Abstract
Radiomics is a relatively new word for the field of radiology, meaning the extraction of a high number of quantitative features from medical images. Artificial intelligence (AI) is broadly a set of advanced computational algorithms that basically learn the patterns in the data provided to make predictions on unseen data sets. Radiomics can be coupled with AI because of its better capability of handling a massive amount of data compared with the traditional statistical methods. Together, the primary purpose of these fields is to extract and analyze as much and meaningful hidden quantitative data as possible to be used in decision support. Nowadays, both radiomics and AI have been getting attention for their remarkable success in various radiological tasks, which has been met with anxiety by most of the radiologists due to the fear of replacement by intelligent machines. Considering ever-developing advances in computational power and availability of large data sets, the marriage of humans and machines in future clinical practice seems inevitable. Therefore, regardless of their feelings, the radiologists should be familiar with these concepts. Our goal in this paper was three-fold: first, to familiarize radiologists with the radiomics and AI; second, to encourage the radiologists to get involved in these ever-developing fields; and, third, to provide a set of recommendations for good practice in design and assessment of future works.
Radiomics is a new word for the field of radiology, deriving from a combination of “radio”, meaning medical images, and “omics”, indicating the various fields like genomics and proteomics that contribute to our understanding of various medical conditions. Radiomics is simply the extraction of a high number of features from medical images (1). The typical radiomic analysis includes the evaluation of size, shape, and textural features that have useful spatial information on pixel or voxel distribution and patterns (1). These radiomic features are further used in creating statistical models with an intent to provide support for individualized diagnosis and management in a variety of organs and systems such as brain (2, 3), pituitary gland (4, 5), lung (6), heart (7), liver (8), kidney (9–12), adrenal gland (13, 14), and prostate (15).
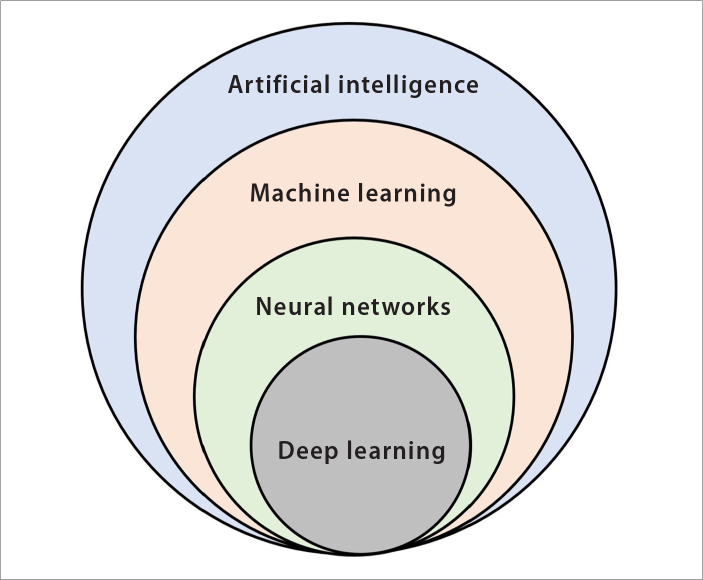
Artificial intelligence (AI) is broadly a set of systems that can accurately perform inferences from a large amount of data, based on advanced computational algorithms (16). Just as in humans, learning is a fundamental need for any intelligent behavior of machines. Hence, the AI is a general concept encompassing different learning algorithms, namely, machine learning (ML) and lately very popular deep learning algorithms (Fig. 1) (17, 18). Although the concept of AI goes back to 1950s, it has gained momentum since 2000 because of the advances in computational power (19–21). Today, AI technology provides numerous indispensable tools for intelligent data analysis for solving several medical problems, particularly for diagnostic issues (17, 18, 21–24).
Figure 1.
Venn diagram of the concepts related to artificial intelligence (AI). AI is the simulation of human intelligence processes like learning, reasoning, and self-correction by the machines, particularly the computer systems. AI is a broad concept that covers many machine learning techniques such as k-nearest neighbors, support vector machine, decision trees, and neural networks. Neural networks include various algorithms ranging from very simple to complex architectures, such as multi-layer perceptron and deep learning or convolutional neural networks.
Relationship between radiomics and AI are mutual. Due to its ever-growing high-dimensional nature, the field of radiomics needs much more powerful analytic tools, and AI appears to be a potential candidate for this purpose, with its extreme capabilities. On the other hand, in medical image analysis, AI applications inevitably need the radiomics because the metrics that are used to train and build the AI models are delivered through radiomic approaches, specifically, feature extraction and feature engineering techniques.
In this paper, we reviewed the radiomics and AI with a rather practical point of view. Our goal was three-fold: first, to familiarize the radiologists with the radiomics and AI; second, to encourage them to get involved in these ever-developing fields; and, third, to provide a set of recommendations and tips for good practice.
Critical questions and answers
Why do we need radiomics?
In conventional radiology practice, except for a few measurements like size and volume, the imaging data sets are generally evaluated visually or qualitatively. This approach not only involves intra- and interobserver variability but also leaves a very large amount of hidden data in the medical images unused. A common clinical scenario for explaining the need for radiomics would be possible with imagining two patients with tumors with rather different qualitative features like size, shape, borders, and heterogeneity. The survival of the patients in this scenario will probably be different even though the tumors have histopathologically similar features. If one could have predicted the prognosis of the patients before any intervention or treatment, the management of the patients would be different. This is actually called precision medicine. In precision medicine, the patients that belong to different subtypes need to be identified for achieving better outcomes. Radiomics can be considered an objective way to achieve these goals. Using either conventional (1) or advanced imaging techniques (25, 26), the primary purpose of the radiomics is to extract as much and meaningful hidden objective data as possible to be used in decision support.
Why do we need AI in radiomics?
The main reason for using AI in radiomics is its better capability of handling a massive amount of data compared with the traditional statistical methods. AI algorithms are essentially used for classification problems. These algorithms basically learn the data provided by analyzing patterns and then make predictions on unseen data sets to check whether these patterns are correct or not. AI algorithms are not only able to analyze the numeric data provided by the predefined or hand-crafted radiomic features but also able to directly analyze the images in order to automatically design its own radiomic features (17, 27–30). This very popular and advanced subset of AI is called deep learning (28). Deep learning algorithms are also able to perform segmentation tasks itself, without any need for human intervention (31).
Is it possible to get involved in radiomics as a radiologist?
Yes, that is perfectly possible. Collective work is of paramount importance because the workflow of radiomics covers a wide range of consecutive steps including preprocessing, segmentation, feature extraction, and data handling (1). Depending on the software used, each step might require a massive amount of time and workload. Authors think that there would be at least three ways to get involved in radiomics in any subfield of medical imaging.
First, the simplest way would be to look for the paid software programs. Those kinds of programs are easy to use because the providers simplified almost all radiomic pipeline. Some of those could provide some statistical tools for further analysis as well.
Second, a little bit harder way would be to use free software programs that allow radiomic feature extraction with a graphical user interface (GUI). Most popular software programs for hand-crafted feature extraction are MaZda (32), LIFEx (33), PyRadiomics (34), and IBEX (35). Nonetheless, even though the authors encourage the radiologists starting with these software programs, they also highly recommend being cautious because the pipeline is not well established in such programs and there are many parameters to be dealt with such as establishing discretization levels, normalization approach, re-sampling, and clearing the non-radiomic data from final feature table. Furthermore, there are also software programs for deep feature extraction with GUI directly from images within the layers of neural network such as Nvidia’s Digits (https://developer.nvidia.com/digits) and Deep Learning Studio (https://deepcognition.ai/).
Third, the hardest way would be to use software programs that allow feature extraction if the user has coding skills or at least familiarity with coding. Most popular platforms for this purpose are MATLAB and Python platforms, which have massive libraries for both hand-crafted and deep feature extraction.
Is it possible to get involved in AI as a radiologist?
Yes, that is perfectly possible as well. Authors think that there would be at least three ways to get involved in AI as a radiologist without formal training about data or computer science.
First, the simplest way would be to find or to be a part of a data science collaboration about medical imaging. Data or computer scientists need meaningful clinical perspectives to provide the unmet need for AI in radiology.
Second, a little bit harder but not the hardest way would be to get some conventional statistical basis and to learn how to use data mining software programs that allow performing AI tasks without knowing how to code. There are many free software programs for this purpose such as Waikato environment for knowledge analysis (WEKA) software (36), Orange data mining software (37), RapidMiner (https://rapidminer.com/), Rattle in R statistics (38), and Deep learning studio (https://deepcognition.ai/). All of these software programs have a GUI to easily implement a wide range of AI tasks covering the very simple to very complex ML algorithms. Also, some of those software programs have options for integration to other common environments (e.g., Python and R) for much more advanced features. Being radiologists, the authors recommend starting first with WEKA or Orange software-like programs considering their simplicity and ease in using the interface. On the other hand, it should be kept in mind that not every software is capable to complete every task. For instance, in our personal experience, WEKA is enough to perform many ML tasks, but it has limited and poor visual capabilities unless it is integrated with the other environments.
Third, the hardest way is, of course, to start with learning how to code. Although it usually seems difficult and daunting to learn to code from scratch, there are very simple languages to start with, such as Python language, which is an object-oriented language with an intuitive and easy to understand syntax, being rather similar to human language. Learning Python language provides various opportunities to use many available AI libraries such as Google’s TensorFlow even for users with low-level programming skills. There are extensive resources to learn to code for AI implementation like books, websites, and online courses (e.g., Coursera, Udemy, edX) at a low cost.
What about the future of radiologists considering the advances in AI?
As it can be seen in recent world-wide annual radiology meetings like RSNA (Radiological Society of North America) and ECR (European Congress of Radiology), there is an evident shift of the overall theme to radiomics and AI, which is much more apparent than any other medical field. Both radiomics and AI have been getting attention for their remarkable success in various radiological tasks, which has been met with anxiety by most of the radiologists due to the fear of replacement by the intelligent machines. Considering ever-developing advances in computational power and availability of large data sets, the marriage of humans and machines in future clinical practice seems inevitable. Therefore, regardless of their feelings, the radiologists should be familiar with these concepts. Authors believe that the radiomics with AI might be helpful for the radiologists by completing or facilitating certain tasks to some extent, reducing the heavy workload of the radiologists, which actually would make the radiologists much more intelligent than ever by providing an opportunity to deal with only the more complex and sophisticated radiological problems in their practice.
Radiomic workflow
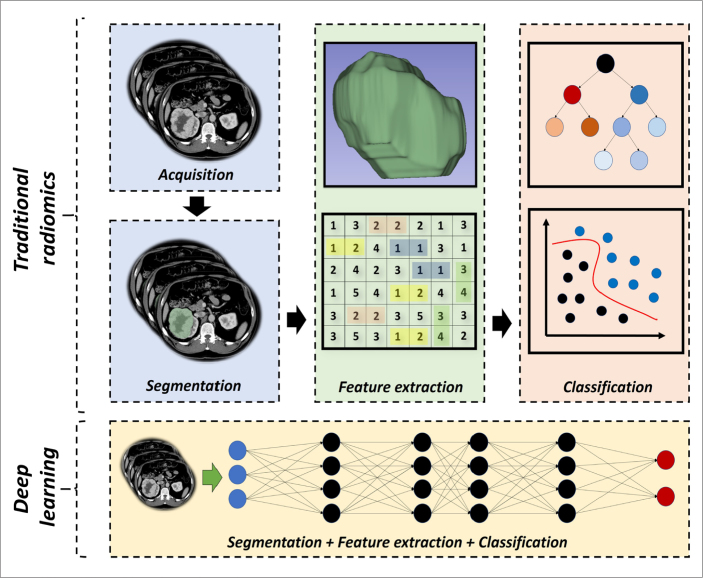
To provide a wider perspective to the readers, over-simplified radiomic pipelines are simply given in Fig. 2 before going into a detailed review of each step.
Figure 2.
Over-simplified representation of traditional and deep learning-based radiomics. Representative CT and MRI images in Fig. 2 and Fig. 3 are obtained from the Cancer Imaging Archive (TCIA), specifically from the collections of TCGA-KIRC (72, 73) and LGG-1p19qDeletion (73–75), which are publicly and freely available.
Image acquisition
Radiomics can be applied to various imaging techniques including computed tomography, magnetic resonance imaging (MRI), positron-emission tomography, X-ray, and ultrasonography. There are a wide variety of acquisition techniques currently in use. Besides, different vendors offer various image reconstruction methods that are customized at each institution depending on the need. This is not only a problem in multi-institutional scale but also a problem in the same institution. Although it is usually underestimated or ignored in visual analysis, the use of different acquisition and image processing techniques might have a great impact in radiomics because it is a process on pixel or voxel level, which may affect image noise and in turn texture, possibly reflecting a different underlying pathology (39, 40). These differences might also lead to inconsistent results in radiomic analyses in independent data sets, which is one of the major problems of the radiomics (39, 40). From a realistic perspective, we should acknowledge that it is not possible to bring all the image acquisition protocols into uniformity. On the other hand, our primary goal should be to find the best technical pipeline to create the most stable and accurate radiomic models that are even applicable to the images obtained with different protocols. To do this, each imaging modality must be taken care of considering their own peculiarities.
Preprocessing
Radiomics has a dependency on some image parameters. The most important of those that need to be dealt with in any imaging modality are the size of the pixel or voxels (41), number of the gray levels (41), and range of gray level values (42). In addition, signal intensity nonuniformity should be removed in MRI (43, 44). There are numerous methods for dealing with these dependencies. For the normalization of the gray level values, the ±3sigma normalization is the most widely used method (45). Pixel resampling can be done using various interpolation methods such as linear and cubic B-spline interpolation (46). Different software programs offer different discretization methods, for instance, fixed bin size and fixed bin number (47). N3 and N4 bias field correction algorithms are widely established techniques for avoiding signal intensity nonuniformity (44). Although some of these preprocessing steps are included in the radiomic software programs, it should be known that many user-friendly open-source tools exist for advanced radiologic imaging data preprocessing such as ImageJ, MIPAV (Medical Image Processing, Analysis, and Visualization), and 3DSlicer.
Segmentation
The most critical step in radiomics is considered to be the segmentation process because the radiomic features are mostly extracted from the segmented areas or volumes. The segmentation process is challenging because of the fact that some tumors have a very unclear margin. The manual segmentation is considered the gold standard provided that it is performed by experts, which is very time-consuming. On the other hand, manual segmentation is subject to intra- and inter-reader variability (48), leading to radiomic feature reproducibility problems. To avoid this variability, a few automatic and semi-automatic methods have been described as follows: active contour (snake) methods (49), level set methods (50), region-based methods (51), graph-based methods (52), and deep learning-based methods (53). Although the automatic segmentation techniques are objective, they are prone to error, especially when images have artifacts and noise and lesions of interest are very heterogeneous.
Feature extraction
Considering the definition of radiomic features, most of them are not part of the radiologists’ lexicon. In this context, it should be kept in mind that radiomics is a hypothesis-free approach. This means that there is no a priori hypothesis made about the clinical relevance of the features, which are computed automatically by image analysis algorithms created by experts. The purpose of the approach is to discover previously unseen image patterns using these agnostic or non-semantic features and to perform classification based on the most discriminative ones, this is also named as the development of radiomic signature. Authors’ view on the subject is also the same. As long as the models are validated on independent data sets, radiomics might be a valid approach, regardless of the individual meaning of the features. In summary, the whole process means that except for some of the histogram or first-order features, if one attempts to define each radiomic feature in a clinical context, it probably results in failure.
There are two categories of radiomic features. The first one is predefined or hand-crafted features, being created by human image processing experts. These are also called as traditional features. Some of the traditional radiomic features (i.e., predefined or hand-crafted features) are presented in Table 1. The second one is deep features, which has gained popularity nowadays because some deep learning algorithms design and select the features themselves for a given task within its layers, without need for any human intervention (28). Some recent works have also suggested the superiority of the deep features to traditional features (54, 55).
Table 1.
Examples of traditional radiomic features
| Feature categories | Example radiomic features |
|---|---|
| Size | Area |
| Volume | |
| Maximum 3D diameter | |
| Major axis length | |
| Minor axis length | |
| Surface area | |
|
| |
| Shape | Elongation |
| Flatness | |
| Sphericity | |
| Spherical disproportion | |
|
| |
| First-order texturea | Energy |
| Entropy | |
| 10th percentile | |
| 90th percentile | |
| Skewness | |
| Kurtosis | |
|
| |
| Second-order textureb | Gray level co-occurrence matrix |
| Gray level run length matrix | |
| Gray level size zone matrix | |
|
| |
| High-order texturec | Autoregressive model |
| Haar wavelet | |
First-order features describe the distribution of intensity within the segmentation.
Second-order features describe the statistical relationships between pixels or voxels.
High-order features are usually based on matrices that consider relationships between three or more pixels or voxels.
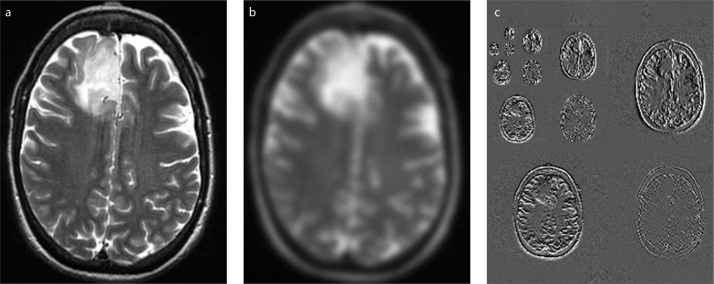
Radiomic features can be extracted using different image types, which contributes to the high-dimensionality of the radiomics. Commonly encountered image types are presented in Fig. 3.
Figure 3 .a–c.
Different image types for radiomic feature extraction: (a), original image; (b), filtered image; (c), wavelet-transformed images. Representative CT and MRI images in Fig. 2 and Fig. 3 are obtained from the Cancer Imaging Archive (TCIA), specifically from the collections of TCGA-KIRC (72, 73) and LGG-1p19qDeletion (73–75), which are publicly and freely available.
Radiomic data handling
Data preparation
Before further analysis of the radiomic data obtained using AI algorithms, certain conditions need to be addressed. Possible data preparation steps would be as follows: feature scaling, discretization, continuization, randomization, over-sampling, under-sampling, and so on.
Considering their major impact in AI-based classification performance, the authors recommend that at least feature scaling and randomization need to be considered in every scientific work.
Radiomic feature values are produced in different scales, which highly interferes with the stability of inner parameters of the AI algorithms, for instance, weights and biases of the artificial neural network. Feature scaling means changing the numeric values to a common scale, avoiding significant distortions in the ranges of values. Feature scaling involves two broad categories: normalization and standardization. The choice of the technique depends on the assumptions about the distribution of the data that AI algorithms make that will be used in further analysis.
Randomization of the data set, on the other hand, is another important factor in creating models because the performance of the ML algorithms is influenced by the initiation or seeding factors. If it is not applied before model creation, some patterns in the data set might strongly influence the results.
Class balance is an important factor to reveal the actual performance of ML classifiers. In the case of significant imbalance, the results might be misleading. To deal with this problem, over-sampling and under-sampling techniques can be used. One of the commonly-used and accepted techniques for balancing the classes is synthetic minority over-sampling technique (SMOTE) (56), which creates new and similar instances from the minority class that are not the exact replications of the actual instances.
Dimension reduction
Radiomic approaches generally lead to high-dimensionality, meaning that they produce a very large number of features to be dealt with. It is a common practice to bring the high-dimensionality to lower levels to optimize the classifier performance, which is basically called dimension reduction (57). The dimension reduction can be done using different approaches such as feature reproducibility analysis (58), collinearity analysis (9), algorithm-based feature selection (57, 59), and cluster analysis.
Feature reproducibility analysis should be done for evaluation of the features that are sensitive to segmentation variabilities (58), particularly the segmentation tasks that need human intervention (10). Furthermore, if possible, this analysis should be extended to evaluate the influence of the different acquisition protocols (60–62). The goal of the reproducibility analysis is to reduce the dimension by excluding the features with relatively poor reproducibility. One of the most common statistical tools for this analysis is the intra-class correlation coefficient (ICC) (63). There are different types of ICC that need to be considered in the analysis (63).
Collinearity analysis is another plausible way of dimension reduction because a very large number of the features have similar information and the extent of which is called the strength of collinearity (64). Pearson’s correlation coefficient can be used to determine redundant features, in other words, the collinear features. If a pair of radiomic features had high collinearity, the one having the highest collinearity with the others should be excluded from the analysis. Of note, there are also some algorithms for this purpose that selects features based on the collinearity status and maximum relevance to the classes, for instance, correlation-based feature selection algorithm (59). These algorithms are useful because it reduces the workload in dimension reduction by doing two techniques at the same time, that is, collinearity analysis and feature selection.
The most widely used dimension reduction technique is algorithm-based feature selection (57). There are various algorithms with different functions such as least absolute shrinkage and selection operator (65), correlation-based feature selection algorithm (59), ReliefF (66), and Gini index (67). The researchers should experiment with these algorithms for achieving the best results.
The most confusing issue in dimension reduction is the final number of features that should be achieved. Although there is no guideline about this, it would be good to reduce the total number of features at least to one-tenth of the total labeled data. However, authors also think that although it is better to keep the number of features as low as possible, it should not be a major concern as long as they are validated on the independent external data with a satisfying performance.
AI-based statistical analysis
Requirements before an AI initiative
There are certain musts that need to be taken care of before an AI initiative: (i), consistent data; (ii), well curation of the data; (iii), expert-driven processing of the data; and (iv) a valid clinical problem or problems to be answered by the AI.
Sample size is also a significant issue to be considered before an AI-based analysis. Although it is usual to encounter AI or ML-based studies with a very small number of patients in the literature, the radiologists should be aware that the sample size is an important factor to avoid some problems in model fitting (Fig. 4) and to improve the generalizability on unseen data. Particularly for very complex algorithms like deep learning, there is absolute need of massive amount of data. Nonetheless, in case of limited or small data, it should be known that there are some well-known augmentation techniques (e.g., image transformation, synthetic minority over-sampling) to be considered as well.
Figure 4.
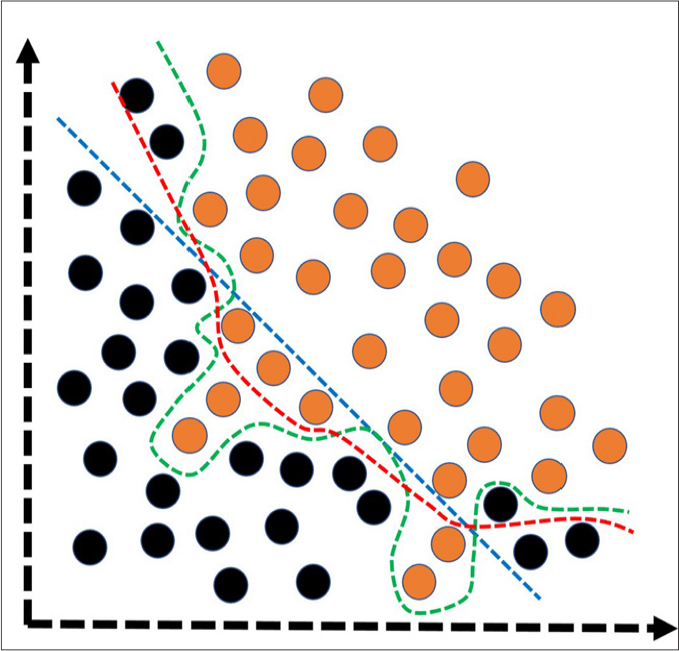
Simplified illustration of the model fitting spectrum. Under-fitting (blue dashed-line) and over-fitting (green dashed-line) are common problems to be solved to create more optimally-fitted (red dashed-line) and generalizable models that are useful on unseen or new data. Under-fitting corresponds to the models having poor performance on both training and test data. In general, the under-fitting problem is not discussed because it is evident in the evaluation of performance metrics. Over-fitting, on the other hand, refers to the models having an excellent performance in training data, but very poor performance on test data. In models with over-fitting, the algorithm learns both the relevant data and the noise that is the primary reason of the over-fitting. In reality, all data sets have noise to some extent. However, in case of small data, the effect of the noise could be much more evident. To reduce the over-fitting, possible steps would be to expand the data size, to use data augmentation techniques, to utilize architectures that generalize well, to use regularization techniques (e.g., L1–L2 regularizations and drop-out), and to reduce to the complexity of the architecture or to use less complex classification algorithms. Black and orange circles represent different classes.
The perception of AI and its training is underestimated by many others in the field. In contrast to the AI systems designed for the distinction of daily life pictures, this task is a little bit more difficult in the field of medicine. Because a nonprofessional or layperson cannot provide reliable processed data for training, experts, in other words, good radiologists and particularly dedicated ones are needed.
Model development
Model development can be done using various algorithms. The most common algorithms are k-nearest neighbors (Fig. 5), naive Bayes (Fig. 6), logistic regression (Fig. 7), support vector machine (Fig. 8), decision tree (Fig. 9a), random forest (Fig. 9b), neural networks, and deep learning (Fig. 10) (18). These algorithms can also be combined with meta-classifiers or ensemble techniques like adaptive boosting and bootstrap aggregation to enhance generalizability (10). Furthermore, there are also other ensemble learning techniques that are composed of more than one algorithm, particularly weak classifiers like k-nearest neighbors, naive Bayes, and C4.5 tree algorithms (68). Although the selection of the algorithm seems to be arbitrary in the literature, the best practice would be the selection of the algorithm with multiple experiments.
Figure 5.

Over-simplified illustration of k-nearest neighbors. This machine learning algorithm classifies the unknown objects or instances (blue triangle) by assigning them to the similar objects of the classes (orange vs. black circles) based on the number of neighbors. For instance, considering 3-nearest neighbors, the class represented with black circles outnumbers the other class (orange circles) so that the unknown object is assigned to the class represented with black circles. On the other hand, in case of 5-nearest neighbors, it is assigned to the class with orange circles because the number of the instances in this class outnumbers the one with black circles.
Figure 6.
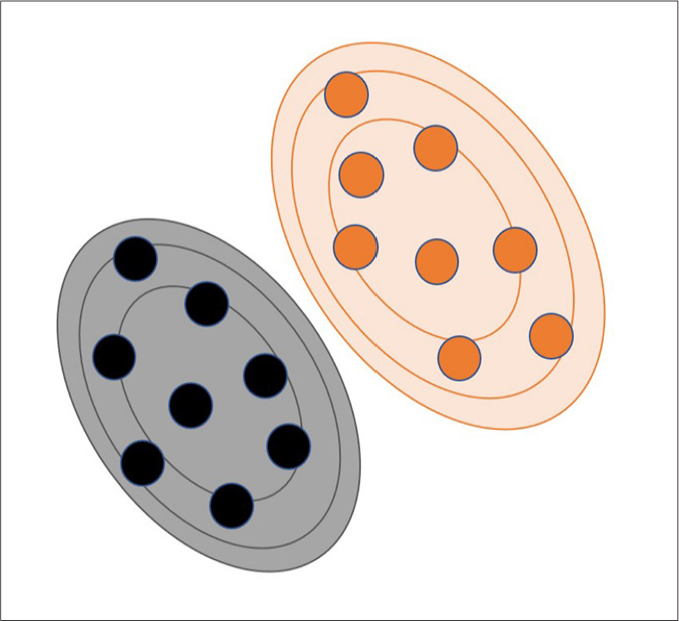
Over-simplified illustration of naive Bayes in a probabilistic space. Naive Bayes is a probabilistic machine learning algorithm and simply based on the strong (naive) independence among the predictor variables (or features). Also, this algorithm assumes that all features equally contribute to the outcome or class prediction. Black and orange circles represent different classes. Black and orange lines represent different probability levels for the instances in different classes.
Figure 7.
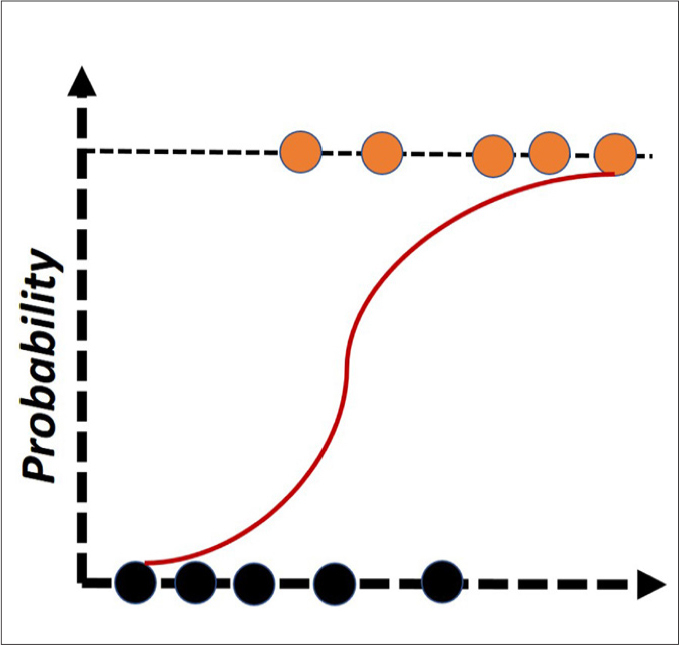
Over-simplified illustration of logistic regression. Even though many extensions of the logistic regression exist, this algorithm simply uses the logistic function to classify the instances to the binary classes. Black and orange circles represent different classes.
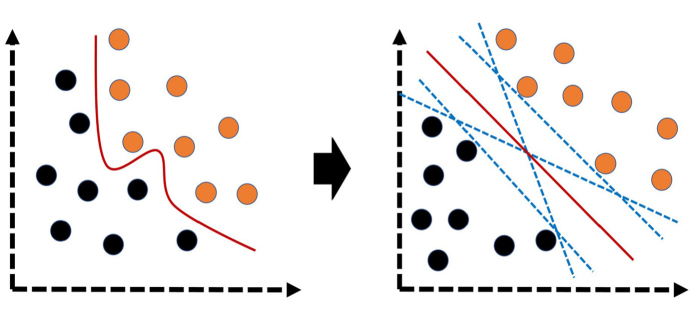
Figure 8.
Over-simplified illustrations of support vector machine. In simple terms, this algorithm transforms the original data (left illustration) to a different space (right illustration) to develop optimal plane or vector (red line) that separates the classes. Black and orange circles represent different classes.
Figure 9. a, b.
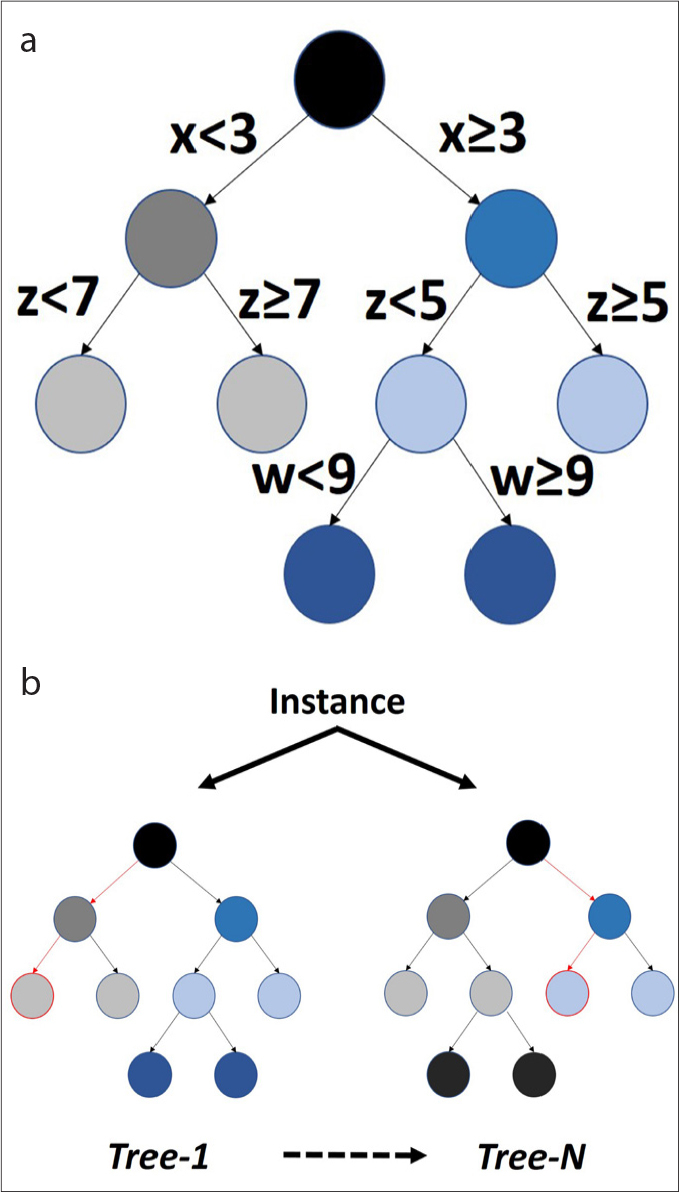
Over-simplified illustrations of decision tree and random forest. In panel (a), decision tree simply creates the most accurate and simple decision points in classification of the instances, providing the most interpretable models for the humans; x, z, and w represent features. In panel (b), to increase the stability and generalizability of the classifications, decision tree algorithm can be iterated several times with various methods. One of the well-known examples is the random forest classifier.
Figure 10.
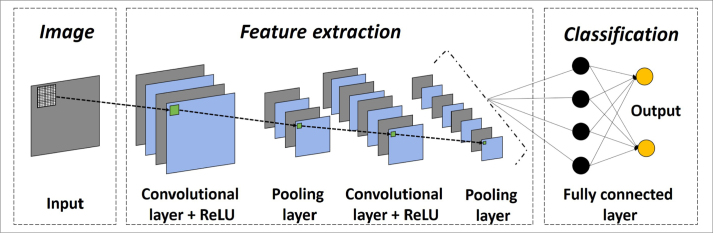
Over-simplified illustration of artificial neural network, particularly deep learning. Neural networks are multi-layer networks of neurons or nodes that are inspired by biological neuronal architecture. Due to computational limitations, early neural networks had very few layers of nodes, generally fewer than 5. Today, it is possible to create useful neural network architectures with many layers. Deep learning or deep neural network generally corresponds to a network with more than 20–25 hidden layers. Variety of deep learning architectures exist in that convolutional neural networks (CNN) are widely used in image analysis. In CNN, image inputs are directly scanned using small-sized filters or kernels, creating transformed images within certain layers like convolutional ones. Convolutional and pooling (or down-sampling) layers are important operations in the CNN architectures, providing the best and most important features of the images (e.g., edges). There are also many important parts of deep learning architectures like activation functions (e.g., rectified linear unit [ReLU], sigmoid function, softmax), regularization (e.g., drop-out layer), and so on. Today, no formula exists to establish the correct number and type of layers for a given classification problem. Therefore, optimal architecture is created with a trial-and-error process. On the other hand, some previously proven architectures and their derivatives are also widely used in similar tasks such as U-Net for segmentation process.
Validation
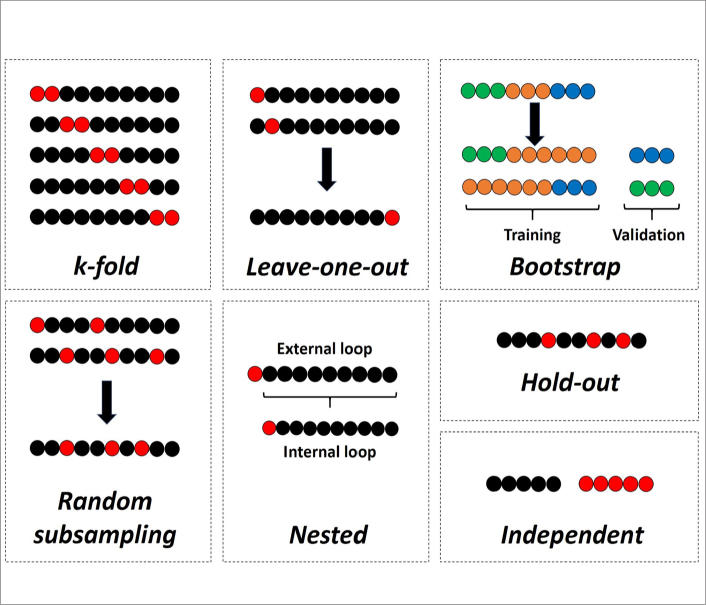
Nowadays, radiomics is considered a mere research area. In order to be accepted in the clinical arena, the results need to be validated using independent data sets, preferably using data from a different institution (1, 69). Hence, the most valuable strategy for the validation of models is considered the independent external validation. However, in small scale pilot or preliminary works, it is not always possible to have such independent validation data. In such cases, internal validation techniques can be used. The most common internal validation techniques that can be encountered in the literature are k-fold, leave-one-out cross-validation, and hold-out. In addition, there are much more sophisticated techniques such as random subsampling, bootstrap cross-validation, and nested cross-validation. Widely used validation techniques are simply presented in Fig. 11 with a didactic approach. Selection of the cross-validation technique mostly depends on the need and capability of the performer in the software along with the specifications of the hardware used. The most important problem in internal validation that must be considered is the possible leakage of the feature selection algorithm in the whole data, which might lead to overly optimistic results. For creating such unseen data sets, although the hold-out technique seems to be the most appropriate internal validation method, there is also nested cross-validation technique that is primarily used for this purpose and might give similar estimates to an independent validation (70).
Figure 11.
Validation techniques with over-simplified illustrations. In k-fold cross-validation, data set is systematically split to k number of folds, with no overlap in validation parts. In leave-one-out cross-validation, data set is systematically divided to a number that is equal to the number of labeled data set, with no overlap in validation parts. In bootstrapping validation, whole data is sampled to create unseen validation parts that are filled or replaced with similar labeled data in the training data set. In random subsampling, data set is randomly sampled many times to create validation parts that may have overlaps in different experiments. In nested cross-validation, the internal loop is used for feature selection along with model optimization; and external loop is used for model validation to simulate an independent process. In hold-out technique, a single split is created with random sampling. In independent validation, validation part corresponds to a completely different data set, preferably an external data set. Except for bootstrapping validation, black and red circles represent training and validation data sets, respectively.
Performance evaluation
Performance evaluation of the classifications is generally done using the area under the receiver operating characteristic curve (AUC) (39). It should be kept in mind that AUC might be a poor performance evaluator if the data set has a class imbalance. For this reason, other performance metrics like accuracy, sensitivity, specificity, precision, recall, F1 measure, and Matthews correlation coefficient should be supplied for further assessment.
Comparison of the validation performance of the AI algorithms can be done by conventional statistical methods (71). Depending on the assumptions of the methods and the number of the classifiers, commonly used statistical tools for comparisons are student t-test, Wilcoxon signed-rank test, analysis of variance, Friedman test, and so on. In multiple comparisons, the multiplicity problem needs to be addressed. The best performing and stable classifier or classifiers are generally selected for the clinical application of interest.
Final recommendations
Radiomics and AI are vast fields with their wide range of different methodologic aspects. This variety leads to a lack of consensus in many steps, which is a challenge that needs to be overcome in the near future. In Table 2, the authors recommend using a checklist of the key features that need to be at least considered and transparently reported in future AI-based radiomic works. Although it is not possible to cover all aspects of radiomics and AI in a review article, we believe the key features included in this paper will be helpful for researchers, reviewers, and the future of the radiomics.
Table 2.
Checklist of the key features that need to be considered and transparently reported in AI-based radiomic studies
| Study parts | Key features |
|---|---|
| Baseline study characteristics | Nature (Retrospective/Prospective) |
| Unmet need for radiomics | |
| Sample size with details of classes | |
| Data source (Single/Multi-institutional/Public) | |
| Data curation by experts | |
| Data overlap | |
| Eligibility criteria | |
|
| |
| Scanning protocol | Acquisition protocol |
| Processing protocol | |
|
| |
| Preprocessing | Pixel or voxel resampling |
| Discretization | |
| Normalization | |
| Bias field correction | |
| Different image types | |
| Registration | |
|
| |
| Segmentation | Manual/Semi-automated/Full-automated |
| 2D/3D/a few slice-based | |
| Excluded/Included regions | |
|
| |
| Feature extraction | Software details |
| Feature types | |
| References for equations | |
| Different image types (Original/Filtered/Transformed) | |
|
| |
| Reliability analysis | Reproducibility analysis to exclude features with poor reproducibility
|
|
| |
| Data handling | Randomization |
| Normalization | |
| Standardization | |
| Class balance | |
| Data augmentation | |
| Collinearity analysis | |
| Feature selection | |
|
| |
| AI-based statistical analysis | Adequacy of sample size considering complexity of AI algorithm |
| Algorithm parameters | |
| Experiments with different algorithms | |
| Validation technique | |
| Precautions for over- and under-fitting | |
| Details for separation of feature selection and model validation | |
| Different performance metrics | |
| Statistical comparison of classification performance | |
AI, artificial intelligence; 2D, two-dimensional; 3D, three-dimensional.
Main points.
Radiomics is simply the extraction of a high number of quantitative features from medical images.
Artificial intelligence is broadly a set of advanced computational algorithms that can accurately perform predictions for decision support.
Primary purpose of radiomics and artificial intelligence is to extract and analyze as much and meaningful hidden quantitative imaging data as possible to be used in objective decision support for any medical condition of interest.
Radiomics and artificial intelligence are vast fields with a wide range of different methodologic aspects, leading to a lack of consensus in many steps, which is a challenge that needs to be overcome in the near future.
Footnotes
Conflict of interest disclosure
The authors declared no conflicts of interest.
References
- 1.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su C, Jiang J, Zhang S, et al. Radiomics based on multicontrast MRI can precisely differentiate among glioma subtypes and predict tumour-proliferative behaviour. Eur Radiol. 2019;29:1986–1996. doi: 10.1007/s00330-018-5704-8. [DOI] [PubMed] [Google Scholar]
- 3.Wang Q, Li Q, Mi R, et al. Radiomics nomogram building from multiparametric MRI to predict grade in patients with glioma: a cohort study. J Magn Reson Imaging. 2019;49:825–833. doi: 10.1002/jmri.26265. [DOI] [PubMed] [Google Scholar]
- 4.Kocak B, Durmaz ES, Kadioglu P, et al. Predicting response to somatostatin analogues in acromegaly: machine learning-based high-dimensional quantitative texture analysis on T2-weighted MRI. Eur Radiol. 2019;29:2731–2739. doi: 10.1007/s00330-018-5876-2. [DOI] [PubMed] [Google Scholar]
- 5.Zeynalova A, Kocak B, Durmaz ES, et al. Preoperative evaluation of tumour consistency in pituitary macroadenomas: a machine learning-based histogram analysis on conventional T2-weighted MRI. Neuroradiology. 2019;61:767–774. doi: 10.1007/s00234-019-02211-2. [DOI] [PubMed] [Google Scholar]
- 6.Yang L, Yang J, Zhou X, et al. Development of a radiomics nomogram based on the 2D and 3D CT features to predict the survival of non-small cell lung cancer patients. Eur Radiol. 2019;29:2196–2206. doi: 10.1007/s00330-018-5770-y. [DOI] [PubMed] [Google Scholar]
- 7.Mannil M, von Spiczak J, Muehlematter UJ, et al. Texture analysis of myocardial infarction in CT: Comparison with visual analysis and impact of iterative reconstruction. Eur J Radiol. 2019;113:245–250. doi: 10.1016/j.ejrad.2019.02.037. [DOI] [PubMed] [Google Scholar]
- 8.Peng J, Zhang J, Zhang Q, Xu Y, Zhou J, Liu L. A radiomics nomogram for preoperative prediction of microvascular invasion risk in hepatitis B virus-related hepatocellular carcinoma. Diagn Interv Radiol. 2018;24:121–127. doi: 10.5152/dir.2018.17467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kocak B, Durmaz ES, Ates E, Kaya OK, Kilickesmez O. Unenhanced CT texture analysis of clear cell renal cell carcinomas: a machine learning-based study for predicting histopathologic nuclear grade. AJR Am J Roentgenol. 2019:W1–W8. doi: 10.2214/AJR.18.20742. [DOI] [PubMed] [Google Scholar]
- 10.Kocak B, Yardimci AH, Bektas CT, et al. Textural differences between renal cell carcinoma subtypes: Machine learning-based quantitative computed tomography texture analysis with independent external validation. Eur J Radiol. 2018;107:149–157. doi: 10.1016/j.ejrad.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 11.Kocak B, Durmaz ES, Ates E, Ulusan MB. Radiogenomics in clear cell renal cell carcinoma: machine learning-based high-dimensional quantitative CT texture analysis in predicting PBRM1 mutation status. AJR Am J Roentgenol. 2019;212:W55–W63. doi: 10.2214/AJR.18.20443. [DOI] [PubMed] [Google Scholar]
- 12.Bektas CT, Kocak B, Yardimci AH, et al. Clear cell renal cell carcinoma: machine learning-based quantitative computed tomography texture analysis for prediction of fuhrman nuclear grade. Eur Radiol. 2019;29:1153–1163. doi: 10.1007/s00330-018-5698-2. [DOI] [PubMed] [Google Scholar]
- 13.Ho LM, Samei E, Mazurowski MA, et al. Can texture analysis be used to distinguish benign from malignant adrenal nodules on unenhanced CT, contrast-enhanced CT, or In-phase and opposed-phase MRI? AJR Am J Roentgenol. 2019;212:554–561. doi: 10.2214/AJR.18.20097. [DOI] [PubMed] [Google Scholar]
- 14.Shi B, Zhang G-M-Y, Xu M, Jin Z-Y, Sun H. Distinguishing metastases from benign adrenal masses: what can CT texture analysis do? Acta Radiol. 2019 doi: 10.1177/0284185119830292. 284185119830292. [DOI] [PubMed] [Google Scholar]
- 15.Min X, Li M, Dong D, et al. Multi-parametric MRI-based radiomics signature for discriminating between clinically significant and insignificant prostate cancer: Cross-validation of a machine learning method. Eur J Radiol. 2019;115:16–21. doi: 10.1016/j.ejrad.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 16.Thrall JH, Li X, Li Q, et al. Artificial intelligence and machine learning in radiology: opportunities, challenges, pitfalls, and criteria for success. J Am Coll Radiol. 2018;15(3 Pt B):504–508. doi: 10.1016/j.jacr.2017.12.026. [DOI] [PubMed] [Google Scholar]
- 17.Chartrand G, Cheng PM, Vorontsov E, et al. Deep learning: a primer for radiologists. Radiographics. 2017;37:2113–2131. doi: 10.1148/rg.2017170077. [DOI] [PubMed] [Google Scholar]
- 18.Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine learning for medical imaging. Radiographics. 2017;37:505–515. doi: 10.1148/rg.2017160130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arimura H, Soufi M, Kamezawa H, Ninomiya K, Yamada M. Radiomics with artificial intelligence for precision medicine in radiation therapy. J Radiat Res. 2019;60:150–157. doi: 10.1093/jrr/rry077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kononenko I. Machine learning for medical diagnosis: history, state of the art and perspective. Artif Intell Med. 2001;23:89–109. doi: 10.1016/S0933-3657(01)00077-X. [DOI] [PubMed] [Google Scholar]
- 21.Auffermann WF, Gozansky EK, Tridandapani S. Artificial intelligence in cardiothoracic radiology. AJR Am J Roentgenol. 2019 Feb;19:1–5. doi: 10.2214/AJR.18.20771. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 22.Harmon SA, Tuncer S, Sanford T, Choyke PL, Türkbey B. Artificial intelligence at the intersection of pathology and radiology in prostate cancer. Diagn Interv Radiol. 2019;25:183–188. doi: 10.5152/dir.2019.19125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le EPV, Wang Y, Huang Y, Hickman S, Gilbert FJ. Artificial intelligence in breast imaging. Clin Radiol. 2019;74:357–366. doi: 10.1016/j.crad.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Bi WL, Hosny A, Schabath MB, et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J Clin. 2019;69:127–157. doi: 10.3322/caac.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harry VN, Semple SI, Parkin DE, Gilbert FJ. Use of new imaging techniques to predict tumour response to therapy. Lancet Oncol. 2010;11:92–102. doi: 10.1016/S1470-2045(09)70190-1. [DOI] [PubMed] [Google Scholar]
- 26.Atri M. New technologies and directed agents for applications of cancer imaging. J Clin Oncol. 2006;24:3299–3308. doi: 10.1200/JCO.2006.06.6159. [DOI] [PubMed] [Google Scholar]
- 27.Savadjiev P, Chong J, Dohan A, et al. Demystification of AI-driven medical image interpretation: past, present and future. Eur Radiol. 2019;29:1616–1624. doi: 10.1007/s00330-018-5674-x. [DOI] [PubMed] [Google Scholar]
- 28.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 29.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 30.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500–510. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moeskops P, Viergever MA, Mendrik AM, de Vries LS, Benders MJNL, Išgum I. Automatic segmentation of MR brain images with a convolutional neural network. IEEE Trans Med Imaging. 2016;35:1252–1261. doi: 10.1109/TMI.2016.2548501. [DOI] [PubMed] [Google Scholar]
- 32.Szczypiński PM, Strzelecki M, Materka A, Klepaczko A. MaZda--a software package for image texture analysis. Comput Methods Programs Biomed. 2009;94:66–76. doi: 10.1016/j.cmpb.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Nioche C, Orlhac F, Boughdad S, et al. LIFEx: A freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res. 2018;78:4786–4789. doi: 10.1158/0008-5472.CAN-18-0125. [DOI] [PubMed] [Google Scholar]
- 34.van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, Fried DV, Fave XJ, Hunter LA, Yang J, Court LE. IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys. 2015;42:1341–1353. doi: 10.1118/1.4908210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Witten IH, Frank E, Hall MA, Pal CJ. Data mining: Practical machine learning tools and techniques. 4th ed. San Francisco: Morgan Kaufmann Publishers Inc; 2016. [Google Scholar]
- 37.Demšar J, Curk T, Erjavec A, et al. Orange: data mining toolbox in Python. J Mach Learn Res. 2013;14:2349–2353. [Google Scholar]
- 38.Williams GJ. Rattle: a data mining GUI for R. R J. 2009;1:45–55. doi: 10.32614/RJ-2009-016. [DOI] [Google Scholar]
- 39.Varghese BA, Cen SY, Hwang DH, Duddalwar VA. Texture analysis of imaging: what radiologists need to know. AJR Am J Roentgenol. 2019;212:520–528. doi: 10.2214/AJR.18.20624. [DOI] [PubMed] [Google Scholar]
- 40.Buckler AJ, Bresolin L, Dunnick NR, Sullivan DC. A collaborative enterprise for multi-stakeholder participation in the advancement of quantitative imaging. Radiology. 2011;258:906–914. doi: 10.1148/radiol.10100799. [DOI] [PubMed] [Google Scholar]
- 41.Shafiq-Ul-Hassan M, Zhang GG, Latifi K, et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys. 2017;44:1050–1062. doi: 10.1002/mp.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shafiq-Ul-Hassan M, Latifi K, Zhang G, Ullah G, Gillies R, Moros E. Voxel size and gray level normalization of CT radiomic features in lung cancer. Sci Rep. 2018;8:10545. doi: 10.1038/s41598-018-28895-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 44.Tustison NJ, Avants BB, Cook PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29:1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collewet G, Strzelecki M, Mariette F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn Reson Imaging. 2004;22:81–91. doi: 10.1016/j.mri.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 46.Parker J, Kenyon RV, Troxel DE. Comparison of interpolating methods for image resampling. IEEE Trans Med Imaging. 1983;2:31–39. doi: 10.1109/TMI.1983.4307610. [DOI] [PubMed] [Google Scholar]
- 47.Duron L, Balvay D, Perre SV, et al. Gray-level discretization impacts reproducible MRI radiomics texture features. PLoS ONE. 2019;14:e0213459. doi: 10.1371/journal.pone.0213459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kocak B, Durmaz ES, Kaya OK, Ates E, Kilickesmez O. Reliability of single-slice-based 2D CT texture analysis of renal masses: influence of intra- and interobserver manual segmentation variability on radiomic feature reproducibility. AJR Am J Roentgenol. 2019:1–7. doi: 10.2214/AJR.19.21212. [DOI] [PubMed] [Google Scholar]
- 49.Kass M, Witkin A, Terzopoulos D. Snakes: Active contour models. Int J Comput Vis. 1988;1:321–331. doi: 10.1007/BF00133570. [DOI] [Google Scholar]
- 50.Suzuki K, Epstein ML, Kohlbrenner R, et al. CT liver volumetry using geodesic active contour segmentation with a level-set algorithm. 2010 Conference Proceedings; SPIE Medical Imaging; San Diego, CA. 2010. p. 7624. [DOI] [Google Scholar]
- 51.Peng J, Hu P, Lu F, Peng Z, Kong D, Zhang H. 3D liver segmentation using multiple region appearances and graph cuts. Med Phys. 2015;42:6840–6852. doi: 10.1118/1.4934834. [DOI] [PubMed] [Google Scholar]
- 52.Wu W, Zhou Z, Wu S, Zhang Y. Automatic liver segmentation on volumetric CT images using supervoxel-based graph cuts. Comput Math Methods Med. 2016;2016 doi: 10.1155/2016/9093721. 9093721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging. 2016;35:1240–1251. doi: 10.1109/TMI.2016.2538465. [DOI] [PubMed] [Google Scholar]
- 54.Ypsilantis P-P, Siddique M, Sohn H-M, et al. Predicting response to neoadjuvant chemotherapy with PET imaging using convolutional neural networks. PLoS ONE. 2015;10:e0137036. doi: 10.1371/journal.pone.0137036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Z, Wang Y, Yu J, Guo Y, Cao W. Deep learning based radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci Rep. 2017;7:5467. doi: 10.1038/s41598-017-05848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over-sampling technique. J Artif Intell Res. 2002;16:321–357. doi: 10.1613/jair.953. [DOI] [Google Scholar]
- 57.Mwangi B, Tian TS, Soares JC. A review of feature reduction techniques in neuroimaging. Neuroinformatics. 2014;12:229–244. doi: 10.1007/s12021-013-9204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kocak B, Ates E, Durmaz ES, Ulusan MB, Kilickesmez O. Influence of segmentation margin on machine learning-based high-dimensional quantitative CT texture analysis: a reproducibility study on renal clear cell carcinomas. Eur Radiol. 2019 doi: 10.1007/s00330-019-6003-8. [DOI] [PubMed] [Google Scholar]
- 59.Hall MA. PhD Thesis. University of Waikato Hamilton; 1999. Correlation-based feature selection for machine learning. [Google Scholar]
- 60.Ahn SJ, Kim JH, Lee SM, Park SJ, Han JK. CT reconstruction algorithms affect histogram and texture analysis: evidence for liver parenchyma, focal solid liver lesions, and renal cysts. Eur Radiol. 2018 doi: 10.1007/s00330-018-5829-9. [DOI] [PubMed] [Google Scholar]
- 61.Hodgdon T, McInnes MDF, Schieda N, Flood TA, Lamb L, Thornhill RE. Can quantitative CT texture analysis be used to differentiate fat-poor renal angiomyolipoma from renal cell carcinoma on unenhanced CT images? Radiology. 2015;276:787–796. doi: 10.1148/radiol.2015142215. [DOI] [PubMed] [Google Scholar]
- 62.Leng S, Takahashi N, Gomez Cardona D, et al. Subjective and objective heterogeneity scores for differentiating small renal masses using contrast-enhanced CT. Abdom Radiol (NY) 2017;42:1485–1492. doi: 10.1007/s00261-016-1014-2. [DOI] [PubMed] [Google Scholar]
- 63.Koo TK, Li MY. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med. 2016;15:155–163. doi: 10.1016/j.jcm.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dormann CF, Elith J, Bacher S, et al. Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography. 2013;36:27–46. doi: 10.1111/j.1600-0587.2012.07348.x. [DOI] [Google Scholar]
- 65.Tibshirani R. Regression shrinkage and selection via the lasso: a retrospective. J R Stat Soc B. 2011;73:273–282. doi: 10.1111/j.1467-9868.2011.00771.x. [DOI] [Google Scholar]
- 66.Kononenko I. Estimating attributes: Analysis and extensions of RELIEF. In: Bergadano F, De Raedt L, editors. Machine Learning: ECML-94. Berlin Heidelberg: Springer; 1994. pp. 171–182. [DOI] [Google Scholar]
- 67.Langs G, Menze BH, Lashkari D, Golland P. Detecting stable distributed patterns of brain activation using Gini contrast. Neuroimage. 2011;56:497–507. doi: 10.1016/j.neuroimage.2010.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shayesteh SP, Alikhassi A, Fard Esfahani A, et al. Neo-adjuvant chemoradiotherapy response prediction using MRI based ensemble learning method in rectal cancer patients. Phys Med. 2019;62:111–119. doi: 10.1016/j.ejmp.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 69.Hayes DF. Biomarker validation and testing. Mol Oncol. 2015;9:960–966. doi: 10.1016/j.molonc.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Varma S, Simon R. Bias in error estimation when using cross-validation for model selection. BMC Bioinformatics. 2006;7:91. doi: 10.1186/1471-2105-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Demšar J. Statistical comparisons of classifiers over multiple data sets. J Mach Learn Res. 2006;7:1–30. [Google Scholar]
- 72.Akin O, Elnajjar P, Heller M, et al. Radiology data from the cancer genome atlas kidney renal clear cell carcinoma [TCGA-KIRC] collection The Cancer Imaging Archive 2016https://wiki.cancerimagingarchive.net/x/woFY. Accessed June 18, 2019 [Google Scholar]
- 73.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Erickson B, Akkus Z, Sedlar J, Korfiatis P. Data from LGG-1p19q deletion. The Cancer Imaging Archive. 2017. [Accessed April 6, 2019]. https://wiki.cancerimagingarchive.net/x/coKJAQ.
- 75.Akkus Z, Ali I, Sedlář J, et al. Predicting deletion of chromosomal arms 1p/19q in low-grade gliomas from MR images using machine intelligence. J Digit Imaging. 2017;30:469–476. doi: 10.1007/s10278-017-9984-3. [DOI] [PMC free article] [PubMed] [Google Scholar]