Cytochrome P450 CYP82Q3 from Tanacetum cinerariifolium is involved in the introduction of the double bond in the side chain of jasmolone to generate pyrethrolone, a component of pyrethrin insecticide.
Abstract
The plant pyrethrum (Tanacetum cinerariifolium) synthesizes highly effective natural pesticides known as pyrethrins. Pyrethrins are esters consisting of an irregular monoterpenoid acid and an alcohol derived from jasmonic acid (JA). These alcohols, referred to as rethrolones, can be jasmolone, pyrethrolone, or cinerolone. We recently showed that jasmolone is synthesized from jasmone, a degradation product of JA, in a single hydroxylation step catalyzed by jasmone hydroxylase (TcJMH). TcJMH belongs to the CYP71 clade of the cytochrome P450 oxidoreductase family. Here, we used coexpression analysis, heterologous gene expression, and in vitro biochemical assays to identify the enzyme responsible for conversion of jasmolone to pyrethrolone. A further T. cinerariifolium cytochrome P450 family member, CYP82Q3 (designated Pyrethrolone Synthase; TcPYS), appeared to catalyze the direct desaturation of the C1–C2 bond in the pentyl side chain of jasmolone to produce pyrethrolone. TcPYS is highly expressed in the trichomes of the ovaries in pyrethrum flowers, similar to TcJMH and other T. cinerariifolium genes involved in JA biosynthesis. Thus, as previously shown for biosynthesis of the monoterpenoid acid moiety of pyrethrins, rethrolones are synthesized in the trichomes. However, the final assembly of pyrethrins occurs in the developing achenes. Our data provide further insight into pyrethrin biosynthesis, which could ultimately be harnessed to produce this natural pesticide in a heterologous system.
Pyrethrins are a group of six similar chemicals, synthesized by the plant pyrethrum to (Tanacetum cinerariifolium), which are very effective at immobilizing and killing flying insects (McLaughlin, 1973). These compounds fall into two groups. The first, called Type-I pyrethrins, includes jasmolin I, pyrethrin I, and cinerin I, and each contain the monoterpene acid transchrysanthemic acid, bound to one of three so-called rethrolones alcohols—respectively, jasmolone, pyrethrolone, and cinerolone (Fig. 1). Although there is some variability between different cultivars, Type-I pyrethrins generally account for approximately half or more of the total pyrethrins in the flower, with pyrethrin I being the most abundant Type-I pyrethrin (Head, 1973). The second class of pyrethrins, called Type II, are esters containing one of the same three alcohols, but linked to pyrethric acid instead of to chrysanthemic acid, and are respectively called jasmolin II, pyrethrin II, and cinerin II (Fig. 1). Pyrethrins pose no danger to humans and most mammals. They are biodegradable and photolabile, properties that slow the evolution of resistance in insects (Mitra et al., 1987). The flowers of pyrethrum, where pyrethrin biosynthesis is maximal, produce all six possible esters in varying concentrations (Head, 1973; Ramirez et al., 2012). For that reason, pyrethrum flowers serve as the source for commercial extraction (Jones, 1973). However, cost considerations limit the use of natural pyrethrins, and cheaper, synthetic pyrethrins, called pyrethroids, are more heavily used, even though they are less biodegradable and some pose more danger to mammals and fish (Barthel, 1973; Sheets et al., 1994; DeMicco et al., 2010).
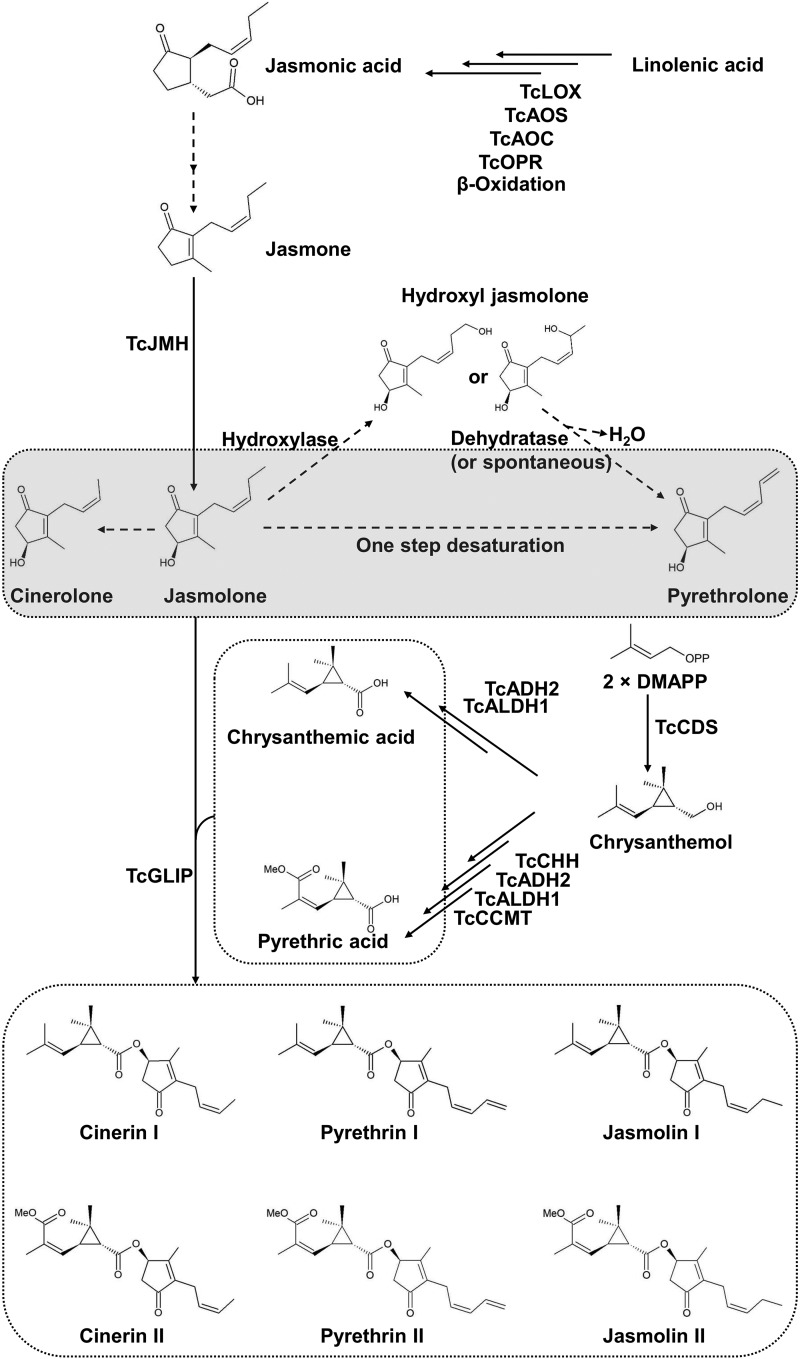
Figure 1.
The biosynthetic pathway of pyrethrins as presently known, and proposed step(s) for the synthesis of pyrethrolone in T. cinerariifolium. The synthesis of JA from linolenic acid is catalyzed by lipoxygenase (TcLOX), allene oxidase synthase (TcAOS), allene oxide cyclase (TcAOC), cis (+)-12-oxo-phytodienenic acid reductase (TcOPR), and three rounds of β-oxidation. JA is metabolized to jasmone in a set of as-yet unidentified reactions. Jasmone is converted to jasmolone in a reaction catalyzed by TcJMH (Li et al., 2018). Pyrethrolone differs from jasmolone by the presence of a double bond between ω1-carbon and the penultimate carbon of the side chain in the former. The introduction of this double bond in jasmolone could be accomplished by a single reaction or by first hydroxylation and then dehydration. Chrysanthemic acid is generated from two molecules of DMAPP in sequential reactions catalyzed by TcCDS, TcADH, and TcALDH (Xu et al., 2018). Pyrethric acid is generated from two molecules of DMAPP by the action of these three enzymes plus TcCHH and TcCCMT (Xu et al., 2019). The pyrethrin esters are formed in a reaction catalyzed by the GDSL lipase-like protein (TcGLIP), which links the acid moiety to the alcohol moiety (Kikuta et al., 2012). Pyrethrins with chrysanthemic acid as the acid moiety are called “type-I” pyrethrins; pyrethrins with pyrethric acid as the acid moiety are called “type-II” pyrethrins.
The details of the enzymatic steps leading to the biosynthesis of the monoterpenoid acids chrysanthemic acid and pyrethric acid were recently elucidated (Rivera et al., 2001; Xu et al., 2018, 2019). Chrysanthemyl diphosphate synthase (TcCDS) condenses two molecules of the universal terpene precursor dimethylallyl diphosphate (DMAPP) to produce chysanthemyl diphosphate. This product, considered an irregular terpene because of the “head to middle” condensation reaction of the two DMAPP molecules, is then converted to chrysanthemol by hydrolysis of the diphosphate group, a reaction that can be catalyzed by TcCDS or by other, as yet unidentified, phosphatases (Rivera et al., 2001; Yang et al., 2014). Chrysanthemol is converted to chrysanthemic acid by the sequential activities of two dehydrogenases, alcohol dehydrogenase2 (TcADH2) and aldehyde dehydrogenase1 (TcALDH1; Xu et al., 2018). To obtain pyrethric acid from chrysanthemol, the activities of TcADH2 and TcALDH1, which form the carboxylic group at C1, are combined with the activity of a cytochrome P450 oxidoreductase, named chrysanthemol 10-hydroxylase (TcCHH), which catalyzes three consecutive oxidation reactions of carbon 10 to form 10-carboxychrysanthemic acid; 10-carboxychrysanthemic acid is then converted to pyrethric acid by the methylation of the 10-carboxyl group by the SABATH-class methyltransferase named 10-carboxychrysanthemic acid 10-methyltransferase (TcCCMT; Xu et al., 2019).
Some progress has also been made in elucidating the synthesis of the alcohol moieties of pyrethrins. Labeling experiments showed that they are derived from jasmonic acid (JA) via jasmone (Matsuda et al., 2005), and we recently demonstrated that jasmolone is produced by hydroxylation of jasmone in a reaction catalyzed by jasmone hydroxylase (TcJMH; Li et al., 2018). Furthermore, feeding pyrethrum flowers with jasmone led not only to a large increase in the concentration of free jasmolone and jasmolin I, but also to a lesser increase in the concentration of free pyrethrolone and pyrethrin I and an even smaller but still statistically significant increase in the concentration of cinerin I (Li et al., 2018). These results suggested that pyrethrolone as well as cinerolone are derived from jasmolone, although the route was not determined.
The genes involved in the biosynthesis of chrysanthemic acid, 10-carboxychrysanthemic acid, and jasmolone are active mostly in the trichomes of the ovaries (Ramirez et al., 2012; Li et al., 2018; Xu et al., 2018, 2019). In contrast, TcCCMT, responsible for the final step in the synthesis of pyrethric acid, and TcGLIP, the gene encoding the GDSL lipase-like protein responsible for linking the acid moiety to the alcohol moiety to form the final pyrethrin molecules, are maximally expressed in the pericarp of the ovaries (Kikuta et al., 2012; Xu et al., 2019). Pyrethrins are also constitutively made in the leaves at lower levels than in the flowers, but their synthesis in the leaves is induced by wounding or treatment with methyl jasmonate (MeJA: Ueda and Matsuda, 2011).
Pyrethrolone differs from jasmolone solely by the presence of a double bond between C1 and C2 in the pentyl side chain of the former (Fig. 1). This desaturation could come about by various mechanisms, including a direct extraction of two electrons or by hydroxylation of either C1 or C2 followed by dehydration (Fig. 1). One class of enzymes that is known to carry out both desaturation and hydroxylation is the cytochrome P450 oxidoreductase superfamily (Morikawa et al., 2006; Field and Osbourn, 2008; Mizutani and Ohta, 2010). To identify such enzymes involved in pyrethrin biosynthesis, we previously performed Pearson coexpression analysis on the RNA-sequencing and metabolic data bases from different stages of pyrethrum flowers, leaves, and stems to identify cytochrome P450 oxidoreductase genes whose expression pattern in pyrethrum flowers positively correlated to some degree with the expression of TcCDS and the synthesis of pyrethrins. A total of 12 gene candidates were selected whose coefficients were higher than 0.75 using TcCDS as a reference (Li et al., 2018; Xu et al., 2018, 2019). We subsequently showed that one of them, designated as CYP71BZ1, encoded TcCHH, the enzyme involved in pyrethric acid biosynthesis (Xu et al., 2019). Another was shown to be CYP71AT148, or TcJMH, the gene encoding the enzyme that catalyzes the formation of jasmolone from jasmone (Li et al., 2018). TcJMH activity was demonstrated by transiently expressing it in Nicotiana benthamiana leaves while feeding the leaves with jasmone, and showing the production of jasmolone (Li et al., 2018). Subsequently, in vitro assays with microsomal preparations from N. benthamiana plants expressing TcJMH directly demonstrated the conversion of jasmone to jasmolone (Li et al., 2018). Here, we extend the coexpression analysis of the cytochrome P450 oxidoreductase genes expressed in pyrethrum flowers to identify the gene pyrethrolone synthase (TcPYS), which encodes the cytochrome P450 oxidoreductase CYP82Q3, the enzyme that catalyzes the formation of pyrethrolone from jasmolone.
RESULTS
Identification of TcPYS by Transient Expression of Candidate Genes in N. benthamiana Leaves
We previously identified 12 cytochrome P450 oxidoreductase genes by coexpression analysis as possibly involved in pyrethrin biosynthesis (Li et al., 2018; Xu et al., 2018, 2019). We transiently expressed each one of them separately in N. benthamiana leaves and fed jasmone to the leaves. Whereas the plants expressing TcJMH produced jasmolone, no production of pyrethrolone was observed in any of the plant lines expressing these 12 cytochrome P450 oxidoreductase candidate genes, suggesting that pyrethrolone cannot be directly produced from jasmone by a P450 enzyme. Furthermore, in previous experiments in which jasmone was fed to pyrethrum stage-3 flowers, we observed that the levels of free jasmolone in the flowers increased by 328%, whereas the concentration of free pyrethrolone increased by 49% (Li et al., 2018). These results suggest that pyrethrolone may be a downstream product of jasmolone.
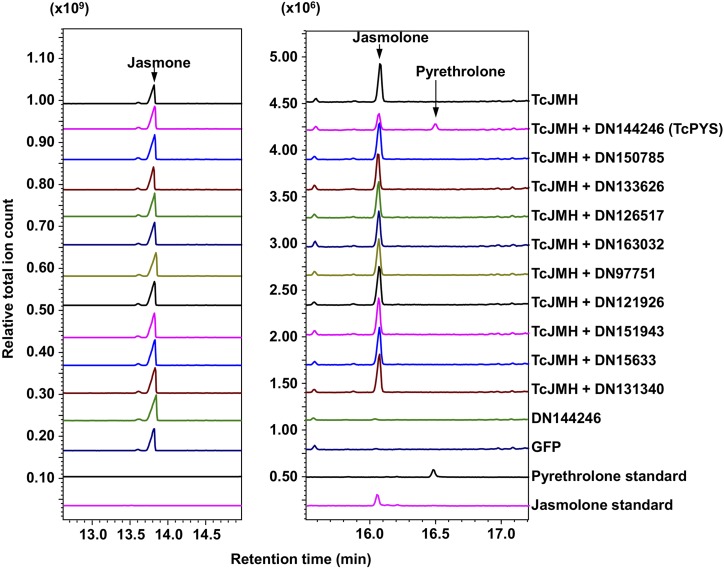
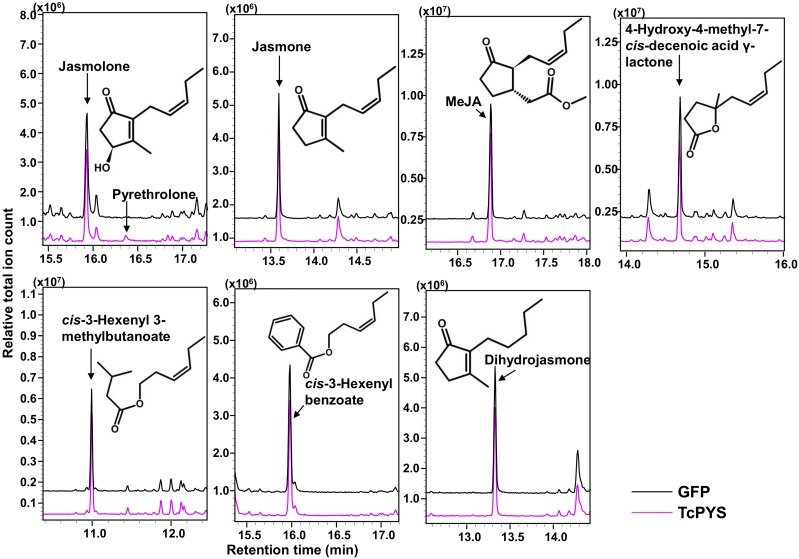
To test if pyrethrolone is produced from jasmolone by the action of a cytochrome P450 oxidoreductase, we transiently coexpressed TcJMH with each of the remaining 10 functionally unassigned cytochrome P450 oxidoreductase genes (i.e. in pairwise combinations) in N. benthamiana leaves immersed in a jasmone solution. After coexpression for 3 d with the leaves immersed in a jasmone solution for the last 24 h of this period, the leaves were ground and extracted with methyl tert-butyl ether (MTBE) and the extract analyzed by gas chromatography-mass spectromery (GC-MS). Pyrethrolone was detected only in leaves in which TcJMH was coexpressed with the cytochrome P450 oxidoreductase gene initially annotated as DN144246 (Fig. 2). As expected, jasmolone production was observed in all gene combinations due to the activity of TcJMH (Fig. 2). GC-MS analysis of leaves expressing TcJMH with DN144246 did not reveal peaks indicating additional new products besides jasmolone and pyrethrolone (Fig. 2; Supplemental Fig. S1). To identify possible intermediates that might be glycosylated or simply less volatile, we further analyzed the plant material in two ways. First, we hydrolyzed the extracts with β-glucosidase and then analyzed the de-glycosylated samples by GC-MS. No additional peaks were detected and there was no change in the concentration amounts of jasmolone and pyrethrolone (Supplemental Fig. S2). Second, we derivatized the hydrolyzed extract with n-tert-butyldimethylsilyl-n-methyltrifluoroacetamide to protect all possible hydroxyl groups from dehydration. By checking the specific ion fragment m/z = 194.0, only silylated jasmolone and pyrethrolone were detected. In contrast, no peaks representing single or double silylation products of possible hydroxyl intermediates were detected (Supplemental Fig. S3).
Figure 2.
Assay for TcPYS activity of the candidate cytochrome P450 oxidoreductases identified in our coexpression analysis. The assays were performed by transiently expressing the candidate genes in N. benthamiana for 3 d via agrobacterium infiltration and immersing their leaves in 2 mm of jasmone for 24 h, after which the leaves were ground and extracted with MTBE. Detection of jasmolone and pyrethrolone in the MTBE extracts were by GC-MS (total ion mode). The jasmolone and pyrethrolone standards were obtained as described in Li et al. (2018). Pyrethrolone was detected only in leaves coexpressing candidate gene DN144246 with TcJMH.
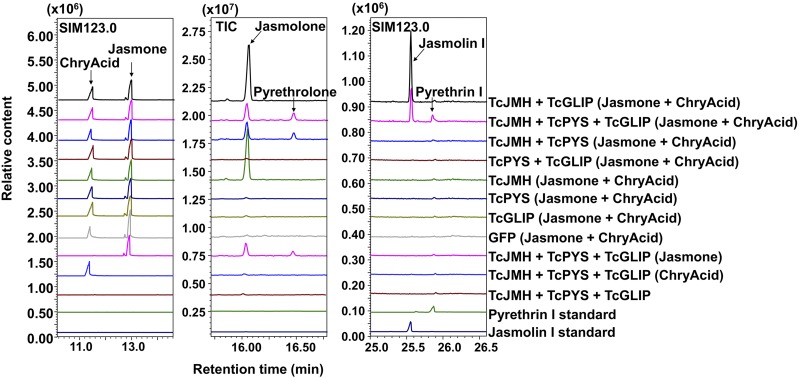
We then transiently coexpressed the three genes TcJMH, DN144246, and TcGLIP in N. benthamiana leaves fed with both jasmone and chrysanthemic acid. GC-MS analysis of extract of these leaves identified pyrethrin I in addition to jasmolin I (Fig. 3; Supplemental Fig. S4). Based on these combined results, we designated gene DN144246 as encoding TcPYS. TcPYS belongs to the CYP82 family, and was given the official catalog designation CYP82Q3 by Dr. David Nelson (https://drnelson.uthsc.edu/CytochromeP450.html; Nelson et al., 1996).
Figure 3.
Generation of pyrethrin I in leaves of N. benthamiana. Plants transiently coexpressed TcJMH, TcPYS, and TcGLIP for 3 d via agrobacteria infiltration and were immersed in a solution containing 2 mm of jasmone and 200 μM of chrysanthemic acid, after which the leaves were ground and extracted with MTBE. Detection of jasmolone and pyrethrolone in the MTBE extracts was by GC-MS (total ion mode). The jasmolin I and pyrethrin I standards were obtained as described in Li et al. (2018). Pyrethrin I was detected only in leaves expressing candidate gene DN144246, along with TcJMH and TcGLIP, and only when fed with both chrysanthemic acid and jasmone.
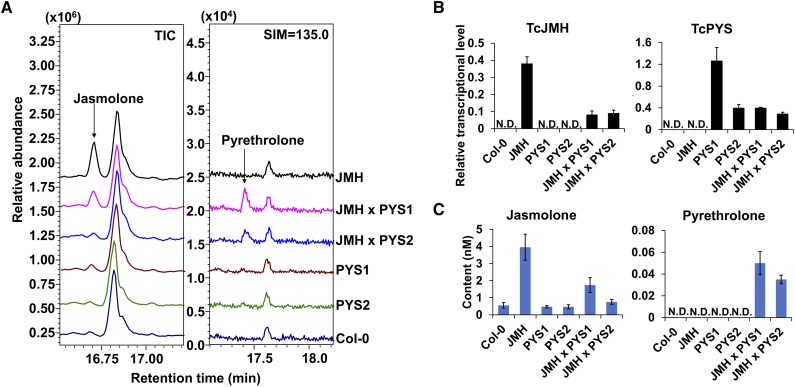
To further confirm TcPYS function, we stably coexpressed TcPYS with TcJMH in Arabidopsis (Arabidopsis thaliana), with expression of both genes independently driven by the 35S promoter. Resulting transgenic plants were tested for the production of jasmolone and pyrethrolone (Fig. 4). Surprisingly, a small amount of jasmolone, but no pyrethrolone, was detected in extracts from leaves of control plants (i.e. Col-0 nontransgenic plants) fed with 50 μM of jasmone (Fig. 4). However, plants expressing TcJMH alone contained 7.5-fold greater levels of jasmolone than control plants (and no pyrethrolone), whereas plants expressing both TcJMH and TcPYS contained pyrethrolone in addition to jasmolone (Fig. 4). These results are consistent with TcPYS having a desaturase activity on jasmolone.
Figure 4.
Coexpression of TcJMH and TcPYS in Arabidopsis. A, GC-MS detection of jasmolone and pyrethrolone in a line stably expressing TcJMH (JMH), two lines expressing TcPYS (line PYS1 and PYS2), and two lines expressing both genes (JMH × PYS1 and JMH × PYS2). Wild-type Col-0 line was used as control. TIC, total ion chromatogram; SIM, single ion monitoring. B, RT-qPCR analysis of relative TcJMH and TcPYS transcript levels in lines analyzed in (A). C, Quantification of jasmolone and pyrethrolone levels in the respective lines; N.D., not detected. Data are presented as means ± sd (n = 3).
Biochemical Characterization of TcPYS
Microsomes of N. benthamiana leaves expressing TcPYS were prepared and tested for activity with purified jasmolone prepared from hydrolyzed jasmolin I as described in Li et al. (2018). After incubation overnight, the reaction solutions were extracted with MTBE and the extract analyzed by GC-MS. This analysis detected pyrethrolone as the sole product (Fig. 5). By incubating microsomes with different concentrations of jasmolone for 2 h, the Km value for TcPYS with jasmolone was calculated to be 34.3 ± 2.6 μM (means ± sd, n = 3; Supplemental Fig. S5). To test the substrate specificity of TcPYS, we selected several available chemicals with similar structures, including jasmone, MeJA, 4-hydroxy-4-methyl-7-cis-decenoic acid γ-lactone, cis-3-Hexenyl 3-methylbutanoate, cis-3-hexenyl benzoate, and dihydrojasmone. TcPYS microsomal preparations did not catalyze the oxidation of any of these compounds, even after overnight incubations (Fig. 5).
Figure 5.
In vitro activity and substrate specificity assays for TcPYS. In vitro conversion of jasmolone to pyrethrolone in microsomes prepared from N. benthamiana leaves transiently expressing TcPYS. Substrate specificity was tested on chemicals with similar structures including jasmone, MeJA, hydroxy-4-methyl-7-cis-decenoic acid γ-lactone, cis-3-Hexenyl 3-methylbutanoate, cis-3-hexenyl benzoate, and dihydrojasmone. The reaction was carried out overnight, after which the solution was extracted with MTBE and the extract analyzed by GC-MS.
Subcellular Localization of TcPYS
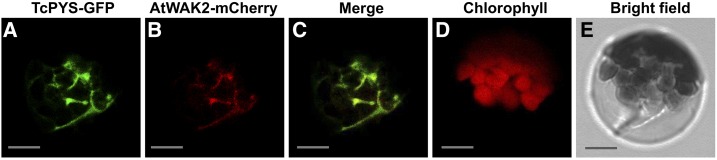
Because TcJMH was previously localized to the endoplasmic reticulum (ER), we checked the subcellular localization of TcPYS by fusing the open reading frame of TcPYS with GFP and transiently expressing this construct in Arabidopsis protoplasts under the control of the 35S promoter. Images obtained via confocal microscopy showed the green signal to be associated with the ER, as indicated by the overlap with the coexpressed ER marker AtWAK2-mCherry (Zhou et al., 2017; Fig. 6).
Figure 6.
Representative results showing subcellular localization of TcPYS. All images are of the same cell. A, Detection of green fluorescence from TcPYS-GFP fusion protein. B, Detection of red fluorescence from the ER marker AtWAK2-mCherry fusion protein. C, Overlay of GFP and red fluorescence from (A) and (B). D, Detection of autofluorescence from chloroplasts. E, Bright-field microscopy of the cell under transmitted white light. Scale bars = 10 μm.
Tissue-Specific Expression of TcPYS
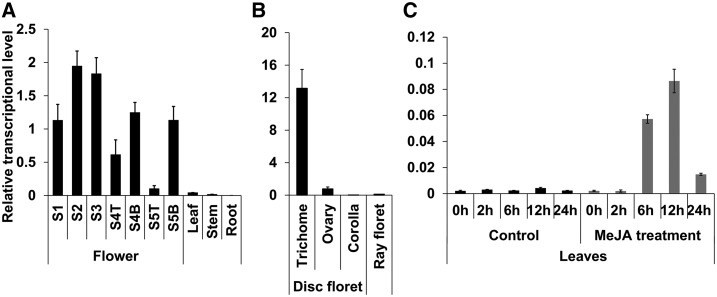
We previously showed that TcJMH converts jasmone to jasmolone in the trichomes of the ovaries (Li et al., 2018). To determine the tissue-specific expression pattern of TcPYS, reverse transcription quantitative PCR (RT-qPCR) experiments were performed with transcripts obtained from five floral developmental stages (Xu et al., 2018) as well from leaf, stem, and root tissues. Disk and ray florets were separated in stage-4 and -5 florets. The transcript analysis showed that TcPYS had a similar expression pattern to that of TcCDS in flowers, with the transcription levels peaking at stages 2 and 3, and transcript levels being barely detectable in leaf, stem, or roots (Fig. 7A).
Figure 7.
TcPYS transcription abundance in different tissues and in leaf under MeJA treatment. A, RT-qPCR analysis of TcPYS transcripts in different developmental stages of the flower (“T” and “B” represent ray florets and disk florets, respectively), leaf, stem, and root. B, RT-qPCR analysis of TcPYS transcripts in different parts of stage-3 flowers. C, RT-qPCR analysis of 2-week–old leaves treated with MeJA. Data are presented as means ± sd (n = 3 or 4).
To further identify in which parts of the flower TcPYS is expressed, we extracted RNA from stage-3 flower trichomes, ovaries, and corollas from disc florets and the entire ray florets for RT-qPCR analysis. RT-qPCR experiments indicated that TcPYS transcripts are found almost exclusively in the trichomes (Fig. 7B).
As our previous work showed that MeJA treatment of leaves induces the transcription of genes involved in pyrethrin biosynthesis (Li et al., 2018), we tested TcPYS for its inducibility by MeJA. RT-qPCR experiments on RNA collected from pyrethrum leaves treated with MeJA indeed showed a characteristic induction of TcPYS by MeJA, with transcript increases peaking at 6 h to 12 h after MeJA application and decreases observed at 24 h (Fig. 7C).
Phylogenetic Tree of Cytochrome P450 CYP82 Family
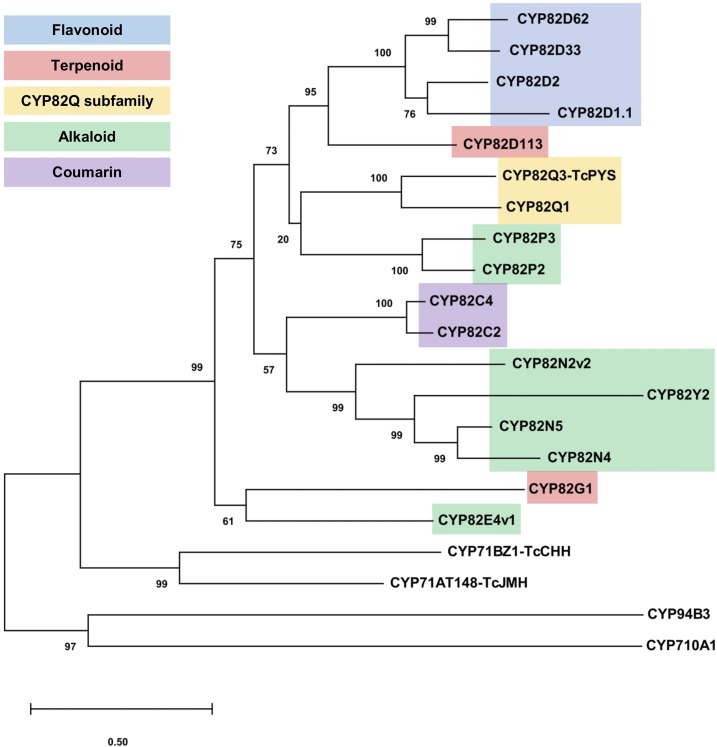
Phylogenetic analysis indicated that TcPYS belongs to the CYP82 clade (Fig. 8), and thus it was named accordingly as CYP82Q3. TcPYS is the first protein in the CYP82Q subfamily that has been biochemically characterized. TcPYS shares 61.7% sequence identity with its closest relative, CYP82Q1 from Stevia rebaudiana, a protein that has not yet been functionally characterized. More distantly related, but functionally characterized, proteins in the CYP82 clade act on flavonoids, terpenes, alkaloids, and coumarins (Siminszky et al., 2005; Kruse et al., 2008; Lee et al., 2010; Beaudoin and Facchini, 2013; Winzer et al., 2015; Hori et al., 2018; Tian et al., 2018; Fig. 8).
Figure 8.
A Maximum Likelihood phylogenetic tree of TcPYS (CYP82Q3) and the proteins most closely related to it from CYP82 family with functional assignments. The tree also includes TcCHH and TcJMH, which are involved in pyrethrin biosynthesis, and CYP94B3, which works on jasmonoyl-l-Ile and CYP710A1 as representative P450 enzymes with desaturase activities. Sources and functions are as follows: CYP82D62, flavone-6-hydroxylase (Ocimum basilicum); CYP82D33, flavone-6-hydroxylase (O. basilicum); CYP82D2, 4′-deoxyflavones hydroxylase (Scutellaria baicalensis); CYP82D1.1, 4′-deoxyflavones hydroxylase (S. baicalensis); CYP82D113, 7-keto-δ-cadinene 18-hydroxylase (Gossypium raimondii); CYP82Q3, TcPYS (T. cinerariifolium); CYP82Q1 (S. rebaudiana); CYP82P3, dihydrosanguinarine 10-hydroxylase (Eschscholzia californica sspp. californica); CYP82P2, dihydrosanguinarine 10-hydroxylase (E. californica sspp. californica); CYP82C4, fraxetin 5-hydroxylase (Arabidopsis); CYP82C2, fraxetin 5-hydroxylase (Arabidopsis); CYP82N2, protopine 6-monooxygenase (E. californica); CYP82Y2, 1,2-dehydroreticulinium reductase (Papaver somniferum); CYP82N5 n-methystylopine hydroxylase (E. californica sspp. californica); CYP82N4, methyltetrahydroprotoberberine 14-monooxygenase (P. somniferum); CYP82G1, (3E)-4,8-dimethyl-1,3,7-nonatriene synthase (Arabidopsis); CYP82E4v1, nicotine demethylase (Nicotiana tabacum); CYP71BZ1, chrysanthemol 10-hydroxylase (T. cinerariifolium); CYP71AT148, TcJMH (T. cinerariifolium); CYP94B3, jasmonoyl-l-Ile hydroxylase (Arabidopsis); CYP710A1, C22-sterol desaturase (Arabidopsis). The tree is drawn to scale (per the number of substitutions per site).
DISCUSSION
Coexpression Analysis, Heterologous Expression, and In Vitro Biochemical Assays Identify Pyrethrum CYP82Q3 as TcPYS
Our previous work suggested that jasmolone is produced in a one-step enzymatic reaction from jasmone, and that pyrethrolone is probably produced from jasmone in a multistep pathway, likely including jasmolone as an intermediate (Li et al., 2018). Here, we employed the N. benthamiana heterologous expression system to coexpress TcJMH, the CYP protein that converts jasmone to jasmolone, together with candidate CYP genes identified by coexpression analysis. This led to identification of TcPYS, which encodes an enzyme that converts jasmolone to pyrethrolone. N. benthamiana leaves coexpressing TcJMH and TcPYS were shown to produce pyrethrolone upon feeding with jasmone. TcPYS is CYP82Q3, a member of the plant-specific CYP82 clade, which includes proteins known to catalyze the modification of other terpenes as well as flavonoids, coumarins, and alkaloids—all plant-specific specialized compounds. Additional evidence for the enzymatic activity of TcPYS in converting jasmolone to pyrethrolone was obtained by coexpression of TcJMH and TcPYS in transgenic Arabidopsis as well as by in vitro assays of N. benthamiana microsomes expressing TcPYS with jasmolone as a substrate. The in vitro assays of TcPYS with jasmolone and related compounds also indicated that the ER-localized enzyme is highly specific for jasmolone. Finally, N. benthamiana leaves coexpressing TcJMH, TcPYS, and TcGLIP produced pyrethrin I upon being fed both jasmone and chrysanthemic acid.
TcPYS May Act as a Desaturase
The oxidative conversion of jasmolone to pyrethrolone could proceed via a hydroxyl intermediate followed by dehydration or directly by the removal of two electrons and two protons (Fig. 1). The various analyses of the products of the in vitro reaction (Figs. 3–5; Supplemental Figs. S2 and S3) identified only pyrethrolone. Whereas it is not possible to rule out production of short-lived, hard-to-detect hydroxylated intermediates and an endogenous N. benthamiana enzyme that acts on such an intermediate, these results suggest that TcPYS catalyzes the direct formation of a C–C double bond between the terminal carbons of the jasmolone pentyl side chain. If so, it would be the first characterized member of the CYP82 subfamily that acts as a desaturase. Most of these proteins catalyze hydroxylation reactions, with some exceptions: CYP82N2/N4 catalyzes a cyclization (Beaudoin and Facchini, 2013; Fujiwara and Ito, 2017), CYP82Y2 catalyzes a regiospecific isomerization (Winzer et al., 2015), CYP82G1 catalyzes oxidative degradation, and CYP82E4v1 catalyzes demethylation (Siminszky et al., 2005; Lee et al., 2010).
Direct desaturation catalyzed by CYP P450 enzymes is uncommon. In most reactions catalyzed by these enzymes, the activated enzyme-bound oxygen-iron complex ([FeO]3+) attacks a carbon (or N or S) atom and abstracts a hydrogen from it, forming a paired [FeOH]3+–carbon radical. This is followed by the newly formed hydroxyl rebounding to the carbon, generating a C-OH group. In some rare cases, the abstraction of a hydrogen might be followed by the abstraction of a second hydrogen from an adjacent carbon atom, followed by the formation of a double bond between two atoms, along with the release of a water molecule (Guengerich, 2001). One such example is desaturation at C22 of sterols in fungi and plants, which is catalyzed by CYP61 and CYP710 enzymes, respectively (Kelly et al., 1997; Morikawa et al., 2006). However, C22 of sterols, while on a side chain, is not a terminal or penultimate carbon. The direct desaturation of valproic acid catalyzed by rat CYP3A1 is a more similar reaction to the formation of the double bond between the ω1-carbon and the ω2-carbon on the side chain of jasmolone by TcPYS (Fisher et al., 1998). Note that in valproic acid oxidation, both hydroxylation and desaturation happen when the initial attack by the enzyme occurs at ω2-carbon, but when the initial attack occurs on the ω1-carbon, the result is 100% hydroxylation (Fisher et al., 1998).
Pyrethrin Biosynthesis Is a Multiorganellar and Multicellular Process
Maximal production of pyrethrins occurs in the pyrethrum flower, and more specifically in the developing achenes (McLaughlin, 1973). The ovaries contain trichomes, where chrysanthemic acid, 10-carboxychrysanthemic acid, and at least the alcohol jasmolone are produced (Rivera et al., 2001; Li et al., 2018; Xu et al., 2019). The final methylation reaction in the synthesis of pyrethric acid as well as the ester formation of the pyrethrins occurs mostly in the pericarp of the ovaries, and the assembled pyrethrins accumulate throughout the achenes and the seeds, evidently for protection (Ramirez et al., 2012; Xu et al., 2019). Even in the trichomes, the synthesis of these components is divided into multiple organelles. Chrysanthemol is produced in the plastid, whereas the oxidation steps leading to the monoterpene acids occur in the cytosol (Yang et al., 2014; Xu et al., 2018). JA, the precursor of the rethrolones (i.e. jasmolone, pyrethrolone, and cinerolone), is produced from linolenic acid partly in the plastid, partly in the ER, and partly in the peroxisome, whereas the production of jasmolone from jasmone occurs on the ER (Song et al., 1993; Schaller et al., 2000; Ziegler et al., 2000; Matsuda et al., 2005; Ramirez et al., 2013; Li et al., 2018). Here we show that TcPYS is also predominantly expressed in pyrethrum flowers of developmental stages 2 and 3 as found for other pyrethrin biosynthetic genes such as TcCDS and TcJMH (Fig. 7A). Furthermore, TcPYS exhibits a pattern of cellular expression (high levels of transcripts in trichomes) and subcellular protein localization (ER localization) that is very similar to that of the TcJMH gene and its protein, which generates jasmolone, the substrate of TcPYS (Figs. 6 and 7).
Pyrethrins are also synthesized in pyrethrum leaves, although at a low level compared to that in flowers. However, the biosynthesis of pyrethrins in leaves is induced by wounding as well as by the wounding hormone jasmonate (Ueda and Matsuda, 2011; Kikuta et al., 2012). Here we showed that TcPYS expression is also jasmonate-inducible (Fig. 7C), similar to the expression of other pyrethrin biosynthetic genes (Li et al., 2018).
Overall, the data presented here suggest that TcPYS may be an unusual cytochrome P450 oxidoreductase capable of directly introducing a double bond in the side chain of jasmolone to produce pyrethrolone. With the recent discovery of TcJMH, the enzyme involved in the synthesis of jasmolone from jasmone, and the set of enzymes involved in the conversion of chrysanthemol to chrysanthemic acid and pyrethric acid, only the synthesis of cinerolone remains unsolved in the biosynthetic pathway of pyrethrins.
MATERIALS AND METHODS
Plant Growth Condition
Pyrethrum (Tanacetum cinerariifolium) plants were grown on soil in growth chambers under the following conditions: 25°C, 16 h, 400 μmol m−2 s−1 light and 20°C, 8-h dark. Nicotiana benthamiana and Arabidopsis (Arabidopsis thaliana) plants were grown on soil in growth chambers at 22°C, 16 h, 150 μmol m−2 s−1 light and 20°C, 8-h dark in a growth room.
Transient Expression in N. benthamiana and Jasmone Feeding
Cytochrome P450 oxidoreductase gene candidates were inserted into vector pEAQ-HT and mobilized into Agrobacterium tumefaciens strain GV3101 as described in Li et al. (2018). Agrobacterium cells were cultured in LB media overnight until the OD value reached 1.0, then collected by centrifugation and resuspended in equal volume of buffer containing 10 mm of MES at pH 6.8, 10 mm of MgCl2, and 100 μM of Acetosyringone (MMA buffer). Agrobacterium cells were injected into 4-week–old N. benthamiana leaves via the abaxial surface. When performing coexpression of several genes, equal volumes of suspensions of different Agrobacterium cells were mixed together for injection. After 3 d, the leaves were harvested for feeding experiment or preparation of microsomes.
In chemical feeding, two N. benthamiana leaves were immersed in 20 mL of 2-mm jasmone, placed under vacuum at 12.5 psi for 10 min, and incubated for 24 h at room temperature. After incubation, leaves were rinsed twice with water, blotted dry with filter paper, and the leaves put into a 15-mL centrifuge tube. MTBE (0.5 mL) was added to the tube and the leaves homogenized with a micro homogenizer to break the tissues. The homogenate was left at room temperature for 1 h, and the sample was centrifuged at 12,000g for 5 min and the supernatant moved to a 2-mL bottle with a 0.25-mL glass insert for GC-MS analysis.
For the hydrolysis and derivatization experiments, 36 leaves were first soaked in an aqueous solution containing 2 mm of jasmone for 24 h. Afterward, the leaves were homogenized and 40 mL of extract buffer (acetonitrile/isopropanol/water: 3:3:2) was added and the solution and incubated overnight at 4°C. The solution was next centrifuged at 20,000g for 10 min, and supernatant was lyophilized and resuspended in 1 mL of a solution of 0.1-m sodium acetate at pH 5.0. Four mg (≥8 units, one unit liberates 1 μmol of Glc from salicin per min at pH 5.0 at 37°C) of almond β-glucosidase (cat. no. G0395; Sigma-Aldrich), which is known to efficiently hydrolyze β-d-glycoside from a wide range of substrates, was added to 250-μL extract, and incubated for 4 h at 37°C. The lysate was next extracted with 250 μL of MTBE for GC-MS analysis. For derivatization, 50 μL of hydrolyzed extract was dried in a speed vacuum device, and 100 μL of n-tert-butyldimethylsilyl-n-methyltrifluoroacetamide was added to the dry material, which was then incubated at 80°C for 4 h, and analyzed directly on GC-MS.
Arabidopsis Stable Expression
The sequences of TcJMH and TcPYS were inserted into the vector pSAT4A, which contains a CaMV 35S promoter and a CaMV 35S terminator, by double digest with EcoRI and BamHI and ligated with T4 DNA ligase. Next, the complete constructs (promoter–gene–terminator) were obtained by cutting with I-SceI and integrating the fragment into the binary vector pPZP-RSCII. The binary vectors were moved into Agrobacterium GV3101 for Arabidopsis floral dip transformation. TcJMH and TcPYS were transferred into Arabidopsis independently, and the two overexpressing lines were crossed to obtain the coexpression lines. Arabidopsis gene AtActin8 was used as internal reference gene to confirm transcription level of TcJMH and TcPYS in transgenic plants. Primers used are listed in Supplemental Table S1.
GC-MS Analysis and Km Value Measurement
GC-MS analysis was performed on GCMS-QP5000 (Shimadzu) with a TR-5MS column. The column temperature was programmed as follows: 50°C hold for 2 mins, 50°C to 330°C at 10°C min−1, then hold 10 min. See our previous work for pyrethrolone, jasmolone, jasmolin I, and pyrethrin I purification (Li et al., 2018).
Purification of microsomes was performed using methods published in Schaller (2017). Kinetic parameter measurements were obtained from 50-μL reactions containing 100 mm of Tris at pH 7.5, 300 μM of NADPH, 40 μL of microsome, and varying concentration of jasmolone (0/25/50/100/200 μM) or 100 μM of substrate for specificity test. After incubation for 2 h (for Km value determination) or overnight (to determine substrate specificity) at room temperature, products were extracted with 100 μL of MTBE for GC-MS analysis and their concentrations plotted to verify enzyme saturation. The Km value was calculated by hyperbolic regression analysis method with software Hyper32 (https://hyper32.software.informer.com/). Data are presented as means ± sd (n = 3).
RT-qPCR of Floral Transcripts and MeJA Induction of Transcripts in Leaves
RNA was extracted with an E.Z.N.A. Plant RNA kit (cat. no. R6827-02; Omega Bio-tek). Reverse transcription reaction was carried out with a high-capacity cDNA reverse transcription kit (cat. no. 4368814; Thermo Fisher Scientific). RT-qPCR was performed with Power SYBR Green PCR master mix (cat. no. 436759; Thermo Fisher Scientific). Different stages of Pyrethrum flower were assessed as described in Xu et al. (2018). Isolation of floral trichomes was performed using published methods from Ramirez et al. (2012) and MeJA induction was done as we reported in Li et al. (2018). Transcriptional level of TcPYS was determined by comparison with internal reference genes TcGAPDH, TcActin7, and TcTubulin3. Primers used are listed in Supplemental Table S1.
Subcellular Localization
Full-length TcPYS gene sequence was integrated into the expression vector pEZS-NL using the primers listed in Supplemental Table S1. Arabidopsis protoplasts preparation, transformation, and confocal microscopy were performed as described in Zhou et al. (2017).
Phylogenetic Analysis
Amino acid sequences of functionally elucidated CYP82 family proteins were downloaded from the National Center for Biotechnology Information. A Maximum Likelihood phylogenetic tree was produced by the software program MEGA7, which is based on the Jones–Taylor–Thornton matrix model (Kumar et al., 2016).
Accession Numbers
The sequence of TcPYS has been submitted to the National Center for Biotechnology Information under accession no. MG874675.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Comparisons of mass spectra of pyrethrolone generated from different sources.
Supplemental Figure S2. Analysis of possible glycosylation of rethrolones in N. benthamiana leaves expressing TcJMH and TcPYS.
Supplemental Figure S3. GC-MS analysis of derivatized rethrolones in N. benthamiana leaves expressing TcJMH and TcPYS.
Supplemental Figure S4. Comparisons of mass spectra of pyrethrin I from different sources.
Supplemental Figure S5. An in vitro enzyme saturation curve for the conversion of jasmolone to pyrethrolone by purified microsome preparations from N. benthamiana leaves expressing TcPYS.
Supplemental Table S1. Primers used in this investigation.
ACKNOWLEDGMENTS
We thank Dr. David Nelson from University of Tennessee for naming TcPYS as CYP82Q3. We also thank Dr. A. Daniel Jones and Dr. Anthony Schilmiller from Michigan State University for derivatization suggestions.
Footnotes
This work was supported by the National Science Foundation (collaborative research grant no. 1565355 to E.P. and no. 1565232 to R.L.L.) and the National Institute of General Medical Sciences of the National Institutes of Health (predoctoral training award grant no. T32-GM110523 to D.B.L.).
Articles can be viewed without a subscription.
References
- Barthel WF. (1973) Toxicity of pyrethrum and its constituents to mammals In Casida JE, ed, Pyrethrum. Academic Press, Cambridge, MA, pp 123–142 [Google Scholar]
- Beaudoin GAW, Facchini PJ (2013) Isolation and characterization of a cDNA encoding (S)-cis-n-methylstylopine 14-hydroxylase from opium poppy, a key enzyme in sanguinarine biosynthesis. Biochem Biophys Res Commun 431: 597–603 [DOI] [PubMed] [Google Scholar]
- DeMicco A, Cooper KR, Richardson JR, White LA (2010) Developmental neurotoxicity of pyrethroid insecticides in zebrafish embryos. Toxicol Sci 113: 177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B, Osbourn AE (2008) Metabolic diversification—independent assembly of operon-like gene clusters in different plants. Science 320: 543–547 [DOI] [PubMed] [Google Scholar]
- Fisher MB, Thompson SJ, Ribeiro V, Lechner MC, Rettie AE (1998) P450-catalyzed in-chain desaturation of valproic acid: Isoform selectivity and mechanism of formation of Δ3-valproic acid generated by baculovirus-expressed CYP3A1. Arch Biochem Biophys 356: 63–70 [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Ito M (2017) Molecular cloning and characterization of a Perilla frutescens cytochrome P450 enzyme that catalyzes the later steps of perillaldehyde biosynthesis. Phytochemistry 134: 26–37 [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (2001) Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol 14: 611–650 [DOI] [PubMed] [Google Scholar]
- Head SW. (1973) Composition of pyrethrum extract and analysis of pyrethrins In Casida JE, ed, Pyrethrum. Academic Press, Cambridge, MA, pp 25–53 [Google Scholar]
- Hori K, Yamada Y, Purwanto R, Minakuchi Y, Toyoda A, Hirakawa H, Sato F (2018) Mining of the uncharacterized cytochrome P450 genes involved in alkaloid biosynthesis in California poppy using a draft genome sequence. Plant Cell Physiol 59: 222–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones GDG. (1973) Pyrethrum production In Casida JE, ed, Pyrethrum. Academic Press, Cambridge, MA, pp 17–22 [Google Scholar]
- Kelly SL, Lamb DC, Baldwin BC, Corran AJ, Kelly DE (1997) Characterization of Saccharomyces cerevisiae CYP61, sterol Δ22-desaturase, and inhibition by azole antifungal agents. J Biol Chem 272: 9986–9988 [DOI] [PubMed] [Google Scholar]
- Kikuta Y, Ueda H, Takahashi M, Mitsumori T, Yamada G, Sakamori K, Takeda K, Furutani S, Nakayama K, Katsuda Y, et al. (2012) Identification and characterization of a GDSL lipase-like protein that catalyzes the ester-forming reaction for pyrethrin biosynthesis in Tanacetum cinerariifolium—a new target for plant protection. Plant J 71: 183–193 [DOI] [PubMed] [Google Scholar]
- Kruse T, Ho K, Yoo HD, Johnson T, Hippely M, Park JH, Flavell R, Bobzin S (2008) In planta biocatalysis screen of P450s identifies 8-methoxypsoralen as a substrate for the CYP82C subfamily, yielding original chemical structures. Chem Biol 15: 149–156 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Badieyan S, Bevan DR, Herde M, Gatz C, Tholl D (2010) Herbivore-induced and floral homoterpene volatiles are biosynthesized by a single P450 enzyme (CYP82G1) in Arabidopsis. Proc Natl Acad Sci USA 107: 21205–21210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhou F, Pichersky E (2018) Jasmone hydroxylase, a key enzyme in the synthesis of the alcohol moiety of pyrethrin insecticides. Plant Physiol 177: 1498–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Kikuta Y, Haba A, Nakayama K, Katsuda Y, Hatanaka A, Komai K (2005) Biosynthesis of pyrethrin I in seedlings of Chrysanthemum cinerariaefolium. Phytochemistry 66: 1529–1535 [DOI] [PubMed] [Google Scholar]
- McLaughlin GA. (1973) History of pyrethrum In Casida JE, ed, Pyrethrum. Academic Press, Cambridge, MA, pp 3–15 [Google Scholar]
- Mitra RB, Kulkarni GH, Khanna PN (1987) A novel approach to synthesis of 1R-cis-caronaldehyde, a key intermediate for photostable pyrethroid insecticides. Synth Commun 17: 1089–1094 [Google Scholar]
- Mizutani M, Ohta D (2010) Diversification of P450 genes during land plant evolution. Annu Rev Plant Biol 61: 291–315 [DOI] [PubMed] [Google Scholar]
- Morikawa T, Mizutani M, Ohta D (2006) Cytochrome P450 subfamily CYP710A genes encode sterol C-22 desaturase in plants. Biochem Soc Trans 34: 1202–1205 [DOI] [PubMed] [Google Scholar]
- Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW, et al. (1996) P450 superfamily: Update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics 6: 1–42 [DOI] [PubMed] [Google Scholar]
- Ramirez AM, Stoopen G, Menzel TR, Gols R, Bouwmeester HJ, Dicke M, Jongsma MA (2012) Bidirectional secretions from glandular trichomes of pyrethrum enable immunization of seedlings. Plant Cell 24: 4252–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez AM, Yang T, Bouwmeester HJ, Jongsma MA (2013) A trichome-specific linoleate lipoxygenase expressed during pyrethrin biosynthesis in pyrethrum. Lipids 48: 1005–1015 [DOI] [PubMed] [Google Scholar]
- Rivera SB, Swedlund BD, King GJ, Bell RN, Hussey CE Jr., Shattuck-Eidens DM, Wrobel WM, Peiser GD, Poulter CD (2001) Chrysanthemyl diphosphate synthase: Isolation of the gene and characterization of the recombinant non-head-to-tail monoterpene synthase from Chrysanthemum cinerariaefolium. Proc Natl Acad Sci USA 98: 4373–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller F, Biesgen C, Müssig C, Altmann T, Weiler EW (2000) 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta 210: 979–984 [DOI] [PubMed] [Google Scholar]
- Schaller GE. (2017) Isolation of endoplasmic reticulum and its membrane. Methods Mol Biol 1511: 119–129 [DOI] [PubMed] [Google Scholar]
- Sheets LP, Doherty JD, Law MW, Reiter LW, Crofton KM (1994) Age-dependent differences in the susceptibility of rats to delta-methrin. Toxicol Appl Pharmacol 126: 186–190 [DOI] [PubMed] [Google Scholar]
- Siminszky B, Gavilano L, Bowen SW, Dewey RE (2005) Conversion of nicotine to nornicotine in Nicotiana tabacum is mediated by CYP82E4, a cytochrome P450 monooxygenase. Proc Natl Acad Sci USA 102: 14919–14924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WC, Funk CD, Brash AR (1993) Molecular cloning of an allene oxide synthase: A cytochrome P450 specialized for the metabolism of fatty acid hydroperoxides. Proc Natl Acad Sci USA 90: 8519–8523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Ruan JX, Huang JQ, Yang CQ, Fang X, Chen ZW, Hong H, Wang LJ, Mao YB, Lu S, et al. (2018) Characterization of gossypol biosynthetic pathway. Proc Natl Acad Sci USA 115: E5410–E5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Matsuda K (2011) VOC-mediated within-plant communications and nonvolatile systemic signals upregulate pyrethrin biosynthesis in wounded seedlings of Chrysanthemum cinerariaefolium. J Plant Interact 6: 89–91 [Google Scholar]
- Winzer T, Kern M, King AJ, Larson TR, Teodor RI, Donninger SL, Li Y, Dowle AA, Cartwright J, Bates R, et al. (2015) Plant science. Morphinan biosynthesis in opium poppy requires a P450-oxidoreductase fusion protein. Science 349: 309–312 [DOI] [PubMed] [Google Scholar]
- Xu H, Li W, Schilmiller AL, van Eekelen H, de Vos RCH, Jongsma MA, Pichersky E (2019) Pyrethric acid of natural pyrethrin insecticide: Complete pathway elucidation and reconstitution in Nicotiana benthamiana. New Phytol 223: 751–765 [DOI] [PubMed] [Google Scholar]
- Xu H, Moghe GD, Wiegert-Rininger K, Schilmiller AL, Barry CS, Last RL, Pichersky E (2018) Coexpression analysis identifies two oxidoreductases involved in the biosynthesis of the monoterpene acid moiety of natural pyrethrin insecticides in Tanacetum cinerariifolium. Plant Physiol 176: 524–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Gao L, Hu H, Stoopen G, Wang C, Jongsma MA (2014) Chrysanthemyl diphosphate synthase operates in planta as a bifunctional enzyme with chrysanthemol synthase activity. J Biol Chem 289: 36325–36335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Wang CY, Gutensohn M, Jiang L, Zhang P, Zhang D, Dudareva N, Lu S (2017) A recruiting protein of geranylgeranyl diphosphate synthase controls metabolic flux toward chlorophyll biosynthesis in rice. Proc Natl Acad Sci USA 114: 6866–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J, Stenzel I, Hause B, Maucher H, Hamberg M, Grimm R, Ganal M, Wasternack C (2000) Molecular cloning of allene oxide cyclase. The enzyme establishing the stereochemistry of octadecanoids and jasmonates. J Biol Chem 275: 19132–19138 [DOI] [PubMed] [Google Scholar]