ABSTRACT
Objectives: This review summarizes the spectroscopic results, which will provide useful suggestions for future research. In addition, the fields that urgently need more information are also advised.
Background: Nitrite-NO-cGMP has been considered as an important signaling pathway of NO in human cells. To date, all the four known human molybdenum-containing enzymes, xanthine oxidase, aldehyde oxidase, sulfite oxidase, and mitochondrial amidoxime-reducing component, have been shown to function as nitrite reductases under hypoxia by biochemical, cellular, or animal studies. Various spectroscopic techniques have been applied to investigate the structure and catalytic mechanism of these enzymes for more than 20 years.
Methods: We summarize the published data on the applications of UV-vis and EPR spectroscopies, and X-ray crystallography in studying nitrite reductase activity of the four human molybdenum-containing enzymes.
Results: UV-vis has provided useful information on the redox active centers of these enzymes. The utilization of EPR spectroscopy has been critical in determining the coordination and redox status of the Mo center during catalysis. Despite the lack of substrate-bound crystal structures of these nitrite reductases, valuable structural information has been obtained by X-ray crystallography.
Conclusions: To fully understand the catalytic mechanisms of these physiologically/pathologically important nitrite reductases, structural studies on substrate-redox center interaction are needed.
KEYWORDS: Nitrite reductase, Molybdenum-containing enzymes, Nitric oxide, UV–vis, EPR, X-ray
Introduction
Molybdenum plays a vital role in the catalysis of important redox reactions of nitrogen, sulfur, and carbon in mammals.1,2 Apart from participating in several cellular processes, including metabolism of purine, sulfite, aldehyde, and certain types of drugs, molybdenum-containing enzymes gained significant attention due to their capability to function as nitrite reductases in recent years.3–6 Nitrite reductase catalyzes one-electron reduction of nitrite to generate nitric oxide (NO) (equation (1)), which is a critical gas signaling molecule and is involved in many physiological/pathological processes including vasodilation, platelet aggregation, neurotransmission, immune response, apoptosis, and gene expression.7–12
| (1) |
There are two known pathways of NO formation in mammals: oxidation of arginine under normoxia or aerobically, and reduction of nitrite under hypoxia or anaerobically (Fig. 1). These two pathways are similar to the ‘ying’ and ‘yang’ philosophy in Asian culture, which complement and interlink with each other.13 In addition to this normoxic pathway of arginine oxidation catalyzed by nitric oxide synthase (NOS), numerous reports have showed that inorganic nitrite (NO2−) can serve as a NO reservoir, stimulating interest in nitrite reductases.3–6,14–19 Currently, identified mammalian molybdenum-containing nitrite reductases include xanthine oxidase (XO), aldehyde oxidase (AO), sulfite oxidase (SO), and mitochondrial amidoxime-reducing component (mARC), which are also the only known molybdenum proteins discovered in human.3–6 Deficiency of Mo proteins would cause severe genetic diseases in babies with characteristic dysmorphic features and mental retardation. Without efficient treatment, children die in 1 or 2 years.3,4
Figure 1.
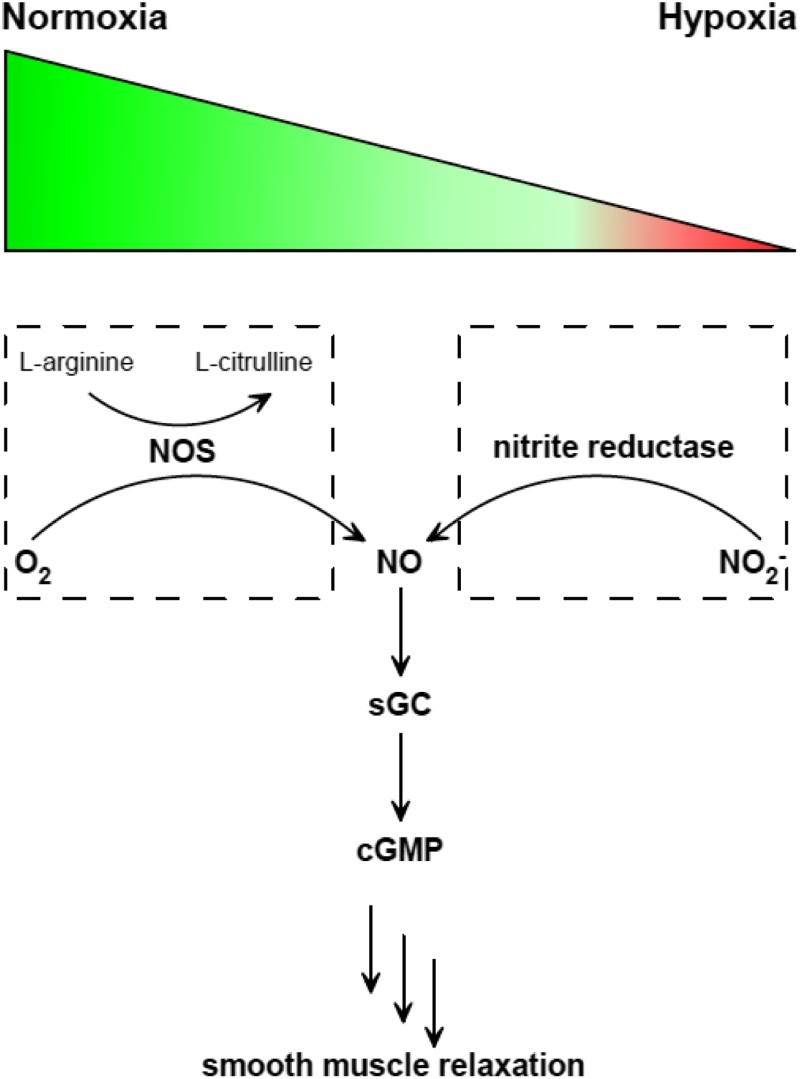
Two pathways of NO generation under normoxia or hypoxia and the sequential signaling pathway afterward. NOS catalyzes the oxidation of l-arginine to l-citrulline in the presence of O2 (left). Nitrite reductase catalyzes NO2− reduction to generate NO. Then NO activates sGC to produce cGMP, which triggers smooth muscle relaxation.
Both XO and AO belong to the xanthine oxidase family.20,21 They are homodimeric molybdenum flavoproteins with each monomer containing two iron–sulfur [2Fe–2S] centers, one molybdenum center, and one FAD cofactor.21 Both enzymes have a molecular weight of approximately 290 kDa.20,21 XO catalyzes the oxidative hydroxylation of hypoxanthine or xanthine to uric acid, making it a crucial enzyme in the catabolism of purines. AO has a similar structure to XO. AO is located in the cytosolic compartment of tissues and facilitates drug metabolism. It catalyzes oxidation of aldehyde into carboxylic acid as well as hydroxylation of many non-aldehydic heterocyclic compounds.22,23 In mammals, SO is a homodimeric 110 kDa enzyme consisting of a C-terminal molybdenum domain and a N-terminal heme domain.1,24,25 SO belongs to the SO family, which also includes assimilatory nitrate reductase. It is located in the mitochondrial intermembrane space where it catalyzes the detoxifying oxidation of sulfite to sulfate. This is the final step in the oxidative degradation of sulfur-containing amino acids, cysteine and methionine.1,24 mARC is the latest discovered molybdenum enzyme in mammals, which does not have other cofactors except the Mo domain.3,26,27 mARC coordinates with two other proteins, cytochrome b5 and cytochrome b5 reductase, to catalyze electron transfer from NADH to amidoxime group. mARC has been known to catalyze the reduction of N-hydroxylated prodrug, but its physiological role is still unclear.3
Although various functions of these four molybdenum enzymes have been thoroughly reviewed,2,21,28–30 there has not been any report summarizing the key spectroscopic features of these proteins, especially from the perspective of their nitrite reductase activity. In this review, we will systematically summarize the spectroscopic data and the proposed redox mechanisms of XO, AO, SO, and mARC as a nitrite reductase, by focusing on ultraviolet–visible (UV–vis) spectroscopies, electron paramagnetic resonance (EPR) spectroscopy, and X-ray crystallography.
UV–vis spectroscopy
Molybdenum-containing nitrite reductases have long been investigated by using UV–vis spectroscopy.31,32 However, due to the low absorption coefficients of Mo center, which is responsible for the reduction of the nitrite to NO in these four Mo enzymes, UV–vis spectroscopy has limited applications in direct study of the Mo center. Furthermore, the characterization of the Mo centers in XO and AO is challenging due to the interference of the much stronger absorbance originated from the [2Fe–2S] and FAD centers.33 Instead, UV–vis spectroscopy has been mainly applied to monitor the changes of other active centers in XO and AO.
Catalytic reduction of nitrite by XO and AO needs coordination of different centers inside these enzymes. The reductive half reaction occurs at the Mo center accompanying nitrite reduction, while the oxidative half reaction takes place at the FAD center upon oxidation of a substrate such as NADH.34,35 The [2Fe–2S] centers serve to mediate the intramolecular electron transfer between the Mo and FAD centers.36
Redox events on the FAD center were investigated in detail using UV–vis spectroscopy.32 Because FAD (λmax = 450 nm) has distinctive UV–vis absorbance from FADH2-bound (λmax = 550 nm) enzyme, the redox reaction can be monitored and quantitated for XO.32
The Mo center of XO is generally analyzed by employing the double-difference spectrum, which allows detecting the change around 425 nm between the oxidized and reduced forms (MoVI–MoIV) of the enzyme, although the raw spectra are dominated by the [2Fe–2S] (λmax = 420 and 470 nm) and FAD (λmax = 360 and 450 nm) centers.33,37 At 420 nm, XO exhibits approximately 3000 M−1 cm−1 increase in extinction coefficient following oxidation (pH 8.5).33 Furthermore, the spectral changes observed during redox events are affected by pH changes presumably due to the presence of ionizable groups close to the Mo center.33 These pH effects include a slight red shift in the absorption maximum and a decrease in the overall intensity upon varying pH from 10 to 6.33 Similarly, the sulfo (Mo = S) and the desulfo (Mo = O) forms of XO can be distinguished by its UV–vis spectrum.37
UV–vis stopped-flow kinetic experiments are also widely utilized to investigate the catalytic mechanism of XO with various substrates.38–40 These studies include the detection of charge transfer complexes involving Mo6+ or Mo4+ in the presence of lumazine (2,4-dihydroxypteridine), Mo6+·lumazine (Eox·S), and Mo4+·violapterin (Ered·P), as well as spectral intermediates in the presence of xanthine.38–40 In the former case, both complexes show an intense band between 550 and 700 nm with an absorption maximum of 650 nm.39 Similarly, these distinct bands are observed upon analysis of the enzyme by circular dichroism spectroscopy.39 The identity of Mo4+·violapterin complex has been also confirmed by resonance Raman studies.41 Furthermore, two long wavelength-absorbing intermediates (λmax = 470 and 540 nm), formed during the reaction of Mo center with 2-hydroxy-6-methylpurine, were detected.31 Other classes of substrates, which produce long wavelength-absorbing intermediates during catalysis, include pyrazolopyrimidines and pteridines.38,42 Early stopped-flow studies involving XO inhibitor, allopurinol, and its product alloxanthine, reveal that the enzyme is trapped at Mo4+·alloxanthine complex.43 The majority of these mechanistic investigations are conducted alongside EPR studies, which is discussed in the next section.
For SO and mARC, UV–vis spectroscopy is also a powerful technique for qualification and quantitation. SO is a homodimer with each monomer containing one cytochrome b5-type heme and one molybdenum domain. A flexible polypeptide chain connects the b5-type heme and the molybdenum domains. The oxidation state of the heme center can be monitored due to the absorbance difference between Fe(II) (λmax = 413 nm) and Fe(III) (λmax = 424 nm).44 Since the heme center has much stronger absorbance than the molybdenum center, usually UV–vis is not eligible to directly detect the Mo center of mammalian SO. However, since plant SO lacks a heme center, it has λmax at 360 nm and a shoulder at ∼480 nm attributing to cysteine-to-molybdenum and enedithiolate-to-molybdenum charge transfers, respectively. These two absorption bands are very similar to the spectra reported for the Mo domain of tryptically cleaved rat SO and of the recombinant Mo domain of human SO.24 When plant SO is reduced, these two bands disappear. When nitrite gets reduced by SO, UV–vis cannot detect changes in the Mo and heme centers, for intraelectron transfer takes place between these two metal centers. mARC has only one active metal domain, the molybdenum domain, which is part of the catalytic reductive chain that requires two other proteins, cytochrome b5 and cytochrome b5 reductase. This chain catalyzes electron transfer from NADH to the terminal oxidase, such as nitrite or molecular oxygen. The physiological function is mARC is still unclear, though it is capable to catalyze reduction of nitrite to NO at its Mo center. Due to the weak absorbance of the Mo center in mARC, UV–vis spectroscopy is usually used to monitor changes at cytochrome b5 (λmax = 414 nm) or cytochrome b5 reductase (λmax = 462 nm). Though the absorbance of the Mo center of these four enzymes are ‘suppressed’ by other metal centers or proteins, UV–vis spectroscopy has been widely employed for activity assays, including nitrite reduction to generate NO.
EPR spectroscopy
EPR spectroscopy is extensively utilized to characterize intermediates of molybdenum-containing nitrite reductases, in order to understand their catalytic mechanisms.21 Mo(V) is EPR-visible, but both Mo(VI) and Mo(IV) are EPR-silent. Low-temperature EPR studies under various conditions demonstrated that XO and AO have very similar EPR spectra with characteristic signals for the two distinct [2Fe–2S] centers, Mo(V), and FADH·.45–49 The corresponding g values are as summarized in Table 1. For both XO and AO, one of the [2Fe–2S] centers (Fe/S I) has near axial symmetry with readily detectable EPR spectrum at 70–100 K.47,50 The second [2Fe–2S] center (Fe/S II) possesses rhombic symmetry and becomes observable only at Τ ≤ 60 K.47 To gain mechanistic information involving Mo(V) center, a common approach is to utilize Mo-specific substrates or inhibitors, such as purine, allopurinol, and ethylene glycol. In the presence of excess substrate, three distinct EPR signals were detected for the Mo(V) center throughout the reaction: ‘slow’, ‘rapid 1’, and ‘rapid 2’, which is based on the time scales they were observed.32,40,51–53 The role of the sulfur atom in the Mo(V) center was investigated in detail by employing various methods such as its removal by cyanide or enrichment with 33S.54,55 Replacement of the essential Mo = S with Mo = O (desulfo-XO or desulfo-AO) results in a ‘slow’ signal.47,56 The enzyme with this Mo = O form is catalytically nonfunctional.32,40,51–54 Depending on the experimental conditions such as substrate identity or concentration the features of the ‘rapid’ signal vary.57,58 Two inequivalent protons are coupled to the ‘rapid type 1’ signals in contrast to the ‘rapid type 2’ signals, which are coupled to two equivalent protons.59–61
Table 1. UV–vis of active centers in XO, AO, SO, mARC, cyt b5, and cyt b5R.
| Mo center | Heme center | [2Fe–2S] | FAD/FADH | Others | |||
|---|---|---|---|---|---|---|---|
| Oxidized | Reduced | Oxidized | Reduced | ||||
| XO | — | — | — | — | 450 (br) | 550 (sh) | — |
| AO | — | — | — | — | 450 (br) | 550 (sh) | — |
| mARC | 450 (sh) 370–400 (br, sh) | 350 (sh) | — | — | — | — | — |
| SO | 360, 480 (sh) | 400 (sh) | 413, 520, 560 | 424, 527, 557 | — | — | — |
| cyt b5 | — | — | 413, 520, 560 | 423, 527, 557 | — | — | — |
| cyt b5R (oxidized) | — | — | — | — | — | — | 391, 462, 480 (sh) |
| cyt b5R (reduced) | — | — | — | — | — | — | 320 (br) 520 (br) |
cyt b5, Cytochrome b5; cyt b5R, cytochrome b5 reductase; br, broad; sh, shoulder.
The mechanistic aspects of XO and AO nitrite reductase activities were also investigated.45 In the presence of nitrite, the characteristic signals of the enzyme (Mo(V)) were decreased due to oxidation.45 This process was accompanied by the emergence of one axial signal (gT = 2.04 and g// = 2.015), which was attributed to a well-known dinitrosyl–Fe/S complex (Fe/S–NO).45,62 Moreover, it was demonstrated that the generation of Fe/S–NO signal requires the presence of an active Mo center for both enzymes.45 EPR studies conducted with the desulfo-XO or desulfo-AO showed that the sulfido group is needed for the formation of Fe/S–NO only in XO, but not in AO.45
EPR spectroscopy has been utilized to study SO since 1980s. Three distinctive EPR forms of mammalian SO are known, including low-pH, high-pH, and phosphate-inhibited forms.63 Experiments were performed for both liquid samples at ambient temperature (295 K) or frozen sample at low temperature (120 K). EPR spectra at 295 K were consistent with spectra recorded at 120 K for both high-pH and low-pH species. This means that the state of ionization of EPR-sensitive species in the mM range stayed the same. Chloride (Cl−) was believed to be an integral part of the low-pH signal-given species. Chloride concentration influenced the interconversation between low-pH and high-pH Mo(V) species. Depending on the concentration of chloride and phosphate (H2PO4−), one, two, or all three forms of EPR species may be present. Under certain conditions, the phosphate form may replace part or all of the low-pH form. Equilibrium among these three forms was as shown in Fig. 2, depending on pH, concentrations of chloride and phosphate anions.63 A cyclic structure was suggested for both the phosphate species, as well as the bisulfite species, which was similar to what was found in xanthine oxidase.
Figure 2.
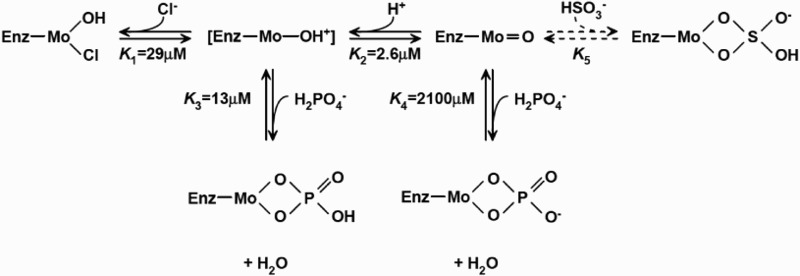
Proposed low pH, high pH, bisulfite, and phosphate-binding to the Mo domain of SO. Values of the equilibrium were the ones which gave the best fit to the experimental data.
At high pH, the EPR spectrum of chicken liver SO (g1,2,3 = 1.987, 1.9641, 1.953) is essential identical to that of plant SO (g1,2,3 = 1.991, 1.965, 1.958).24,64 At low pH, the EPR spectrum of chicken liver SO (g1,2,3 = 2.004, 1.972, 1.966) is shifted (Table 2).
Table 2. EPR comparison among XO, AO, SO, mARC, and other Mo enzymes.
| Enzyme | Signal | g1,2,3 | gav | ||
|---|---|---|---|---|---|
| hmARC-1 | — | 1.9983 | 1.9691 | 1.9585 | 1.9753 |
| C246S hmARC-1 | — | 1.9990 | 1.9693 | 1.9587 | 1.9757 |
| hmARC-2 | — | 1.9994 | 1.9658 | 1.9616 | 1.9756 |
| Chicken SO | Low pH | 2.0037 | 1.9720 | 1.9658 | 1.9805 |
| C207S human SO | — | 1.9789 | 1.9654 | 1.9545 | 1.9663 |
| Spinach nitrate reductase | — | 1.9957 | 1.9692 | 1.9652 | 1.9767 |
| SO | Low pH | 2.0037 | 1.9720 | 1.9658 | 1.9805 |
| High pH | 1.9872 | 1.9641 | 1.9531 | 1.9681 | |
| PO43– | 1.9917 | 1.9692 | 1.9614 | 1.9741 | |
| Nitrate reductase | Low pH | 1.9989 | 1.9855 | 1.9628 | 1.9824 |
| High pH | 1.9870 | 1.9805 | 1.9612 | 1.9762 | |
| Xanthine oxidase | Rapid type 1 | 1.9893 | 1.9703 | 1.9657 | 1.9751 |
| Rapid type 2 | 1.9951 | 1.9712 | 1.9616 | 1.9760 | |
| Slow | 1.9719 | 1.9671 | 1.9551 | 1.9647 | |
| Aldehyde oxidase | Slow (Bicine/Na2S2O4) | 1.9720 | 1.9670 | 1.9558 | 1.9649 |
| Aldehyde oxidase | Rapid type 2 (purine) | 1.9876 | 1.9687 | 1.9607 | 1.9723 |
Studies with isolated molybdenum domain of SO has shown that excess sulfite reduces Mo(VI) to Mo(IV), which is oxidized by excess nitrite only to Mo(V). However, Mo(V) cannot be reduced by sulfite, a strict two-electron reductant, but vulnerable to be reduced by phenosafranine (a one- or two-electron reductant) forming EPR-silent Mo(IV).4 In the holo SO enzyme, Mo(V) is always detected upon reduction by sulfite, due to an intraelectron transfer between heme and Mo domains. Reduction of the holo SO enzyme by phenosafranine leads to EPR-silent Mo(IV), due to an exhaustive reduction of both Mo domain and heme domain by phenosafranine.64,65
EPR spectroscopy studies on human mARC have shown similar signals as shown in low-pH form of SO (Table 2). Mo(V) state was generated with partial reduction with NADH/cyt b5R/cyt b5. However, replacement of the cysteine residue by serine at the active site of mARC does not significantly change the EPR signal (Table 2). Recent studies using EPR and computation together had provided more information about the coordination at the active sites of mARC. When nitrite meets reduced Mo(IV), the metal gets oxidized to Mo(V), which has very similar EPR spectra to that of SO at low pH. It has proposed that an intermediate Mo(IV)–O–N–O was formed first, enabling the one-electron transfer between Mo and nitrite.66
Besides continuous wave EPR, pulsed EPR spectroscopy has also been used to understand the molybdenum center of SO and mARC, which provides information about electron transfer and ligand exchange. 2H, 17O, 33S labeling have been used together with pulse EPR investigation, which confirms seven-coordination or six-coordination of the Mo center.67
X-ray crystallography
Crystal structure can provide insights into the substrate/product binding site of Mo-containing nitrite reductases. To date, there is no resolved structure for nitrite- or NO-binding Mo enzyme, but quite a few structures are available for XO, AO, and SO to help understand the catalytic reduction of nitrite.
The structure of XO has been investigated with X-ray crystallography in the presence or absence of various substrates and inhibitors.68–73 These studies indicate that each XO monomer consists five domains: a N-terminal domain composed of mainly β-sheets and a [2Fe–2S] cluster (Fe/S II) followed by a mainly α-helical domain containing the other [2Fe–2S] center (Fe/S I).68,69 The third domain contains FAD, and the two C-terminal domains that bind the Mo center at the interface.68,69 The only contacts between the two monomers are at the Mo-binding part of the enzyme.68,69 Investigation of its active site has led to the determination of several key residues (e.g. Glu802, Gln767, Arg880/881,Glu1261), intermediates, and mechanistic aspects.68–70,72–74 Moreover, recent findings point to the importance of C-terminal on the structure and activity of XO.71 The deletion of the C-terminal amino acids lead to generation of an intermediate form, which exhibits both oxidase and dehydrogenase activities.71
Recently, the crystal structure of a mammalian AO was obtained.75 Comparison of the structure with XO has shown that although the two structures are similar, there are significant differences around FAD.75 Furthermore, several conserved (Gln772, Phe919, Phe1014, Glu1266) and non-conserved (Ala807, Tyr885, Lys889, Pro1015, Tyr1019, Arg717, Asp878, Glu880, Leu881, Thr1081) key residues were found in the active site as well as in the access channel, shining light onto the differences between the substrate specificities of AO and XO.75 In accordance with these, studies show that E803 V and R881M mutants of XO have significantly different preference toward purine substrates, and display aldehyde oxidase activity.73
So far there is no crystal structure for human and rat SO, but wild-type chicken SO and its variants have provided valuable information.76–78 SO purified from chicken liver contains one C-terminal dimerization domain, one central Mo domain, and one N-terminal cytochrome b5 domain for each subunit. The distance between the Mo and heme center is 32 Å.76 Some patients who suffered from SO deficiency were diagnosed with Arg160 mutation.79 This mutation results in increased Km and decreased kcat; thus, about 1000-fold decrease in its second-order rate constant (kcat/Km). For chicken SO R138Q (R160Q in human) mutant, the substrate-binding pocket is significantly altered.80 Arg138 forms a hydrogen bond with the water/hydroxo ligand of molybdenum. In addition, the active site residue Arg450 adopts different conformation when sulfate is present or absent.
Plant SO from Arabidopsis thaliana has similar folding and coordination of Mo domain as mammalian SO, but it does not have a heme center. The Mo and dimerization domains show 48 and 43% identity between plant SO and chicken.81 Arg374 is an important residue for substrate binding. Up to today, there is no reported crystal structure of mARC.
Catalytic mechanisms
Based on the data collected with UV–vis, EPR, X-ray crystallography spectroscopies, the mechanism of nitrite reduction by Mo-containing enzymes has been proposed as Fig. 3. UV–vis has provided useful information about the redox active centers in these four nitrite reductases. EPR spectroscopy helps us understand coordination and redox states of the Mo center when nitrite or reducing substrate interacts with Mo. Though there is no available nitrite-binding or NO-binding crystal structure of XO, AO, SO, and mARC, reported X-ray crystallography structures can still enlighten our understanding of the redox process. However, there is plenty of spectroscopic information still missing, including how nitrite or NO binds to the Mo center, how other ligands influence nitrite binding, what are the intermediates, etc. We are looking forward to more spectroscopic data to clarify this physiologically/pathologically important nitrite reduction to NO process.82,83
Figure 3.
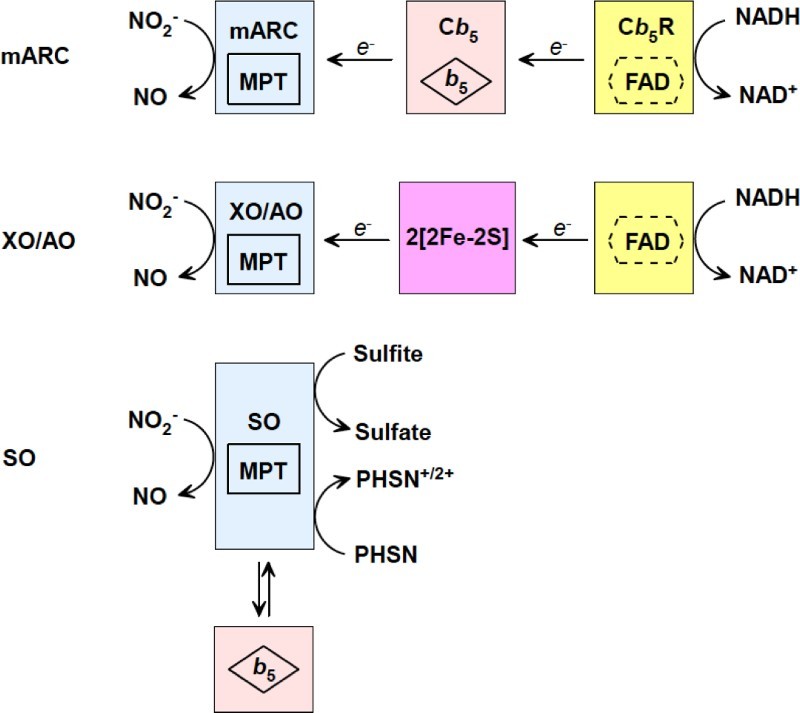
Proposed mechanisms of nitrite reduction catalyzed by XO, AO, SO, and mARC.
Funding Statement
Biotechnology and Biological Sciences Research Council.
Disclaimer statements
Contributors Jun Wang and Gizem Keceli contribute to this paper equally.
Conflict of interest There are no conflicts of interest.
Ethics approval Yes.
ORCiD
Gizem Keceli http://orcid.org/0000-0002-9562-7994
Rui Cao http://orcid.org/0000-0002-0316-0152
Jiangtao Su http://orcid.org/0000-0003-0708-1168
References
- [1].Kisker C, Schindelin H, Rees DC. Molybdenum-cofactor-containing enzymes: structure and mechanism. Annu Rev Biochem 1997;66:233–67. doi: 10.1146/annurev.biochem.66.1.233 [DOI] [PubMed] [Google Scholar]
- [2].Ott G, Havemeyer A, Clement B. The mammalian molybdenum enzymes of mARC. J Biol Inorg Chem 2015;20:265–75. doi: 10.1007/s00775-014-1216-4 [DOI] [PubMed] [Google Scholar]
- [3].Sparacino-Watkins CE, Tejero J, Sun B, Gauthier MC, Thomas J, Ragireddy V, et al. Nitrite reductase and nitric-oxide synthase activity of the mitochondrial molybdopterin enzymes mARC1 and mARC2. J Biol Chem 2014;289(15):10345–58. doi: 10.1074/jbc.M114.555177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wang J, Krizowski S, Fischer-Schrader K, Niks D, Tejero J, Sparacino-Watkins CE, et al. Sulfite oxidase catalyzes single-electron transfer at molybdenum domain to reduce nitrite to nitric oxide. Antioxid Redox Signal 2015;23(4):283–94. doi: 10.1089/ars.2013.5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li H, Samouilov A, Liu X, Zweier JL. Characterization of the magnitude and kinetics of xanthine oxidase-catalyzed nitrite reduction. Evaluation of its role in nitric oxide generation in anoxic tissues. J Biol Chem 2001;276(27):24482–9. doi: 10.1074/jbc.M011648200 [DOI] [PubMed] [Google Scholar]
- [6].Li H, Kundu TK, Zweier JL. Characterization of the magnitude and mechanism of aldehyde oxidase-mediated nitric oxide production from nitrite. J Biol Chem 2009;284(49):33850–8. doi: 10.1074/jbc.M109.019125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zuckerbraun BS, Shiva S, Ifedigbo E, Mathier MA, Mollen KP, Rao J, et al. Nitrite potently inhibits hypoxic and inflammatory pulmonary arterial hypertension and smooth muscle proliferation via xanthine oxidoreductase-dependent nitric oxide generation. Circulation 2010;121:98–109. doi: 10.1161/CIRCULATIONAHA.109.891077 [DOI] [PubMed] [Google Scholar]
- [8].Webb AJ, Patel N, Loukogeorgakis S, Okorie M, Aboud Z, Misra S, et al. Acute blood pressure lowering, vasoprotective, and antiplatelet properties of dietary nitrate via bioconversion to nitrite. Hypertension 2008;51:784–90. doi: 10.1161/HYPERTENSIONAHA.107.103523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wiklund CU, Olgart C, Wiklund NP, Gustafsson LE. Modulation of cholinergic and substance P-like neurotransmission by nitric oxide in the guinea-pig ileum. Br J Pharmacol 1993;110(2):833–9. doi: 10.1111/j.1476-5381.1993.tb13888.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bogdan C. Nitric oxide and the immune response. Nat Immunol 2001;2(10):907–16. doi: 10.1038/ni1001-907 [DOI] [PubMed] [Google Scholar]
- [11].Albina JE, Cui S, Mateo RB, Reichner JS. Nitric oxide-mediated apoptosis in murine peritoneal macrophages. J Immunol 1993;150(11):5080–5. [PubMed] [Google Scholar]
- [12].Durante W, Kroll MH, Christodoulides N, Peyton KJ, Schafer AI. Nitric oxide induces heme oxygenase-1 gene expression and carbon monoxide production in vascular smooth muscle cells. Circ Res 1997;80(4):557–64. doi: 10.1161/01.RES.80.4.557 [DOI] [PubMed] [Google Scholar]
- [13].Lundberg JO, Weitzberg E, Gladwin MT. The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat Rev Drug Discov 2008;7(2):156–67. doi: 10.1038/nrd2466 [DOI] [PubMed] [Google Scholar]
- [14].Cosby K, Partovi KS, Crawford JH, Patel RP, Reiter CD, Martyr S, et al. Nitrite reduction to nitric oxide by deoxyhemoglobin vasodilates the human circulation. Nat Med 2003;9(12):1498–505. doi: 10.1038/nm954 [DOI] [PubMed] [Google Scholar]
- [15].Jayaraman T, Tejero J, Chen BB, Blood AB, Frizzell S, Shapiro C, et al. 14-3-3 binding and phosphorylation of neuroglobin during hypoxia modulate six-to-five heme pocket coordination and rate of nitrite reduction to nitric oxide. J Biol Chem 2011;286(49):42679–89. doi: 10.1074/jbc.M111.271973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shiva S, Huang Z, Grubina R, Sun JH, Ringwood LA, MacArthur PH, et al. Deoxymyoglobin is a nitrite reductase that generates nitric oxide and regulates mitochondrial respiration. Circ Res 2007;100(5):654–61. doi: 10.1161/01.RES.0000260171.52224.6b [DOI] [PubMed] [Google Scholar]
- [17].Sturms R, DiSpirito AA, Hargrove MS. Plant and cyanobacterial hemoglobins reduce nitrite to nitric oxide under anoxic conditions. Biochemistry 2011;50(19):3873–8. doi: 10.1021/bi2004312 [DOI] [PubMed] [Google Scholar]
- [18].Tejero J, Gladwin MT. The globin superfamily: functions in nitric oxide formation and decay. Biol Chem 2014;395:631–9. doi: 10.1515/hsz-2013-0289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tiso M, Tejero J, Basu S, Azarov I, Wang X, Simplaceanu V, et al. Human neuroglobin functions as a redox-regulated nitrite reductase. J Biol Chem 2011;286(20):18277–89. doi: 10.1074/jbc.M110.159541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Huber R, Hof P, Duarte RO, Moura JJ, Moura I, Liu MY, et al. A structure-based catalytic mechanism for the xanthine oxidase family of molybdenum enzymes. Proc Natl Acad Sci USA 1996;93(17):8846–51. doi: 10.1073/pnas.93.17.8846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hille R, Hall J, Basu P. The mononuclear molybdenum enzymes. Chem Rev 2014;114(7):3963–4038. doi: 10.1021/cr400443z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Johns DG. Human liver aldehyde oxidase: differential inhibition of oxidation of charged and uncharged substrates. J Clin Invest 1967;46(9):1492–505. doi: 10.1172/JCI105641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Garattini E, Terao M. The role of aldehyde oxidase in drug metabolism. Expert Opin Drug Metab Toxicol 2012;8(4):487–503. doi: 10.1517/17425255.2012.663352 [DOI] [PubMed] [Google Scholar]
- [24].Eilers T, Schwarz G, Brinkmann H, Witt C, Richter T, Nieder J, et al. Identification and biochemical characterization of Arabidopsis thaliana sulfite oxidase: a new player in plant sulfur metabolism. J Biol Chem 2001;276(50):46989–94. doi: 10.1074/jbc.M108078200 [DOI] [PubMed] [Google Scholar]
- [25].Cohen HJ, Fridovich I. Hepatic sulfite oxidase: purification and properties. J Biol Chem 1971;246(2):359–66. [PubMed] [Google Scholar]
- [26].Klein JM, Busch JD, Potting C, Baker MJ, Langer T, Schwarz G. The mitochondrial amidoxime-reducing component (mARC1) is a novel signal-anchored protein of the outer mitochondrial membrane. J Biol Chem 2012;287(51):42795–803. doi: 10.1074/jbc.M112.419424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Krompholz N, Krischkowski C, Reichmann D, Garbe-Schönberg D, Mendel RR, Bittner F, et al. The mitochondrial amidoxime reducing component (mARC) is involved in detoxification of N-hydroxylated base analogues. Chem Res Toxicol 2012;25(11):2443–50. doi: 10.1021/tx300298m [DOI] [PubMed] [Google Scholar]
- [28].Maia LB, Moura JJG. Nitrite reduction by molybdoenzymes: a new class of nitric oxide-forming nitrite reductases. J Biol Inorg Chem 2015;20(2):403–33. doi: 10.1007/s00775-014-1234-2 [DOI] [PubMed] [Google Scholar]
- [29].Havemeyer A, Lang J, Clement B. The fourth mammalian molybdenum enzyme mARC: current state of research. Drug Metab Rev 2011;43(4):524–39. doi: 10.3109/03602532.2011.608682 [DOI] [PubMed] [Google Scholar]
- [30].Romão MJ. Molybdenum and tungsten enzymes: a crystallographic and mechanistic overview. Dalton Trans 2009:4053–68. doi: 10.1039/b821108f [DOI] [PubMed] [Google Scholar]
- [31].McWhirter RB, Hille R. The reductive half-reaction of xanthine oxidase. J Biol Chem 1991;266(35):23724–31. [PubMed] [Google Scholar]
- [32].Olson JS, Ballou DP, Palmer G, Massey V. The mechanism of action of xanthine oxidase. J Biol Chem 1974;249(14):4363–82. [PubMed] [Google Scholar]
- [33].Ryan MG, Ratnam K, Hille R. The molybdenum centers of xanthine oxidase and xanthine dehydrogenase. J Biol Chem 1995;270(33):19209–12. doi: 10.1074/jbc.270.33.19209 [DOI] [PubMed] [Google Scholar]
- [34].Komai H, Massey V, Palmer G. The preparation and properties of deflavo xanthine oxidase. J Biol Chem 1969;244(7):1692–700. [PubMed] [Google Scholar]
- [35].Bray RC, Palmer G, Beinert H. Direct studies on the electron transfer sequence in xanthine oxidase by electron paramagnetic resonance spectroscopy: II. Kinetic studies employing rapid freezing. J Biol Chem 1964;239(8):2667–76. [PubMed] [Google Scholar]
- [36].Hille R, Anderson RF. Electron transfer in milk xanthine oxidase as studied by pulse radiolysis. J Biol Chem 1991;266(9):5608–15. [PubMed] [Google Scholar]
- [37].Maiti NC, Tomita T, Kitagawa T, Okamoto K, Nishino T. Resonance Raman studies on xanthine oxidase: observation of MoVI-ligand vibrations. J Biol Inorg Chem 2003;8(3):327–33. [DOI] [PubMed] [Google Scholar]
- [38].Davis MD, Olson JS, Palmer G. The reaction of xanthine oxidase with lumazine. Characterization of the reductive half-reaction. J Biol Chem 1984;259(6):3526–33. [PubMed] [Google Scholar]
- [39].Davis MD, Olson JS, Palmer G. Charge transfer complexes between pteridine substrates and the active center molybdenum of xanthine oxidase. J Biol Chem 1982;257(24):14730–7. [PubMed] [Google Scholar]
- [40].Kim JH, Hille R. Reductive half-reaction of xanthine oxidase with xanthine. J Biol Chem 1993;268(1):44–51. [PubMed] [Google Scholar]
- [41].Oertling TA, Hille R. Resonance-enhanced Raman scattering from the molybdenum center of xanthine oxidase. J Biol Chem 1990;265(29):17446–50. [PubMed] [Google Scholar]
- [42].Massey V, Komai H, Palmer G, Elion GB. On the mechanism of inactivation of xanthine oxidase by allopurinol and other pyrazolo[3,4-d]pyrimidines. J Biol Chem 1970;245(11):2837–44. [PubMed] [Google Scholar]
- [43].Massey V, Komai H, Palmer G, Ellion GB. On the mechanism of inactivation of xanthine oxidase by allopurinol and other pyrazolo[3,4-d]pyrimidines. J Biol Chem 1970;245(11):2837–44. [PubMed] [Google Scholar]
- [44].Toghrol F, Southerland WM. Purification of Thiobacillus novellus sulfite oxidase. Evidence for the presence of heme and molybdenum. J Biol Chem 1983;258(11):6762–6. PubMed PMID: WOS:A1983QT65000024. English. [PubMed] [Google Scholar]
- [45].Maia LB, Moura JJG. Nitrite reduction by xanthine oxidase family enzymes: a new class of nitrite reductases. J Biol Inorg Chem 2011;16(3):443–60. doi: 10.1007/s00775-010-0741-z [DOI] [PubMed] [Google Scholar]
- [46].Bray RC, George GN, Gutteridge S, Norlander L, Stell JGP, Stubley C. Studies by electron-paramagnetic-resonance spectroscopy of the molybdenum centre of aldehyde oxidase. Biochem J 1982;203(1):263–7. doi: 10.1042/bj2030263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Barber MJ, Coughlan MP, Rajagopalan KV, Siegel LM. Properties of the prosthetic groups of rabbit liver aldehyde oxidase: a comparison of molybdenum hydroxylase enzymes. Biochemistry 1982;21(15):3561–8. doi: 10.1021/bi00258a006 [DOI] [PubMed] [Google Scholar]
- [48].Rajagopalan KV, Handler P, Palmer G, Beinert H. Studies of aldehyde oxidase by electron paramagnetic resonance spectroscopy. J Biol Chem 1968;243(14):3797–806. [PubMed] [Google Scholar]
- [49].Rajagopalan KV, Handler P, Palmer G, Beinert H. Studies of aldehyde oxidase by electron paramagnetic resonance spectroscopy. J Biol Chem 1968;243(14):3784–96. [PubMed] [Google Scholar]
- [50].Bray RC. In: Boyer PD, (ed.) The enzymes. 12, pt. B 3rd ed New York, NY: Academic Press; 1975. p. 303–88. [Google Scholar]
- [51].McGartoll MA, Pick FM, Swann JC, Bray RC. Properties of xanthine oxidase preparations dependent on the proportions of active and inactivated enzyme. Biochim Biophys Acta 1970;212(3):523–6. doi: 10.1016/0005-2744(70)90264-0 [DOI] [PubMed] [Google Scholar]
- [52].Rosenberry TL, Chang HW, Chen YT. Purification of acetylcholinesterase by affinity chromatography and determination of active site stoichiometry. J Biol Chem 1972;247(5):1555–65. [PubMed] [Google Scholar]
- [53].Bray RC. The inorganic biochemistry of molybdoenzymes. Q Rev Biophys 1988;21(3):299–329. doi: 10.1017/S0033583500004479 [DOI] [PubMed] [Google Scholar]
- [54].Malthouse JPG, George GN, Lowe DJ, Bray RC. Coupling of [33S]sulphur to molybdenum(V) in different reduced forms of xanthine oxidase. Biochem J 1981;199(3):629–37. doi: 10.1042/bj1990629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Coughlan MP, Johnson JL, Rajagopalan KV. Mechanisms of inactivation of molybdoenzymes by cyanide. J Biol Chem 1980;255(7):2694–9. [PubMed] [Google Scholar]
- [56].Gutteridge S, Tanner SJ, Bray RC. Comparison of the molybdenum centres of native and desulpho xanthine oxidase. The nature of the cyanide-labile sulphur atom and the nature of the proton-accepting group. Biochem J 1978;175(3):887–97. doi: 10.1042/bj1750887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Gutteridge S, Tannerdi SJ, Bray RC. The molybdenum centre of native xanthine oxidase. Evidence for proton transfer from substrates to the centre and for existence of an anion-binding site. Biochem J 1978;175(3):869–78. doi: 10.1042/bj1750869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Bray RC, Barber MJ, Lowe DJ. Electron-paramagnetic-resonance spectroscopy of complexes of xanthine oxidase with xanthine and uric acid. Biochem J 1978;171(3):653–8. doi: 10.1042/bj1710653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Edmondson D, Ballou D, Heuvelen AV, Palmer G, Massey V. Kinetic studies on the substrate reduction of xanthine oxidase. J Biol Chem 1973;248(17):6135–44. [PubMed] [Google Scholar]
- [60].Tanner SJ, Bray RC, Bergmann F. 13C hyperfine splitting of some molybdenum electron-paramagnetic-resonance signals from xanthine oxidase. Biochem Soc Trans 1978;6(6):1328–30. doi: 10.1042/bst0061328 [DOI] [PubMed] [Google Scholar]
- [61].Bray RC, George GN. Electron-paramagnetic-resonance studies using pre-steady-state kinetics and substitution with stable isotopes on the mechanism of action of molybdoenzymes. Biochem Soc Trans 1985;13(2):560–7. doi: 10.1042/bst0130560 [DOI] [PubMed] [Google Scholar]
- [62].Ichimori K, Fukahori M, Nakazawa H, Okamoto K, Nishino T. Inhibition of xanthine oxidase and xanthine dehydrogenase by nitric oxide: nitric oxide converts reduced xanthine-oxidizing enzymes into the desulfo-type inactive form. J Biol Chem 1999;274(12):7763–8. doi: 10.1074/jbc.274.12.7763 [DOI] [PubMed] [Google Scholar]
- [63].Bray RC, Gutteridge S, Lamy MT, Wilkinson T. Equilibria amongst different molybdenum (V)-containing species from sulphite oxidase. Evidence for a halide ligand of molybdenum in the low-pH species. Biochem J 1983;211(1):227–36. PubMed PMID: WOS:A1983QK54500024. English. doi: 10.1042/bj2110227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lamy MT, Gutteridge S, Bray RC. Electron-paramagnetic-resonance parameters of molybdenum(V) in sulphite oxidase from chicken liver. Biochem J 1980;185(2):397–403. PubMed PMID: WOS:A1980JE41900012. English. doi: 10.1042/bj1850397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wahl B, Reichmann D, Niks D, Krompholz N, Havemeyer A, Clement B, et al. Biochemical and spectroscopic characterization of the human mitochondrial amidoxime reducing components hmARC-1 and hmARC-2 suggests the existence of a new molybdenum enzyme family in eukaryotes. J Biol Chem 2010;285(48):37847–59. PubMed PMID: WOS:000284424000075. English. doi: 10.1074/jbc.M110.169532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Yang J, Giles LJ, Ruppelt C, Mendel RR, Bittner F, Kirk ML. Oxyl and hydroxyl radical transfer in mitochondrial amidoxime reducing component-catalyzed nitrite reduction. J Am Chem Soc 2015. Apr;137(16):5276–9. PubMed PMID: WOS:000353931500009. English. doi: 10.1021/jacs.5b01112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Rajapakshe A, Astashkin AV, Klein EL, Reichmann D, Mendel RR, Bittner F, et al. Structural studies of the molybdenum center of mitochondrial amidoxime reducing component (mARC) by pulsed EPR spectroscopy and O-17-labeling. Biochemistry 2011;50(41):8813–22. PubMed PMID: WOS:000295661200005. English. doi: 10.1021/bi2005762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Enroth C, Eger BT, Okamoto K, Nishino T, Nishino T, Pai EF. Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion. Proc Natl Acad Sci USA 2000;97(20):10723–8. doi: 10.1073/pnas.97.20.10723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Cao H, Pauff JM, Hille R. X-ray crystal structure of a xanthine oxidase complex with the flavonoid inhibitor quercetin. J Nat Prod 2014;77(7):1693–9. doi: 10.1021/np500320g [DOI] [PubMed] [Google Scholar]
- [70].Cao H, Hall J, Hille R. X-ray crystal structure of arsenite-inhibited xanthine oxidase: μ-sulfido, μ-oxo double bridge between molybdenum and arsenic in the active site. J Am Chem Soc 2011;133(32):12414–7. doi: 10.1021/ja2050265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Nishino T, Okamoto K, Kawaguchi Y, Matsumura T, Eger BT, Pai EF, et al. The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase. FEBS J 2015;282(16):3075–90. doi: 10.1111/febs.13277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Pauff JM, Cao H, Hille R. Substrate orientation and catalysis at the molybdenum site in xanthine oxidase: crystal structures in complex with xanthine and lumazine. J Biol Chem 2009;284(13):8760–7. doi: 10.1074/jbc.M804517200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Yamaguchi Y, Matsumura T, Ichida K, Okamoto K, Nishino T. Human xanthine oxidase changes its substrate specificity to aldehyde oxidase type upon mutation of amino acid residues in the active site: roles of active site residues in binding and activation of purine substrate. J Biochem 2007;141(4):513–24. doi: 10.1093/jb/mvm053 [DOI] [PubMed] [Google Scholar]
- [74].Cao H, Pauff JM, Hille R. Substrate orientation and catalytic specificity in the action of xanthine oxidase: the sequential hydroxylation of hypoxanthine to uric acid. J Biol Chem 2010;285(36):28044–53. doi: 10.1074/jbc.M110.128561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Coelho C, Mahro M, Trincao J, Carvalho ATP, Ramos MJ, Terao M, et al. The first mammalian aldehyde oxidase crystal structure: insights into substrate specificity. J Biol Chem 2012;287(48):40690–702. doi: 10.1074/jbc.M112.390419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kisker C, Schindelin H, Pacheco A, Wehbi WA, Garrett RM, Rajagopalan KV, et al. Molecular basis of sulfite oxidase deficiency from the structure of sulfite oxidase. Cell 1997;91(7):973–83. PubMed PMID: WOS:000071281400014. English. doi: 10.1016/S0092-8674(00)80488-2 [DOI] [PubMed] [Google Scholar]
- [77].George GN, Pickering IJ, Kisker C. X-ray absorption spectroscopy of chicken sulfite oxidase crystals. Inorg Chem 1999;38(10):2539–40. PubMed PMID: WOS:000080459200045. English. doi: 10.1021/ic981451b [DOI] [Google Scholar]
- [78].Qiu JA, Wilson HL, Rajagopalan KV. Structure-based alteration of substrate specificity and catalytic activity of sulfite oxidase from sulfite oxidation to nitrate reduction. Biochemistry 2012;51(6):1134–47. PubMed PMID: WOS:000300132900009. English. doi: 10.1021/bi201206v [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Garrett RM, Johnson JL, Graf TN, Feigenbaum A, Rajagopalan KV. Human sulfite oxidase R160Q: identification of the mutation in a sulfite oxidase-deficient patient and expression and characterization of the mutant enzyme. Proc Natl Acad Sci USA 1998;95(11):6394–8. PubMed PMID: WOS:000073852600097. English. doi: 10.1073/pnas.95.11.6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Karakas E, Wilson HL, Graf TN, Xiang S, Jaramillo-Buswuets S, Rajagopalan KV, et al. Structural insights into sulfite oxidase deficiency. J Biol Chem 2005;280(39):33506–15. PubMed PMID: WOS:000232058100050. English. doi: 10.1074/jbc.M505035200 [DOI] [PubMed] [Google Scholar]
- [81].Schrader N, Fischer K, Theis K, Mendel RR, Schwarz G, Kisker C. The crystal structure of plant sulfite oxidase provides insights into sulfite oxidation in plants and animals. Structure 2003;11(10):1251–63. PubMed PMID: WOS:000185758500012. English. doi: 10.1016/j.str.2003.09.001 [DOI] [PubMed] [Google Scholar]
- [82].Kosaka T, Takazawa T, Kubota K, Watanabe N, Nakamura T. Identification of rat liver aldehyde oxidase across species by accurate peptide-mass fingerprinting and sequence-tagging with Fourier transform ion cyclotron resonance mass spectrometry. J Mass Spectrom Soc Jpn 2000;48(3):179–86. doi: 10.5702/massspec.48.179 [DOI] [Google Scholar]
- [83].Barr JT, Jones JP, Joswig-Jones CA, Rock DA. Absolute quantification of aldehyde oxidase protein in human liver using liquid chromatography-tandem mass spectrometry. Mol Pharm 2013;10(10):3842–9. doi: 10.1021/mp4003046 [DOI] [PMC free article] [PubMed] [Google Scholar]


