Introduction
Transceptors are nutrient transporters that act as receptors to regulate downstream signaling pathways. Ammonium transceptors regulate fungal morphology in response to limiting levels of ammonium, which is utilized as a nitrogen source. Few studies have addressed to what extent ammonium transceptors are conserved in pathogenic fungi. Plant and animal fungal pathogens undergo a variety of developmental transitions during infection, and work on some plant fungal pathogens establishes the importance of ammonium transporters during host infection [1, 2]. More widespread studies are needed to determine if ammonium transceptors have a general role in regulating the virulence of fungal pathogens.
Ammonium transceptors
Fungal ammonium transceptors belong to a large family of ammonium transporters that are found throughout biology, including the human Rhesus proteins [3]. Individual fungal species contain more than one member of this protein family, and in the model fungi that have been studied to date, only one of these acts as an ammonium sensor. In response to limiting ammonium, transceptors regulate morphological change in the yeasts Saccharomyces cerevisiae (ScMep2), Candida albicans (CaMep2), Ustilago maydis (Ump2), and Cryptococcus neoformans (Amt2) [4–7]. Ammonium transporters from filamentous fungi also act as transceptors when expressed in S. cerevisiae, suggesting that this form of signal transduction may be conserved in divergent fungi [8, 9]. Of importance to the classification of these proteins as nutrient receptors, amino acid substitutions have been identified that separate their transport and signaling functions, establishing that this form of ammonium sensing does not relate to nitrogen assimilation but involves some aspect of ammonium conductance through the transceptor [10–13]. Although fungal ammonium transporters are readily identified through homology searches, it is not possible to predict which of these are potential receptors as it is not yet clear what differentiates a transporter that only imports ammonium from one that also senses ammonium availability. Identifying transceptors therefore requires the phenotypic analysis of fungal mutants that lack individual ammonium transporters.
The structure of the Mep2 transceptor
The high-resolution structures of several ammonium transporters have been determined [14–19]. The basic architecture of these proteins is similar, as they all form stable trimers with each monomer containing a channel through which ammonium is transported. Each monomer contains an extracellular ammonium binding site, a pair of conserved phenylalanine residues that gate the channel, and a twin-histidine motif lining the narrow hydrophobic pore. While the bacterial and human proteins have open conformations, the yeast transceptors as purified are closed due to changes in the positions of their first and third intracellular loops [19]. The yeast transceptors, therefore, appear to need to undergo significant conformational changes to open the transport channel and allow ammonium import. The closed structures make sense as ammonium is potentially toxic, and its import must be carefully regulated. In bacteria, this is achieved by the regulated binding of a PII-like signal transduction protein that inhibits ammonium import via binding to the transporter and blocking the pore [20]. Similarly, ammonium transporters from S. cerevisiae that are not transceptors are controlled by the binding of a target of rapamycin-regulated protein [21]. It is therefore possible that transceptor-mediated ammonium import may be regulated by a different mechanism that controls conformational changes that open the pore of the transporter. Support for this comes from the position of the transceptor C-terminal cytoplasmic domain (CTD), which makes few contacts with the main body of the protein when compared with the CTD of the bacterial transporters [19]. The transceptor CTD may therefore be important to maintain the protein in its closed conformation.
The mechanism of ammonium sensing
Several questions remain that relate to the mechanisms of both ammonium transport and ammonium signaling by ammonium transceptors. Early studies suggested that ammonium transport by the Amt/Mep/Rh proteins involves the passive diffusion of ammonia gas [15, 16]. Although this likely remains true for the Rhesus proteins, more recent studies on the bacterial and plant transporters are consistent with electrogenic uniport of ammonium ions (NH4+) or symport of ammonia gas and protons (NH3/H+), indicating that different mechanisms of ammonium transport are used within this family of proteins [22–28]. Crystal structures and computer simulations of ammonium transport by the Escherichia coli AmtB transporter are consistent with the ammonium ion being recruited at an extracellular site. Subsequent stages are less clear, but a plausible scenario is that the ammonium ion passes through the phenylalanine gate and is then deprotonated at a second ammonium binding site [29, 30]. The resulting ammonium gas diffuses through the largely hydrophobic pore, while proton symport most likely occurs via a relay involving the twin-histidine motif [29]. The critical E. coli AmtB residues for ammonium recruitment and transport are conserved in the ScMep2 and CaMep2 transceptors and occupy identical positions within their ammonium-translocating pore, suggesting a conserved transport mechanism [19]. Remarkably, amino acid substitutions that block ammonium signaling but not ammonium transport by ScMep2 and CaMep2 involve residues that lie within or close to this central pore [10–13]. Presumably, these amino acid substitutions either affect the conformational changes that Mep2 undergoes during ammonium transport or the nature of the transported substrate, in accordance with the two mechanisms that have been proposed to explain the signaling function of ammonium transceptors (Fig 1). The first involves the transceptors acting analogous to G-protein-coupled receptors where the transceptor interacts with a signaling partner that responds to conformational changes in the transporter to regulate a downstream signal transduction pathway [4, 31, 32]. Consistent with this model, the Mep2 CTD undergoes large conformational changes in response to the phosphorylation of a regulatory serine residue within the CTD that activates the transporter [19] (Fig 2). An alternative signaling model proposes that ammonium transport causes changes in cytosolic pH that is then sensed by an internal pH-responsive mechanism [11]. Indeed, a link between pH and polarized growth has been identified in various fungal systems [33–37]. Moreover, a global screen has identified subunits of the vacuolar H+-ATPase as being essential for the induction of pseudohyphal growth by S. cerevisiae [38]. Identifying the mechanism of transport (electrogenic versus electroneutral) by transceptors and their nonsignaling homologues will determine if pH sensing is a potential mechanism that underpins transceptor function.
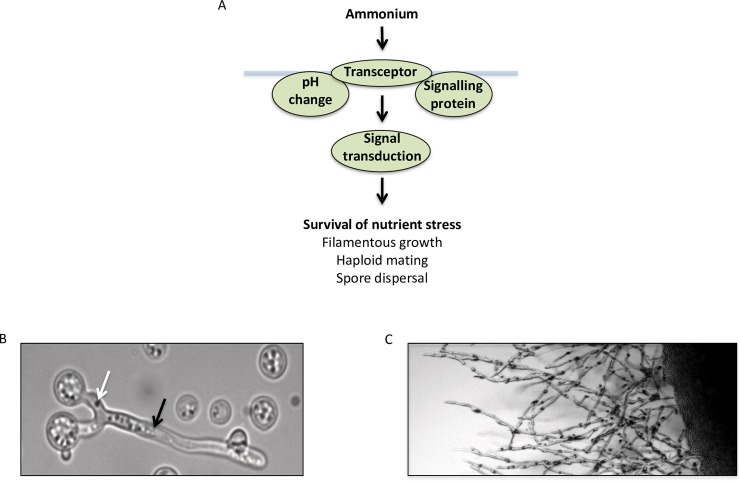
Fig 1.
(A) Studies in model dimorphic yeasts support two possible mechanisms of ammonium transceptor function. In the first, ammonium uptake results in cytosolic changes in pH that acts as the signal to initiate morphological change. A different mechanism involves the transceptor physically interacting with a signaling partner to regulate development. The role of the transceptor may be to signal that the levels of ammonium entering the cell are low but sufficient enough to support growth. For saprobic yeasts, this induces processes that promote survival during nutrient stress, such as the induction of filamentous growth. In fungal pathogens, similar mechanisms may regulate morphological changes involved in virulence. (B, C) Low-ammonium conditions can induce mating and filamentous growth. (B) Haploid strains of Cryptococcus neoformans mate by producing a conjugation tube (white arrow), followed by the growth of a dikayon filament (black arrow). (C) On low-ammonium agar, haploid strains of C. neoformans (serotype D) produce filaments that grow away from the main body of the colony. This process is enhanced when the colony is grown near a colony consisting of yeast of the opposite mating type.
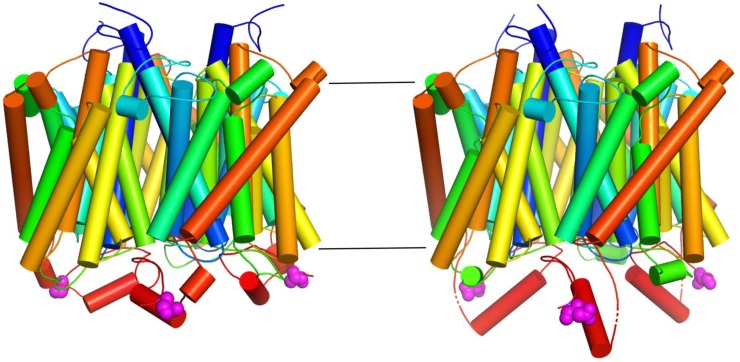
Fig 2. The Mep2 transceptor undergoes a phosphorylation-dependent conformational change.
Trimer cartoon models viewed from the side for (left) wild-type Candida albicans Mep2 (CaMep2) and (right) a CaMep2 variant that mimics the phosphorylation of a regulatory serine residue (S453) within the cytoplasmic CTD that activates the transporter. The cartoons are in rainbow representation. The phosphorylation-mimicking CaMep2 variant has undergone a large conformational change, resulting in the formation of a 12-residue-long α-helix (red) in the CTD [19]. Changes in transceptor conformation may regulate its interaction with a signaling partner. CTD, C-terminal cytoplasmic domain.
Ammonium transceptors and the response to nitrogen starvation
Ammonium transceptors are transcriptionally regulated in response to nitrogen availability so that they are induced when ammonium is absent or at low concentrations within the environment. Transceptor signaling, however, does not occur in the absence of ammonium, as mutants that fail to transport but that are correctly localized to the plasma membrane fail to initiate a dimorphic switch [10, 39]. Therefore, ammonium import when environmental levels of ammonium are low is the signal to induce morphological change. This is consistent with the view that ammonium transceptors regulate processes that promote survival when ammonium levels are limiting but sufficient to support slow growth. These survival strategies involve the switch to filamentous growth by diploid yeast and the mating between compatible haploid yeast, followed by the formation of dikaryon filaments. The diploid form of the baker’s yeast S. cerevisiae forms chains of elongated cells known as pseudohyphae in response to low-ammonium conditions [40]. Pseudohyphal formation requires a significant change in growth and involves a switch from bipolar to unipolar budding, an increase in apical growth, and the continued adhesion of mother and daughter cells. These pseudohyphal filaments grow away from the area of ammonium limitation, allowing the colony to grow towards new sources of nitrogen [40]. The diploid human pathogenic yeast C. albicans undergoes a similar transceptor-mediated response by producing branching filaments that grow away from the colony [6]. Although the molecular details of transceptor signaling are not understood, transceptors may regulate the protein kinase A (PKA) or mitogen-activated protein kinase pathways as constitutively active components of these pathways restore the dimorphic switch in transceptor-lacking mutants [4, 6, 10]. Haploid yeast can form filaments to survive limiting nitrogen conditions but will also initiate mating if they are in the presence of a mating partner. Both C. neoformans and U. maydis undergo ammonium responsive mating and dikaryon formation, a process that is dependent on an ammonium transceptor [1, 7]. Presumably, the generation of a diploid organism with its potential to produce spores for further dispersal provides an additional strategy to survive nutrient stress.
Perspectives
Fungal pathogens undergo significant morphological changes that are required for the successful infection of their hosts. These include the dimorphic transition between yeast and filamentous forms and the development of specialized infection structures. The formation and dispersal of spores also allow the spread of infection and can promote genetic variability that results in antifungal resistance. Fungal pathogens can experience nitrogen limitation, and regulatory overlap between pathogenicity and nitrogen starvation genes has been identified, including the induction of ammonium transporter genes during infection [41–45]. Similar to their role in saprobic fungi, ammonium transceptors may therefore regulate morphological change in pathogenic fungi to induce processes that promote their dispersal within or from their hosts. There are a few examples that suggest that this may be the case. A mutant of the broad host range plant pathogen Colletotrichum gloeosporioides that lacks the MepB ammonium transporter has reduced levels of PKA activity and appressoria formation and is less virulent than a wild-type strain [2]. Secondary metabolite production by the rice pathogen Fusarium fuikuroi is induced in a mutant lacking the MepB ammonium transporter, which may have a transceptor-like role [8]. A more definitive transceptor role during fungal infection has been assigned to the Ump2 ammonium transceptor in the maize pathogen U. maydis. A mutant lacking Ump2 fails to filament in response to low ammonium, exhibits reduced mating, and is significantly less virulent [1, 5]. Together, these studies support a potential link between ammonium transceptor function and fungal pathogenicity. It is widely accepted that fungal pathogens have a negative impact on human health and agricultural productivity worldwide. Due to increasing pathogen resistance to antifungal agents, there is a timely need for the identification of new antifungal targets. Ammonium transceptors may fall within this category as they support survival during nutrient stress and their accessibility as cell surface proteins makes them ideal antifungal targets. An analogy can be made with human G-protein-coupled receptors that are cell membrane proteins that are the targets of approximately 35% of approved drugs [46]. Further studies are therefore needed to identify the role that ammonium transceptors may have in plant and animal disease and any small molecules that inhibit their signaling function.
Funding Statement
SL was supported by a UK Biotechnology and Biological Sciences Research Council studentship (https://bbsrc.ukri.org). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Paul JA, Wallen RM, Zhao C, Shi T, Perlin MH. Coordinate regulation of Ustilago maydis ammonium transporters and genes involved in mating and pathogenicity. Fungal Biol. 2018;122(7):639–50. 10.1016/j.funbio.2018.03.011 [DOI] [PubMed] [Google Scholar]
- 2.Shnaiderman C, Miyara I, Kobiler I, Sherman A, Prusky D. Differential activation of ammonium transporters during the accumulation of ammonia by Colletotrichum gloeosporioides and its effect on appressoria formation and pathogenicity. Mol Plant Microbe Interact. 2013;26(3):345–55. 10.1094/MPMI-07-12-0170-R [DOI] [PubMed] [Google Scholar]
- 3.Andrade SL, Einsle O. The Amt/Mep/Rh family of ammonium transport proteins. Mol Membr Biol. 2007;24(5–6):357–65. 10.1080/09687680701388423 [DOI] [PubMed] [Google Scholar]
- 4.Lorenz MC, Heitman J. MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 1998;17(5):1236–47. 10.1093/emboj/17.5.1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith DG, Garcia-Pedrajas MD, Gold SE, Perlin MH. Isolation and characterization from pathogenic fungi of genes encoding ammonium permeases and their roles in dimorphism. Mol Microbiol. 2003;50(1):259–75. 10.1046/j.1365-2958.2003.03680.x [DOI] [PubMed] [Google Scholar]
- 6.Biswas K, Morschhäuser J. The Mep2 ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol Microbiol. 2005;56(3):649–69. 10.1111/j.1365-2958.2005.04576.x [DOI] [PubMed] [Google Scholar]
- 7.Rutherford JC, Lin X, Nielsen K, Heitman J. Amt2 permease is required to induce ammonium responsive invasive growth and mating in Cryptococcus neoformans. Eukaryot Cell. 2008;7(2):237–46. 10.1128/EC.00079-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teichert S, Rutherford JC, Wottawa M, Heitman J, Tudzynski B. Impact of ammonium permeases mepA, mepB, and mepC on nitrogen-regulated secondary metabolism in Fusarium fujikuroi. Eukaryot Cell. 2008;7(2):187–201. 10.1128/EC.00351-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Javelle A, Morel M, Rodríguez-Pastrana BR, Botton B, André B, Marini AM, et al. Molecular characterization, function and regulation of ammonium transporters (Amt) and ammonium-metabolizing enzymes (GS, NADP-GDH) in the ectomycorrhizal fungus Hebeloma cylindrosporum. Mol Microbiol. 2003;47(2):411–30. 10.1046/j.1365-2958.2003.03303.x [DOI] [PubMed] [Google Scholar]
- 10.Rutherford JC, Chua G, Hughes T, Cardenas ME, Heitman J. A Mep2-dependent transcriptional profile links permease function to gene expression during pseudohyphal growth in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19(7):3028–39. 10.1091/mbc.E08-01-0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boeckstaens M, André B, Marini AM. Distinct transport mechanisms in yeast ammonium transport/sensor proteins of the Mep/Amt/Rh family and impact on filamentation. J Biol Chem. 2008;283(31):21362–70. 10.1074/jbc.M801467200 [DOI] [PubMed] [Google Scholar]
- 12.Van Nuland A, Vandormael P, Donaton M, Alenquer M, Lourenço A, Quintino E, et al. Ammonium permease based sensing mechanism for rapid ammonium activation of the protein kinase A pathway in yeast. Mol Microbiol. 2006;59(5):1485–505. 10.1111/j.1365-2958.2005.05043.x [DOI] [PubMed] [Google Scholar]
- 13.Dabas N, Schneider S, Morschhäuser J. Mutational analysis of the Candida albicans ammonium permease Mep2p reveals residues required for ammonium transport and signaling. Eukaryot Cell. 2009;8(2):147–60. 10.1128/EC.00229-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruswitz F, Chaudhary S, Ho JD, Schlessinger A, Pezeshki B, Ho CM, et al. Function of human Rh based on structure of RhCG at 2.1 A. Proc Natl Acad Sci U S A. 2010;107(21):9638–43. 10.1073/pnas.1003587107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khademi S, O'Connell J 3rd, Remis J, Robles-Colmenares Y, Miercke LJ, Stroud RM. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 A. Science. 2004;305(5690):1587–94. 10.1126/science.1101952 [DOI] [PubMed] [Google Scholar]
- 16.Zheng L, Kostrewa D, Bernèche S, Winkler FK, Li XD. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc Natl Acad Sci U S A. 2004;101(49):17090–5. 10.1073/pnas.0406475101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrade SL, Dickmanns A, Ficner R, Einsle O. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc Natl Acad Sci U S A. 2005;102(42):14994–9. 10.1073/pnas.0506254102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Javelle A, Lupo D, Zheng L, Li XD, Winkler FK, Merrick M. An unusual twin-his arrangement in the pore of ammonia channels is essential for substrate conductance. J Biol Chem. 2006;281(51):39492–8. 10.1074/jbc.M608325200 [DOI] [PubMed] [Google Scholar]
- 19.van den Berg B, Chembath A, Jefferies D, Basle A, Khalid S, Rutherford JC. Structural basis for Mep2 ammonium transceptor activation by phosphorylation. Nat Commun. 2016;7:11337 10.1038/ncomms11337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas G, Coutts G, Merrick M. The glnKamtB operon. A conserved gene pair in prokaryotes. Trends Genet. 2000;16(1):11–4. 10.1016/s0168-9525(99)01887-9 [DOI] [PubMed] [Google Scholar]
- 21.Boeckstaens M, Merhi A, Llinares E, Van Vooren P, Springael JY, Wintjens R, et al. Identification of a Novel Regulatory Mechanism of Nutrient Transport Controlled by TORC1-Npr1-Amu1/Par32. PLoS Genet. 2015;11(7):e1005382 10.1371/journal.pgen.1005382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baday S, Orabi EA, Wang S, Lamoureux G, Bernèche S. Mechanism of NH4(+) Recruitment and NH3 Transport in Rh Proteins. Structure. 2015;23(8):1550–7. 10.1016/j.str.2015.06.010 [DOI] [PubMed] [Google Scholar]
- 23.Wacker T, Garcia-Celma JJ, Lewe P1, Andrade SL. Direct observation of electrogenic NH4(+) transport in ammonium transport (Amt) proteins. Proc Natl Acad Sci U S A. 2014;111(27):9995–10000. 10.1073/pnas.1406409111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neuhäuser B, Ludewig U. Uncoupling of ionic currents from substrate transport in the plant ammonium transporter AtAMT1;2. J Biol Chem. 2014;289(17):11650–5. 10.1074/jbc.C114.552802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludewig U, von Wirén N, Frommer WB. Uniport of NH4+ by the root hair plasma membrane ammonium transporter LeAMT1;1. J Biol Chem. 2002;277(16):13548–55. 10.1074/jbc.M200739200 [DOI] [PubMed] [Google Scholar]
- 26.Mayer M, Dynowski M, Ludewig U. Ammonium ion transport by the AMT/Rh homologue LeAMT1;1. Biochem J. 2006;396(3):431–7. 10.1042/BJ20060051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayer M, Ludewig U. Role of AMT1;1 in NH4+ acquisition in Arabidopsis thaliana. Plant Biol (Stuttg). 2006;8(4):522–8. [DOI] [PubMed] [Google Scholar]
- 28.Søgaard R, Alsterfjord M, Macaulay N, Zeuthen T. Ammonium ion transport by the AMT/Rh homolog TaAMT1;1 is stimulated by acidic pH. Pflugers Arch. 2009;458(4):733–43. 10.1007/s00424-009-0665-z [DOI] [PubMed] [Google Scholar]
- 29.Wang S, Orabi EA, Baday S, Bernèche S, Lamoureux G. Ammonium transporters achieve charge transfer by fragmenting their substrate. J Am Chem Soc. 2012;134(25):10419–27. 10.1021/ja300129x [DOI] [PubMed] [Google Scholar]
- 30.Ariz I, Boeckstaens M, Gouveia C, Martins AP, Sanz-Luque E, Fernández E, et al. Nitrogen isotope signature evidences ammonium deprotonation as a common transport mechanism for the AMT-Mep-Rh protein superfamily. Sci Adv. 2018;4(9):eaar3599 10.1126/sciadv.aar3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thevelein JM, Voordeckers K. Functioning and evolutionary significance of nutrient transceptors. Mol Biol Evol. 2009;26(11):2407–14. 10.1093/molbev/msp168 [DOI] [PubMed] [Google Scholar]
- 32.Holsbeeks I, Lagatie O, Van Nuland A, Van de Velde S, Thevelein JM. The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem Sci. 2004;29(10):556–64. 10.1016/j.tibs.2004.08.010 [DOI] [PubMed] [Google Scholar]
- 33.Bowman EJ, O'Neill FJ, Bowman BJ. Mutations of pma-1, the gene encoding the plasma membrane H+-ATPase of Neurospora crassa, suppress inhibition of growth by concanamycin A, a specific inhibitor of vacuolar ATPases. J Biol Chem. 1997;272(23):14776–86. 10.1074/jbc.272.23.14776 [DOI] [PubMed] [Google Scholar]
- 34.Vylkova S, Carman AJ, Danhof HA, Collette JR, Zhou H, Lorenz MC. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. MBio. 2011;2(3):e00055–11. 10.1128/mBio.00055-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minc N, Chang F. Electrical control of cell polarization in the fission yeast Schizosaccharomyces pombe. Curr Biol. 2010;20(8):710–6. 10.1016/j.cub.2010.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martínez-Espinoza AD, Ruiz-Herrera J, León-Ramírez CG, Gold SE. MAP kinase and cAMP signaling pathways modulate the pH-induced yeast-to-mycelium dimorphic transition in the corn smut fungus Ustilago maydis. Curr Microbiol. 2004;49(4):274–81. 10.1007/s00284-004-4315-6 [DOI] [PubMed] [Google Scholar]
- 37.Rane HS, Bernardo SM, Raines SM, Binder JL, Parra KJ, Lee SA. Candida albicans VMA3 is necessary for V-ATPase assembly and function and contributes to secretion and filamentation. Eukaryot Cell. 2013;12(10):1369–82. 10.1128/EC.00118-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan O, Shapiro RS, Kurat CF, Mayhew D, Baryshnikova A, Chin B, et al. Global gene deletion analysis exploring yeast filamentous growth. Science. 2012;337(6100):1353–6. 10.1126/science.1224339 [DOI] [PubMed] [Google Scholar]
- 39.Marini AM, Boeckstaens M, Benjelloun F, Chérif-Zahar B, André B. Structural involvement in substrate recognition of an essential aspartate residue conserved in Mep/Amt and Rh-type ammonium transporters. Curr Genet. 2006;49(6):364–74. 10.1007/s00294-006-0062-5 [DOI] [PubMed] [Google Scholar]
- 40.Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68(6):1077–90. 10.1016/0092-8674(92)90079-r [DOI] [PubMed] [Google Scholar]
- 41.Oh Y, Robertson SL, Parker J, Muddiman DC, Dean RA. Comparative proteomic analysis between nitrogen supplemented and starved conditions in Magnaporthe oryzae. Proteome Sci. 2017;15:20 10.1186/s12953-017-0128-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.López-Berges MS, Rispail N, Prados-Rosales RC, Di Pietro A. A nitrogen response pathway regulates virulence functions in Fusarium oxysporum via the protein kinase TOR and the bZIP protein MeaB. Plant Cell. 2010;22(7):2459–75. 10.1105/tpc.110.075937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang J, Zhao J, Duan W, Tian S, Wang X, Zhuang H, et al. TaAMT2;3a, a wheat AMT2-type ammonium transporter, facilitates the infection of stripe rust fungus on wheat. BMC Plant Biol. 2019;19(1):239 10.1186/s12870-019-1841-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang F, Li W, Jørgensen HJ. Transcriptional reprogramming of wheat and the hemibiotrophic pathogen Septoria tritici during two phases of the compatible interaction. PLoS ONE. 2013;8(11):e81606 10.1371/journal.pone.0081606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lanver D, Müller AN, Happel P, Schweizer G, Haas FB, Franitza M, et al. The Biotrophic Development of Ustilago maydis Studied by RNA-Seq Analysis. Plant Cell. 2018;30(2):300–23. 10.1105/tpc.17.00764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sriram K, Insel PA. G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs? Mol Pharmacol. 2018;93(4):251–8. 10.1124/mol.117.111062 [DOI] [PMC free article] [PubMed] [Google Scholar]