Abstract
Two machine learning platforms were successfully used to provide diagnostic guidance in the differentiation between common cancer conditions in veteran populations.
Artificial intelligence (AI), first described in 1956, encompasses the field of computer science in which machines are trained to learn from experience. The term was popularized by the 1956 Dartmouth College Summer Research Project on Artificial Intelligence.1 The field of AI is rapidly growing and has the potential to affect many aspects of our lives. The emerging importance of AI is demonstrated by a February 2019 executive order that launched the American AI Initiative, allocating resources and funding for AI development.2 The executive order stresses the potential impact of AI in the health care field, including its potential utility to diagnose disease. Federal agencies were directed to invest in AI research and development to promote rapid breakthroughs in AI technology that may impact multiple areas of society.
Machine learning (ML), a subset of AI, was defined in 1959 by Arthur Samuel and is achieved by employing mathematic models to compute sample data sets.3 Originating from statistical linear models, neural networks were conceived to accomplish these tasks.4 These pioneering scientific achievements led to recent developments of deep neural networks. These models are developed to recognize patterns and achieve complex computational tasks within a matter of minutes, often far exceeding human ability.5 ML can increase efficiency with decreased computation time, high precision, and recall when compared with that of human decision making.6
ML has the potential for numerous applications in the health care field.7–9 One promising application is in the field of anatomic pathology. ML allows representative images to be used to train a computer to recognize patterns from labeled photographs. Based on a set of images selected to represent a specific tissue or disease process, the computer can be trained to evaluate and recognize new and unique images from patients and render a diagnosis.10 Prior to modern ML models, users would have to import many thousands of training images to produce algorithms that could recognize patterns with high accuracy. Modern ML algorithms allow for a model known as transfer learning, such that far fewer images are required for training.11–13
Two novel ML platforms available for public use are offered through Google (Mountain View, CA) and Apple (Cupertino, CA).14,15 They each offer a user-friendly interface with minimal experience required in computer science. Google AutoML uses ML via cloud services to store and retrieve data with ease. No coding knowledge is required. The Apple Create ML Module provides computer-based ML, requiring only a few lines of code.
The Veterans Health Administration (VHA) is the largest single health care system in the US, and nearly 50 000 cancer cases are diagnosed at the VHA annually.16 Cancers of the lung and colon are among the most common sources of invasive cancer and are the 2 most common causes of cancer deaths in America.16 We have previously reported using Apple ML in detecting non-small cell lung cancers (NSCLCs), including adenocarcinomas and squamous cell carcinomas (SCCs); and colon cancers with accuracy.17,18 In the present study, we expand on these findings by comparing Apple and Google ML platforms in a variety of common pathologic scenarios in veteran patients. Using limited training data, both programs are compared for precision and recall in differentiating conditions involving lung and colon pathology.
In the first 4 experiments, we evaluated the ability of the platforms to differentiate normal lung tissue from cancerous lung tissue, to distinguish lung adenocarcinoma from SCC, and to differentiate colon adenocarcinoma from normal colon tissue. Next, cases of colon adenocarcinoma were assessed to determine whether the presence or absence of the KRAS proto-oncogene could be determined histologically using the AI platforms. KRAS is found in a variety of cancers, including about 40% of colon adenocarcinomas.19 For colon cancers, the presence or absence of the mutation in KRAS has important implications for patients as it determines whether the tumor will respond to specific chemotherapy agents.20 The presence of the KRAS gene is currently determined by complex molecular testing of tumor tissue.21 However, we assessed the potential of ML to determine whether the mutation is present by computerized morphologic analysis alone. Our last experiment examined the ability of the Apple and Google platforms to differentiate between adenocarcinomas of lung origin vs colon origin. This has potential utility in determining the site of origin of metastatic carcinoma.22
METHODS
Fifty cases of lung SCC, 50 cases of lung adenocarcinoma, and 50 cases of colon adenocarcinoma were randomly retrieved from our molecular database. Twenty-five colon adenocarcinoma cases were positive for mutation in KRAS, while 25 cases were negative for mutation in KRAS. Seven hundred fifty total images of lung tissue (250 benign lung tissue, 250 lung adenocarcinomas, and 250 lung SCCs) and 500 total images of colon tissue (250 benign colon tissue and 250 colon adenocarcinoma) were obtained using a Leica Microscope MC190 HD Camera (Wetzlar, Germany) connected to an Olympus BX41 microscope (Center Valley, PA) and the Leica Acquire 9072 software for Apple computers. All the images were captured at a resolution of 1024 x 768 pixels using a 60x dry objective. Lung tissue images were captured and saved on a 2012 Apple MacBook Pro computer, and colon images were captured and saved on a 2011 Apple iMac computer. Both computers were running macOS v10.13.
Creating Image Classifier Models Using Apple Create ML
Apple Create ML is a suite of products that use various tools to create and train custom ML models on Apple computers.15 The suite contains many features, including image classification to train a ML model to classify images, natural language processing to classify natural language text, and tabular data to train models that deal with labeling information or estimating new quantities. We used Create ML Image Classification to create image classifier models for our project (Appendix A).
Creating ML Modules Using Google Cloud AutoML Vision Beta
Google Cloud AutoML is a suite of machine learning products, including AutoML Vision, AutoML Natural Language and AutoML Translation.14 All Cloud AutoML machine learning products were in beta version at the time of experimentation. We used Cloud AutoML Vision beta to create ML modules for our project. Unlike Apple Create ML, which is run on a local Apple computer, the Google Cloud AutoML is run online using a Google Cloud account. There are no minimum specifications requirements for the local computer since it is using the cloud-based architecture (Appendix B).
Experiment 1
We compared Apple Create ML Image Classifier and Google AutoML Vision in their ability to detect and subclassify NSCLC based on the histopathologic images. We created 3 classes of images (250 images each): benign lung tissue, lung adenocarcinoma, and lung SCC.
Experiment 2
We compared Apple Create ML Image Classifier and Google AutoML Vision in their ability to differentiate between normal lung tissue and NSCLC histopathologic images with 50/50 mixture of lung adenocarcinoma and lung SCC. We created 2 classes of images (250 images each): benign lung tissue and lung NSCLC.
Experiment 3
We compared Apple Create ML Image Classifier and Google AutoML Vision in their ability to differentiate between lung adenocarcinoma and lung SCC histopathologic images. We created 2 classes of images (250 images each): adenocarcinoma and SCC.
Experiment 4
We compared Apple Create ML Image Classifier and Google AutoML Vision in their ability to detect colon cancer histopathologic images regardless of mutation in KRAS status. We created 2 classes of images (250 images each): benign colon tissue and colon adenocarcinoma.
Experiment 5
We compared Apple Create ML Image Classifier and Google AutoML Vision in their ability to differentiate between colon adenocarcinoma with mutations in KRAS and colon adenocarcinoma without the mutation in KRAS histopathologic images. We created 2 classes of images (125 images each): colon adenocarcinoma cases with mutation in KRAS and colon adenocarcinoma cases without the mutation in KRAS.
Experiment 6
We compared Apple Create ML Image Classifier and Google AutoML Vision in their ability to differentiate between lung adenocarcinoma and colon adenocarcinoma histopathologic images. We created 2 classes of images (250 images each): colon adenocarcinoma lung adenocarcinoma.
RESULTS
Twelve machine learning models were created in 6 experiments using the Apple Create ML and the Google AutoML (Table). To investigate recall and precision differences between the Apple and the Google ML algorithms, we performed 2-tailed distribution, paired t tests. No statistically significant differences were found (P = .52 for recall and .60 for precision).
TABLE.
Results Summary for the Apple Create ML and the Google AutoML Machine Learning Models
| Machine Learning Model | Classes | Recall | Precision | ||
|---|---|---|---|---|---|
| Apple, % | Google, % | Apple, % | Google, % | ||
| Model 1 | Lung normal | 100.0 | 100.0 | 98.0 | 100.0 |
|
| |||||
| Lung adenocarcinoma | 86.0 | 85.7 | 97.7 | 81.8 | |
|
| |||||
| Lung squamous cell carcinoma | 98.0 | 89.9 | 89.1 | 90.6 | |
|
| |||||
| Model 2 | Lung normal | 100.0 | 100.0 | 98.0 | 100.0 |
|
| |||||
| Non-small cell lung cancer | 98.0 | 100.0 | 100.0 | 100.0 | |
|
| |||||
| Model 3 | Lung adenocarcinoma | 90.0 | 82.4 | 91.8 | 87.5 |
|
| |||||
| Lung squamous cell carcinoma | 92.0 | 92.3 | 90.2 | 88.9 | |
|
| |||||
| Model 4 | Colon normal | 98.0 | 95.8 | 100.0 | 100.0 |
|
| |||||
| Colon adenocarcinoma | 100.0 | 100.0 | 98.0 | 96.0 | |
|
| |||||
| Model 5 | Colon adenocarcinoma KRAS+ | 72.0 | 88.2 | 69.2 | 71.4 |
|
| |||||
| Colon adenocarcinoma KRAS− | 68.0 | 50.0 | 70.8 | 75.0 | |
|
| |||||
| Model 6 | Lung adenocarcinoma | 96.0 | 91.3 | 96.0 | 100.0 |
|
| |||||
| Colon adenocarcinoma | 96.0 | 100.0 | 96.0 | 93.9 | |
Overall, each model performed well in distinguishing between normal and neoplastic tissue for both lung and colon cancers. In subclassifying NSCLC into adenocarcinoma and SCC, the models were shown to have high levels of precision and recall. The models also were successful in distinguishing between lung and colonic origin of adenocarcinoma (Figures 1–4). However, both systems had trouble discerning colon adenocarcinoma with mutations in KRAS from adenocarcinoma without mutations in KRAS.
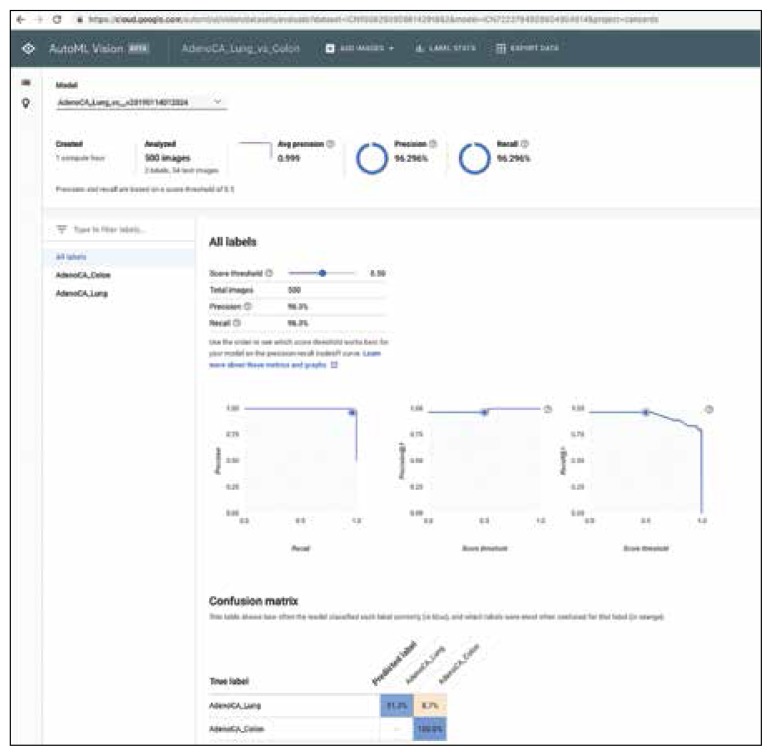
FIGURE 1.
Results Using Google AutoML to Differentiate Lung Adenocarcinoma From Colon Adenocarcinoma With Overall Precision and Recall Data (Experiment 6)
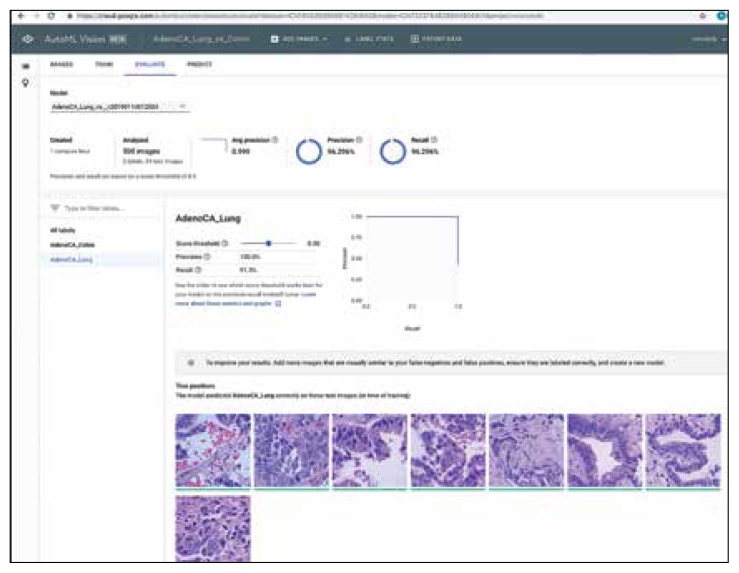
FIGURE 2.
Results Using Google AutoML to Differentiate Lung Adenocarcinoma From Colon Adenocarcinoma With Precision and Recall Data for Lung Adenocarcinoma Label (Experiment 6)
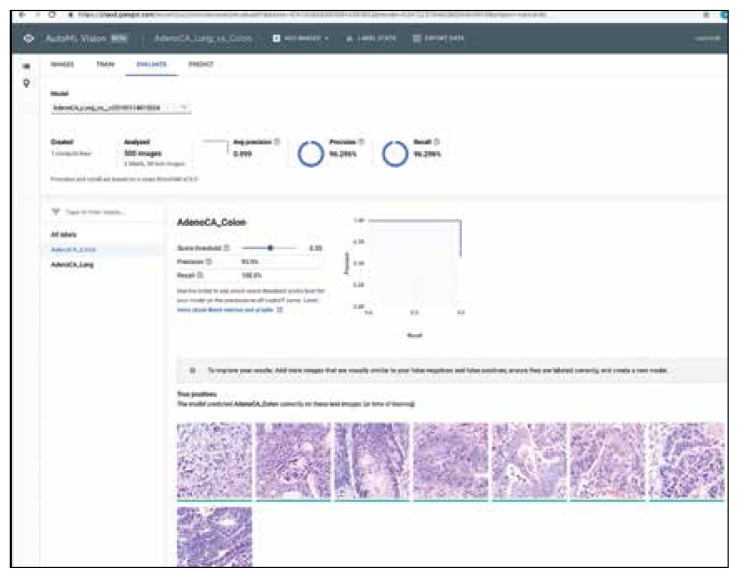
FIGURE 3.
Results Using Google AutoML to Differentiate Lung Adenocarcinoma From Colon Adenocarcinoma With Precision and Recall Data for Colon Adenocarcinoma Label (Experiment 6)
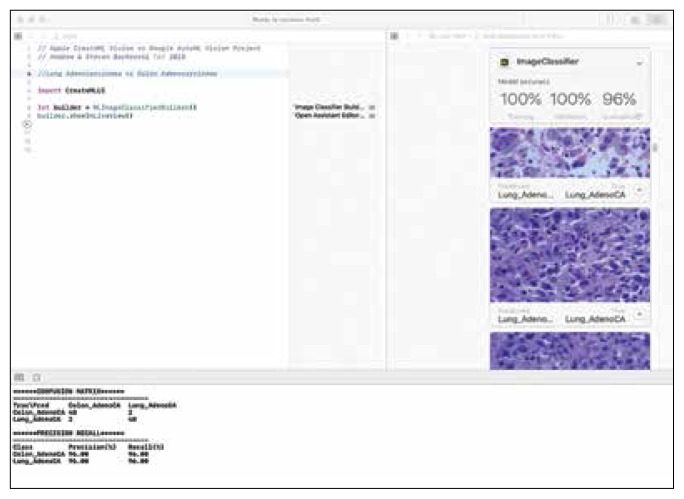
FIGURE 4.
Results Using Apple Create ML Demonstrating Precision and Recall Data Differentiating Lung Adenocarcinoma From Colon Adenocarcinoma
DISCUSSION
Image classifier models using ML algorithms hold a promising future to revolutionize the health care field. ML products, such as those modules offered by Apple and Google, are easy to use and have a simple graphic user interface to allow individuals to train models to perform humanlike tasks in real time. In our experiments, we compared multiple algorithms to determine their ability to differentiate and subclassify histopathologic images with high precision and recall using common scenarios in treating veteran patients.
Analysis of the results revealed high precision and recall values illustrating the models’ ability to differentiate and detect benign lung tissue from lung SCC and lung adenocarcinoma in ML model 1, benign lung from NSCLC carcinoma in ML model 2, and benign colon from colonic adenocarcinoma in ML model 4. In ML model 3 and 6, both ML algorithms performed at a high level to differentiate lung SCC from lung adenocarcinoma and lung adenocarcinoma from colonic adenocarcinoma, respectively. Of note, ML model 5 had the lowest precision and recall values across both algorithms demonstrating the models’ limited utility in predicting molecular profiles, such as mutations in KRAS as tested here. This is not surprising as pathologists currently require complex molecular tests to detect mutations in KRAS reliably in colon cancer.
Both modules require minimal programming experience and are easy to use. In our comparison, we demonstrated critical distinguishing characteristics that differentiate the 2 products.
Apple Create ML image classifier is available for use on local Mac computers that use Xcode version 10 and macOS 10.14 or later, with just 3 lines of code required to perform computations. Although this product is limited to Apple computers, it is free to use, and images are stored on the computer hard drive. Of unique significance on the Apple system platform, images can be augmented to alter their appearance to enhance model training. For example, imported images can be cropped, rotated, blurred, and flipped, in order to optimize the model’s training abilities to recognize test images and perform pattern recognition. This feature is not as readily available on the Google platform. Apple Create ML Image classifier’s default training set consists of 75% of total imported images with 5% of the total images being randomly used as a validation set. The remaining 20% of images comprise the testing set. The module’s computational analysis to train the model is achieved in about 2 minutes on average. The score threshold is set at 50% and cannot be manipulated for each image class as in Google AutoML Vision.
Google AutoML Vision is open and can be accessed from many devices. It stores images on remote Google servers but requires computing fees after a $300 credit for 12 months. On AutoML Vision, random 80% of the total images are used in the training set, 10% are used in the validation set, and 10% are used in the testing set. It is important to highlight the different percentages used in the default settings on the respective modules. The time to train the Google AutoML Vision with default computational power is longer on average than Apple Create ML, with about 8 minutes required to train the machine learning module. However, it is possible to choose more computational power for an additional fee and decrease module training time. The user will receive e-mail alerts when the computer time begins and is completed. The computation time is calculated by subtracting the time of the initial e-mail from the final e-mail.
Based on our calculations, we determined there was no significant difference between the 2 machine learning algorithms tested at the default settings with recall and precision values obtained. These findings demonstrate the promise of using a ML algorithm to assist in the performance of human tasks and behaviors, specifically the diagnosis of histopathologic images. These results have numerous potential uses in clinical medicine. ML algorithms have been successfully applied to diagnostic and prognostic endeavors in pathology,23–28 dermatology,29–31 ophthalmology,32 cardiology,33 and radiology.34–36
Pathologists often use additional tests, such as special staining of tissues or molecular tests, to assist with accurate classification of tumors. ML platforms offer the potential of an additional tool for pathologists to use along with human microscopic interpretation.37,38 In addition, the number of pathologists in the US is dramatically decreasing, and many other countries have marked physician shortages, especially in fields of specialized training such as pathology.39–42 These models could readily assist physicians in underserved countries and impact shortages of pathologists elsewhere by providing more specific diagnoses in an expedited manner.43
Finally, although we have explored the application of these platforms in common cancer scenarios, great potential exists to use similar techniques in the detection of other conditions. These include the potential for classification and risk assessment of precancerous lesions, infectious processes in tissue (eg, detection of tuberculosis or malaria),24,44 inflammatory conditions (eg, arthritis subtypes, gout),45 blood disorders (eg, abnormal blood cell morphology),46 and many others. The potential of these technologies to improve health care delivery to veteran patients seems to be limited only by the imagination of the user.47
Regarding the limited effectiveness in determining the presence or absence of mutations in KRAS for colon adenocarcinoma, it is mentioned that currently pathologists rely on complex molecular tests to detect the mutations at the DNA level.21 It is possible that the use of more extensive training data sets may improve recall and precision in cases such as these and warrants further study. Our experiments were limited to the stipulations placed by the free trial software agreements; no costs were expended to use the algorithms, though an Apple computer was required.
CONCLUSION
We have demonstrated the successful application of 2 readily available ML platforms in providing diagnostic guidance in differentiation between common cancer conditions in veteran patient populations. Although both platforms performed very well with no statistically significant differences in results, some distinctions are worth noting. Apple Create ML can be used on local computers but is limited to an Apple operating system. Google AutoML is not platform-specific but runs only via Google Cloud with associated computational fees. Using these readily available models, we demonstrated the vast potential of AI in diagnostic pathology. The application of AI to clinical medicine remains in the very early stages. The VA is uniquely poised to provide leadership as AI technologies will continue to dramatically change the future of health care, both in veteran and nonveteran patients nationwide.
Acknowledgments
The authors thank Paul Borkowski for his constructive criticism and proofreading of this manuscript. This material is the result of work supported with resources and the use of facilities at the James A. Haley Veterans’ Hospital.
APPENDIX A. Apple Create ML Test Procedure
For each model, we used the following procedure. We opened the Apple Xcode macOS playground v10 on a 2018 Apple MacBook Pro (macOS v10.14, 2.3 GHz Intel Core I5, 8 GB 2133 MHz LPDDR3, Intel Iris Plus Graphics 655 GPU 1536 MB, 256 SSD) (Standard configuration) to create image classifier model with following lines of code (Apple Swift programming language):
import CreateMLUI let builder = MLImageClassifierBuilder() builder.showInLiveView()
We then opened the assistant editor in Xcode and ran the code. The live view displayed the image classifier UI. We then dragged in the training folder for training and validating the model. Once training and validation were complete, we dragged in the testing folder to evaluate the model performance on the indicated locations in live view. All training, validation, and testing were done at the default setting.
When handling images, Apple Create ML Image Classifier randomly assigned 80% of the images to a training folder and 20% to a testing folder. The training folder included class-labeled subfolders with training images (80% of total). The testing folder included class-labeled subfolders with testing images (20% of total). The Apple program randomly assigned 5% of the images from the training folder for validation.
APPENDIX B. Google AutoML Test Procedure
For each machine model, we used the following procedure. We created and named new datasets in AutoML Vision Datasets and imported images using the graphic user interface. We left the classification type box unchecked because each of our image classes had only a single label. We created class labels and imported images for each of the classes.
Training a new model took about 8 to 9 minutes. The following information was available: average precision (area under the precision-recall tradeoff curve), overall precision and recall, score threshold slider, precision-recall graphs, and confusion matrix. Confusion matrix table, provided by the AutoML, displays how often trained model classifies each label correctly and how often incorrectly. When images were uploaded, Google AutoML Vision randomly assigned 80% for training, 10% for validation, and 10% for testing the module.
Footnotes
Author disclosures
The authors report no actual or potential conflicts of interest with regard to this article.
Disclaimer
The opinions expressed herein are those of the authors and do not necessarily reflect those of Federal Practitioner, Frontline Medical Communications Inc., the US Government, or any of its agencies.
References
- 1.Moor J. The Dartmouth College artificial intelligence conference: the next fifty years. AI Mag. 2006;27(4):87–91. [Google Scholar]
- 2.Trump D. Accelerating America’s leadership in artificial intelligence. [Accessed September 4, 2019]. https://www.whitehouse.gov/articles/accelerating-americas-leadership-in-artificial-intelligence. Published February 11, 2019.
- 3.Samuel AL. Some studies in machine learning using the game of checkers. IBM J Res Dev. 1959;3(3):210–229. [Google Scholar]
- 4.SAS Users Group International. Neural networks and statistical models. In: Sarle WS, editor. Proceedings of the Nineteenth Annual SAS Users Group International Conference. SAS Institute; Cary, North Carolina: 1994. [Accessed September 16, 2019]. pp. 1538–1550. http://www.sascommunity.org/sugi/SUGI94/Sugi-94-255%20Sarle.pdf. [Google Scholar]
- 5.Schmidhuber J. Deep learning in neural networks: an overview. Neural Networks. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 6.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 7.Jiang F, Jiang Y, Li H, et al. Artificial intelligence in healthcare: past, present and future. Stroke Vasc Neurol. 2017;2(4):230–243. doi: 10.1136/svn-2017-000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine learning for medical imaging. Radiographics. 2017;37(2):505–515. doi: 10.1148/rg.2017160130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deo RC. Machine learning in medicine. Circulation. 2015;132(20):1920–1930. doi: 10.1161/CIRCULATIONAHA.115.001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J Pathol Inform. 2016;7(1):29. doi: 10.4103/2153-3539.186902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oquab M, Bottou L, Laptev I, Sivic J. Learning and transferring mid-level image representations using convolutional neural networks. Presented at: IEEE Conference on Computer Vision and Pattern Recognition; 2014; [Accessed September 4, 2019]. http://openaccess.thecvf.com/content_cvpr_2014/html/Oquab_Learning_and_Transferring_2014_CVPR_paper.html. [Google Scholar]
- 12.Shin HC, Roth HR, Gao M, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35(5):1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tajbakhsh N, Shin JY, Gurudu SR, et al. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging. 2016;35(5):1299–1312. doi: 10.1109/TMI.2016.2535302. [DOI] [PubMed] [Google Scholar]
- 14.Cloud AutoML. [Accessed September 4, 2019]. https://cloud.google.com/automl.
- 15.Create ML.https://developer.apple.com/documentation/createml. Accessed September 4, 2019
- 16.Zullig LL, Sims KJ, McNeil R, et al. Cancer incidence among patients of the U.S. Veterans Affairs Health Care System: 2010 Update. Mil Med. 2017;182(7):e1883–e1891. doi: 10.7205/MILMED-D-16-00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borkowski AA, Wilson CP, Borkowski SA, Deland LA, Mastorides SM. Using Apple machine learning algorithms to detect and subclassify non-small cell lung cancer. [Accessed September 4, 2019]. https://arxiv.org/ftp/arxiv/papers/1808/1808.08230.pdf.
- 18.Borkowski AA, Wilson CP, Borkowski SA, Thomas LB, Deland LA, Mastorides SM. Apple machine learning algorithms successfully detect colon cancer but fail to predict KRAS mutation status. [Accessed September 4, 2019]. http://arxiv.org/abs/1812.04660. Revised January 15, 2019.
- 19.Armaghany T, Wilson JD, Chu Q, Mills G. Genetic alterations in colorectal cancer. Gastrointest Cancer Res. 2012;5(1):19–27. [PMC free article] [PubMed] [Google Scholar]
- 20.Herzig DO, Tsikitis VL. Molecular markers for colon diagnosis, prognosis and targeted therapy. J Surg Oncol. 2015;111(1):96–102. doi: 10.1002/jso.23806. [DOI] [PubMed] [Google Scholar]
- 21.Ma W, Brodie S, Agersborg S, Funari VA, Albitar M. Significant improvement in detecting BRAF, KRAS, and EGFR mutations using next-generation sequencing as compared with FDA-cleared kits. Mol Diagn Ther. 2017;21(5):571–579. doi: 10.1007/s40291-017-0290-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greco FA. Molecular diagnosis of the tissue of origin in cancer of unknown primary site: useful in patient management. Curr Treat Options Oncol. 2013;14(4):634–642. doi: 10.1007/s11864-013-0257-1. [DOI] [PubMed] [Google Scholar]
- 23.Bejnordi BE, Veta M, van Diest PJ, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318(22):2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong Y, Ba X, Hou A, Zhang K, Chen L, Li T. Automatic detection of mycobacterium tuberculosis using artificial intelligence. J Thorac Dis. 2018;10(3):1936–1940. doi: 10.21037/jtd.2018.01.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruz-Roa A, Gilmore H, Basavanhally A, et al. Accurate and reproducible invasive breast cancer detection in whole-slide images: a deep learning approach for quantifying tumor extent. Sci Rep. 2017;7:46450. doi: 10.1038/srep46450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24(10):1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ertosun MG, Rubin DL. Automated grading of gliomas using deep learning in digital pathology images: a modular approach with ensemble of convolutional neural networks. AMIA Annu Symp Proc. 2015;2015:1899–1908. [PMC free article] [PubMed] [Google Scholar]
- 28.Wahab N, Khan A, Lee YS. Two-phase deep convolutional neural network for reducing class skewness in histopathological images based breast cancer detection. Comput Biol Med. 2017;85:86–97. doi: 10.1016/j.compbiomed.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 29.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han SS, Park GH, Lim W, et al. Deep neural networks show an equivalent and often superior performance to dermatologists in onychomycosis diagnosis: automatic construction of onychomycosis datasets by region-based convolutional deep neural network. PLoS One. 2018;13(1):e0191493. doi: 10.1371/journal.pone.0191493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujisawa Y, Otomo Y, Ogata Y, et al. Deep-learning-based, computer-aided classifier developed with a small dataset of clinical images surpasses board-certified dermatologists in skin tumour diagnosis. Br J Dermatol. 2019;180(2):373–381. doi: 10.1111/bjd.16924. [DOI] [PubMed] [Google Scholar]
- 32.Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316(22):2402–2010. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 33.Weng SF, Reps J, Kai J, Garibaldi JM, Qureshi N. Can machine-learning improve cardiovascular risk prediction using routine clinical data? PLoS One. 2017;12(4):e0174944. doi: 10.1371/journal.pone.0174944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng J-Z, Ni D, Chou Y-H, et al. Computer-aided diagnosis with deep learning architecture: applications to breast lesions in US images and pulmonary nodules in CT scans. Sci Rep. 2016;6(1):24454. doi: 10.1038/srep24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Yang W, Weinreb J, et al. Searching for prostate cancer by fully automated magnetic resonance imaging classification: deep learning versus non-deep learning. Sci Rep. 2017;7(1):15415. doi: 10.1038/s41598-017-15720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284(2):574–582. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]
- 37.Bardou D, Zhang K, Ahmad SM. Classification of breast cancer based on histology images using convolutional neural networks. IEEE Access. 2018;6(6):24680–24693. [Google Scholar]
- 38.Sheikhzadeh F, Ward RK, van Niekerk D, Guillaud M. Automatic labeling of molecular biomarkers of immunohistochemistry images using fully convolutional networks. PLoS One. 2018;13(1):e0190783. doi: 10.1371/journal.pone.0190783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metter DM, Colgan TJ, Leung ST, Timmons CF, Park JY. Trends in the US and Canadian pathologist workforces from 2007 to 2017. JAMA Netw Open. 2019;2(5):e194337. doi: 10.1001/jamanetworkopen.2019.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benediktsson H, Whitelaw J, Roy I. Pathology services in developing countries: a challenge. Arch Pathol Lab Med. 2007;131(11):1636–1639. doi: 10.5858/2007-131-1636-PSIDCA. [DOI] [PubMed] [Google Scholar]
- 41.Graves D. The impact of the pathology workforce crisis on acute health care. Aust Health Rev. 2007;31(suppl 1):S28–S30. doi: 10.1071/ah070s28. [DOI] [PubMed] [Google Scholar]
- 42.NHS pathology shortages cause cancer diagnosis delays. [Accessed September 4, 2019]. https://www.gmjournal.co.uk/nhs-pathology-shortages-are-causing-cancer-diagnosis-delays. Published September 18, 2018.
- 43.Abbott LM, Smith SD. Smartphone apps for skin cancer diagnosis: Implications for patients and practitioners. Australas J Dermatol. 2018;59(3):168–170. doi: 10.1111/ajd.12758. [DOI] [PubMed] [Google Scholar]
- 44.Poostchi M, Silamut K, Maude RJ, Jaeger S, Thoma G. Image analysis and machine learning for detecting malaria. Transl Res. 2018;194:36–55. doi: 10.1016/j.trsl.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orange DE, Agius P, DiCarlo EF, et al. Identification of three rheumatoid arthritis disease subtypes by machine learning integration of synovial histologic features and RNA sequencing data. Arthritis Rheumatol. 2018;70(5):690–701. doi: 10.1002/art.40428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodellar J, Alférez S, Acevedo A, Molina A, Merino A. Image processing and machine learning in the morphological analysis of blood cells. Int J Lab Hematol. 2018;40(suppl 1):46–53. doi: 10.1111/ijlh.12818. [DOI] [PubMed] [Google Scholar]
- 47.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]