Overexpressed Tomosyn protein inhibits secretion in pollen by competitively binding to syntaxins, the proteins that control secretory vesicle fusion with its target compartment.
Abstract
SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptors) complex formation is necessary for intracellular membrane fusion and thus has a key role in processes such as secretion. However, little is known about the regulatory factors that bind to Qa-SNAREs, which are also known as syntaxins (SYPs) in plants. Here, we characterized Arabidopsis (Arabidopsis thaliana) Tomosyn protein (AtTMS) and demonstrated that it is a conserved regulator of SYPs in plants. AtTMS binds strongly via its R-SNARE motif-containing C terminus to the Qa domain of PM-resident, pollen-expressed SYP1s (SYP111, SYP124, SYP125, SYP131, and SYP132), which were narrowed down from 12 SYPs. AtTMS is highly expressed in pollen from the bicellular stage onwards, and overexpression of AtTMS under the control of the UBIQUITIN10, MSP1, or LAT52 promoter all resulted in defective pollen after the microspore stage in which secretion was inhibited, leading to the failure of intine deposition and cell plate formation during pollen mitosis I. In tobacco (Nicotiana benthamiana) leaf epidermal cells, overexpression of AtTMS inhibited the secretion of secreted GFP. The defects were rescued by mCherry-tagged SYP124, SYP125, SYP131, or SYP132. In vivo, SYP132 partially rescued the pMSP1:AtTMS phenotype. In addition, AtTMS, lacking a transmembrane domain, was recruited to the plasma membrane by SYP124, SYP125, SYP131, and SYP132 and competed with Vesicle-Associated Membrane Protein721/722 for binding to, for example, SYP132. Together, our results demonstrated that AtTMS might serve as a negative regulator of secretion, whereby active secretion might be fine-tuned during pollen development.
Male gametophyte development is a key process during plant reproduction. A number of events, such as the deposition of the intine, the expansion of the cell plate during pollen mitosis I (PMI), and pollen tube growth, require active membrane traffic to the plasma membrane (PM; Kato et al., 2010; Twell, 2011; Shi et al., 2015). It is generally recognized that SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptors) proteins control the ultimate fusion of a secretory vesicle with its target compartment (Jahn and Scheller, 2006). They have been grouped into Q-SNAREs (Qa, Qb, Qc, or Qbc) or R-SNAREs based on the conserved residues within the SNARE domain (Fasshauer et al., 1998). Different sets of Q- and R-SNAREs in two opposing membranes associate into trans-SNARE complexes through SNARE domains, which drive membrane fusion and cargo secretion (Jahn and Scheller, 2006).
In recent years, several SNARE proteins have been shown to play roles in male gametophyte development. The PM-resident Qa-SNAREs SYP124, SYP125, and SYP131 (Uemura et al., 2004) are expressed specifically in pollen (Enami et al., 2009), and the syp124 syp125 syp131 triple mutant shows defective pollen tube growth (Slane et al., 2017). SYP111 and SYP132 are expressed in the early stages of pollen development and are possible candidates to mediate fusion processes during the mitotic divisions that give rise to tricellular pollen (Enami et al., 2009; Slane et al., 2017). Unlike SYP111, SYP132 was essential for secretory trafficking to the PM (Karnahl et al., 2018; Park et al., 2018). The R-SNARE SEC22 localized at the endoplasmic reticulum, and sec22-2 pollen development was defective (El-Kasmi et al., 2011). Overexpressing the pollen-specific R-SNARE PiVAMP726 in Petunia inflata inhibited growth and caused the formation of novel membrane compartments within the tip of the pollen tube (Guo and McCubbin, 2012). Apparently, more components of the vesicle fusion machinery and regulators remain to be studied in pollen.
Compared with the characterization of the fusion machinery itself, little is known about factors modulating SNARE complex assembly that ensure the correct vesicle fusion spatially and temporally (Toonen and Verhage, 2003; Pobbati et al., 2004). In plants, the best-studied SNARE regulator is the SM/SEC1 family protein KEULE. KEULE binds to the open conformation of Qa-SNARE SYP111/KNOLLE, thus stabilizing KNOLLE and promoting trans-SNARE formation (Park et al., 2012). As a result, keule and knolle single and keule knolle double mutants had abnormal vesicle accumulation and defective cell plate formation during somatic cytokinesis (Jürgens, 2005).
Tomosyn (TMS) was first identified as a binding protein for the Qa-SNARE domain of Syntaxin1 from rat cerebra (Fujita et al., 1998) and is conserved in mammals. Overexpression of TMS inhibits exocytosis in a number of neuroendocrine secretory cells (Widberg et al., 2003; Gladycheva et al., 2007) and in neurons (Yizhar et al., 2004, 2007). Moreover, genetic studies with a Caenorhabditis elegans tom-1 mutant (Gracheva et al., 2007) along with a tomosyn knockout mouse (Sakisaka et al., 2008) have demonstrated enhanced synaptic transmission.
In this study, we provide evidence that AtTMS might act as a negative regulator of secretion in pollen development by competitively binding to a set of SYP1s.
RESULTS
AtTMS Is an R-SNARE Motif-Containing Protein Homologous to Animal TMSs
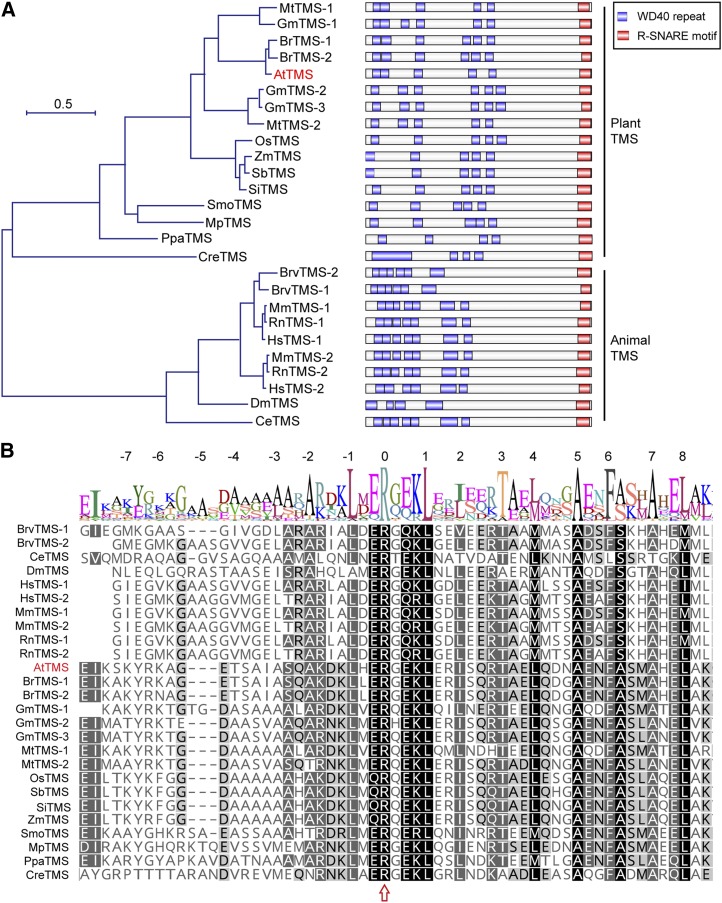
AtTMS (AT5G05570) encodes a 1,124-amino acid protein containing a small C terminus harboring a coiled-coil R-SNARE motif and a large N terminus containing WD40 repeats (Fig. 1A). A maximum-likelihood phylogenetic tree analysis showed that TMSs are highly conserved in animals and plants (Fig. 1A). The R-SNARE motif in each TMS homolog bears 16 fully conserved hydrophobic amino acids and the conserved Arg in layer 0 of the central heptad repeat of the α-helix (Fig. 1B). This motif shows very high sequence similarity to that of the R-SNARE synaptobrevin and serves as the main interaction site with Syntaxin1a (Hatsuzawa et al., 2003; Pobbati et al., 2004). These data indicate that AtTMS is a conserved TMS protein in plants and may have a Qa-SNARE-binding capacity like its animal counterparts.
Figure 1.
AtTMS is homologous to animal TMSs. A, Left, Phylogenetic relationship among AtTMS, other plant TMS homologs, and animal TMS proteins. The phylogenic tree was constructed using MEGA version 7.0. Species abbreviations are as follows: Mt, Medicago truncatula; Gm, Glycine max; Br, Brassica rapa; At, Arabidopsis thaliana; Os, Oryza sativa; Zm, Zea mays; Sb, Sorghum bicolor; Si, Setaria italica; Smo, Selaginella moellendorfii; Mp, Marchantia polymorpha; Ppa, Physcomitrella patens; Cre, Chlamydomonas reinhardtii; Mm, Mus musculus; Rn, Rattus norvegicus; Hs, Homo sapiens; Brv, Brachydanio rerio var; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans. The sequence alignment used for this analysis is available as Supplemental Data Set S1. Right, The domain structure of each TMS family member is presented next to the corresponding protein. B, Sequence alignment of R-SNARE motif of TMS proteins. Numbers at the top indicate helical layers formed during SNARE complex assembly. The arrow indicates the Arg residue (R) at layer 0.
AtTMS Localizes to the Trans-Golgi Network, PM, and Cytosol
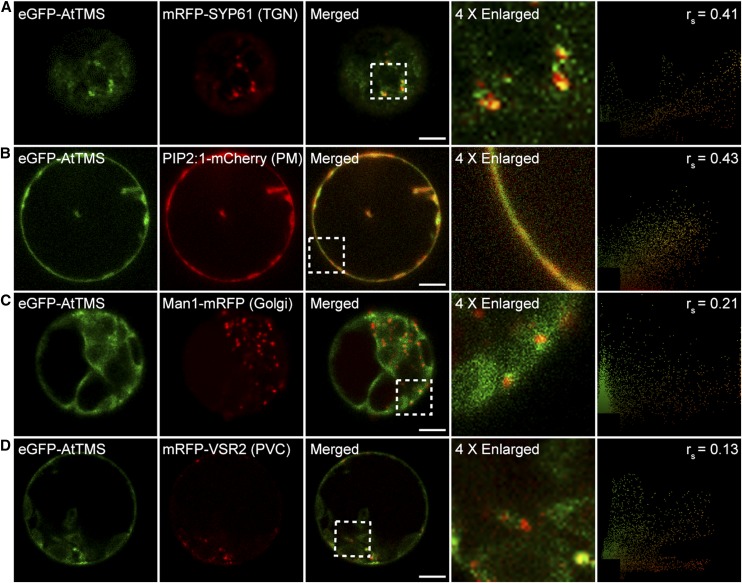
To narrow down the possible AtTMS-interacting Qa-SNAREs, subcellular localization of eGFP-tagged AtTMS was studied in Arabidopsis (Arabidopsis thaliana) protoplasts. eGFP-AtTMS partially overlapped with the trans-Golgi network (TGN; Fig. 2A; Lam et al., 2007) and PM (Fig. 2B; Hou et al., 2014) but not with the Golgi (Fig. 2C; Tse et al., 2004) and the prevacuolar compartment (Fig. 2D; Miao et al., 2006). A similar AtTMS localization pattern was demonstrated in tobacco (Nicotiana benthamiana) leaf epidermal cells (Supplemental Fig. S1). These data demonstrated that AtTMS disperses in the cytosol and also localizes to the TGN and the PM.
Figure 2.
Subcellular localization of AtTMS. eGFP-AtTMS was coexpressed with known organelle markers in Arabidopsis protoplasts. Images were collected by confocal microscopy after 16 to 18 h of incubation. The nonlinear Spearman’s rank (rs) correlation coefficient between eGFP-AtTMS and each marker is shown in the right column. Bars = 10 µm.
AtTMS Interacts with PM-Resident SYP111/124/125/131/132 and TGN-Resident SYP41/43
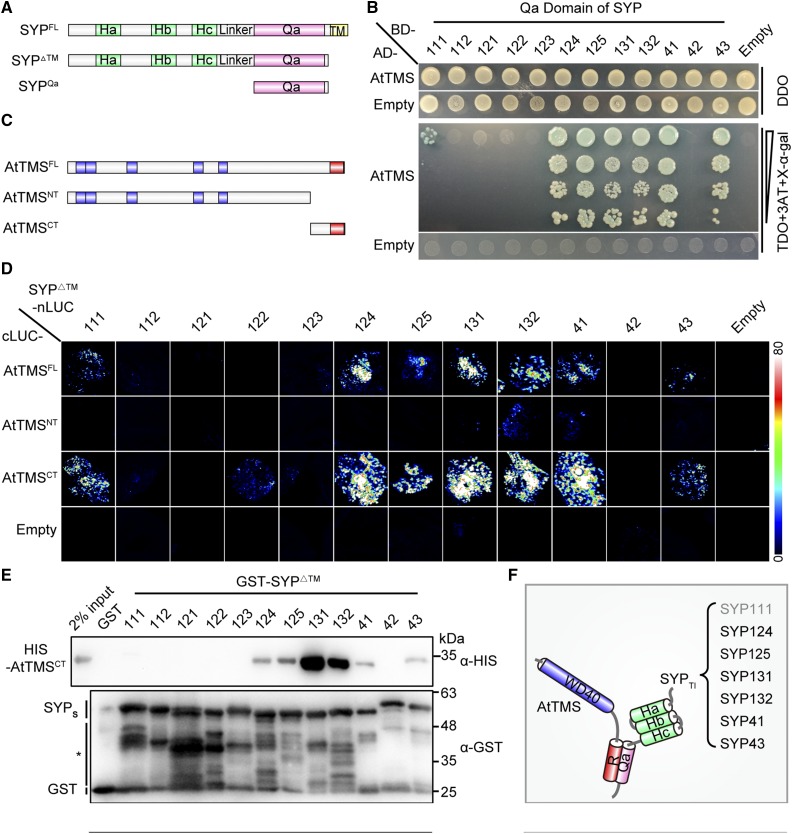
The subcellular localization pattern (Fig. 2) suggested that AtTMS-interacting Qa-SNAREs should be of TGN and/or PM origin. There are nine PM (SYP1 subfamily) and three TGN (SYP4 subfamily) Qa-SNAREs in Arabidopsis (Uemura et al., 2004). The interaction between AtTMS and the Qa domain of 12 individual SYPs was then tested. In a yeast-two-hybrid assay, AtTMS interacted strongly with the Qa domain of SYP124/125/131/132/41/43 (forward slash means and), weakly with that of SYP111, but not at all with SYP112/121/122/123/42 (Fig. 3, A and B).
Figure 3.
The C terminus of AtTMS and the SNARE domain of SYPs mediate mutual binding between TMS-interacting SYP (SYPsTI) and AtTMS. A, Schematic presentation of truncations for SYP proteins. B, A yeast two-hybrid assay showed that AtTMS interacts with the Qa domain of SYP111/124/125/131/132/41/43. Transformed yeast cells able to grow and turn blue on triple dropout medium (-Leu-Trp-His; TDO) + 5 µm 3-aminotriazole (3AT) + 200 ng mL−1 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (X-α-gal) indicates positive interactions. DDO, Double dropout medium (-Leu-Trp). C, Schematic representation of the size and position of each AtTMS truncation. The WD40 domain (blue) and R-SNARE motif (red) are indicated. D, SYP111/124/125/131/132/41/43ΔTM interact with both the AtTMSFL and the AtTMSCT in firefly LCI assays. Different pairs of the indicated plasmids were cotransformed in tobacco leaves. A pair of cLUC and nLUC empty vectors was used as negative controls (Empty). The luciferase signal was captured using a low-light cooled CCD camera, and the color scale at right shows the range of luminescence intensity. E, Only SYP111/124/125/131/132/41/43ΔTM can interact with the AtTMSCT. GST and each GST-SYPΔTM protein coupled to beads were incubated with His-tagged AtTMSCT. Bound proteins were detected using mouse anti-His antibody. The blot was stripped and reblotted with mouse anti-GST antibody to ensure the quality and coupling of the bait proteins. The asterisk indicates nonspecific or degraded protein bands. F, Diagram of the mode of interaction between AtTMS and SYPsTI. Gray text for SYP111 indicates a weak interaction.
Next, each of the 12 SYPs without a transmembrane domain (SYPΔTM) was tested for its binding capacity to AtTMS full length (AtTMSFL; amino acids 1–1,124), N terminus-containing WD40 repeats (AtTMSNT; amino acids 1–921), and C terminus-containing R-SNARE motif (AtTMSCT; amino acids 922–1,124; Fig. 3C) using a firefly luciferase complementation imaging (LCI) assay (Chen et al., 2008). Only SYP111/124/125/131/132/41/43ΔTM-nLUC, but not SYP112/121/122/123/42ΔTM-nLUC, reconstituted a high luciferase activity with either cLUC-AtTMSFL or cLUC-AtTMSCT, respectively (Fig. 3D). Similarly, glutathione S-transferase (GST)-SYP111/124/125/131/132/41/43ΔTM, but neither GST alone nor GST-SYP112/121/122/123/42ΔTM, were able to pull down His-tagged AtTMSCT (Fig. 3E).
Taken together, these results demonstrated that among 12 SYPs, AtTMS binds specifically to only seven of them (SYPsTI; i.e. PM-resident SYP111/124/125/131/132 [SYP1sTI] and TGN-resident SYP41/43 [SYP4sTI]). Furthermore, the Qa domain of each SYPsTI and the R-SNARE motif containing AtTMSCT were demonstrated to mediate the reciprocal bindings. Notably, AtTMS interacted only weakly with SYP111 in two assays (Fig. 3, B and E). These interaction studies are summarized in a schematic drawing (Fig. 3F).
Overexpression of AtTMS Impairs Pollen Development after the Microspore Stage
To analyze the function of AtTMS, attms-1/+ and attms-2/+ mutants were generated by CRISPR/Cas9-mediated gene editing, and homozygotes were obtained. Sequence analysis revealed a single base A and T deletion at the eighth exon in attms-1 and attms-2, respectively, and both mutations caused premature translational termination (Supplemental Fig. S2). However, attms-1 and attms-2 mutants exhibited no obvious abnormality in growth under the standard conditions.
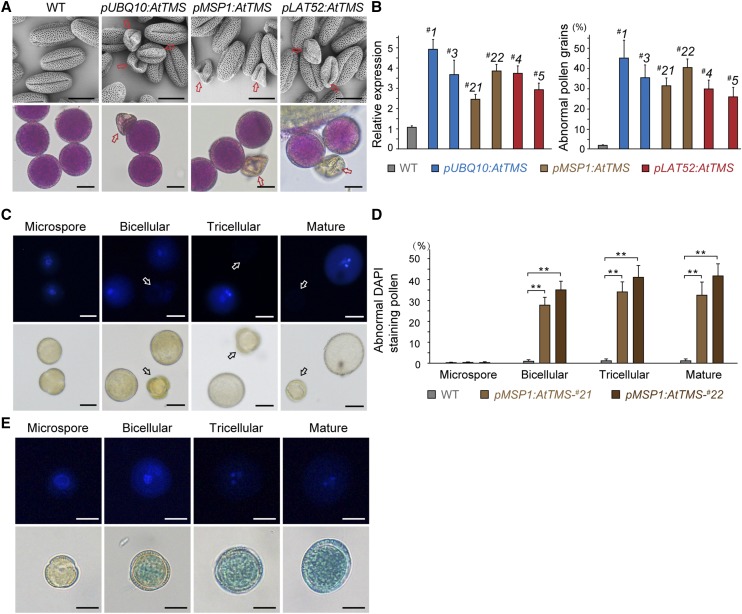
The UBIQUITIN10 promoter (pUBQ10), which facilitates moderate gene expression in nearly all tissues, including pollen (Norris et al., 1993), was first used to drive the expression of AtTMS. Three independent T0 lines of pUBQ10:AtTMS showed significantly increased shriveled and nonviable pollen grains (Fig. 4A). To verify this result, two additional pollen-specific promoters, MICROSPORE-SPECIFIC (pMSP1; Honys et al., 2006) and LAT52 (Twell et al., 1989; Yao et al., 2018), were used. Similar pollen defects were noticed in 10 pMSP1:AtTMS and nine pLAT52:AtTMS T0 lines, respectively (Fig. 4A). This phenotype could be inherited stably, and T2 generation plants were used for further observation. In two randomly selected lines of each transformation, the level of AtTMS expression was found to correlate positively with the ratio of pollen abortion (Fig. 4B). Consistently, endogenous AtTMS was highly expressed in pollen and pollen tube, as indicated by GUS assays performed with pAtTMS:GUS plants (pAtTMS; 1,965 bp in relation to ATG; Supplemental Fig. S3). It is worth noting that the abnormal pollen grains in three AtTMS overexpression (AtTMS-OE) transgenics were not caused by the possible variations of growth conditions, since other pLAT52-bearing transgenic plants cultured in the same growth room developed normal pollen (Tan et al., 2016; Li et al., 2017).
Figure 4.
AtTMS-OE impairs pollen development after the microspore stage. A, Scanning electron microscopy (top row) and Alexander staining (bottom row) micrographs of pollen grains from wild-type (WT) and AtTMS-OE plants using different promoters. The arrows indicate the shrunken and nonviable pollen grains. B, #1 and #3 of pUBQ10:AtTMS, #21 and #22 of pMSP1:AtTMS, and #4 and #5 of pLAT52:AtTMS transgenic lines were chosen randomly for analysis. Left graph, mRNA used for real time quantitative PCR (RT-qPCR) was extracted from flower buds. AtTMS expression relative to that of the wild type is shown for each line; right graph, n > 200 pollen was counted for each line. Values represent means ± sd of three independent experiments. C, pMSP1:AtTMS-#22 pollen was stained with DAPI and visualized under fluorescence (top row) and bright-field (bottom row) optics. Arrows indicate abnormal pollen. D, Quantification of abnormal DAPI-stained pollen. n > 200 pollen was counted for each line. Values represent means ± sd of three independent experiments. **, P < 0.001 by Student’s t test. E, AtTMS expression starts from the bicellular stage. GUS-stained pollen from pAtTMS:GUS plants in the bottom row was further stained with DAPI (top row) before observation. Bars = 10 µm in A, C, and E.
Phenotypic analysis was performed in two randomly chosen pMSP1:AtTMS lines: #21 and #22. Wild-type pollen development proceeds through well-defined stages that can be easily followed by counting 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei (Backues et al., 2010). pMSP1:AtTMS pollen was normal at the microspore stage (Fig. 4C); however, some pollen began to display only one or no nucleus from the bicellular stage (Fig. 4, C and D), associated with its endogenous expression pattern (Fig. 4E). Our results demonstrated that AtTMS-OE affected pollen development after the microspore stage.
AtTMS-OE Leads to Defective PMI Cell Plate and Intine Formation
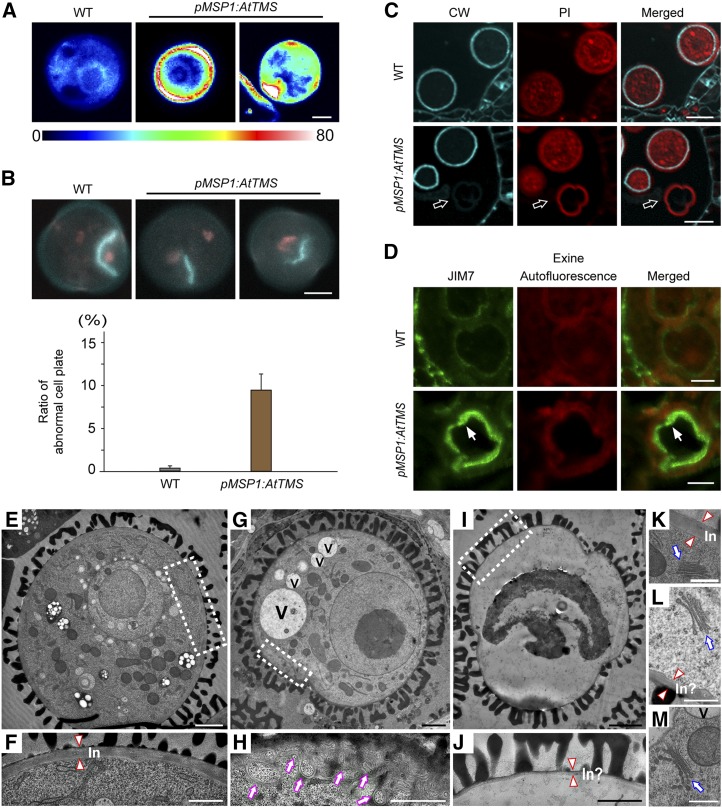
Ectopic callose deposition was noticed in pMSP1:AtTMS bicellular pollen (Fig. 5A), indicating a defective membrane trafficking and cell wall deposition (Backues et al., 2010). During PMI, a distinct hemispherical cell plate forms around the generative nucleus in wild-type pollen (Twell, 2011; Fig. 5B). However, in pMSP1:AtTMS, ∼9% of PMI pollen displayed incomplete cell wall stubs between two separating nuclei (Fig. 5B). In pMSP1:AtTMS tricellular pollen, cellulose was barely detectable by Calcofluor White staining (Fig. 5C). Further examination of pMSP1:AtTMS bicellular stage pollen showed that the secretion of JIM7-labeled highly methylesterified homogalacturonan was blocked (Fig. 5D).
Figure 5.
The phenotype of pMSP1:AtTMS pollen. A, Ectopic callose deposition found in bicellular pMSP1:AtTMS pollen. The color scale at bottom shows the range of callose intensity. WT, Wild type. B, Cell wall stubs were detected at PMI in pMSP1:AtTMS pollen. DAPI-stained (red) and Aniline Blue-stained (blue) nuclei and callose, respectively, are visible. The quantification of abnormal cell plate formation is shown underneath the micrographs; n > 100. Values represent means ± sd of three independent experiments. C, Cellulose deposition is defective in pMSP1:AtTMS bicellular pollen. Anthers were fixed in a formalin-acetic acid-alcohol solution overnight, embedded, and sectioned in 1-µm-thick slices. The blue fluorescence represents cellulose labeled by Calcofluor White (CW). Red fluorescence signals denote cytoplasm stained by propidium iodide (PI). Arrows indicate the arrested pMSP1:AtTMS pollen. D, JIM7-positive signals were blocked inside pMSP1:AtTMS bicellular pollen. Highly methylesterified homogalacturonan labeled by mouse JIM7 monoclonal antibodies (green, left), exine autofluorescence (red, middle), and merged images (right) are shown. E, TEM micrograph of a normal bicellular pollen in pMSP1:AtTMS anthers. F, Magnified view of the boxed area in E. Intine (In) is indicated between two triangles. G, TEM micrograph of abnormal bicellular pollen in pMSP1:AtTMS anthers. V, Vacuole. H, Magnified view of the boxed area in G. Purple arrows indicate abnormal membrane structures. I, TEM micrograph of a shriveled bicellular pollen in pMSP1:AtTMS anthers. J, Magnified view of the boxed area in I. In?, Abnormal intine formation. K to M, Golgi stacks from normal (K) and defective (L and M) bicellular stage pollen in pMSP1:AtTMS. Two triangles point to normal or abnormal intine. Blue arrows indicate the Golgi stacks. Bars = 5 µm in A, B, and D; 10 µm in C; 2 μm in E, G, and I; 1 μm in F, H, and J; and 500 nm in K to M.
Consistent with this, as revealed by transmission electron microscopy (TEM), the intine was not deposited properly in the same stage pollen (Fig. 5, E–J); in some cases, irregular membrane structures were seen to accumulate just beneath the PM (Fig. 5, G and H). However, Golgi stacks looked largely normal (Fig. 5, K–M), indicating a post-Golgi trafficking defect. In summary, AtTMS-OE leads to defective cell plate and intine formation by inhibiting the secretion of cell wall components.
AtTMS-OE Causes SecGFP Blockage in Tobacco Leaf Epidermal Cells That Is Rescued by Overexpression of SYP124, SYP125, SYP131 and SYP132
The syp41 syp43 double mutant displayed normal plant and pollen development (Uemura et al., 2012), indicating that the AtTMS-OE effect might not be due to an inhibition of TGN-resident SYP4sTI; therefore, they were not included in the following assays performed in tobacco leaf epidermal cells.
In AtTMS-OE cells, vacuolar transport of sporamin was not affected (Supplemental Fig. S4). SecGFP, the secreted form of GFP, was then used as a secretory marker that accumulates poorly and exhibits weak fluorescence in the apoplast (Zheng et al., 2005). Notably, intracellular SecGFP signals accumulated when AtTMS was coexpressed (Fig. 6B), whereas cells exhibited very weak SecGFP fluorescence with an empty vector (Fig. 6A). This provided an excellent system to determine which SYP1sTI (Fig. 3) could rescue the AtTMS-OE defects, thus acting as potential AtTMS-regulated SYP1s in vivo.
Figure 6.
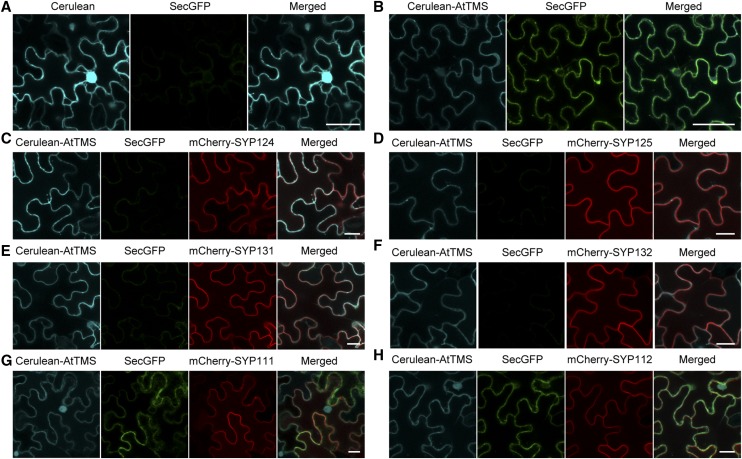
Intracellular SecGFP accumulation caused by AtTMS-OE is released by coexpression of SYP124/125/131/SYP132. A and B Overexpression of Cerulean-AtTMS (B), but not Cerulean alone (A), inhibits the trafficking of SecGFP to the apoplast of tobacco leaf epidermal cells. Cerulean fluorescent protein was used to create the Cerulean-AtTMS fusion protein. In A, Cerulean highlights nonspecifically the nucleus as a bright blue spot. C to H, Different combinations of plasmids were coinfiltrated into tobacco leaf epidermal cells and assayed after 72 h. Cerulean protein alone (A) or Cerulean-AtTMS fusion protein (B–H) are presented in cyan, SecGFP protein in green, and mCherry-SYP1s in red. Bars = 50 µm in A and B and 20 µm in C to H.
First, we showed that individual mCherry-SYP1sTI under their respective native regulatory sequence (Enami et al., 2009) imparted no inhibition on SecGFP secretion (Supplemental Fig. S5). Different mCherry-SYP1sTI were then coinfiltrated with SecGFP and Cerulean-AtTMS into the cells. Our results showed that SYP124, SYP125, SYP131, and SYP132 (Fig. 6, C–F), but not SYP111 (weakly interacting with AtTMS; Fig. 6G) or SYP112 (not interacting with AtTMS; Fig. 6H), rescued SecGFP secretion defects.
AtTMS Is Recruited to the PM by SYP124/125/131/132
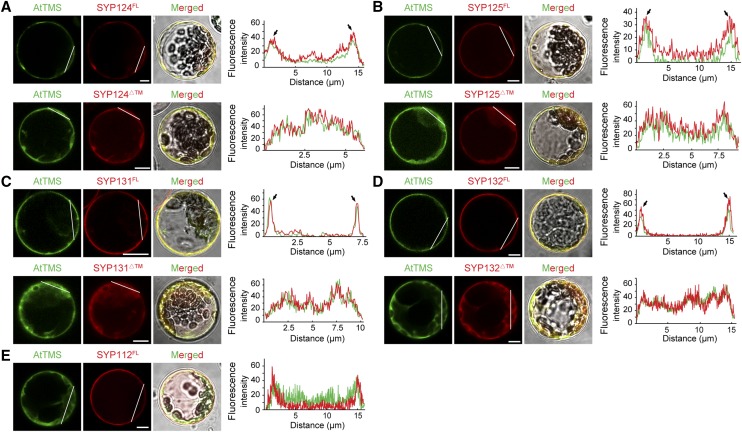
AtTMS bears no signal peptide and lacks a transmembrane domain (http://octopus.cbr.su.se/); thus, it might be recruited by PM-localized SYP1sTI (Uemura et al., 2004). Indeed, AtTMS displayed a sharp PM localization when coexpressed with full-length SYP124/125/131/132FL and a largely cytosolic pattern when coexpressed with SYP124/125/131/132ΔTM (Figs. 3A and 7, A–D–D). In addition, AtTMS could not be recruited by noninteracting SYP112 (Fig. 7E). These data suggested that AtTMS could be recruited to the PM specifically by SYP124/125/131/132.
Figure 7.
AtTMS is recruited to the PM by SYP124/125/131/132. A, AtTMS displayed a sharp PM localization when coexpressed with mCherry-SYP124FL (first row) and a largely cytosolic pattern with mCherry-SYP124ΔTM (second row). At far right for each row, the fluorescence intensity along the white lines is measured by a plot profile plugin for ImageJ. PM localization is indicated by arrows. B to E, AtTMS could be recruited by SYP125 (B), SYP131 (C), and SYP132 (D) but not by SYP112 (E). Bars = 5 µm.
AtTMS Competes with VAMP721/722 for the Binding to SYP132, and SYP132 Partially Rescues Pollen Defects in pMSP1:AtTMS Plants
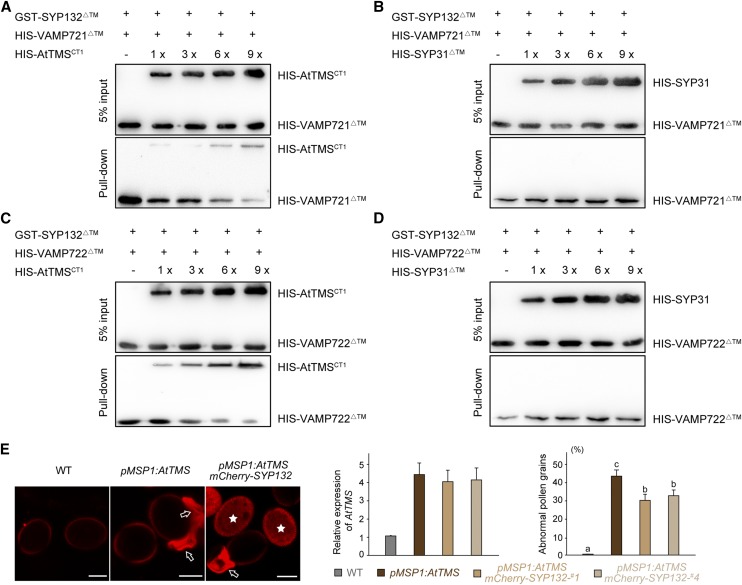
As AtTMS and R-SNAREs VAMP721/722 all bind to the Qa domain of SYP132 (Fig. 3; Park et al., 2018), we wanted to know if AtTMS and VAMP721/722 bind to SYP132 sequentially or if their association with SYP132 is mutually exclusive. For this purpose, equal amounts of GST-SYP132ΔTM immobilized on GST-Bind resin were incubated with His-VAMP721ΔTM (∼30 kD) in the presence of increasing concentrations of HIS-AtTMSCT1 (amino acids 793–1124, including an R-SNARE motif, ∼ 46 kD) or HIS-SYP31ΔTM (∼41 kD), which is a Golgi-resident Qa-SNARE. As shown in Figure 8, A and B, the interaction between VAMP721ΔTM and SYP132ΔTM was substantially significantly reduced in the presence of increasing amounts of HIS-AtTMSCT1 but not HIS-SYP31ΔTM. Therefore, AtTMS and VAMP721 specifically compete for the binding site in SYP132. Moreover, the binding of AtTMS and VAMP722 to SYP132 is mutually exclusive as well (Fig. 8, C and D).
Figure 8.
AtTMS competes with VAMP721/722 for the binding to SYP132, SYP132 partially rescues defects of pMSP1:AtTMS pollen. A and B, SYP132 partially rescues defects of pMSP1:AtTMS pollen (A), and AtTMSCT1 competes with VAMP721ΔTM for binding to SYP132ΔTM (B). In each lane, 6 µg of GST-SYP132ΔTM proteins coupled to beads was incubated with 2 µg of VAMP721ΔTM and increasing amounts (1, 3, 6, and 9 µg) of HIS-AtTMSCT1 (A) or 2 µg of VAMP721ΔTM with increasing amounts of Golgi resident Qa-SNARE HIS-SYP31ΔTM (B). After washing, bound proteins were detected using anti-His antibodies. C and D The same method used in A and B was adopted. Similarly, AtTMSCT1 was shown to compete with VAMP722ΔTM for the binding to SYP132ΔTM. E, Confocal images of pollen grains from the wild type (WT), pMSP1:AtTMS, and pMSP1:AtTMS + pSYP132:mCherry-SYP132. Arrows and asterisks indicate collapsed pollen and mCherry-SYP132-expressing pollen, respectively. Note that collapsed pollen grains display specific red fluorescence. In the left-hand graph, mRNA used for RT-qPCR was extracted from flower buds, and AtTMS expression relative to that of the wild type is presented. In the right-hand graph, one pMSP1::AtTMS line showed approximately 45% abnormal pollen grains, whereas two lines of pSYP132::mCherry-SYP132 + pMSP1::AtTMS showed about 30% abnormal pollen grains. n > 200 pollen was counted for each line. Values represent means ± sd of three independent experiments. Statistical significance using Duncan’s ANOVA is indicated by lowercase letters (P < 0.05). Bars = 10 μm.
The syp124 syp125 syp131 triple mutant showed normal male gametogenesis but pollen tube growth arrest (Slane et al., 2017). Therefore, we choose SYP132 among SYP124/125/131/132 to see whether SYP132 could rescue the AtTMS-OE defects in Arabidopsis. Two independent transgenic lines bearing pSYP132:mCherry-SYP132 and pMSP1:AtTMS were generated by crossing, and we found that SYP132 could reduce the amount of defective pollen by about 15%, thereby partially rescuing the AtTMS-OE phenotype (Fig. 8E).
Dexamethasone-Induced AtTMS Expression Causes a Pleiotropic Phenotype Related to Secretion Blockage
To see the possible effects of AtTMS-OE in sporophytic growth, plants expressing AtTMS under the control of the dexamethasone (DEX)-inducible promoter were generated. Upon DEX treatment, AtTMS-OE seedlings became dwarfed with short root hairs. Root cells showed cytokinesis defects with multiple nuclei and cell wall stubs, and JIM7 antigen secretion was blocked in the root hairs (Supplemental Fig. S6). These phenotypes suggested a negative role of AtTMS in secretory vesicle fusion during vegetative growth as well.
DISCUSSION
AtTMS-OE Inhibits Exocytosis in Developing Pollen
AtTMS is a TMS homolog that contains the highly conserved R-SNARE motif (Fig. 1). In this study, the physiological function of AtTMS in plant development was explored. Overexpression of AtTMS driven by three different promoters, pUBQ10, pMSP1, and pLAT52, caused a high percentage of shriveled mature pollen. Notably, the ratio of abnormal pollen is positively correlated with the AtTMS expression level in all three independent transgenic events (Fig. 4). The abnormal pollen development was only detectable from the bicellular stage onward, when the endogenous AtTMS starts expressing.
Pollen development involves intensive secretion that is required for intine development and cytokinesis during PMI (Shi et al., 2015). In pMSP1:AtTMS bicellular pollen, the highly methylesterified homogalacturonan was retained inside the cell (Fig. 5) and the intine structures were not detected. Instead, large amounts of irregular membranes accumulated beneath the PM (Fig. 5); meanwhile, the expansion of the cell plate during PMI, a redirected secretion event (Jürgens et al., 2015), was affected (Fig. 5). These data implied that AtTMS plays a predominantly inhibitory role in secretory trafficking in developing pollen.
The attms mutants have no detectable phenotype (Supplemental Fig. S2). There is an AtTMS-like (AtTMSL) protein in Arabidopsis that bears an R-SNARE-like motif without the conserved Arg residue. It will be interesting to explore the attmsl mutants in the future.
AtTMS Interacts with Pollen-Expressed PM-Localized SYP1sTI, and Four SYP1sTI Could Rescue AtTMS-OE Secretion Defects
In this study, we found several AtTMS binding partners. AtTMS binds to pollen-expressed PM-localized SYP1sTI, which is consistent with its subcellular localization (Figs. 2 and 3). These interactions are highly specific, since AtTMS did not interact with non-pollen-expressed SYP112/121/122/123 (Fig. 3), implying a specific, endogenous role of AtTMS in pollen development.
SYP124/125 were first detected during PMI, while SYP131 was detected at advanced stages of floral development (Ichikawa et al., 2015). Although the syp124 syp125 syp131 triple mutant showed normal male gametogenesis (Slane et al., 2017), they might have an overlapping function with SYP132, which displayed a clear PM localization in many tissues, including developing pollen (Enami et al., 2009; Karnahl et al., 2018). In agreement with this, overexpressing SYP124, SYP125, SYP131, or SYP132 rescued SecGFP trafficking defects caused by AtTMS-OE (Fig. 6). In addition, SYP132 transgene could partially rescue the pMSP1:AtTMS pollen phenotype (Fig. 8). Although SYP111 localizes to the PMI cell plate and maybe participates in cell plate expansion (Lee et al., 2007; Slane et al., 2017), it only weakly interacted with AtTMS (Fig. 3), and SYP111 could not rescue SecGFP defects (Fig. 6). Therefore, in pollen, AtTMS-OE inhibits secretion probably through binding to the SNARE domain of SYP124/125/131/132.
Brefeldin A (BFA) blocks the functions of BFA-sensitive ARF-GEFs, and this aggregates Golgi and post-Golgi compartments, such as the TGN and endosomes (Dettmer et al., 2006). SYP124/125 mainly localized to the BFA compartment, suggesting that SYP124/125 functions in both recycling and secretion (Slane et al., 2017), whereas SYP131/132 mainly serves the secretion of newly synthesized proteins (Karnahl et al., 2018; Park et al., 2018). As AtTMS also presents at the TGN, AtTMS might be able to inhibit the recycling pathway as well during pollen development.
DEX-inducible AtTMS-OE seedlings display a range of phenotypes that could be attributed to secretion defects, suggesting that AtTMS plays a similar role in vegetative growth.
AtTMS Might Participate in the Formation of Nonfusogenic Complexes
Animal TMS contains an R-SNARE motif that shares sequence similarity with that of the R-SNARE synaptobrevin and has been shown to bind Syntaxin and SNAP-25, forming a SNARE-like complex precluding synaptobrevin (Ashery et al., 2009). Our data suggested that a similar model of AtTMS action might hold true in plants. AtTMS lacks a membrane anchor and cannot act as a true, fusion-active R-SNARE. In the AtTMS-OE plants, each pair of AtTMS-SYP1 (AtTMS with SYP124, SYP125, SYP131, or SYP132) is nonfusogenic, preventing genuine R-SNAREs (VAMP721 or VAMP722) from entering the SNARE complexes whereby the secretion events are blocked, resulting in defective pollen development. Therefore, AtTMS might serve to fine-tune the active secretion events in pollen development.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was used in the experiments. Seeds were surface sterilized for 1 min in 70% (v/v) ethanol, followed by 10 min of incubation in 2% (v/v) NaClO with occasional mixing, and washed five times with sterile distilled water. Seeds were germinated and grown on Murashige and Skoog plates containing 1% (w/v) agar. AtTMS-OE seeds were germinated on the same medium supplemented with 2.5 × 10−2 mg mL−1 hygromycin. After 7 d, the seedlings were transferred to soil and grown at 22°C with a 16-h/8-h light/dark photoperiod in a growth room. Tobacco plants (Nicotiana benthamiana) were grown at 25°C with a 16-h/8-h light/dark photoperiod in a growth chamber.
Phylogenetic Analysis
Homologs of Arabidopsis AtTMS were identified using the BLASTP search program of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). The phylogenetic tree was constructed using MEGA 7.0 (http://www.megasoftware.net), based on the neighbor-joining method with the following parameters: p-distance model, pairwise deletion, and bootstrap (1,000 replicates; random seed).
Subcellular Localization Assay in Arabidopsis Protoplasts
To study the subcellular localization of AtTMS protein, two transient expression systems were used. First, the coding sequence of AtTMS was cloned into pAN580 to create p35S:eGFP-AtTMS, which was transformed into Arabidopsis protoplasts with or without fluorescent markers. Second, the coding sequence of AtTMS was cloned into pCAMBIA1305 to obtain a pUBQ10:eGFP-AtTMS construct, which was introduced into Agrobacterium tumefaciens (EHA105), infiltrated into tobacco leaves, and observed after 72 h (Wu et al., 2017). To study the recruitment of AtTMS by various SYPs, the coding sequences of SYP112/124/125/131/132 and SYP124/125/131/132ΔTM were cloned into pAN583 vector to obtain N-terminal mCherry fusion constructs. Pairs of constructs were transformed into Arabidopsis protoplasts for observation.
Yeast Two-Hybrid Assay
Yeast-two hybrid analysis was performed using the Matchmaker GAL4 Two-Hybrid System according to the supplier’s instructions (Clontech) and was performed as described previously (Tan et al., 2016). In the assay, the open reading frame of AtTMS were subcloned into pGADT7, while the Qa domain of SYP111/112/121/122/123/124/125/131/132/41/42/43 was cloned into pGBKT7.
Firefly LCI Assay
For the firefly LCI assay, the open reading frame, NT, and CT of AtTMS were cloned into vector pCAMBIA1300-cLuc to generate cLUC-AtTMS. Meanwhile, the cytoplasmic fragments of SYP111/112/121/122/123/124/125/131/SYP132//41//42/43 were ligated into pCAMBIA1300-nLUC to generate SYPsΔTM-nLUC. The firefly LCI assay was performed as previously described (Chen et al., 2008; Wu et al., 2017).
GST Pull-Down Assay
The C terminus of AtTMS containing the R-SNARE motif was cloned into pET28a to generate HIS-AtTMS-CT (2,766–3,372 bp). The cytoplasmic portions of SYP/112/121/122/123/124/125/131/132/41/42/43 were ligated into pGEX4T-1 to generate GST-SYPsΔTM. The GST pull-down assays were performed as previously described (Wu et al., 2013).
Generation of AtTMS-OE and Knockout Lines
To overexpress the AtTMS gene, the coding sequence of AtTMS was cloned into pCAMBIA1300 (which contained a UBQ10, MSP1, or Lat52 promoter) at the XbaI site to generate pUBQ10:AtTMS, MSP1:AtTMS, or Lat52:AtTMS, and the coding sequence of AtTMS was also cloned into pTA7002 (which contained a DEX-inducible promoter) at XhoI and SpeI. To create AtTMS knockout mutants, an RNA targeting the sequence within the eighth exon (ATCTATGGTGGTGATATCAT) was cloned into the pAtU6-26:sgRNA-2×p35S:Cas9 pBlunt vector. The resulting constructs were transformed into Columbia-0 using floral dipping (Clough and Bent, 1998).
RT-qPCR Analysis
Total RNA was isolated using the RNA prep pure plant kit (Tiangen). The first-strand cDNA was synthesized using oligo(dT)18 as the primer and PrimeScript reverse transcriptase (TaKaRa). Arabidopsis gene Tubulinß8 was used as an internal control. Quantitative analysis was performed using an ABI 7500 real-time qPCR system with the SYBR Green Mix (Bio-Rad) and three biological repeats (Tan et al., 2016).
Phenotype Characterization in Pollen
To visualize nuclei, pollen grains were collected into a DAPI staining solution (0.1 m sodium phosphate, pH 7, 1 mm EDTA, 0.1% [v/v] Triton X-100, and 0.5 mg mL−1 DAPI) and examined 15 min after staining. Pollen viability was examined using Alexander staining (Tan et al., 2016). Cell plate and deposited callose in pollen were revealed using Aniline Blue staining. To observe cellulose deposition, anthers were fixed in a formalin-acetic acid-alcohol solution overnight, embedded, and sectioned to 1-µm-thick slices with a Leica EM UC7 microtome. After 10 mg mL−1 propidium iodide and 35 mg mL−1 Calcofluor White staining, the sections were observed using confocal microscopy (LSM780; Zeiss). To observe JIM7 antigen distribution, wild-type and pMSP1:AtTMS anthers were fixed in 0.1 m phosphate buffer and embedded in LR White resin. Sections (1 µm thick) were blocked in a buffer (3% [w/v] BSA in phosphate-buffered saline, 4.3 mm dibasic sodium phosphate, 1.4 mm potassium phosphate monobasic, 138 mm NaCl, and 2.7 mm KCl, pH 7.3) for 30 min, washed with phosphate-buffered saline, and incubated with the JIM7 antibodies (Carbo Souse) for 1 h. After washing, the sections were labeled with Alexa-488 conjugated secondary antibodies (Life Technologies; 1322327) for 30 min before analysis.
GUS Staining
Histochemical staining for GUS expression in transgenic plant materials was performed according to the method of Tan et al. (2016).
Transient Expression in Tobacco Leaf Epidermal Cells
Each construct for transient expression was transformed into A. tumefaciens. For subcellular localization studies, individual bacterial strains (OD600 = 0.1) bearing AtTMS or various fluorescent markers were infiltrated together into tobacco leaf epidermal cells. For functional studies, different combinations of an A. tumefaciens strain bearing Cerulean-AtTMS, SecGFP, Spo-mCherry, mCherry-SYP111, mCherry-SYP124, mCherry-SYP125, mCherry-SYP131, mCherry-SYP132, and mCherry-SYP112 constructs were infiltrated. The OD600 of bacteria used for these infiltrations was 0.05 for SecGFP, 0.5 for Cerulean-AtTMS, mCherry-SYP131, mCherry-SYP132, and mCherry-SYP112, and 0.1 for Spo-mCherry, 0.08 for mCherry-SYP111, mCherry-SYP124, and mCherry-SYP125. After 72 h of infiltration, the leaves were observed using confocal microscopy.
Scanning Electron Microscopy and TEM
For scanning electron microscopy observation, mature pollen grains were coated with gold particles (EIKOIB-3) and observed using a Hitachi S-2000N scanning electron microscope. For TEM observation, anthers were immediately frozen in a high-pressure freezer (EM HPM100; Leica), followed by subsequent freeze substitution in 100% (v/v) acetone containing 0.1% (w/v) uranyl acetate at −85°C in an AFS freeze substitution unit (Leica), and substituted with 2% (w/v) OsO4 in 100% acetone and infiltrated with Epon resin as previously described (Tan et al., 2016). Sections were examined using a transmission electron microscope (H-7650; Hitachi) with a CCD camera (Hitachi High-Technologies) operating at 80 kV.
All primers used in this study are listed in Supplemental Table S1.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource under the following accession numbers: AtTMS (AT5G05570), SYP111/KNOLLE (AT1G12360), SYP112 (AT2G18260), SYP121 (AT3G11820), SYP122 (AT3G52400), SYP123 (AT4G03330), SYP124 (AT1G61290), SYP125 (AT1G11250), SYP131 (AT3G03800), SYP132 (AT5G08080), SYP41 (AT5G26980), SYP42 (AT4G02195), SYP43 (AT3G05710), and AtTMSL (AT4G35560).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Localization of AtTMS in tobacco leaf epidermal cells.
Supplemental Figure S2. Generation of attms mutants by CRISPR/Cas9-mediated gene editing.
Supplemental Figure S3. Expression pattern of AtTMS:GUS in Arabidopsis.
Supplemental Figure S4. Overexpression of Cerulean-AtTMS does not inhibit vacuolar trafficking of sporamin in tobacco leaf epidermal cells.
Supplemental Figure S5. mCherry-tagged SYP111, SYP124, SYP125, SYP131, SYP132, and SYP112 alone under their respective promoters do not affect SecGFP secretion in tobacco leaf epidermal cells.
Supplemental Figure S6. Overexpression of AtTMS under the control of a DEX-inducible promoter affected vegetative growth of Arabidopsis.
Supplemental Table S1. List of primers used in this study.
Supplemental Data Set S1. Text file of sequence alignment used for the phylogenetic analysis in Figure 1A.
ACKNOWLEDGMENTS
We thank Dr. Liwen Jiang (Chinese University of Hong Kong) for providing subcellular compartment markers, Dr. Huanquan Zheng (McGill University) for the SecGFP marker, and Dr. Jianmin Zhou (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for the pCAMBIA1200-nLuc and pCAMBIA1200-cLuc vectors.
Footnotes
This work was supported by the National Science Foundation of China (31770202) and the Ministry of Agriculture of China for Transgenic Research (2018ZX08009-20B).
References
- Ashery U, Bielopolski N, Barak B, Yizhar O (2009) Friends and foes in synaptic transmission: The role of tomosyn in vesicle priming. Trends Neurosci 32: 275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backues SK, Korasick DA, Heese A, Bednarek SY (2010) The Arabidopsis dynamin-related protein2 family is essential for gametophyte development. Plant Cell 22: 3218–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K (2006) Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kasmi F, Pacher T, Strompen G, Stierhof YD, Müller LM, Koncz C, Mayer U, Jürgens G (2011) Arabidopsis SNARE protein SEC22 is essential for gametophyte development and maintenance of Golgi-stack integrity. Plant J 66: 268–279 [DOI] [PubMed] [Google Scholar]
- Enami K, Ichikawa M, Uemura T, Kutsuna N, Hasezawa S, Nakagawa T, Nakano A, Sato MH (2009) Differential expression control and polarized distribution of plasma membrane-resident SYP1 SNAREs in Arabidopsis thaliana. Plant Cell Physiol 50: 280–289 [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Sutton RB, Brunger AT, Jahn R (1998) Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA 95: 15781–15786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Shirataki H, Sakisaka T, Asakura T, Ohya T, Kotani H, Yokoyama S, Nishioka H, Matsuura Y, Mizoguchi A, et al. (1998) Tomosyn: A syntaxin-1-binding protein that forms a novel complex in the neurotransmitter release process. Neuron 20: 905–915 [DOI] [PubMed] [Google Scholar]
- Gladycheva SE, Lam AD, Liu J, D’Andrea-Merrins M, Yizhar O, Lentz SI, Ashery U, Ernst SA, Stuenkel EL (2007) Receptor-mediated regulation of tomosyn-syntaxin 1A interactions in bovine adrenal chromaffin cells. J Biol Chem 282: 22887–22899 [DOI] [PubMed] [Google Scholar]
- Gracheva EO, Burdina AO, Touroutine D, Berthelot-Grosjean M, Parekh H, Richmond JE (2007) Tomosyn negatively regulates both synaptic transmitter and neuropeptide release at the C. elegans neuromuscular junction. J Physiol 585: 705–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, McCubbin AG (2012) The pollen-specific R-SNARE/longin PiVAMP726 mediates fusion of endo- and exocytic compartments in pollen tube tip growth. J Exp Bot 63: 3083–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsuzawa K, Lang T, Fasshauer D, Bruns D, Jahn R (2003) The R-SNARE motif of tomosyn forms SNARE core complexes with syntaxin 1 and SNAP-25 and down-regulates exocytosis. J Biol Chem 278: 31159–31166 [DOI] [PubMed] [Google Scholar]
- Honys D, Oh SA, Renák D, Donders M, Solcová B, Johnson JA, Boudová R, Twell D (2006) Identification of microspore-active promoters that allow targeted manipulation of gene expression at early stages of microgametogenesis in Arabidopsis. BMC Plant Biol 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S, Wang X, Chen D, Yang X, Wang M, Turrà D, Di Pietro A, Zhang W (2014) The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7. PLoS Pathog 10: e1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa M, Iwano M, Sato MH (2015) Nuclear membrane localization during pollen development and apex-focused polarity establishment of SYP124/125 during pollen germination in Arabidopsis thaliana. Plant Reprod 28: 143–151 [DOI] [PubMed] [Google Scholar]
- Jahn R, Scheller RH (2006) SNAREs: Engines for membrane fusion. Nat Rev Mol Cell Biol 7: 631–643 [DOI] [PubMed] [Google Scholar]
- Jürgens G. (2005) Plant cytokinesis: Fission by fusion. Trends Cell Biol 15: 277–283 [DOI] [PubMed] [Google Scholar]
- Jürgens G, Park M, Richter S, Touihri S, Krause C, El Kasmi F, Mayer U (2015) Plant cytokinesis: A tale of membrane traffic and fusion. Biochem Soc Trans 43: 73–78 [DOI] [PubMed] [Google Scholar]
- Karnahl M, Park M, Krause C, Hiller U, Mayer U, Stierhof YD, Jürgens G (2018) Functional diversification of Arabidopsis SEC1-related SM proteins in cytokinetic and secretory membrane fusion. Proc Natl Acad Sci USA 115: 6309–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N, He H, Steger AP (2010) A systems model of vesicle trafficking in Arabidopsis pollen tubes. Plant Physiol 152: 590–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SK, Siu CL, Hillmer S, Jang S, An G, Robinson DG, Jiang L (2007) Rice SCAMP1 defines clathrin-coated, trans-Golgi-located tubular-vesicular structures as an early endosome in tobacco BY-2 cells. Plant Cell 19: 296–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YR, Li Y, Liu B (2007) Two Arabidopsis phragmoplast-associated kinesins play a critical role in cytokinesis during male gametogenesis. Plant Cell 19: 2595–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tan X, Wang M, Li B, Zhao Y, Wu C, Rui Q, Wang J, Liu Z, Bao Y (2017) Exocyst subunit SEC3A marks the germination site and is essential for pollen germination in Arabidopsis thaliana. Sci Rep 7: 40279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Yan PK, Kim H, Hwang I, Jiang L (2006) Localization of green fluorescent protein fusions with the seven Arabidopsis vacuolar sorting receptors to prevacuolar compartments in tobacco BY-2 cells. Plant Physiol 142: 945–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris SR, Meyer SE, Callis J (1993) The intron of Arabidopsis thaliana polyubiquitin genes is conserved in location and is a quantitative determinant of chimeric gene expression. Plant Mol Biol 21: 895–906 [DOI] [PubMed] [Google Scholar]
- Park M, Krause C, Karnahl M, Reichardt I, El Kasmi F, Mayer U, Stierhof YD, Hiller U, Strompen G, Bayer M, et al. (2018) Concerted action of evolutionarily ancient and novel SNARE complexes in flowering-plant cytokinesis. Dev Cell 44: 500–511.e4 [DOI] [PubMed] [Google Scholar]
- Park M, Touihri S, Müller I, Mayer U, Jürgens G (2012) Sec1/Munc18 protein stabilizes fusion-competent syntaxin for membrane fusion in Arabidopsis cytokinesis. Dev Cell 22: 989–1000 [DOI] [PubMed] [Google Scholar]
- Pobbati AV, Razeto A, Böddener M, Becker S, Fasshauer D (2004) Structural basis for the inhibitory role of tomosyn in exocytosis. J Biol Chem 279: 47192–47200 [DOI] [PubMed] [Google Scholar]
- Sakisaka T, Yamamoto Y, Mochida S, Nakamura M, Nishikawa K, Ishizaki H, Okamoto-Tanaka M, Miyoshi J, Fujiyoshi Y, Manabe T, et al. (2008) Dual inhibition of SNARE complex formation by tomosyn ensures controlled neurotransmitter release. J Cell Biol 183: 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Cui M, Yang L, Kim YJ, Zhang D (2015) Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci 20: 741–753 [DOI] [PubMed] [Google Scholar]
- Slane D, Reichardt I, El Kasmi F, Bayer M, Jürgens G (2017) Evolutionarily diverse SYP1 Qa-SNAREs jointly sustain pollen tube growth in Arabidopsis. Plant J 92: 375–385 [DOI] [PubMed] [Google Scholar]
- Tan X, Cao K, Liu F, Li Y, Li P, Gao C, Ding Y, Lan Z, Shi Z, Rui Q, et al. (2016) Arabidopsis COG complex subunits COG3 and COG8 modulate Golgi morphology, vesicle trafficking homeostasis and are essential for pollen tube growth. PLoS Genet 12: e1006140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toonen RFG, Verhage M (2003) Vesicle trafficking: Pleasure and pain from SM genes. Trends Cell Biol 13: 177–186 [DOI] [PubMed] [Google Scholar]
- Tse YC, Mo B, Hillmer S, Zhao M, Lo SW, Robinson DG, Jiang L (2004) Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16: 672–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell D. (2011) Male gametogenesis and germline specification in flowering plants. Sex Plant Reprod 24: 149–160 [DOI] [PubMed] [Google Scholar]
- Twell D, Wing R, Yamaguchi J, McCormick S (1989) Isolation and expression of an anther-specific gene from tomato. Mol Gen Genet 217: 240–245 [DOI] [PubMed] [Google Scholar]
- Uemura T, Kim H, Saito C, Ebine K, Ueda T, Schulze-Lefert P, Nakano A (2012) Qa-SNAREs localized to the trans-Golgi network regulate multiple transport pathways and extracellular disease resistance in plants. Proc Natl Acad Sci USA 109: 1784–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH (2004) Systematic analysis of SNARE molecules in Arabidopsis: Dissection of the post-Golgi network in plant cells. Cell Struct Funct 29: 49–65 [DOI] [PubMed] [Google Scholar]
- Widberg CH, Bryant NJ, Girotti M, Rea S, James DE (2003) Tomosyn interacts with the t-SNAREs syntaxin4 and SNAP23 and plays a role in insulin-stimulated GLUT4 translocation. J Biol Chem 278: 35093–35101 [DOI] [PubMed] [Google Scholar]
- Wu C, Tan L, van Hooren M, Tan X, Liu F, Li Y, Zhao Y, Li B, Rui Q, Munnik T, et al. (2017) Arabidopsis EXO70A1 recruits Patellin3 to the cell membrane independent of its role as an exocyst subunit. J Integr Plant Biol 59: 851–865 [DOI] [PubMed] [Google Scholar]
- Wu J, Tan X, Wu C, Cao K, Li Y, Bao Y (2013) Regulation of cytokinesis by exocyst subunit SEC6 and KEULE in Arabidopsis thaliana. Mol Plant 6: 1863–1876 [DOI] [PubMed] [Google Scholar]
- Yao X, Tian L, Yang J, Zhao YN, Zhu YX, Dai X, Zhao Y, Yang ZN (2018) Auxin production in diploid microsporocytes is necessary and sufficient for early stages of pollen development. PLoS Genet 14: e1007397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O, Lipstein N, Gladycheva SE, Matti U, Ernst SA, Rettig J, Stuenkel EL, Ashery U (2007) Multiple functional domains are involved in tomosyn regulation of exocytosis. J Neurochem 103: 604–616 [DOI] [PubMed] [Google Scholar]
- Yizhar O, Matti U, Melamed R, Hagalili Y, Bruns D, Rettig J, Ashery U (2004) Tomosyn inhibits priming of large dense-core vesicles in a calcium-dependent manner. Proc Natl Acad Sci USA 101: 2578–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Camacho L, Wee E, Batoko H, Legen J, Leaver CJ, Malhó R, Hussey PJ, Moore I (2005) A Rab-E GTPase mutant acts downstream of the Rab-D subclass in biosynthetic membrane traffic to the plasma membrane in tobacco leaf epidermis. Plant Cell 17: 2020–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]