Multi-omics analyses reveal metabolic pathway activities induced by two synthetic compounds that increase lipid synthesis during microalgal growth.
Abstract
Microalgae accumulate lipids during stress such as that of nutrient deprivation, concomitant with cessation of growth and depletion of chloroplasts. By contrast, certain small chemical compounds selected by high-throughput screening in Chlamydomonas reinhardtii can induce lipid accumulation during growth, maintaining biomass. Comprehensive pathway analyses using proteomics, transcriptomics, and metabolomics data were acquired from Chlamydomonas cells grown in the presence of one of two structurally distinct lipid activators. WD10784 stimulates both starch and lipid accumulation, whereas WD30030-treated cells accumulate only lipids. The differences in starch accumulation are largely due to differential effects of the two compounds on substrate levels that feed into starch synthesis and on genes encoding starch metabolic enzymes. The compounds had differential effects on photosynthesis, respiration, and oxidative stress pathways. Cells treated with WD10784 showed slowed growth over time and reduced abundance of photosynthetic proteins, decreased respiration, and increased oxidative stress proteins, glutathione, and reactive oxygen species specific to this compound. Both compounds maintained central carbon and nitrogen metabolism, including the tricarboxylic acid cycle, glycolysis, respiration, and the Calvin–Benson–Bassham cycle. There were few changes in proteins and transcripts related to fatty acid biosynthesis, whereas proteins and transcripts for triglyceride production were elevated, suggesting that lipid synthesis is largely driven by substrate availability. This study reports that the compound WD30030 and, to a lesser extent WD10784, increases lipid and lipid droplet synthesis and storage without restricting growth or biomass accumulation by mechanisms that are substantially different from nutrient deprivation.
Green algae hold great promise as a feedstock for biofuel production due to their ability to synthesize and store triacylglycerides (TAGs) in lipid droplets (LDs; Guschina and Harwood, 2006; Li-Beisson et al., 2019). However, the relatively low lipid accumulation per unit biomass produced in standard growth conditions has rendered them economically impractical for biofuel production (Wijffels and Barbosa, 2010). Therefore, much effort has gone into devising methods to increase both biomass and lipid yield, and to reduce the induction of stress response pathways. The unicellular green alga Chlamydomonas reinhardtii has emerged as a valuable model system to study both photosynthesis and metabolic processes that are important for the biological production of economically valuable products. The annotated genome sequence in Phytozome facilitates gene-based comparative studies to understand lipid biochemistry. Although several strategies have been attempted to improve lipid production, such as targeted strain design and improvements in growth and harvesting techniques, in most cases the green algae accumulate lipids only when subjected to severe stresses such as nutrient depletion, particularly nitrogen starvation (Wang et al., 2009; Miller et al., 2010; Schmollinger et al., 2014). However, nutrient starvation arrests growth, which is disadvantageous for large-scale production. Therefore, to improve lipid yield for biofuel production, it is important to design methods to decouple lipid accumulation from growth arrest.
Numerous previous studies have examined lipid accumulation in green algae for cells grown under a variety of stressors such as nitrogen, sulfur, phosphorus, or iron deprivation (Wang et al., 2009; Miller et al., 2010; Nguyen et al., 2011; Lee et al., 2012; Blaby et al., 2013; Schmollinger et al., 2014; Wase et al., 2014; Ajjawi et al., 2017). A subset of these studies employed data-rich omics technologies to identify key alterations in expression of metabolic networks or regulatory components that might contribute to lipid production (Boyle et al., 2012; Blaby et al., 2013; Goodenough et al., 2014; Schmollinger et al., 2014; Wase et al., 2014; Ajjawi et al., 2017; Gargouri et al., 2017). Classical hallmarks of nitrogen starvation include downregulation of protein synthesis, degradation of the photosynthetic apparatus, and the movement of primary carbon into long-term storage molecules as starch and TAGs (Blaby et al., 2013; Alboresi et al., 2016). This is correlated with increased expression of diglyceride acyltransferase (DGTT1) and the major lipid droplet protein (MLDP). However, it is unclear if these factors and processes are necessary to induce lipid accumulation or are merely expressed coincident with the nutrient deprivation response.
A novel approach to increase lipid yield is to select chemical inducers of TAG production during phototrophic or mixotrophic growth conditions (for review, see Wase et al., 2018). Franz et al. (2013) employed microplate-based phenotypic screening of a small library of bioactive molecules (∼50) in four strains of oleaginous microalgae (Nannochloropsis salina, Nannochloropsis oculata, Nannochloris sp, and Phaeodactylum tricornutum) to select compounds that increased lipid accumulation by at least 2-fold without terminating growth.Among the hits were forskolin, which increases cAMP levels, and quinacrine, which inhibits NF-κB and facilitates p53 activator, in mammalian systems. A similar study identified the MAP kinase inhibitor U0126 (Promega) and another cAMP modulator IBMX (cat. no. I5879; Sigma-Aldrich) in Chlamydomonas (Choi et al., 2015).
Although counterintuitive, compounds known to inhibit important enzymes of fatty acid (FA) and glycerolipid biosynthesis were also found to increase storage lipids including the fop herbicides thiolactomycin, cerulenin, and triclosan. The fop herbicides inhibit cytosolic acetyl-CoA carboxylase (Xiang et al., 2009), whereas cerulenin and triclosan inhibit the type-II FA, synthase (Chevalier et al., 2016). Thus, in these cases, induction of storage lipids is most likely due to turnover of other lipid species as de novo synthesis is inhibited. The inhibitor of Arabidopsis (Arabidopsis thaliana) monogalactosyldiacylglycerol synthase Galvestine-1 was identified using a similar screening effort (Botté et al., 2011), again suggesting inhibition of one arm of lipid synthesis funnels substrates or metabolites into different complex lipid species.
The most comprehensive high throughput screen for lipid-inducing compounds during algal growth in the absence of stress conditions such as nutrient deprivation was conducted by this laboratory using Chlamydomonas (Wase et al., 2017). More than 43,000 compounds were screened, and 243 active compounds were identified and verified using dose-response curves to estimate lipid accumulation levels during growth. Among the primary hits were two compounds called “WD10784” and “WD30030,” which represented two different chemical structural classes that act to increase lipid storage in algae. Secondary screens confirmed both compounds increased lipid storage 4- to 5-fold over 72 h of compound treatment in a dose-dependent manner. Importantly, lipid induction also occurred during growth in three additional freshwater algae including Chlorella vulgaris UTEX395, Chlorella sorokiniana UTEX1230, and Tetrachlorella alterns UTEX2453 (Wase et al., 2017). The two compounds induced both similar and unique profiles regarding cellular physiology and metabolism. Briefly, both induced FA and triglyceride accumulation without altering galactolipid levels (Wase et al., 2017). WD30030 did not influence starch storage, whereas WD10784 increased starch 2.5-fold. Additionally, the WD10784 compound reduced chlorophyll (Chl) a and b levels and total carotenoids and the WD30030 compound did not affect these pigments. In this work, proteomics, transcriptomics, and metabolomics profiling were conducted to provide further insight into pathway adjustments leading to lipid synthesis and storage induced by these two compounds. The goal of this work was to define similarities and differences between each compound and to contrast the mechanistic impact of the compounds on cellular physiology and biochemistry, particularly in comparison with well-characterized abiotic stress conditions, such as nitrogen deprivation, which have been used extensively to increase lipid synthesis and storage. Importantly, this work supports and extends previous studies demonstrating that cells treated with WD30030 maintain proteins involved in photosynthesis, protein synthesis, and central carbon metabolism that support growth and lipid synthesis. In contrast, treatment of cells with WD10784 after 48 h in culture, induced some stress response pathways and resulted in limited chlorosis. Thus, WD30030 in particular represents an important reagent in our arsenal to provide economically feasible biofuel feedstocks and other high-value products.
RESULTS
Comparative Proteomics, Transcriptomics, and Metabolomics of Chlamydomonas after Treatment with WD30030 or WD10784
WD30030 and WD10784 were identified in a high throughput screen that selected compounds that increased triglyceride accumulation during growth (Wase et al., 2017). The core structures of the compounds are different, and each represents the most potent in inducing lipid accumulation among that compound class. Each passed secondary screens that confirmed the growth and lipid phenotypes but also distinguished the impact of the two compounds in terms of levels of starch, Glc, specific amino acids, and tricarboxylic acid cycle intermediates. This study was carried out to further understand and contrast the impacts of WD30030 and WD10784 on algal metabolism leading to lipid production and storage using a multiomics approach as has proven effective to understand biochemical and physiological shifts that occur in nutrient deprivation conditions (Blaby et al., 2013; Schmollinger et al., 2014; Wase et al., 2014; Park et al., 2015). Our ultimate goal is to find the most effective reagent for maximizing lipid storage without inducing stress responses or limiting biomass yield.
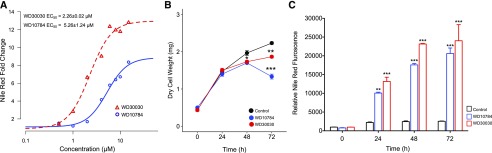
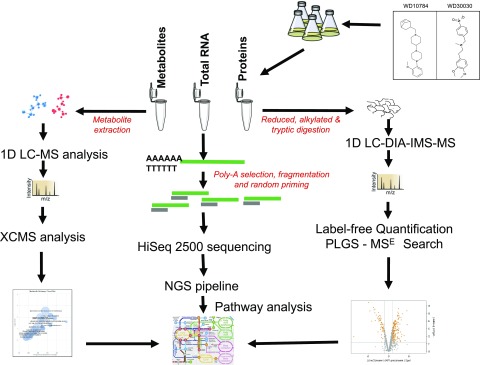
We compared the response of Chlamydomonas to treatment with each compound at a final concentration of 5 µM, which approximates the concentration at which a half-maximal response (i.e. lipid accumulation) occurs (EC50; Fig. 1A). The compounds were added to a single well containing 500,000 cells in 200 μL of culture media (∼OD600 0.15) and growth was continued for up to 72 h. Estimates of dry weight showed that compound-treated cells accumulated approximately the same dry cell mass as controls for up to 48 h, after which growth slows and differences between compound-treated and controls become discernible (Fig. 1B). Lipid accumulation is increased to 3-fold in the first 24 hours of growth after treatment and continues to accumulate over 72 h (Fig. 1C). From these data, it is also apparent that compound WD30030 promotes higher lipid accumulation and higher biomass accumulation compared with WD10784. These results further demonstrate lipid accumulation can be induced to high levels without introducing a severe stress that rapidly leads to growth termination as occurs during nutrient limitation (Hu et al., 2008) or in response to previously identified chemical inducers of TAG accumulation including Brefeldin A and fenpropimorph (Kim et al., 2015; Wase et al., 2015). Employing these data, triplicate cultures were treated with the compounds, harvested after 72 h, and then proteins, RNA, and metabolites were prepared for global screening and comparison with untreated control cultures as detailed in “Materials and Methods” and illustrated in Figure 2. The data were then compared to provide mechanistic insight into metabolic and regulatory changes that lead to lipid storage.
Figure 1.
Estimates of lipid accumulation and biomass after compound treatment. A, Log dose response curves with varying concentrations of compounds by comparison with controls (vehicle alone). B, Biomass estimated as dry cell weight over time at 5 μm of compound or vehicle alone (Control) as indicated. C, Estimation of lipid concentration in cells over time after compound treatment; n = 3 biological replicates, error bars indicate mean ± se. Two way-ANOVA with Bonferroni posttest was performed to determine significant differences between compound treatment and controls over time. Statistical significance is indicated by *P ≤ 0.05 , **P ≤ 0.01, ***P ≤ 0.001.
Figure 2.
Outline of the workflow adopted for multiomics experiments to characterize the metabolic changes induced by lipid activators in Chlamydomonas. NGS, Next-Generation Sequencing, PLGS-MSE,ProteinLynx Global SERVER (Waters Corp), XCMS, https://bioconductor.org/packages/release/bioc/.
Label-free comparative proteomics was applied to three biological replicates for each compound treatment and independently compared to the control sample. A total of 45,384 features comprising charges of 2+, 3+, and 4+ were identified. Of these, 8,590 features were assigned to specific peptide matches by an MSE search resulting in identification and quantification of 884 proteins across all sample groups (Supplemental Dataset S1). Differentially abundant proteins were identified and analyzed to compare the concordance and differences in protein abundance for those significantly changed after compound treatment (Tables 1 and 2; Supplemental Fig. S1). Differentially abundant proteins were defined by a log2-fold change (log2FC) > 0.5 or < −0.5 with a P value ≤ 0.05. In the subset of proteins with increased abundance, 63 were unique to the WD10784 treatment and 23 proteins were unique to the WD30030 treatment, whereas 123 were shared between the two treatments. Similarly, 69 proteins with reduced abundance relative to controls were unique to compound WD10784, whereas 23 proteins were unique to compound WD30030 treatment, and 48 proteins were shared between the two treatment conditions. Enriched pathway clusters were generated from the data resulting in identification of 13 that were significantly altered after WD10784 treatment and 11 after WD30030 treatment. There was more concordance between the two compound treatments for proteins that increased in abundance in the pathway clusters of carbon metabolism, biosynthesis of secondary metabolites, and amino acid and sugar metabolism compared to those that were reduced. In this case, WD10784 decreased abundance of some proteins that mapped to carbon fixation, synthesis of secondary metabolites, pyruvate metabolism, and the pentose P pathway, whereas WD30030 did not.
Table 1. Selected proteins with increased abundance after compound treatment (from Supplemental Datasets S1 and S5–S13).
Significant fold change and P values are indicated in bold face.
| Protein | Protein Name | Peptides | Gene Symbol | Fold Change | P Value | Pathway | ||
|---|---|---|---|---|---|---|---|---|
| WD30030 | WD10784 | WD30030 | WD10784 | |||||
| Cre09.g396650.t1.2 | Phosphate acetyltransferase | 2 | PAT2 | 3.49 | 8.67 | 0.0172 | 0.0025 | Acetate utilization and formation |
| Cre06.g268750.t1.2 | NAD malic enzyme | 10 | NAD-ME2 | 3.74 | 4.31 | 0.0019 | 0.0013 | Tricarboxylic acid cycle |
| Cre10.g423250.t1.2 | Malate dehydrogenase | 12 | MDH2 | 2.53 | 2.25 | 0.0095 | 0.0151 | |
| Cre12.g514750.t1.2 | Citrate synthase | 14 | CIS1 | 1.54 | 2.16 | 0.0439 | 0.0068 | |
| Cre05.g241850.t1.2 | ATP citrate lyase, subunit A | 3 | ACLA1 | 8.67 | 3.42 | 0.0029 | 0.0211 | |
| Cre02.g088600.t1.2 | ATP citrate lyase, subunit B | 11 | ACLA2 | 2.29 | 2.34 | 0.0444 | 0.0414 | |
| Cre12.g526800.t1.2 | 6-Phosphogluconate dehydrogenase | 13 | 6PGD | 1.73 | 2.64 | 0.0455 | 0.0115 | Glycolysis/gluconeogenesis |
| Cre12.g485150.t1.2 | Glyceraldehyde 3-P dehydrogenase | 12 | GAP1a | 10.18 | 14.15 | 0.0018 | 0.0011 | |
| Cre12.g553250.t1.2 | Phosphofructokinase | 3 | PFK2 | 3.06 | 3.71 | 0.0285 | 0.0173 | |
| Cre17.g721500.t1.2 | Starch synthase, chloroplastic | 17 | STA2 | 1.23 | 2.41 | 0.2031 | 0.0029 | Starch and Suc metabolism |
| Cre07.g336950.t1.1 | Alpha-1,4 glucan phosphorylase | 19 | PHOA | 1.86 | 2.86 | 0.033 | 0.005 | |
| Cre12.g552200.t1.2 | Glycogen/starch/alpha-glucan phosphorylase | 6 | PHOB | 1.40 | 1.82 | 0.173 | 0.044 | |
| Cre06.g278210.t1.1 | Phosphoglucomutase | 22 | GPM1 | 2.41 | 2.61 | 0.0043 | 0.0032 | |
| Cre13.g566650.t2.1 | Putative long-chain acyl-CoA synthetase | 10 | LCS2 | 3.10 | 2.59 | 0.0036 | 0.0068 | FA and lipid metabolism |
| Cre16.g687350.t1.2 | Acyl-CoA oxidase | 4 | ACX3 | 2.51 | 4.18 | 0.0327 | 0.0076 | |
| Cre17.g723650.t1.2 | Acetyl-CoA Acyltransferase | 20 | ATO1 | 5.31 | 2.29 | 0.0072 | 0.0657 | |
| Cre01.g053000.t1.2 | Glycerol-3-P dehydrogenase | 12 | GDPH | 2.03 | 3.67 | 0.0520 | 0.0072 | |
| Cre01.g000300.t1.1 | 1-acylglycerol-3-P O-acyltransferase | 6 | GDP8 | 2.94 | 6.69 | 0.0177 | 0.0024 | |
| Cre09.g405500.t1.1 | Major LD protein | 3 | MLDP1 | 3.16 | 1.99 | 0.022 | 0.046 | |
| Cre09.g407700.t1.2 | Phospholipase A2 | 8 | CEP1 | 9.03 | 1.72 | 0.032 | 0.471 | |
| Cre16.g695050.t1.2 | Enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase | 15 | ECH1 | 1.79 | 0.99 | 0.039 | 0.957 | |
| NP_958385.1 | PSII reaction center protein H (PSII-H) | 4 | PsbH | 1.51 | 0.99 | 0.0018 | 0.8800 | Photosynthesis |
| Cre12.g508750.t1.2 | Chl a/b binding protein domain | 2 | Lhca2 | 4.14 | 1.04 | 0.02 | 0.93 | |
| Cre06.g284200.t1.2 | Chl a/b binding protein, chloroplastic | 7 | Lhcbm9 | 0.92 | 3.90 | 0.7670 | 0.0067 | |
| Cre13.g568800.t1.2 | NADH-ubiquinone oxidoreductase | 3 | NUOB12 | 2.59 | 5.10 | 0.1268 | 0.0294 | Respiration and oxidative phosphorylation |
| Cre16.g691552.t1.1 | Pyridine nucleotide-disulphide oxidoreductase | 2 | NDA1 | 1.28 | 7.32 | 0.7203 | 0.0371 | |
| Cre10.g458450.t1.1 | Glutathione peroxidase | 15 | GPXH | 2.05 | 3.00 | 0.0321 | 0.0080 | Redox homeostasis |
| Cre09.g391900.t1.1 | Thioredoxin h1 | 15 | TRXH | 5.53 | 4.39 | 0.0008 | 0.0013 | |
| Cre16.g688550.t1.2 | Glutathione-s-transferase | 18 | GSTS1 | 9.68 | 17.29 | 0.0004 | 0.0002 | |
| Cre03.g145787.t1.1 | HSP 22C | 8 | HSP22C | 1.38 | 10.38 | 0.3935 | 0.0021 | Heat shock proteins |
| Cre16.g677000.t1.2 | HSP 70E | 23 | HSP70E | 2.08 | 3.29 | 0.0034 | 0.0006 | |
| Cre06.g250100.t1.2 | HSP 70B | 38 | HSP70B | 1.54 | 2.25 | 0.0107 | 0.0011 | |
| Cre01.g029650.t1.1 | Subunit of retromer complex | 5 | VPS35 | 3.60 | 4.66 | 0.0018 | 0.0009 | Endocytosis |
| Cre17.g728150.t1.2 | R-SNARE protein, YKT6-family | 3 | YKT6 | 13.87 | 12.18 | 0.0046 | 0.0055 | SNARE interactions in vesicular transport |
| Cre06.g269950.t1.2 | Flagellar-associated protein | 22 | CDC48 | 2.16 | 3.16 | 0.0342 | 0.0092 | Protein processing in ER |
| Cre06.g278163.t1.1 | Acetyl-Orn aminotransferase | 2 | ARG9 | 1.45 | 7.69 | 0.3754 | 0.0054 | Biosynthesis of amino acids |
| Cre06.g252650.t1.2 | Isopropylmalate dehydratase, small subunit | 2 | LEU1S | 2.23 | 3.42 | 0.0245 | 0.0056 | |
Table 2. Selected proteins with decreased abundance after compound treatment (from Supplemental Datasets S1 and S5–S13).
Significant fold change and P values are indicated in bold face.
| Protein | Protein Name | Peptides | Gene Symbol | Fold Change | P Value | Pathway | ||
|---|---|---|---|---|---|---|---|---|
| WD30030 | WD10784 | WD30030 | WD10784 | |||||
| Cre03.g158900.t1.2 | Dihydrolipoamide acetyltransferase | 21 | DLA2 | 0.41 | 0.36 | 0.0137 | 0.0084 | Carbon metabolism |
| Cre01.g010900.t1.2 | Glyceraldehyde-3-P dehydrogenase | 26 | GAP3 | 1.08 | 0.23 | 0.7135 | 0.0018 | |
| Cre02.g141400.t1.2 | Phosphoenolpyruvate carboxykinase 1 | — | PCK1 | 0.84 | 0.49 | 0.5162 | 0.0430 | |
| Cre12.g511900.t1.2 | Ribulose-P 3-epimerase | 4 | RPE1 | 0.24 | 0.19 | 0.0025 | 0.0014 | |
| Cre03.g185550.t1.2 | Sedoheptulose-1,7-bisphosphatase | 14 | SEBP1 | 0.58 | 0.29 | 0.0598 | 0.0041 | |
| Cre14.g630847.t1.1 | Transaldolase | 11 | TAL2 | 0.55 | 0.23 | 0.0844 | 0.0048 | |
| Cre02.g080200.t1.2 | Transketolase | 19 | TRK1 | 0.58 | 0.25 | 0.0487 | 0.0020 | |
| Cre06.g282800.t1.2 | Isocitrate lyase | 22 | ICL1 | 0.72 | 0.15 | 0.0472 | 0.0002 | Glyoxylate cycle |
| Cre03.g144807.t1.1 | Malate synthase | — | MAS1 | 1.02 | 0.44 | 0.9388 | 0.0249 | |
| Cre13.g567950.t1.2 | Glc-1-P adenylyltransferase | 17 | STA1 | 0.65 | 0.44 | 0.104 | 0.016 | Starch metabolism |
| Cre03.g188250.t1.2 | Glc-1-P adenylyltransferase | 3 | STA6 | 0.72 | 0.36 | 0.203 | 0.003 | |
| Cre08.g359350.t1.2 | Biotin carboxylase, acetyl-CoA carboxylase component | 7 | BCR1 | 0.72 | 0.36 | 0.287 | 0.020 | FAs and lipids |
| Cre06.g284250.t1.2 | Chl a/b binding protein, chloroplastic | 14 | LHCBM8 | 0.07 | 0.57 | 0.0047 | 0.3003 | |
| Cre02.g077300.t1.2 | Nucleolar protein | 6 | NOP1 | 0.27 | 0.13 | 0.0063 | 0.0011 | Ribosomes |
| Cre06.g264350.t1.2 | Plastid ribosomal protein L13 | 6 | PRPL13 | 0.19 | 0.12 | 0.0035 | 0.0013 | |
| Cre06.g299000.t1.2 | Plastid ribosomal protein L21 | 2 | PRPL21 | 0.25 | 0.09 | 0.0097 | 0.0013 | |
| Cre06.g272850.t1.2 | Plastid ribosomal protein L10 | 2 | PRPL10 | 0.45 | 0.26 | 0.0480 | 0.0086 | |
| Cre02.g091100.t1.2 | Ribosomal protein L15 | 2 | RPL15 | 0.70 | 0.20 | 0.2988 | 0.0059 | |
| Cre02.g102250.t1.2 | Ribosomal protein S3 | 6 | RPS3 | 0.35 | 0.35 | 0.0053 | 0.0051 | |
| Cre06.g272800.t1.2 | 40S ribosomal protein S8 | 16 | RPS8 | 0.54 | 0.52 | 0.0032 | 0.0024 | |
| Cre06.g259850.t1.2 | Mitochondrial ribosomal protein L29 | 2 | MRPL29 | 0.12 | 0.05 | 0.0021 | 0.0005 | |
| Cre17.g734450.t1.2 | Plastid ribosomal protein L19 | 3 | PRPL19 | 1.13 | 0.52 | 0.4259 | 0.0077 | |
| Cre13.g581650.t1.2 | Plastid ribosomal protein L7/L12 | 18 | PRPL7/L12 | 0.46 | 0.37 | 0.0216 | 0.0093 | |
| Cre16.g659950.t1.1 | Plastid ribosomal protein S5 | 7 | PRPS5 | 0.47 | 0.33 | 0.0041 | 0.0010 | |
| Cre02.g083950.t1.1 | Plastid-specific ribosomal protein 3 | 6 | PSRP-3 | 0.57 | 0.44 | 0.0170 | 0.0047 | |
| Cre01.g039250.tl. | Cytosolic 80S ribosomal protein S2 | 17 | RPS2 | 0.60 | 0.36 | 0.0828 | 0.0099 | |
To assess compound impacts on gene expression, whole transcriptome sequencing by RNA sequencing (RNA-Seq) was performed. From a total of 15,965 mapped genes, 1,816 differentially expressed genes (DEGs; log2FC > 2 or < −2 and P ≤ 0.01) were detected with compound WD30030 treatment (Supplemental Dataset S2; Supplemental Fig. S2), of which 910 were significantly increased, whereas 926 were decreased. By comparison, compound WD10784 treatment resulted in a total of 4,364 transcripts significantly different from controls of which 2,266 were increased, whereas 2,098 transcripts were decreased. Of these, 722 transcripts were upregulated by both compounds and 664 were reduced by both compound treatments. These data indicated a more pronounced impact of WD10784 treatment on transcription by comparison with WD30030. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the DEGs with reduced abundance from WD30030-treated cells showed enrichment in only three categories including ribosomes, photosynthesis, and photosynthetic antenna proteins, whereas transcripts with increased expression mapped to nine categories, the most abundant of which were genes involved in DNA replication, purine metabolism, and protein processing in the endoplasmic reticulum (ER; Supplemental Fig. S3). Treatment with compound WD10784 had a pronounced effect on genes in 25 metabolic pathways. Most of the transcripts with decreased expression were in the categories “ribosomes,” “biosynthesis of secondary metabolites,” “carbon metabolism,” and “photosynthesis and photosynthetic antenna proteins,” whereas transcripts with increased expression were distributed in the categories “DNA replication” and “glutathione (GSH) metabolism,” and others related primarily to lipid and carbon metabolism. To evaluate the reliability of the RNA-Seq results, we performed reverse transcription quantitative PCR (RT-qPCR) on 12 genes (acetyl-CoA synthetase [ACS1], Peroxisomal acyl-coenzyme A oxidase 1 [ACX1], acetyl-COA biotin carboxyl carrier protein1 [BCC1], citrate synthase [CIS1], diacylglycerol acyltransferase, DAGAT Type 2 [DGTT1], 3-ketoacyl-ACP-synthase [KAS2], lecithin:cholesterol acyltransferase [LCA1], mitochondrial pyruvate dehydrogenase complex, E1 component, alpha subunit [PDC1], phosphofructokinase [PFK1], glycerol-3-phosphate acyltransferase [PLSB1], pyruvate Kinase [PYK1], and ADP-glucose pyrophosphorylase small subunit [STA6]) using RNA from cells prepared using the same treatment conditions used for RNA-Seq (Supplemental Dataset S3; Supplemental Fig. S4). These genes were selected to represent a range of targets identified to be important for lipid accumulation in Chlamydomonas. For all 12 genes, the expression patterns from RNA-Seq and RT-qPCR were comparable as demonstrated by correlation coefficients near 1.0 (WD10784 R2 = 0.96; WD30030 R2 = 0.9).
To further understand the metabolic outcomes of pathway changes identified through proteomic and transcriptomic analyses, an untargeted high-resolution metabolomics approach was employed to assess levels of low-molecular–weight polar metabolites. The differentially abundant features in each treatment group were compared with the untreated controls, features with intensity <1,000 were filtered out, and then statistical analysis was performed on the remaining features in the set. Differentially abundant features were defined as those having a log2FC change in abundance ≥ 2 or ≤ −2 with P values ≤ 0.01. A total of 11,752 aligned features were identified, of which 939 (8%) and 1,124 (9.56%) features were significantly changed by compound WD30030 and WD10784 treatments, respectively (Supplemental Dataset S4; Supplemental Fig. S5). Of these, 371 were unique to WD30030, 556 to WD10784, and 568 were common to both compound treatments.
To ascribe metabolite identifications to the features whose abundance was significantly different between treatments, Mummichog (http://mummichog.org/) analysis was performed using the Metaboanalyst server (http://www.metaboanalyst.ca/) using mass accuracies of 0.1 ppm and a P value cutoff of 0.0001. Masses were searched in “positive ionization” mode using the Arabidopsis metabolic model as background. This analysis identified 134 unique metabolites significantly increased after WD30030 treatment, whereas 125 were increased after WD10784; 257 metabolites were shared between the two. Similarly, 26 metabolites were decreased in abundance by WD30030 alone, 75 decreased by WD10784, and 204 were shared between the two treatments (Supplemental Fig. S5).
Effect of Compound Treatments on Acetate Utilization, acetyl-CoA Generation, and Central Carbon Metabolism
In this study, Chlamydomonas was grown mixotrophically on acetate as a source of carbon. Acetate assimilation occurs by two pathways: It is either directly converted to acetyl-CoA upon import in a reaction catalyzed by ACS1, ACS2, or ACS3, or it is metabolized to acetyl-CoA via a two-step reaction catalyzed by acetate kinase (ACK1 or ACK2) and P acetyltransferase (PAT1 or PAT2). Alternatively, a recent study proposed a potential acetate permease GFY 3-5 may be involved in acetate uptake (Goodenough et al., 2014). Only three proteins were detected in proteomics experiments and among these, neither ACS3 nor ACK2 protein levels were significantly affected by compound treatments (Supplemental Dataset S5), whereas PAT2 was increased by both. Based on RNA-Seq data, transcript levels of ACK1, ACK2, and PAT1 were significantly higher than controls after treatment with either compound. These data suggest that the two-step pathway to acetyl-CoA is induced to potentiate acetate assimilation under compound treatment. Similar results were observed during nitrogen starvation (Wase et al., 2014; Park et al., 2015).
Once formed, acetyl-CoA may be oxidized in the tricarboxylic acid cycle to produce NADH, FADH2, ATP, and CO2 (Johnson and Alric, 2013). NADH and FADH2 are further oxidized by the electron transport chain to yield ATP through oxidative phosphorylation (OxPhos). At the protein level, the abundance of six of 14 tricarboxylic acid cycle–associated proteins identified and quantified were significantly increased by either WD30030 or WD10784 (Table 1; Supplemental Dataset S5). Transcripts encoding both glyoxylate cycle enzymes, isocitrate lyase and malate synthase, were reduced by either compound treatment although, for protein abundance, only WD10784 treatment resulted in reduced enzyme levels (Table 2). The reduction in abundance of the glyoxylate cycle enzymes is in contrast to results obtained after nitrogen starvation of starchless strains where both were increased (Blaby et al., 2013).
Among the most significant alterations in enzymes that lead to FA and lipid synthesis were increases in the abundance of citrate synthase (both CIS1 and CIS2), malic enzyme (NAD_ME2), and ATP citrate lyases (ACL1 and ACL2; Table 1; Supplemental Dataset S5). These critical enzymes promote the production of cytosolic acetyl-CoA to provide substrates and reducing power to FA synthesis. Previously, citrate levels were determined to be unchanged by WD10784 treatment, but were increased significantly by WD30030 (Wase et al., 2017).
Gluconeogenesis and the Calvin–Benson–Bassham cycle pathways are responsible for generating 6-C backbones that provide substrates to the oxidative pentose P pathway and monosaccharide synthesis. It has been reported that the 6-C backbones can also be catabolized via glycolysis leading to acetyl-CoA production for FA synthesis (Johnson and Alric, 2013). There was no effect on protein levels of Rubisco, the rate-limiting enzyme of the Calvin–Benson–Bassham cycle after either compound treatment. However, RNA levels were reduced, particularly by WD10784, suggesting a future impact as the protein turns over. It was noted that the transcript and protein expression of additional key enzymes in these interrelated pathways including 6-phosphofructokinase (PFK2), glyceraldehyde-3-P dehydrogenase (GAP1), and 6-phosphogluconate dehydrogenase (6PGD) were significantly increased by both treatment conditions (Table 1; Supplemental Dataset S2). Protein levels of pyruvate kinase (PYK2) were unchanged.
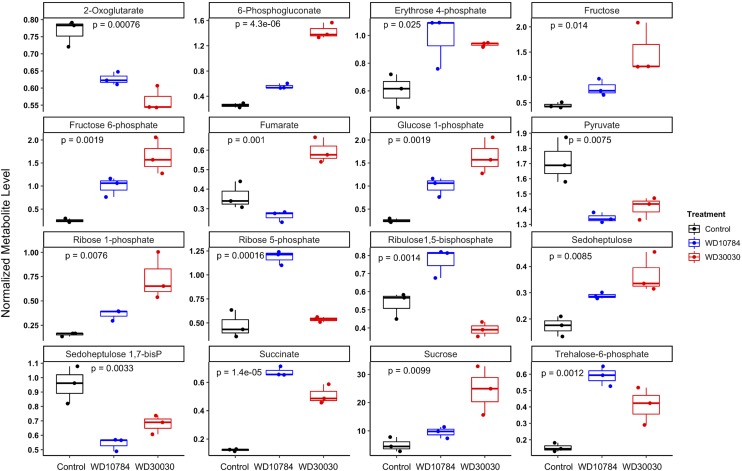
Fru-1,6 bisphosphate aldolase (FBA) generally functions in both gluconeogenesis and the Calvin–Benson–Bassham cycle and catalyzes the synthesis of 6-C Fru-bisphosphate from two 3-C sugars, dihydroxyacetone P, and glyceraldehyde-3-P (G3P). None of the FBA proteins were identified in label-free proteomics analyses, but at the transcript level, three genes encoding FBA were reduced, whereas FBA4 transcript abundance was significantly increased by both treatment conditions. The next enzyme in the pathway, Fru-1,6-bisphosphatase (FBP1), had reduced transcript abundance but was increased at the protein level ∼2-fold after either compound treatment. Metabolite analysis indicated reduction in Fru-1,6-bisphosphate and elevation of Fru-6-P (17-fold by WD30030 and 300-fold by WD10784). This subsequently resulted in elevation of Glc-6-P and Glc-1-P levels, substrates for starch synthesis.
An additional 12 proteins of enzymes in the Calvin–Benson–Bassham cycle were identified and compared (Supplemental Dataset S5). Of these, only the abundance of one, Ribulose-P 3-epimerase (RPE1), was reduced by both compounds; all others were not significantly different from controls. WD10784 treatment also reduced glyceraldehyde/erythrose P dehydrogenase family, transketolase (TRK1), RPE1, and sedoheptulose-1,7-bisphosphatase (SEBP1). This marks a key difference in the metabolic response to WD30030 compared with WD10784, demonstrating the latter compound could eventually reduce the function of the Calvin–Benson–Bassham cycle and production of hexose products from carbon dioxide. When metabolites were examined, erythrose 4-P was increased after either treatment, whereas ribose 5-P and d-xylulose 5-P were found to be significantly increased only after WD10784 treatment. The 6-phosphogluconate, an intermediate in the pentose phosphate and Entner–Doudoroff glycolytic pathway, was significantly increased by either treatment (Fig. 3; Supplemental Dataset S4).
Figure 3.
Boxplot illustration of differential expression of selected metabolites. One-way ANOVA was performed for multiple group comparison. Adjusted P value was calculated using Bonferroni–Hochberg (n = 3 biological replicates).
An important consideration for glycerolipid synthesis is the transport of glycerol-3-P from the chloroplast to the cytosol to fuel lipid synthesis in the ER. Two predicted triose phosphate transporters and four nucleotide-sugar transport members were identified in RNA-Seq experiments (Supplemental Dataset S5). All were elevated by both compound treatments. Among these one, Cre16.g663800.t1.2, is predicted to localize to the chloroplast. It was increased 6- to 7-fold by either WD30030 or WD10784.
Starch Metabolism
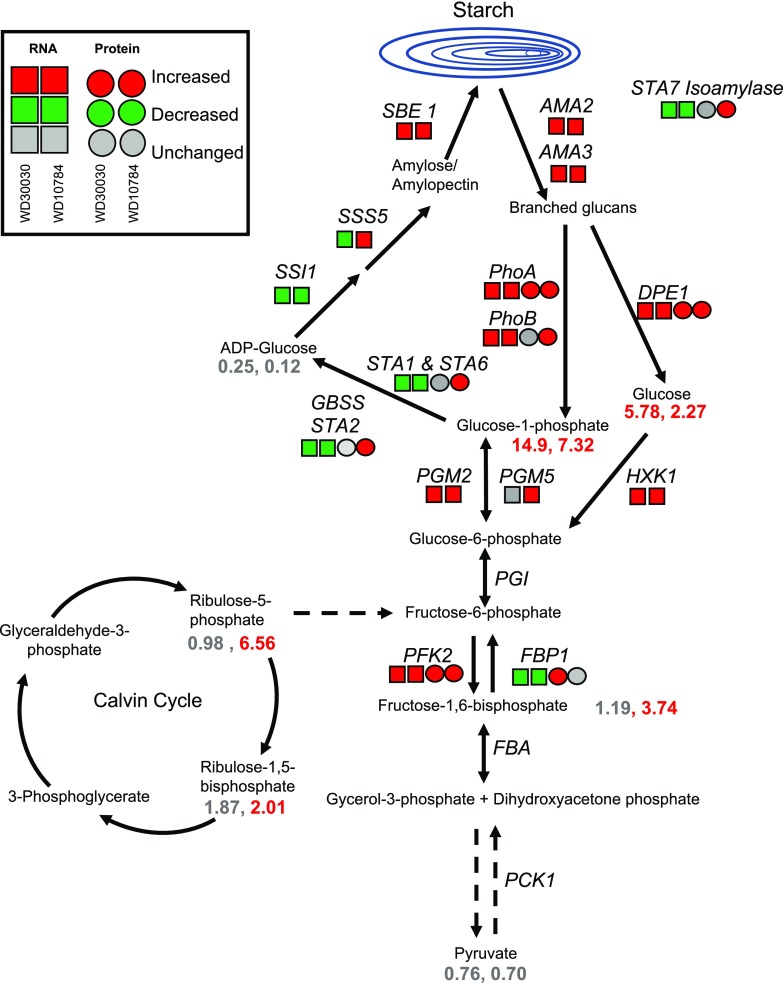
Wase et al. (2017) previously reported that WD10784 increases starch accumulation whereas WD30030 does not, therefore we sought to understand this diversification of storage outcomes using the multiomics data. Metabolite analysis demonstrated that levels of substrates for early steps in starch synthesis were highly elevated after either compound treatment including Fru-6-P, Glc-6-P, and Glc-1-P (Fig. 4; Supplemental Dataset S4; Wase et al., 2017). Glc-1-P adenylyltransferase catalyzes the rate-limiting step in the starch synthesis pathway, which controls the flux of carbon into starch. The regulatory architecture is composed of the large regulatory STA1 enzyme (Cre13.g567950.t1.2) and small catalytic STA6 enzyme (Cre03.g188250.t1.2). In nitrogen starvation, starch levels were increased coincident with a reduction in both the STA1 and STA6 enzyme levels (Miller et al., 2010; Msanne et al., 2012; Schmollinger et al., 2014; Wase et al., 2014). WD10784 treatment resulted in a significant reduction in STA1 and STA6 proteins, but an increase in STA2 protein; protein levels were not significantly different from controls after treatment with WD30030 (Tables 1 and 2; Fig. 4; Supplemental Dataset S5). Expression of a number of genes of starch biosynthesis were increased only in cells treated with WD10784. These included, for example, phosphoglycerate mutase1 (PGM5) and glycogen/starch/α-glucan phosphorylase (PhoA and PhoB). The abundance of several other enzymes was increased by both compound treatments, although impact of WD10784 was more substantial in each case, resulting in high levels of RNA encoding GAP1, ATP-dependent PFK2, starch branching enzyme (SBE1), FBA4, and UTP-Glc-1-P uridylyltransferase (UGP1). Also significantly increased by treatment with either compound were two transcripts that were identified as encoding α-amylase, an enzyme required for starch breakdown. The balance between starch synthesis and breakdown has been invoked as important for the potentiation of lipid synthesis in nitrogen deprivation whereby inhibition of starch synthesis results in lipid accumulation (Li et al., 2010).
Figure 4.
Changes in transcript (squares) and protein (circles) levels for enzymes in the starch biosynthesis and degradation pathways after treatment with lipid-inducing compounds. Dashed arrows indicate multiple enzymatic steps. Fold change values of metabolite levels are given as numbers (see also Supplemental Datasets S1, S2, and S5). Enzyme abbrevations: SBE1, starch branching enzyme; SSS5, soluble starch synthase; STA1 and 6, glucose-1-phosphate adenylyltransferase; AMA2, 3, amylase DPE:disproportionating enzyme; HXK1, hexokinase; PGM2, 5, phosphoglucomutase; PFK1, phosphofructokinase; FBP1, fructose 1,6-bisphosphatase; FBA, fructose-bisphosphate aldolase; PCK1, phosphoenolpyruvate carboxykinase; PHOA, B, α-1,4 glucan phosphorylase
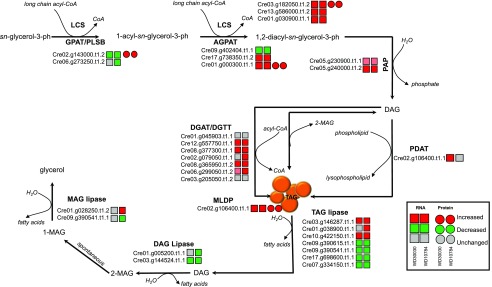
Lipid Metabolism
A primary goal of this work was to identify factors and mechanisms that impact lipid synthesis and storage in response to the small molecule activators. The increase in lipid accumulation could be due to induction of lipid synthesis and storage pathways, decrease in degradation pathway components, or increased substrate availability and utilization. Notably, a prior strong case has been made for increased citrate levels leading to FA biosynthesis and ultimately storage in complex lipids, particularly TAGs (Wase et al., 2014, 2017). As mentioned above in the "Effect of Compound Treatments on Acetate Utilization, acetyl-CoA Generation, and Central Carbon Metabolism" section, this is supported in part by increased levels of CIS and ACL at both the protein and RNA levels after treatment with either compound.
Activity of the FA biosynthetic pathway might be expected to increase to provide FA substrates for TAG synthesis in response to the lipid-inducing compounds. Acyl-carrier protein1 (ACP1) was not detected in the proteomics analysis and ACP1 transcripts were not affected by either treatment. ACP2 protein abundance was unchanged whereas transcript abundance was significantly decreased by both compounds (Supplemental Dataset S6). Acetyl-CoA biotin carboxyl carrier protein2 (BCC2) was increased 5.6- and 15.8-fold by WD30030 and WD10784 treatments, respectively, but the corresponding transcripts were decreased in abundance (Table 1; Supplemental Dataset S6). There was no evidence that other FA biosynthetic enzymes were increased in abundance by either compound. Notably increased at the RNA level were two long-chain acyl-CoA synthetases LCS1 and LCS3. These proteins are required for complex lipid synthesis because acyl-CoAs are the activated substrate for the synthetic pathways. Among components of the FA desaturase family, no proteins were detected. However, RNA levels were substantially increased for fatty acid desaturase2.a (FAD2.a) and FAD5c, whereas fatty acid desaturase3 (FAD3), monogalactosyldiacylglycerol synthase Galvestine-1-specific palmitate delta-7 desaturase (FAD5a), and chloroplast glycerolipid omega-3 fatty acid desaturase (FAD7) were significantly decreased.
Several enzymes and proteins required for TAG synthesis and storage were induced at the transcript level (Supplemental Dataset S6; Fig. 5). These included the diacylglycerol acyltransferases (DGATs) DGTT1, DGTT3, and DGTT5, as well as three other acyltransferases. Among these was PDAT1, encoding phospholipid:DGAT, whose expression was increased in response to WD30030, whereas it remained at baseline after WD10784 treatment. There were increases in transcripts encoding three enzymes involved in sulphoquinovosyl diacylglycerols, which are essential for structure and function of thylakoid membranes. Additionally, transcripts for LD-associated proteins including major lipid droplet protein (MLDP), CDP-ethanolamine:DAG ethanolamine phosphotransferase, and putative lysophosphatidylcholine acyltransferase, were all elevated compared with controls in response to either compound. Of these, only MLDP1 protein was identified in the proteomics experiment and it was found to be induced by both compounds (Table 1). These data follow a trend seen for other lipid-inducing conditions, including nitrogen starvation, in which the abundance of FA biosynthetic enzymes were either unchanged or decreased, while complex lipid synthetic enzymes were induced (Miller et al., 2010; Alboresi et al., 2016).
Figure 5.
Changes in transcript (squares) and protein (circles) abundance relative to controls for enzymes in the TAG biosynthesis and degradation pathways after treatment with lipid-inducing compounds (see also Supplemental Datasets S1, S2, and S6). AGPAT, 1-acylglycerol-3-phosphate-o-acyltransferase; DAG, diacylglycerol; PLSB,Glycerol-3-phosphate acyltransferase, chloroplastic; LCS, long chain acyl-CoA synthetase; MAG, Monoacylglycerol; PDAT, phospholipid:diacylglycerol acyltransferase.
Several TAG lipases that degrade storage lipids had lower abundance in compound treatment conditions, which may have also contributed to TAG accumulation. These included LIP1 and two other lipases encoded by transcripts Cre17.g698600.t1.1 and Cre07.g334150.t1.1. The transcripts of each were greatly reduced under either treatment condition whereas other lipases including phospholipase A2 (Cre08.g381250.t1.2) and ELT3 (Cre03.g146287.t1.1) were induced by both the treatments.
Amino Acid and Nitrogen Metabolism
A notable impact of treatment with the selected lipid-inducing compounds profiled by our group previously and herein by untargeted metabolomics was an increase in the abundance of intermediates and products of amino acid biosynthetic pathways (Wase et al., 2017; Supplemental Data S4). Arg was significantly increased by both compounds. l-His accumulated 8-fold with compound WD30030 and 4-fold with WD10784 treatments. Levels of aliphatic amino acids l-Leu and l-Val were both significantly increased as a result of compound treatments. Of the two sulfur-containing amino acids, l-cystine and l-Met, only Met levels were significantly increased. Of the aromatic amino acids, l-Tyr, l-Phe, and l-Trp levels were significantly elevated by both compounds. By contrast, nitrogen starvation results in a reduction in Glu, Gln, Asp, Put, Gly, and Ala (Wase et al., 2014; Park et al., 2015).
Nineteen proteins categorized into amino acid metabolism pathways were identified in these analyses; of these, six were significantly different in response to either compound treatment and three by WD10784 alone (Supplemental Dataset S7). Among these proteins, isopropyl malate isomerase (LEU1L; Cre01.g004500.t1.2), required for Leu biosynthesis, was increased in abundance by 3-fold in response to WD10784 treatment. Recently, it was suggested that Leu may be involved in the regulation of lipid metabolism in Yarrowia lipolytica because supplementation of culture medium with Leu resulted in higher lipid accumulation (Blazeck et al., 2014). It was hypothesized that elevated Leu levels repress amino acid biosynthesis and redirect carbon flux toward lipid metabolism (Kerkhoven et al., 2016). Our observations do not support this hypothesis, as amino acid biosynthesis was largely potentiated by the lipid-inducing compound treatments and there was no evidence that carbon surplus from amino acid and protein degradation served as a source of substrates for lipid synthesis and storage.
For the KEGG pathway termed “nitrogen metabolism,” only five proteins were identified while 62 transcripts were identified (Supplemental Dataset S7). Among the transcripts, 56 were differentially abundant. Among the proteins, Glu dehydrogenase and Glu synthetase were increased ∼2-fold by WD30030 treatment at the protein level, but there was no effect elicited by WD10784 treatment. Overall, WD10784 increased the abundance of most transcripts identified. WD30030 treatment decreased the abundance of the transcripts for several transporters, including the ammonium transporter AMT5, sodium transporter DUR5, and formate/nitrate transporter NAR1-2. In nitrogen starvation, there is a notable induction of transcripts and proteins involved in ammonia assimilation including periplasmic l-amino acid oxidase (LAO), ammonia transporters, and enzymes of the glutamine synthetase and glutamate synthase (GS/GOGAT) system (Schmollinger et al., 2014; Wase et al., 2014; Kerkhoven et al., 2016). In contrast, neither lipid-inducing compound treatment altered LAO1, LAO2, or LAO3 transcript levels, whereas NRT2.1 and NRT2.4 were repressed, and NRT2.5 was increased, especially in response to WD10784. Transcripts encoding enzymes of the GS/GOGAT system that are normally induced during nitrogen starvation were repressed by either compound treatment. These results are in direct opposition to those reported in nitrogen deprivation and further emphasize the unique opportunity to modify metabolism to enhance lipid production without starvation (Miller et al., 2010; Schmollinger et al., 2014; Wase et al., 2014).
Differential Impacts of the Compounds on Protein and RNA Levels for Components of Photosynthesis and Photorespiration
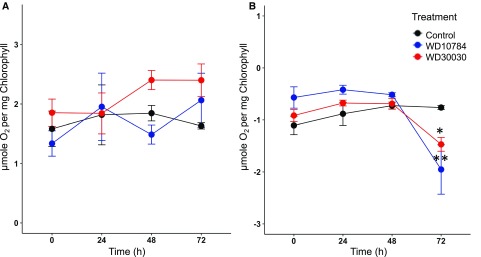
During nitrogen limitation, which is frequently used to induce lipid synthesis and storage, chlorosis of the culture is observed primarily due to downregulation of PSI and PSII proteins, the light-harvesting complexes (LHCs) cytochrome b6f and ATP synthase (Lee et al., 2012; Schmollinger et al., 2014; Wase et al., 2014; Park et al., 2015). Previously, it was reported that WD30030 treatment did not cause chlorosis and there was no reduction in Chl a or b, nor was there a reduction in total carotenoids (Wase et al., 2017). By comparison, there was a 50% reduction in pigment levels when cells were treated with WD10784 after 72 h, which led to concerns that photosynthesis would be reduced over time with this compound. These findings were extended to demonstrate that the maximal photosynthetic rates were similar between control and cells treated with either compound up to 72 h (Fig. 6A). The relative quantum yield per milligram of CHL (i.e. the initial slope of the light saturation curves) did not vary markedly between the different treatment conditions. Similarly, the respiratory rates of control and compound-treated cells were not statistically different up to 48 h, after which they started to decline (Fig. 6B). These data substantiate the modest impact of the compound treatments on growth and cellular function.
Figure 6.
Photosynthetic and respiratory rates during compound treatments. A, Photosynthetic rates of compound-treated cells measured at 24-h intervals for 72 h using a Clark oxygen electrode. Rates are expressed as O2 evolved per milligram of Chl (n = 3 biological replicates; ±sd). B, Respiratory rates were similarly determined by measuring oxygen consumption. Two way-ANOVA with Bonferroni posttest was applied to determine changes in experimental group over time (n = 3 ± sd). Statistically significant differences are indicated by *P ≤ 0.05, **P ≤ 0.01.
Assessment of both gene and protein expression profiles provided further insight into the impact of the compounds on the photosynthetic proteins and genes. In previous studies, proteins of PSI and PSII were found to be significantly reduced during nitrogen starvation (Longworth et al., 2012; Wase et al., 2014). In contrast, protein levels were relatively stable during WD30030 or WD10784 treatments; only PSI reaction center subunit IX (NP_958417.1) was significantly reduced by both the treatment conditions at 72 h (Supplemental Dataset S8). A differential response was observed after compound treatment for the subunits of the LHCs. LHCA2 was increased 4-fold by WD30030 (P < 0.05), whereas the abundance of this protein was not affected by WD10784 (Table 1). Several LHC proteins such as LHCA4, LHCBM2, LHCBM3, LHCBM5, and LHCM6 were decreased after WD30030 treatment, but remained unchanged by WD10784. The subunits of the oxygen-evolving complex that are generally reduced during nitrogen starvation retained their abundance (Supplemental Dataset S8). Although not identified in RNA-Seq experiments, 16 proteins encoded by the plastid genome were identified by label-free proteomics (Supplemental Dataset S8). Thirteen remained unaffected by either compound treatment. PsbH increased in response to WD30030; PetB encoding Cytochrome b6f was reduced ∼50% by WD10784; and both compounds had reduced PSI reaction center subunit IX.
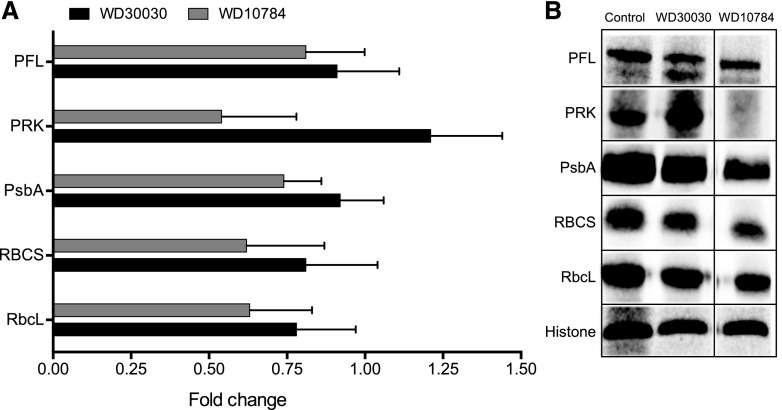
To verify the impact of the compound treatments on proteins involved in photosynthesis, western blot analysis was performed on six selected proteins (Fig. 7). WD30030 treatment did not affect protein abundance of the large and small subunits of Rubisco, PFL, phosphoribulose kinase (PRK), PsbA, or histone, which was included as a loading control. In contrast, WD10784 decreased the abundance of PRK. These results verify findings from untargeted, global proteomic assessment that showed only WD10784 had a significantly adverse effect on the photosynthetic machinery. WD30030 had no impact on proteins linked to the antenna proteins and photosynthesis, whereas WD10784 reduced the abundance of proteins in both categories. Reduction in the antenna apparatus has been suggested as one way to limit excess excitation energy and increase algal biomass (Nama et al., 2015; de Mooij et al., 2016; Girolomoni et al., 2017).
Figure 7.
Verification of the impact of compound treatment on selected proteins identified in label-free proteomic analysis using western blotting. A, Data from label-free proteomic analysis expressed as fold-change compared with control samples. B, Expression of the selected proteins in cell extracts was visualized using targeted antibodies. (Note: All lanes are from a single gel, but column 3 was two lanes distal from lane 2.)
At the transcript level, most genes encoding proteins of Rubisco, PSI, PSII, and the LHCs were significantly reduced by both compounds and, in keeping with the more severe impact on growth, treatment with WD10784 was more repressive than WD30030 (Supplemental Dataset S8). Therefore, it is presumed that, over time (i.e. beyond 72 h), compound treatment would reduce the levels of these proteins, most likely eventually leading to a reduction in photosynthetic capacity. Importantly, these results point to significant differences in the metabolic status of the algal cells between nutrient deprivation conditions compared with treatment with our lipid-inducing compounds, which supports both growth and lipid accumulation.
Protein Abundance of OxPhos and Respiratory Pathway Components Are Largely Unaltered by Compound Treatment
In previous proteomics analyses of nitrogen-starved cells, many proteins and enzymes of oxidative phosphorylation were significantly increased in abundance, suggesting protein expression was induced to compensate for energy deficiencies (Wase et al., 2014). In this study, among a large number of proteins (41) involved in OxPhos and respiration identified and quantified, only three proteins were significantly altered by WD10784 treatment and none were altered by WD30030 treatment (Supplemental Dataset S9). Of the four, in complex I, one protein (NUOB12) was significantly increased (P < 0.05) by WD10784 at both the protein and transcript levels. In complex II, SDH2 succinate dehydrogenase [ubiquinone] iron–sulfur subunit (Cre06.g264200.t1.2) was significantly reduced in abundance. In addition to the proton pumping complex I, Chlamydomonas contains several type-II NAD(P)H dehydrogenases. These enzymes are located on the mitochondrial inner membrane surface and facilitate electron transport from NAD(P)H to ubiquinone. Two proteins were profiled, of which only one (Cre16.g691552.t1.1; NDA1) was increased 7-fold by compound WD10784 treatment.
RNA-Seq data reflected the minimal impact on the expression of genes encoding proteins involved in these pathways (Supplemental Dataset S9). The abundance of very few transcripts was significantly different for proteins encoding complex I proteins. For other OxPhos complexes, a number of transcripts were increased in abundance, particularly after treatment with WD10784. This is more similar to the changes in transcript abundance after nitrogen deprivation (Miller et al., 2010). Transcript analysis showed that some components of complex IV were increased including COX16, COX17, and mitochondrion-associated cytochrome c oxidase assembly factor, especially after WD10784 treatment. Five proteins in this complex were identified, but none were significantly different from controls.
Evaluation of Cellular Redox Status
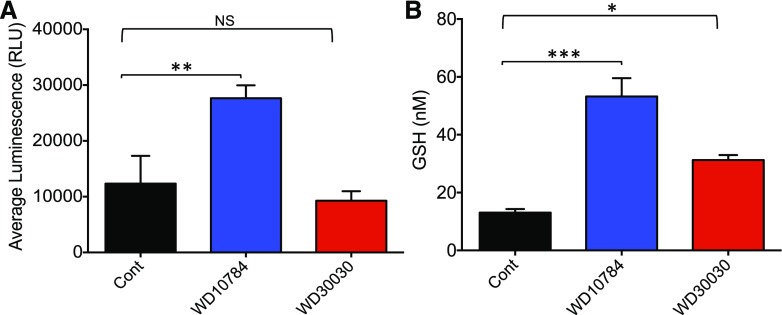
Reactive oxygen species (ROS), which are by-products of aerobic metabolism, may act as signaling molecules or indicators of stress in various algal, plant, and animal systems (Szivák et al., 2009; Menon et al., 2013). GSH is an efficient scavenger of ROS, and levels of reduced GSH rise in response to various culture and stress conditions to compensate for elevated ROS. Compound WD10784 increased both ROS and GSH levels significantly (2- and 4-fold, respectively), whereas WD30030 increased only GSH (2-fold; Fig. 8). Cells mitigate increased oxidative stress by inducing the expression of scavenging enzymes such as superoxide dismutase, catalase, and glutathione reductase (GR). Proteomics and transcriptomics analyses identified 22 proteins and 34 transcripts in the redox homeostasis pathways after treatment with either compound (Supplemental Dataset S10). Overall, these data further reflect the stronger increase in redox stress response by treatment with WD10784 as compared with WD30030. However, both compounds induced a manganese superoxide dismutase (MSD1), encoded by Cre02.g096150.t1.2 (increased 2- to 2.5-fold by both compounds at the protein level and 3-fold at the RNA level). Likewise, a glutathione reductase (GSHR2; Cre09.g396252.t1.1) was increased 4- and 11-fold, respectively, for WD30030 and WD10784 at the protein level. However, this enzyme was only modestly increased at the RNA level (2-fold each). Catalase (CAT1; Cre09.g417150.t1.2) protein levels were not affected by either compound, and RNA levels were modestly reduced.
Figure 8.
Evaluation of redox stress after compound treatment. A, Levels of ROS estimated using an R-Glo H2O2 Assay Kit (Promega). B, Levels of reduced GSH assessed using the GSH-Glo Assay. Bar heights indicate the mean of three biological replicates, and error bars indicate the sd. Significance was determined by Student’s t test comparing the compound-treated samples with control samples within the same experiment. Statistical significance is indicated by NS, no significance; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. RLU, relative luminescence unit.
Impacts on Ribosomal Protein Components
Reduced expression of ribosomes under stress conditions including nitrogen starvation was first reported in early work using ultrastructure studies (Martin et al., 1976). This was more recently confirmed using quantitative proteomics and transcriptomics (Martin et al., 1976; Schmollinger et al., 2014; Wase et al., 2014). In this study, 84 cytoplasmic, plastidic, and mitochondrial ribosomal proteins were identified and quantified (Table 2; Supplemental Dataset S11). Few cytosolic ribosomal proteins were significantly downregulated in the cells treated with either compound. Of these, the average reduction was 35% to 40% with WD30030 and 55% to 60% for WD10784 as compared with untreated control cells. Interestingly, ribosomal protein L29 (Cre03.g207050.t1.2; RPL29) was significantly increased in cells treated with compound WD10784. This protein is suggested to be necessary for the stable assembly of the large subunit of chloroplast ribosome (Myers et al., 1984).
Mitochondrial and plastidic ribosomal components were more profoundly impacted by the compound treatments (Supplemental Dataset S11). Most mitochondrial ribosomal protein transcripts were increased by either compound treatment and, of two proteins identified, neither were significantly changed in abundance. Among the 17 plastidic ribosomal proteins identified and compared to untreated controls, 10 were significantly reduced by WD30030 treatment and 11 by WD10784 treatment. The most severely affected were PRPL11, PRPL13, and PRPL21 (70% to 85% reduced). It was noted that WD10784 reduced plastidic ribosomal proteins and transcripts, in most cases, more severely than WD30030, which further supports the contention that this compound induces more metabolic dysfunction than WD30030 at the same concentration and over the same culturing period.
Compound Treatment has Limited Impacts on Heat Shock Response Factors and Chaperones
Chlamydomonas is generally grown at 25°C to optimize growth rate. Elevated temperatures (35°C to 41°C) result in the induction of heat shock proteins (HSPs) and chaperonins that assist protein folding and assembly (Kobayashi et al., 2014; Maikova et al., 2016). Other stresses may induce the same proteins and similar responses (Schroda et al., 1999; Schulz-Raffelt et al., 2007; Maikova et al., 2016). The accumulation of heat shock proteins (HSPs) correlates with heat shock factor (HSF) hyperphosphorylation (Schulz-Raffelt et al., 2007). Treatment of cells with either WD30030 or WD10784 resulted in different impacts on HSP family members (Supplemental Dataset S12). HSF1 was only detected at the transcript level and was increased 1.7-fold by WD10784 but was not affected by WD30030. Six small HSFs of the HSP22 family were identified. Of these, five were significantly increased in expression by WD10784, whereas all were decreased by WD30030 treatment. The HSP60 family members are type I chaperonins localized to plastid and mitochondria (Bertsch et al., 1992). Of 19 transcripts and 11 proteins identified, none were significantly altered by WD30030. After WD10784 treatment, the transcripts of only three GroEL-like proteins were induced and no proteins levels were significantly increased. Similar results were found for the HSP70s. These results indicate neither compound significantly induced the heat shock stress response.
Effects of Lipid-Accumulating Compounds on Factors Involved in ER Stress, Autophagy, and Vesicle-Mediated Lipid Transport
Recent evidence has indicated that LDs are synthesized along an ER pathway involving vesicular trafficking and are catabolized via autophagic processes (Stordeur et al., 2014; Welte and Gould, 2017; Garcia et al., 2018). Autophagy of LDs, sometimes called “lipophagy,” plays an important role in lipid and protein homeostasis critical to energy control pathways. Therefore, we examined these pathways to provide insight into LD assembly and utilization after compound treatment.
To test whether the lipid-accumulating compounds induce ER stress in Chlamydomonas, we examined the transcript and protein expression profile of selected ER stress markers. For example, a previous study showed that Brefeldin A, a compound that induces lipid accumulation in algae, induces an ER stress response as indicated by increased expression of classical ER stress marker genes including Binding Protein1(BIP1) and BIP2 and Sar-type small GTPase1 (SAR1; Kim et al., 2013). WD10784 induced expression of BIP2 protein 2.3-fold but had no effect on Bip1 or SAR1. Twenty transcripts were identified and categorized as part of the autophagy pathway. Two, APG18 and APG20, were induced by both compound treatments 2- to 3-fold. Another, DIN1, was decreased 80% to 95% by both. Among autophagy proteins, only DIN6 was detected, and its abundance was not significantly altered by either compound. Therefore, it does not appear that either compound induces lipophagy.
The ARF1/COPI protein machinery, known for its role in vesicle trafficking, is also implicated in the synthesis, maintenance, and catabolism of LDs through a mechanism that is poorly understood (Beller et al., 2010). Only one protein was identified as a component of this pathway, VPS26, but its abundance was not affected by either treatment. Twenty-three transcripts were identified that were categorized to the ARF/COPII system (Supplemental Dataset S13). While both compounds induced many of these transcripts, the strongest induction was by WD10784 treatment. Transcripts increased in abundance included ADP-ribosylation factor (ARF family), ARF-like small GTPase (ARL1), coatomer subunits (COPII α, β, γ, and ε), vacuolar sorting proteins of the VPS family, SEC31, two syntaxins, and a Septin 9 homolog. The increase in components of this pathway is consistent with a physiological state induced by the compounds that favor complex lipid and protein synthesis involving the ER and secretory apparatus necessary to produce LDs as storage vesicles.
Further Comparison of Proteomic and Transcriptomic Results after Compound Treatment with Nitrogen Deprivation Using Published Data
To further compare and contrast alterations in the proteomic and transcriptomic profiles of Chlamydomonas after treatment with either compound and the nitrogen starvation response, we compared results from this study with assessment of protein levels after nitrogen starvation previously reported by this lab (Wase et al., 2014) and transcript levels from the work of Miller et al. (2010). In making these comparisons, it must be kept in mind that there are significant differences in experimental conditions for each study, including for example, length of treatment (72-h compound treatment and 48-h nitrogen starvation), as well as different controls employed (addition of vehicle in the case of the compounds versus nitrogen-replete media for the starvation experiments) and different analytical methods. For this illustrative assessment, the raw transcript data available in published archives was employed in the comparative analyses. For comparison of protein expression, the results of the analyses were used after processing because the raw data files were acquired using different analytical methods. The results of the proteomics comparisons can be found in Supplemental Dataset S14 and the transcriptomics data comparisons in Supplemental Dataset S15. The methods of analyses are detailed in Supplemental Methods S1.
For treatment with WD30030 or WD10784, 747 proteins were identified and quantified with at least two peptides, whereas in the nitrogen starvation study of Wase et al. (2014), 794 proteins were identified and quantified, of which 572 were identified with two peptides. Only proteins that were common to both studies were considered for comparison here. For WD30030 treatment, 45 proteins were increased in abundance; 55 proteins were increased by WD10784; and 18 were increased 48 h after nitrogen starvation. A Venn diagram illustrating these comparative results can be found in Supplemental Figure S6. A list of specific proteins that have significantly altered abundance after at least one of the three treatments is given in Table 3. Only five proteins with increased abundance were common to all treatments, whereas 33 were shared between WD30030 and WD10784. For proteins with significantly decreased abundance, 21 were detected after WD30030 treatment, 44 with WD10784, and 31 after nitrogen starvation. Of these, 17 were common to both compound treatments and only two to the compounds and nitrogen starvation. Among the unique expression patterns distinguishing compound treatments from nitrogen limitations is RPE1 of the oxidative pentose phosphate pathway (OPPP) that was reduced by either compound treatment but not significantly different after nitrogen starvation. As mentioned above, CIS1 was increased in all treatments, possibly contributing acetyl-CoA for FA synthesis. Of three photosynthetic proteins, all were significantly reduced by nitrogen starvation but none were significantly changed after treatment with either lipid-inducing compound. These data, while limited, further support the unique impacts of WD30030 and WD10784 compared with nitrogen starvation response.
Table 3. Comparisons of the abundance of selected proteins for compound-treated cells versus nitrogen starvation based on this work (Supplemental Dataset S14) and Wase et al. (2014).
Significant fold change and P values are indicated in bold face. NS, not significant.
| Protein | Name | Gene Symbol | Fold Change | P Values | ||||
|---|---|---|---|---|---|---|---|---|
| WD30030 | WD10784 | Nitrogen, 48 h | WD30030 | WD10784 | Nitrogen, 48 h | |||
| Carbon metabolism | ||||||||
| Cre06.g264200.t1.2 | Succinate dehydrogenase 2-2 | SDH2 | 0.93 | 0.38 | 1.36 | 0.822 | 0.034 | 0.043 |
| Cre02.g080200.t1.2 | Transketolase | TRK1 | 0.58 | 0.25 | 0.61 | 0.049 | 0.002 | 0.000 |
| Cre12.g510650.t1.2 | Fru-1,6-bisphosphatase I | FBP1 | 1.90 | 1.58 | 0.59 | 0.036 | 0.091 | 0.067 |
| Cre12.g511900.t1.2 | d-Ribulose-5-P-3-epimerase | RPE1 | 0.24 | 0.19 | 0.97 | 0.003 | 0.001 | 0.890 |
| Cre12.g554800.t1.2 | Phosphoribulokinase | PRK1 | 1.21 | 0.54 | 0.63 | 0.491 | 0.072 | 0.031 |
| Cre12.g526800.t1.2 | 6-Phosphogluconate dehydrogenase family protein | 6PGD | 1.73 | 2.64 | 1.63 | 0.045 | 0.011 | 0.014 |
| Cre03.g149100.t1.2 | Citrate synthase 2 | CIS2 | 1.32 | 1.06 | 0.46 | 0.088 | 0.674 | 0.000 |
| Cre12.g514750.t1.2 | Citrate synthase family protein 1 | CIS1 | 1.54 | 2.16 | 1.56 | 0.044 | 0.007 | 0.006 |
| Cre06.g272050.t1.2 | Phosphoglycerate mutase, 2,3-bisphosphoglycerate-independent | PGM1a| PGM1b | 1.19 | 1.55 | 1.62 | 0.621 | 0.246 | 0.001 |
| Cre12.g485150.t1.2 | Glyceraldehyde-3-P dehydrogenase of plastid 1 | GAP1a| GAP1b | 10.18 | 14.15 | 4.03 | 0.002 | 0.001 | 0.000 |
| Cre12.g513200.t1.2 | Enolase | PGH1 | 1.16 | 0.99 | 1.50 | 0.624 | 0.977 | 0.000 |
| Fatty acid and lipid metabolism | ||||||||
| Cre13.g577100.t1.2 | Acyl carrier protein | ACP2 | 0.69 | 0.63 | 1.23 | 0.165 | 0.102 | 0.323 |
| Cre08.g359350.t1.2 | Acetyl coenzyme, a carboxylase biotin carboxylase subunit | BCR1 | 0.27 | 0.13 | 0.93 | 0.006 | 0.001 | 0.732 |
| Cre12.g519100.t1.2 | Acetyl coenzyme, a carboxylase carboxyltransferase alpha-subunit | ACX1 | 0.54 | 0.20 | 1.01 | 0.368 | 0.046 | 0.937 |
| Cre01.g035350.t1.2 | Polyketide synthase, enoylreductase family protein | Not assigned | 3.46 | 5.70 | 1.18 | 0.010 | 0.003 | 0.495 |
| Cre11.g476550.t1.2 | Polyketide synthase, enoylreductase family protein | Not assigned | 2.69 | 2.41 | 1.23 | 0.001 | 0.002 | NS |
| Cre01.g053000.t1.2 | NAD-dependent glycerol-3-P dehydrogenase family protein | Not assigned | 2.03 | 3.67 | 2.92 | 0.052 | 0.007 | 0.028 |
| Starch metabolism | ||||||||
| Cre13.g567950.t1.2 | Glc-1-P adenylyltransferase | STA1 | 0.65 | 0.44 | 0.67 | 0.104 | 0.016 | 0.294 |
| Cre03.g188250.t1.2 | Glc-1-P adenylyltransferase | STA6 | 0.72 | 0.36 | 0.55 | 0.287 | 0.020 | 0.117 |
| Cre03.g181500.t1.2 | Disproportionating enzyme | STA11 | 2.41 | 2.23 | 1.74 | 0.021 | 0.028 | 0.535 |
| Cre07.g336950.t1.1 | Alpha-glucan phosphorylase 2 | PHOA | 1.86 | 2.86 | 2.74 | 0.033 | 0.005 | 0.000 |
| Cre12.g552200.t1.2 | Alpha-glucan phosphorylase 2 | PHOB | 1.40 | 1.82 | 1.14 | 0.173 | 0.044 | 0.131 |
| Photosynthesis | ||||||||
| Cre12.g550850.t1.2 | PSII subunit P-1 | PSBP1 | 0.90 | 0.74 | 0.21 | 0.670 | 0.225 | 0.001 |
| Cre16.g673650.t1.1 | LHC of PSII 5 | IHCB5 | 0.98 | 0.77 | 0.19 | 0.907 | 0.179 | 0.000 |
| Cre17.g720250.t1.2 | LHC PSII | LHCB4 | 0.88 | 0.76 | 0.21 | 0.551 | 0.255 | 0.000 |
| Amino acid biosynthesis | ||||||||
| Cre06.g252650.t1.2 | Aconitase/3-isopropylmalate dehydratase protein | LEU1S | 2.23 | 3.42 | 0.96 | 0.024 | 0.006 | NS |
To assess similarities and differences in the effect of treatment with either chemical inducer and nitrogen limitation on gene expression, a total of 15,965 transcripts common to all treatment conditions were compared (Supplemental Dataset S15; Supplemental Fig. S7). These analyses demonstrated striking contrasts as well. Treatment with WD30030 and WD10784 resulted in 666 transcripts in common that were increased in abundance and 588 transcripts in common with reduced abundance. In contrast, few of these changes were shared with nitrogen limitation: 125 transcripts were increased in expression, whereas 48 were reduced under all three treatment conditions. KEGG enrichment analysis further pointed to unique transcriptomic profiles. Of the top-30 pathways identified as significantly altered during treatment, 16 were increased by WD30030 treatment, 20 by WD10784 treatment, and only five by nitrogen deprivation (Supplemental Dataset S15). Reduction of transcripts encoding pathway components of the categories “photosynthetic antenna proteins,” “photosynthesis,” “metabolic pathways,” and “DNA replication” were the top four altered in nitrogen deprivation. For either compound treatment, reduction in components of the pathway category “ribosome” ranked first, followed by the pathway categories “endocytosis,” “DNA replication,” and the “proteasome,” each of which increased. These differences in transcript abundance after treatment further support unique and extensive differences in impact from either compound treatment (WD10784 or WD30030) versus nitrogen deprivation.
DISCUSSION
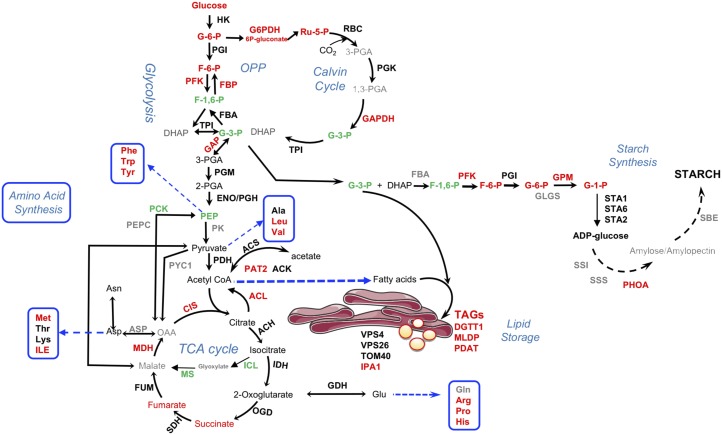
This study was conducted to define similarities and differences in the metabolic response of Chlamydomonas to WD30030 and WD10784 treatments that occur coincident with induction of lipid synthesis and storage. The small compound inducers can be compared with drugs that are administered to induce a specific phenotype, but that may have additional off-target effects that also contribute to their effectiveness and therapeutic application. Thus, the physiological, biochemical, and multiomics analyses reported here provide a comprehensive understanding of the effects of these two compounds on Chlamydomonas during growth. Further, we rationalized that convergence between metabolic pathways enhanced or repressed by both compounds would provide information on common mechanisms leading to TAG synthesis and accumulation. These data have also been compared with nutrient deprivation conditions, notably the well-characterized nitrogen starvation response, to determine similar or unique pathway effects. The most important differences between nutrient deprivation responses and the two compounds are the maintenance of growth and photosynthetic capacity over time, whereas growth stops within one cell doubling when an essential nutrient such as nitrogen is removed from the culture media (Miller et al., 2010; Lee et al., 2012; Wase et al., 2014; Park et al., 2015). The maintenance of growth and photosynthesis is most apparent for cells treated with WD30030, because after 48 h, cultures treated with WD10784 elicit signs of stress and chlorosis coincident with a reduction in expression of photosynthetic protein coding genes. An oversimplified interpretation of the response of microalgae to nutrient deprivation is that excess carbon pools are the direct result of scavenging cellular components (e.g. for nitrogen) to maintain essential cellular functions, thereby generating the need to sequester the excess carbon in storage compounds including starch and lipid. As shown previously and herein, the lipid-activating compounds do not induce the turnover of cellular proteins or membrane lipids associated with starvation stress response (Schmollinger et al., 2014; Wase et al., 2017). Instead, protein synthesis is maintained, amino acid biosynthesis is potentiated, and carbon flow is directed toward storage of only TAG (i.e. WD30030 treatment) or both starch and TAG (i.e. WD10784). The common pathway shifts induced by both WD10784 and WD30030 compared to controls provide insight into alterations in protein and metabolites that contribute to TAG production (Fig. 9), while differences, particularly those related to stress and starch synthesis induced by compound WD10784, help to eliminate nonproductive metabolic perturbations.
Figure 9.
Major changes in proteins and metabolites shared by both WD10784 and WD30030 treatments. The pathways are simplified to emphasize major routes in central carbon metabolism leading to triglyceride and starch storage. Red, increased abundance; green, decreased; black, no change in at least one condition; gray, not detected. For impacts of each compound on starch and triglyceride synthesis, see Figures 4 and 5, respectively. Enzyme abbreviations: ACH, aconitate hydratase, mitochondrial; ACK, acetate kinase; ACL, ATP-citrate lyase; CIS, citrate synthase; DGTT1, diacylglycerol acyltransferase type 2; FBA, fructose-1,6-bisphosphate aldolase; FBP, fructose 1,6-bisphosphatase; FUM, fumarate hydratase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GDH, glutamate dehydrogenase; GPD1, glycerol-3-phosphate dehydrogenase; GPM, phosphoglucomutase; HK, hexokinase; ICL, isocitrate lyase; IDH, isocitrate dehydrogenase; IPA1, importin subunit α; MDH: malate dehydrogenase; MLDP, major lipid droplet protein; MS, malate synthase; OGD, oxoglutarate dehydrogenase E2 component; PAT2, phosphotransacetylase; PCK, phosphoenolpyruvate carboxykinase; PDAT, phospholipid:diacylglycerol acyltransferase; PDH, pyruvate dehydrogenase; PEPC, phosphoenolpyruvate carboxylase; PFK, phosphofructokinase; PGI, phosphoglucose isomerase; PGM, phosphoglucomutase; PHOA, starch phosphorylase; PYC1, pyruvate carboxylase; SBE, starch branching enzyme; SDH, succinate dehydrogenase; SS1, soluble starch synthase; SSS, Starch synthase, chloroplastic/amyloplastic; STA1,2,6, ADP-glucose pyrophosphorylase; TOM40, 40 kDa translocon at mitochondrial outer envelope membrane; TPI, triosephosphate isomerase; VPS26, subunit of retromer complex.
Central Carbon Metabolism in Compound Treatment
In Chlamydomonas, the Calvin–Benson–Bassham cycle, OPPP, and glycolysis, overlap and share several reaction steps in common, but are differentially compartmentalized such that the reactions of glycolysis from Glc to G3P are localized in the plastid and the reactions from 3-PGA to pyruvate are localized in the cytosol (Johnson and Alric, 2013). The OPPP enzymes Glc-6-P dehydrogenase and 6-PG dehydrogenase are found within both compartments. Metabolite analysis indicated elevated levels of several key intermediates including Fru-6-P, ribulose-5-P, ribulose-1,6-bisphosphate, and xylulose-5-P for either compound treatments compared with controls. PFK2 (Cre12.g553250.t1.2) regulates the conversion of Fru-6-P to Fru-1,6-bisphosphate and when ATP concentrations are low, PFK2 is activated and enhances glycolytic flux and starch breakdown (Gibbs et al., 1986) to induce the Pasteur effect characterized by the activation of glycolysis in response to inhibition of mitochondrial ATP production (Rebeille and Gans, 1988; Gans and Rebeille, 1990). In previous work, Fru-6-P was found to be elevated 17-fold by WD30030 treatment and 360-fold by WD108784, whereas Fru-1,6-P levels were reduced 0.47- and 0.71-fold, respectively, most likely due to metabolic conversion to G3P (Wase et al., 2017). G3P can provide the backbone to glycerol lipids including storage TAG as catalyzed by glycerol-3-P acyltransferase (PLSB). In this studies, PFK2 levels were significantly increased at both the protein and RNA levels, as was glyceraldehyde-3-P dehydrogenase (GAPDH), which catalyzes the conversion of G3P to 3-PGA. GAPDH encoded within GAP1a and GAP2 was highly upregulated in cells treated with either compound. In Chlamydomonas, GAPDH forms a complex with PRK1. Although WD30030 had no significant effect on PRK1 abundance, the protein level was reduced ∼35% and RNA ∼93% by WD10784 treatment, which may ultimately affect 3-PGA levels available for the Calvin–Benson–Bassham cycle.
In the final reaction of glycolysis, pyruvate dehydrogenase catalyzes the formation of acetyl-CoA for use in the tricarboxylic acid cycle. The abundance of this protein was increased over time after nitrogen removal and continued for up to 144 h and, concomitantly, pyruvate levels were reduced (Wase et al., 2014). Neither WD30030 nor WD10784 affected pyruvate dehydrogenase protein or transcript abundance, supporting the unique impact of these compounds in preserving the integrity of central carbon metabolism.
The amphibolic tricarboxylic acid cycle is required both to produce energy as ATP and to supply critical intermediates for anabolic processes including amino acid, nucleotide, and sugar biosynthesis. Overall, there was little impact on the expression of protein components of the tricarboxylic acid cycle, suggesting regulatory impact of the compounds on this pathway most likely occur at the level of metabolite flux. At the metabolite level, the compounds elevate isocitrate, succinate, and fumarate levels, concomitant with an elevation of various amino acids that are synthesized in anabolic reactions utilizing tricarboxylic acid cycle intermediates (herein and Wase et al., 2017). This generally only occurs when there is sufficient ATP and substrates for the anabolic reactions. The increases in ATP-isocitrate lyase and CIS were notable in that these enzymes are critical for the formation of cytosolic acetyl-CoA for FA biosynthesis and are only active when ATP and citrate are abundant.
Starch biosynthesis was examined critically to help understand why WD10784 but not WD30030 increases this storage compound. The answer seems to lie in the relative impact of this compound on the precursor metabolites, as well as transcripts and proteins. G-6-P and F-6-P levels are 15- and 20-fold higher, respectively, when cells are treated with WD10784 relative to WD30030. Some key proteins are also elevated including STA2, PhoA, and PhoB. Likewise, transcripts for many starch-synthesizing enzymes accumulate to higher levels compared with controls. Thus, it appears the cumulative effects of these unique patterns of change induced by WD10784 result in elevated starch levels.
Catabolic Enzymes of Lipid Metabolism Are Maintained or Reduced in Expression Potentiating Lipid Accumulation
Reduction in lipid degradation by lipases and FA oxidation enzymes might also contribute to lipid accumulation and storage. Lipases and phospholipases hydrolyze the ester bond between the acyl-chain of TAGs or phospholipids, respectively, and the glycerol backbone, yielding a free FA and a corresponding lyso-lipid (Goold et al., 2015). The released acyl-chain may be further incorporated into glycerolipids or shuttled between the membrane lipids and TAG pool. Lipases are also suggested to maintain the equilibrium during continuous oil synthesis to prevent hyper-accumulation of oil and lipotoxicity in the cell. As noted above, the expression of at least three lipases was reduced in compound-treated cells compared with controls. In terms of β-oxidation enzymes, the expression of the acyl-CoA oxidase gene encoded by ACX1 was reduced by both compounds, whereas ACX3 protein was induced by both. Enoyl-CoA hydratase1, ECH1, protein expression was increased by WD30030 but not affected by compound WD10784. A major reduction was observed in the expression of two transcripts encoding peroxisomal 2,4-dienoyl-CoA reductase and 3-hydroxyisobutyryl-CoA hydrolase. Although mixed, overall these results suggest that reductions in lipolysis and/or FA oxidation are not major contributing factors to lipid accumulation in response to either compound.
LD Formation Is Potentiated
LD formation has been most extensively studied in eukaryotic systems where a model has emerged in which lipid storage molecules including TAGs are synthesized concurrently with modification of the ER membrane to include LD proteins (Chapman et al., 2019). As the lipid particle expands, it is released by a process of budding and scission. It has further been proposed that the ARF1/COPI machinery controls the formation of a membrane bridge between LDs and the ER to target proteins including DGAT, glycerol-3-P acyltransferase, and adipose triglyceride lipase to the maturing LD (Thiam et al., 2013). Additionally, the ARF1/COPI proteins may directly act on the LDs to remove phospholipids from the LD surface by forming nano-sized LDs. This is suggested to result in an increase in the surface tension of the LDs and helps in building a bridge between the LDs and the ER to contribute to LD formation (Wilfling et al., 2014). Both compounds induced the transcript levels of the ADP-ribosylation factor, the coatomer subunits COPII α, β, γ, and ε, and the vacuolar sorting proteins of the VPS family and SEC31. Recently, a number of Chlamydomonas proteins have been proposed as homologs of yeast and mammalian proteins involved in LD dynamics (Huang et al., 2019). Five of these that are proposed to regulate droplet fusion, lipid accumulation, and maturation were increased at the transcript level by either WD10784 or WD30030. These include two syntaxins (Cre16.g692050 and Cre17.g711450), one Rab GTPase (Cre15.g641800), a Septin 9-like protein (Cre12.g556250), and, as mentioned, Arf1 (Cre10.g448200). Together with the induction of TAG biosynthetic genes and proteins including MLDP and DGTT1, these results support the role for WD30030 and WD10784 in inducing LD biosynthesis and accumulation.
Compound Treatment Maintains Photosynthesis and Respiration
An essential distinction between WD30030 and WD10784 treatment to induce lipid synthesis and nutrient deprivation conditions is the maintenance of photosynthetic capacity by WD30030 relative to WD10784. This was measured directly by assessing photosynthetic rates and indirectly using proteomic and transcriptomic analyses. Many proteins of PSI, PSII, and the LHCs were identified and compared using label-free proteomics. These data demonstrated that the protein subunits of the complexes are largely maintained during compound treatment. However, at the level of RNA, most transcripts were reduced in abundance after 72 h of compound treatment, with repression of gene expression in response to WD10784 being more severe compared with WD30030 treatment. Enzymes of the Calvin–Benson–Bassham cycle, especially Rubisco, are generally severely reduced in nutrient starvation and other stresses (Longworth et al., 2012; Schmollinger et al., 2014; Wase et al., 2014; Park et al., 2015), whereas expression of triosephosphate isomerase and FBA3 are increased (Schmollinger et al., 2014). The Rubisco small and large subunit proteins are maintained during either compound treatment. In contrast, the Calvin–Benson–Bassham enzymes RPE1 and TRK1 both were significantly downregulated in both compound treatment conditions. SEBP1 was significantly downregulated at the protein level only during treatment with compound WD10784 (0.29-fold).
The maintenance of respiratory complex proteins coordinated with photosynthetic proteins during compound treatment, in general, suggests cellular energy production is maintained. This is in direct contrast to our previous findings assessing the proteome of cells during nitrogen deprivation that demonstrated proteins involved in OxPhos were significantly elevated, whereas levels of proteins of the photosynthetic and LHCs were reduced (Wase et al., 2017). The results in nitrogen deprivation conditions were interpreted to indicate that cellular metabolism was in energy deficit. The results for treatment with WD30030 and WD10784, in this article, are in direct contrast.
CONCLUSION
Although a specific single target of either WD30030 or WD10784 cannot be identified from these studies, the multiomics analyses of Chlamydomonas cells grown in the presence of these lipid-inducing small molecules strongly support the contention that microalgae can be induced to produce and store triglycerides in LDs without terminating growth, or stimulating a severe stress response, or causing destruction of thylakoid membranes and proteins. By analogy to drugs aimed at a therapeutic endpoint (e.g. reduction in obesity), the importance of these results lies in the broad understanding they provide in the shifts in algal physiology, biochemical pathways, and metabolites that result from compound treatment. Metabolite analysis reported herein and previously (Wase et al., 2017) further demonstrated that these small molecules could be used to produce high levels of Calvin–Benson–Bassham, glycolytic, and tricarboxylic acid cycle intermediates, as well as amino acids and antioxidant compounds to drive both growth and carbon storage. Therefore, these compounds offer the opportunity to delve further into the metabolic shifts required to maximize lipid production as required if microalgae are to be employed as feedstock for biofuels or other high-value products. Additionally, these results provide a database of information that may be used to identify additional pathways and products of interest to the research and commercial communities. WD30030 in particular is being further examined for industrial applications.
MATERIALS AND METHODS
Cell Culture and Compound Treatment
The Chlamydomonas reinhardtii strain CC-125 was used for all studies. Cells were routinely maintained on TAP solid media and were used to inoculate a liquid starter culture. The cells were routinely cultured in liquid TAP media at 25°C under constant illumination with cool-white fluorescent bulbs with a photon flux density of 50 μmol photons m−2 s−1 and continuous shaking (120 rpm). For compound treatment, cells from midlog phase culture were harvested, washed once with fresh sterile TAP media, and inoculated at starting density of 5 × 105 cells/mL. The test compounds WD30030 and WD10784 were then added to a final concentration of 5 μm unless otherwise stated and controls received the vehicle dimethyl sulfoxide. Cell cultures (100 mL) were grown in 250-mL Erlenmeyer flasks with rubber stoppers adapted to facilitate gas exchange in an Innova 43 Orbital Shaking Growth Chamber (New Brunswick) under continuous lighting. Cell numbers were quantified using a model no. TC-10 Automated Cell Counter (Bio-Rad). Harvesting was performed by centrifugation. Three independent biological replicates were employed for RNA, protein, or metabolite extraction. For all experiments, control cultures contained dimethyl sulfoxide instead of compound. For comparison, the protein, RNA, and metabolite data values are presented relative to data obtained for the controls.
Statistical Analysis
Significant differences among treatment groups and controls for assessment of growth, photosynthetic and respiration rates measurements, and GSH and H2O2 assays were determined using either the Student’s t test or one-way ANOVA followed by Tukey’s posthoc test. Comparison of groups over time was carried out with mixed-model, two-way ANOVA with Bonferroni posttest.
Additional Methods
Additional detailed methods related to assessment of growth, photosynthetic, and respiration rates measurements, GSH and H2O2 assay, proteome analysis, mass spectrometry (MS) analysis of the peptides, data processing of the proteomics dataset, western analysis of targeted proteins, transcriptome analysis, gene expression by RT-qPCR, and cellular metabolite analysis by ultra performance liquid chromatography (UPLC)-MS are available in Supplemental Methods S1.
Accession Numbers
All original RNA-Seq data are deposited in the National Center for Biotechnology Information BioProject database archive under accession number PRJNA324451 (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA324451). Raw mass spectrometry data for label-free quantitative proteome profiling is deposited in the Pride ProteomeXchange data repository under accession number PXD006736 (http://www.ebi.ac.uk/pride/archive/projects/PXD006736). Raw mass spectrometry data for untargeted LC-MS metabolomics analysis are archived in the Metabolomics Workbench database (http://metabolomicsworkbench.org/) under the accession number ST000621.
Supplemental Data
The following supplemental materials are available.
Supplemental Methods. Detailed methods of data acquisition and analysis.
Supplemental Figure S1. Differential expression analysis of proteomics datasets.
Supplemental Figure S2. Overview of the transcriptome dataset analysis of RNA-Seq data.
Supplemental Figure S3. KEGG enrichment analysis of differentially expressed transcripts.
Supplemental Figure S4. Correlations between data obtained using RNA-Seq and RT-qPCR for selected genes after each treatment.
Supplemental Figure S5. Assessment of metabolites that are significantly increased or decreased in cells treated with either compound.
Supplemental Figure S6. Comparisons between protein abundance from this compound treatment study and nitrogen starvation from Wase et al. (2014) presented in Venn diagrams.
Supplemental Figure S7. Overview of the transcriptome dataset analyses of RNA-Seq data for comparison of treatment with the two compounds (this study) and nitrogen starvation from Miller et al. (2010).
Supplemental Dataset S1. Quantitative proteomics data and KEGG pathway enrichment.
Supplemental Dataset S2. Transcriptomics data from RNA-Seq and KEGG pathway analysis.
Supplemental Dataset S3. List of primers and comparison of expression of selected target genes using RT-qPCR, RNA-Seq, and quantitative proteomics.
Supplemental Dataset S4. Untargeted LC-MS metabolomics data.
Supplemental Dataset S5. DEGs and proteins related to central carbon metabolism (acetate, glycolysis, Calvin–Benson–Bassham cycle, tricarboxylic acid cycle, and starch metabolism).
Supplemental Dataset S6. DEGs and proteins related to FA and lipid metabolism.
Supplemental Dataset S7. DEGs and proteins related to nitrogen and amino acid metabolism
Supplemental Dataset S8. DEGs and proteins of photosynthesis.
Supplemental Dataset S9. DEGs and proteins of respiration and oxidative photophosphorylation.
Supplemental Dataset S10. DEGs and proteins related to redox homeostasis.
Supplemental Dataset S11. DEGs and proteins related to ribosomes.
Supplemental Dataset S12. DEGs and proteins related to heat shock stress and autophagy.
Supplemental Dataset S13. DEGs and proteins related to protein transport and vesicular trafficking.
Supplemental Dataset S14. Differential protein expression patterns for compound-treated cells or N-starved cells.
Supplemental Dataset S15. Differential transcript expression patterns for compound-treated cells or N-starved cells.
ACKNOWLEDGMENTS
Thanks are due to Dr. Istvan (Steve) Ladunga and Dr. Wayne Reikhof for critically reviewing the article.
Footnotes
This work was supported by the National Science Foundation (EPSCoR EPS-1004094, EPS-1264409, and CBET-1402896) and the Nebraska Center for Energy Science Research.
Articles can be viewed without a subscription.
References
- Ajjawi I, Verruto J, Aqui M, Soriaga LB, Coppersmith J, Kwok K, Peach L, Orchard E, Kalb R, Xu W, et al. (2017) Lipid production in Nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator. Nat Biotechnol 35: 647–652 [DOI] [PubMed] [Google Scholar]
- Alboresi A, Perin G, Vitulo N, Diretto G, Block M, Jouhet J, Meneghesso A, Valle G, Giuliano G, Maréchal E, et al. (2016) Light remodels lipid biosynthesis in Nannochloropsis gaditana by modulating carbon partitioning between organelles. Plant Physiol 171: 2468–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller M, Thiel K, Thul PJ, Jäckle H (2010) Lipid droplets: A dynamic organelle moves into focus. FEBS Lett 584: 2176–2182 [DOI] [PubMed] [Google Scholar]
- Bertsch U, Soll J, Seetharam R, Viitanen PV (1992) Identification, characterization, and DNA sequence of a functional “double” groES-like chaperonin from chloroplasts of higher plants. Proc Natl Acad Sci USA 89: 8696–8700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby IK, Glaesener AG, Mettler T, Fitz-Gibbon ST, Gallaher SD, Liu B, Boyle NR, Kropat J, Stitt M, Johnson S, et al. (2013) Systems-level analysis of nitrogen starvation-induced modifications of carbon metabolism in a Chlamydomonas reinhardtii starchless mutant. Plant Cell 25: 4305–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazeck J, Hill A, Liu L, Knight R, Miller J, Pan A, Otoupal P, Alper HS (2014) Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat Commun 5: 3131. [DOI] [PubMed] [Google Scholar]
- Botté CY, Deligny M, Roccia A, Bonneau AL, Saïdani N, Hardré H, Aci S, Yamaryo-Botté Y, Jouhet J, Dubots E, et al. (2011) Chemical inhibitors of monogalactosyldiacylglycerol synthases in Arabidopsis thaliana. Nat Chem Biol 7: 834–842 [DOI] [PubMed] [Google Scholar]
- Boyle NR, Page MD, Liu B, Blaby IK, Casero D, Kropat J, Cokus SJ, Hong-Hermesdorf A, Shaw J, Karpowicz SJ, et al. (2012) Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J Biol Chem 287: 15811–15825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KD, Aziz M, Dyer JM, Mullen RT (2019) Mechanisms of lipid droplet biogenesis. Biochem J 476: 1929–1942 [DOI] [PubMed] [Google Scholar]
- Chevalier F, Carrera LC, Nussaume L, Maréchal E (2016) Chemical genetics in dissecting membrane glycerolipid functions. Subcell Biochem 86: 159–175 [DOI] [PubMed] [Google Scholar]
- Choi YE, Rhee JK, Kim HS, Ahn JW, Hwang H, Yang JW (2015) Chemical genetics approach reveals importance of cAMP and MAP kinase signaling to lipid and carotenoid biosynthesis in microalgae. J Microbiol Biotechnol 25: 637–647 [DOI] [PubMed] [Google Scholar]
- de Mooij T, Schediwy K, Wijffels RH, Janssen M (2016) Modeling the competition between antenna size mutant and wild type microalgae in outdoor mass culture. J Biotechnol 240: 1–13 [DOI] [PubMed] [Google Scholar]
- Franz AK, Danielewicz MA, Wong DM, Anderson LA, Boothe JR (2013) Phenotypic screening with oleaginous microalgae reveals modulators of lipid productivity. ACS Chem Biol 8: 1053–1062 [DOI] [PubMed] [Google Scholar]
- Gans P, Rebeille F (1990) Control in the dark of the plastoquinone redox state by mitochondrial activity in Chlamydomonas reinhardtii. Biochim Biophys Acta 1015: 150–155 [Google Scholar]
- Garcia EJ, Vevea JD, Pon LA (2018) Lipid droplet autophagy during energy mobilization, lipid homeostasis and protein quality control. Front Biosci 23: 1552–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargouri M, Bates PD, Park JJ, Kirchhoff H, Gang DR (2017) Functional photosystem I maintains proper energy balance during nitrogen depletion in Chlamydomonas reinhardtii, promoting triacylglycerol accumulation. Biotechnol Biofuels 10: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs M, Gfeller RP, Chen C (1986) Fermentative metabolism of Chlamydomonas reinhardtii: III. Photoassimilation of acetate. Plant Physiol 82: 160–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girolomoni L, Ferrante P, Berteotti S, Giuliano G, Bassi R, Ballottari M (2017) The function of LHCBM4/6/8 antenna proteins in Chlamydomonas reinhardtii. J Exp Bot 68: 627–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U, Blaby I, Casero D, Gallaher SD, Goodson C, Johnson S, Lee JH, Merchant SS, Pellegrini M, Roth R, et al. (2014) The path to triacylglyceride obesity in the sta6 strain of Chlamydomonas reinhardtii. Eukaryot Cell 13: 591–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goold H, Beisson F, Peltier G, Li-Beisson Y (2015) Microalgal lipid droplets: Composition, diversity, biogenesis and functions. Plant Cell Rep 34: 545–555 [DOI] [PubMed] [Google Scholar]
- Guschina IA, Harwood JL (2006) Lipids and lipid metabolism in eukaryotic algae. Prog Lipid Res 45: 160–186 [DOI] [PubMed] [Google Scholar]
- Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A (2008) Microalgal triacylglycerols as feedstocks for biofuel production: Perspectives and advances. Plant J 54: 621–639 [DOI] [PubMed] [Google Scholar]
- Huang S, Jiang L, Zhuang X (2019) Possible roles of membrane trafficking components for lipid droplet dynamics in higher plants and green algae. Front Plant Sci 10: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Alric J (2013) Central carbon metabolism and electron transport in Chlamydomonas reinhardtii: Metabolic constraints for carbon partitioning between oil and starch. Eukaryot Cell 12: 776–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhoven EJ, Pomraning KR, Baker SE, Nielsen J (2016) Regulation of amino-acid metabolism controls flux to lipid accumulation in Yarrowia lipolytica. NPJ Syst Biol Appl 2: 16005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Jang S, Kim S, Yamaoka Y, Hong D, Song WY, Nishida I, Li-Beisson Y, Lee Y (2015) The small molecule fenpropimorph rapidly converts chloroplast membrane lipids to triacylglycerols in Chlamydomonas reinhardtii. Front Microbiol 6: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim H, Ko D, Yamaoka Y, Otsuru M, Kawai-Yamada M, Ishikawa T, Oh HM, Nishida I, Li-Beisson Y, et al. (2013) Rapid induction of lipid droplets in Chlamydomonas reinhardtii and Chlorella vulgaris by Brefeldin A. PLoS One 8: e81978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Harada N, Nishimura Y, Saito T, Nakamura M, Fujiwara T, Kuroiwa T, Misumi O (2014) Algae sense exact temperatures: Small heat shock proteins are expressed at the survival threshold temperature in Cyanidioschyzon merolae and Chlamydomonas reinhardtii. Genome Biol Evol 6: 2731–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Park JJ, Barupal DK, Fiehn O (2012) System response of metabolic networks in Chlamydomonas reinhardtii to total available ammonium. Mol Cell Proteomics 11: 973–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Han D, Hu G, Dauvillee D, Sommerfeld M, Ball S, Hu Q (2010) Chlamydomonas starchless mutant defective in ADP-glucose pyrophosphorylase hyper-accumulates triacylglycerol. Metab Eng 12: 387–391 [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Thelen JJ, Fedosejevs E, Harwood JL (2019) The lipid biochemistry of eukaryotic algae. Prog Lipid Res 74: 31–68 [DOI] [PubMed] [Google Scholar]
- Longworth J, Noirel J, Pandhal J, Wright PC, Vaidyanathan S (2012) HILIC- and SCX-based quantitative proteomics of Chlamydomonas reinhardtii during nitrogen starvation induced lipid and carbohydrate accumulation. J Proteome Res 11: 5959–5971 [DOI] [PubMed] [Google Scholar]
- Maikova A, Zalutskaya Z, Lapina T, Ermilova E (2016) The HSP70 chaperone machines of Chlamydomonas are induced by cold stress. J Plant Physiol 204: 85–91 [DOI] [PubMed] [Google Scholar]
- Martin NC, Chiang K-S, Goodenough UW (1976) Turnover of chloroplast and cytoplasmic ribosomes during gametogenesis in Chlamydomonas reinhardtii. Dev Biol 51: 190–201 [DOI] [PubMed] [Google Scholar]
- Menon KR, Balan R, Suraishkumar GK (2013) Stress induced lipid production in Chlorella vulgaris: Relationship with specific intracellular reactive species levels. Biotechnol Bioeng 110: 1627–1636 [DOI] [PubMed] [Google Scholar]
- Miller R, Wu G, Deshpande RR, Vieler A, Gärtner K, Li X, Moellering ER, Zäuner S, Cornish AJ, Liu B, et al. (2010) Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol 154: 1737–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Msanne J, Xu D, Konda AR, Casas-Mollano JA, Awada T, Cahoon EB, Cerutti H (2012) Metabolic and gene expression changes triggered by nitrogen deprivation in the photoautotrophically grown microalgae Chlamydomonas reinhardtii and Coccomyxa sp. C-169. Phytochemistry 75: 50–59 [DOI] [PubMed] [Google Scholar]
- Myers AM, Harris EH, Gillham NW, Boynton JE (1984) Mutations in a nuclear gene of Chlamydomonas cause the loss of two chloroplast ribosomal proteins, one synthesized in the chloroplast and the other in the cytoplasm. Curr Genet 8: 369–378 [DOI] [PubMed] [Google Scholar]
- Nama S, Madireddi SK, Devadasu ER, Subramanyam R (2015) High light induced changes in organization, protein profile and function of photosynthetic machinery in Chlamydomonas reinhardtii. J Photochem Photobiol B 152(Pt B): 367–376 [DOI] [PubMed] [Google Scholar]
- Nguyen HM, Baudet M, Cuiné S, Adriano JM, Barthe D, Billon E, Bruley C, Beisson F, Peltier G, Ferro M, et al. (2011) Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: With focus on proteins involved in lipid metabolism. Proteomics 11: 4266–4273 [DOI] [PubMed] [Google Scholar]
- Park JJ, Wang H, Gargouri M, Deshpande RR, Skepper JN, Holguin FO, Juergens MT, Shachar-Hill Y, Hicks LM, Gang DR (2015) The response of Chlamydomonas reinhardtii to nitrogen deprivation: A systems biology analysis. Plant J 81: 611–624 [DOI] [PubMed] [Google Scholar]
- Rebeille F, Gans P (1988) Interaction between chloroplasts and mitochondria in microalgae: Role of glycolysis. Plant Physiol 88: 973–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmollinger S, Mühlhaus T, Boyle NR, Blaby IK, Casero D, Mettler T, Moseley JL, Kropat J, Sommer F, Strenkert D, et al. (2014) Nitrogen-sparing mechanisms in Chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plant Cell 26: 1410–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M, Vallon O, Wollman FA, Beck CF (1999) A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11: 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz-Raffelt M, Lodha M, Schroda M (2007) Heat shock factor 1 is a key regulator of the stress response in Chlamydomonas. Plant J 52: 286–295 [DOI] [PubMed] [Google Scholar]
- Stordeur C, Puth K, Sáenz JP, Ernst R (2014) Crosstalk of lipid and protein homeostasis to maintain membrane function. Biol Chem 395: 313–326 [DOI] [PubMed] [Google Scholar]
- Szivák I, Behra R, Sigg L (2009) Metal-induced reactive oxygen species production in Chlamydomonas reinhardtii (chlorophyceae)(1). J Phycol 45: 427–435 [DOI] [PubMed] [Google Scholar]
- Thiam AR, Antonny B, Wang J, Delacotte J, Wilfling F, Walther TC, Beck R, Rothman JE, Pincet F (2013) COPI buds 60-nm lipid droplets from reconstituted water–phospholipid–triacylglyceride interfaces, suggesting a tension clamp function. Proc Natl Acad Sci USA 110: 13244–13249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZT, Ullrich N, Joo S, Waffenschmidt S, Goodenough U (2009) Algal lipid bodies: Stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot Cell 8: 1856–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wase N, Black P, DiRusso C (2018) Innovations in improving lipid production: Algal chemical genetics. Prog Lipid Res 71: 101–123 [DOI] [PubMed] [Google Scholar]
- Wase N, Black PN, Stanley BA, DiRusso CC (2014) Integrated quantitative analysis of nitrogen stress response in Chlamydomonas reinhardtii using metabolite and protein profiling. J Proteome Res 13: 1373–1396 [DOI] [PubMed] [Google Scholar]
- Wase N, Tu B, Allen JW, Black PN, DiRusso CC (2017) Identification and metabolite profiling of chemical activators of lipid accumulation in green algae. Plant Physiol 174: 2146–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wase N, Tu B, Black PN, DiRusso C (2015) Phenotypic screening identifies Brefeldin A/Ascotoxin as an inducer of lipid storage in the algae Chlamydomonas reinhardtii. Algal Res 11: 74–84 [Google Scholar]
- Welte MA, Gould AP (2017) Lipid droplet functions beyond energy storage. Biochim Biophys Acta Mol Cell Biol Lipids 1862(10 Pt B): 1260–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijffels RH, Barbosa MJ (2010) An outlook on microalgal biofuels. Science 329: 796–799 [DOI] [PubMed] [Google Scholar]
- Wilfling F, Thiam AR, Olarte MJ, Wang J, Beck R, Gould TJ, Allgeyer ES, Pincet F, Bewersdorf J, Farese RV Jr, et al. (2014) Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. eLife 3: e01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Callaghan MM, Watson KG, Tong L (2009) A different mechanism for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by tepraloxydim. Proc Natl Acad Sci USA 106: 20723–20727 [DOI] [PMC free article] [PubMed] [Google Scholar]