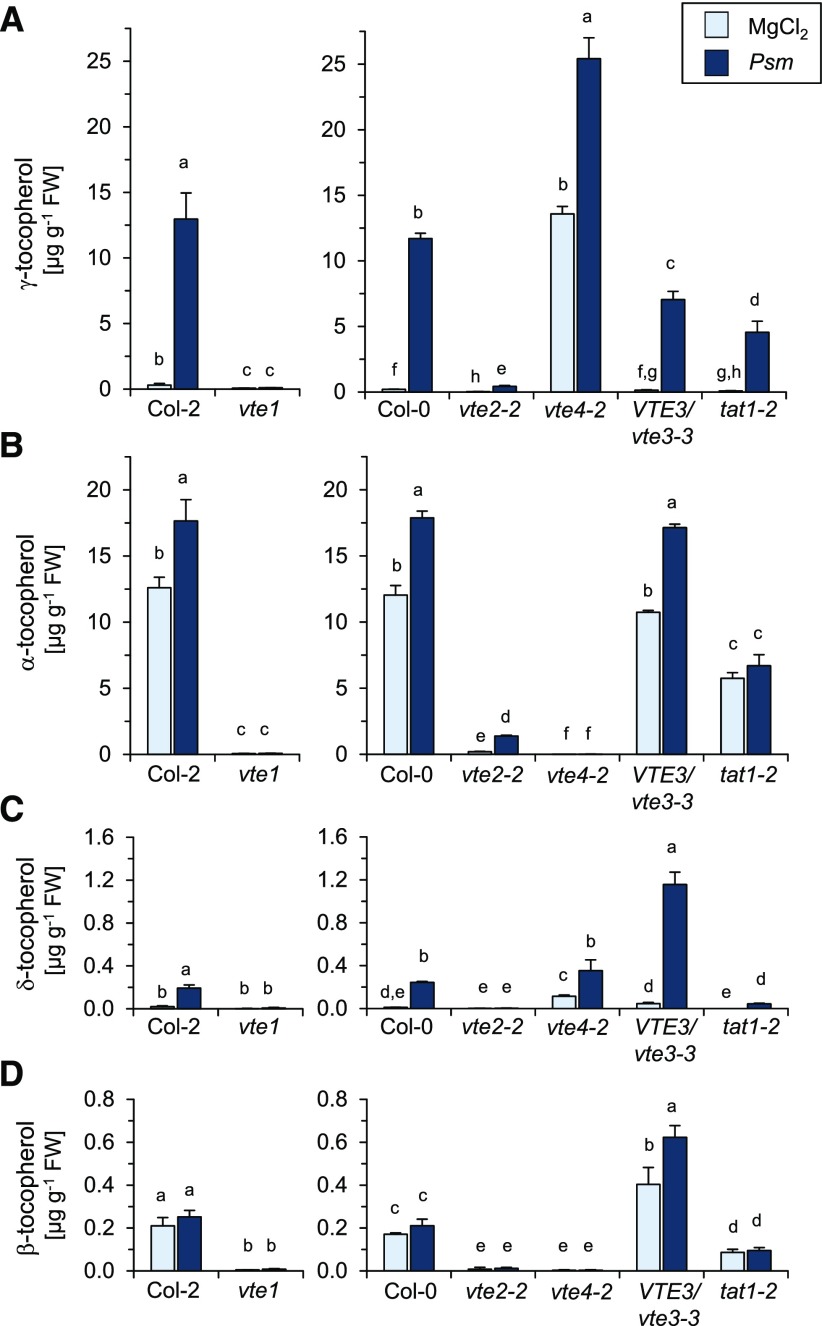
To further characterize the biotic stress-inducible generation of tocopherols in Arabidopsis leaves, we took advantage of the availability of mutant plants impaired in the aforementioned steps of the tocopherol biosynthetic pathway (Fig. 1). The employed mutants included the gene knockout lines vte1 and vte2-2 that were previously obtained from ethyl methanesulfonate mutagenesis (Porfirova et al., 2002; Havaux et al., 2005). Former analyses of different vte1 and vte2 mutants indicated that tocopherols are strongly reduced in their leaves under stress-free conditions (Porfirova et al., 2002; Sattler et al., 2003; Havaux et al., 2005). To examine a possible accumulation of tocopherols in the leaves of vte1 and vte2-2 mutant plants in response to biotic stress, we compared the leaf tocopherol levels at 48 h after Psm and mock treatments in the two mutants with those of the wild-type Col-2 and Col-0 plants. Col-2 and Col-0 plants both showed similar tocopherol accumulation patterns at 48 hpi (Fig. 5), just as described in the previous experiments for Col-0 (Fig. 3). By contrast, the leaves of mock- and Psm-treated vte1 plants only contained faint levels of all four tocopherol forms, indicating that both the basal and inducible biosynthesis of tocopherols is virtually absent in vte1 (Fig. 5). Similar to vte1, the leaves of vte2-2 control plants exhibited strongly reduced basal levels of α-tocopherol and only traces of the other tocopherols. Moreover, upon Psm inoculation, vte2-2 lacked the δ-tocopherol accumulation observed in the wild type and showed strongly diminished accumulation of γ- and α-tocopherol (Fig. 5). Thus, the P. syringae-inducible tocopherol biosynthesis in Arabidopsis leaves essentially requires both intact HGA phytyltransferase VTE2 and tocopherol cyclase VTE1 genes (Fig. 1).
Figure 5.
Psm-triggered tocopherol accumulation in leaves is quantitatively and qualitatively altered in tocopherol biosynthetic mutants. Endogenous levels of tocopherols are shown in leaves of different Arabidopsis lines infiltrated with 10 mm MgCl2 (mock treatment) or inoculated with Psm at 48 h post treatment. Different letters above the bars denote statistically significant differences of the values in each section (P < 0.05, ANOVA and posthoc Tukey’s HSD test). Other details are as described in Figure 3. The vte1 mutant is in the Col-2 background; all the other mutants are in the Col-0 background. A, Levels of γ-tocopherol. B, Levels of α-tocopherol. C, Levels of δ-tocopherol. D, Levels of β-tocopherol. FW, Fresh weight.
We also analyzed tat1-2 mutant plants (SALK_141402), which harbor a T-DNA insertion in the Tyr aminotransferase gene TAT1 and exhibit reduced basal tocopherol levels (Wang et al., 2019). Consistently, tat1-2 showed less than half of the levels of α- and β-tocopherol than the Col-0 wild type in mock-control plants. Moreover, the strong and moderate accumulation of γ- and δ-tocopherol observed for Col-0 plants, respectively, were both attenuated by more than 50% in tat1-2 mutant plants (Fig. 5). This indicates that a large part of inducible tocopherol biosynthesis proceeds via TAT1.
In addition, we examined tocopherol levels in the SALK_031151 line that carries a predicted T-DNA insertion in the MPBQ methyltransferase gene VTE3. For this line, however, we only identified plants heterozygous for the T-DNA integration by PCR-based genotyping (Alonso et al., 2003), which might be related to the previously described viability defects of homozygous vte3 mutants (Cheng et al., 2003). We thus employed heterozygous plants termed VTE3/vte3-3 for tocopherol analyses. The VTE3/vte3-3 plants showed wild type-like levels of α-tocopherol but decreased accumulation of γ-tocopherol. Conversely, δ- and β-tocopherol moderately overaccumulated in VTE3/vte3-3 after Psm inoculation. Finally, we examined vte4-2 mutant plants that carry a T-DNA insertion in the coding sequence of the γ-tocopherol methyltransferase gene VTE4 (Fig. 1; Bergmüller et al., 2003). Consistent with previous findings (Bergmüller et al., 2003), leaves of noninoculated vte4-2 plants lacked basal levels of α- and β-tocopherol and contained strongly and moderately elevated levels of γ- and δ-tocopherol, respectively. Moreover, Psm inoculation resulted in a strong overaccumulation of γ-tocopherol to levels of about 25 µg g−1 fresh weight and a wild type-like δ-tocopherol accumulation, while α- and β-tocopherol remained undetectable. Together, the qualitative changes of the individual tocopherol levels in VTE3/vte3-3 and vte4-2 plants are consistent with the functions of VTE3 and VTE4 as MPBQ methyltransferase and γ-/δ-tocopherol methyltransferase, respectively. Our analyses also demonstrate that the pathogen-inducible tocopherol biosynthetic pathway is channeled to predominantly produce tocopherols with a methyl group at the 7-position of the chromanol ring (mainly γ-tocopherol and occasionally α-tocopherol; Fig. 1).
vte2 Mutant Plants Exhibit Increased Susceptibility to Virulent P. syringae
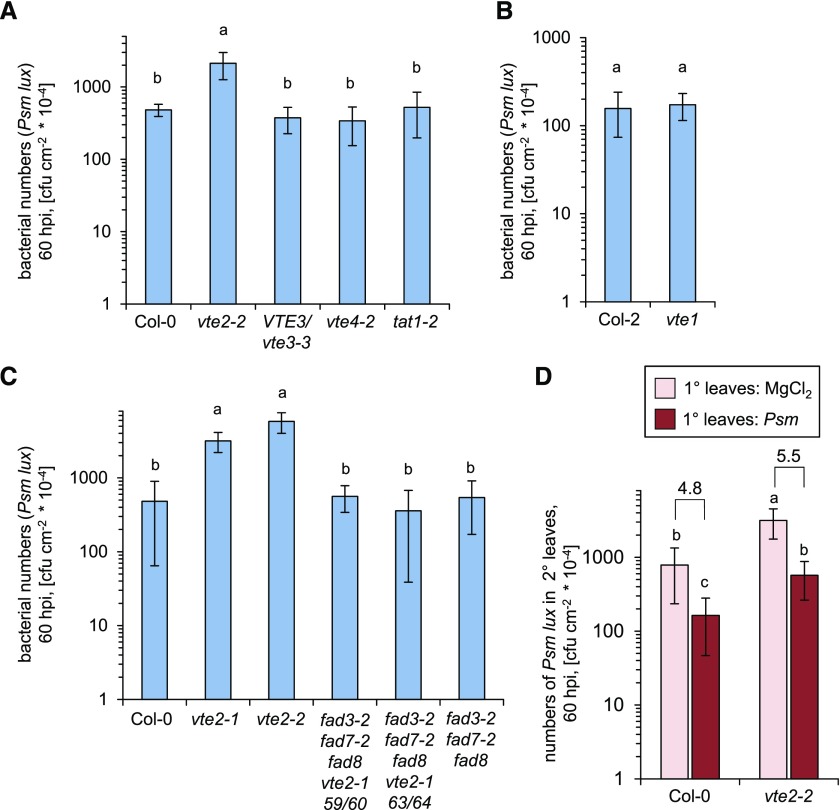
We next examined whether the observed accumulation of tocopherols in P. syringae-inoculated leaves would be functionally relevant for immunity against bacterial infection. To determine plant basal resistance to P. syringae, we leaf inoculated the tocopherol biosynthetic mutants and the corresponding wild-type accessions with a compatible Psm strain that expresses the luxCDABE operon from Photorhabdus luminescens (Psm lux) and assessed bacterial growth via the quantification of bioluminescence (Fan et al., 2008; Gruner et al., 2018; Hartmann et al., 2018). Bacterial growth in the leaves of vte1, VTE3/vte3-3, vte4-2, and tat1-2 was similar to the respective wild type, but the vte2-2 mutant allowed significantly higher bacterial proliferation than the wild-type Col-0 and the other lines under investigation (Fig. 6A). In several independent experiments, the average numbers of Psm lux in the leaves of vte2-2 were fourfold to eightfold higher than the numbers in Col-0 leaves at 60 hpi (Fig. 6, A and C; Supplemental Fig. S4), indicating that vte2-2 suffers from impaired basal resistance to infection by the virulent P. syringae strain. Moreover, vte2-1, another VTE2 loss-of-function mutant, showed a similar deficiency in basal resistance to vte2-2 (Fig. 6C). This impaired basal immunity was also evident on the phenotypic level, since leaves of vte2-2 plants showed more pronounced disease symptoms in terms of leaf yellowing and necrosis than the other lines under investigation (Supplemental Fig. S5). Together, these results indicate that the HGA phytyltransferase gene VTE2 is required for a full basal immune response of Arabidopsis against challenge by compatible P. syringae.
Figure 6.
VTE2 contributes to basal resistance against Psm infection. A to C, Basal resistance to Psm infection of different Arabidopsis lines. Three leaves of naïve plants were inoculated with bioluminescent Psm lux (OD600 = 0.001), and bacterial numbers in leaves were assessed 60 h later. Data represent means ± sd of growth values from at least 10 replicate plants. Different letters above the bars denote statistically significant differences (P < 0.001, ANOVA and posthoc Tukey’s HSD test). The results were similar in three independent experiments (see Supplemental Fig. S4). D, Systemic acquired resistance in Col-0 and vte2-2 plants. Three lower (1°) leaves of a given plant were infiltrated with 10 mm MgCl2 or inoculated with Psm (OD600 = 0.005), and three upper (2°) leaves were challenge inoculated 2 d later with Psm lux (OD600 = 0.001). Growth of Psm lux in 2° leaves was assessed 60 h post 2° challenge inoculation. Data represent means ± sd of growth values from at least nine replicate plants. Different letters above the bars denote statistically significant differences (P < 0.01, ANOVA and posthoc Tukey’s HSD test). The numbers above the brackets in D indicate the fold growth reduction due to the SAR effect. The results were similar in two independent experiments.
A localized leaf inoculation with Psm can induce SAR in the entire Arabidopsis foliage (Mishina and Zeier, 2007). Since common regulatory principles of basal resistance and SAR have been described previously (Shah and Zeier, 2013), we tested whether the defect of vte2 in basal immunity would be accompanied by compromised SAR. To this end, Col-0 and vte2-2 plants were either inoculated with Psm in three lower (1°) leaves or mock treated in 1° leaves with 10 mm MgCl2 (Mishina and Zeier, 2007). Two days later, SAR establishment was assessed by challenging upper (2°) leaves of the differently pretreated plants with Psm lux and determining bacterial growth in 2° leaves 2.5 d later. Bacterial growth was decreased to a similar factor in Col-0 and vte2-2 plants as a consequence of the Psm preinoculation, suggesting that VTE2 and an intact tocopherol biosynthetic pathway are not necessary for SAR activation (Fig. 6D). However, as with naïve or mock-pretreated control plants, SAR-induced vte2-2 mutants allowed higher bacterial growth than SAR-induced Col-0 plants (Fig. 6D), indicating that tocopherols strengthen the levels of resistance to the compatible Psm lux strain under both basal and SAR-induced conditions.
Our finding that tocopherol levels also increased in response to the incompatible Psm avrRpm1 strain prompted us to test whether specific resistance against this avirulent, HR-inducing pathogen is altered in tocopherol biosynthetic mutants. However, resistance against Psm avrRpm1 was similar to the wild type for all the tocopherol mutants under investigation (Supplemental Fig. S6).
vte2 Mutant Plants Are Impaired in Basal and P. syringae-Induced (D)MPBQ Precursor Biosynthesis
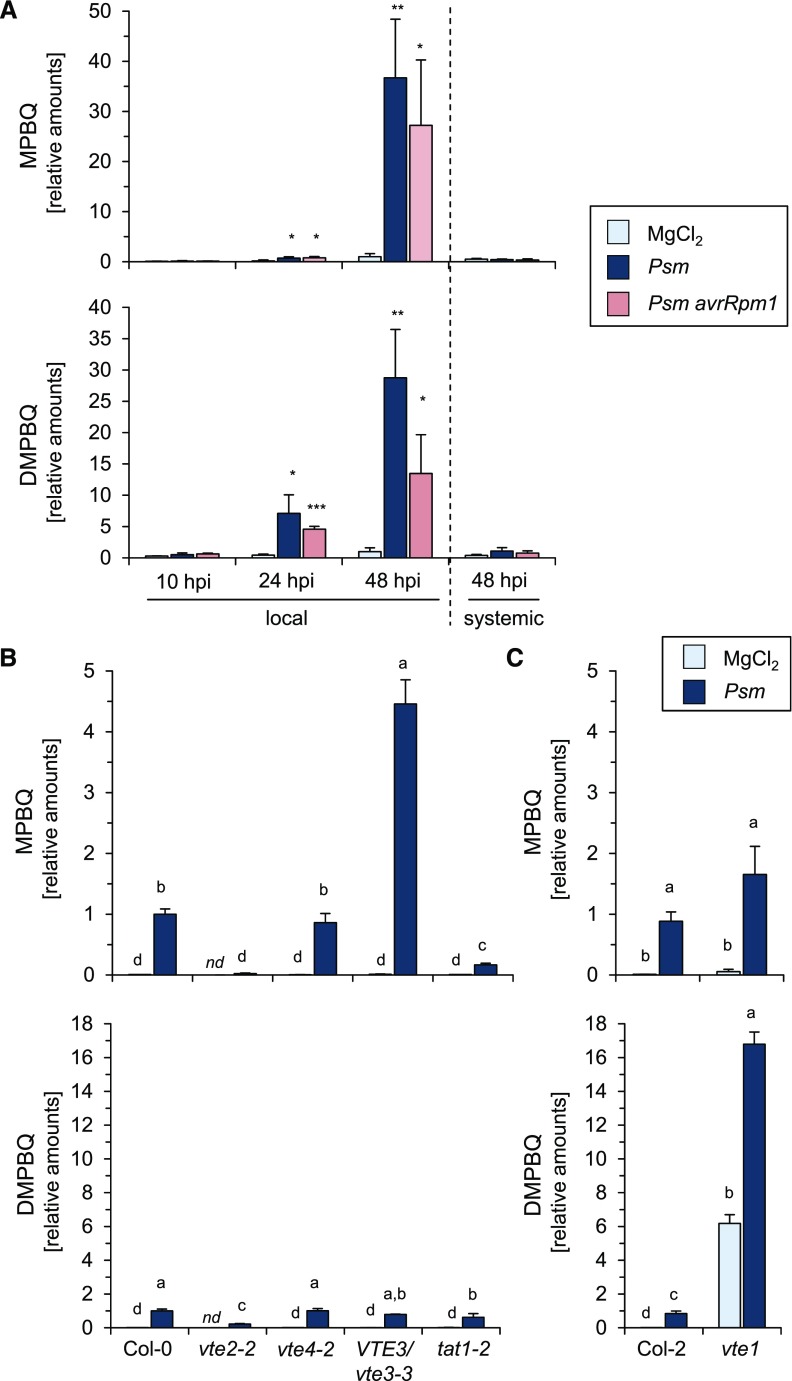
The vte2-2 mutant is strongly deficient in basal and Psm-inducible tocopherol biosynthesis and exhibits attenuated basal resistance, which is in accordance with the assumption that tocopherols could play a role in basal immunity to bacterial infection. However, the metabolic phenotype of the vte1 mutant argued against this hypothesis, because vte1 lacked basal and pathogen-induced tocopherol accumulation and still exhibited wild type-like basal resistance (Figs. 5 and 6). A marked difference between vte1 and vte2 mutant plants concerns the basal levels of the benzoquinol precursors MPBQ and DMPBQ, which are elevated in vte1 and diminished in vte2, respectively (Porfirova et al., 2002; Sattler et al., 2003; Havaux et al., 2005). We detected both tocopherol precursors with our GC-MS-based method in the leaf extracts (Supplemental Fig. S7; Porfirova et al., 2002; Kobayashi and DellaPenna, 2008). Due to the unavailability of authentic standards, we could not determine absolute (D)MPBQ amounts but were able to quantitatively assess the treatment- and genotype-dependent relative levels of the two tocopherol precursors in leaves. In the wild type, the levels of both MPBQ and DMPBQ started to rise at 24 h post Psm or Psm avrRpm1 attack and were strongly increased at 48 hpi in inoculated leaves (Fig. 7A). This accumulation was significantly reduced in tat1-2 and almost absent in vte2-2 (Fig. 7B). Consistent with a role of VTE3 as an MPBQ methyltransferase, MPBQ overaccumulated about fourfold in VTE3/vte3-3 (Fig. 7B). Moreover, the vte1 mutant contained elevated basal levels of DMPBQ and, compared with the Col-2 wild type, overaccumulated this tocopherol precursor to about 20-fold in response to Psm inoculation (Fig. 7C). Due to the relative abundances of the MPBQ, DMPBQ, and γ-tocopherol peaks in the respective ion chromatograms (Supplemental Fig. S7), we carefully estimated the accumulation of MPBQ and DMPBQ at 48 h post Psm attack to be in the low µg/g range for wild-type plants, suggesting that the pathogen-induced DMPBQ levels in vte1 are in a similar quantitative range to the induced γ-tocopherol levels of the wild type. Together, the accumulation patterns of MPBQ and DMPBQ upon bacterial inoculation are consistent with the biosynthetic scenario depicted in Figure 1. Moreover, our analyses show that vte2 is the only tocopherol biosynthetic mutant that is deficient in both tocopherol and benzoquinol precursor accumulation, a characteristic that is associated with enhanced susceptibility to compatible P. syringae attack.
Figure 7.
P. syringae inoculation induces the accumulation of the tocopherol precursors (D)MPBQ, which are absent in vte2 and strongly overaccumulate in vte1. A, Relative amounts of MPBQ (top) and DMPBQ (bottom) in local leaves of Arabidopsis Col-0 plants at 10, 24, and 48 h following MgCl2 treatment, Psm inoculation, or Psm avrRpm1 inoculation (OD600 = 0.005 each) and in systemic leaves at 48 h post treatment. Details are as described for Figure 3. Amounts of (D)MPBQ in leaves are given relative to the mean of the 48-h mock samples, which consequently has a numerical value of 1. MPBQ and DMPBQ were identified according to their mass spectra (Supplemental Fig. S7). B and C, Relative amounts of MPBQ (top) and DMPBQ (bottom) in leaves of different mock-treated or Psm-inoculated Arabidopsis lines at 48 h post treatment. Different letters above the bars denote statistically significant differences of the values of each subfigure (P < 0.05, ANOVA and posthoc Tukey’s HSD test). Other details are as described in Figure 5. Amounts of (D)MPBQ in leaves are given relative to the mean of the Col-0 48-h Psm samples, which as a consequence has a numerical value of 1.
The Enhanced Disease Susceptibility Phenotype of vte2 Is Associated with Attenuated SA Biosynthesis and Increased Production of Oxidation Markers for Triunsaturated Lipids upon Bacterial Infection
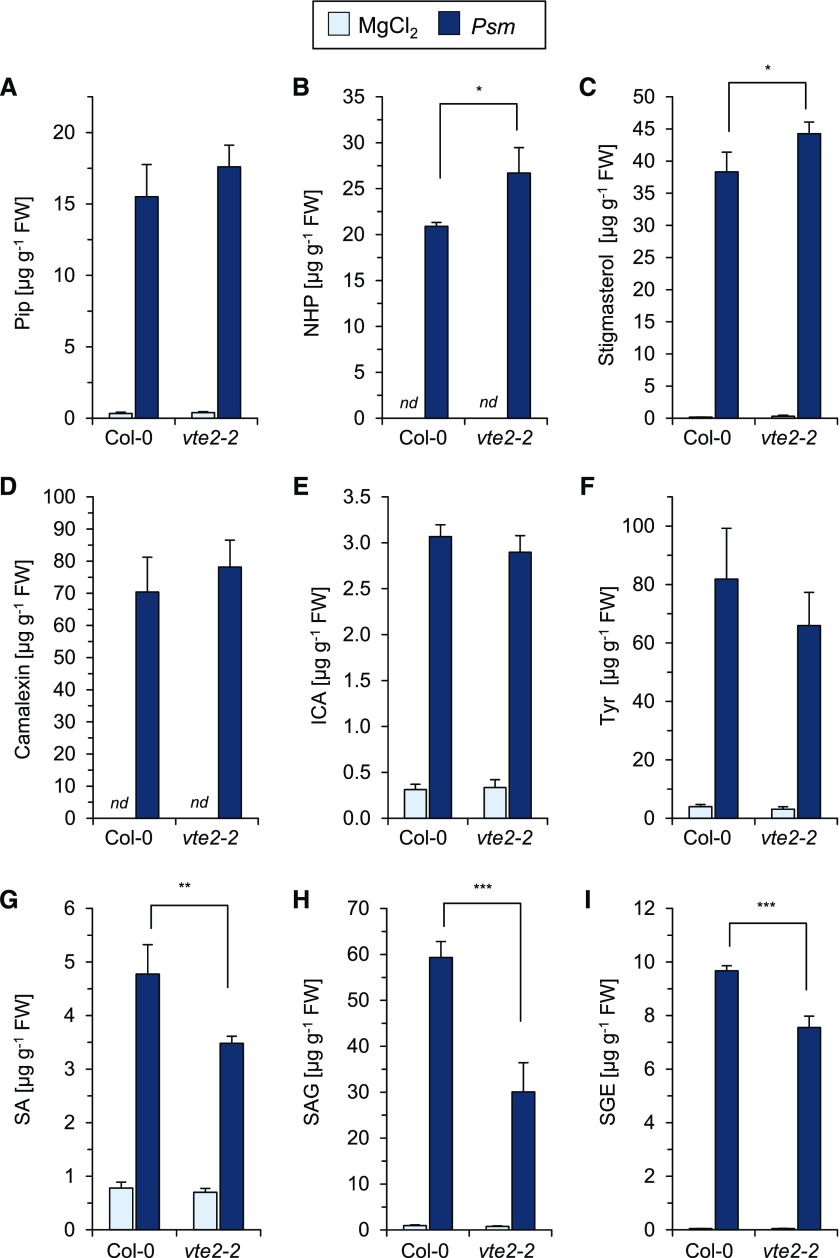
We next examined whether the alleviated basal resistance of vte2 was related to a compromised production of defense-related metabolites in this mutant (Fig. 8). At 24 h post Psm inoculation, the leaves of wild-type Col-0 plants accumulated substantial amounts of the pipecolate pathway products Pip and NHP, the unsaturated sterol stigmasterol, the indolics camalexin and ICA, the tocopherol precursor amino acid Tyr, SA, and the glycosylated SA derivatives SA-β-glucoside (SAG) and SA glucose ester (SGE). While Pip, camalexin, ICA, and Tyr generation was similar in Col-0 and vte2-2, NHP and stigmasterol production was moderately more prominent in the mutant. Remarkably, the pathogen-induced accumulation of SA, SAG, and SGE was significantly lower in vte2-2 than in Col-0, so that the total levels of SA (i.e. the sum of SA, SAG, and SGE) were about 1.8-fold higher in wild-type than in vte2-2 leaves after Psm attack (Fig. 8). Therefore, the attenuated resistance of vte2 to Psm infection is accompanied by a reduced biosynthesis of SA, a main determinant of plant basal immunity.
Figure 8.
The vte2-2 mutant is not generally impaired in the production of defense-related metabolites but exhibits attenuated SA biosynthesis. Endogenous levels are shown for defense-related metabolites at 24 h post Psm inoculation or 10 mm MgCl2 infiltration in leaves of Col-0 and vte2-2 mutant plants. Values given represent means ± sd of four replicate leaf samples. nd, not detected. Other details are as described in Figure 3. Asterisks above the brackets denote statistically significant differences between the Psm samples of Col-0 and vte2-2: ***, P < 0.001; **, P < 0.01; and *, P < 0.05 (two-tailed Student’s t test). A, Pip. B, NHP. C, Stigmasterol. D, Camalexin. E, ICA. F, Tyr. G, SA. H, SAG. I, SGE. FW, Fresh weight.
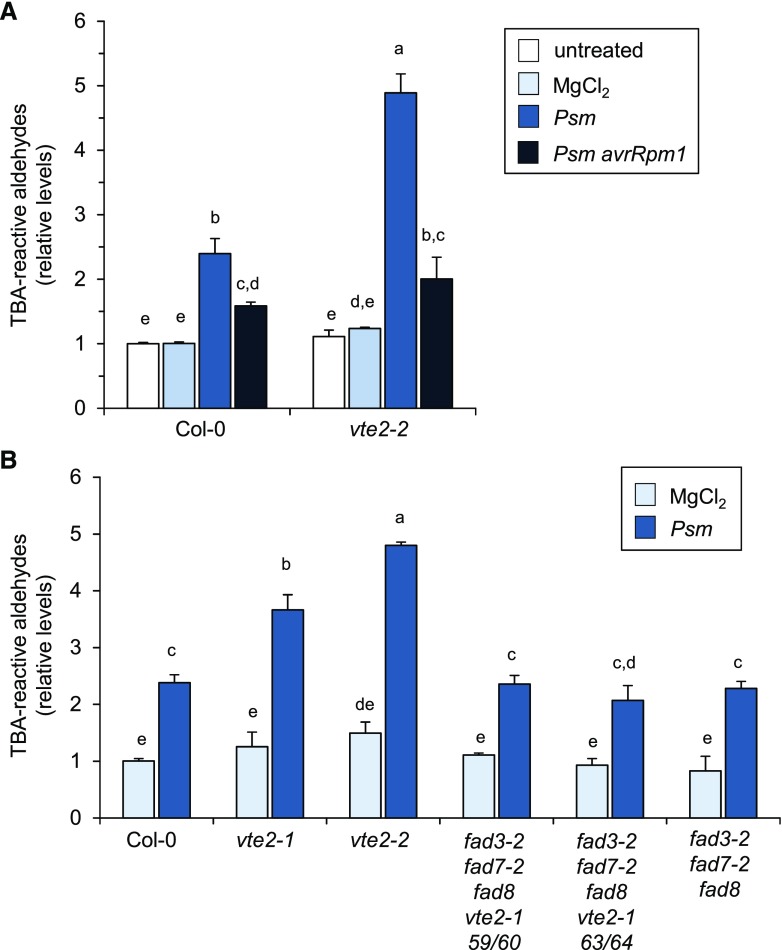
Tocopherols, as well as their benzoquinol precursurs MPBQ and DMPBQ, possess antioxidant activities and can protect polyunsaturated lipids, such as trienoic fatty acids, from oxidative damage (Schneider, 2005; Sattler et al., 2006). For example, previous studies with vte2 mutant plants indicated that tocopherols diminish lipid peroxidation in Arabidopsis during seed and early seedling development (Sattler et al., 2004, 2006). We estimated the degree of lipid peroxidation using the thiobarbituric acid (TBA) assay, which determines the amounts of malondialdehyde and other aldehydic products of the lipid peroxidation process (Hodges et al., 1999; Yin et al., 2010; Ayala et al., 2014). The results of the TBA assay suggested that challenge of wild-type Col-0 plants with Psm is accompanied by increased lipid peroxidation in leaves (Fig. 9). The amount of lipid peroxidation also increased in leaves inoculated with the avirulent Psm avrRpm1 strain, but to a lower degree than in leaves infected with compatible Psm (Fig. 9A). Notably, the levels of estimated lipid peroxidation products after Psm challenge were significantly higher in both vte2-2 and vte2-1 compared with the wild type, indicating increased pathogen-induced lipid peroxidation in the vte2 plants (Fig. 9).
Figure 9.
vte2 mutants exhibit increased oxidation of triunsaturated lipids following Psm infection. A, The generation of TBA-reactive aldehydes was assessed in Arabidopsis Col-0 plants as a marker for lipid peroxidation in untreated leaves, in leaves infiltrated with 10 mm MgCl2, and in leaves inoculated with Psm or Psm avrRpm1 (OD600 = 0.005 each) in both Col-0 and vte2-2 plants at 48 h post treatment. Values represent means ± sd of three replicate leaf samples from different plants. Each replicate sample consisted of six leaves (three leaves from two plants). Peroxidation levels were normalized to the level measured in untreated Col-0 leaves. Different letters above the bars denote statistically significant differences (P < 0.01, ANOVA and posthoc Tukey’s HSD test). B, Assessment of lipid peroxidation in leaves of mock-treated or Psm-inoculated Col-0, vte2-1, vte2-2, and fad3-2 fad7-2 fad8 plants as well as in two different lines (59/60 and 63/64) of the quadruple mutant fad3-2 fad7-2 fad8 vte2-1 at 48 h post treatment. Details are as described in A.
The biosynthesis of the plant trienoic fatty acids 18:3 and 16:3 in Arabidopsis is mediated by the three fatty acid desaturases FAD3, FAD7, and FAD8, and a triple mutant with defects in all three FAD genes (fad3-2 fad7-2 fad8) is deficient of trienoic fatty acid-containing leaf lipids (McConn and Browse, 1996). Previous work has employed a trienoic acid- and tocopherol-deficient fad3-2 fad7-2 fad8 vte2-1 quadruple mutant to estimate the degree of peroxidation of triunsaturated lipids in vte2 mutant plants (Mène-Saffrané et al., 2007). Notably, the overproduction of lipid peroxidation products estimated for vte2-1 in response to Psm attack was absent in the fad3-2 fad7-2 fad8 vte2-1 mutant, and the quadruple mutant plants showed similar Psm-induced peroxidation levels to wild-type and fad3-2 fad7-2 fad8 triple mutant plants (Fig. 9B). This suggests that the assessed overproduction of pathogen-induced TBA-reactive aldehydes in vte2 is due to the oxidation of lipids composed of trienoic fatty acids. Interestingly, the fad3-2 fad7-2 fad8 vte2-1 quadruple mutant exhibited similar resistance to Psm lux infection than the wild type and the fad3-2 fad7-2 fad8 triple mutant and higher basal resistance than vte2-1 and vte2-2 plants (Fig. 6C). Therefore, the attenuated basal resistance phenotype of Arabidopsis in the tocopherol- and benzoquinol precursor-deficient vte2 background is related to the levels of trienoic fatty acid-containing lipids in leaves and might be associated with decreased protection of these triunsaturated lipids from peroxidation in the course of bacterial pathogen attack.
To directly assess the levels of lipid-esterified trienoic fatty acids, we performed lipid extraction from Psm-inoculated and mock-control leaves according to Bligh and Dyer (1959), transesterified extracted lipids with a boron trifluoride/methanol solution, and analyzed the released methyl esters of 18:3 and 16:3 with GC-MS. We observed the tendency that Psm inoculation causes a decrease of lipid-bound 18:3 and 16:3 at 48 hpi and that this tendency appeared more significant in the vte2-2 mutant plants than in the Col-0 wild type (Supplemental Fig. S8, A and B). In addition, we spotted two substances in these analyses that markedly increased upon Psm inoculation of leaves. The mass spectrum of the compound with the higher retention time was virtually identical to the published mass spectrum of the methyl ester of iso-12-oxophytodienoic acid (iso-OPDA; Supplemental Fig. S8, B and C; Dabrowska et al., 2011), an isomer of the 18:3-derived oxylipin OPDA. The mass spectrum of the lower-molecular-weight compound differed in putative M+ and M+-OCH3 ions by 28 mass units from the analogous masses of the iso-OPDA spectrum but otherwise exhibited a closely similar fragmentation pattern, suggesting a putative iso-dinor-OPDA structure for this compound (Supplemental Fig. S8, B and D). Dinor-OPDA, the ET homolog of OPDA, was reported as a 16:3-derived oxylipin (Weber et al., 1997). The accumulation of both iso-OPDA and the putative iso-dinor-OPDA upon Psm inoculation tended to be higher in samples from vte2-2 than Col-0 (Supplemental Fig. S8A), which is in line with our assumption of heightened 18:3 and 16:3 oxidation in the vte2 mutant.
DISCUSSION
Upon biotic stress exposure, plants induce the biosynthesis of a variety of metabolites that may activate plant defense responses, possess antimicrobial activity, or otherwise exert influences on plant immunity. Here, we report that leaf inoculation of Arabidopsis plants with either a virulent or an HR-inducing P. syringae strain results in a significant induction of the tocopherol biosynthetic pathway (Fig. 1). P. syringae challenge resulted in a massive accumulation of γ-tocopherol (up to ∼15 µg g−1) and a moderate biosynthesis of δ-tocopherol in leaves (Figs. 3 and 5; Supplemental Fig. S2). The levels of α-tocopherol, the dominant tocopherol form in unstressed Arabidopsis leaves (DellaPenna, 2005; ∼12–15 µg g−1), remained unchanged in response to bacterial attack in most of the performed experiments (Figs. 3 and 5; Supplemental Fig. S2 and S3), and β-tocopherol levels always were constant at low basal levels (Figs. 3 and 5). Thus, at a stage of bacterial infection that is characterized by the first appearance of chlorotic disease symptoms (Supplemental Fig. S5), Arabidopsis leaves possess about equal levels of α- and γ-tocopherol. In addition to tocopherols, P. syringae inoculation of leaves triggers a significant accumulation of MPBQ and DMPBQ, the direct biosynthetic precursors of tocopherols (Figs. 1 and 7).
Biochemical and Regulatory Aspects of Pathogen-Inducible Tocopherol Biosynthesis in Arabidopsis Leaves
The pathogen-triggered activation of tocopherol biosynthesis begins with an accumulation of the precursor amino acid Tyr, which essentially provides the phenolic ring of the chromanol head group (Fig. 1; DellaPenna and Pogson, 2006). For instance, Tyr accumulated in Arabidopsis leaves inoculated with the compatible Psm strain or the HR-inducing P. syringae pv tomato avrRpm1 strain sevenfold to 25-fold above basal levels (Figs. 4B and 8F; Návarová et al., 2012; Monteoliva et al., 2014). The increased availability of Tyr is coupled with an enhanced expression of the early tocopherol biosynthetic genes TAT1 and HPPD (Fig. 2; Supplemental Table S1), which is supposed to direct the metabolic flow toward the aromatic tocopherol precursor HGA (Fig. 1). The reduced tocopherol and (D)MPBQ accumulation phenotype of the tat1-2 mutant indicates that more than 50% of pathogen-inducible tocopherol biosynthesis is mediated by the Tyr aminotransferase TAT1 (Figs. 5B and 7). To a similar extent, TAT1 is involved in basal tocopherol production in unstressed leaves (Fig. 5B; Riewe et al., 2012; Wang et al., 2019). Sequence comparisons suggest that Arabidopsis possesses nine TAT1 homologs (Wang et al., 2016), some of which might code for Tyr aminotransferases that further contribute to basal and inducible tocopherol biosynthesis. In Arabidopsis, TAT1 and HPPD are localized in the cytosol, while other tocopherol pathway enzymes reside in the chloroplast. Thus, transport processes such as export of plastid-synthesized Tyr to the cytosol or plastidial import of HGA might be involved in the regulation of tocopherol biosynthesis (Bernsdorff et al., 2016; Mène-Saffrané, 2017; Hartmann and Zeier, 2018).
In addition to TAT1 and HPPD, the HGA phytyltransferase gene VTE2 is significantly up-regulated in response to bacterial attack (Fig. 2; Supplemental Table S1). Besides HGA, VTE2-mediated MPBQ biosynthesis requires phytyl-PP as a substrate (Mène-Saffrané, 2017). Phytyl-PP for tocopherol biosynthesis mainly originates from the recycling of phytol moieties released in the course of chlorophyll catabolism by two successive phosphorylation reactions catalyzed by the phytol kinase VTE5 and the phytyl-phosphate kinase VTE6 (Ischebeck et al., 2006; Valentin et al., 2006; Vom Dorp et al., 2015). Chlorophyll is markedly degraded in Arabidopsis leaves suffering P. syringae infection (Mecey et al., 2011; Supplemental Fig. S5), and this might favor chlorophyll-derived phytyl-PP generation for MPBQ and tocopherol biosynthesis. Therefore, transcriptional activation of early pathway genes and metabolic precursor availability supposedly represent a main driving force for the P. syringae-induced accumulation of tocopherols. This assumption is supported by several previous studies. For example, feeding with HGA and/or phytol was sufficient to trigger a significant increase of tocopherol levels in plant cell cultures of different species (Furuya et al., 1987; Caretto et al., 2004; Mène-Saffrané, 2017), and overexpression of the VTE2 gene in transgenic Arabidopsis plants resulted in a marked overaccumulation of tocopherols (Collakova and DellaPenna, 2003a). Since only very small residual tocopherol and (D)MPBQ levels were detected in vte2-2 mutant plants, the pathogen-inducible activation of tocopherol biosynthesis proceeds almost exclusively via the HGA phytyltransferase VTE2 (Figs. 5 and 7).
The expression levels of the (D)MPBQ cyclase gene VTE1 did not alter in response to bacterial inoculation (Fig. 2E). This suggests that the basal expression levels of VTE1 in wild-type plants are sufficient to effectively cyclize the elevated amounts of benzoquinol substrates generated upon P. syringae attack into tocopherols. In spite of that, elevated VTE1 transcription can promote tocochromanol biosynthesis, as indicated previously by the finding that VTE1-overexpressing Arabidopsis lines showed increased tocopherol and PC-8 contents (Kanwischer et al., 2005; Zbierzak et al., 2009). The complete lack of tocopherols in P. syringae-inoculated vte1 mutants confirms that the pathogen-inducible tocopherol biosynthesis entirely proceeds via VTE1 (Fig. 5), like the basal tocopherol production in unstressed plants (Porfirova et al., 2002). Consistently, vte1 strongly overaccumulated the direct γ-tocopherol precursor DMPBQ in response to Psm inoculation (Fig. 7). By contrast, overaccumulation of the δ- and β-tocopherol precursor MPBQ was hardly observed in vte1, suggesting that pathogen-inducible tocopherol biosynthesis involves effective methylation of MPBQ to DMPBQ and a subsequent VTE1-mediated cyclization of DMPBQ to γ-tocopherol. In this way, the metabolic flow is directed toward generation of the 7-methylated tocopherol variant γ-tocopherol (Fig. 1), which is, together with the constitutively synthesized α-tocopherol, the predominant tocopherol form in the P. syringae-inoculated wild-type leaf (Fig. 3; Supplemental Fig. S2).
The methylation of MPBQ to DMPBQ in Arabidopsis is catalyzed by the methyltransferase VTE3 (Cheng et al., 2003). In line with the involvement of VTE3 in the conversion of MPBQ to DMPBQ in P. syringae-inoculated leaves, heterozygous VTE3/vte3-3 plants overaccumulated MPBQ and δ-tocopherol upon bacterial attack (Figs. 5 and 7). However, both our expression analyses and the evaluated microarray data indicate that the VTE3 transcript levels in leaves become significantly reduced following P. syringae inoculation (Figs. 1 and 2D; Supplemental Table S1). But the microarray data also show that the absolute levels of VTE3 transcription are high under basal and still noticeable under biotic stress conditions (Supplemental Table S1), which might explain that MPBQ-to-DMPBQ conversion is not impaired in infected leaves. In addition to VTE3, the δ-/γ-tocopherol methyltransferase gene VTE4 is down-regulated in response to bacterial attack. This is in line with our findings that P. syringae inoculation primarily results in the accumulation of γ- instead of α-tocopherol and that, on the level of the less prominent 7-unsubstituted tocopherols, δ- but not β-tocopherol is increased upon inoculation (Figs. 1 and 3; Supplemental Fig. S2). Moreover, several studies show that VTE4 activity may become limiting when the metabolic flow through the tocopherol biosynthetic pathway is high, as is the case in leaves challenged with P. syringae. For example, transgenic Arabidopsis plants expressing a bifunctional bacterial chorismate mutase/prephenate dehydratase and thus overproducing HGA exhibited increased γ- but not α-tocopherol contents (Tzin et al., 2009). In addition, tocopherol accumulation in response to different abiotic stress stimuli was invariably associated with significant increases in γ-tocopherol contents (Bergmüller et al., 2003; Collakova and DellaPenna, 2003b), and the induction of leaf senescence was associated with a marked accumulation of γ-tocopherol (Mishina et al., 2007). Arabidopsis seed tissue exhibits very low VTE4 transcript levels, and the seed oil contains γ-tocopherol as the major tocopherol form. However, when VTE4 was overexpressed in Arabidopsis, the seed oil composition shifted in favor of α-tocopherol (Shintani and DellaPenna, 1998; Mène-Saffrané, 2017). Together, these findings indicate that γ-tocopherol methyltransferase activity is limiting in wild-type Arabidopsis seeds and in leaves of plants that suffer from biotic and abiotic stress conditions, which in either case results in a strong production of γ-tocopherol in the respective tissue.
The compatible Psm and the HR-inducing Psm avRpm1 strain triggered tocopherol accumulation, suggesting that both PTI and ETI might be involved in the activation of tocopherol biosynthesis (Fig. 3). Analyses of Arabidopsis mutants impaired in biotic stress-related hormonal pathways indicate that the P. syringae-triggered induction of tocopherol biosynthesis largely proceeds independently of the SA-, NHP-, JA-, and ET-induced defense pathways (Fig. 4A), although JA and ET signaling have been previously implicated in UV light- and drought stress-induced tocopherol production, respectively (Sandorf and Holländer-Czytko, 2002; Cela et al., 2009). However, the pathogen-triggered production of γ-tocopherol and the precursor amino acid Tyr is strongly potentiated by the immune regulators EDS1 and PAD4 (Fig. 4, A and B). The EDS1 and PAD4 proteins physically interact with each other and build a signaling node essential for basal resistance to (hemi)biotrophic pathogen attack (Wiermer et al., 2005). Following pathogen inoculation, EDS1/PAD4 positively regulates the expression of a battery of defense-related genes, the biosynthesis of SA, and the accumulation of the phytoalexin camalexin (Jirage et al., 1999; Feys et al., 2001; Wang et al., 2008). Moreover, the EDS1/PAD4 node boosts Pip and NHP biosynthesis in the course of SAR induction (Návarová et al., 2012; Hartmann et al., 2018). Immune responses activated by EDS1/PAD4 are separable into SA-dependent and SA-independent branches, and regulation of expression of the NHP biosynthesis gene FLAVIN-DEPENDENT MONOOXYGENASE1 falls into the second, SA-independent signaling sector (Bartsch et al., 2006; Mishina and Zeier, 2006). Since the accumulation of γ-tocopherol is not compromised in the SA pathway mutants sid2 and npr1 (Fig. 4A), an SA-independent branch of EDS1/PAD4 signaling also seems to operate in the regulation of pathogen-induced tocopherol biosynthesis (Fig. 10).
Figure 10.
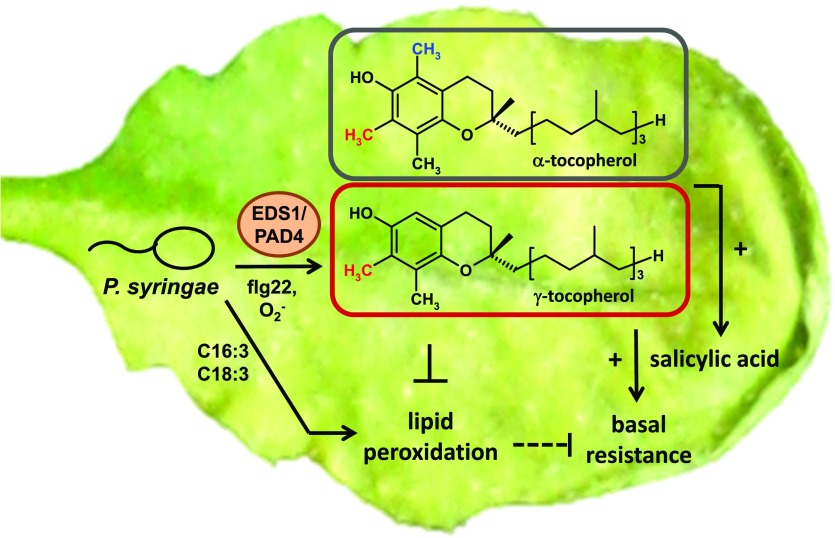
Proposed model for the role of tocopherols in Arabidopsis basal resistance against compatible P. syringae. Leaf inoculation with Psm triggers strong generation of γ-tocopherol in addition to the constitutively synthesized α-tocopherol homolog, resulting in high levels of both α- and γ-tocopherol in the course of infection. The pathogen-induced biosynthesis of tocopherols proceeds essentially via the biosynthetic scheme established previously for uninfected conditions (Fig. 1), is promoted by the EDS1/PAD4 signaling node, and is induced by flagellin perception and ROS generation. The elevated tocopherol levels in infected leaves (presumably together with elevated levels of their benzoquinol precursors) protect from pathogen-induced peroxidation of triunsaturated fatty acid-containing lipids, ensure effective SA biosynthesis, and contribute to basal resistance against P. syringae.
γ-Tocopherol also accumulated in leaves upon treatment with the flagellin peptide flg22 (Fig. 4B), whose perception is sufficient to trigger defense responses such as ROS production (Gómez-Gómez et al., 1999). Likewise, exogenous O2− generation induced γ-tocopherol accumulation in leaves, and the Arabidopsis cpr5-2 mutant that exhibits constitutively heightened ROS levels contained elevated basal contents of γ-tocopherol (Fig. 4, A and D). Together, these findings indicate that ROS have a triggering function in the pathogen-inducible biosynthesis of tocopherols. Our mutant analyses show that γ-tocopherol accumulation is independent of RbohD (Fig. 4A), the Arabidopsis NADPH oxidase isoform primarily responsible for the early oxidative burst triggered by avirulent bacterial pathogens (Torres et al., 2002). This and the relatively slow kinetics of tocopherol accumulation (Fig. 3) support a scenario in which bacteria proliferate in the leaf apoplast in the course of a plant-bacterium interaction, thereby exposing a rising number of pathogen-associated molecular patterns, which results in increased PTI signaling, ROS generation, and finally tocopherol accumulation (Fig. 10). Overall, the regulatory pattern of tocopherol accumulation is reminiscent of the regulation of stigmasterol biosynthesis in P. syringae-challenged leaves (Griebel and Zeier, 2010). Interestingly, stigmasterol also accumulated in Arabidopsis plants that overproduced hydrogen peroxide specifically in the chloroplast due to overexpression of a glycolate oxidase, indicating that the redox status of the chloroplast can control immune-related metabolic alterations (Sewelam et al., 2014).
Tocopherol Biosynthesis Is Required for Effective Basal Resistance of Arabidopsis to P. syringae
ROS production is a common molecular response of plants exposed to distinct biotic and abiotic stressors (Hasanuzzaman et al., 2012; Camejo et al., 2016). Consistent with our finding that elevated ROS can trigger the accumulation of γ-tocopherol (Fig. 4C), previous studies have described several abiotic stress situations that stimulate tocopherol biosynthesis in Arabidopsis. For example, plants exposed to UV light, high light, heat, or cold significantly accumulated both α- and γ-tocopherol in leaves (Sandorf and Holländer-Czytko, 2002; Bergmüller et al., 2003), while combined nutrient deficiency and high light exposure led to foliar increases in all four tocopherol forms (Collakova and DellaPenna, 2003b). In addition, accumulation of α-tocopherol was detected in heavy metal-treated and drought-stressed plants (Collin et al., 2008; Cela et al., 2009). Mutant analyses indicate that tocopherols contribute to the tolerance of plants to at least some of these abiotic stresses. For instance, vte2, and to a lesser extend vte1, exhibited diminished growth, reduced seed production, and increased anthocyanin accumulation under extended low-temperature treatment (Maeda et al., 2006). In addition, oxidative stress, lipid peroxidation, and deficiency symptoms induced by exposure to Cu and Cd were higher in tocopherol-deficient vte1 than in wild-type plants (Collin et al., 2008), and vte1 also exhibited increased sensitivity to salinity stress (Ellouzi et al., 2013). Together with additional mechanisms such as zeaxanthin biosynthesis and nonphotochemical energy dissipation, tocopherols also contribute to the tolerance to long-term high-light exposure (Havaux et al., 2005).
The results of this study show that tocopherol biosynthesis is not only required for abiotic stress tolerance but also for plant basal immunity to bacterial pathogen infection, because Arabidopsis vte2 mutants exhibited compromised resistance to infection by compatible Psm (Fig. 6, A and C; Supplemental Fig. S4). The decreased resistance phenotype of vte2 is associated with a significantly attenuated pathogen-inducible biosynthesis of SA and its direct derivatives SAG and SGE, while other defense-related metabolites are produced to at least wild type-like levels in vte2 (Fig. 8). SA accumulation is critical for basal resistance against distinct biotrophic and hemibiotrophic pathogens, including P. syringae (Nawrath and Métraux, 1999; Wildermuth et al., 2001; Bernsdorff et al., 2016; Klessig et al., 2018). The partially compromised biosynthesis of SA might thus be causative for the observed susceptibility phenotype of vte2.
The triunsaturated fatty acids 18:3 and 16:3, which are both susceptible to nonenzymatic oxidation (Mène-Saffrané et al., 2009), typically account for the majority of chloroplast membrane-bound fatty acids and for more than 90% of the fatty acids of the major thylakoid lipid monogalactosyldiacylglycerol (Routaboul et al., 2000). By GC-MS analyses of transesterified leaf lipid extracts, we found that membrane-bound 18:3 and 16:3 tend to decrease after Psm inoculation. This was associated with increases in presumable 18:3- and 16:3-derived oxylipins, which were detected to a higher degree in vte2 than in the Col-0 wild type (Supplemental Fig. S8). Moreover, as estimated by the quantification of TBA-reactive aldehydic products, we observed augmented lipid peroxidation in leaves following Psm challenge. Lipid peroxidation was significantly higher in vte2 than in wild-type plants, indicating that tocopherol biosynthesis attenuates the Psm-induced peroxidation of leaf lipids (Fig. 9). Trienoic acid- and tocopherol-deficient fad3-2 fad7-2 fad8 vte2-1 quadruple mutants showed similar Psm-induced lipid peroxidation to fad3-2 fad7-2 fad8 and wild-type plants, suggesting that the increased peroxidation levels observed in vte2 result from oxidation of lipids that contain triunsaturated fatty acids (Fig. 9).
The vte2 mutant plants showed increased susceptibility to Psm infection, while fad3-2 fad7-2 fad8 vte2-1 and fad3-2 fad7-2 fad8 plants showed wild type-like resistance (Fig. 6C). Taking these results together, we propose a model in which tocopherols contribute to basal immunity by protecting trienoic fatty acid-containing chloroplast lipids from oxidative damage in the course of a bacterial infection (Fig. 10). In this way, they might support the integrity of chloroplast membranes and of the entire organelle. Chloroplasts represent important cellular sites of plant defense (Serrano et al., 2016). For example, the key enzyme of stress-inducible SA biosynthesis in Arabidopsis, ISOCHORISMATE SYNTHASE1 (ICS1), resides in the chloroplast (Wildermuth et al., 2001). Protection of chloroplasts from oxidative damage by tocopherols might thus positively influence the generation of prodefense signals, as observed here for the immune-regulatory metabolite SA (Fig. 8).
In contrast to vte2, the tocopherol biosynthetic mutants tat1-2, VTE3/vte3-3, vte4-2, and vte1 exhibited wild type-like basal resistance to Psm infection (Fig. 6, A and B). The tat1-2 mutant contained decreased but still substantial levels of tocopherols that might be sufficient for a defense-related function (Fig. 5). Although the tocopherol profiles of VTE3/vte3-3 and vte4-2 were qualitatively different, the levels of total tocopherols in these lines were similar to the total tocopherol content in the wild type (Fig. 5). Thus, the qualitative nature of a given tocopherol mixture present in attacked leaves appears to be subordinate for the execution of basal immunity, although specific physiological roles for α- and γ-tocopherol have been proposed previously (Abbasi et al., 2007). While vte1 and vte2 were both severely compromised in basal and Psm-inducible tocopherol biosynthesis (Fig. 5), only vte2 essentially lacked accumulation of the benzoquinol precursors (D)MPBQ. In fact, vte1 even heavily overaccumulated DMPBQ upon bacterial attack (Fig. 7). Thus, the failure of both tocopherol and benzoquinol precursor accumulation appears to determine the enhanced susceptibility phenotype of vte2. As suggested previously, the tocopherol precursors (D)MPBQ might possess antioxidant activity and functionally compensate for the deficiency of tocopherols in vte1 (Sattler et al., 2004). The differences observed for vte2 and vte1 in plant basal resistance observed here are reminiscent of the phenotypic differences reported by Sattler et al. (2004, 2006) in context with early seedling development: vte2 but not vte1 plants exhibited marked seedling growth defects after germination and contained strongly elevated levels of lipid peroxidation products such as TBA-reactive aldehydes, hydroxy fatty acids, and phytoprostanes.
Arabidopsis is also able to produce PC-8, another type of tocochromanol with a long, solanesyl diphosphate-derived terpenoid side chain that is synthesized by the VTE1-mediated cyclization of its benzoquinol precursor plastoquinol-9 (PQ-9; Mène-Saffrané, 2017). vte1 mutants are thus impaired in PC-8 biosynthesis but not in PQ-9 production. Since an HGA prenyltransferase different from VTE2 is involved in the generation of PQ-9, vte2 mutants can still synthesize the redox-active PQ-9 and PC-8 molecules. Due to their relatively high Mr, PQ-9 and PC-8 are not amenable to our GC-MS analysis, and we have therefore not determined whether these compounds accumulate in response to bacterial infection. The resistance phenotypes of the vte1 and vte2 mutants, however, do not necessarily support a role for PC-8 in basal immunity. Seedlings of vte1 vte2 double mutants possess a severe growth inhibition phenotype, suggesting that PC-8 functions as a lipid antioxidant in early plant development (Mène-Saffrané et al., 2010).
While vte2 mutant plants show attenuated basal resistance to the compatible Psm strain, they exhibit, as all other examined tocopherol biosynthetic mutants, wild type-like specific resistance to an avirulent, HR-inducing Psm strain that expresses the avrRpm1 avirulence gene (Supplemental Fig. S6). Consistently, a recent study reported that vte1, vte4, and wild-type plants exhibit similar resistance to avirulent P. syringae pv tomato avrRpm1 (Cela et al., 2018). We found that Psm avrRpm1 inoculation caused markedly weaker lipid peroxidation in Arabidopsis leaves than infection by the compatible Psm strain. Moreover, the Psm avrRpm1-triggered increase in leaf lipid peroxidation was not significantly different between the wild type and vte2-2 mutants (Fig. 9A). This suggests that tocopherol biosynthesis is functionally irrelevant for the studied incompatible plant-bacterium interaction that is associated with an HR and an early distinct ROS burst around 4 to 6 h after inoculation (Lamb and Dixon, 1997; Zeier, 2005). By comparison, ROS production in response to the compatible Psm strain starts several hours later, essentially coinciding with the onset of Psm-induced tocopherol accumulation (Fig. 3; Hamdoun et al., 2013).
The vte2 mutant was not compromised in the Psm-inducible activation of the pipecolate pathway that produces the SAR-associated metabolites Pip and NHP following Psm inoculation (Fig. 8, A and B). The accumulation of NHP is necessary and sufficient for SAR induction (Hartmann et al., 2018), and, congruently, the degree of SAR induced in wild-type and vte2 plants was similar (Fig. 6D). Thus, the biosynthesis of tocopherols is not required to trigger SAR. However, just as in noninduced plants, the SAR-induced vte2 displayed significantly weaker resistance to Psm than the wild type (Fig. 6D). We have previously proposed a model for SAR establishment in which the pipecolate pathway (i.e. NHP) acts as a switch that induces SAR and the salicylate pathway (i.e. SA) influences the strength of SAR (Bernsdorff et al., 2016; Hartmann and Zeier, 2018; Hartmann et al., 2018). The observed metabolite pattern and SAR phenotype of vte2 are consistent with this model (Figs. 6D and 8).
In summary, we have shown that tocopherols and their benzoquinol precursor are required to ensure proper SA accumulation and basal resistance to infection by the hemibiotrophic bacterium P. syringae. In addition, different disease susceptibility phenotypes of Arabidopsis wild-type, vte1, and vte4 plants following Botrytis cinerea inoculation indicate that the tocopherol content and composition can influence plant resistance to infection by necrotrophic pathogens (Cela et al., 2018). We propose that the pathogen-inducible tocopherol biosynthetic pathway protects Arabidopsis plants from oxidation of triunsatured fatty acid-containing lipids during bacterial leaf infection. Thus, the antioxidant activity of tocopherols is necessary for an optimal performance of plants in biotic, abiotic, and developmental stress situations.
MATERIALS AND METHODS
Plant Material and Cultivation
Arabidopsis (Arabidopsis thaliana) plants were cultivated in pots containing a mixture of soil (Klasmann-Deilmann, Substrat BP3), vermiculite, and sand (8:1:1) in a controlled environmental chamber with a 10-h-day (9 am–7 pm; photon flux density of 100 μmol m−2 s−1)/14-h-night cycle and a relative humidity of 60%. Temperatures during the day and night periods were 21°C and 18°C, respectively. Experiments were generally conducted with 5-week-old plants.
The tocopherol biosynthetic mutants used in this study included vte1 (Porfirova et al., 2002), vte2-1 (Mène-Saffrané et al., 2007), vte2-2 (Havaux et al., 2005), vte4-2 (Bergmüller et al., 2003), and fad3-2 fad7-2 fad8 vte2-1 (Mène-Saffrané et al., 2007). The VTE3/vte3-3 and tat1-2 mutants, which correspond to the SALK T-DNA insertion lines SALK_031151C and SALK_141402C (Wang et al., 2019), respectively, were identified by the PCR-based protocol described by Alonso et al. (2003). Other lines used in this study were etr1 (Bleecker et al., 1988), dde2 (von Malek et al., 2002), sid2-1 (Nawrath and Métraux, 1999), npr1-3 (Cao et al., 1997 [Nottingham Arabidopsis Stock Centre identifier N3802]), eds1-2 (Bartsch et al., 2006), pad4-1 (Jirage et al., 1999), aba2-1 (Léon-Kloosterziel et al., 1996), cpr5-2 (Bowling et al., 1997), coi1-35 (Staswick and Tiryaki, 2004), ald1 (Návarová et al., 2012), rbohD (Torres et al., 2002), fad3-2 fad7-2 fad8 (McConn and Browse, 1996), and NahG (Lawton et al., 1995). All lines are in the Col-0 background except for vte1, which has a Col-2 background.
Cultivation and Inoculation of Bacteria
Pseudomonas syringae pv maculicola strain ES4326, Psm carrying the avrRpm1 avirulence gene (Psm avrRpm1), and Psm carrying the luxCDABE operon from Photorhabdus luminescens under the control of a constitutive promoter (Psm lux) were cultivated in King’s B medium containing the appropriate antibiotics (Zeier et al., 2004; Fan et al., 2008). Overnight cultures were grown at 28°C under permanent shaking, washed three times with a 10 mm MgCl2 solution, and diluted to different OD600 values. The bacterial solutions were infiltrated, between 10 and 12 am, from the abaxial side into the leaves with a needleless syringe. For metabolite determination, RT-qPCR analysis, and the assessment of lipid peroxidation, three leaves per plant were inoculated with a suspension of Psm or Psm avrRpm at OD600 = 0.005. Control plants were mock infiltrated with a 10 mm MgCl2 solution. Leaves were harvested at different times after treatment.
Determination of Local Resistance to P. syringae and SAR Assays
To determine plant basal resistance to P. syringae, three leaves per plant were infiltrated with a suspension of Psm lux at OD600 = 0.001 between 10 and 12 am. Sixty hours later, leaf discs (10 mm in diameter) were punched out from inoculated leaves and bacterial numbers were assessed by determining relative light units of the bioluminescent Psm lux strain with a Sirius luminometer (Berthold Detection Systems). Colony-forming units, which linearly correlate with relative light unit values, were calculated by an experimentally determined calibration line (Gruner et al., 2018).
To assess SAR establishment, three lower (1°) leaves of a given plant were either infiltrated with a suspension of Psm at OD600 = 0.005 (SAR induction) or with a 10 mm MgCl2 solution (mock treatment). Two days after the primary treatment, three upper leaves (2°) of all plants were infiltrated with a suspension of Psm lux at OD600 = 0.001 and the growth of bacteria in 2° leaves was estimated as described above.
The assessment of local resistance to the HR-inducing Psm avrRpm1 strain was performed by bacterial plating assay following leaf tissue homogenization in 10 mm MgCl2 solution at 60 h post Psm avrRpm1 inoculation as described previously (Zeier et al., 2004).
Flg22 and X/XO Treatments
Three leaves of a given plant were infiltrated with flg22 peptide at a concentration of 100 nm or with deionized water as a control treatment. The flg22 peptide was synthesized by Mimotopes. To investigate the effect of superoxide, the O2−-producing combination of X and XO was applied as described previously (Griebel and Zeier, 2010). X and XO were applied at concentrations of 0.5 mm X and 0.5 units mL−1 XO in 20 mm sodium phosphate buffer (pH 6.5). X and XO were obtained from Sigma-Aldrich. Control infiltration was performed with 0.5 mm X in 20 mm sodium phosphate buffer without XO.
RT-qPCR Analysis
RNA isolation, cDNA synthesis, and RT-qPCR analysis were performed as described previously (Návarová et al., 2012). The POLYPYRIMIDINE TRACT-BINDING PROTEIN1 (PTB1) gene (At3g01150), which is nonresponsive to P. syringae infection, was used as a reference gene (Czechowski et al., 2005). The gene-specific primers used in this study are listed in Supplemental Table S2. The expression levels were normalized to those of the reference gene and were expressed relative to the value of the gene in MgCl2-infiltrated leaves.
GC-MS Analyses of Tocopherols, (D)MPBQ, Defense-Related Metabolites, and Lipid-Bound Fatty Acids
Five-week-old Arabidopsis plants treated in three full-grown leaves as described above (see “Cultivation and Inoculation of Bacteria”) were used for metabolite analyses. At least six plants per treatment and plant genotype were analyzed, whereby individual replicate leaf samples (n ≥ 3) were pooled from two plants. The experiments depicted in the figures were repeated at least three times with similar results.
The determination of endogenous levels of tocopherols and their biosynthetic precursors MPBQ and DMPBQ in Arabidopsis leaves was performed according to a GC-MS-based analytical procedure described previously by Hartmann et al. (2018). Minor modifications to this procedure concerned the use of the extraction buffer and the internal standardization. In brief, 50 mg of freshly pulverized, frozen leaf tissue was extracted twice with 1 mL of CHCl3:MeOH/H2O (1:2.5:1, v/v/v). Tocol (1,000 ng) was used as an internal standard (IST). Six hundred microliters of the extract was evaporated to dryness using a ScanSpeed vacuum centrifuge (Labogene). To convert free hydroxyl groups of the analytes into their trimethylsilyl derivatives, 20 µL of pyridine, 20 µL of N-methyl-N-trimethylsilyltrifluoroacetamide containing 1% (v/v) trimethylchlorosilane, and 60 µL of hexane were added and the reaction mixture was incubated at 70°C for 30 min. An aliquot of the cooled sample was transferred to a GC vial and diluted fivefold with hexane. Two microliters of the sample mixture was separated on a gas chromatograph (GC 7890A; Agilent Technologies) equipped with a fused silica capillary column (Phenomenex ZB-35; 30 m × 0.25 mm × 0.25 µm). Mass spectra were recorded in the range between mass-to-charge ratio (m/z) 50 and 750 with a 5975C mass spectrometric detector (Agilent Technologies) in the electron ionization mode. The following temperature program was used for GC separation: 70°C for 2 min, with 10°C min−1 to 320°C, and 320°C for 5 min. For quantification of metabolites, peaks emanating from selected ion chromatograms were integrated using MSD ChemStation software version E.02.01.1177 (Agilent Technologies): m/z 502 for α-tocopherol, m/z 488 for β- and γ-tocopherol, m/z 474 for δ-tocopherol, m/z 546 for MPBQ, and m/z 560 for DMPBQ (Supplemental Figs. S1 and S7). The corresponding peak areas were related to the peak area of the IST tocol (m/z 460). The absolute contents of the four tocopherols were calculated by considering correction factors experimentally determined using authentic substances (Supelco; catalog nos. 47783 [α-tocopherol], 46401 [β-tocopherol], 47785 [γ-tocopherol], and 47784 [δ-tocopherol]) and are generally given in µg g−1 fresh weight. Due to the unavailability of authentic MPBQ and DMPBQ standards, the relative endogenous contents of (D)MPBQ were given. To relate tocopherol contents to leaf dry weight (Supplemental Fig. S3C), frozen and homogenized plant samples were freeze dried (Alpha 1-2 LD plus; CHRIST), followed by extraction, derivatization, and GC-MS analysis as described above.
For the analysis of other defense-related metabolites, the protocol of Hartmann et al. (2018) was followed as described. SA (selected ion chromatogram m/z 267) was determined relative to D4-SA as IST (m/z 271), Pip (m/z 156) relative to D9-Pip as IST (m/z 165), NHP (m/z 172) relative to D9-NHP as IST (m/z 181), camalexin (m/z 272) and ICA (m/z 246) relative to indole-3-propionic acid (m/z 202) as IST, Tyr (m/z 218) relative to nor-Val (m/z 144) as IST, stigmasterol (m/z 484) relative to tocol (m/z 460) as IST, and both SAG (m/z 267) and SGE (m/z 193) relative to salicin (m/z 268) as IST. Authentic SAG and SGE standards for the determination of analytical correction factors were kindly provided by Patrick Saindrenan (Paris-Sud University; Chong et al., 2002).
For the analysis of lipid-bound fatty acids (Supplemental Fig. S8), 50 mg of pulverized, frozen leaf tissue was extracted with 1 mL of CHCl3:MeOH:H2O (2:2:1, v/v/v) according to Bligh and Dyer (1959) using added tetracosane (3 µg) as an IST. After centrifugation and phase separation, the organic phase was removed and the solvent was evaporated in a stream of nitrogen. Fatty acids were released as methyl esters from the extracted lipids by transesterification with a solution of 10% (w/w) boron trifluoride in methanol (Fluka) at 70°C overnight (Zeier, 1998). Two milliliters of CHCl3 was added, and the solution was washed twice with 1 mL of a saturated sodium chloride solution. The organic phase was separated, dried over Na2SO4, and evaporated to dryness. The residue was redissolved in 300 µL of CH2Cl2, and fatty acid methyl esters were analyzed by GC-MS as described above for tocopherol analysis. The following ion chromatograms were used for the relative quantification of compounds: m/z 292 (18:3), m/z 264 (16:3), m/z 177 (iso-OPDA and putative iso-dnOPDA), and m/z 338 (tetracosane, IST).
Assessment of Lipid Peroxidation
The degree of lipid peroxidation was estimated by assessing the amount of TBA-reactive aldehydes according to a previously published protocol (Hodges et al., 1999). The plant treatments were performed as described under “Cultivation and Inoculation of Bacteria” and “GC-MS Analyses of Tocopherols, (D)MPBQ, Defense-Related Metabolites, and Lipid-Bound Fatty Acids.” The frozen leaf material was homogenized, mixed with 2 mL of 0.1% (w/v) TCA, and centrifuged at 10,000g for 15 min. One milliliter of the supernatant was supplemented with 2 mL of 20% (w/v) TCA and 2 mL of 0.5% (w/v) TBA, and the mixture was incubated at 95°C for 30 min. Thereby, TBA-aldehyde complexes with a specific A532 were formed. The specific A532 and a nonspecific A600 were measured with a UV-VIS spectrophotometer (UVmini-1240; Shimadzu), and the nonspecific absorbance was subtracted from the specific absorbance. The concentrations of TBA-reactive aldehydes were calculated using Beer-Lambert’s equation with an extinction coefficient of 155 mm−1 cm−1 (Heath and Packer, 1968) and related to the sample fresh weight. The aldehyde levels estimated for the different samples were expressed relative to the means of the samples for untreated Col-0 leaves.
Reproducibility of Experiments and Statistical Analyses
The results presented are generally derived from one representative experiment. Given data points show means ± sd of values from multiple biological replicates, each replicate consisting of samples from different plants. Bacterial growth values originated from nine or more biological replicates. Each replicate value was determined from three leaf discs of one given plant. For metabolite, gene expression, and TBA analyses, three to four biological replicates were used. Each replicate sample was derived from six leaves harvested from two plants. For the statistical evaluation of the data, ANOVA or Student’s t test was used as described (Bernsdorff et al., 2016). For ANOVA, log10-transformed values were analyzed. Student’s t test was applied to data values without prior log transformation. The reported tendencies were generally observed in at least three independent experiments. The tendencies for the fatty acid analysis depicted in Supplemental Figure S8 were only observed twice.
Accession Numbers
Sequence data from genes described in this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: TAT1 (At5g53970), HPPD (At1g06570), VTE1 (At4g32770), VTE2 (At2g18950), VTE3 (At3g63410), VTE4 (At1g64970), ETR1 (At1g66340), DDE2 (At5g42650), SID2/ICS1 (At1g74710), ABA2 (At1g52340), CPR5 (At5g64930), COI1 (At2g39940), ALD1 (At2g13810), RBOHD (At5g47910), FAD3 (At2g29980), FAD7 (At3g11170), FAD8 (At5g05580), and PTB (At3g01150).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. GC-MS-based analysis of tocopherols.
Supplemental Figure S2. Time-dependent accumulation of tocopherols upon Psm inoculation.
Supplemental Figure S3. Levels of α-tocopherol in Psm-inoculated leaves.
Supplemental Figure S4. vte2 mutant plants are compromised in basal resistance to P. syringae.
Supplemental Figure S5. Disease symptoms of leaves of Col-0, vte2-2, VTE3/vte3-3, vte4-2, and tat1-2 plants inoculated with compatible Psm.
Supplemental Figure S6. Resistance against avirulent Psm avrRpm1 of tocopherol biosynthetic mutants is similar to the wild type.
Supplemental Figure S7. GC-MS-based analysis of MPBQ and DMPBQ.
Supplemental Figure S8. Levels of lipid-esterified triunsaturated fatty acids and related oxylipins in the leaves of Psm-inoculated and control plants.
Supplemental Table S1. Expression patterns of tocopherol biosynthetic genes in P. syringae-inoculated Arabidopsis leaves according to publicly available microarray data sets.
Supplemental Table S2. List of primers used in this study.
ACKNOWLEDGMENTS
We thank Peter Dörmann for allocating seeds of vte1, vte2-2, and vte4-2 and Eric Kemen for providing seeds of fad3-2 fad7-2 fad8. We are also indebted to Laurent Mène-Saffrané for allocating seeds of vte2-1 and fad3-2 fad7-2 fad8 vte2-1 mutants as well as for critically discussing the data.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) under Germany’s Excellence Strategy (EXC 2048/1-390686111) and by DFG grant ZE467/6-2.
Articles can be viewed without a subscription.
E.S. performed most of the experiments, analyzed the corresponding data, and contributed to article preparation and writing; M.H. completed bacterial growth assays, significantly contributed to the estimation of lipid peroxidation and defense metabolite analyses, and performed (D)MPBQ analyses; N.S. completed bacterial growth assays; J.Z. conceived, initialized, and supervised the project, evaluated data sets, and wrote the article.
References
- Abbasi AR, Hajirezaei M, Hofius D, Sonnewald U, Voll LM (2007) Specific roles of alpha- and gamma-tocopherol in abiotic stress responses of transgenic tobacco. Plant Physiol 143: 1720–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja I, Kissen R, Bones AM (2012) Phytoalexins in defense against pathogens. Trends Plant Sci 17: 73–90 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Attaran E, Rostás M, Zeier J (2008) Pseudomonas syringae elicits emission of the terpenoid (E,E)-4,8,12-trimethyl-1,3,7,11-tridecatetraene in Arabidopsis leaves via jasmonate signaling and expression of the terpene synthase TPS4. Mol Plant Microbe Interact 21: 1482–1497 [DOI] [PubMed] [Google Scholar]
- Attaran E, Zeier TE, Griebel T, Zeier J (2009) Methyl salicylate production and jasmonate signaling are not essential for systemic acquired resistance in Arabidopsis. Plant Cell 21: 954–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala A, Muñoz MF, Argüelles S (2014) Lipid peroxidation: Production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev 2014: 360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE (2006) Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek P, Osbourn A (2009) Plant-microbe interactions: Chemical diversity in plant defense. Science 324: 746–748 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Bergmüller E, Porfirova S, Dörmann P (2003) Characterization of an Arabidopsis mutant deficient in γ-tocopherol methyltransferase. Plant Mol Biol 52: 1181–1190 [DOI] [PubMed] [Google Scholar]
- Bernsdorff F, Döring AC, Gruner K, Schuck S, Bräutigam A, Zeier J (2016) Pipecolic acid orchestrates plant systemic acquired resistance and defense priming via salicylic acid-dependent and -independent pathways. Plant Cell 28: 102–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H (1988) Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241: 1086–1089 [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Block A, Schmelz E, Jones JB, Klee HJ (2005) Coronatine and salicylic acid: The battle between Arabidopsis and Pseudomonas for phytohormone control. Mol Plant Pathol 6: 79–83 [DOI] [PubMed] [Google Scholar]
- Bohlmann J, Keeling CI (2008) Terpenoid biomaterials. Plant J 54: 656–669 [DOI] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camejo D, Guzmán-Cedeño Á, Moreno A (2016) Reactive oxygen species, essential molecules, during plant-pathogen interactions. Plant Physiol Biochem 103: 10–23 [DOI] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Caretto S, Bray Speth E, Fachechi C, Gala R, Zacheo G, Giovinazzo G (2004) Enhancement of vitamin E production in sunflower cell cultures. Plant Cell Rep 23: 174–179 [DOI] [PubMed] [Google Scholar]
- Cela J, Falk J, Munné-Bosch S (2009) Ethylene signaling may be involved in the regulation of tocopherol biosynthesis in Arabidopsis thaliana. FEBS Lett 583: 992–996 [DOI] [PubMed] [Google Scholar]
- Cela J, Tweed JKS, Sivakumaran A, Lee MRF, Mur LAJ, Munné-Bosch S (2018) An altered tocopherol composition in chloroplasts reduces plant resistance to Botrytis cinerea. Plant Physiol Biochem 127: 200–210 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Sattler S, Maeda H, Sakuragi Y, Bryant DA, DellaPenna D (2003) Highly divergent methyltransferases catalyze a conserved reaction in tocopherol and plastoquinone synthesis in cyanobacteria and photosynthetic eukaryotes. Plant Cell 15: 2343–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Baltz R, Schmitt C, Beffa R, Fritig B, Saindrenan P (2002) Downregulation of a pathogen-responsive tobacco UDP-Glc:phenylpropanoid glucosyltransferase reduces scopoletin glucoside accumulation, enhances oxidative stress, and weakens virus resistance. Plant Cell 14: 1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collakova E, DellaPenna D (2003a) Homogentisate phytyltransferase activity is limiting for tocopherol biosynthesis in Arabidopsis. Plant Physiol 131: 632–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collakova E, DellaPenna D (2003b) The role of homogentisate phytyltransferase and other tocopherol pathway enzymes in the regulation of tocopherol synthesis during abiotic stress. Plant Physiol 133: 930–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin VC, Eymery F, Genty B, Rey P, Havaux M (2008) Vitamin E is essential for the tolerance of Arabidopsis thaliana to metal-induced oxidative stress. Plant Cell Environ 31: 244–257 [DOI] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE (2015) Effector-triggered immunity: From pathogen perception to robust defense. Annu Rev Plant Biol 66: 487–511 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabrowska P, Shabab M, Brandt W, Vogel H, Boland W (2011) Isomerization of the phytohormone precursor 12-oxophytodienoic acid (OPDA) in the insect gut: A mechanistic and computational study. J Biol Chem 286: 22348–22354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D. (2005) Progress in the dissection and manipulation of vitamin E synthesis. Trends Plant Sci 10: 574–579 [DOI] [PubMed] [Google Scholar]
- DellaPenna D, Pogson BJ (2006) Vitamin synthesis in plants: Tocopherols and carotenoids. Annu Rev Plant Biol 57: 711–738 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Xia Y, Dixon RA, Lamb C (1998) Nitric oxide functions as a signal in plant disease resistance. Nature 394: 585–588 [DOI] [PubMed] [Google Scholar]
- de Torres Zabala M, Bennett MH, Truman WH, Grant MR (2009) Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J 59: 375–386 [DOI] [PubMed] [Google Scholar]
- de Torres Zabala M, Littlejohn G, Jayaraman S, Studholme D, Bailey T, Lawson T, Tillich M, Licht D, Bölter B, Delfino L, et al. (2015) Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nat Plants 1: 15074. [DOI] [PubMed] [Google Scholar]
- Ding Y, Dommel M, Mou Z (2016) Abscisic acid promotes proteasome-mediated degradation of the transcription coactivator NPR1 in Arabidopsis thaliana. Plant J 86: 20–34 [DOI] [PubMed] [Google Scholar]
- Dörmann P. (2007) Functional diversity of tocochromanols in plants. Planta 225: 269–276 [DOI] [PubMed] [Google Scholar]
- Ellouzi H, Hamed KB, Cela J, Müller M, Abdelly C, Munné-Bosch S (2013) Increased sensitivity to salt stress in tocopherol-deficient Arabidopsis mutants growing in a hydroponic system. Plant Signal Behav 8: e23136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban R, Olano JM, Castresana J, Fernández-Marín B, Hernández A, Becerril JM, García-Plazaola JI (2009) Distribution and evolutionary trends of photoprotective isoprenoids (xanthophylls and tocopherols) within the plant kingdom. Physiol Plant 135: 379–389 [DOI] [PubMed] [Google Scholar]
- Falk J, Munné-Bosch S (2010) Tocochromanol functions in plants: Antioxidation and beyond. J Exp Bot 61: 1549–1566 [DOI] [PubMed] [Google Scholar]
- Fan J, Crooks C, Lamb C (2008) High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J 53: 393–399 [DOI] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J 20: 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya T, Yoshikawa T, Kimura T, Kaneko H (1987) Production of tocopherols by cell culture of safflower. Phytochemistry 26: 2741–2747 [Google Scholar]
- Glawischnig E. (2007) Camalexin. Phytochemistry 68: 401–406 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Felix G, Boller T (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18: 277–284 [DOI] [PubMed] [Google Scholar]
- Griebel T, Zeier J (2010) A role for β-sitosterol to stigmasterol conversion in plant-pathogen interactions. Plant J 63: 254–268 [DOI] [PubMed] [Google Scholar]
- Grun G, Berger S, Matthes D, Mueller MJ (2007) Early accumulation of non-enzymatically synthesized oxylipins in Arabidopsis thaliana after infection with Pseudomonas syringae. Funct Plant Biol 34: 65–71 [DOI] [PubMed] [Google Scholar]
- Gruner K, Griebel T, Návarová H, Attaran E, Zeier J (2013) Reprogramming of plants during systemic acquired resistance. Front Plant Sci 4: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner K, Zeier T, Aretz C, Zeier J (2018) A critical role for Arabidopsis MILDEW RESISTANCE LOCUS O2 in systemic acquired resistance. Plant J 94: 1064–1082 [DOI] [PubMed] [Google Scholar]
- Hamdoun S, Liu Z, Gill M, Yao N, Lu H (2013) Dynamics of defense responses and cell fate change during Arabidopsis-Pseudomonas syringae interactions. PLoS ONE 8: e83219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M, Kim D, Bernsdorff F, Ajami-Rashidi Z, Scholten N, Schreiber S, Zeier T, Schuck S, Reichel-Deland V, Zeier J (2017) Biochemical principles and functional aspects of pipecolic acid biosynthesis in plant immunity. Plant Physiol 174: 124–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M, Zeier J (2018) l-Lysine metabolism to N-hydroxypipecolic acid: An integral immune-activating pathway in plants. Plant J 96: 5–21 [DOI] [PubMed] [Google Scholar]
- Hartmann M, Zeier J (2019) N-Hydroxypipecolic acid and salicylic acid: A metabolic duo for systemic acquired resistance. Curr Opin Plant Biol 50: 44–57 [DOI] [PubMed] [Google Scholar]
- Hartmann M, Zeier T, Bernsdorff F, Reichel-Deland V, Kim D, Hohmann M, Scholten N, Schuck S, Bräutigam A, Hölzel T, et al. (2018) Flavin monooxygenase-generated N-hydroxypipecolic acid is a critical element of plant systemic immunity. Cell 173: 456–469.e16 [DOI] [PubMed] [Google Scholar]
- Hasanuzzaman M, Hossain MA, Teixeira da Silva JA, Fujita M (2012) Plant response and tolerance to abiotic oxidative stress: Antioxidant defense is a key factor In Venkateshwarulu B, Shanker AK, Shanker C, and Mandapaka M, eds, Crop Stress and Its Management: Perspectives and Strategies. Springer, Berlin, pp 261–316 [Google Scholar]
- Havaux M, Eymery F, Porfirova S, Rey P, Dörmann P (2005) Vitamin E protects against photoinhibition and photooxidative stress in Arabidopsis thaliana. Plant Cell 17: 3451–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125: 189–198 [DOI] [PubMed] [Google Scholar]
- Hodges DM, DeLong JM, Forney CF, Prange RK (1999) Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207: 604–611 [DOI] [PubMed] [Google Scholar]
- Ischebeck T, Zbierzak AM, Kanwischer M, Dörmann P (2006) A salvage pathway for phytol metabolism in Arabidopsis. J Biol Chem 281: 2470–2477 [DOI] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA 96: 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwischer M, Porfirova S, Bergmüller E, Dörmann P (2005) Alterations in tocopherol cyclase activity in transgenic and mutant plants of Arabidopsis affect tocopherol content, tocopherol composition, and oxidative stress. Plant Physiol 137: 713–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klessig DF, Choi HW, Dempsey DA (2018) Systemic acquired resistance and salicylic acid: Past, present, and future. Mol Plant Microbe Interact 31: 871–888 [DOI] [PubMed] [Google Scholar]
- Kobayashi N, DellaPenna D (2008) Tocopherol metabolism, oxidation and recycling under high light stress in Arabidopsis. Plant J 55: 607–618 [DOI] [PubMed] [Google Scholar]
- Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Lawton K, Weymann K, Friedrich L, Vernooij B, Uknes S, Ryals J (1995) Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol Plant Microbe Interact 8: 863–870 [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JAD, Koornneef M (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10: 655–661 [DOI] [PubMed] [Google Scholar]
- Lewis LA, Polanski K, de Torres-Zabala M, Jayaraman S, Bowden L, Moore J, Penfold CA, Jenkins DJ, Hill C, Baxter L, et al. (2015) Transcriptional dynamics driving MAMP-triggered immunity and pathogen effector-mediated immunosuppression in Arabidopsis leaves following infection with Pseudomonas syringae pv tomato DC3000. Plant Cell 27: 3038–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, DellaPenna D (2007) Tocopherol functions in photosynthetic organisms. Curr Opin Plant Biol 10: 260–265 [DOI] [PubMed] [Google Scholar]
- Maeda H, Song W, Sage TL, DellaPenna D (2006) Tocopherols play a crucial role in low-temperature adaptation and phloem loading in Arabidopsis. Plant Cell 18: 2710–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo A, Funck D, Mühlenbock P, Kular B, Mullineaux PM, Karpinski S (2006) Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. J Exp Bot 57: 1795–1807 [DOI] [PubMed] [Google Scholar]
- McConn M, Browse J (1996) The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J (1997) Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA 94: 5473–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecey C, Hauck P, Trapp M, Pumplin N, Plovanich A, Yao J, He SY (2011) A critical role of STAYGREEN/Mendel’s I locus in controlling disease symptom development during Pseudomonas syringae pv tomato infection of Arabidopsis. Plant Physiol 157: 1965–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mène-Saffrané L. (2017) Vitamin E biosynthesis and its regulation in plants. Antioxidants 7: E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mène-Saffrané L, Davoine C, Stolz S, Majcherczyk P, Farmer EE (2007) Genetic removal of tri-unsaturated fatty acids suppresses developmental and molecular phenotypes of an Arabidopsis tocopherol-deficient mutant: Whole-body mapping of malondialdehyde pools in a complex eukaryote. J Biol Chem 282: 35749–35756 [DOI] [PubMed] [Google Scholar]
- Mène-Saffrané L, Dubugnon L, Chételat A, Stolz S, Gouhier-Darimont C, Farmer EE (2009) Nonenzymatic oxidation of trienoic fatty acids contributes to reactive oxygen species management in Arabidopsis. J Biol Chem 284: 1702–1708 [DOI] [PubMed] [Google Scholar]
- Mène-Saffrané L, Jones AD, DellaPenna D (2010) Plastochromanol-8 and tocopherols are essential lipid-soluble antioxidants during seed desiccation and quiescence in Arabidopsis. Proc Natl Acad Sci USA 107: 17815–17820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina TE, Lamb C, Zeier J (2007) Expression of a nitric oxide degrading enzyme induces a senescence programme in Arabidopsis. Plant Cell Environ 30: 39–52 [DOI] [PubMed] [Google Scholar]
- Mishina TE, Zeier J (2006) The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol 141: 1666–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina TE, Zeier J (2007) Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J 50: 500–513 [DOI] [PubMed] [Google Scholar]
- Monteoliva MI, Rizzi YS, Cecchini NM, Hajirezaei MR, Alvarez ME (2014) Context of action of proline dehydrogenase (ProDH) in the hypersensitive response of Arabidopsis. BMC Plant Biol 14: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Návarová H, Bernsdorff F, Döring AC, Zeier J (2012) Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24: 5123–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Métraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porfirova S, Bergmüller E, Tropf S, Lemke R, Dörmann P (2002) Isolation of an Arabidopsis mutant lacking vitamin E and identification of a cyclase essential for all tocopherol biosynthesis. Proc Natl Acad Sci USA 99: 12495–12500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Wang J, Gong Z, Zhou JM (2017) Apoplastic ROS signaling in plant immunity. Curr Opin Plant Biol 38: 92–100 [DOI] [PubMed] [Google Scholar]
- Rajniak J, Barco B, Clay NK, Sattely ES (2015) A new cyanogenic metabolite in Arabidopsis required for inducible pathogen defence. Nature 525: 376–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riewe D, Koohi M, Lisec J, Pfeiffer M, Lippmann R, Schmeichel J, Willmitzer L, Altmann T (2012) A tyrosine aminotransferase involved in tocopherol synthesis in Arabidopsis. Plant J 71: 850–859 [DOI] [PubMed] [Google Scholar]
- Routaboul JM, Fischer SF, Browse J (2000) Trienoic fatty acids are required to maintain chloroplast function at low temperatures. Plant Physiol 124: 1697–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandorf I, Holländer-Czytko H (2002) Jasmonate is involved in the induction of tyrosine aminotransferase and tocopherol biosynthesis in Arabidopsis thaliana. Planta 216: 173–179 [DOI] [PubMed] [Google Scholar]
- Sattler SE, Cahoon EB, Coughlan SJ, DellaPenna D (2003) Characterization of tocopherol cyclases from higher plants and cyanobacteria: Evolutionary implications for tocopherol synthesis and function. Plant Physiol 132: 2184–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SE, Gilliland LU, Magallanes-Lundback M, Pollard M, DellaPenna D (2004) Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell 16: 1419–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SE, Mène-Saffrané L, Farmer EE, Krischke M, Mueller MJ, DellaPenna D (2006) Nonenzymatic lipid peroxidation reprograms gene expression and activates defense markers in Arabidopsis tocopherol-deficient mutants. Plant Cell 18: 3706–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. (2005) Chemistry and biology of vitamin E. Mol Nutr Food Res 49: 7–30 [DOI] [PubMed] [Google Scholar]
- Serrano I, Audran C, Rivas S (2016) Chloroplasts at work during plant innate immunity. J Exp Bot 67: 3845–3854 [DOI] [PubMed] [Google Scholar]
- Sewelam N, Jaspert N, Van Der Kelen K, Tognetti VB, Schmitz J, Frerigmann H, Stahl E, Zeier J, Van Breusegem F, Maurino VG (2014) Spatial H2O2 signaling specificity: H2O2 from chloroplasts and peroxisomes modulates the plant transcriptome differentially. Mol Plant 7: 1191–1210 [DOI] [PubMed] [Google Scholar]
- Shah J, Zeier J (2013) Long-distance communication and signal amplification in systemic acquired resistance. Front Plant Sci 4: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani D, DellaPenna D (1998) Elevating the vitamin E content of plants through metabolic engineering. Science 282: 2098–2100 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Dong X (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12: 89–100 [DOI] [PubMed] [Google Scholar]
- Stahl E, Bellwon P, Huber S, Schlaeppi K, Bernsdorff F, Vallat-Michel A, Mauch F, Zeier J (2016) Regulatory and functional aspects of indolic metabolism in plant systemic acquired resistance. Mol Plant 9: 662–681 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16: 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticher L, Mauch-Mani B, Métraux JP (1997) Systemic acquired resistance. Annu Rev Phytopathol 35: 235–270 [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Penninckx IAMA, Broekaert WF, Cammue BPA (2001) The complexity of disease signaling in Arabidopsis. Curr Opin Immunol 13: 63–68 [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JDG (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA 99: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzin V, Malitsky S, Aharoni A, Galili G (2009) Expression of a bacterial bi-functional chorismate mutase/prephenate dehydratase modulates primary and secondary metabolism associated with aromatic amino acids in Arabidopsis. Plant J 60: 156–167 [DOI] [PubMed] [Google Scholar]
- Valentin HE, Lincoln K, Moshiri F, Jensen PK, Qi Q, Venkatesh TV, Karunanandaa B, Baszis SR, Norris SR, Savidge B, et al. (2006) The Arabidopsis vitamin E pathway gene5-1 mutant reveals a critical role for phytol kinase in seed tocopherol biosynthesis. Plant Cell 18: 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick BA, Zimmerman DC (1984) Biosynthesis of jasmonic acid by several plant species. Plant Physiol 75: 458–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vom Dorp K, Hölzl G, Plohmann C, Eisenhut M, Abraham M, Weber APM, Hanson AD, Dörmann P (2015) Remobilization of phytol from chlorophyll degradation is essential for tocopherol synthesis and growth of Arabidopsis. Plant Cell 27: 2846–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Malek B, van der Graaff E, Schneitz K, Keller B (2002) The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216: 187–192 [DOI] [PubMed] [Google Scholar]
- Wang L, Mitra RM, Hasselmann KD, Sato M, Lenarz-Wyatt L, Cohen JD, Katagiri F, Glazebrook J (2008) The genetic network controlling the Arabidopsis transcriptional response to Pseudomonas syringae pv. maculicola: Roles of major regulators and the phytotoxin coronatine. Mol Plant Microbe Interact 21: 1408–1420 [DOI] [PubMed] [Google Scholar]
- Wang M, Toda K, Block A, Maeda HA (2019) TAT1 and TAT2 tyrosine aminotransferases have both distinct and shared functions in tyrosine metabolism and degradation in Arabidopsis thaliana. J Biol Chem 294: 3563–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Toda K, Maeda HA (2016) Biochemical properties and subcellular localization of tyrosine aminotransferases in Arabidopsis thaliana. Phytochemistry 132: 16–25 [DOI] [PubMed] [Google Scholar]
- Weber H, Vick BA, Farmer EE (1997) Dinor-oxo-phytodienoic acid: A new hexadecanoid signal in the jasmonate family. Proc Natl Acad Sci USA 94: 10473–10478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M, Feys BJ, Parker JE (2005) Plant immunity: The EDS1 regulatory node. Curr Opin Plant Biol 8: 383–389 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Yin L, Mano J, Wang S, Tsuji W, Tanaka K (2010) The involvement of lipid peroxide-derived aldehydes in aluminum toxicity of tobacco roots. Plant Physiol 152: 1406–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbierzak AM, Kanwischer M, Wille C, Vidi PA, Giavalisco P, Lohmann A, Briesen I, Porfirova S, Bréhélin C, Kessler F, et al. (2009) Intersection of the tocopherol and plastoquinol metabolic pathways at the plastoglobule. Biochem J 425: 389–399 [DOI] [PubMed] [Google Scholar]
- Zeier J (1998) Pfanzliche Abschlussgewebe der Wurzel: Chemische Zusammensetzung und Feinstruktur der Endodermis in Abhängigkeit von Entwicklung und äußeren Einfüssen. PhD thesis. University of Würzburg, Wuerzburg, Germany.
- Zeier J. (2005) Age-dependent variations of local and systemic defense responses in Arabidopsis leaves towards an avirulent strain of Pseudomonas syringae. Physiol Mol Plant Pathol 66: 30–39 [Google Scholar]
- Zeier J. (2013) New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ 36: 2085–2103 [DOI] [PubMed] [Google Scholar]
- Zeier J, Pink B, Mueller MJ, Berger S (2004) Light conditions influence specific defence responses in incompatible plant-pathogen interactions: Uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta 219: 673–683 [DOI] [PubMed] [Google Scholar]