Based on transcriptome analysis, the WRKY1 transcription factor simultaneously mediates regulation of nitrogen and light signaling pathways in a potential energy conservation mechanism.
Abstract
Plant responses to multiple environmental stimuli must be integrated to enable them to adapt their metabolism and development. Light and nitrogen (N) are two such stimuli whose downstream signaling pathways must be intimately connected to each other to control plant energy status. Here, we describe the functional role of the WRKY1 transcription factor in controlling genome-wide transcriptional reprogramming of Arabidopsis (Arabidopsis thaliana) leaves in response to individual and combined light and N signals. This includes a cross-regulatory network consisting of 724 genes regulated by WRKY1 and involved in both N and light signaling pathways. The loss of WRKY1 gene function has marked effects on the light and N response of genes involved in N uptake and assimilation (primary metabolism) as well as stress response pathways (secondary metabolism). Our results at the transcriptome and at the metabolite analysis level support a model in which WRKY1 enables plants to activate genes involved in the recycling of cellular carbon resources when light is limiting but N is abundant and upregulate amino acid metabolism when both light and N are limiting. In this potential energy conservation mechanism, WRKY1 integrates information about cellular N and light energy resources to trigger changes in plant metabolism.
Plants perceive multiple stimuli and dynamically respond to complex environmental challenges to survive. Responses to stimuli or stresses occur via signal transduction pathways that initiate transcriptional responses. Signal response pathways do not often act alone, but instead interact with other signaling pathways within a cell or tissue, ultimately resulting in emergent properties in the underlying gene regulatory networks (GRNs). These pathways are likely connected via integrator molecules that mediate some common effects (Seo and Park, 2010; Matiolli et al., 2011; Chen et al., 2013). Light and nitrogen (N) signaling pathways are closely connected (Reed et al., 1983; Riens and Heldt, 1992; Oliveira and Coruzzi, 1999; Oliveira et al., 2001; Chen et al., 2016), and increasing evidence supports the notion that the transcriptional cross talk between light and N signaling pathways enables plants to fine-tune plant energy status (Jonassen et al., 2008; Krouk et al., 2009; Nunes-Nesi et al., 2010; Obertello et al., 2010). Indeed, this coordinated regulation is physiologically important, as nitrogen assimilation is dependent on reducing power and carbon (C) skeletons derived from photosynthesis, while the photosynthetic apparatus is dependent on N availability to support the formation of chlorophyll and other components necessary for biomass accumulation (Matt et al., 2001a, 2001b; Bläsing et al., 2005; Lillo, 2008).
Knowledge about the regulation of genes common to light and N signaling pathways by transcription factors (TFs) is limited. Whole transcriptome analysis of the Arabidopsis (Arabidopsis thaliana) response to combinations of C, N, and light treatments revealed a change in expression of several genes, including a few known TFs (Krouk et al., 2009). That previous study also revealed that 35% of the genome is controlled by one of either light or N signals or their combination (Krouk et al., 2009). A handful of components shared between the light and N signal transduction pathways have been identified by studying the Arabidopsis basic Leu-zipper (bZIP) TFs, including bZIP1 (Obertello et al., 2010) and ELONGATED HYPOCOTYL 5/HY5-HOMOLOG (HY5/HYH; Jonassen et al., 2008). Genome-wide analysis of bzip1 mutant seedlings revealed 33 genes with a significant interaction term for genotype (G), light, and N treatments, indicating that bZIP1 regulates a small group of genes involved in both light and N sensing (Obertello et al., 2010). HY5 and HYH were essential for light-activated/phytochrome-mediated expression of nitrate reductase (Jonassen et al., 2008), in which the enhancement of NIA2 expression by light is dependent on HY5/HYH (Jonassen et al., 2009). It was also shown that HY5 is a shoot-to-root mobile TF that mediates the light-activated N uptake by inducing expression of the NO3− transporter NRT2.1 (Chen et al., 2016). Thus, despite the many and varied interactions between N and light signaling pathways, to our knowledge, only TFs from the bZIP gene family have been experimentally validated to common regulatory N and light signal transduction pathways to date.
In our previous work on Arabidopsis gene regulation by N status, we used network analysis to predict regulatory connections between genes and associated TFs (Gutiérrez et al., 2008). In that study, our networks identified several TFs involved in either positive or negative regulation of organic N metabolism and catabolism. Three regulatory hubs of an organic N regulatory network identified were CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), GOLDEN2-LIKE 1 (GLK1), and bZIP1 (Gutiérrez et al., 2008), each of which have been implicated in N and/or light signaling pathways (Wang and Tobin, 1998; Waters et al., 2009; Obertello et al., 2010; Dietrich et al., 2011; Maekawa et al., 2015). Indeed, independent experiments revealed that CCA1, GLK1 (Gutiérrez et al., 2008), and bZIP1 (Baena-González et al., 2008; Gutiérrez et al., 2008; Obertello et al., 2010; Dietrich et al., 2011; Para et al., 2014) are involved in the regulation of genes in response to N and/or light signals. Our subsequent expanded network analysis of the N-regulated genes described in Gutiérrez et al. (2008) revealed an additional TF hub, WRKY1, which was also previously shown to be regulated by light (Krouk et al., 2009) and is the focus of our current study.
WRKY1 is a member of a family of TFs that have diverse regulatory functions in response to biotic and abiotic stresses (Wei et al., 2008; Jia et al., 2015). WRKY TFs activate or repress transcription and in some instances have dual activator/repressor functions (e.g. rice [Oryza sativa] OsWRKY72 and OsWRKY77 activate abscisic acid [ABA] signaling and repress GA signaling; Xie et al., 2005). Recent studies show that AtWRKY1 plays a key role in the direct and indirect regulation of genes involved in ABA signaling and drought response (Qiao et al., 2016). Prior studies also showed that other WRKY family member TFs respond to and also regulate gene response to light signals, where AtWRKY22 is activated by light and repressed by dark (Nozue et al., 2011; Zhou et al., 2011), while AtWRKY40 and AtWRKY63 repress and activate, respectively, genes involved in high light signaling (Van Aken et al., 2013). Additionally, WRKY TFs have been implicated in nutrient deficiency response signaling pathways, where AtWRKY75 is induced by Pi starvation (Devaiah et al., 2007) and AtWRKY45 and AtWRKY65 are induced by C starvation (Contento et al., 2004). Likewise, previous studies in Arabidopsis (ecotype Columbia 0 [Col-0]) revealed that WRKY1 expression is repressed by organic N treatment (Gutiérrez et al., 2008) and induced by N starvation (Krapp et al., 2011). Here, we investigate the role of WRKY1 in coordinating responses to light and N signaling. We show that WRKY1 participates in genome-wide transcriptional reprogramming of Arabidopsis leaves in response to individual and combined light and N signals, and our metabolite studies support its potential role as an integrator of light and N signaling pathways toward the fine-tuning of plant energy status.
RESULTS
GRN Analysis Reveals WRKY1 Is a Hub in the N Assimilation Pathway
Our previous studies of N-regulatory networks in Arabidopsis identified a subnetwork of 367 connected nodes, including WRKY1 (Supplemental Data Set 1; Gutiérrez et al., 2008). In that initial N-regulatory network, protein-DNA interactions were predicted based on an overrepresentation of the regulatory motif for that TF, and the expression of the TF and putative target gene was highly (≥0.7 or ≤−0.7) and significantly (P ≤ 0.01) correlated (Gutiérrez et al., 2008). Subsequent chromatin immunoprecipitation analysis of a top hub (CCA1) in the network, in combination with bioinformatic cis-regulatory element (CRE)-motif analysis, revealed that the presence of a single binding site was sufficient for direct regulation of the target gene by the TF (Gutiérrez et al., 2008). Based on these experimental results, we reanalyzed the N-response data in Gutiérrez et al. (2008) by relaxing the predicted protein-DNA interaction to require a minimum of a single regulatory motif for the TF in the promoter of a putative target, rather than an overrepresentation of cis-binding sites for that TF. This resulted in an expanded N-regulatory subnetwork, which increased the number of regulatory edges from WRKY1 to putative target genes (Supplemental Data Set 1). In this expanded N-regulatory network, WRKY1 was one of the most highly connected TFs (eight edges) directly associated with metabolic genes involved in N assimilation, such as GLU DEHYDROGENASE 1 (GDH1), NITRATE REDUCTASE 1 (NIA1) and NIA2, and ASN SYNTHETASE 1 (ASN1). WRKY1 was predicted to activate GDH1, NIA1, and NIA2, and to repress ASN1 (Supplemental Fig. S1; Supplemental Table S1).
WRKY1 Target Genes Are Involved in Nitrogen and Light Signaling Pathways
As discussed above, our expanded network analysis shows WRKY1 is predicted to be a major hub of an organic N regulatory network (Gutiérrez et al., 2008) and to transcriptionally repress expression of ASN1, a gene regulated in response to light, N and C signaling (Thum et al., 2003). Here, we validate the involvement of WRKY1 as a hub of an N and light signaling network by exposing wrky1 mutant plants to N and light signals and performing genome-wide analysis. To this end, we compared three T-DNA alleles of WRKY1 (wrky1-1 [SALK_070989], wrky1-2 [SALK_016954], and wrky1-3 [SALK_136009]) to wild-type Col-0 (the genetic background of the mutants). The wrky1 mutant phenotype (SALK_016954) was previously described by Qiao et al. (2016). We performed RNA analysis and found that WRKY1 expression was altered in the three wrky1 transfer DNA (T-DNA) mutant alleles, ranging from below the level of detection in wrky1-1 to 2% and 24% of wild-type WRKY1 expression levels in wrky1-2 and wrky1-3, respectively (Supplemental Figs. S2–S4).
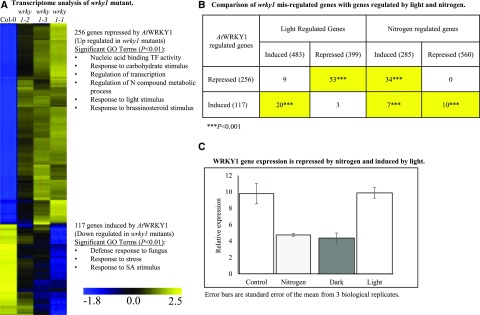
To determine the effect of the wrky1 T-DNA mutations on genome-wide expression, we grew wrky1 plants under “steady-state” conditions or in transient light and/or nitrogen treatments. For the steady-state experiments we grew the three wrky1 T-DNA alleles and Col-0 on basal Murashige and Skoog (MS) media under 16 h/8 h light/dark regime for 14 d. Shoot tissue was extracted for mRNA analysis by reverse-transcription quantitative PCR (RT-qPCR), and microarray analysis was performed with the Arabidopsis Genome ATH1 array (Affymetrix/Thermo Fisher Scientific) to identify genes misregulated in the wrky1 mutants using rank product (Breitling et al., 2004) statistical analysis (see “Materials and Methods”). We identified the “core set” of WRKY1-regulated genes as those that are misregulated in the most severe knockdown mutant, wrky1-1, and also either in wrky1-2 or wrky1-3. This analysis identified 256 genes upregulated and 117 genes downregulated in the wrky1 mutants (Fig. 1A; Supplemental Table S2). The 117 genes downregulated in the wrky1 mutants (i.e. genes induced by WRKY1) include a significant overrepresentation (P < 0.01) of genes enriched in gene ontology (GO) terms involved in secondary metabolic processes, such as defense response and response to stress (using the BioMaps function in VirtualPlant 1.3; Katari et al., 2010; Fig. 1A). This role of WRKY TFs has also been reported for several members of the WRKY family of TFs (Rushton et al., 2010; Agarwal et al., 2011; Chen et al., 2012; Qiao et al., 2016). By contrast, the 256 genes upregulated in the wrky1 mutants (i.e. genes repressed by WRKY1) were significantly enriched for GO terms involved in primary metabolic processes (P = 0.0001), response to carbohydrate stimulus (P = 2.3e-05), regulation of the nitrogen compound metabolic process (P = 4.9e-05), and response to light stimulus (P = 0.0003; Fig. 1A), revealing potential new regulatory roles for WRKY1 as a transcriptional repressor.
Figure 1.
Downregulation of WRKY1 results in misregulation of genes involved in light, nitrogen, and stress response pathways. A, Transcriptome analysis of wild-type (Col-0) and mutant wrky1-1 (SALK_070989), wrky1-2 (SALK_016954), and wrky1-3 (SALK_136009) seedlings. The heat map of transcriptome data includes genes with significant (P < 0.01; FDR 5%) change in expression from the wild type in wrky1-1, wrky1-2, or wrky1-3. The top significantly (P < 0.01) overrepresented GO terms are listed. B, Significance of overlaps (P < 0.001) of WRKY1-regulated, light-regulated (Nozue et al., 2011), and nitrogen-regulated (Gutiérrez et al., 2008) gene sets calculated using the GeneSect (R)script with the microarray as background. The total numbers of genes are in parentheses; the numbers of overlapping genes are shown in boxes. Boxes highlighted in yellow have P < 0.001, indicating that the size of the intersection is higher than expected. C, Relative expression levels of WRKY1 in wild-type (Col-0) seedlings in response to N (20 mm KCl in the control group, 20 mm NH4NO3 + 20 mm KNO3− in the treatment group) and light treatments (normal long day for the control group, extended dark for the treatment group). Error bars represent the mean ± se for three biological replicates.
To further explore the underlying mechanism, we searched for known CREs in the putative promoter regions (2 kb 5ʹ upstream of transcription start site sequences) of the genes misregulated in the wrky1 mutants. This cis analysis uncovered a statistical overrepresentation of the W-box promoter motif (e-value = 5.17e-05) among the 117 genes downregulated in the wrky1 mutants. Although the W-box motif was present in 245 of 256 promoters of genes upregulated in the wrky1 mutants (on average 4.4 W-box elements per promoter), it was not statistically overrepresented. Instead, the I-box (e-value = 1.15e-72), GATA (e-value = 1.23e-46), ABA response element-like (e-value = 3.27e-28), and G-box (e-value = 2.06e-22) motifs were the most statistically overrepresented among the 256 genes upregulated in the wrky1 mutants. It is possible that these genes are either indirect targets of WRKY1 or that WRKY1 is part of a TF complex that represses their expression in the wild type. Interestingly, many protein-protein interactions (PPIs) have been shown among WRKYs and other protein families (Chi et al., 2013), including 14-3-3 proteins. A search for PPIs involving WRKY1 using the Arabidopsis Interactions Viewer (Geisler-Lee et al., 2007) revealed an interaction (Proteomics Standard Initiative Common Query Interface confirmed by affinity chromatography) with the 14-3-3 protein GENERAL REGULATORY FACTOR 1 (GRF1; AT4G09000), which is a G-box factor whose native form is a heterodimer. GRF1 is expressed in both wild-type and wrky1 mutant plants; thus, one hypothesis these results imply is that WRKY1 interacts with GRF1 as a heterodimer to downregulate the expression of these genes in the wild type.
Investigation of the core genes involved in the nitrogen-assimilation pathway revealed that downregulation of WRKY1 expression in response to light affected the expression of genes involved in both N uptake and organic nitrogen metabolism and catabolism (Fig. 1A; Supplemental Table S2). In wild-type plants, the expression of genes encoding several nitrate transporters, as well as genes directly involved in Gln biosynthesis, are upregulated, while genes involved in Gln catabolism, such as ASN1 (ASN SYNTHETASE 1/DARK INDUCIBLE6), are downregulated during the light period. However, in wrky1 mutant plants, the nitrate transporters NRT1.7 and NRT3.1 and the Glu receptor GLR1.1 are downregulated, while ASN1 is upregulated in the light. Thus, these transcriptome results provide support for the predicted edge between WRKY1 and ASN1 from the network analysis described above (Supplemental Fig. S1). The observed overall reprogramming of the nitrogen network and the presence of the W-box motif in the promoter of N-related genes also support that WRKY1 is likely a regulatory molecule for this N-uptake/assimilation pathway.
We next investigated the involvement of WRKY1 in mediating temporal responses to light and N signals by intersecting the list of wrky1 misregulated genes with lists of genes previously identified as responsive to N treatments (Gutiérrez et al., 2008) or light treatments (Fig. 1B; Nozue et al., 2011). We found that genes normally repressed by WRKY1 in the wild type (i.e. the 256 genes upregulated in the wrky1 mutants) share a significant overlap with genes repressed by light treatments (P < 0.001; Fig. 1B). By contrast, genes induced by WRKY1 in the wild type (i.e. the 117 genes downregulated in the wrky1 mutant) share a significant overlap with genes induced by light treatments (P < 0.001; Fig. 1B). With regard to a potential role for WRKY1 in N signaling, we found that genes normally repressed by WRKY1 in the wild type (i.e. induced in the wrky1 mutant) are induced by N treatments (P < 0.001), while genes induced by WRKY1 (i.e. repressed in the wrky1 mutants) are either repressed or induced by N treatments (P < 0.001; Fig. 1B).
These reciprocal patterns of expression of genes regulated by WRKY1 in light and N treatment datasets support our hypothesis that WRKY1 is an integrator of light and N signaling pathways. This hypothesis is further supported by our finding that WRKY1 expression itself is independently and reciprocally regulated by light and N treatments. Specifically, WRKY1 expression is induced by light treatment and repressed by N treatment (Fig. 1C). To further investigate the regulatory role of WRKY1 in N and/or light signaling and the possible crosstalk, we exposed the wild type and wrky1 mutants to three treatments: (1) light treatment only; (2) N treatment only; and (3) combined N and light treatments, as described below (Table 1). Three separate treatments were performed, as opposed to a single combined treatment, to eliminate omitted-variable bias and accurately determine the role of WRKY1 in the independent light and N signaling pathways and in regulating crosstalk between pathways, since transcriptional changes in response to double abiotic stress treatments are not predictable from responses to single-stress treatments (Prasch and Sonnewald, 2013; Rasmussen et al., 2013).
Table 1. Experimental design for the different treatment groups.
L, Light only; N, nitrogen only; LN, combined light and nitrogen
| Experiment | Light | Nitrogen |
|---|---|---|
| L1 | + | + |
| L2 | − | + |
| N1 | + | − |
| N2 | + | + |
| LN1 | − | − |
| LN2 | − | + |
| LN3 | + | − |
| LN4 | + | + |
WRKY1 Mediates the Light Repression of Genes Involved in Organic Resource Catabolism
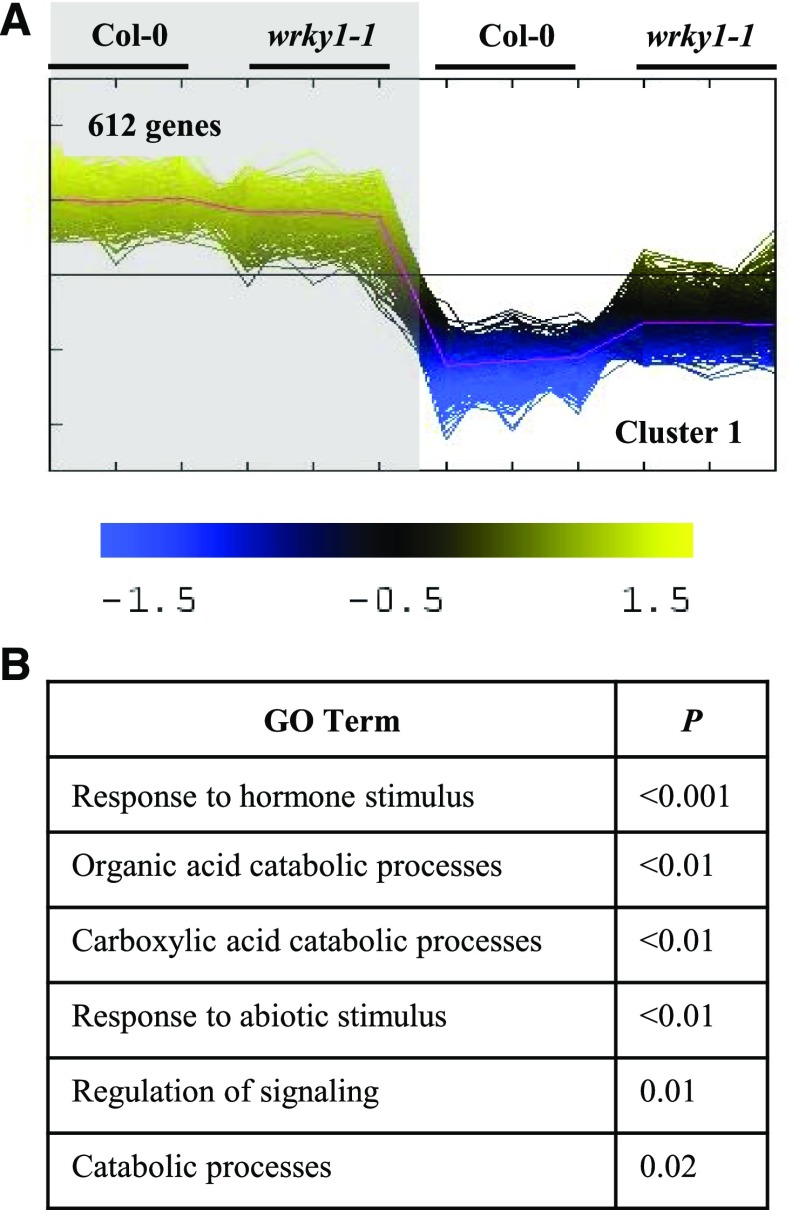
Our finding that WRKY1 regulates genes implicated in light and N signaling (e.g. ASN1) inspired us to further examine the role of WRKY1 in the regulation of genes involved in the light response. To do this, we compared light-regulated gene expression in the null wrky1-1 mutant and wild-type Col-0 (Fig. 2). For this experiment, we grew seedlings on MS media to 13 d after planting (DAP) and either maintained the seedlings in the normal light/dark cycle or moved them to extended darkness for 24 h prior to harvest. We used two-way ANOVA of transcriptome data followed by false discovery rate (FDR) correction (P < 0.01 for the ANOVA model) to identify 1,110 genes with a significant light × G interaction term (P < 0.01 for the coefficient of the term). Intersection of the 1,110 genes regulated by a light × G interaction with the 373 genes misregulated in a knockdown of WRKY1 (wrky1) under steady-state light conditions (Fig. 1) revealed a 35% overlap at a high level of significance (P < 0.001). The large number of affected genes and highly significant overlap with genes misregulated in the wrky1 mutant at steady-state conditions suggest a strong involvement of WRKY1 in light signaling.
Figure 2.
Cluster analysis of WRKY1-dependent genes with significant G × light interaction reveals loss of light repression for some dark-inducible genes. A, Cluster analysis of genes with significant (P < 0.02, FDR 5%) G × light interaction effect (1,567 genes). The shaded area indicates dark conditions, and the nonshaded area indicates light conditions. Only cluster 1 is shown. The full cluster analysis can be viewed in Supplemental Fig. S5. B, GO-term analysis of gene cluster 1 with significant G × light effect.
To identify patterns of genes misregulated by light in the wrky1 mutant, we performed cluster analysis of the microarray data. Specifically, we used gene expression cluster analysis (Multiple Expression Viewer [MEV], quality threshold clustering [QTC] analysis) and Tukey’s honestly significant difference (HSD) mean-separation test on the genes with significant light × G interaction. Cluster analysis of the genes misregulated in response to a light × G interaction resulted in 11 distinct gene expression clusters regulated by WRKY1 (Fig. 2; Supplemental Fig. S5A). Cluster 1 was the largest (612 genes) and contained a set of genes that had partially lost light repression in the wrky1 mutant (Fig. 2A). For this set of genes, expression is normally repressed by light in the wild type but was upregulated in the wrky1-1 mutant (Fig. 2A). Genes in cluster 1 include the dark-inducible genes DIN1, DIN4, DIN6/ASN1, and DIN10 (Fujiki et al., 2001), which all have an overrepresentation of W-box CRE-motifs in their promoters. The intersection of cluster 1 WRKY1-regulated genes with previously identified light-induced or light-repressed genes (Nozue et al., 2011) revealed a significant overlap with the light-repressed genes (P < 0.001; 50 genes). Cluster 1 WRKY1-regulated genes comprised GO-term enrichments (BioMaps) for organic acid and carboxylic acid catabolic processes (P < 0.01; Fig. 2B). This observation suggests that WRKY1 plays a large regulatory role in the light repression of genes involved in catabolism of organic resources, which are specifically required in plants exposed to extended darkness. Two other highly significantly overrepresented GO terms in the WRKY1-controlled genes include “response to abscisic acid stimulus” (P = 0.01) and “regulation of abscisic acid mediated signaling pathway” (P = 0.07). These results are compatible with the recent finding that WRKY1 regulates ABA signaling in response to drought (Qiao et al., 2016).
Unique and significant GO-term enrichments (BioMaps) were also uncovered for the other clusters of genes regulated by a light × G interaction in the wrky1 mutants (Fig. 2; Supplemental Fig. S5B). These include nitrogen-compound metabolic processes (cluster 2), disaccharide biosynthetic processes (cluster 4), generation of precursor metabolites and energy (cluster 5), ATP biosynthetic process (cluster 10), and carbohydrate metabolic process (cluster 11).
Finally, Tukey’s HSD mean separation test of the light × G interaction term revealed a larger G effect in the light (901 genes) than in the dark (499 genes; P < 0.01, Tukey’s HSD mean-separation test), while the light effect was 89% similar between the wild type and the wrky1 mutant. These results support a role for WRKY1 in the regulation of light-responsive genes (Fig. 2; Supplemental Fig. S6).
To summarize, in the light, WRKY1 specifically (1) repressed a network of genes that are required to catabolize cellular resources when light (i.e. in the form of C) is limited (cluster 1); and (2) activated a subset of genes involved in the biosynthesis of energy-dependent metabolites synthesized during the day (clusters 4 and 5). By contrast, in the dark, WRKY1 (1) activated a subset of genes involved in processes of respiration and the production of energy metabolites; and (2) repressed genes involved in energy-expensive, secondary metabolic processes.
To explore the potential direct regulation of these genes by WRKY1, we performed analysis of known cis elements in the promoter of WRKY1-regulated genes. This analysis showed that the W-box motif is the most enriched CRE in genes that are upregulated in the wrky1-1 mutant in both the light (1.00e-∞) and the dark (5.03e-29). Although the W-box is not statistically overrepresented in promoters of genes downregulated in the wrky1-1 mutant in light or dark, a promoter scanning algorithm (Find Individual Motif Occurrences; Grant et al., 2011) uncovered at least one W-box canonical sequence (TTGAC) that was present in 446 of the 456 genes downregulated in the light and 365 of the 367 genes downregulated in the dark. Thus, nearly all the genes misregulated in the wrky1-1 mutant in the light and dark have at least one W-box CRE-motif in their putative promoter regions, suggesting potential direct transcriptional regulation by WRKY1.
WRKY1 Mediates Transcriptional Reprogramming in Response to Nitrogen Treatment
Our analysis of steady-state levels of mRNA in the wrky1 mutant revealed misregulation of genes involved in the N-assimilation pathway (Fig. 1A; Supplemental Table S2). However, to test whether WRKY1 is involved in the regulation of the plant N response, it was necessary for us to explore changes in expression of genes involved in the N signaling pathway to a transient N treatment in both the most severe T-DNA mutant, wrky1-1 (SALK_070989), and wild-type (Col-0) seedlings. To do this, we grew wrky1-1 and wild-type seedlings on basal MS media supplemented with 1 mm KNO3− for 14 DAP under a long-day cycle. At the start of day, seedlings were transferred to either the N-levels present in MS media (20 mm KNO3− + 20 mm NH4NO3) or 20 mm KCl (control) for 2 h prior to harvest. This N treatment was previously shown to elicit both inorganic and organic N-response in 2-week-old Arabidopsis seedlings (Gutiérrez et al., 2008). A two-way ANOVA of genome-wide transcriptome data followed by FDR correction of the ANOVA model (P < 0.01) uncovered 123 genes with a significant N × G interaction term (P < 0.02). Of these 123 genes, 11 had a significant overlap (P < 0.05) with nitrogen-regulated genes in a N-regulatory network previously identified by Gutiérrez et al. (2008), including nitrate reductase 1 (NIA1). This result indicates that a different network of genes respond to transient N treatment when WRKY1 is absent compared to when it is present.
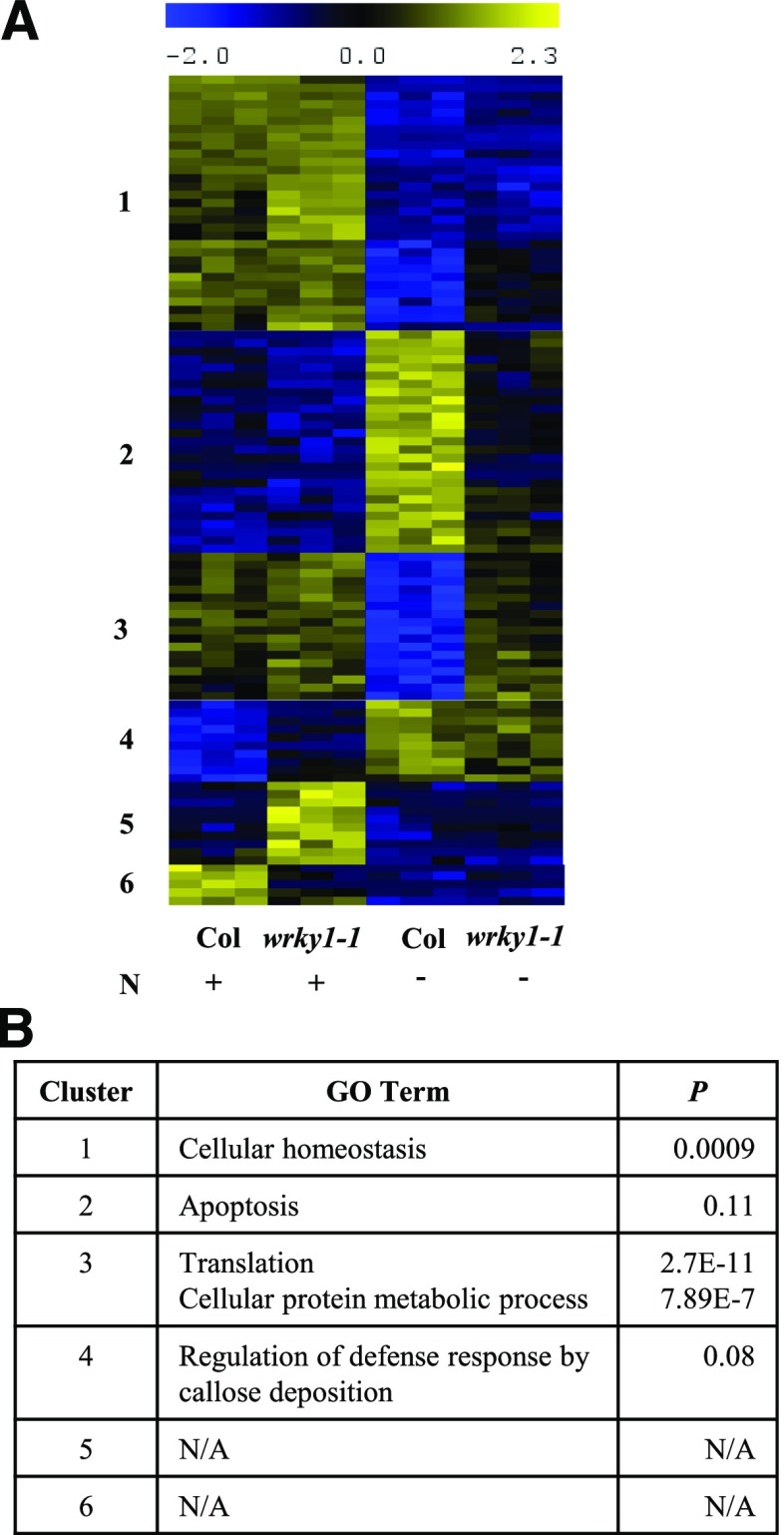
To better understand the biological role of the 123 genes with expression regulated by WRKY1 and a significant N × Ginteraction, we performed gene expression cluster analysis (QTC function in MEV) and Tukey’s HSD mean-separation test. This cluster analysis resulted in six distinct gene expression clusters of WRKY1-regulated genes (Fig. 3A) involved in different biological processes (Fig. 3B). WRKY1-regulated genes in clusters 1 and 3 contained genes with the most significant overrepresentation of GO terms (BioMaps), in which genes in cluster 1 are predominantly involved in cellular homeostasis (P = 0.0009), while genes in cluster 3 are involved in translation (P = 2.7e-11) and the cellular protein metabolic process (P = 7.89e-07; Fig. 3B).
Figure 3.
Cluster analysis of genes with significant N × G effect reveal that WRKY1 participates in plant response to N limitation. A, Cluster analysis of genes with significant (P < 0.02, FDR 5%) G × N interaction effect (123 genes). B, GO-term analysis of gene clusters with significant G × N effect.
Further inspection of the WRKY1-regulated gene expression clusters 2 and 3 revealed that a subset of genes do not respond to N limitation in the wrky1 mutant as they do in plants with wild-type WRKY1 function (Fig. 3; Supplemental Table S3). This finding suggests that the role of WRKY1 in the N signaling pathway may be as a transcriptional regulator in the control of nitrogen-starvation signaling processes. According to this hypothesis, the subset of WRKY1-regulated genes must meet two criteria: (1) in the presence of N, gene expression is the same between the wild type and wrky1-1; and (2) in the absence of N, gene expression is different between the wild type and wrky1-1. To identify a full subset of WRKY1-regulated genes that meet these criteria, we compared Tukey’s HSD mean-separation test results from the two-way interaction terms for “N effect in wrky1-1 mutant plants” and for “G effect in the absence of N”. This analysis uncovered 38 WRKY1-regulated genes that met our criteria, of which 18% are involved in C compound and carbohydrate metabolism, including O-Glycosyl hydrolases family 17 protein (At5g58090); UDP-Glycosyltransferase superfamily protein (At2g36970); Phosphofructokinase family protein (At1g20950); XTH6_xyloglucan endotransglucosylase/hydrolase 6 (At5g65730); and others (Supplemental Table S3). This result provides further support that WRKY1 is involved in N- and energy-related signaling pathways. Additionally, Tukey’s HSD mean-separation test results revealed that the N response was significantly altered in the wrky1 mutant for 63 genes (P < 0.01). Of these, 43 genes responded to N in the wild type but not in the wrky1-1 mutant, while 18 genes had a significant N response only in the wrky1 mutant seedlings.
Cumulatively, these analyses reveal that (1) WRKY1 regulates a different transcriptional program of genes in response to transient N treatment compared to steady-state N conditions; (2) fewer genes are misregulated by knockdown of WRKY1 expression in response to transient N treatment compared to steady-state N conditions; and (3) the N response is altered in wrky1 mutants compared to the wild type, in which WRKY1 represses genes involved in defense response in the presence of N. However, in the absence of N, WRKY1 activates genes involved in apoptosis (Fig. 3, cluster 2) and represses genes involved in translation and protein metabolic processes (Fig. 3, cluster 3) that require N. This last result further supports the hypothesis that WRKY1 is involved in energy conservation, where under N-limiting conditions WRKY1 activates genes involved in recycling of cellular resources while simultaneously suppressing genes involved in energy-expensive protein biosynthesis.
To predict whether WRKY1 directly or indirectly regulates the differentially expressed genes (DEGs), we performed promoter analysis of known cis elements and found different combinations of enriched CREs in the promoters of genes up- and downregulated in response to N treatment in wild-type and wrky1 mutant plants. Only genes upregulated in the wrky1-1 mutant in the presence of N had a statistical overrepresentation of W-box motifs (3.12e-03). Promoter scanning using FIMO revealed that all 20 genes downregulated in the wrky1-1 mutant in the presence of N have at least one W-box motif. Likewise, 45 of 47 genes upregulated and all 40 genes downregulated in the mutant in the absence of N have at least one W-box motif. A number of these genes (25 upregulated and 12 downregulated) are involved in N-related processes, suggesting that WRKY1 may have a direct role in their transcriptional regulation in response to an N signal.
Combined Light and Nitrogen Treatments Reveal that WRKY1 Regulates Cross Talk between Light and Nitrogen Signaling Pathways
The above studies collectively support the hypothesis that WRKY1 is a regulatory node in the Arabidopsis light and N interaction network. This implication, along with evidence from previous research (Jonassen et al., 2008; Krouk et al., 2009; Nunes-Nesi et al., 2010; Obertello et al., 2010), reinforces the notion that transcriptional cross talk occurs between light and N signaling pathways to fine-tune plant energy status. This hypothesis was further investigated by performing combined treatments with light and N on wild-type and wrky1-1 null mutant (SALK_070989) seedlings. Here, we aimed to determine if combined light and N treatments will reveal different transcriptional reprogramming by WRKY1 than is observed in response to individual light or N treatments, as has been observed for 10 Arabidopsis ecotypes in response to single- and double-stress treatments (Rasmussen et al., 2013).
To test this hypothesis, we grew seedlings on basal MS media supplemented with 1 mm KNO3− under a long-day light cycle for 13 d. For dark treatment, wild-type and mutant seedlings were moved to continuous dark for 24 h prior to N treatment. For N treatment, seedlings were transferred to basal MS media supplemented with the N-levels in MS media (20 mm KNO3− plus 20 mm NH4NO3) at the start of the light cycle (or the putative light cycle for dark-treated seedlings) for 2 h. RNA was extracted from shoot tissue for expression analysis by microarrays, and the data were analyzed by three-way ANOVA followed by FDR correction of the ANOVA model (P < 0.01). Three-way ANOVA revealed significant main effects, two-way interaction effects, and a three-way interaction effect (Table 2), which identified 724 genes with significant G × light × N three-way interaction. Gene network analysis was used to organize these 724 WRKY1-dependent genes into a cross talk network, revealing predicted interactions among nodes based on coexpression and protein-DNA regulatory interactions (Supplemental Fig. S7). Analysis of promoters for known cis elements identified the W-box motif in 706 of 724 promoters of WRKY1-regulated genes at an average of 4.4 cis motifs/promoter.
Table 2. Results of three-way ANOVA for individual and interaction terms.
The number of genes is the number of ATH1-genechip identifiers (probes).
| Effect | No. Genes |
|---|---|
| G | 2,356 |
| N | 5,062 |
| L | 10,158 |
| G × N | 1,022 |
| G × L | 1,459 |
| N × L | 2,114 |
| G × N × L | 700 |
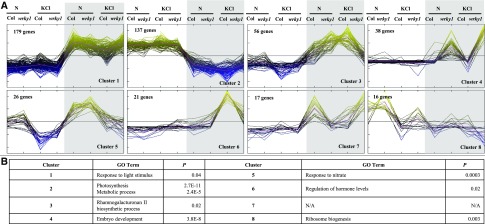
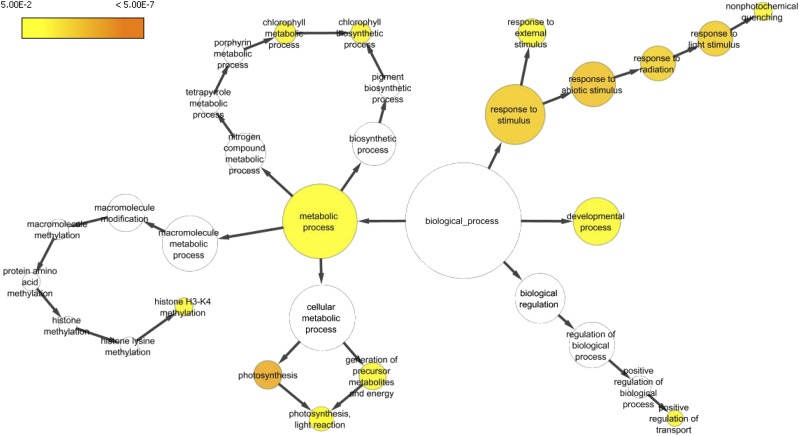
To identify groups of WRKY1-regulated genes with similar expression patterns within the 724 genes whose expression is affected by a G × light × N interaction, we performed cluster analysis, using the QTC function in MEV, for genes with a significant three-way interaction term (P < 0.01), which resulted in eight distinct WRKY1-regulated gene clusters (Fig. 4A). GO-term analysis (BioMaps) revealed unique and significant biological functions for WRKY1-regulated genes in clusters 1–6 and 8 (Fig. 4B), including response to light stimulus (cluster 1; P = 0.04); photosynthesis (cluster 2; P = 2.7E-11); embryo development (cluster 4; P = 3.8e-8); response to nitrate (cluster 5; P = 0.0003); and regulation of hormone levels (cluster 6; P = 0.02). We next performed gene network analysis, which resulted in a cross talk network in which WRKY1-regulated genes were grouped by significant (P < 0.05) overrepresentation of shared biological processes (BinGO Plugin Cytoscape; Fig. 5). The largest WRKY1-regulated gene clusters had overrepresented GO terms for “metabolic processes” (177 genes); “response to stimulus” (106 genes); and “developmental process” (60 genes; Fig. 5). This analysis provides insight into the biological processes influenced by the cross talk between N and light signaling pathways in which WRKY1 is a regulatory node.
Figure 4.
Combinatorial treatment of wrky1 mutants and the wild type with N and light results in a significant three-way G × N × light interaction. Cluster analysis of genes with significant (P < 0.01, FDR 5%) G × N × light interaction effect (724 genes). B, GO-term analysis of gene clusters with significant G × N × light effect. The shaded area indicates dark conditions. KCl, control treatment; wrky1, wrky1-1.
Figure 5.
Genes commonly regulated by WRKY1, N, and light comprise a network of diverse biological functions. Shown is the N of statistically overrepresented GO terms in the set of 724 genes with significant G × N × light interaction effect. The node area is proportional to the number of genes within the functional category (i.e. more genes equals a larger node). Colored nodes are significantly overrepresented (see color legend), while white nodes are not significantly overrepresented (BinGO; Maere et al., 2005).
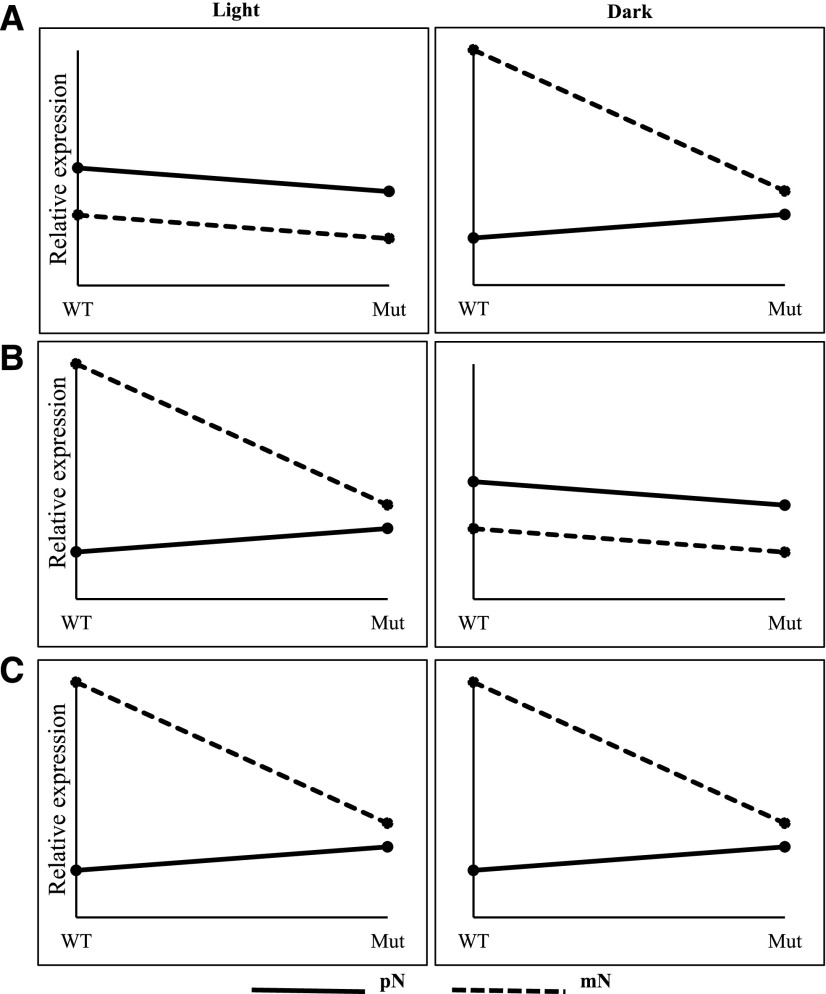
To fully interpret the three-way interaction term (G × light × N), we used a sequential ANOVA approach to investigate genes with significant three-way interaction to statistically determine how the various two-way interactions differed across the levels of the third variable (Fig. 6). Sequential ANOVA is a commonly used post hoc analysis to investigate the interaction term from ANOVA models (UCLA Statistical Consulting Group, 2019). Principal component analysis (PCA) of all single and combined treatments revealed that light was the dominant effect (PC1), accounting for 49% of the variance, while nitrogen corresponded to PC2, explaining 30% of the total variance (Supplemental Fig. S8). Therefore, we performed the first iteration of sequential ANOVA across levels of the light variable, and the second iteration across levels of the nitrogen variable. Two-way ANOVAs of genes with significant three-way interaction under each light condition revealed significant N × G interaction exclusively in the dark for 78% of genes (P < 0.05); exclusively in the light for 12% of genes; and in both dark and light conditions for 10% of genes (Fig. 6).
Figure 6.
Graphical representation of the sequential ANOVA to interpret the three-way interaction term. These plots represent the general observations for the 724 genes with significant three-way interaction. This example shows the three scenarios we observed for how gene expression changes in the light and the dark in response to the nitrogen variable across levels of the genotype variable. In our study, 78% of the genes only have a significant G × N interaction in the dark (A), 12% of the genes only have a significant G × N interaction in the light (B), and 10% of the genes have a significant G × N interaction in both dark and light conditions (C). mN, 20 mM KCl; Mut, wrky1-1; pN, 20 mM KNO3 + 20mM NH4NO3; WT, Col-0 wild type.
These results indicate that the N × Ginteraction was most significant in dark conditions, which was also observed visually from the cluster analysis of WRKY1-regulated genes (Fig. 4). WRKY1-regulated genes with significant N × Ginteraction in the dark were uniquely and significantly enriched in GO terms (BioMaps) for photosynthesis (P = 0.002), response to light stimulus (P = 0.004), and Gln metabolic process (P = 0.05). Alternatively, genes with significant N × Ginteraction exclusively in the light (12% of genes with significant three-way interaction) were uniquely enriched in GO terms for the lignin metabolic process (P = 0.02). Genes with significant two-way interaction (P < 0.05) in both light and dark conditions were enriched in GO terms for the mRNA catabolic process (P = 0.04) and intracellular transport (P = 0.04).
To better understand the two-way N × G interaction term, the next step in our sequential ANOVA analysis was to perform one-way ANOVAs under each N condition while holding the light variable as either “dark” or “light”. Here, we focused on one-way ANOVA results in dark conditions, since it was revealed as the dominant effect from the previous step. Our comparison of one-way ANOVA models of genes with significant two-way interaction terms in the dark revealed that there was a significant G effect for 16% of genes exclusively in the presence of N, 47% of the genes exclusively in the absence of N, and 37% of genes in both the presence and absence of N.
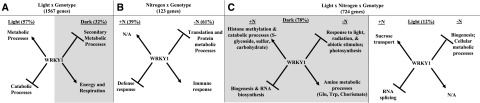
The genes with significant genotype effect in the dark exclusively in the presence of N have significant enrichment of GO terms for the S-glycoside catabolic process (P = 0.04) and carbohydrate catabolic process (P = 0.05), where in response to transient N treatment, WRKY1 activates a network of genes involved in the remobilization of cellular carbon resources and represses genes involved in biogenesis (Fig. 7C; Supplemental Fig. S9). However, genes with significant G effect in the dark exclusively in the absence of N are uniquely enriched in GO terms for light stimulus (P = 0.0004); photosynthesis (P = 0.0016); and amine metabolic processes (P = 0.0088), in which WRKY1 activates genes involved in metabolic and biosynthetic processes for production of Gln, Trp, and chorismate and represses genes that respond to light stimulus (Fig. 7C). Additionally, genes with a significant G effect (P < 0.05) under both N regimes are enriched in GO terms for the chlorophyll biosynthetic process (P = 0.004), the reproductive process (P = 0.006), and embryo development ending in seed dormancy (P = 0.02).
Figure 7.
WRKY1 regulates genes in light and nitrogen pathways and is an integrator of light and N signaling. Shown is the putative mechanism by which WRKY1 regulates different transcriptional programs under three conditions: light treatment (A) N treatment (B), and combined light and N treatment. The most significantly overrepresented GO terms for the biological process are shown. Arrows indicate activation and lines and bars indicate repression. Percentages indicate the number of genes from a given group that adhere to the proposed mechanism for that group. Shaded areas indicate dark conditions. +N, nitrogen treatment; −N, control treatment.
These results indicate that the G effect is weakest in the presence of nitrogen. This finding supports our earlier hypothesis that a less significant G effect is observed between the wild type and wrky1 mutants when N is present. Ultimately, the sequential ANOVA analysis indicates that the G effect caused by mutation of WRKY1 is revealed most significantly in the dark and in the absence of N (Fig. 7C; Supplemental Fig. S9). This finding suggests a mechanism by which WRKY1 regulates a transcriptional program of genes in response to light and N limitation. Moreover, analysis of the three-way interaction term provides support for WRKY1 being a regulatory node connecting N and light signaling pathways. In this model of transcriptional regulation, WRKY1 modulates the expression of a new network of genes in response to simultaneous N and light signals compared to the transcriptional programs controlled by WRKY1 in response to either N or light signaling alone (Fig. 7, A–C).
Phenotypic Analysis Reveals the Regulatory Role of WRKY1 in Nitrogen Metabolism
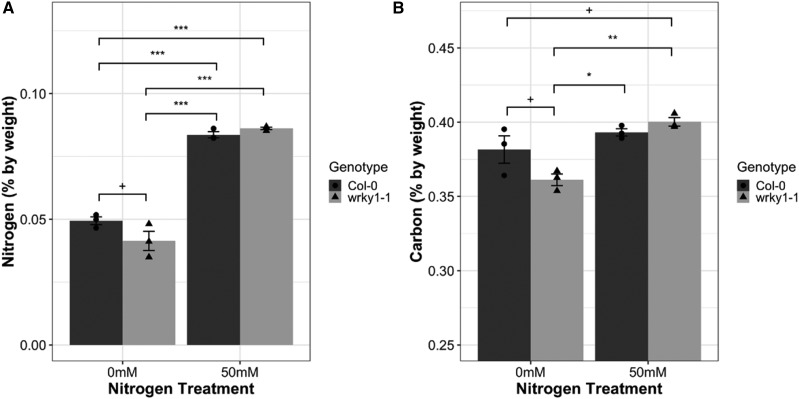
Our transcriptional and bioinformatics analysis above suggests a role for WRKY1 in the regulation of light and N signaling. To investigate the physiological effect of the wrky1 mutation on plants, we grew wrky1-1 and wild-type lines on soil under low (0 mm N supplement) and high (50 mm N supplement) N fertilization regimes and then subjected shoot tissues to elemental analysis to assess total C and N. Total N analysis revealed a significant (P = 0.041) N × G effect (Fig. 8A; Supplemental Table S4) in which there was more total N in the wild type than in the mutant under low N conditions. Likewise, there was a significant (P = 0.033) N × G effect for total C content (Fig. 8B; Supplemental Table S4), where there was higher C content in the wild type under low N conditions, and there was similar C content between the wild type and mutant under high N content.
Figure 8.
Total N and C contents in wild-type and wrky1-1 plants. A, Total N by percent weight (mean ± sd percent by weight) in Col-0 and wrky1-1 under 0 mM and 50 mM N supplement. B, Total C by percent weight (mean ± sd percent by weight) in Col-0 and wrky1-1 under 0 mM and 50 mM N supplement (Tukey’s HSD mean-separation test, +P < 0.15, *P < 0.05, **P < 0.01, ***P < 0.001). Circles represent individual sample values G = Col-0; triangles represent individual sample values G = wrky1-1; n = 3.
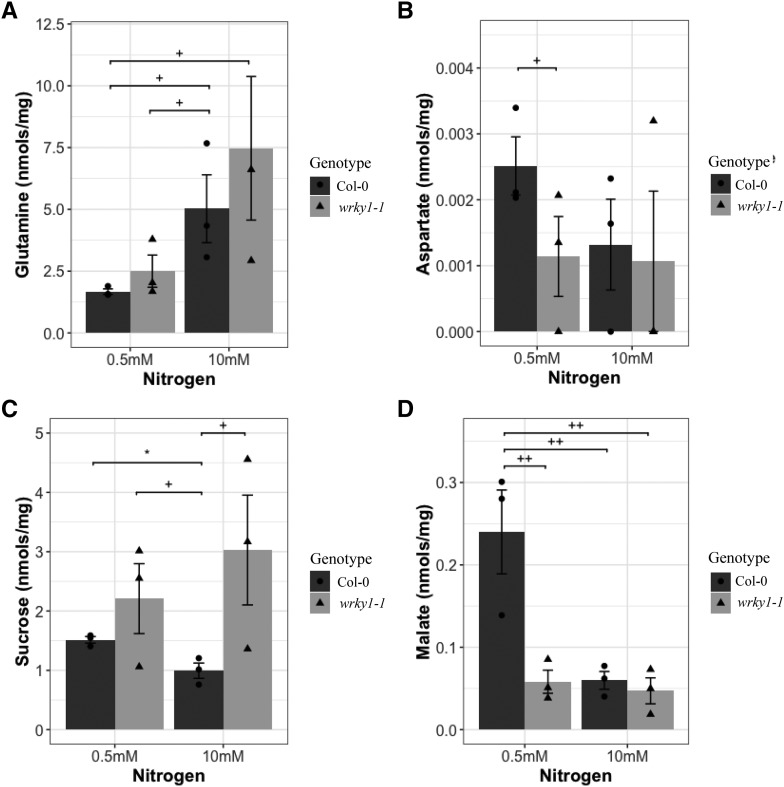
We further investigated the underlying changes in plant metabolism by analyzing free amino acids and carbohydrates using gas chromatography (GC)-mass spectrometry to determine the underlying cause for the change in total nitrogen and carbon content in wrky1 mutants compared to the wild type. To do this, we grew plants for 2 weeks on MS media supplemented with either 0.5 mm or 10 mm KNO3−. The majority of free amino acids were not significantly different between wild-type and mutant plants (Supplemental Fig. S10; Supplemental Table S4). However, the average concentration of Gln was higher in wrky1-1 mutants under both low and high [NO3−] compared to the wild type. Likewise, there was a higher accumulation of Asp in the wild type than in the mutant under low [NO3−] (P = 0.147; Fig. 9, A and B; Supplemental Table S4). Upon examining carbohydrates, the wrky1-1 mutant had higher concentrations of Suc (G effect, P = 0.039; Fig. 9C), and its products Glc (N effect, P = 0.141) and Fru (G effect, P = 0.163) under both low and high NO3− conditions. However, wild-type plants had higher concentrations of the dicarboxylic acid malate (N effect, P = 0.00932; G effect, P = 0.00854; N × G effect, P = 0.01682) only under low NO3− conditions (Fig. 9D; Supplemental Table S4).
Figure 9.
Measured metabolite levels (mean ± sd [nmol/mg]) in Col-0 and wrky1-1. Metabolites Gln (A), Asp (B), Suc (C), and malate (D) were measured under nitrogen treatment of 0.5 mM KNO3− and 10.0 mM KNO3− (Student’s t-test; +P < 0.2, ++P < 0.1, * P < 0.05). Circles represent individual sample values G = Col-0; triangles represent individual sample values G = wrky1-1; n = 3.
Overall, these reciprocal patterns of Gln/Asp and Suc/malate suggest a reprogramming of central C and N metabolism in wrky1 mutant plants that results in lower overall C and N content when N is limiting. The wild-type function of WRKY1 may be to regulate genes involved in the redirection of flux through the tricarboxylic acid cycle away from Gln biosynthesis and toward malate/Asp synthesis under nitrogen-limiting conditions as part of a resource conservation mechanism.
DISCUSSION
WRKY1 Regulates Different Transcriptional Programs Depending on the Signal or Combination of Signals Perceived
Our data on the wrky1 T-DNA mutants reveal a defect in genome-wide expression that is dependent on light, N, and a combination of light and N. This suggests that WRKY1 mediates cross talk between light and N signaling. Vert and Chory (2011) established two criteria for cross talk to exist between two signaling pathways: (1) “the combinatorial signal from both pathways should produce a different response than that triggered by each pathway alone”; and (2) “the two pathways must be connected directly or indirectly.” Our PCA analysis of the wrky1 mutant revealed that light is the dominant effect among single and combined treatments (Supplemental Fig. S8). However, to explain the variance in gene expression, our ANOVA analyses revealed that a different transcriptional program is activated in response to concurrent N and light treatments in wrky1 mutant and wild-type seedlings compared to the transcriptional response to individual N and light treatments. In addition, ∼80% of the 724 WRKY1 regulated genes shared between the light and N pathways (those with significant three-way interaction) are unique to the combined light and N treatment compared to individual treatments, indicating a direct connection between these pathways.
Our results for WRKY1 are similar to those reported by a recent study that revealed that 61% of transcriptional changes in 10 Arabidopsis ecotypes in response to double abiotic stress treatments were not predictable from responses to single-stress treatments (Rasmussen et al., 2013). Therefore, it is reasonable to suggest that WRKY1 is a regulator of cross talk between light and N signaling pathways. Moreover, the resulting cross talk network of WRKY1 controlled genes contains 29 downstream TFs (Supplemental Table S5), of which 15 have significant (P = 5.37e-10) overrepresentation of the GO term “regulation of N compound metabolic process,” further indicating a direct connection between WRKY1 control of the N and light signaling pathways. The sequential ANOVA performed here effectively deconstructed the three-way interaction term to reveal dominant effects among the interactions to define the plant status under which WRKY1 mediates cross talk between N and light signaling pathways.
Our analysis revealed a potential mechanism by which WRKY1 functions to repress genes involved in plant response to light stimulus and activate genes involved in amine metabolic processes when both light and exogenous N are limiting (Fig. 7C). This example may be extrapolated as a mechanism by which plant TFs influence the partitioning of cellular resources in response to complex environmental signals. The intensive statistical analysis presented herein can be used to decipher multifaceted interactions arising from similar or even more complex combinatorial experiments.
Although genome-wide transcriptional changes were observed between wild-type and WRKY1 mutant plants under a variety of experimental conditions, it is less clear whether the majority of these changes are due to the direct regulation of genes by WRKY1. WRKY TFs are known to be involved in the regulation of a number of biotic and abiotic stress response pathways (Wei et al., 2008; Jia et al., 2015; Qiao et al., 2016). Thus, it is important to consider whether WRKY1 is involved in shared or unique responses to N and/or light signaling. Evidence from the literature suggests that WRKY TFs help fine-tune plant response to specific stresses (Chen et al., 2019). For example, mitogen-activated protein kinase (MAPK) cascades are activated early following the perception of stress stimuli, and many WRKY family TFs have been identified as substrates for MAPKs. MAPKs phosphorylate different WRKY proteins to either positively or negatively modulate their DNA-binding ability (Chi et al., 2013; Chen et al., 2019). AtWRKY33 is phosphorylated by AtMPK3/AtMPK6 in response to Botrytis cinerea infection, which specifically influences pathogen-induced camalexin accumulation (Mao et al., 2011). Likewise, during senescence, AtMEKK both activates expression of AtWRKY53 and phosphorylates the protein to enhance its DNA-binding activity, which in turn promotes senescence (Miao et al., 2007). It is possible that WRKY1 is part of MAPK signaling cascades that fine-tune activation by N and/or light stimuli.
Promoter analysis of DEGs in the three WRKY1 mutant lines under “steady-state” conditions revealed that all the genes downregulated in wrky1 mutants had a highly significant overrepresentation of W-box motifs, while genes upregulated in the mutants did not, even though at least one W-box motif was located in the promoter of every gene. This is consistent with results from a recent study that used “Network Walking” to determine direct and indirect targets of 33 TFs (Brooks et al., 2019). That study found that all 33 TFs act as both inducers and repressors but that cis-binding motifs for a TF are specific to a certain direction of regulation (induced or repressed). For example, a TF can either directly repress targets and indirectly activate targets, or directly induce targets and indirectly repress targets through partner TFs (Brooks et al., 2019).
Additional promoter analysis of DEGs from the light, N, and combined treatments revealed that a W-box motif appears in the promoters of the majority of DEGs, and specifically in those involved in N-related processes. A previous study by Gutiérrez et al. (2008) shows that a single CRE motif is sufficient for transcriptional activation. Based on this information, we relaxed the original (Gutiérrez et al., 2008) nitrogen GRN, which led to the identification of WRKY1 as a regulatory hub in the N-network. However, the remaining DEGs with no detectable W-box motif in their putative promoter region suggest indirect regulation by WRKY1.
WRKY family TFs are known to have PPI with chromatin remodeling, calmodulin, and 14-3-3 proteins (Chi et al., 2013; Chen et al., 2019), which could result in “indirect” transcriptional regulation in response to light and/or N signals in the WRKY1 mutants. Interestingly, one of the most significant CREs found in the promoters of the up-regulated genes in the WRKY mutants was the G-box, and the only confirmed protein-protein interaction for AtWRKY1 is with a 14-3-3 protein, GRF1. Our combined gene expression and promoter analysis suggest mechanisms for both direct and indirect regulation of genes by WRKY1 in response to light and/or nitrogen signals that can be experimentally tested in future studies.
WRKY1 Is Likely Involved in an Energy Conservation Mechanism in Response to Low Energy Signaling
ANOVA revealed that the majority of the 724 genes misregulated in the wrky1-1 mutant had significant G × N interaction in the dark compared to the light. This result, in combination with GO-term analysis, generates the hypothesis that WRKY1 is part of an energy conservation mechanism by which targets of WRKY1 remobilize C resources in the dark when N is abundant but upregulates N metabolism in the dark when N is limiting. In this mechanism, WRKY1 integrates information about cellular N and energy resources to trigger processes necessary for plant metabolism in response to a transient N signal. For example, when both light (i.e. C) and nitrogen resources are limiting, genes involved in light response and photosynthesis were significantly upregulated in the wrky1 mutant. By contrast, genes involved in Gln and Trp metabolic and biosynthetic processes were significantly downregulated in the wrky1 mutant (Fig. 7C). These results are supported by research by Urbanczyk-Wochniak and Fernie (2005), who uncovered the surprising result that several amino acid pools, including Arg, Asn, and Glu, are higher under N-deficient conditions compared to N-saturated conditions. Specifically, the authors discovered that N-deficient plants in low light conditions have increased carbohydrate content, and that Glu and Trp metabolite pools increase initially in response to N-deficiency in both high- and low-light conditions. Our model of WRKY1-mediated regulation of genes in the dark provides transcriptional support for the observed changes in metabolite pools in response to N-deficiency in low-light conditions (Urbanczyk-Wochniak and Fernie, 2005). In our own experiments, we observed that under normal light conditions, but with N limitation, there is a decrease in total N and C content in the wrky1 mutant but a higher concentration of Gln and a lower concentration of malate and Asp compared to the wild type (Fig. 9, B and D). This analysis of free metabolite pools suggests that the wrky1 mutant fails to redirect metabolism toward Asp biosynthesis and instead maintains Gln biosynthesis even when C and N resources are limiting (Fig. 9). In future studies, it would be interesting to examine protein-bound amino acids to enhance our understanding of the underlying metabolism that contributes to the altered total C and N contents in mutant plants.
Our proposed energy conservation mechanism regulated by WRKY1 is also apparent in the regulation of a suite of dark-inducible genes (DIN1, DIN4, DIN6/ASN1, and DIN10), in which WRKY1 represses these genes in the light. AtDIN6/ASN1 in particular has been associated with both C and N signaling networks and energy conservation mechanisms in response to abiotic stress (Baena-Gonzáles et al., 2007; Lam et al., 1998). The influence of WRKY1 on ASN1 under light and N stress is similar to the “low energy syndrome” (LES) described by (Baena-González and Sheen, 2008; Tomé et al., 2014). The LES syndrome plays a role in plant adaptation to stressful conditions in which nonspecific stresses cause common energy deprivation responses. LES causes substantial perturbation of cellular processes, including the arrest of metabolism and sugar storage and induction of catabolism, photosynthesis, and remobilization of sugar (Baena-González et al., 2008; Tomé et al., 2014). AtKIN10, an SNF1-related protein kinase, has been implicated as a factor controlling LES (Baena-González et al., 2007). Comparison of genes up- and downregulated by AtKIN10 (Baena-González et al., 2007) with genes up- and downregulated in wrky1 mutant plants revealed a unique and highly significant overlap (P < 0.001) between 81 genes upregulated by AtKIN10 and genes downregulated by WRKY1 (Supplemental Fig. S11). GO-term analysis of the 81 overlapping genes found a significant overrepresentation for the term “trehalose metabolic/biosynthetic processes” (P = 0.009). This is of particular interest, since an association between trehalose metabolism and sugar sensing in plants has recently been shown (Tsai and Gazzarrini, 2014), in which it is hypothesized that trehalose acts as a signal of Suc availability (Schluepmann et al., 2003) and is shown to inhibit activity of the AtSnRK1-KIN10 complex. Although there were no observable differences in free Asn levels in 2-week-old mutant and wild-type plants, it is likely that there are differences in either protein-bound Asn levels or in free Asn levels, but at a later stage, since Asn is a known storage form of N (Lea et al., 2007; Gaufichon et al., 2016). Together, these results suggest that WRKY1 may play a role in mediating the LES syndrome in plants, having a potentially inverse but complementary role to the SnRK family of protein kinases.
CONCLUSION
TFs in the WRKY superfamily exist uniquely in plants and are primarily associated with biotic and abiotic stress response (Rushton et al., 2010; Chen et al., 2013; Jia et al., 2015). Our extended network analysis of N-regulatory networks predicted that WRKY1 is a regulatory hub in the Arabidopsis N-assimilation pathway, a component of primary metabolism. Here, we found that downregulation of this single TF in wrky1-1 null mutants resulted in genome-wide transcriptional reprogramming of both N and light signaling pathways, two essential plant response pathways. The phenotype of the wrky-1 null mutant shows it plays a nonfunctionally redundant role compared to WRKY family members. Our transcriptome and metabolite assays show that wrky1 mutants are affected in key metabolites of N assimilation, including Gln, Asp, and Gly. Our results for C limitation (via light) suggest that WRKY1 is involved in the low-energy response pathways in Arabidopsis and possibly other plant species. Our results further support that WRKY1 is involved in mediating other abiotic stress responses, as was recently shown by Qiao et al. (2016). Our results at the transcriptome and the metabolite analysis levels support a model in which WRKY1 enables plants to activate genes involved in the recycling of cellular C resources when light is limiting but N is abundant and to upregulate amino acid metabolism when both light and N are limiting. In this potential energy conservation mechanism, WRKY1 integrates information about cellular N and light-energy resources to trigger changes in plant metabolism.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) wild-type (ecotype Col-0) seeds were obtained from Lehle Seeds, while wrky1 T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center. Homozygous mutants were identified by PCR genotyping using gene-specific primers in combination with the T-DNA-specific primer LBb1.3 (Supplemental Table S6). The lines SALK_016954 and SALK_136009 have a single polymorphism in the WRKY1 gene (At2g04880) in an intron in the 5ʹ untranslated region (UTR) and promoter, respectively. The SALK_070989 line was recently shown by SALKSEQ to contain multiple polymorphisms: the insertion site for SALKSEQ_070989.0 lies within the 5ʹ UTR of At2g04880; SALK_070989.56.00.x lies within 300 bases of the 5ʹ end of At2g04880; both are present in the SALK_070989 line used in this study (Supplemental Fig. S3). SALKSEQ_070989.1 is a T-DNA insertion in the exon sequence of AT3G20460, a major facilitator superfamily protein; however, this insertion was not present in the SALK_070989 line used (Supplemental Fig. S3C), and the gene was not expressed based on microarray analysis. SALKSEQ_070989.2 is a T-DNA insertion in the intron of AT4G20300, a putative Ser/Thr-kinase. This insertion was present in the SALK_070989 line used (Supplemental Fig. S3D); however, the expression of this gene was not statistically significantly different between the wild type and mutant line (Supplemental Table S2).
For steady-state or no-treatment experiments, wild-type (Col-0) and homozygous mutant (SALK_070989; SALK_016954; SALK_136009) seeds were vapor-phase sterilized, vernalized for 3 d, then grown on basal MS media (Sigma M5524-1L), with 0.5 g/L MES hydrate (Sigma-Aldrich), 0.1% (w/v) Suc, 1% (w/v) agar at pH 5.7. Plants were grown vertically on plates for 14 d in an Intellus environment controller (Percival Scientific), under long-day (16 h light/8 h dark) conditions with light intensity of 50 μmol m−2 s−1 at constant temperature of 22°C. Seedlings were harvested 2 h after the start of the light period and flash-frozen in liquid N. For light treatments, Col-0 and SALK_070989 seedlings were grown in exactly the same way as no-treatment seedlings; however, at 13 DAP at the start of the light period, half of the seedling plates were wrapped in a double layer of foil to extend darkness then placed back in the same chamber. On 14 DAP, seedlings were harvested 2 h after the start of the light period, or putative start of the light period for dark treated seedlings, and immediately placed in liquid N. Dark-treated seedlings were harvested at the same time as long-day seedlings, but in complete darkness, and flash-frozen in liquid N.
For nitrogen treatments, Col-0 and SALK_070989 seedlings were grown on basal MS media without N (custom GIBCO) supplemented with 1 mm KNO3−, 0.5 g/L MES hydrate (Sigma-Aldrich), 0.1% (w/v) Suc, and 1% (w/v) agar at pH 5.7. Seedlings were grown under the same conditions as the no-treatment seedlings for 14 d. Then at the start of the light period, wild type and mutant seedlings were transferred to either N-rich media (basal MS media without N [Phytotech] supplemented with 20 mm NH4NO3 plus 20 mm KNO3−, 0.5 g/L MES hydrate [Sigma-Aldrich], 0.1% [w/v] Suc, 1% [w/v] agar at pH 5.7) or control media (basal MS media without N [Phytotech] supplemented with 20 mm KCl [molar equivalent for potassium in KNO3−], 0.5 g/L MES hydrate [Sigma-Aldrich], 0.1% [w/v] Suc, 1% [w/v] agar at pH 5.7) for 2 h and then harvested and flash-frozen in liquid N. The level of N for growing and treatment conditions was chosen based on previous studies showing that this level of N prevents N-starvation stress but facilitates both the organic and inorganic N response (Gutiérrez et al., 2008).
For combined light and N treatments, half of the seedlings received extended dark treatment at 13 DAP, as done for the light treatments, while the other half remained under the normal light/dark regime. Nitrogen treatments were performed as before on 14 DAP in both light and dark conditions at the start of the light period. For all treatments, shoots and roots were harvested separately, and subsequent analyses were performed on shoot tissue only.
Under the conditions of our experiments, there were no detectable morphological differences among the wild-type and wrky1 mutant lines.
RNA Isolation, RT-qPCR, and Microarray
RNA from three biological replicates from each experiment was extracted from shoots using an RNeasy Mini Kit with RNase-free DNaseI Set (QIAGEN) and quantified on both a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific) and a Bioanalyzer RNA Nano Chip (Agilent Technologies). RNA was converted to cDNA (Thermoscript kit, Invitrogen) then analyzed by RT-qPCR using LightCycle FastStart DNA MasterPLUS SYBR Green I kit (Roche) with a LightCycler 480 (Roche). RT-qPCR primers are listed in Supplemental Table S6. Then, a 100 ng aliquot of total RNA was converted into cDNA, amplified and labeled with GeneChip 3ʹ IVT Express Kit Assay (Affymetrix). The labeled complementary DNA (cDNA) was hybridized, washed, and stained on an ATH1-121501 Arabidopsis Genome Array using a Hybridization Control Kit, a GeneChip Hybridization, Wash, and Stain Kit, a GeneChip Fluidics Station 450 and a GeneChip Scanner (all from Affymetrix).
Analysis and Clustering of Microarray Data
Microarray intensities were normalized using the GCRMA (http://mev.tm4.org and http://geneontology.org/) package in R (http://www.r-project.org/; Wu et al., 2019). For the steady-state experiment, DEGs for each mutant genotype were determined by rank product (Breitling et al., 2004), and raw P values were adjusted by FDR with a cutoff of 5%. For l-only and N-only experiments, DEGs were determined by two-way ANOVA with G and light or N as factors. A gene was identified as differentially expressed if the FDR corrected P value of ANOVA models was <0.01 and the P value of the interaction coefficient (G × N or G × light), was <0.02. Tukey’s HSD mean-separation test was used for multiple comparison to identify interaction-term means that were significantly different from each other and greater than the expected se. Only unambiguous probes were included. Multiple Experiment Viewer software (TIGR; http://mev.tm4.org) was used to create heat maps and perform cluster analysis using QTC with Pearson correlation, Hierarchical Clustering: average linkage method, and diameter 0.1. The significance of overlaps of gene sets were calculated using the GeneSect (R)script (15) using the microarray as background. The significance of overrepresented GO-term analysis was performed with BioMaps (VirtualPlant 1.3; Katari et al., 2010) using genes that are represented on the microarray as background in which the P value of overrepresentation was measured by Fisher’s exact test with FDR correction and P value cutoff of 0.01 or as otherwise indicated in figures. All microarray data have been deposited into Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under GSE76278.
Three wrky1 mutant T-DNA insertion lines were used to understand the core regulatory role of WRKY1 in response to N and light perturbations (see “Plant Material and Growth Conditions”). Microarray analysis was done for all three mutant lines plus the wild type for “steady-state” and individual light and nitrogen treatments. Probes were normalized using the GCRMA method. Probes with more than one Present Call (P) in at least one group of replicates and sd > 0 were kept for further analysis. ANOVA with model simplification followed by Tukey’s HSD mean-separation test was performed for these three experiments in R using reshape2 (Wickham, 2007) and tidyverse (Wickham, 2007) packages and the GCRMA (Wu et al., 2019), affy (Gautier et al., 2004), and BiocGenerics packages from Bioconductor. This analysis revealed that the three mutants lines respond in the same way to light and/or N perturbations, which is different from the wild type response (Supplemental Figs. S6 and S12), meaning that the same genes are either up- or downregulated across mutant genotypes in response to the treatment. Based on this analysis, results were presented from the most severe wrky1 mutant (wrky1-1) and wild-type control plants.
Sequential ANOVA for Combined Experiment
For the combined N and light experiment, DEGs were determined by a three-way ANOVA with G, N, and light as factors. The ANOVA model was adjusted by FDR at a cutoff of 1%, and genes significantly regulated by the interaction of G × light × N were selected with a P value (ANOVA after FDR correction) cutoff of 0.01. Genes with significant three-way interaction were subjected to sequential ANOVA starting with a two-way ANOVA, in which G × N interactions were explored across levels of the light variable, resulting in Dark and Light ANOVA models. Sequential ANOVA is a commonly used post hoc analysis to investigate the interaction term from ANOVA models (UCLA Statistical Consulting Group, 2019). Genes with significant two-way interactions (ANOVA, P < 0.05 after model FDR correction, cutoff 5%) from Dark and Light ANOVA models were subjected to one-way ANOVA in which G factor was explored across levels of the N variable, resulting in “Dark Nitrogen,” “Dark Control,” “Light Nitrogen,” and “Light Control” ANOVA models. Genes with P < 0.05 (ANOVA after model FDR correction, cutoff 5%) were considered to have a significant G effect. Heat maps, cluster analysis, GO-term analysis, and gene-set overlap analysis were all performed as described above.
Gene Network Analysis
Analysis of the N-regulatory subnetwork was performed as described in Gutiérrez et al. (2008), except that only one regulatory binding site was required for protein-DNA edges. The N × light cross talk network was constructed from the 724 genes with a significant three-way interaction term for G × N × light. The Arabidopsis multinetwork (VirtualPlant 1.3) was queried with this list of genes, and only significant correlation and protein-DNA regulatory edges were included in the network using the Pearson correlation (cutoffs 1 to 0.7 or −1 to −0.7, with P ≤ 0.01). Networks were generated using the “Gene Networks” tool in the VirtualPlant system (www.virtualplant.org). Networks are visualized in Cytoscape 3.2.1.
Promoter Analysis
Significantly regulated genes (Supplemental Tables S2, S3, S7, and S8) were clustered using QTC with the default parameters in the Multiple Experiment Viewer software (TIGR; http://mev.tm4.org). Using the GO Enrichment Analysis tool at GeneOntolgy (http://geneontology.org/; Mi et al., 2017), we performed a GO-term analysis to identify genes involved in nitrogen-specific processes. Top level GO terms containing “nitrogen” and “amino acid” were selected (Supplemental Table S9). The 2-kb 5ʹ end upstream of the transcription start site were considered the putative promoter regions of genes of interest and were obtained using the associated tool with Elefinder (http://stan.cropsci.uiuc.edu/prom2.php). These regions were analyzed for known CRE overrepresentation within a group of genes using Elefinder (http://stan.cropsci.uiuc.edu/cgi-bin/elefinder/compare.cgi), which returns an e-value that indicates the likelihood of the result being returned by chance based on the binomial distribution. Using the same clusters of genes and the same promoter sequences from the analysis with Elefinder, a promoter scanning algorithm, FIMO (Grant et al., 2011), was used to find individual matches of the W-box motif obtained from PlantTFDB. The P-value cutoff was set to 1, with other parameters left at their default settings in order to fine-tune the presence/absence of the canonical W-box motif sequence (TTGAC). From the output, exact matches containing the canonical sequence were tallied for each promoter in Microsoft Excel.
Principal Component Analysis
Forty experiments (22 from light × N × G, 12 from light × G, and six from steady-state experiments) were renormalized together using GCRMA. The N × G hybridizations are the same as light treatments in the light × N × G interaction. Normalized expression values were centered and used for PCA using the prcomp function in R. The summary function in R was used to obtain the information regarding the percentage variance explained.
Elemental Analysis
Arabidopsis lines Col-0 and SALK_070989 were grown on autoclaved soil (Sunshine Mix LC1) and fertilized with either one-half strength MS media minus C and N or one-half strength MS media supplemented with 50 mm NH4NO3. Total C, hydrogen, and N were determined by elemental analysis using an Exeter Analytical CHN Analyzer (model CE440). Dried samples (30 mg fresh weight, which is ∼1.5 mg dry weight) were weighed in consumable tin capsules and purged with helium prior to combustion in pure oxygen under static conditions. Results were statistically analyzed using two-way ANOVA using R 3.5.2 (Supplemental Table S4).
Metabolite Analysis
Arabidopsis lines Col-0 and SALK_070989 were grown on 50 mL of MS modified basal-salt mixture (Phytotech Labs, M531) containing 1% (w/v) Suc and either 0.5 mm or 10 mm NH4NO3 solution containing 20 g/L BD Bacto agar (BD Biosciences) in a 100 × 100 × 15-mm square petri dish with a grid (Light Labs, D210-16), with three biological replicates each. Plants were grown for 14 d in a Percival growth chamber (Percival Scientific) under long-day (16 h light/8 h dark) conditions with a light intensity of 120 μmol m−2 s−1 and at a constant temperature of 22°C. Seedlings were harvested 2 h after the start of the light period on the day 14. The shoots from each plate were cut off, placed in an Eppendorf tube, and flash-frozen in liquid nitrogen. The samples were then stored at −80°C.
Metabolites were extracted based on the method outlined by Fiehn et al. (2008). The extraction solvent was prepared by mixing isopropanol, acetonitrile, and water at a volume ratio of 3:3:2. For amino acid analysis, the concentrated samples were fractionated as outlined by Orlova et al. (2006). One mL of water was added to each sample and vortexed until the residue was resuspended, and 25 μL of 10 mm ribitol and 10 mM α-aminobutyric acid were added as internal standards.
Samples were derivatized as outlined by Fiehn et al. (2008). An Agilent 7890B/7693 GC-mass spectrometry system was used with a fused silica capillary column SPB-35 column (30 m × 320 μm × 0.25 μm; Supelco 24094). One μL of each sample was injected using a splitless mode at 230°C. Helium (ultra high purity) was applied as the carrier gas using constant flow mode. The mass spectrometer transfer line, ion source and quadrupole were kept at 250°C, 250°C, and 150°C, respectively. The GC oven was set to an initial temperature of 80°C and held for 2 min. The temperature then increased at a rate of 5°C/min to a maximal temperature of 275°C and held for 6 min. The mass spectrometer was set to scan mode, and set to detect compounds eluting from 50 to 600 m/z.
Agilent MassHunter Qualitative Analysis B.07.00 was used to obtain peak areas. Metabolite peaks were normalized to the internal standard and quantified as nmol/g fresh weight. Pairwise statistical analysis (Students t-test) was performed using R 3.5.2 (Supplemental Table S4).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number At2g04880 (WRKY1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. WRKY1 regulatory subnetwork (VirtualPlant 1.1).
Supplemental Figure S2. Relative expression of WRKY1 in the wild type (Col-0) and wrky1 T-DNA mutants measured by RT-qPCR.
Supplemental Figure S3. SALK_070989 genotyping gels.
Supplemental Figure S4. SALK_016954 and SALK_136009 genotyping gels.
Supplemental Figure S5. Full cluster analysis of genes with significant G × light interaction.
Supplemental Figure S6. QTC of DEGs under dark and light treatments.
Supplemental Figure S7. Cross talk network of genes with significant three-way interaction (G × N × light).
Supplemental Figure S8. PCA of all experiments.
Supplemental Figure S9. Representative genes of the WRKY1 mechanism in the dark.
Supplemental Figure S10. Measured metabolite levels in Col-0 and wrky1-1.
Supplemental Figure S11. Overlapping WRKY1- and AtKIN10-regulated genes.
Supplemental Figure S12. QT clustering of DEGs under low and high N treatments.
Supplemental Table S1. Putative targets of WRKY1 identified from gene network analysis.
Supplemental Table S2. Differentially-expressed genes from the steady-state (no treatment) conditions.
Supplemental Table S3. Differentially-expressed genes from the nitrogen treatment conditions.
Supplemental Table S4. Two-way ANOVA and Tukey;s HSD mean-separation test results for metabolite and HCN analyses.
Supplemental Table S5. BioMaps output for 29 transcription factors in the crosstalk network.
Supplemental Table S6. Primers used for genotyping germplasms and for RT-qPCR.
Supplemental Table S7. Differentially-expressed genes from the light treatment conditions.
Supplemental Table S8. Differentially-expressed genes from the combined nitrogen and light treatment conditions.
Supplemental Table S9. Top level nitrogen process GO terms used for promoter analysis.
Supplemental Data Set S1. Relaxed network regulatory predictions from Gutierrez et al., 2008.
Acknowledgments
The authors gratefully acknowledge Ying Li and Daniel Tranchina for advice on data analysis, and Gabriel Krouk for advice.
Footnotes
This work was supported by the National Institutes of Health (NIH) (grant no. R01-GM032877 to G.C.) and a National Research Service Award (grant no. GM095273 to A.M.-C.) and the National Science Foundation (grant no. MCB-0929338 to G.C.).
Articles can be viewed without a subscription.
References
- Agarwal P, Reddy MP, Chikara J (2011) WRKY: Its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol Biol Rep 38: 3883–3896 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Sheen J (2008) Convergent energy and stress signaling. Trends Plant Sci 13: 474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Sheen J (2008) KIN10/11 are master regulators of the convergent stress transcriptome. In Photosynthesis. Energy from the Sun: 14th International Congress on Photosynthesis. Allen JF, Gantt E, Golbeck JH, Osmond B, eds, Springer, Dordrecht, The Netherlands, pp 1331–1337 [Google Scholar]
- Bläsing OE, Gibon Y, Günther M, Höhne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, Stitt M (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P (2004) Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573: 83–92 [DOI] [PubMed] [Google Scholar]
- Brooks MD, Cirrone J, Pasquino AV, Alvarez JM, Swift J, Mittal S, Juang C-L, Varala K, Gutiérrez RA, Krouk G, et al. (2019) Network Walking charts transcriptional dynamics of nitrogen signaling by integrating validated and predicted genome-wide interactions. Nat Commun 10: 1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta 1819: 120–128 [DOI] [PubMed] [Google Scholar]
- Chen L, Zhang L, Li D, Wang F, Yu D (2013) WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA 110: E1963–E1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Yao Q, Gao X, Jiang C, Harberd NP, Fu X (2016) Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr Biol 26: 640–646 [DOI] [PubMed] [Google Scholar]
- Chen X, Li C, Wang H, Guo Z (2019) WRKY transcription factors: Evolution, binding, and action. Phytopathol Res 1: 13 [Google Scholar]
- Chi Y, Yang Y, Zhou Y, Zhou J, Fan B, Yu JQ, Chen Z (2013) Protein–protein interactions in the regulation of WRKY transcription factors. Mol Plant 6: 287–300 [DOI] [PubMed] [Google Scholar]
- Contento AL, Kim SJ, Bassham DC (2004) Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol 135: 2330–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143: 1789–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich K, Weltmeier F, Ehlert A, Weiste C, Stahl M, Harter K, Dröge-Laser W (2011) Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell 23: 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee DY, Lu Y, Moon S, Nikolau B (2008) Quality control for plant metabolomics: Reporting MSI-compliant studies. Plant J 53: 691–704 [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Yoshikawa Y, Sato T, Inada N, Ito M, Nishida I, Watanabe A (2001) Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol Plant 111: 345–352 [DOI] [PubMed] [Google Scholar]
- Gaufichon L, Rothstein SJ, Suzuki A (2016) Asparagine metabolic pathways in Arabidopsis. Plant Cell Physiol 57: 675–689 [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J, O’Toole N, Ammar R, Provart NJ, Millar AH, Geisler M (2007) A predicted interactome for Arabidopsis. Plant Physiol 145: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS (2011) FIMO: Scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, et al. (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Wang C, Wang F, Liu S, Li G, Guo X (2015) GhWRKY68 reduces resistance to salt and drought in transgenic Nicotiana benthamiana. PLoS One 10: e0120646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen EM, Lea US, Lillo C (2008) HY5 and HYH are positive regulators of nitrate reductase in seedlings and rosette stage plants. Planta 227: 559–564 [DOI] [PubMed] [Google Scholar]
- Jonassen EM, Sévin DC, Lillo C (2009) The bZIP transcription factors HY5 and HYH are positive regulators of the main nitrate reductase gene in Arabidopsis leaves, NIA2, but negative regulators of the nitrate uptake gene NRT1.1. J Plant Physiol 166: 2071–2076 [DOI] [PubMed] [Google Scholar]
- Katari MS, Nowicki SD, Aceituno FF, Nero D, Kelfer J, Thompson LP, Cabello JM, Davidson RS, Goldberg AP, Shasha DE, et al. (2010) VirtualPlant: A software platform to support systems biology research. Plant Physiol 152: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Berthomé R, Orsel M, Mercey-Boutet S, Yu A, Castaings L, Elftieh S, Major H, Renou JP, Daniel-Vedele F (2011) Arabidopsis roots and shoots show distinct temporal adaptation patterns toward nitrogen starvation. Plant Physiol 157: 1255–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Tranchina D, Lejay L, Cruikshank AA, Shasha D, Coruzzi GM, Gutiérrez RA (2009) A systems approach uncovers restrictions for signal interactions regulating genome-wide responses to nutritional cues in Arabidopsis. PLOS Comput Biol 5: e1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Hsieh MH, Coruzzi G (1998) Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis thaliana. Plant J 16: 345–353 [DOI] [PubMed] [Google Scholar]
- Lea PJ, Sodek L, Parry MAJ, Shewry PR, Halford NG (2007) Asparagine in plants. Ann Appl Biol 150: 1–26 [Google Scholar]
- Lillo C. (2008) Signalling cascades integrating light-enhanced nitrate metabolism. Biochem J 415: 11–19 [DOI] [PubMed] [Google Scholar]
- Maekawa S, Takabayashi A, Huarancca Reyes T, Yamamoto H, Tanaka A, Sato T, Yamaguchi J (2015) Pale-green phenotype of atl31 atl6 double mutant leaves is caused by disruption of 5-aminolevulinic acid biosynthesis in Arabidopsis thaliana. PLoS One 10: e0117662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matiolli CC, Tomaz JP, Duarte GT, Prado FM, Del Bem LE, Silveira AB, Gauer L, Corrêa LG, Drumond RD, Viana AJ, et al. (2011) The Arabidopsis bZIP gene AtbZIP63 is a sensitive integrator of transient abscisic acid and glucose signals. Plant Physiol 157: 692–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matt P, Geiger M, Walch-Liu P, Engels C, Krapp A, Stitt M (2001a) Elevated carbon dioxide increases nitrate uptake and nitrate reductase activity when tobacco is growing on nitrate, but increases ammonium uptake and inhibits nitrate reductase activity when tobacco is growing on ammonium nitrate. Plant Cell Environ 24: 1119–1137 [Google Scholar]
- Matt P, Geiger M, Walch-Liu P, Engels C, Krapp A, Stitt M (2001b) The immediate cause of the diurnal changes of nitrogen metabolism in leaves of nitrate-replete tobacco: A major imbalance between the rate of nitrate reduction and the rates of nitrate uptake and ammonium metabolism during the first part of the light period. Plant Cell Environ 24: 177–190 [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD (2017) PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45(D1): D183–D189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Laun TM, Smykowski A, Zentgraf U (2007) Arabidopsis MEKK1 can take a short cut: It can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Mol Biol 65: 63–76 [DOI] [PubMed] [Google Scholar]
- Nozue K, Harmer SL, Maloof JN (2011) Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol 156: 357–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Fernie AR, Stitt M (2010) Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol Plant 3: 973–996 [DOI] [PubMed] [Google Scholar]
- Obertello M, Krouk G, Katari MS, Runko SJ, Coruzzi GM (2010) Modeling the global effect of the basic-leucine zipper transcription factor 1 (bZIP1) on nitrogen and light regulation in Arabidopsis. BMC Syst Biol 4: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira IC, Coruzzi GM (1999) Carbon and amino acids reciprocally modulate the expression of glutamine synthetase in Arabidopsis. Plant Physiol 121: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira IC, Brenner E, Chiu J, Hsieh MH, Kouranov A, Lam HM, Shin MJ, Coruzzi G (2001) Metabolite and light regulation of metabolism in plants: Lessons from the study of a single biochemical pathway. Braz J Med Biol Res 34: 567–575 [DOI] [PubMed] [Google Scholar]
- Orlova I, Marshall-Colón A, Schnepp J, Wood B, Varbanova M, Fridman E, Blakeslee JJ, Peer WA, Murphy AS, Rhodes D, et al. (2006) Reduction of benzenoid synthesis in petunia flowers reveals multiple pathways to benzoic acid and enhancement in auxin transport. Plant Cell 18: 3458–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Para A, Li Y, Marshall-Colón A, Varala K, Francoeur NJ, Moran TM, Edwards MB, Hackley C, Bargmann BO, Birnbaum KD, et al. (2014) Hit-and-run transcriptional control by bZIP1 mediates rapid nutrient signaling in Arabidopsis. Proc Natl Acad Sci USA 111: 10371–10376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasch CM, Sonnewald U (2013) Simultaneous application of heat, drought, and virus to Arabidopsis plants reveals significant shifts in signaling networks. Plant Physiol 162: 1849–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Z, Li CL, Zhang W (2016) WRKY1 regulates stomatal movement in drought-stressed Arabidopsis thaliana. Plant Mol Biol 91: 53–65 [DOI] [PubMed] [Google Scholar]
- Rasmussen S, Barah P, Suarez-Rodriguez MC, Bressendorff S, Friis P, Costantino P, Bones AM, Nielsen HB, Mundy J (2013) Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol 161: 1783–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed AJ, Canvin DT, Sherrard JH, Hageman RH (1983) Assimilation of [15N]nitrate and [15N]nitrite in leaves of five plant species under light and dark conditions. Plant Physiol 71: 291–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riens B, Heldt HW (1992) Decrease of nitrate reductase activity in spinach leaves during a light-dark transition. Plant Physiol 98: 573–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Schluepmann H, Pellny T, van Dijken A, Smeekens S, Paul M (2003) Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 6849–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Park CM (2010) MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol 186: 471–483 [DOI] [PubMed] [Google Scholar]
- Thum KE, Shasha DE, Lejay LV, Coruzzi GM (2003) Light- and carbon-signaling pathways. Modeling circuits of interactions. Plant Physiol 132: 440–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomé F, Nägele T, Adamo M, Garg A, Marco-Llorca C, Nukarinen E, Pedrotti L, Peviani A, Simeunovic A, Tatkiewicz A, et al. (2014) The low energy signaling network. Front Plant Sci 5: 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AY, Gazzarrini S (2014) Trehalose-6-phosphate and SnRK1 kinases in plant development and signaling: The emerging picture. Front Plant Sci 5: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UCLA Statistical Consulting Group (2019) FAQ: How can I understand a three-way interaction in ANOVA? https://stats.idre.ucla.edu/spss/faq/how-can-i-explain-a-three-way-interaction-in-anova-2/ (August 22, 2019)
- Urbanczyk-Wochniak E, Fernie AR (2005) Metabolic profiling reveals altered nitrogen nutrient regimes have diverse effects on the metabolism of hydroponically-grown tomato (Solanum lycopersicum) plants. J Exp Bot 56: 309–321 [DOI] [PubMed] [Google Scholar]
- Van Aken O, Zhang B, Law S, Narsai R, Whelan J (2013) AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiol 162: 254–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Chory J (2011) Crosstalk in cellular signaling: Background noise or the real thing? Dev Cell 21: 985–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Zhang Y, Han L, Guan Z, Chai T (2008) A novel WRKY transcriptional factor from Thlaspi caerulescens negatively regulates the osmotic stress tolerance of transgenic tobacco. Plant Cell Rep 27: 795–803 [DOI] [PubMed] [Google Scholar]
- Wickham H. (2007) Reshaping data with the reshape package. J Stat Softw 21: 1–20 [Google Scholar]
- Wu J, Gentry J, MacDonald J. 2019. gcrma: Background Adjustment Using Sequence Information. R package version 2.56.0. https://rdrr.io/bioc/gcrma/
- Xie Z, Zhang ZL, Zou X, Huang J, Ruas P, Thompson D, Shen QJ (2005) Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol 137: 176–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Jiang Y, Yu D (2011) WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol Cells 31: 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]