The high-resolution cryo-EM structure of a synthetic β-carboxysome shell reveals the specific protein interactions and assists in future study of permeability and encapsulation properties.
Abstract
Carboxysomes are capsid-like, CO2-fixing organelles that are present in all cyanobacteria and some chemoautotrophs and that substantially contribute to global primary production. They are composed of a selectively permeable protein shell that encapsulates Rubisco, the principal CO2-fixing enzyme, and carbonic anhydrase. As the centerpiece of the carbon-concentrating mechanism, by packaging enzymes that collectively enhance catalysis, the carboxysome shell enables the generation of a locally elevated concentration of substrate CO2 and the prevention of CO2 escape. A functional carboxysome consisting of an intact shell and cargo is essential for cyanobacterial growth under ambient CO2 concentrations. Using cryo-electron microscopy, we have determined the structure of a recombinantly produced simplified β-carboxysome shell. The structure reveals the sidedness and the specific interactions between the carboxysome shell proteins. The model provides insight into the structural basis of selective permeability of the carboxysome shell and can be used to design modifications to investigate the mechanisms of cargo encapsulation and other physiochemical properties such as permeability. Notably, the permeability properties are of great interest for modeling and evaluating this carbon-concentrating mechanism in metabolic engineering. Moreover, we find striking similarity between the carboxysome shell and the structurally characterized, evolutionarily distant metabolosome shell, implying universal architectural principles for bacterial microcompartment shells.
Carboxysomes are bacterial microcompartments (BMCs) that encapsulate Rubisco and carbonic anhydrase (CA) in a selectively permeable protein shell. They are found in some chemoautotrophs and all cyanobacteria and are an important part of global carbon fixation (Garcia-Pichel et al., 2003). Bacteria that contain carboxysomes actively accumulate cytosolic HCO3−, which diffuses across the carboxysome shell. Inside the carboxysome, CA converts HCO3− to CO2, the substrate of Rubisco, to enhance CO2 fixation (for overview, see Yeates et al., 2008; Kerfeld et al., 2018). The carboxysome was the first BMC identified, evident as ∼200-nm polyhedral particles, based on electron microscopy (Drews and Niklowitz, 1956) and subsequent isolation and biochemical characterization (Shively et al., 1973; Holthuijzen et al., 1987). More recently, the availability of genomic sequence data and bioinformatics analysis have shown that compartmentalization of metabolism in BMCs is widespread across the Bacterial Kingdom (Axen et al., 2014). Whereas the encapsulated enzymes differ widely, the basic building blocks of the shell are conserved. Two different folds compose the BMC shell architecture, the mixed α-helix/β-sheet pfam00936 domain that forms regular hexagons consisting of either six BMC-H chains (Kerfeld et al., 2005) or three BMC-T chains (each BMC-T chain contains two pfam00936 domains; Heldt et al., 2009; Klein et al., 2009; Sagermann et al., 2009; Crowley et al., 2010) and the pure β-sheet pfam03319 domain that forms pentagonal shaped truncated pyramids consisting of five BMC-P chains (Tanaka et al., 2008). Structural characterization of shells made of proteins of an uncharacterized catabolic BMC from the myxobacterium Haliangium ochraceum (HO shell) shows that proteins from these two folds can form icosahedral structures with the BMC-H and BMC-T on the facets and the BMC-P located at the vertices (Sutter et al., 2017).
There are two types of carboxysomes (α and β) that perform the same biological function but differ in composition, assembly, and evolution (Kerfeld and Melnicki, 2016). Our model shell is of the β-type, and the prefix of the protein names Ccm originates from carbon-concentrating mechanism, starting at CcmK because the extent of the operon was unknown at the time (Price et al., 1993). The conserved carboxysomal genomic loci generally consist of the shell proteins CcmK1 and CcmK2 (BMC-H), CcmL (BMC-P), CcmO (a BMC-T of unknown structure), as well as the lumenal proteins CcmM and CcmN. There are additional proteins in satellite chromosomal locations, such as the BMC-T type CcmP and the BMC-H proteins CcmK3 and CcmK4 (Sommer et al., 2017). A recent study shows in detail how Rubisco interacts with parts of the CA CcmM to form networks (Wang et al., 2019) around which the β-carboxysome shell is thought to assemble (Cameron et al., 2013). The α-carboxysome shell has been shown to provide a barrier to CO2 leakage (Cai et al., 2009), contributing to the carbon-concentrating mechanism of carboxysomes.
We have previously shown the formation of shells from the β-carboxysome proteins CcmK1, CcmK2, CcmL, and CcmO from Halothece sp. PCC 7418 and demonstrated their potential for engineering purposes (Cai et al., 2016). Here, we optimized the purification of this particle and determined 3D structures definitively composed of CcmK1 and CcmK2 and CcmL with cryo-electron microscopy (cryo-EM) single-particle analysis. The shell proteins self-assemble predominantly into two families of shells with pseudo-T = 3 and pseudo-T = 4 icosahedral symmetry with diameters of 210 and 245 Å, respectively. The resolution is sufficient to identify the specific amino acid interactions among the various shell proteins, which are likely the same as in native carboxysomes. The complete structural description of this synthetic β-carboxysome shell provides a foundational system for the design of cargo-loading strategies and mutational studies that will enable direct measurement of the carboxysome shell permeability to gases and metabolites.
RESULTS
Cryo-EM of Purified Synthetic Carboxysome Shells
A synthetic β-carboxysome shell operon encoding BMC-H proteins CcmK1 and CcmK2, BMC-T type CcmO, and C-terminally Strep-tagged BMC-P/CcmL was recombinantly expressed in Escherichia coli, and self-assembled shells were purified by affinity and anion-exchange chromatography. The composition of the purified sample was assessed by SDS-PAGE and negative-stain transmission electron microscopy prior to vitrification for cryo-EM imaging (Supplemental Fig. S1, A and B).
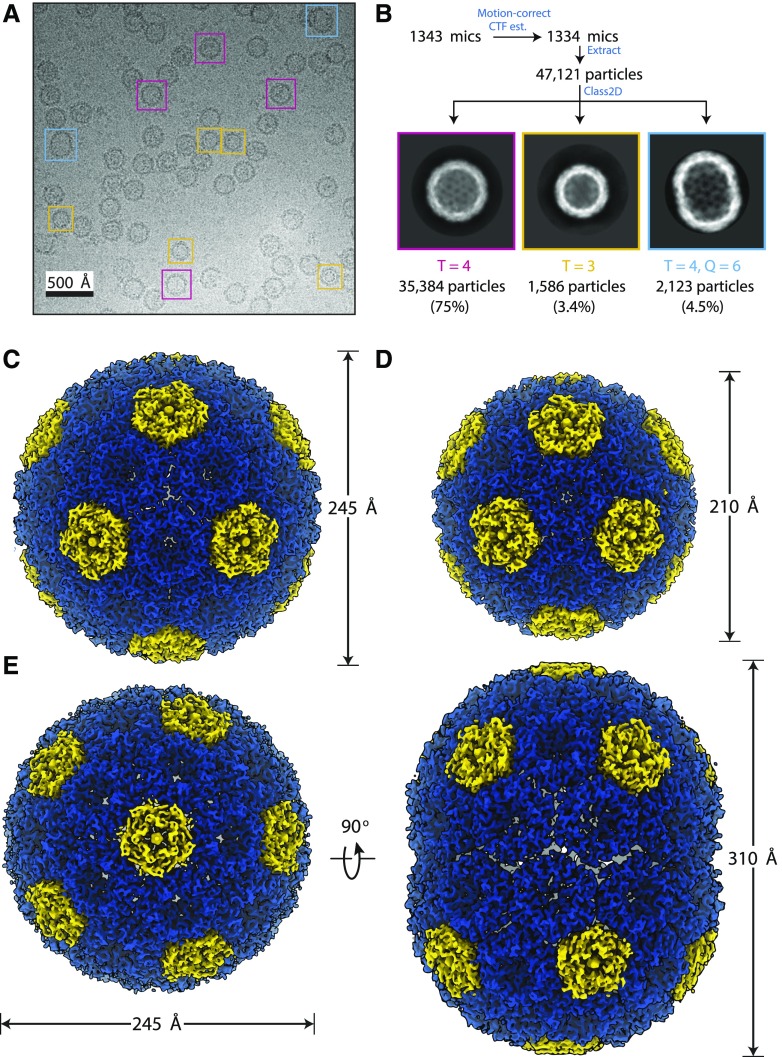
Cryo-EM imaging and subsequent single-particle analysis revealed that multiple classes of shell assemblies were present in the sample (Fig. 1A; Supplemental Fig. S2). We obtained 3D reconstructions of three classes (Fig. 1B; Supplemental Fig. S3). The most prominent and highest resolution class (75% picked particles, 2.6 Å) was a pseudo-T = 4 icosahedral shell (designated pseudo because not all subunits are identical), with a diameter of approximately 245 Å (Fig. 1, B and C). A smaller, pseudo-T = 3 icosahedral shell with a diameter of 210 Å was also found in the sample at a lower occurrence of 3.4%, but we were still able to obtain good resolution (3 Å) due to the high inherent particle symmetry (Fig. 1, B and D). An elongated shell (Fig. 1E), similar to prolate capsids of some bacteriophages (Luque and Reguera, 2010), was also found at low occurrence (4.5%). It has dimensions of 245 × 310 Å, and we were only able to obtain a low-resolution structure (Fourier shell correlation [FSC0.143] = 4.1 Å), likely due to the lower symmetry and possibly structural variability. There were also some larger shells (greater than 300 Å diameter) that frequently appeared filled (Supplemental Fig. S2), indicating that cargo encapsulation (albeit nonspecific, because the shell was not produced in the native organism) might favor larger shells that are closer to the full carboxysome size. However, density can also be seen in pseudo-T = 3 and pseudo-T = 4 shells occasionally (Fig. 1A; Supplemental Fig. S2, A and B), indicating that E. coli cytosolic cargo is commonly packaged.
Figure 1.
Cryo-EM reconstruction of synthetic shells. A, Example cryo-EM micrograph used in reconstruction with example particles of different shell types boxed (T = 4 in magenta, T = 3 in yellow, and T = 4, Q = 6 prolate in cyan). All particles were used for reconstruction, but only selected ones are highlighted for clarity. B, Single-particle analysis preprocessing workflow with example 2D class averages for the different shell types. Particle counts and percentage of input particles for each class are indicated. C to E, 3D reconstructions for the T = 4 (C), T = 3 (D), and prolate (E) shell types with dimensions indicated.
Structural Analysis of the Carboxysome Shell Assemblies
Atomic modeling into the density was facilitated by available high-resolution crystal structures of homologous carboxysome shell protomers (Kerfeld et al., 2005; Tanaka et al., 2008). The T = 3 and T = 4 icosahedral shell reconstructions were of exceptional quality, and atomic models could be readily built and refined into the density (Supplemental Table S1; Supplemental Fig. S4). For the prolate shell, shell proteins could be reliably docked but not refined because of the poor quality of the density. In all cases, the shell reconstructions are composed entirely of the BMC-H protomers CcmK1/CcmK2 and the BMC-P protomer CcmL. The BMC-H paralogs CcmK1 and CcmK2 share 92% sequence identity over 101 aligned residues (CcmK1 has a 12-residue C-terminal extension that is not visible in the density) and could not be distinguished in the T = 4 reconstruction at 2.6 Å resolution. However, ambiguous density at the side chain positions where the two proteins differ possibly indicates that the two proteins are used interchangeably in those shells (Supplemental Fig. S5). For simplicity, all BMC-H protomers are modeled as CcmK1 (hereafter, CcmK refers to either unless specified). Density corresponding to the BMC-T protomer CcmO was not observed in any of the 3D reconstructions. It is possible that CcmO is binding to particles on the inside or outside, or it could be a structural part of particles with low occurrence (e.g. some of the larger particles shown in Supplemental Fig. S2, A and B).
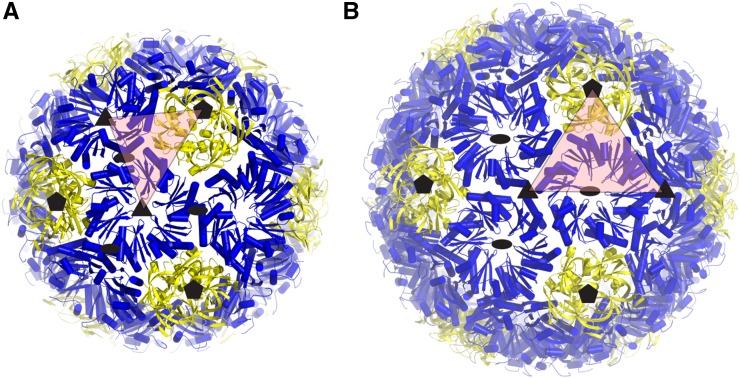
The T = 3 shell icosahedral asymmetric unit consists of one CcmL protomer and two CcmK protomers, with the complete shell composed of 20 CcmK hexamers and 12 CcmL pentamers at the vertices (Fig. 2A). The T = 4 shell icosahedral asymmetric unit contains an additional CcmK protomer relative to the T = 3 shell for a total of 30 CcmK hexamers and 12 CcmL pentamers (Fig. 2B). In the prolate capsid, the asymmetric unit consists of six CcmL protomers and 24 CcmK protomers, with the complete shell composed of 40 CcmK hexamers and 12 CcmL pentamers.
Figure 2.
Geometrical description of the regular icosahedral shell types. Closeup of the cartoon model for each shell type is shown with the asymmetric unit indicated by red triangles. Subunits corresponding to the T = 3 and T = 4 lattices are indicated by solid lines for the icosahedral shells in A and B, respectively.
In all of the shells, the orientation of the subunits was consistent. For the CcmL pentamers, which are shaped like truncated pyramids, the broader side (pyramid base) and C termini face outward. The CcmK proteins have distinct concave and convex surfaces. In all shells, the concave side, which contains both termini, faces outward (Fig. 2). The sidedness of the carboxysome shell proteins is therefore consistent with the previously characterized shell of a metabolosome (Sutter et al., 2017; Greber et al., 2019).
Structural Details of the Shell
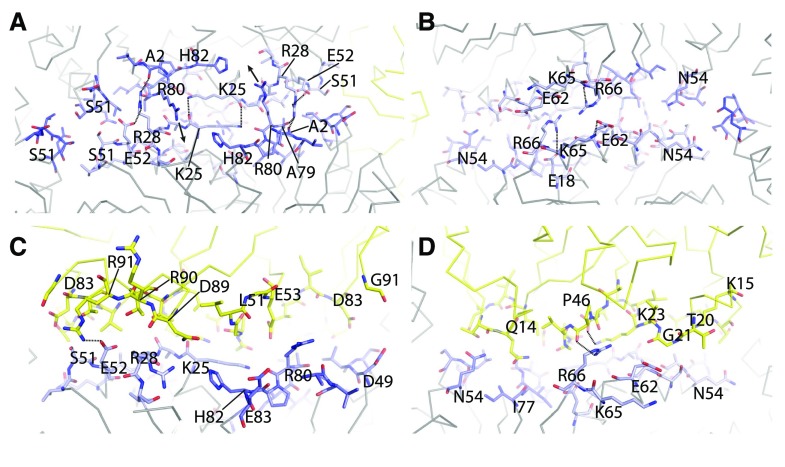
The interfaces between the individual proteins for the T = 3 and the T = 4 shell are almost perfectly identical; we show only the T = 4 interfaces for clarity. The residues at the interfaces between the BMC-H:BMC-H and BMC-P:BMC-H are highly conserved across different microcompartment types (Sutter et al., 2017). For the CcmK-CcmK interface, we could recognize the absolutely conserved K25 making an antiparallel interaction with its counterpart (Fig. 3A). Another highly conserved interaction is R80 of the (A/P)-R-P-H motif inserting into a pocket formed by H82 and backbone oxygens of the neighboring CcmK (Fig. 3A). This CcmK-CcmK interface is at an angle of 30° and, as observed previously for the HO shell, the same interface can probably also adopt a planar configuration where the R80 is extending all the way into the pocket. An interaction that seems to be specific to β-carboxysomes is the hydrogen bonds formed between the R28 side chain and the side chain of E52, its own chain, as well as the backbone oxygen of A79 and A2 across the interface (Fig. 3A). Looking at the CcmK-CcmK interface from the interior of the shell, we noticed additional intrachain interactions between residues E62 and K65 as well as R66 and E18 (Fig. 3B); the E62 and R66 residues are positioned such that they could possibly interact across the interface in a planar configuration. Interestingly, this is a position where Halothece sp. PCC 7418 CcmK1 and CcmK2 differ (E62/K65 in CcmK1, D62/N65 in CcmK2), which could cause a preference of one shell subunit over another for angled/planar interfaces. This residue pattern is not consistent, though, for all β-carboxysomal CcmK1 and CcmK2; there is no distinguishing feature that separates CcmK1 from CcmK2 other than the C-terminal extension (Sommer et al., 2017).
Figure 3.
Protein subunit interfaces. A and B, View onto the CcmK-CcmK interface from the outside (A) and from the inside (B). C and D, View of the CcmK-CcmL interface from the inside (C) and outside (D). Interface residues are shown as sticks, CcmK in shades of blue and CcmL in yellow. Important residues are labeled, and dashed lines indicate specific interactions.
The composition of our T = 4 shell enables visualization for the first time, in the context of a complete BMC shell, of the interface where three BMC-H proteins meet; in the HO shell, a BMC-T is always one of the three oligomers at this interface. The small side chain of residue S51 seems to prevent steric clashes (Fig. 3A) and explains why the residues at this position are generally Gly, Ala, or Ser.
The interface between CcmL and CcmK seems to rely mainly on shape complementarity (Fig. 3, C and D). There is a notable interaction between K23 of CcmL and the backbone oxygen of P46 (Fig. 3D), which, similar to the antiparallel Lys residues of CcmK, creates a flat surface. There are a few charged interactions, including CcmK’s E52 side chain and CcmL’s R91 as well as CcmK’s R66 side chain to the backbone oxygen of P46.
The Structural Basis of Permeability to Carboxysome Metabolites 3-Phosphoglyceric Acid and Bicarbonate
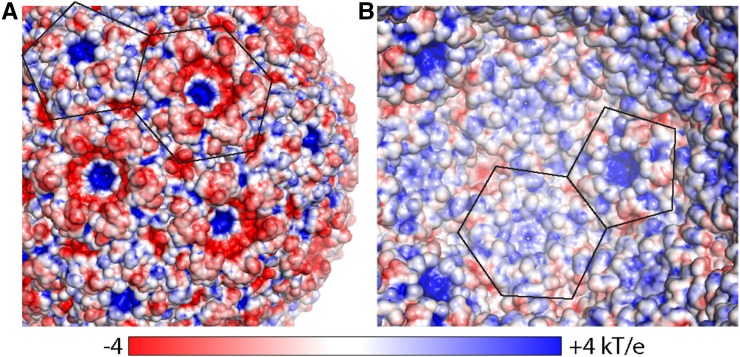
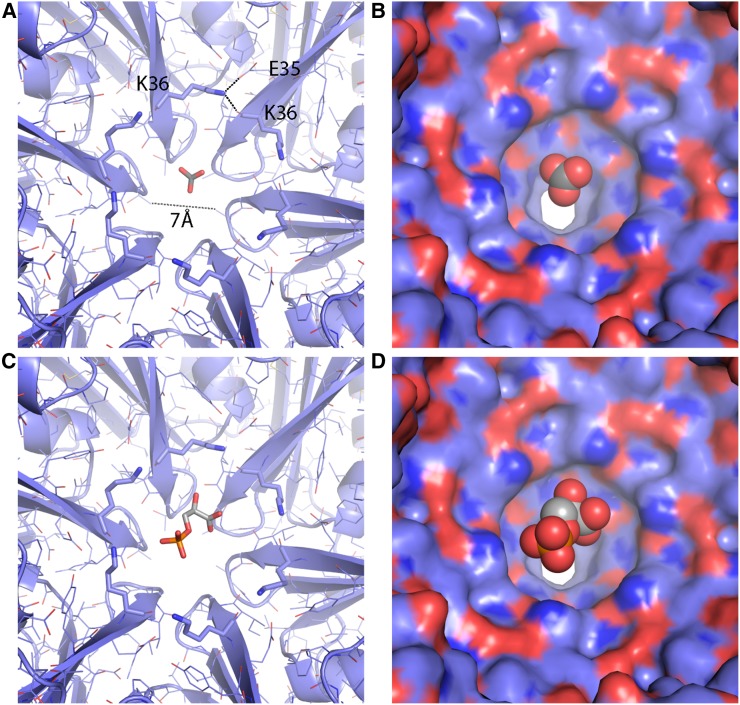
The CcmK proteins are the major building blocks of the facets of the β-carboxysome shell. Protein-protein interactions in the facets completely seal the synthetic shell from bulk solvent, and the pores formed by the cyclic axes of symmetry provide the only conduits into and out of the shell. The EM density for the residues around the CcmK pore of the T = 4 shell was of good quality and allowed placement of the backbone and side chains for all residues (Supplemental Fig. S4C). CcmK1 and CcmK2 are identical in their pore-forming residues, and therefore even if the observed CcmK proteins are heterohexamers (i.e. mixtures or CcmK1 and CcmK2), the diameter of the pore is unlikely to be affected. The pores in the CcmK hexamers are about 7 Å in diameter, consistent with the sizes estimated from the hexamers in isolation (Kerfeld et al., 2005). Electrostatic surface analyses of the shells revealed a generally more negatively charged exterior and positively charged interior (Fig. 4). The pores of both CcmK and CcmL are positively charged, especially pronounced on the exterior surface, possibly favorable to interaction with the negatively charged metabolites that must cross the shell.
Figure 4.
Electrostatics of the T = 4 shell. A, Surface as seen from the outside colored by electrostatic potential (red, negative; blue, positive), with outlines of CcmK and CcmL indicated. B, Same as A, but for the inside view. The scale bar below indicates the range from −4 to +4 kT/e.
There is some residual density at the center of the CcmK pore (Supplemental Fig. S4C), but it is weaker than the surrounding protein density, unlike the very strong density observed at the center of CcmL (Supplemental Fig. S6A). At the center of the pentamer pore, we observed a large spherical density on the concave, outside-facing side of the shell. The density is approximately 8 Å in diameter and appears relatively smooth and featureless. Asymmetric reconstructions still resulted in featureless density occupying the CcmL pore (Supplemental Fig. S6B); therefore, the density likely corresponds to an average of a molecule in multiple orientations. The density is surrounded by positively charged residues K39 and R59, and it is possible that a negatively charged molecule from the E. coli cytosol was bound and retained throughout the purification process.
Comparison with Other Shell Proteins and the Metabolosome (HO) Shell
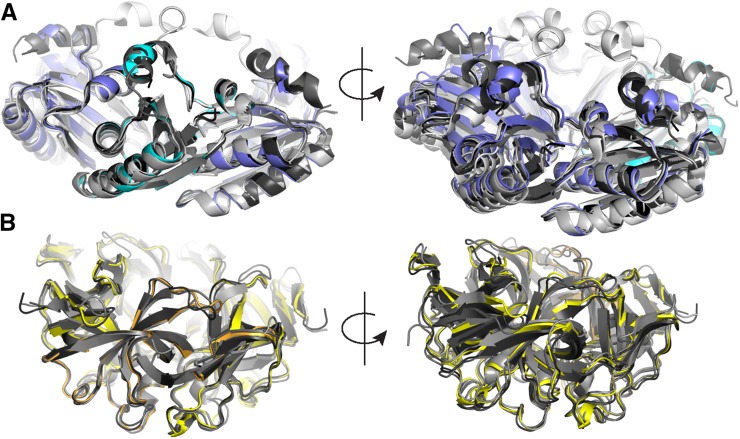
There are a number of crystal structures available for both CcmK1 and CcmK2 and CcmL. When we aligned the structure of one chain of the CcmK hexamer extracted from the shell to other available β-carboxysomal CcmK1 and CcmK2 structures (Fig. 5A, left), the root-mean-square deviation (rmsd) values were very low (0.3–0.5 Å over 80–90 aligned C-α atoms). However, when looking at the opposite side of the locally aligned hexamer, we noted that the hexamer extracted from the shell is more compact than its isolated counterparts (Fig. 5A, right). This indicates that the BMC-H monomer fold is quite rigid but that there is some flexibility in the interactions between individual chains that allow the hexamer to adopt slightly different overall shapes. The difference could be due to the fact that the hexamer in the shell is more constrained by surrounding proteins relative to a single type of hexamer in the context of only crystal-packing interactions. However, the flexibility could also have a role in the native particles, where CcmK presumably interacts with itself and CcmL and also CcmO and CcmP. The CcmL monomer also aligned very well to structures of available β-carboxysomal counterparts (Fig. 5B, left; 0.4–0.55 Å over 75–85 aligned C-α atoms), but, in contrast to the CcmK hexamer, the whole pentamer also aligned very well (Fig. 5B, right), indicating that the shape of the pentamer is quite rigid.
Figure 5.
Structural alignment of Halothece sp. PCC 7418 BMC-H and BMC-P from the shell with corresponding isolated structures of other β-carboxysomes. A, Alignment of Halothece sp. PCC 7418 CcmK1 and CcmK2 (blue), with available crystal structures of CcmK1 and CcmK2 (Protein Data Bank identifiers 2A1B, 3BN4, 3CIM, 3SSQ, 3SSS, and 4OX7) in different shades of gray. Views of the aligned chain in cyan (left) and opposite side (right) are shown. B, Alignment of Halothece sp. PCC 7418 CcmL (yellow), with homologous structures (Protein Data Bank identifiers 2QW7, 4JVZ, 4JW0, and 4N8X) in different shades of gray.
Prolate Shells
The peculiar architecture of the prolate shell consists of two hemispheres similar to the T = 4 shell connected by an additional belt of hexamer subunits, described by an elongation number of six (Q = 6; Luque and Reguera, 2010). Due to the additional hexamer belt, the prolate shell lacks the threefold symmetry that would otherwise be present if the shell were a regular icosahedron, thus the structure is D5 symmetric. The low abundance of this class necessitated enforcement of this D5 symmetry during 3D classification and refinement to yield sufficient resolution reconstructions to discern the identity of the constitutive shell protomers. Whereas symmetric refinement of the entire complex resulted in density maps of sufficient quality to resolve secondary structure elements, the density on the circumference of the hemisphere regions appeared as a partial mix of subunit density and central belt that was very weak relative to the hemisphere regions.
To address the structural heterogeneity and improve the resolution of the reconstruction, we performed symmetry expansion on the prolate particle images and treated the artificially increased data set without enforcing symmetry. Classification on the expanded particles with a soft mask about the vertices and without angular searches revealed a mix of pentamer- and hexamer-centered particles. This result indicated a mixture of prolate shells in which the T = 4 hemispheres are rotated 36° relative to one another (Supplemental Fig. S4C). The angular assignments for hexamer-centered particles were appropriately rotated, combined with the pentamer-centered class, and refined to yield a 3.7 Å reconstruction. However, the central belt hexamers are still much lower quality than the remainder of the reconstruction. Nevertheless, despite the low quality, we could identify the shell protomers as CcmK and CcmL. The low quality of parts of the structure and low abundance indicate that this is likely an incorrectly assembled particle rather than a relevant structural form.
DISCUSSION
The structures we have determined for the synthetic β-carboxysome shells are formed by the major carboxysome shell proteins CcmK1 and CcmK2 and the pentamer protein CcmL (Kerfeld et al., 2005; Tanaka et al., 2008; Rae et al., 2012). Despite coexpression of CcmO and its presence throughout the purification, we could not unambiguously identify this protein in the final structure. It is possible that it is bound to the shell randomly or present in particles that are not ordered well enough to enable classification. Purified CcmO has so far eluded structural characterization (C.A. Kerfeld group, unpublished data), and it is possible that only in native (enzymes + shell) carboxysomes does CcmO play a functional role and is even properly folded. We observe two main types of structures, corresponding to pseudo- (because not all subunits are identical) T = 3 and T = 4 icosahedral particles as well as an elongated prolate-type structure that is likely a result of misassembly. Native β-carboxysomes are typically much larger (greater than 150 nm diameter; Rae et al., 2012), but it has been shown that the assembly of functional carboxysomes starts with an aggregation of the interior components (Cameron et al., 2013), which might influence their size. Larger particles presumably have extended sheets of planar CcmK1 and CcmK2, but the pentamer caps and the surrounding hexamers are expected to be the same as in our model, because the angles are conserved among all sizes of icosahedral particles.
The permeability of carboxysome shells has not been measured directly; it is an essential parameter in estimating the CO2 fixation efficiency of natural and designed carboxysomes, the latter of which includes the goal of enhancing CO2 fixation in plants by heterologous expression of carboxysomes. The surface of the synthetic β-carboxysome shell is tightly sealed, perforated only by 7-Å openings at the cyclic symmetry axis of each CcmK hexamer. These openings are large enough to readily allow the passage of bicarbonate/HCO3− (Fig. 6, A and B) and potentially 3-phosphoglyceric acid (PGA; Fig. 6, C and D), but they are unlikely to conduct ribulose 1,5-bisphosphate. Moreover, by revealing the sidedness of the shell proteins, our model provides new structural information about the orientation of the pores and the electrostatic potential on the interior and exterior surfaces of the shell (Fig. 4); these provide constraints that now can be used to inform theoretical models of the influence of shell permeability (i.e. CO2 barrier, substrate, and product permeability) on CO2 fixation efficiency (Mangan and Brenner, 2014; McGrath and Long, 2014; Hinzpeter et al., 2017; Tsitkov and Hess, 2019). Notably, the exterior surface (Fig. 4A) is predominantly negative, with relatively strong positive charges focused at the pores. The metabolites (HCO3−, PGA, and ribulose 1,5-bisphosphate) that cross the carboxysome shell all are negatively charged. The overall global charge distribution is likely important for electrostatic steering of HCO3− to the openings in the shell. The synthetic shell system and its atomic resolution model provide an ideal system to probe shell permeability. CcmK1 and CcmK2 represent the major shell protein of the facets (Rae et al., 2012) and, as such, are prime targets to obtain estimates for shell permeability, one of the parameters still missing for accurate theoretical modeling (Mangan and Brenner, 2014; Hinzpeter et al., 2017; Tsitkov and Hess, 2019). Knowing the structure will facilitate the design of modifications, such as the SpyTag/Catcher system that has been used to program synthetic metabolosome shells (Hagen et al., 2018) to reliably encapsulate cargo. Since the CcmK structures are highly conserved, a substitution of some subunits with homologs is possible, for example to test the effect of capping of CcmK4 subunits with CcmK3-CcmK4 heterohexamers, as was shown recently (Sommer et al., 2019). Our detailed structure of a synthetic carboxysome shell, together with our previous biochemical characterization (Cai et al., 2016), makes this an attractive model system to study aspects like encapsulation and shell permeability.
Figure 6.
A, Closeup of the CcmK pore of the Halothece sp. PCC 7418 shell with a bicarbonate modeled in the center. K36 lines the concave side of the pore, and hydrogen bonds to the E35 side chain and the backbone oxygen of K36 of the adjacent chain. B, Surface representation of A. C and D, Same as A and B, but with PGA.
We recently reported the structure of a synthetic catabolic BMC shell derived from the phylogenetically remote BMC from H. ochraceum. This HO shell is constructed from a BMC-P protein, a single type of hexamer (there are no BMC-H paralogs in the genome), and three BMC-T proteins. The carboxysome and HO BMC are functionally distinct and are found in organisms remote from one another on a phylogenetic tree (Δ-proteobacteria versus cyanobacteria; Schulz et al., 2017). The amino acid sequences of HO-BMC-P and CcmL are 49% identical, and a structural alignment yields an rmsd of 0.7 Å over 72 aligned C-α atoms. Similar values are observed when comparing the HO-BMC-H and CcmK1 and CcmK2; the sequences share 53% sequence identity, and the structures align with an rmsd of 0.65 Å over 78 aligned C-α atoms. We also compared the interfaces between two hexamers and between the hexamer and pentamer for the β-carboxysomal shell and the HO shell. Despite a number of amino acid substitutions, the backbones of the subunits involved in both the hexamer-hexamer and hexamer-pentamer interfaces align almost perfectly (Supplemental Fig. S7, A and B). In a phylogenetic tree of BMC-P proteins (BMC-H trees are much more difficult to generate and interpret due to permutation and duplication events), the Halothece sp. PCC 7418 CcmL and H. ochraceum BMC-P are in very distinct clades and separated almost all the way to the base of the tree (Supplemental Fig. S8). In this context, the structural conservation between these functionally distinct BMC types is remarkable and suggests that this is a very stable architectural solution for capping interaction between the pentamer and its surrounding hexamers, one that has persisted through evolution and functional diversification. Indeed, BMC-H and BMC-P seem to form an optimal structure that allows the formation of the vertices, where a BMC-P pentamer is surrounded by five BMC-H hexamers, while allowing the BMC-H to form specific interactions between each other. The CcmK-CcmK interaction around the pentamer is at a 30° angle, whereas a planar interaction (as observed in the HO shell) is necessary to form sheets for larger facets. These findings provide detailed structural information on key intersubunit interfaces such as BMC-P:BMC-H and BMC-H:BMC-H and subunit sidedness, providing a foundation for the direct study of carboxysome shell permeability.
MATERIALS AND METHODS
Expression and Purification of Synthetic Carboxysome Shells
The plasmid Halo-1 as described (Cai et al., 2016) was modified using Gibson cloning to include a Strep-II tag (WSHPQFEK) at the C terminus of CcmL connected by a GS linker. Escherichia coli (DE3) cells were transformed with this plasmid and grown in Studier’s autoinducing medium at 25°C overnight. The cell pellet was resuspended in 100 mm Tris, pH 8, 150 mm NaCl, and 1 mm EDTA (buffer A), after which Benzonase and Lysozyme were added (EMD Millipore, Novagen; manufacturer’s instructions) and cells were lysed using a French press. The lysate was centrifuged in an SS-34 rotor at 20,000 rpm for 20 min. The soluble lysate was applied to a 5-mL GE StrepTrap equilibrated in buffer A, the column was then washed with 6 column volumes of buffer A, and the protein was eluted with 5 column volumes of 20 mm Tris, pH 8, 50 mm NaCl, 1 mm EDTA, and 2.5 mm d-desthiobiotin. The eluate was then applied on a Suc cushion (30% [w/v] Suc in 20 mm Tris, pH 8, and 50 mm NaCl) and centrifuged in a Ti-70 rotor for 16 h at 42,000 rpm. The glassy pellet was then resuspended in 20 mL of 20 mm Tris, pH 8, and 50 mm NaCl and applied to a MonoQ 10/100 anion-exchange column. Shells eluted at around 35% of a 50 to 1,000 mm NaCl gradient in 20 mm Tris, pH 8.
Cryo-EM Grid Preparation and Data Collection
In the chamber of a Vitrobot Mark IV at 4°C and 100% humidity, 3 µL of sample at ∼0.6 mg mL−1 was applied to a freshly glow-discharged R1/2 Cu 300 grid (Quantifoil) and immediately blotted with Whatman No. 1 filter paper for 6 s prior to plunging into liquid ethane contained in a liquid nitrogen bath. Data were collected on a Titan Krios G2 transmission electron microscope operated in EFTEM mode at 300 kV and equipped with a K2 summit direct electron detector and Quantum LS Imaging Filter (slit width of 25 eV). Movies were recorded in superresolution mode at an effective pixel size of 0.4454 Å with a cumulative exposure of 50 e− Å−2 distributed uniformly over a 6-s exposure into 25 frames. Data acquisition was performed using SerialEM (Mastronarde, 2018) with one exposure per hole and focusing for each exposure to a target defocus range of −0.5 to −2 µm. A total of 1,343 movies were collected.
Single-Particle Analysis Image Processing
Preprocessing was performed concurrently with data collection through Focus (Biyani et al., 2017). Superresolution movies were drift corrected, dose weighted, and Fourier cropped to 0.8908 Å per pixel using MotionCor2-v1.1.0 (Zheng et al., 2017), and contrast transfer function (CTF) parameters were estimated using Gctf-v1.06 (Zhang, 2016). Aligned micrographs were manually curated, and 1,334 were selected for further processing. Nontemplated particle picking was performed using Gautomatch-v0.56 (https://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch), and results were curated to remove empty picks, resulting in 47,121 particles. Particles were extracted in a 512-pixel box and binned by a factor of 2 using RELION-v3.0.b2 (Scheres, 2012; Zivanov et al., 2018), subjected to reference-free 2D classification, and the different assembly types were selected individually for further processing. All reported global map resolutions correspond to the gold-standard FSC at 0.143 between independently refined half-maps, with phase randomization to correct for masking artifacts (Scheres and Chen, 2012). Local resolution maps were generated using RELION.
For the T = 4 assembly, 35,383 particles were reextracted in a 424-pixel box and imported into cryoSPARC-v2.2 (Punjani et al., 2017) for ab initio reference generation and subsequent homogenous refinement with icosahedral symmetry enforced, resulting in a 3.1-Å map. The cryoSPARC refinement metadata were converted to RELION STAR format for further processing using Daniel Asarnow’s pyem scripts (unpublished data; https://github.com/asarnow/pyem). CTF parameter refinement followed by 3D autorefine improved the global resolution to 2.8 Å. Bayesian polishing (Zivanov et al., 2019) further improved the map to 2.6 Å. Further 3D classification without alignment into four classes resulted in a class of 15,354 particles that yielded a 2.6-Å map with marginal improvements.
The T = 3 assembly was processed similarly to the T = 4 for 1,586 particles extracted in a 384-pixel box and achieving final global resolution of 3 Å. Further classification of the particles did not yield an improved map.
For the prolate (T = 4, Q = 6) assembly, 2,123 particles were extracted in a 484-pixel box and binned by a factor of 2 (Nyquist limit ∼ 3.6 Å). An ab initio reference could not successfully be generated using either cryoSPARC or RELION. Nevertheless, based on the known dimensions of the shell protomers and 2D class averages, a coordinate model was built and converted to a pseudo-map using Chimera-v1.12 (Goddard et al., 2007). The pseudo-map was low-pass filtered to 12 Å and used as an initial reference for autorefinement in RELION using D5 symmetry, which resulted in a 4.4-Å map. Refinement of CTF parameters and Bayesian polishing improved the map to 4.1 Å. To further improve the resolution, the particle images were expanded with D5 symmetry, resulting in 21,230 expanded particles. Focused classification of the expanded particles was performed into five classes, using C1 symmetry and applying a soft mask encompassing an asymmetric unit of the complex. This resulted in two dominant classes, one a pentamer-centered face (58%) and the other hexamer centered (22%). One-tenth of a rotation was applied to the hexamer-centered particle metadata and particles merged with the pentamer-centered class, resulting in 16,941 particles that yielded a 3.7-Å map after autorefinement. The necessity to invert the handedness of the map suggests low initial reference bias, and FSC curves dropping to around zero indicate the absence of duplicate particles.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AFZ45643.1 (ccmK1), AFZ45642.1 (ccmK2), and AFZ45644.1 (ccmL) for Halothece sp. PCC 7418 shell proteins. Density maps were deposited with the Electron Microscopy Data Bank with accession codes EMD-20207 (T = 4 shell), EMD-20208 (T = 3 shell), EMD-20209 (prolate shell), and EMD-20210 (symmetry-expanded prolate shell). Raw frames were deposited with the Electron Microscopy Public Image Archive with accession code EMPIAR-10275. Coordinate models were deposited with the Protein Data Bank with accession codes 6OWF (T = 3 shell) and 6OWG (T = 4 shell).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Purification of synthetic carboxysome shells.
Supplemental Figure S2. Larger particles observed in the cryo-EM data.
Supplemental Figure S3. Cryo-EM reconstruction workflow.
Supplemental Figure S4. Cryo-EM map and model quality.
Supplemental Figure S5. CcmK1 versus CcmK2 comparison.
Supplemental Figure S6. Density occupying the CcmL pore.
Supplemental Figure S7. Alignment of the interfaces with the corresponding HO shell proteins.
Supplemental Figure S8. Phylogenetic tree of BMC-P sequences.
Supplemental Table S1. Cryo-EM data collection, refinement, and validation statistics.
ACKNOWLEDGMENTS
We thank Basil Greber for advice with cryo-EM data processing, Daniel Toso and Elizabeth Montabana for assistance with training and cryo-EM data collection, Paul Tobias for computation support, Fei Cai for help with the initial purifications of Halothece sp. PCC 7418 shells for cryo-EM, Andrew Hagen for help with cloning construct design, and Sylvia Choi for initial EM work. Cryo-EM samples were optimized at the Lawrence Berkeley National Laboratory cryo-EM resource, and image data were collected at the Bay Area Cryo-EM facility at the University of California, Berkeley.
Footnotes
This work was supported by the Office of Science of the U.S. Department of Energy (DE-FG02-91ER20021) with infrastructure support from MSU AgBio Research (to C.A.K.) and the Early Career Research Program (DE-AC02-O5CH11231 to K.M.D.), by a National Science Foundation Graduate Research Fellowship and The Molecular Basis of Cell Function National Institutes of Health predoctoral training grant (NIGMS project 5T32GM007232-38 to T.G.L.), by the Human Frontier Science Program (fellowship LT000234/2018-L to D.S.), and by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (5 R01 AI114975-05 to C.A.K., N.B.S., and M.S.).
Articles can be viewed without a subscription.
References
- Axen SD, Erbilgin O, Kerfeld CA (2014) A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLOS Comput Biol 10: e1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biyani N, Righetto RD, McLeod R, Caujolle-Bert D, Castano-Diez D, Goldie KN, Stahlberg H (2017) Focus: The interface between data collection and data processing in cryo-EM. J Struct Biol 198: 124–133 [DOI] [PubMed] [Google Scholar]
- Cai F, Bernstein SL, Wilson SC, Kerfeld CA (2016) Production and characterization of synthetic carboxysome shells with incorporated luminal proteins. Plant Physiol 170: 1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F, Menon BB, Cannon GC, Curry KJ, Shively JM, Heinhorst S (2009) The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS ONE 4: e7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA (2013) Biogenesis of a bacterial organelle: The carboxysome assembly pathway. Cell 155: 1131–1140 [DOI] [PubMed] [Google Scholar]
- Crowley CS, Cascio D, Sawaya MR, Kopstein JS, Bobik TA, Yeates TO (2010) Structural insight into the mechanisms of transport across the Salmonella enterica Pdu microcompartment shell. J Biol Chem 285: 37838–37846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews G, Niklowitz W (1956) Beitrage zur Cytologie der Blaualgen. II. Zentroplasma und granuläre Einschlüsse von Phormidium uncinatum. Arch Mikrobiol 24: 147–162 [PubMed] [Google Scholar]
- Garcia-Pichel F, Belnap J, Neuer S, Schanz F (2003) Estimates of global cyanobacterial biomass and its distribution. Algol Stud 109: 213–227 [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE (2007) Visualizing density maps with UCSF Chimera. J Struct Biol 157: 281–287 [DOI] [PubMed] [Google Scholar]
- Greber BJ, Sutter M, Kerfeld CA (2019) The plasticity of molecular interactions governs bacterial microcompartment shell assembly. Structure 27: 749–763.e744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen A, Sutter M, Sloan N, Kerfeld CA (2018) Programmed loading and rapid purification of engineered bacterial microcompartment shells. Nat Commun 9: 2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldt D, Frank S, Seyedarabi A, Ladikis D, Parsons JB, Warren MJ, Pickersgill RW (2009) Structure of a trimeric bacterial microcompartment shell protein, EtuB, associated with ethanol utilization in Clostridium kluyveri. Biochem J 423: 199–207 [DOI] [PubMed] [Google Scholar]
- Hinzpeter F, Gerland U, Tostevin F (2017) Optimal compartmentalization strategies for metabolic microcompartments. Biophys J 112: 767–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuijzen YA, Kuenen JG, Konings WN (1987) Activity of ribulose-1,5-bisphosphate carboxylase in intact and disrupted carboxysomes of Thiobacillus neapolitanus. FEMS Microbiol Lett 42: 121–124 [Google Scholar]
- Kerfeld CA, Aussignargues C, Zarzycki J, Cai F, Sutter M (2018) Bacterial microcompartments. Nat Rev Microbiol 16: 277–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld CA, Melnicki MR (2016) Assembly, function and evolution of cyanobacterial carboxysomes. Curr Opin Plant Biol 31: 66–75 [DOI] [PubMed] [Google Scholar]
- Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO (2005) Protein structures forming the shell of primitive bacterial organelles. Science 309: 936–938 [DOI] [PubMed] [Google Scholar]
- Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA (2009) Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J Mol Biol 392: 319–333 [DOI] [PubMed] [Google Scholar]
- Luque A, Reguera D (2010) The structure of elongated viral capsids. Biophys J 98: 2993–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan N, Brenner M (2014) Systems analysis of the CO2 concentrating mechanism in cyanobacteria. eLife 3: e02043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. (2018) Advanced data acquisition from electron microscopes with serialEM. Microsc Microanal 24(Suppl S1): 864–865 [Google Scholar]
- McGrath JM, Long SP (2014) Can the cyanobacterial carbon-concentrating mechanism increase photosynthesis in crop species? A theoretical analysis. Plant Physiol 164: 2247–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Howitt SM, Harrison K, Badger MR (1993) Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. strain PCC7942 involved in carboxysome assembly and function. J Bacteriol 175: 2871–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA (2017) cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14: 290–296 [DOI] [PubMed] [Google Scholar]
- Rae BD, Long BM, Badger MR, Price GD (2012) Structural determinants of the outer shell of β-carboxysomes in Synechococcus elongatus PCC 7942: Roles for CcmK2, K3-K4, CcmO, and CcmL. PLoS ONE 7: e43871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagermann M, Ohtaki A, Nikolakakis K (2009) Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment. Proc Natl Acad Sci USA 106: 8883–8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW. (2012) RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180: 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW, Chen S (2012) Prevention of overfitting in cryo-EM structure determination. Nat Methods 9: 853–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz F, Eloe-Fadrosh EA, Bowers RM, Jarett J, Nielsen T, Ivanova NN, Kyrpides NC, Woyke T (2017) Towards a balanced view of the bacterial tree of life. Microbiome 5: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shively JM, Ball F, Brown DH, Saunders RE (1973) Functional organelles in prokaryotes: Polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 182: 584–586 [DOI] [PubMed] [Google Scholar]
- Sommer M, Cai F, Melnicki M, Kerfeld CA (2017) β-Carboxysome bioinformatics: Identification and evolution of new bacterial microcompartment protein gene classes and core locus constraints. J Exp Bot 68: 3841–3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer M, Sutter M, Gupta S, Kirst H, Turmo A, Lechno-Yossef S, Burton RL, Saechao C, Sloan NB, Cheng X, et al. (2019) Heterohexamers formed by CcmK3 and CcmK4 increase the complexity of beta carboxysome shells. Plant Physiol 179: 156–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter M, Greber B, Aussignargues C, Kerfeld CA (2017) Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell. Science 356: 1293–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO (2008) Atomic-level models of the bacterial carboxysome shell. Science 319: 1083–1086 [DOI] [PubMed] [Google Scholar]
- Tsitkov S, Hess H (2019) Design principles for a compartmentalized enzyme cascade reaction. ACS Catal 9: 2432–2439 [Google Scholar]
- Wang H, Yan X, Aigner H, Bracher A, Nguyen ND, Hee WY, Long BM, Price GD, Hartl FU, Hayer-Hartl M (2019) Rubisco condensate formation by CcmM in β-carboxysome biogenesis. Nature 566: 131–135 [DOI] [PubMed] [Google Scholar]
- Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM (2008) Protein-based organelles in bacteria: Carboxysomes and related microcompartments. Nat Rev Microbiol 6: 681–691 [DOI] [PubMed] [Google Scholar]
- Zhang K. (2016) Gctf: Real-time CTF determination and correction. J Struct Biol 193: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA (2017) MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14: 331–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJH, Lindahl E, Scheres SHW (2018) New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7: e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Scheres SHW (2019) A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]