Abstract
Background
A DNA extraction and preservation protocol that yields sufficient and qualitative DNA is pivotal for the success of any nucleic acid amplification test (NAAT), but it still poses a challenge for soil-transmitted helminths (STHs), including Ascaris lumbricoides, Trichuris trichiura and the two hookworms (Necator americanus and Ancylostoma duodenale). In the present study, we assessed the impact of different DNA extraction and preservativation protocols on STH-specific DNA amplification from stool.
Methodology and principal findings
In a first experiment, DNA was extracted from 37 stool samples with variable egg counts for T. trichiura and N. americanus applying two commercial kits, both with and without a prior bead beating step. The DNA concentration of T. trichiura and N. americanus was estimated by means of qPCR. The results showed clear differences in DNA concentration across both DNA extraction kits, which varied across both STHs. They also indicated that adding a bead beating step substantially improved DNA recovery, particularly when the FECs were high. In a second experiment, 20 stool samples with variable egg counts for A. lumbricoides, T. trichiura and N. americanus were preserved in either 96% ethanol, 5% potassium dichromate or RNAlater and were stored at 4°C for 65, 245 and 425 days. DNA was extracted using the DNeasy Blood & Tissue kit with a bead beating step. Stool samples preserved in ethanol proved to yield higher DNA concentrations as FEC increased, although stool samples appeared to be stable over time in all preservatives.
Conclusions
The choice of DNA extraction kit significantly affects the outcome of NAATs. Given the clear benefit of bead beating and our validation of ethanol for (long-term) preservation, we recommend that these aspects of the protocol should be adopted by any stool sampling and DNA extraction protocol for downstream NAAT-based detection and quantification of STHs.
Author summary
DNA-based tools are increasingly being used for the diagnosis of intestinal worm infections in both clinical and research laboratories. However, recovering DNA from intestinal worm eggs in stool remains a challenge since this DNA is protected by a very rigid egg shell. Furthermore, stool contains inhibitors that can affect test results and these should be removed during DNA extraction. Prior to DNA extraction, samples are often preserved, but the impact of the type of preservatives and the duration of preservation remains poorly studied. In the present study, we assessed the impact of four DNA extraction and three preservation protocols on the downstream performance of a DNA-based diagnostic tool for intestinal worms. We found significant differences in DNA recovery across the DNA and preservation protocols, but DNA from worm eggs in stool proved to be stable over time in all preservatives.
Introduction
Soil-transmitted helminth (STH) infections are among the most common parasitic infections globally and affect more than a quarter of the world’s population, mainly poor populations in (sub)tropical regions. These infections are caused by intestinal helminths (worms) which excrete eggs through human stool, that contaminate soil in areas where sanitation is poor and in turn infect the human host orally or through skin contact. The main STH species are the giant roundworm (Ascaris lumbricoides), the whipworm (Trichuris trichiura), and the two hookworms (Necator americanus and Ancylostoma duodenale). Today, STHs are responsible for nearly two million disability-adjusted life years [1, 2].
To control the burden caused by these worms, the World Health Organization (WHO) recommends preventive chemotherapy (PC) programs, during which anthelmintic drugs are administered to at-risk populations (i.e., pre-school-aged children, school-aged children and women of reproductive age). WHO recommends a bi-annual PC when the prevalence of any STHs exceeds 50% and an annual PC when the prevalence is between 20% and 50%. For a prevalence below 20%, it is not recommended to initiate a PC program [3].
Diagnostic tools play a pivotal role in these PC programs as they provide information on the population prevalence and infection intensity distribution that ultimately guides PC program decisions. Today, microscopy of Kato-Katz thick smears is the most widely used tool for the detection and enumeration of STH eggs in stool [3, 4].
More recently, nucleic-acid amplification tests (NAATs) are being used increasingly in clinical [5] and research settings [6–10]. Although these NAATs are associated with a considerable cost for both pre-analytical procedures (DNA extraction), equipment, reagents and training, they have some important advantages which make them an attractive diagnostic tool for STH PC programs. First, NAATs are much more sensitive, which makes them more appropriate to monitor the progress of PC programs when both infection intensity and prevalence of STHs declined after multiple rounds of PC [11]. Second, next to detecting STH DNA, they allow for the simultaneous detection of a variety of other co-endemic pathogens such as Strongyloides stercoralis, another important STH, for which large-scale epidemiological data is lacking due to the poor diagnostic performance of the Kato-Katz method for this helminth [12–14]. Third, NAATs allow differentiation of STH species that cannot be distinguished using microscopy, such as the hookworms. This is important because the human hookworms N. americanus and A. duodenale have a clear different impact on health [15]. Furthermore, there is an increasing number of studies that indicate that animal STHs can infect humans. Important zoonotic species that are known to cause patent infections in humans are Ascaris suum and Trichuris suis from pigs [16] and Ancylostoma ceylanicum [17–18], Ancylostoma caninum [19] and Trichuris vulpis from dogs [20]. Finally, NAATs also allow for the detection of mutations in genes that have been associated with anthelmintic resistance [21, 22]. Despite these advantages, the diagnostic performance of NAATs is highly dependent on an effective DNA extraction. This is a particular challenge for STH eggs in stool, as the shell of the STH eggs are hard to lyse [10,23]. Moreover, stool is a complex matrix containing numerous compounds that may inhibit the amplification of nucleic-acids [24]. Moreover, when NAATs are to be applied in STH PC programs, samples will be collected usually from remote rural areas and transported to a centralized laboratory for analysis, necessitating stool preservation [25]. In the past a variety of common (ethanol and potassium dichromate) and commercial preservatives (e.g., RNAlater and PAXgene) have been used [9,26–28], but the impact of these preservatives and the duration of preservation on DNA recovery remains poorly studied. In this study, we compared the performance of four DNA extraction protocols (DNA extraction experiment), three preservatives and three storage periods (preservation experiment) for the downstream quantitative PCR (qPCR) based detection and quantification of STHs in human clinical stool samples.
Methods
Ethical statements
For the two experiments (the DNA extraction and the preservation experiment), stool samples were collected from primary school children. The protocols for the two experiments were separately approved by the Institutional Review Board of Jimma University, Ethiopia (DNA extraction experiment: reference RPGC/478/2014; preservation experiment: reference RPGC/547/2016). The school administrators, teachers, parents/legal guardians and children were informed about the objectives of the study. Children that appeared in overall healthy condition, whose parents or legal guardians signed an informed consent and who volunteered to provide a sufficient amount of stool sample were included. Children who were found excreting any of the STH eggs were treated with a single oral dose of 400 mg albendazole (GlaxoSmithKline).
DNA extraction experiment
Selection of samples
Based upon previous STH prevalence data from Jimma Town (Ethiopia) [29], three primary schools were strategically chosen for stool sample collection, with the ultimate aim to enroll STH positive subjects excreting different levels of eggs concentration in stool. A total of 195 stool samples from children 5–14 years of age were microscopically screened using the McMaster egg counting method at the Neglected Tropical Disease Laboratory of Jimma University (Ethiopia) using a previously described protocol [30,31]. Briefly, 2 grams of stool was suspended in 30 ml of flotation solution (saturated sodium chloride; specific density = 1.2). The suspension was sieved using a plastic tea strainer to withhold the large debris. This sieved stool suspension was then mixed by pouring 10 times from one cup to the other. Finally, the suspension was transferred to both chambers of a McMaster slide using a Pasteur pipette. STH eggs were allowed to float for 2 min and they were subsequently microscopically counted in both chambers (= 2 x 0.15 ml) using a 100x magnification. To obtain the fecal egg counts (FECs) expressed as number of eggs per gram of stool (EPG), the number of eggs for each STH was multiplied by 50. Based on the FECs, the intensity of infection was classified as low (T. trichiura: FEC <1,000 EPG; hookworm: FEC <2,000) and as moderate-to-heavy (T. trichiura: FEC ≥1,000 EPG; hookworm: FEC ≥2,000 EPG) [32]. We aimed at including a minimum of 10 stool samples of each level of infection intensity for both T. trichiura and hookworms. We restricted the analysis of these two STHs, as their eggs are the most difficult (T. trichiura) and the easiest to lyse (hookworm). In addition, we also included 10 negative samples. Finally, 3 grams of the selected stool samples were preserved in 96% ethanol to make a total volume of 10 ml and stored at room temperature until DNA extraction. All samples were preserved within 6 hours after the stool collection.
DNA extraction protocols
We compared two commercially available DNA extraction kits, i.e. the QIAamp DNA Stool Mini kit and DNeasy Blood & Tissue kit (both Qiagen, Germany). The Qiagen DNeasy Blood & Tissue kit was chosen because it was our in-house DNA extraction protocol, while the QIAamp DNA Stool Mini kit was selected because it is Qiagen’s recommended kit for stool samples. Both kits were tested with and without a preceding bead beating step. This step aims to mechanically rupture the egg shells [32], but this is not included in both commercial Qiagen protocols. For each of the four extraction protocols, an aliquot of approximately 666 μl of the 96% ethanol preserved stool sample was transferred into an Eppendorf tube. This volume corresponds with the 0.2 g of stool recommended by the manufacturer of the QIAamp DNA Stool Mini kit. As there is no recommended amount of stool mentioned in the DNeasy Blood & Tissue kit manual, we used the same amount of stool as for the QIAamp DNA Stool Mini kit protocols. To avoid any systematical error (e.g., the first aliquot systematically being assigned to the same DNA extraction protocol), we randomized the aliquots across the four DNA extraction protocols. Subsequently, the ethanol was removed from all stool aliquots. To this end, all aliquots were centrifuged at 10,000 rpm (8,944 g) for 1 min (Heraeus™ Pico™ 17 Microcentrifuge, Thermo scientific, Germany), after which the supernatant was discarded by pipetting. The remaining pellet was further washed by adding 1 ml of phosphate buffer saline (PBS), after which it was mixed by vortexing and centrifuged at 10,000 rpm (8,944 g) for 1 min. Subsequently, the supernatant was discarded. The pellet was then resuspended in 200 μl of PBS. To enhance egg shell rupture and minimize inhibition, a freeze-thaw-boiling step was included in all extraction protocols (not included in the manufacturer’s protocols). This step consisted of a freeze phase at -80°C for 30 min followed by a thaw-boiling phase in a preheated shaking heating block at 100°C for 10 min. For the two bead beating extraction protocols, the samples were transferred to Green Bead tubes (Roche, Germany) and subjected to bead beating by manual vortexing (VM-300, Jemmy Industrial Corp., Taiwan) at maximum speed (3,150 rpm) for 5 min.
Thereafter, we further followed the manufacturer’s protocol for all four extraction protocols. Briefly, for both QIAamp DNA Stool Mini kit extraction protocols (with and without preceding bead beating), 2 ml of buffer ASL was added to each sample tube and thoroughly mixed by vortexing. Subsequently, 1.6 ml of this suspension was transferred to a 2 ml Eppendorf tube and heated at 70°C for 5 min. After centrifugation at 10,000 rpm (8,944 g) for 2 min, 1.2 ml of supernatant was transferred to a new 2 ml Eppendorf tube containing 1 InhibitEX tablet, which adsorbs inhibitors from the suspension. After centrifugation at 10,000 rpm (8,944 g) for 3 min, 200 μl of supernatant was added to an Eppendorf tube, to which 15 μl of Proteinase K and 200 μl AL lysis buffer was added. Tubes were then incubated at 70°C for 10 min. The complete lysate was added to QIAamp spin column and centrifuged until the lysate completely passed through the spin column membrane enabling nucleic acids to attach to the column membrane. Subsequently, the column was washed with buffers AW1 and AW2. Finally, bound DNA was eluted in 200 μl of AE buffer.
For both DNeasy Blood & Tissue kit extraction protocols (with and without preceding bead beating), 180 μl of buffer ATL containing 20 μl of Proteinase K was added to the sample and incubated at 55°C for 2 h. Subsequently, 400 μl of buffer AL was added and samples where incubated at 70°C for 10 min. Then, the suspension was centrifuged at 10,000 rpm (8,944 g) for 30 seconds, and the supernatant was transferred to an Eppendorf containing 400 μl of 96% ethanol. A total of 600 μl of the mixture was pipetted to a spin column supported by a 2 ml collecting tube and centrifuged at 10,000 rpm (8,944 g) until the lysate completely passed through spin column and the same step applied for rest of the lysate. Subsequently, the column was washed with buffers AW1 and AW2. Finally, bounded DNA was eluted in 200 μl of buffer AE. All DNA extracts were stored at -20°C until shipment on dry ice to the Laboratory for Medical Microbiology and Immunology, Elisabeth-TweeSteden Hospital, Tilburg (The Netherlands) for qPCR analysis.
qPCR
DNA of T. trichiura and hookworms (N. americanus and A. duodenale) was detected using qPCR assays that are routinely used at the Laboratory for Medical Microbiology and Immunology, Elisabeth-TweeSteden Hospital, Tilburg (The Netherlands). The primers and probes used in this qPCR analysis are described in Table 1. The assays were performed on a RotorGene amplification platform using the following cycling conditions: initial denaturation of 15 min at 95°C, followed by 45 cycles of 10 s at 95°C, 15 s at 60°C and 15 s at 72°C. The qPCR results were expressed as the number of genome equivalents per ml of DNA extract (GE/ml) as described by Cools and co-workers [11]. A detailed standard operating procedure used to assess the linkage between Ct and GE/ml is provided in S1 File.
Table 1. Primers and probes used for the qPCR assays.
| Species | Primer/ probe |
Sequence (5’-3’) | Target region | References |
|---|---|---|---|---|
| Ascaris lumbricoides | Fwd | GTAATAGCAGTCGGCGGTTTCTT | ITS-1 | [34] |
| Rev | GCCCAACATGCCACCTATTC | [34] | ||
| Probe | Texas Red-TTGGCGGACAATTGCATGCGAT-BHQ2 | [35] | ||
| Trichuris trichiura | Fwd | TTGAAACGACTTGCTCATCAACTT | 18S | [36] |
| Rev | CTGATTCTCCGTTAACCGTTGTC | [36] | ||
| Probe | Yakima Yellow-CGATGGTACGCTACGTGCTTACCATGG-BHQ1 | [36] | ||
| Ancylostoma duodenale | Fwd | GAATGACAGCAAACTCGTTGTTG | ITS-2 | [37] |
| Rev | ATACTAGCCACTGCCGAAACGT | [37] | ||
| Probe* | Cy5-ATCGTTTACCGACTTTAG- BHQ2 | [37] | ||
| Necator americanus | Fwd | CTGTTTGTCGAACGGTACTTGC | ITS-2 |
[37] |
| Rev | ATAACAGCGTGCACATGTTGC | [37] | ||
| Probe* | FAM-CTGTACTACGCATTGTATAC-BHQ1 | [37] |
*Minor groove binding probes; Fwd: forward primer; Rev: reverse primer; BHQ1: black hole quencher 1; BHQ2: black hole quencher 2; ITS-1: internal transcribed spacer 1; 18S: 18S ribosomal RNA gene; ITS-2: internal transcribed spacer 2
Stool preservation experiment
Selection of samples
In April 2016, a total of 140 stool samples were collected from children of two primary schools in Jimma Town, with the ultimate aim to have 20 stool samples with at least 150 EPG for two STH species. Stool samples were microscopically screened applying a single Kato-Katz thick smear as previously described (WHO, 1992). To avoid clearance of hookworm eggs, all smears were examined within 30–60 min for the presence of STH eggs. The number of STH eggs was counted as per STH species basis and multiplied by 24 to obtain the FECs in EPG.
Stool preservation
After homogenization, three aliquots of 0.5 gram from each of the selected samples were preserved in 1 ml of 96% ethanol, three aliquots in 1 ml of 5% potassium dichromate and three aliquots in 1 ml of RNAlater (Invitrogen). All samples were preserved within 6 hours after the stool collection. Except during the shipment of the samples, all aliquots were stored at 4°C until DNA extraction. To avoid any systematic error (e.g., the first aliquot being assigned to the same preservative and duration of preservation), we randomized the aliquots to one of the three preservatives and time points of DNA extraction. At 65, 245 and 425 days of preservation, one of the three aliquots was subjected to DNA extraction.
DNA extraction and qPCR
DNA of preserved stool samples were extracted using the in-house protocol of the Laboratory for Medical Microbiology and Immunology, Elisabeth-TweeSteden Hospital, Tilburg (The Netherlands). Briefly, 250 μl of stool suspension was placed in an Eppendorf tube and washed by centrifugation at 8,500 g for 1 min. Subsequently, the supernatant was aspirated and discarded. A total volume of 1 ml PBS was added to the pellet, which was again vortexed and centrifuged at 8,500 g for 1 min. Then the supernatant was once more discarded. Subsequently, 500 μl of 2% polyvinylpyrrolidone (PVPP, Sigma) was added to prevent inhibition in downstream qPCR steps, and the suspension was transferred to Green Bead tubes (Roche). A freeze-thaw cycle was performed by placing the tubes at -80°C for 30 min. After thawing, mechanical disruption of the sample was performed by placing the tubes in the MagNA Lyser (Roche) for 1 min at 3,000 rpm. After a short spin, 500 μl of ATL buffer (Qiagen) containing 50 μl of Proteinase K (Qiagen) was added and placed at 55°C for 2 h. Subsequently, samples were placed in the automated QiaSymphony platform for purification. Finally, extracted DNA from each preservative and each time point was used as a template in qPCR assays for the quantification of DNA of A. lumbricoides, T. trichiura and hookworms (N. americanus and A. duodenale) using the primers described in Table 1.
Data analysis
DNA extraction protocols
The performance of the different extraction protocols was assessed by comparing the sensitivity and the DNA concentration of the different STHs expressed as GE/ml. The sensitivity was determined using the principle of the composite reference standard method [38] as the gold standard, which in our case classifies a stool sample as positive for a STH species if at least one egg was detected applying microscopy or one of the four extraction protocols has a downstream positive qPCR for that particular STH species. For the DNA concentration, the geometric mean was applied as a summary statistic. We explored the sensitivity and DNA concentration for each of the four DNA extraction protocols separately. We determined the sensitivity across the different levels of infection intensity and the Pearson’s coefficient between the log transformed DNA concentration and the log transformed FECs.
Significant differences in sensitivity and DNA concentration across DNA extraction protocols were assessed applying generalized linear mixed effect models. For the sensitivity, the ‘glmer’ function in R was used, incorporating the binary qPCR test (positive or negative for that particular target) as dependent variable and STH species (2 levels: T. trichiura and N. americanus), the DNA extraction kit (2 levels: QIAamp DNA Stool Mini kit and DNeasy Blood & Tissue kit), the inclusion of a bead beating step (2 levels: yes and no) and log transformed FECs as potential predicting variables. For the DNA concentration, the ‘lmer’ function in R was used, incorporating the log transformed DNA concentration of that particular target as dependent variable and the aforementioned potential predicting variables. In both analyses, only those samples that were found positive based on the composite reference standard method were incorporated in the analysis. In addition, samples containing DNA of multiple STHs were considered as separate samples, one for each STH species. The level of significance was set at p <0.05.
Stool preservation
We assessed the impact of different preservatives, duration of preservation and FECs by comparing the sensitivity and the DNA concentration of the different STHs expressed as GE/ml applying the aforementioned methodologies. For the sensitivity, a stool sample was classified as positive for a STH species if at least one egg was detected applying microscopy or one of the preservatives had a downstream positive qPCR at least one time point of DNA extraction for that particular STH species. For the generalized linear mixed effect models, the STH species (3 levels: A. lumbricoides, T. trichiura and N. americanus), the preservative (3 levels: 96% ethanol, RNAlater and 5% potassium dichromate), duration of preservation expressed in days (continuous variable) and the log transformed FECs were evaluated as potential predicting variables. The level of significance was set at p <0.05. The raw data of the two different experiments is made available in S2 File.
Results
Comparison of the four DNA extraction protocols
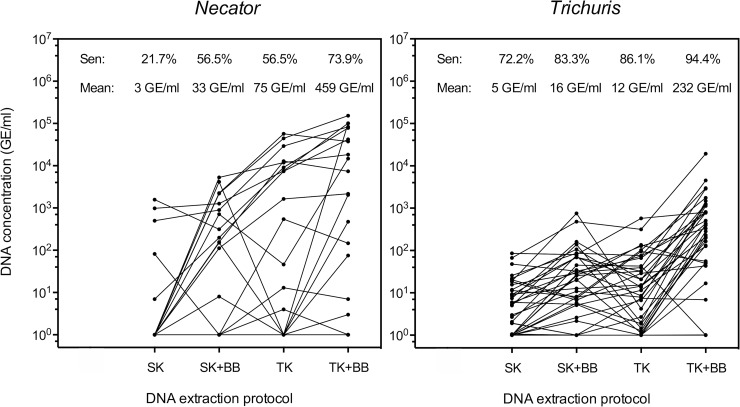
Eggs and/or DNA of T. trichiura and N. americanus were detected in 36 and 23 samples, respectively. Fig 1 illustrates the sensitivity and DNA concentration for each of the four DNA extraction protocols for both T. trichiura and N. americanus. The sensitivity when applying the DNeasy Blood & Tissue kit was higher compared to the QIAamp Stool Mini kit for both STH species, and this independent of the inclusion of bead beating. Similar trends were observed for DNA concentration. The measured DNA concentrations were higher when the DNeasy Blood & Tissue kit was used and when a bead beating step was applied. For both STHs, we also explored the variation in sensitivity across the FECs (See Table 2). In general, the sensitivity increased when the FECs increased and this increase in sensitivity was proportionally more pronounced when QIAamp Stool Mini kit or no bead beating was applied.
Fig 1. The sensitivity and DNA concentration across four DNA extraction protocols for Trichuris and Necator.
The left panel represents the sensitivity (sen) and geometric mean of DNA concentration expressed as genome equivalents per ml of DNA extract (mean; GE/ml) for 20 Necator americanus samples preserved in 96% ethanol and extracted by four DNA extraction protocols. The DNA extraction protocols include the QIAamp DNA Stool Mini kit without (SK) and with bead beating (SK + BB), DNeasy Blood & Tissue kit without (TK) and without bead beating (TK + BB). The right panel represents the same parameters across 36 Trichuris trichiura samples preserved in 96% ethanol. Each line represents a sample.
Table 2. The sensitivity across four DNA extraction protocols for different levels of Trichuris and Necator infections.
The intensity of infection was classified as low (T. trichiura: fecal egg count (FEC) <1,000 eggs per gram of stool (EPG); N. americanus: FEC <2,000 EPG) and as moderate-to-heavy (T. trichiura: FEC ≥1,000 EPG; N. americanus: FEC ≥2,000 EPG). The zero FECs, represent subjects for which no eggs were found applying McMaster.
| N | QIAamp DNA Stool Mini kit | DNeasy Blood & Tissue kit | ||||
|---|---|---|---|---|---|---|
| Without bead beating (%) | With bead beating (%) | Without bead beating (%) | With bead beating (%) | |||
| Trichuris trichiura | ||||||
| Zero FECs | 15 | 66.7 | 73.3 | 73.3 | 86.7 | |
| Low | 11 | 54.5 | 81.8 | 90.9 | 100 | |
| Moderate-to-heavy | 10 | 100 | 100 | 100 | 100 | |
| Necator americanus | ||||||
| Zero FECs | 9 | 22.2 | 44.4 | 44.4 | 66.7 | |
| Low | 14 | 21.4 | 64.3 | 64.3 | 78.6 | |
For T. trichiura, there was a positive correlation between the FECs and the DNA concentration for each of the four DNA extraction protocols (See S3 File), the correlation being higher when a bead beating step was applied (QIAamp DNA Stool Mini kit: 0.39 (p = 0.03) vs. 0.47 (p = 0.004); DNeasy Blood & Tissue kit: 0.32 (p = 0.07) vs. 0.64 (p <0.001)). Similar patterns were observed for N. americanus (QIAamp DNA Stool Mini kit: 0.01 (p = 0.96) vs. 0.35 (p = 0.10); DNeasy Blood & Tissue kit: 0.23 (p = 0.28) vs. 0.31 (p = 0.14); S4 File), but the correlation coefficients were not significant for all DNA extraction protocols.
The outcome of the general linear mixed effect regression models confirmed significant differences in sensitivity and DNA recovery across both STHs, DNA extraction protocols and FECs. The odds of detecting a case of N. americanus was approximately 18.1 (95% confidence intervals (95%CI): 3.9–84.6, p <0.001) times lower than the odds for T. trichiura. Applying the DNeasy Blood & Tissue kit increased the odds with a factor 5.6 (95%CI: 2.2–14.1, p <0.001) compared to the QIAamp DNA Stool Mini kit. Inclusion of a bead beating step further improved the detection of cases, the odds being 4.8 times higher (95%CI: 1.9–11.8, p <0.001) than when bead beating was not included. An increase in FEC of 1 EPG increased the odds ratios 1.3 times (95%CI: 1.0–1.6, p = 0.02). The two-way interactions between STH, DNA extraction kit, bead beating and FECs were not significant.
The DNA concentration increased approximately 6.3 (95%CI: 3.5–11.5) times when the DNeasy Blood & Tissue kit was used instead of the QIAamp DNA Stool Mini kit. A bead beating step increased the DNA concentration 3.6 (96%CI: 1.8–7.4) times. There was no significant difference in DNA recovery across N. americanus and T. trichiura, but the impact of the DNA extraction kit on DNA recovery was different for these STHs. Compared to T. trichiura, the DNA recovery gain when applying the DNeasy Blood & Tissue kit instead of QIAamp DNA Stool Mini kit was 2.8 (96%CI: 1.1–7.4) times higher for N. americanus. An interaction was also observed between FECs and the bead beating. When the FEC increases with 1 EPG, bead beating resulted in the DNA concentration that was 1.2 (95%CI: 1.1–1.4) times higher compared to when no bead beating was done.
Comparison of the different stool preservatives and period of preservation
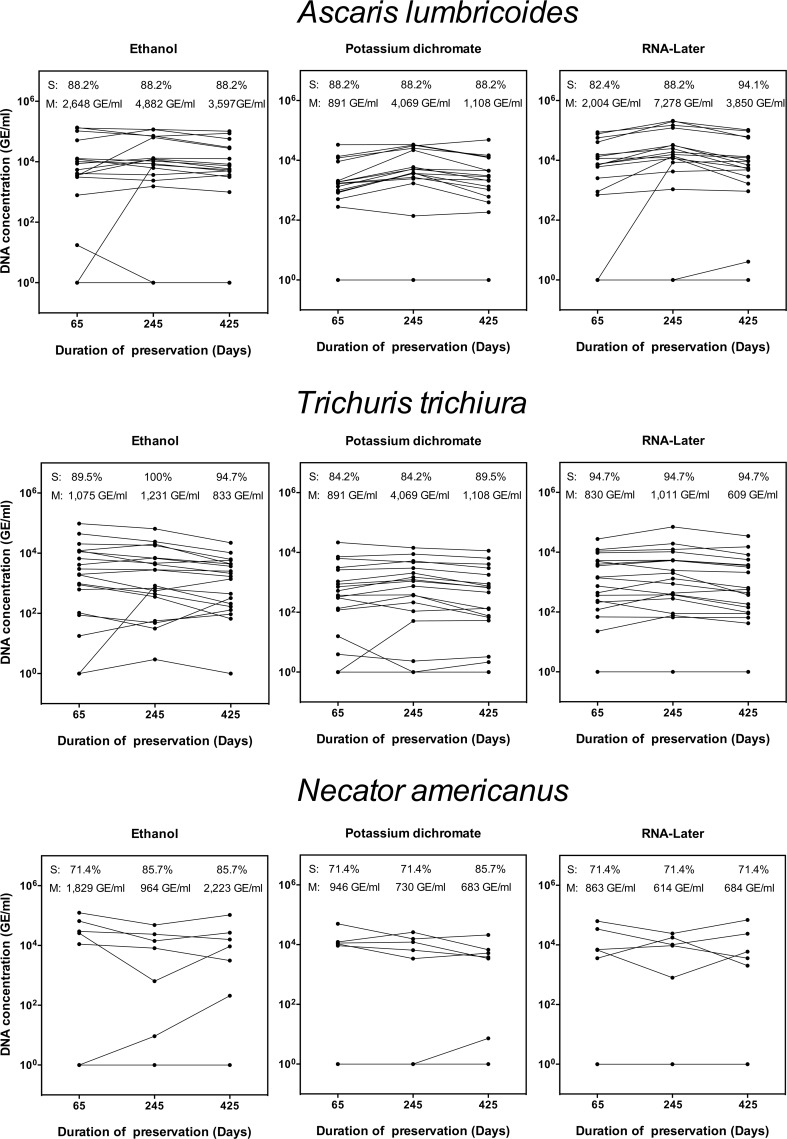
Eggs and/or DNA of A. lumbricoides, T. trichiura and N. americanus were detected in 17, 19 and 7 samples, respectively. Fig 2 summarizes the sensitivity and DNA concentration for each of the preservatives for A. lumbricoides, T. trichiura and N. americanus over time. In general, there were only minor differences in sensitivity and DNA concentration over time and DNA concentration in samples preserved in potassium dichromate where lower compared to that found in sample preserved in ethanol and RNAlater. As illustrated in S5 File, the sensitivity generally increased with increasing FECs for the three STH species. The gain in detecting cases was more pronounced for T. trichiura and potassium dichromate. For all three STH species, there was a significant correlation between the FECs and the DNA concentration for each of the three preservatives (See S6 File), the correlation being high (Pearson’s correlation coefficient (R) >0.79) for each of the preservatives and STH species.
Fig 2. The sensitivity and DNA concentration across three preservatives and three preservation times.
The line graphs represent the sequential differences in sensitivity (Sen) and geometric mean (mean) of DNA concentration expressed genome equivalents/ml (GE/ml) of DNA over time (65, 125 and 425 days) for 17 Ascaris lumbricoides, 19 Trichuris trichiura and 7 Necator americanus samples stored in three preservatives. The preservatives include 96% ethanol (left plot), 5% potassium dichromate (middle graph) and RNAlater (right graph). Each line represents a sample.
The outcome of the regression models indicated a significant difference in sensitivity between STHs and FECs, but did not reveal any significant difference between preservatives and duration of preservation. Compared to A. lumbricoides (reference), the odds of correctly classifying a case were 2.1 times (95%CI: 1.0; 4.5, p = 0.06) lower for N. americanus, though only marginally significant. For T. trichiura, there was no significant difference in odds ratio. A significant difference in DNA concentration was observed between STHs, preservatives and FECs, but not for duration of preservation. Compared to A. lumbricoides (reference), the DNA concentration for N. americanus was 8.8 times (95%CI: 1.5–52.7) lower. The DNA concentration increased with a factor 1.5 (95%CI: 1.2–1.8) per increase of 1 EPG. In addition, there was a significant interaction between FECs, and both STHs and preservatives, indicating that the gain in DNA concentration for an equal increase FEC is different across STHs and preservatives. When compared to A. lumbricoides (reference), an increase in FEC with 1 EPG (reference) resulted in 1.4 (95%CI: 1.0–1.9) higher DNA concentration for N. americanus and 1.4 (95%CI: 1.1–1.7) times lower DNA concentration for T. trichiura. When compared to the DNA concentration of samples preserved ethanol (reference), an increase in FEC with 1 EPG resulted in 1.3 less DNA when preserved in potassium dichromate (95%CI: 1.0–1.7) and RNA-later (95%CI: 1.1–1.6).
Discussion
Research and clinical laboratories are increasingly using NAATs for both detection and quantification of STHs in stool. However, any DNA based examination requires DNA extraction method that yields sufficient and qualitative DNA. In the current study, we assessed the performance of the different extraction and preservation protocols by comparing the sensitivity and the DNA concentration (expressed as GE/ml) of the different STHs. To our knowledge, we are the first to compare the preservation of stool samples over a period of 14 months in three widely used preservatives for STHs, i.e. ethanol, potassium dichromate and RNAlater.
DNeasy Blood & Tissue kit outcompetes QIAamp DNA Stool Mini
The DNA extraction experiment indicated that extracting DNA applying the DNeasy Blood & Tissue kit improved the sensitivity and DNA concentration for both T. trichiura and N. americanus. Moreover, the gain in DNA concentration significantly varied across kits, with the DNA recovery when using the DNeasy Blood & Tissue kit being higher for N. americanus than for T. trichiura. The relatively poor performance of the QIAamp DNA Stool Mini kit is rather unexpected, particularly when this kit is designed for extracting DNA from stool, for example by inclusion of a inhibitor removal step. Although it remains difficult to identify any other differences in both protocols (e.g. recipe of different buffers is not known; ASL in stool kit vs. ATL buffer in tissue kit), there is a clear difference in both time and temperature at which the samples are lysed (stool kit: 10 min at 70°C vs. tissue kit: 2 hours at 55°C). In addition to a lesser performance, the QIAamp DNA Stool Mini kit is also more expensive than the DNeasy Blood & Tissue kit (€322 vs. €201 for 50 samples, prices obtained from the company website) [39,40] and requires more operational steps (e.g. adding and removal of the inhibitEX tablet). In the present study we only tested two kits from the same company, but there is a plethora of commericial DNA extractions kits (e.g. DNease PowerSoil kit (fomer PowerSoil kit), FastDNA SPIN Kit and MagnaPure Roche kit; (Cools et al., under review), and hence it remains unclear whether these kits outcompete the DNeasy Blood & Tissue kit.
Bead beating is a crucial step in any DNA extraction protocol
The DNA extraction experiment clearly showed that a bead beating step too has an important impact on the sensitivity and DNA recovery. This is in line with previous studies that compared DNA extraction protocols with and without bead beating for the detection of parasites, including but not limited to human STHs (A. lumbricoides [33], A. suum [41], A. duodenale [33], Necator americanus [33], T. trichiura [33,41], Ostertagia ostertagi [42], Echinococcus multilocularis [43,44], Toxocara canis, Toxocara catis and Toxoascaris leonina [45]). In addition, the benefit of bead beating was more pronounced when FECs were higher, which might be explained by the increased likelihood of contact of beads with eggs (being higher when more eggs are present for same number of beads), and suggests that a higher concentration of beads might be recommended when FECs are low. The current and previous studies have used different types of beads such as glass [46], garnet [33], zirconium [41], ceramic [42] and steel [45], but the impact of these on the sensitivity of and DNA concentration too remains to be elucidated.
Ethanol is a cheap and reliable preservative for long-term storage for STHs
We found that ethanol and RNAlater preserved stool samples yielded higher DNA concentrations of A. lumbricoides, T. trichiura and N. americanus compared to potassium dichromate although there were no significant differences in sensitivity across preservatives. Furthermore, for each of the preservatives we demonstrated limited variation in DNA concentration over the evaluated time period. This justifies the use of both ethanol, potassium dichromate and RNAlater for long time storage. These findings are in line with previous studies comparing preservation of N. americanus (ethanol, potassium dichromate and RNAlater over 60 days) [28] and Giardia duodenalis cysts (ethanol and potassium dichromate over three months) [47]. On the contrary, Kuk and Cetinkaya (2012) suggested that potassium dichromate was a better preservative for G. duodenalis trophozoites compared to 75% ethanol (over four weeks), but conclusions were based on a small sample size (n = 5) [48]. Ethanol has some important advantages over the other two preservatives. First, the results highlight that the net gain in DNA concentration compared to other preservatives increased when FEC are higher, suggesting that it is better preservative. Second, ethanol is easily accessible in resource-limited settings, where e.g. the commercial RNAlater and potassium dichromate can be difficult to obtain. Moreover, ethanol is approximately two times and over 100 times cheaper compared to potassium dichromate (5%) and RNAlater, respectively. The cost of 100 ml ethanol, 100 ml potassium dichromate (5%) and 100 ml RNAlater were estimated to be €1, €1.8 and €150, respectively.
Limitations of the study
This study has six important limitations, which needs to be considered when both interpreting extrapolating the results. First, we only evaluated a restricted number of DNA extraction (DNA extraction kit and beads) and preservative protocols, focusing on DNA extraction protocols previously used by our laboratory and widely used preservatives. Consequentially, extrapolation to any other combination of DNA and preservation protocol should be done with care. Second, the two experiments were conducted on two different sets of stool samples at different time points applying a different microscopic method for screening cases (DNA extraction experiment: McMaster and preservation experiment: Kato-Katz thick smear) and a slightly adapted DNA extraction protocol on the selected samples (e.g. manual vortexing vs. automated bead beating). Although this study is indeed not standardized across both experiments, this difference in methodology across experiments has no impact on the conclusions drawn from the separate experiments. Third, we did not include a non-preservation control (extraction of fresh samples at time of collection) or a standardized reference (e.g. freezing of samples) in our experiments. Hence, we cannot exclude any DNA degradation with absolute certainty. Inclusion of a non-preservative control or a standard reference was logistically difficult. All samples needed to be shipped to The Netherlands for further molecular analysis, which impeded extraction of all samples at collection (fresh samples would have been extracted with a slightly different DNA extraction protocol), required strict measurements to adhere to safety regulations and to ensure cold chain throughout the transfer of material. Moreover, it is important to highlight that previous studies indicated that analysis of samples preserved in ethanol provide a level of performance that was at least equal to that of samples immediately frozen after collection [33]. This underscores that ethanol-preserved samples too could serve as valid standard reference, which on top is more feasible under field conditions. Fourth, we stored the preserved samples at 4°C under controlled conditions (except during shipment of the samples). Although there is an increased storage capacity in STH endemic countries, these conditions and thus the results might not be representative for all laboratory settings. Fifth, due to the low endemicity we were not able to include sufficient cases of N. americanus, and hence more research is required to confirm our findings. In addition to this, we are not able to draw any conclusions on the other hookworm species (A. duodenale and A. ceylanicum).
In conclusion, DNeasy Blood & Tissue kit DNA extraction protocol with preceded bead beating maximized the extraction of STH DNA in stool in our study. Ethanol was found to be the most cost-effective preservative. Given the clear benefit of bead beating and our validation of ethanol for (long-term) preservation, we recommend that these aspects of the protocol should be adopted by any stool sampling and DNA extraction protocol for downstream NAAT-based detection and quantification of STHs.
Supporting information
(PDF)
The raw data of the DNA and preservation experiment can be found in the worksheets ‘DNA extraction’ and ‘Preservation’, respectively. The worksheet ‘Legend’ explains the headers (variables) in across both data sets.
(XLSX)
The fecal egg counts are expressed as eggs per gram of stool (EPG), whereas the DNA concentration is expressed as genome equivalents per ml (GE/ml). ‘R’ represents the Pearson’s correlation coefficient.
(PDF)
The fecal egg counts are expressed as eggs per gram of stool (EPG), whereas the DNA concentration is expressed as genome equivalents per ml (GE/ml). ‘R’ represents the Pearson’s correlation coefficient.
(PDF)
The intensity of infection was classified as low (Ascaris lumbricoides: fecal egg count (FEC) <5,000 eggs per gram of stool (EPG); Trichuris trichiura: FEC <1,000 EPG; Necator americanus: FEC <2,000 EPG) and as moderate-to-heavy (Ascaris lumbricoides: FEC ≥5,000 EPG; Trichuris trichiura: FEC ≥1,000 EPG; Necator americanus: FEC ≥2,000 EPG). The zero FECs, represent subjects for which no eggs were found applying Kato-Katz thick smear, but for which at least on preservation protocol resulted in a positive qPCR. The sample size equals to the number (N) of subjects multiplied by the number of time points at which DNA is extracted (3 time points).
(DOCX)
The scatterplot illustrate the agreement in fecal egg counts (FECs; expressed as eggs per gram of stool (EPG) and the DNA concentration (expressed as genome equivalents per ml (GE/ml)) across ethanol, potassium dichromate and RNAlater for Ascaris lumbricoides (top graphs), Trichuris trichiura (middle graphs) and Necator americanus (bottom graphs).’R’ represents the Pearson’s correlation coefficient.
(PDF)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
PC is financially supported through a grant from the Bill & Melinda Gates foundation (OPP1120972, principal investigator is BL) (www.starworms.org). JV is financially supported through an International Coordination Action of the Flemish Research Foundation (www.fwo. BL is a postdoctoral fellow of the Research Foundation (www.fwo.be). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jourdan PM, Lamberton PH, Fenwick A, Addiss DG. Soil-transmitted helminth infections. The Lancet. 2018;391(10117):252–65. [DOI] [PubMed] [Google Scholar]
- 2.GBD 2017 DALYs and HALE Collaborators. Global, regional, and national disability-adjusted life-years (DALYs) for 359 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1859–922. 10.1016/S0140-6736(18)32335-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Preventive chemotherapy to control soil-transmitted helminth infections in at-risk population groups. Geneva, Switzerland: World Health Organization, 2017. [PubMed] [Google Scholar]
- 4.World Health Organization. Basic laboratory methods in medical parasitology. Geneva, Switzerland: World Health Organization, 1991. [Google Scholar]
- 5.Verweij JJ, Stensvold CR. Molecular testing for clinical diagnosis and epidemiological investigations of intestinal parasitic infections. Clin Microbiol Rev. 2014;27(2):371–418. 10.1128/CMR.00122-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Easton AV, Oliveira RG, O’Connell EM, Kepha S, Mwandawiro CS, Njenga SM, et al. Multi-parallel qPCR provides increased sensitivity and diagnostic breadth for gastrointestinal parasites of humans: field-based inferences on the impact of mass deworming. Parasit Vectors. 2016;9(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Llewellyn S, Inpankaew T, Nery SV, Gray DJ, Verweij JJ, Clements AC, et al. Application of a multiplex quantitative PCR to assess prevalence and intensity of intestinal parasite infections in a controlled clinical trial. PloS Neglect Trop Dis. 2016;10(1):e0004380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staudacher O, Heimer J, Steiner F, Kayonga Y, Havugimana JM, Ignatius R, et al. Soil-transmitted helminths in southern highland Rwanda: associated factors and effectiveness of school‐based preventive chemotherapy. Trop Med Int Health. 2014;19(7):812–24. 10.1111/tmi.12321 [DOI] [PubMed] [Google Scholar]
- 9.Nery SV, Qi J, Llewellyn S, Clarke NE, Traub R, Gray DJ, et al. Use of quantitative PCR to assess the efficacy of albendazole against Necator americanus and Ascaris spp. in Manufahi District, Timor-Leste. Parasit Vectors. 2018;11(1):373 10.1186/s13071-018-2838-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Connell EM, Nutman TB. Molecular diagnostics for soil-transmitted helminths. Am J of Trop Med Hyg. 2016;95(3):508–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cools P, Vlaminck J, Albonico M, Ame S, Ayana M, Cringoli G, et al. Diagnostic performance of qPCR, Kato-Katz thick smear, Mini-FLOTAC and FECPAKG2 for the detection and quantification of soil-transmitted helminths in three endemic countries. PLoS Negl Trop Dis. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schär F, Trostdorf U, Giardina F, Khieu V, Muth S, Marti H, et al. Strongyloides stercoralis: global distribution and risk factors. Plos Neglect Trop D. 2013;7(7):e2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buonfrate D, Formenti F, Perandin F, Bisoffi Z. Novel approaches to the diagnosis of Strongyloides stercoralis infection. Clin Microbiol Infect. 2015;21(6):543–52. 10.1016/j.cmi.2015.04.001 [DOI] [PubMed] [Google Scholar]
- 14.Krolewiecki AJ, Lammie P, Jacobson J, Gabrielli A-F, Levecke B, Socias E, et al. A public health response against Strongyloides stercoralis: time to look at soil-transmitted helminthiasis in full. PloS Neglect Trop Dis. 2013;7(5):e2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bethony J, Brooker S, Albonico M, Geiger SM, Loukas A, Diemert D, et al. Soil-transmitted helminth infections: ascariasis, trichuriasis, and hookworm. Lancet. 2006;367(9521):1521–32. 10.1016/S0140-6736(06)68653-4 [DOI] [PubMed] [Google Scholar]
- 16.Nejsum P, Betson M, Bendall R, Thamsborg SM, Stothard J. Assessing the zoonotic potential of Ascaris suum and Trichuris suis: looking to the future from an analysis of the past. J Helminthol. 2012;86(2):148–55. 10.1017/S0022149X12000193 [DOI] [PubMed] [Google Scholar]
- 17.Ngui R, Mahdy MA, Chua KH, Traub R, Lim YA. Genetic characterization of the partial mitochondrial cytochrome oxidase c subunit I (cox 1) gene of the zoonotic parasitic nematode, Ancylostoma ceylanicum from humans, dogs and cats. Acta Trop. 2013;128(1):154–7. 10.1016/j.actatropica.2013.06.003 [DOI] [PubMed] [Google Scholar]
- 18.Traub RJ, Inpankaew T, Sutthikornchai C, Sukthana Y, Thompson RA. PCR-based coprodiagnostic tools reveal dogs as reservoirs of zoonotic ancylostomiasis caused by Ancylostoma ceylanicum in temple communities in Bangkok. Vet Parasitol. 2008;155(1–2):67–73. 10.1016/j.vetpar.2008.05.001 [DOI] [PubMed] [Google Scholar]
- 19.George S, Levecke B, Kattula D, Velusamy V, Roy S, Geldhof P, et al. (2016) Molecular Identification of Hookworm Isolates in Humans, Dogs and Soil in a Tribal Area in Tamil Nadu, India. PLoS Negl Trop Dis. 10(8): e0004891 10.1371/journal.pntd.0004891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Areekul P, Putaporntip C, Pattanawong U, Sitthicharoenchai P, Jongwutiwes S. Trichuris vulpis and T. trichiura infections among schoolchildren of a rural community in northwestern Thailand: the possible role of dogs in disease transmission. Asian Biomed. 2010;4(1):49–60. [Google Scholar]
- 21.Diawara A, Drake LJ, Suswillo RR, Kihara J, Bundy DA, Scott ME, et al. Assays to detect β-tubulin codon 200 polymorphism in Trichuris trichiura and Ascaris lumbricoides. PloS Neglect Trop Dis. 2009;3(3):e397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diawara A, Halpenny CM, Churcher TS, Mwandawiro C, Kihara J, Kaplan RM, et al. Association between response to albendazole treatment and β-tubulin genotype frequencies in soil-transmitted helminths. PloS Neglect Trop Dis. 2013;7(5):e2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verweij JJ. Application of PCR-based methods for diagnosis of intestinal parasitic infections in the clinical laboratory. Parasitology. 2014;141(14):1863–72. 10.1017/S0031182014000419 [DOI] [PubMed] [Google Scholar]
- 24.Schrader C, Schielke A, Ellerbroek L, Johne R. PCR inhibitors–occurrence, properties and removal. J Appl Microbiol. 2012;113(5):1014–26. 10.1111/j.1365-2672.2012.05384.x [DOI] [PubMed] [Google Scholar]
- 25.Ten Hove R, van Esbroeck M, Vervoort T, van den Ende J, Van Lieshout L, Verweij J. Molecular diagnostics of intestinal parasites in returning travellers. Eur J Clin Microbiol Infect Dis. 2009;28(9):1045–53. 10.1007/s10096-009-0745-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Gruijter JM, van Lieshout L, Gasser RB, Verweij JJ, Brienen EA, Ziem JB, et al. Polymerase chain reaction‐based differential diagnosis of Ancylostoma duodenale and Necator americanus infections in humans in northern Ghana. Trop Med Int Health. 2005;10(6):574–80. 10.1111/j.1365-3156.2005.01440.x [DOI] [PubMed] [Google Scholar]
- 27.Nsubuga AM, Robbins MM, Roeder AD, Morin PA, Boesch C, Vigilant L. Factors affecting the amount of genomic DNA extracted from ape faeces and the identification of an improved sample storage method. Mol Ecol. 2004;13(7):2089–94. 10.1111/j.1365-294X.2004.02207.x [DOI] [PubMed] [Google Scholar]
- 28.Papaiakovou M, Pilotte N, Baumer B, Grant J, Asbjornsdottir K, Schaer F, et al. A comparative analysis of preservation techniques for the optimal molecular detection of hookworm DNA in a human fecal specimen. PloS Negl Trop Dis. 2018;12(1):e0006130 10.1371/journal.pntd.0006130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mekonnen Z, Levecke B, Boulet G, Bogers J, Vercruysse J. Efficacy of different albendazole and mebendazole regimens against heavy-intensity Trichuris trichiura infections in school children, Jimma Town, Ethiopia. Pathog Glob Health. 2013;107(4):207–9. 10.1179/2047773213Y.0000000092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levecke B, Behnke JM, Ajjampur SS, Albonico M, Ame SM, Charlier J, et al. A comparison of the sensitivity and fecal egg counts of the McMaster egg counting and Kato-Katz thick smear methods for soil-transmitted helminths. PloS Negl Trop Dis. 2011;5(6):e1201 10.1371/journal.pntd.0001201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.http://www.starworms.org/tools/overview/visual-tutorials, Accessed July 10th 2019.
- 32.World Health Organization. Guidelines for the evaluation of soil-transmitted helminthiasis and schistosomiasis at community level A guide for control programme managers. Geneva, Switzerland: World Health Organization, 1998. [Google Scholar]
- 33.Kaisar MMM, Brienen EAT, Djuardi Y, Sartono E, Yazdanbakhsh M, Verweij JJ, et al. Improved diagnosis of Trichuris trichiura by using a bead-beating procedure on ethanol preserved stool samples prior to DNA isolation and the performance of multiplex real-time PCR for intestinal parasites. Parasitology. 2017;144(7):965–74. 10.1017/S0031182017000129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Gratz J, Amour C, Nshama R, Walongo T, Maro A, et al. Optimization of quantitative PCR methods for enteropathogen detection. PloS One. 2016;11(6):e0158199 10.1371/journal.pone.0158199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiria AE, Prasetyani MA, Hamid F, Wammes LJ, Lell B, Ariawan I, et al. Does treatment of intestinal helminth infections influence malaria? Background and methodology of a longitudinal study of clinical, parasitological and immunological parameters in Nangapanda, Flores, Indonesia (ImmunoSPIN Study). BMC Infect Dis. 2010;10(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Gratz J, Amour C, Kibiki G, Becker S, Janaki L, et al. A laboratory-developed TaqMan Array Card for simultaneous detection of 19 enteropathogens. J Clin Microbiol. 2013;51(2):472–80. 10.1128/JCM.02658-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verweij JJ, Brienen EA, Ziem J, Yelifari L, Polderman AM, Van Lieshout L. Simultaneous detection and quantification of Ancylostoma duodenale, Necator americanus, and Oesophagostomum bifurcum in fecal samples using multiplex real-time PCR. Am J Trop Med Hyg. 2007;77(4):685–90. [PubMed] [Google Scholar]
- 38.Alonzo TA, Pepe MS. Using a combination of reference tests to assess the accuracy of a new diagnostic test. Stat in Med. 1999;18(22):2987–3003. [DOI] [PubMed] [Google Scholar]
- 39.https://www.qiagen.com/be/shop/sample-technologies/dna/genomic-dna/dneasy-blood-and-tissue-kit/. Accessed July 10th 2019.
- 40.https://www.qiagen.com/be/shop/sample-technologies/dna/genomic-dna/qiaamp-fast-dna-stool-mini-kit/. Accessed July 10th 2019.
- 41.Andersen LOB, Röser D, Nejsum P, Nielsen HV, Stensvold CR. Is supplementary bead beating for DNA extraction from nematode eggs by use of the NucliSENS easyMag protocol necessary? J Clin Microbiol. 2013;51(4):1345–7. 10.1128/JCM.03353-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harmon AF, Zarlenga DS, Hildreth MB. Improved methods for isolating DNA from Ostertagia ostertagi eggs in cattle feces. Vet Parasitol. 2006;135(3–4):297–302. 10.1016/j.vetpar.2005.10.014 [DOI] [PubMed] [Google Scholar]
- 43.Irie T, Ito T, Kouguchi H, Yamano K, Uraguchi K, Yagi K, et al. Diagnosis of canine Echinococcus multilocularis infections by copro-DNA tests: comparison of DNA extraction techniques and evaluation of diagnostic deworming. Parasitol Res. 2017;116(8):2139–44. 10.1007/s00436-017-5514-y [DOI] [PubMed] [Google Scholar]
- 44.Maksimov P, Schares G, Press S, Fröhlich A, Basso W, Herzig M, et al. Comparison of different commercial DNA extraction kits and PCR protocols for the detection of Echinococcus multilocularis eggs in faecal samples from foxes. Vet Parasitol. 2017;237:83–93. 10.1016/j.vetpar.2017.02.015 [DOI] [PubMed] [Google Scholar]
- 45.Mikaeili F, Kia E, Sharbatkhori M, Sharifdini M, Jalalizand N, Heidari Z, et al. Comparison of six simple methods for extracting ribosomal and mitochondrial DNA from Toxocara and Toxascaris nematodes. Exp Parasitol. 2013;134(2):155–9. 10.1016/j.exppara.2013.02.008 [DOI] [PubMed] [Google Scholar]
- 46.Babaei Z, Oormazdi H, Rezaie S, Rezaeian M, Razmjou E. Giardia intestinalis: DNA extraction approaches to improve PCR results. Experimental Parasitol. 2011;128(2):159–62. [DOI] [PubMed] [Google Scholar]
- 47.Wilke H, Robertson LJ. Preservation of Giardia cysts in stool samples for subsequent PCR analysis. Journal Microbiolog Meth. 2009;78(3):292–6. [DOI] [PubMed] [Google Scholar]
- 48.Kuk S, Cetinkaya U. Stool sample storage conditions for the preservation of Giardia intestinalis DNA. Mem Inst Oswaldo Cruz. 2012;107(8):965–8. 10.1590/s0074-02762012000800001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
The raw data of the DNA and preservation experiment can be found in the worksheets ‘DNA extraction’ and ‘Preservation’, respectively. The worksheet ‘Legend’ explains the headers (variables) in across both data sets.
(XLSX)
The fecal egg counts are expressed as eggs per gram of stool (EPG), whereas the DNA concentration is expressed as genome equivalents per ml (GE/ml). ‘R’ represents the Pearson’s correlation coefficient.
(PDF)
The fecal egg counts are expressed as eggs per gram of stool (EPG), whereas the DNA concentration is expressed as genome equivalents per ml (GE/ml). ‘R’ represents the Pearson’s correlation coefficient.
(PDF)
The intensity of infection was classified as low (Ascaris lumbricoides: fecal egg count (FEC) <5,000 eggs per gram of stool (EPG); Trichuris trichiura: FEC <1,000 EPG; Necator americanus: FEC <2,000 EPG) and as moderate-to-heavy (Ascaris lumbricoides: FEC ≥5,000 EPG; Trichuris trichiura: FEC ≥1,000 EPG; Necator americanus: FEC ≥2,000 EPG). The zero FECs, represent subjects for which no eggs were found applying Kato-Katz thick smear, but for which at least on preservation protocol resulted in a positive qPCR. The sample size equals to the number (N) of subjects multiplied by the number of time points at which DNA is extracted (3 time points).
(DOCX)
The scatterplot illustrate the agreement in fecal egg counts (FECs; expressed as eggs per gram of stool (EPG) and the DNA concentration (expressed as genome equivalents per ml (GE/ml)) across ethanol, potassium dichromate and RNAlater for Ascaris lumbricoides (top graphs), Trichuris trichiura (middle graphs) and Necator americanus (bottom graphs).’R’ represents the Pearson’s correlation coefficient.
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.