Abstract
The influence of acetic and propionic acids on baker's yeast was investigated in order to expand our understanding of the effect of weak organic acid food preservatives on eukaryotic cells. Both acids decreased yeast survival in a concentration-dependent manner, but with different efficiencies. The acids inhibited the fluorescein efflux from yeast cells. The inhibition constant of fluorescein extrusion from cells treated with acetate was significantly lower in parental strain than in either PDR12 (ABC-transporter Pdr12p) or WAR1 (transcriptional factor of Pdr12p) defective mutants. The constants of inhibition by propionate were virtually the same in all strains used. Yeast exposure to acetate increased the level of oxidized proteins and the activity of antioxidant enzymes, while propionate did not change these parameters. This suggests that various mechanisms underlie the yeast toxicity by acetic and propionic acids. Our studies with mutant cells clearly indicated the involvement of Yap1p transcriptional regulator and de novo protein synthesis in superoxide dismutase up-regulation by acetate. The up-regulation of catalase was Yap1p independent. Yeast pre-incubation with low concentrations of H2O2 caused cellular cross-protection against high concentrations of acetate. The results are discussed from the point of view that acetate induces a prooxidant effect in vivo, whereas propionate does not.
Keywords: ABC-transporter Pdr12p, Acetic acid, Antioxidant enzymes, Propionic acid, Protein carbonyls, Yeast cross-protection
Introduction
Weak organic acids (WOAs) are naturally occurring compounds that inhibit or prevent the growth of many microorganisms. Monocarboxylic WOAs are the most widely used acid preservatives in industrial food and beverage production. The major WOA food preservatives include acetic acid (AA) and propionic acid (PA), the concentration of which usually ranges from 0.1 to 1.0 M. Because WOAs may be potentially harmful to humans, the effects of these compounds on eukaryotic cells have been extensively studied. The yeast Saccharomyces cerevisiae is a good model system to investigate acid stress and defense against it in eukaryotes, since its protective mechanisms are generally similar to those of higher organisms.1–6
It was found that WOAs were mutagenic towards the mitochondrial genome in aerobically grown baker's yeast.2 Accumulation of acid anions in the yeast cells also resulted in energetic stress,7 programmed cell death,8–12 and inhibition of essential metabolic pathways.13–15 On the other hand, baker's yeast is shown to be relatively resistant to monocarboxylic WOAs.14,16
One of the most important mechanisms protecting yeast against acid stress is prevention of intracellular accumulation of acid anions to high, potentially toxic levels. Adenosine-5′-triphosphate (ATP) binding cassette Pdr12p appears to be acting as an efflux pump for WOA anions. It is well documented that S. cerevisiae is capable of growing in the presence of sorbate due to induction of Pdr12p via War1p transcriptional factor-dependent pathway.17 Pdr12p is responsible for the active extrusion of benzoate, propionate, fluorescein, catabolic products of amino acids, and multicyclic compounds such as caffeine.18–20 However, possible role of the involvement of the pump in acetate efflux from S. cerevisiae cells is still debated.20–22 In vivo, the activity of Pdr12p can be readily monitored as fluorescein extrusion from the cell.18,20 Here we measured fluorescein efflux from yeast cells in the presence of acetate and propionate with the aim to compare the effects of these acid food preservatives on the Pdr12p transport system.
The effects of WOAs are rather complex and yeast response involves other defense mechanisms. For example, antioxidant system was shown to be involved in yeast adaptation to AA.18,20 There seems to be little information available on the possible role of antioxidant enzymes in yeast defense against PA. In order to expand our understanding of S. cerevisiae response and adaptation to WOA-induced stress, the influence of AA and PA on the level of oxidized proteins and the activity of antioxidant enzymes was investigated in this work.
It is widely believed that cell exposure to mild stress results in the acquisition of cellular resistance to lethal stress, what is called ‘adaptive response’. The phenomenon has been observed in various organisms, including S. cerevisiae. For example, yeast exposed to a mild dose of oxidative stress could subsequently survive an otherwise lethal dose of the same or another stress.5,6,23–25 The latter is known as ‘cross-adaptation’ or ‘cross-protection’. In the present work, we examined the influence of cell pre-adaptation by sublethal concentrations of hydrogen peroxide on yeast survival under acid stress. Possible mechanisms of yeast resistance to WOAs and coordination of antioxidant system response are discussed.
Materials and methods
Yeast strains
The S. cerevisiae strains used were as follows: W303-1A (wild type, MATa ura3-1 leu2-3,112 his3-11,15 trp1-1 ade2-1 can1-100) and its isogenic derivatives ΔPDR12 (W303-1A pdr12Δ::hisG-URA3-hisG) and ΔWAR1 (W303-1A war1Δ::HIS3) kindly provided by Prof. K Kuchler (Medical University of Vienna, Austria); YPH250 (wild type, MATa trp1-Δ1 his3-Δ200 lys2-801 leu2-Δ1 ade2-101 ura3-52) and its isogenic derivatives ΔCTT1ΔCTA1 (YPH250 ctt1Δ::URA3 cta1Δ::TRP1), ΔYAP1 (YPH250 yap1Δ::HIS3) earlier described.26,27
Construction of ΔSOD1ΔSOD2 strain
The strain ΔSOD1ΔSOD2 (sod1Δ::kanMX sod2Δ::TRP1) was constructed from YPH250. SOD1 gene was disrupted using pUC-sod1Δ::URA3 plasmid donated by Dr E Gralla (University of California at Los Angeles).28 To disrupt SOD2, the pUCsod2Δ::TRP1 plasmid was constructed as follows: SOD2 gene was amplified using the primers SOD2-F1 (5′-TGGCTGAGATTCTCCTTGATGGCGAAGCAA-3′) and SOD2-R1 (5′-TCGGGAATAAAGGCTGAGCTCATTTTCGTA-3′). The SacI site was designed in the SOD2-R1 primer (underlined). The amplified fragment was digested with BamHI and SacI, and the resultant fragment was cloned into the BamHI–SacI site of pUC19. The resultant plasmid (pUCSOD2) was digested with StyI followed by Klenow fragment treatment, and the StyI–StyI fragment in SOD2 was replaced with TRP1 gene further treated with Klenow fragment. The resultant plasmid (pUCsod2Δ::TRP1) was digested with BamHI and SacI, and the sod2Δ::TRP1 fragment was introduced to the SOD2 loci of wild type and sod1Δ::URA3 strains of YPH250. Disruption of SOD2 was confirmed by polymerase chain reaction.
Growth conditions, WOA stress, and cell pre-treatment with H2O2
Yeast cells were grown at 28 °C with shaking at 175 rpm to early stationary phase (24 hours) in a liquid medium containing 1% yeast extract, 2% peptone, and 1% dextrose (YPD). Aliquots of experimental culture were incubated with different AA or PA concentrations at pH 3.0 and 28 °C for 120 minutes. At low pH, AA (pKa 4.75) and PA (pKa 4.88) exist mainly in the undissociated state, in which they enter the cell rather easily. In order to reach maximum penetration of acids into cells, the pH values of YPD medium was adjusted to 3.0 with HCl.21,29 Control cells were incubated in YPD medium at pH 6.75 or 3.0 without organic acids. At рН 3.0–4.5, YPD medium has been shown to act as a buffer system and recommended to be used for yeast incubation under WOA-induced stress.29,30
To study pre-adaptation effect on cell survival under weak organic stress, aliquots of experimental culture were pre-incubated with 0.05, 0.10, and 0.25 mM H2O2 at pH 6.75 for 30 minutes, followed by incubation with 200 mM AA at pH 3.0 for 120 minutes.
Cell survival was evaluated as colony-forming units. To evaluate respiratory-deficient petites, triphenyltetrazolium chloride was used.2
Fluorescein distribution and efflux from whole cells
Cell suspensions (∼108 cells/ml) were incubated with 50 µM fluorescein diacetate (FDA) as described previously.20 Briefly, after loading with FDA the yeast cells were: (1) pelleted and cell-free supernatant was used for measurement of extracellular fluorescein fluorescence; (2) vortexed in the incubation medium to measure total fluorescence of intra- and extracellular fluorescein; or (3) harvested, washed, and vortexed for intracellular fluorescence measurement in cell-free extract. The intensity of fluorescence was determined using λexcitation = 435 nm and λemission = 525 nm with a SpectraMAX GeminiEM 96-well plate spectrafluorometer and Soft Max Pro 4.7 software (both from Molecular Devices, Sunnyvale, CA, USA).
Assay of superoxide dismutase and catalase activities and protein carbonyls levels
The parameters were measured spectrophotometrically with a Specoll 211 spectrophotometer (Carl Zeiss, Germany) and CΦ-46 (ЛOMO, USSR). The activity of superoxide dismutase (SOD) and catalase was assayed in cell lysates as described previously.31 The content of carbonyl proteins (CPs) was measured by determining the amount of dinitrophenylhydrazone formed upon reaction with 2,4-dinitrophenylhydrazine.31 To inhibit de novo protein synthesis, yeast cells were pre-exposed to 15 µM cycloheximide.
Protein measurement and statistics
Protein concentration was determined by the Coomassie brilliant blue G-250 dye-binding method32 with bovine serum albumin as the standard. Experimental data were expressed as the mean value of 3–6 independent experiments ± the standard error of the mean (SEM), and statistical analysis was performed using analysis of variance (ANOVA) followed by a Student–Newman–Keuls test.
Results
Toxicity of acetate and propionate
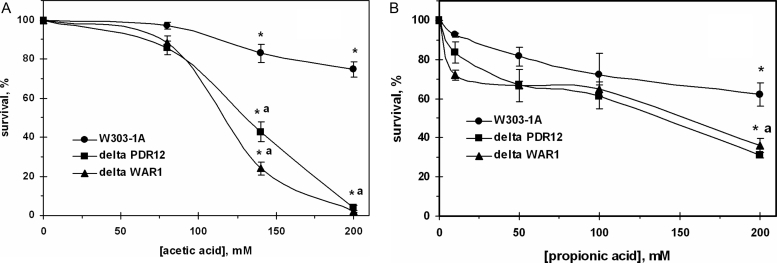
The survival of the wild-type yeast decreased with increasing acid concentrations, accounting for 75% at 200 mM acetate (Fig. 1A) and 62% at 200 mM propionate (Fig. 1B) relative to control. Mutant cells defective in either PDR12 (ABC-transporter Pdr12p) or WAR1 (transcriptional factor of Pdr12p) demonstrated higher sensitivity to both acids compared to the parental strain. However, the susceptibilities of two knockouts to the acids differed: at concentrations up to 100 mM yeast demonstrated higher sensitivity to propionate compared with acetate. Parental strain was less tolerant to low concentrations of propionate than acetate. In contrast, very few of the mutant cells were able to survive after treatment with 200 mM AA (Fig. 1A), whereas about 30–35% of defective cells survived stress induced by 200 mM PA (Fig. 1B).
Figure 1.
Survival of S. cerevisiae W303-1A wild type and its derivatives ΔPDR12 and ΔWAR1 after treatment with AA (A) and PA (B). Data are mean ± SEM (n = 4–6). Significantly different from respective values obtained: *under treatment with 80 mM AA (A) with P < 0.005 or 20 mM PA (B) with P < 0.05 and for aW303-1A wild strain with P < 0.05.
Piper2 found that sorbate and benzoate were mutagenic towards the mitochondrial genome in aerobically grown baker's yeast. In the present study, we used the same test with triphenyltetrazolium chloride to evaluate the amount of respiratory-deficient petites. Neither acetate nor propionate increased the petites amounts (not shown), which might reflect the absence of mutagenic effects towards mitochondrial DNA.
Acetate and propionate effects on fluorescein efflux from yeast cells
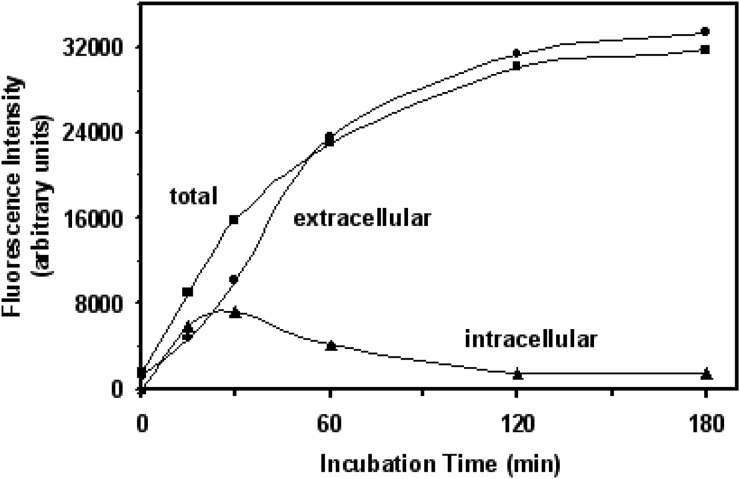
In the present study, yeast cells were incubated with FDA freely diffusing into cytosol, where it is cleaved by intracellular esterases to fluorescein. The latter can be extruded from the cell by Pdr12p.18,20 Fig. 2 shows the dynamics of intra- and extracellular fluorescein distribution in cell suspension during yeast incubation with FDA. The accumulation of fluorescein in the cells increased in parallel to its extrusion during the first 20 minutes of yeast exposure to FDA. Then the fluorescence intensity of extracellular fluorescein exceeded the intracellular one. Although extra- and intracellular fluorescence demonstrated different dynamics, the total fluorescence corresponded well to their sum. Recently, we obtained very similar data with another yeast strain.20 Because the incubation for 20–30 minutes yielded maximum fluorescein accumulation, further experiments were performed using 20-minute incubation with FDA.
Figure 2.
Time course of intra- and extracellular fluorescein distribution in S. cerevisiae YPH250. Data are from representative experiment.
To characterize the influence of acetate and propionate on Pdr12 transport system in S. cerevisiae, we measured the efflux of fluorescein from wild type and in either PDR12 or WAR1 defective yeast cells. Exogenously added WOAs linearly inhibited fluorescein extrusion from the yeast cells. Half-maximal inhibition of fluorescein efflux was virtually the same for both acids in the wild type (Table 1). The efflux of fluorescein from the mutant cells defective in PDR12 or WAR1 was also inhibited by AA, but inhibition constants were ∼3-fold higher than those in the parental strain. It seems that Pdr12p in some way affects fluorescein transport system in the presence of AA, but not PA (Table 1). Neither the defect in PDR12 nor the defect in WAR1 changed inhibition constant in the experiments, where PA was used.
Table 1.
Inhibition constants (mM) of fluorescein efflux from different strains of S. cerevisiae in the presence of 25–400 mM AA and 10–200 mM PA
| Strain | Half-maximal inhibition constant | |
|---|---|---|
| AA | PA | |
| W303-1A (wild type) | 18.6 ± 4.0 | 15.3 ± 4.7 |
| ΔPDR12 | 53.9 ± 2.8* | 18.3 ± 2.5** |
| ΔWAR1 | 55.0 ± 2.8* | 15.2 ± 2.8** |
Data are mean ± SEM (n = 3–4). Significantly different from respective values for: *W303-1A wild type with P < 0.005 and **AA with P < 0.005.
Acetate and propionate effects on CP content and activity of antioxidant enzymes
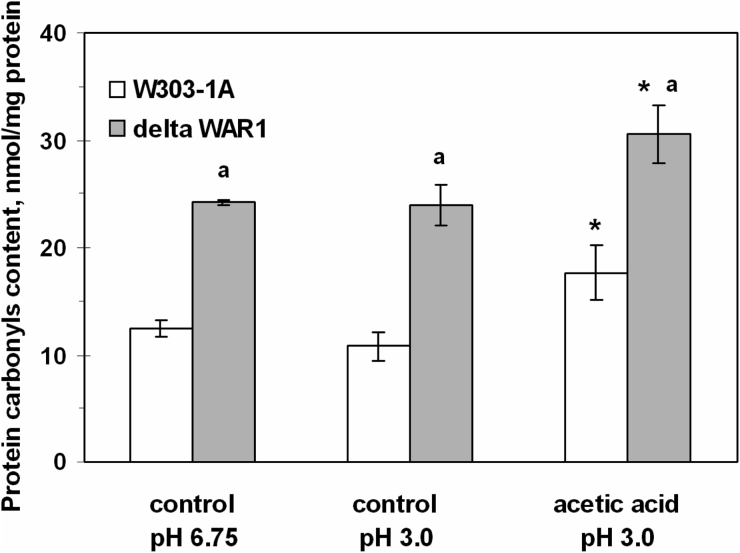
Fig. 3 shows that under control conditions (without AA) the mutant strain defective in War1p (ΔWAR1) had a 2-fold higher CP content than the wild type (W303-1A). Exposure to AA increased CP levels by 1.4-fold in the parental strain and by 1.3-fold in ΔWAR1. It should be added that yeast incubation at pH 3.0 without acetate did not affect CP content in both strains (Fig. 3). In contrast to acetate, PA did not change CP level in the yeast cells (Table 2).
Figure 3.
CP content in S. cerevisiae W303-1A wild type and its derivative ΔWAR1 under exposure to AA. Data are mean ± SEM (n = 3–4). Significantly different from respective values for: *controls with P < 0.005 and aW303-1A wild strain with P < 0.005.
Table 2.
CP levels and activities of SOD and catalase in S. cerevisiae W303-1A cells under PA treatment
| Incubation conditions | CP level (nmol/mg protein) | SOD activity (U/mg protein) | Catalase activity (U/mg protein) |
|---|---|---|---|
| Control (pH 6.75) | 12.8 ± 0.5 | 3167 | 34.3 ± 4.4 |
| Control (pH 3.0) | 12.5 ± 0.9 | 323 ± 11 | 33.0 ± 4.3 |
| PA (pH 3.0) | |||
| 10 mM | ND | 310 ± 23 | 34.3 ± 3.2 |
| 50 mM | ND | 292 ± 34 | 33.0 ± 2.5 |
| 100 mM | 10.1 ± 1.0 | 348 ± 18 | 39.0 ± 4.7 |
ND, not determined.
Data are mean ± SEM (n = 3).
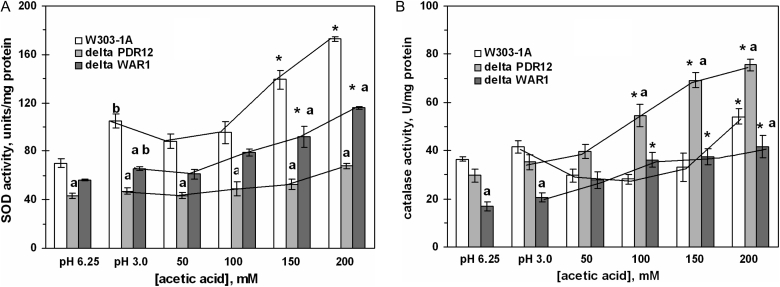
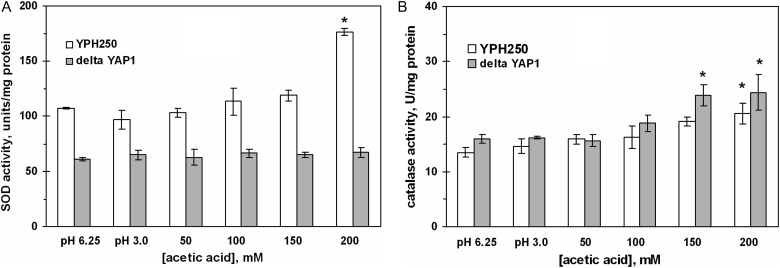
The activity of SOD was significantly higher in the wild strain and the ΔWAR1 mutant after cell treatment with AA high concentrations (Fig. 4A). Both defective strains also demonstrated concentration-dependent increase in catalase activity (Fig. 4B). In the wild type, 50–150 mM AA was either inhibitory or not effective, while 200 mM AA increased catalase activity by 40% relative to pH 3.0 control, and by 60% relative to neutral control. In contrast to acetate, PA did not change SOD and catalase activities in the wild-type yeast (Table 2).
Figure 4.
Activities of SOD (A) and catalase (B) in S. cerevisiae W303-1A and its derivatives ΔPDR12 and ΔWAR1 under AA treatment. Data are mean ± SEM (n = 4–6). Significantly different from respective values for: *control (pH 3.0) with P < 0.05, aW303-1A wild strain with P < 0.01, and bcontrol (pH 6.75) with P < 0.05.
Role of SOD, Yap1p, and catalase in yeast susceptibility to AA
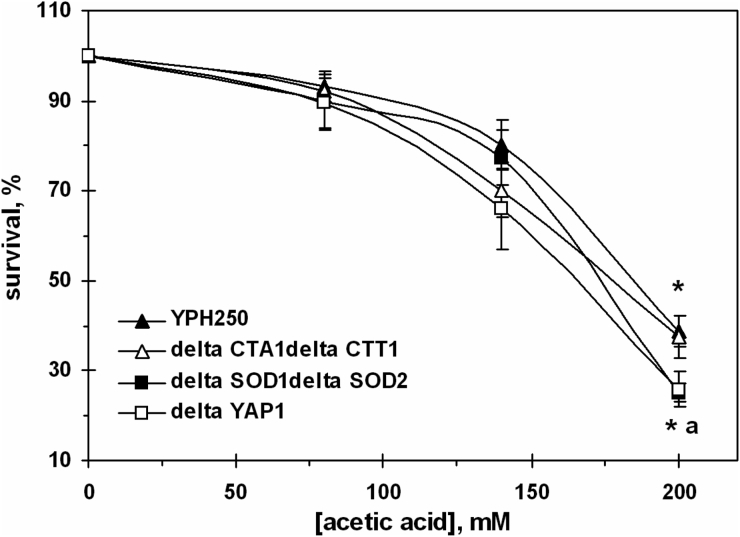
In order to clarify the role of antioxidant enzymes in yeast survival under AA-induced stress, we used strains derived from YPH250: ΔCTA1ΔCTT1 (defective in both catalases), ΔSOD1ΔSOD2 (defective in both SODs), and ΔYAP1 (lacking Yap1p transcriptional regulator of catalases and SODs). Incubation of YPH250 cells with AA caused concentration-dependent killing (Fig. 5). It should be noted that YPH250 strain demonstrated somewhat higher sensitivity to 200 mM AA as compared to W303-1A (Fig. 1A) and virtually the same sensitivity to PA (not shown). Inactivation of genes encoding S. cerevisiae catalases did not affect cell susceptibility to 200 mM AA. However, the defects in genes encoding SODs increased the sensitivity to 200 mM AA by 32%. The Yap1p deletion mutant behaved similarly to the mutant defective in both SODs (Fig. 5).
Figure 5.
Survival of S. cerevisiae YPH250 and its derivatives ΔCTA1ΔCTT1, ΔSOD1ΔSOD2, and ΔYAP1 under AA treatment. Data are mean ± SEM (n = 4–6). Significantly different from respective values obtained: *under treatment with 80 mM AA with P < 0.005 and afor YPH250 with P < 0.05.
Similarly to W303-1A strain (Fig. 4), yeast exposure to 200 mM AA increased the activity of SOD and catalase in YPH250 (Fig. 6). However, in the ΔYAP1 mutant, cell incubation with 50–200 mM AA did not change SOD activity (Fig. 6A), while catalase activity increased (Fig. 6B) in a similar way as in the wild-type strain. In addition, cycloheximide, an inhibitor of protein synthesis in eukaryotes, prevented the AA effect on SOD, but not catalase (not shown).
Figure 6.
Activities of SOD (A) and catalase (B) in S. cerevisiae YPH250 and its derivative ΔYAP1 under AA treatment. Data are mean ± SEM (n = 5–6).*Significantly different from respective values for control (pH 3.0) with P < 0.05.
Cross-protection by H2O2 against AA high concentrations
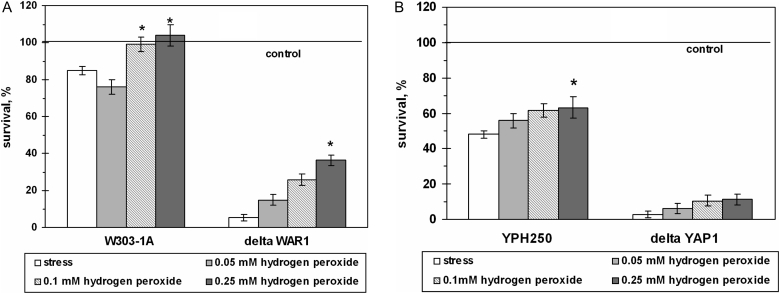
Yeast pre-adaptation by sublethal concentrations of hydrogen peroxide increased cell viability under AA-induced stress in dose- and strain-dependent manners (Fig. 7). For example, pre-incubation with 0.10 and 0.25 mM H2O2 elevated by 1.3-fold cell viability after exposure to 200 mM AA in W303-1A and YPH250 wild strains. Survival of the ΔWAR1 mutant cells after pre-incubation increased by 6.6-fold. However, H2O2 pre-adaptation did not improve the survival of the ΔYAP1 mutant significantly.
Figure 7.
Survival of S. cerevisiae cells pre-treated with sublethal doses of H2O2 under exposure to AA. Strains used: W303-1A and ΔWAR1 (A), and YPH250 and ΔYAP1 (B). Data are mean ± SEM (n = 5–6). *Significantly different from respective values for cells exposed to 200 mM AA with P < 0.01.
Discussion
In this study we have demonstrated different effects of AA and PA on baker's yeast S. cerevisiae. First of all, we examined yeast survival under exposure to AA or PA (Fig. 1A and B). It is well known that WOA toxicity is essentially determined by their hydrophobicity and pKa.5 Although both AA and PA are hydrophilic and have quite close pKa, PA was more toxic to yeast at low concentrations and less toxic at high concentrations relative to AA. Earlier it was also found that S. cerevisiae demonstrated higher sensitivity to low doses of PA comparing with AA.7,33 In general, baker's yeast cells are known to be relatively resistant to monocarboxylic WOAs.14,16 At least three different mechanisms are supposed to be responsible for that: (1) prevention of diffusion of exogenous acid molecules into the cell, (2) extrusion of intracellular acid molecules from the cell, and (3) acid metabolism. Thus it can be suggested that the difference in yeast sensitivities to AA and PA is due to some difference in at least one of the abovementioned mechanisms.
At low pH, WOAs with low pKa exist mainly in the undissociated state; therefore, they rather easily enter the cell by passive diffusion as neutral molecules. It has been reported that AA can also enter the cell by Fps1p-mediated facilitated diffusion.34–36 However, no information on the relation between aquaglyceroporin Fps1p and PA could be found.
At neutral intracellular pH acid molecules dissociate.16,37,38 Yeast can extrude acid anions through the anion efflux pump Pdr12p, which has been found to mediate resistance to both AA and PA.18 However, later works by the same research group reported that acetate did not induce expression of PDR12 gene, while PA did.17,39 It was believed that expression of Pdr12p is controlled by the sole War1p transcriptional factor.17 To date, it is well documented that weak acid stress-induced response depends on the following transcription factors: War1p, Msn2/Msn4p, Rim101p, and Haa1p,5,38 and the sole War1p-target gene, PDR12, was identified among the genes activated by Haa1p in response to AA.40 Analysis of these controversial data demonstrates that possible role of the Pdr12p involvement in acetate efflux is still an open problem.
The common approach to study physiological role of any protein is an investigation of physiology of cells lacking the activity of this protein. Two different ways can be used: knockout isogenic strains and specific protein inhibitors. Both of these ways were applied earlier in our laboratory in the in vivo investigation of dual antioxidant/prooxidant role of SOD.31,41
To the best of our knowledge, no specific inhibitor of Pdr12p has been found; therefore, most researchers use defective mutant cells. However, auxotrophic markers present in mutant strain may lead to incorrect evaluation of gene function. Several years ago it was reported that previous experiments, in which Pdr12p has been identified as the main system responsible for AA extrusion from yeast cells, were experimental artifacts.21 Authors observed inability of the mutant lacking Pdr12p to grow in the presence of acetate only for yeast auxotrophic for tryptophan and concluded that WOAs inhibited uptake of tryptophan from the medium.21 It should be noted that all strains used in this study, parental and isogenic derivatives, are trp1-1 auxotrophs. Thus, the effects observed do not seem to be associated with tryptophan metabolism. In one of the latest works by Mira et al.40, a 4.2-fold increase in PDR12 gene expression by acetate is reported, which is not in agreement with the previous suggestion about artifacts.21 It is possible that other targets than tryptophan metabolism may exist for acetate and be responsible for its toxicity.
One more experimental approach for in vivo evaluation of Pdr12p-mediated anion transport is measurement of fluorescein extrusion from yeast cells.18,21 Therefore, next we compared AA and PA effects on fluorescein efflux from yeast cells (Table 1). Both acids inhibited fluorescein extrusion in all strains used. Earlier it was shown that orthovanadate, an inhibitor of ATPases, and cyanide, an inhibitor of oxidative phosphorylation, did not affect fluorescein efflux.20 At the same time, iodoacetate, a glycolytic inhibitor, significantly suppressed fluorescein extrusion showing that glycolysis plays a critical role in this case.20 It is possible that glycolytic inhibition caused by WOAs may be responsible for lower activity of Pdr12p. It cannot be excluded that certain proteins, e.g. Pdr10p42 or Fps1p36,43, involved in membrane transport and found to modulate Pdr12 activity, can also be affected by AA and PA. Taking into account that quite recently 650 genes were identified as determinants of tolerance to AA,37 complex investigation is needed to clarify the role of Pdr12p efflux pump in yeast response to acid stress. One can conclude that Pdr12p affected the inhibition of fluorescein transport by acetate, but not propionate, since the constants of inhibition by acetate and propionate were different for the mutants (Table 1).
It is widely believed that AA toxicity is associated with reactive oxygen species (ROS) generation and programmed cell death induction in the yeast.8–12 To study and compare possible mechanisms of acetate and propionate effects on yeast, we monitored CP accumulation and the activities of selected antioxidant enzymes. These parameters are used as markers of oxidative stress.44,45 Recently, both SOD and catalase were claimed to play an important role in yeast defense against AA-induced programmed cell death via ROS detoxification.11 Here we found that acetate elevated SOD and catalase activities and CP levels (Figs 3 and 4), while PA did not change these parameters (Table 2). Therefore, one may suggest that different mechanisms underlie the yeast toxicity caused by AA and PA. Based on the present results, it can be assumed that AA indicates a prooxidant effect in vivo, while PA does not.
Earlier, Piper2 reported that the ΔPDR12 mutant exposed to benzoate and sorbate demonstrated increased superoxide production. Recently, Mollapour et al.16 suggested that reduction of endogenous oxidative stress is one of the major advantages of Pdr12p. Our study shows that CP level in the yeast lacking War1p is significantly higher compared to the parental strain (Fig. 3). These data together with the results by others support the idea on oxidative stress development in yeast defective in the Pdr12 transport system.
It was shown that Pdr12p was not under control of either Yap1p or other yeast regulators responsive to oxidative stress.14 Recent works reported that both Pdr12p and Yap1p are members of the Haa1p-regulon responsive to AA.5,40 Results presented in Fig. 5 indicate that Yap1p is at least partially responsible for yeast survival under stress induced by AA. Yap1p is also involved in SOD activation by AA via protein synthesis (Fig. 6A). However, the up-regulation of catalase by AA seems to be Yap1p independent (Fig. 6B). As earlier demonstrated, the expression of the CTT gene encoding cytosolic catalase T was enhanced by sorbic acid treatment via Msn2p/4p-dependent way.46 Msn2p/4p was found to be involved in acetate-stress response also.5,38,40 Therefore, the up-regulation of catalase by regulators other than Yap1p under acetate-induced stress (Figs 4B and 6B) cannot be excluded. As another possible explanation, acetate did not change the expression of catalase genes, but instead activated pre-existing non-active molecules. Earlier we suggested that cells treated with low sublethal concentrations of hydrogen peroxide accumulated non-active stress-protective molecules, further activation of which made microorganisms resistant to lethal concentrations of H2O2.25,47
Overall, microorganism exposure to mild stress leads to the acquisition of cellular resistance to lethal stress.5,6,23–25 Here we found that H2O2 pre-incubation increased the survival of yeast exposed to 200 mM AA (Fig. 7). The highest protective effect was observed for cells lacking War1p (Fig. 7A) and not found in the ΔYAP1 mutant (Fig. 7B). Earlier Hatzixanthis et al.39 demonstrated that H2O2 did not affect PDR12 expression. Piper2 also suggested that PDR12 induction by WOAs was not a direct response to oxidative stress in sorbate-exposed yeast cells. Interestingly, in an experiment of Papadimitriou et al.,22 pre-inducing Pdr12p to maximal levels by subjecting cells to a mild sorbic acid stress did not lead to cells with an acquired resistance. The authors concluded that induction of the pump by sorbate is not sufficient to acquire resistance to acid stress. Therefore, the effects of low concentrations of H2O2 found in the present study seem not associated with a direct activation of the acid extrusion system (Fig. 7A), but rather depend on Yap1p transcriptional regulator (Fig. 7B). It suggests that cells possess complex mechanisms sensing various stress conditions via intracellular redox status, which in turn can be changed by such oxidant as H2O2.
In summary, several interesting conclusions can be drawn: (1) PA was more toxic to yeast at low concentrations and less toxic at high concentrations relative to AA; (2) Pdr12p in some way affects fluorescein transport system in the presence of acetate, but not propionate; (3) yeast exposure to AA increases the level of oxidatively modified proteins and activity of antioxidant enzymes, whereas PA does not change these parameters; (4) Yap1p and protein synthesis de novo is involved in SOD activation by AA, but not catalase; (5) yeast pre-incubation with low concentrations of hydrogen peroxide causes Yap1p-dependent cellular cross-protection against toxic effect of high concentrations of AA. Finally, it is possible that different mechanisms underlie the yeast toxicity by AA and PA: acetate indicates a prooxidant effect in vivo, whereas propionate does not.
Acknowledgments
The authors are grateful to Professor K Kuchler for providing us with yeast strains. HS and VL were partially supported by visiting professor grants from Jagiellonian University, and OA was partially supported by the Queen Jadwiga Fund from Jagiellonian University.
References
- 1.Jamieson DJ. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 1998;14:1511–27. [DOI] [PubMed] [Google Scholar]
- 2.Piper P. Yeast superoxide dismutase mutants reveal a pro-oxidant action of weak organic acid food preservatives. Free Radic Biol Med 1999;17:1219–27. [DOI] [PubMed] [Google Scholar]
- 3.Hazan R, Levine A, Abeliovich H. Benzoic acid, a weak organic acid food preservative, exerts specific effects on intracellular membrane trafficking pathways in Saccharomyces cerevisiae. Appl Environ Microbiol 2004;70:4449–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sá-Correia I, Tenreiro S. The multidrug resistance transporters of the major facilitator superfamily, 6 years after disclosure of Saccharomyces cerevisiae genome sequence. J Biotechnol 2002;98:215–26. [DOI] [PubMed] [Google Scholar]
- 5.Mira NP, Teixeira MC, Sá-Correia I. Adaptive response and tolerance to weak acids in Saccharomyces cerevisiae: a genome-wide view. OMICS 2010;14:525–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lushchak VI. Adaptive response to oxidative stress: bacteria, fungi, plants and animals. Comp Biochem Physiol 2011;153:175–90. [DOI] [PubMed] [Google Scholar]
- 7.Epstein CB, Waddle JA, Hale IVW, Davé V, Thornton J, Macatee TL, et al.. Genome-wide responses to mitochondrial dysfunction. Mol Biol Cell 2001;12:297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ludovico P, Sousa MJ, Silva MT, Leão C, Côrte-Real M. Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology 2001;147:2409–15. [DOI] [PubMed] [Google Scholar]
- 9.Knorre DA, Smirnova EA, Severin FF. Natural conditions inducing programmed cell death in the yeast Saccharomyces cerevisiae. Biochemistry (Moscow) 2005;70:264–6. [DOI] [PubMed] [Google Scholar]
- 10.Giannattasio S, Atlante A, Antonacci L, Guaragnella N, Lattanzio P, Passarella S, et al.. Cytochrome c is released from coupled mitochondria of yeast en route to acetic acid-induced programmed cell death and can work as an electron donor and a ROS scavenger. FEBS Lett 2008;582:1519–25. [DOI] [PubMed] [Google Scholar]
- 11.Guaragnella N, Antonacci L, Giannattasio S, Marra E, Passarella S. Catalase T and Cu,Zn-superoxide dismutase in the acetic acid-induced programmed cell death in Saccharomyces cerevisiae. FEBS Lett 2008;582:210–4. [DOI] [PubMed] [Google Scholar]
- 12.Almeida B, Ohlmeier S, Almeida AJ, Madeo F, Leão C, Rodrigues F, et al.. Yeast protein expression profile during acetic acid-induced apoptosis indicates causal involvement of the TOR pathway. Proteomics 2009;9:720–32. [DOI] [PubMed] [Google Scholar]
- 13.Pearce AK, Booth IR, Brown AJ. Genetic manipulation of 6-phosphofructo-1-kinase and fructose 2,6-bisphosphate levels affects the extent to which benzoic acid inhibits the growth of Saccharomyces cerevisiae. Microbiology 2001;147:403–10. [DOI] [PubMed] [Google Scholar]
- 14.Piper P, Calderon CO, Hatzixanthis K, Mollapour M. Weak acid adaptation: the stress response that confers yeasts with resistance to organic acid food preservatives. Microbiology 2001;147:2635–42. [DOI] [PubMed] [Google Scholar]
- 15.Abrat O, Semchyshyn H, Lushchak V. Acid stress in the yeast Sacccharomyces cerevisiae. Ukrainian Biochem J 2008;80:19–31. [PubMed] [Google Scholar]
- 16.Mollapour M, Shepherd A, Piper PW. Novel stress responses facilitate Saccharomyces cerevisiae growth in the presence of the monocarboxylate preservatives. Yeast 2008;25:169–77. [DOI] [PubMed] [Google Scholar]
- 17.Kren A, Mamnun IM, Bauer BE, Schuller C, Wolfger H, Hatzixanthis K, et al.. War1p, a novel transcription factor controlling weak acid stress response in yeast. Mol Cell Biol 2003;23:1775–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holyoak CD, Bracey D, Piper PW, Kuchler K, Coote PJ. The Saccharomyces cerevisiae weak-acid inducible ABC transporter Pdr12 transports fluorescein and preservative anions from the cytosol by an energy-dependent mechanism. J Bacteriol 1999;181:4644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hazelwood LA, Tai SL, Boer VM, de Winde JH, Pronk JT, Daran JM. A new physiological role for Pdr12p in Saccharomyces cerevisiae: exportofaromatic and branched-chain organic acids produced in amino acid catabolism. FEMS Yeast Res 2006;6:937–45. [DOI] [PubMed] [Google Scholar]
- 20.Lushchak V, Abrat O, Miedzobrodzki J, Semchyshyn H. Pdr12p-dependent and -independent fluorescein extrusion from baker's yeast cells. Acta Biochim Pol 2008;55:595–601. [PubMed] [Google Scholar]
- 21.Bauer B, Rossington D, Mollapour M, Mamnun Y, Kuchler K, Piper P. Weak organic acid stress inhibits aromatic amino acid uptake by yeast, causing a strong influence of amino acid auxotrophies on the phenotypes of membrane transporter mutants. Eur J Biochem 2003;270:3189–95. [DOI] [PubMed] [Google Scholar]
- 22.Papadimitriou MN, Resende C, Kuchler K, Brul S. High Pdr12 levels in spoilage yeast (Saccharomyces cerevisiae) correlate directly with sorbic acid levels in the culture medium but are not sufficient to provide cells with acquired resistance to the food preservative. Int J Food Microbiol 2007;113:173–9. [DOI] [PubMed] [Google Scholar]
- 23.Collinson LP, Dawes IW. Inducibility of the response of yeast cells to peroxide stress. J Gen Microbiol 1992;138:329–35. [DOI] [PubMed] [Google Scholar]
- 24.Berry DB, Gasch AP. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol Biol Cell 2008;19:4580–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semchyshyn H. Hydrogen peroxide-induced response in E. coli and S. cerevisiae: different stages of the flow of the genetic information. Cent Eur J Biol 2009;4:142–53. [Google Scholar]
- 26.Izawa S, Inoue Y, Kimura A. Importance of catalase in the adaptive response to hydrogen peroxide: analysis of acatalasaemic Saccharomyces cerevisiae. Biochem J 1996;320:61–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inoue Y, Matsuda T, Sugiyama K, Izawa S, Kimura A. Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae. J Biol Chem 1999;274:27002–9. [DOI] [PubMed] [Google Scholar]
- 28.Gralla EB, Valentine JS. Null mutants of Saccharomyces cerevisiae Cu, Zn superoxide dismutase: characterization and spontaneous mutation rates. J Bacteriol 1991;173:5918–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abrat O, Semchyshyn H, Miedzobrodzki J, Lushchak V. Fluorescein transport and antioxidant systems in the yeast Saccharomyces cerevisiae under acid stress. Ukrainian Biochem J 2008;80:70–7. [PubMed] [Google Scholar]
- 30.Thomas KC, Hynes SH, Ingledew WM. Influence of medium buffering capacity on inhibition of Saccharomyces cerevisiae growth by acetic and lactic acids. Appl Environ Microbiol 2002;68:1616–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lushchak V, Semchyshyn H, Lushchak O, Mandryk S. Diethyldithiocarbamate inhibits in vivo Cu,Zn-superoxide dismutase and perturbs free radical processes in the yeast Saccharomyces cerevisiae cells. Biochem Biophys Res Commun 2005;338:1739–44. [DOI] [PubMed] [Google Scholar]
- 32.Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:289–92. [DOI] [PubMed] [Google Scholar]
- 33.Fernandes AR, Mira NP, Vargas RC, Canelhas I, Sá-Correia I. Saccharomyces cerevisiae adaptation to weak acids involves the transcription factor Haa1p and Haa1p-regulated genes. Biochem Biophys Res Commun 2005;337:95–103. [DOI] [PubMed] [Google Scholar]
- 34.Mollapour M, Piper PW. Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol Cell Biol 2007;27:6446–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casal M, Paiva S, Queirós O, Soares-Silva I. Transport of carboxylic acids in yeasts. FEMS Microbiol Rev 2008;32:974–94. [DOI] [PubMed] [Google Scholar]
- 36.Mollapour M, Shepherd A, Piper PW. Presence of the Fps1p aquaglyceroporin channel is essential for Hog1p activation, but suppresses Slt2(Mpk1)p activation, with acetic acid stress of yeast. Microbiology 2009;155:3304–11. [DOI] [PubMed] [Google Scholar]
- 37.Mira NP, Palma M, Guerreiro JF, Sá-Correia I. Genome-wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid. Microb Cell Fact 2010;9:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teixeira MC, Mira NP, Sá-Correia I. A genome-wide perspective on the response and tolerance to food-relevant stresses in Saccharomyces cerevisiae. Curr Opin Biotechnol 2010, in press. DOI:10.1016/j.copbio.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 39.Hatzixanthis K, Mollapour M, Seymour I, Bauer BE, Krapf G, Schuller C, et al.. Moderately lipophilic carboxylate compounds are the selective inducers of the Saccharomyces cerevisiae Pdr12p ATP-binding cassette transporter. Yeast 2003;20:575–85. [DOI] [PubMed] [Google Scholar]
- 40.Mira NP, Becker JD, Sá-Correia I. Genomic expression program involving the Haa1p-regulon in Saccharomyces cerevisiae response to acetic acid. OMICS 2010;14:587–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lushchak V, Semchyshyn H, Mandryk S, Lushchak O. Possible role of superoxide dismutases in the yeast Saccharomyces cerevisiae under respiratory conditions. Arch Biochem Biophys 2005;441:35–40. [DOI] [PubMed] [Google Scholar]
- 42.Rockwell NC, Wolfger H, Kuchler K, Thorner J. ABC transporter Pdr10 regulates the membrane microenvironment of Pdr12 in Saccharomyces cerevisiae. J Membr Biol 2009;229:27–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang JG, Liu XY, He XP, Guo XN, Lu Y, Zhang BR. Improvement of acetic acid tolerance and fermentation performance of Saccharomyces cerevisiae by disruption of the FPS1 aquaglyceroporin gene. Biotechnol Lett 2011;33:277–84. [DOI] [PubMed] [Google Scholar]
- 44.Dalle-Donne I, Rossi R, Giustarini D, Milzani A, Colombo R. Protein carbonyl groups as biomarkers of oxidative stress. Clin Chim Acta 2003;329:23–38. [DOI] [PubMed] [Google Scholar]
- 45.Lushchak V. Free radical oxidation of proteins and its relationship with functional state of organisms. Biochemistry (Moscow) 2007;72:809–995. [DOI] [PubMed] [Google Scholar]
- 46.Schuller C, Mamnun YM, Mollapour M, Krapf G, Schuster M, Bauer BE, et al.. Global phenotypic analysis and transcriptional profiling defines the weak acid stress response regulon in Saccharomyces cerevisiae. Mol Biol Cell 2004;15:706–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bayliak MM, Semchyshyn HM, Lushchak VI. Possible accumulation of non-active molecules of catalase and superoxide dismutase in S. cerevisiae cells under hydrogen peroxide induced stress. Cent Eur J Biol 2007;2:326–36. [Google Scholar]