Abstract
Background
Autism Spectrum Disorder (ASD) is behaviorally and biologically heterogeneous and likely represents a series of conditions arising from different underlying genetic, metabolic, and environmental factors. There are currently no reliable diagnostic biomarkers for ASD. Based on evidence that dysregulation of branch chain amino acids (BCAA) may contribute to the behavioral characteristics of ASD, we tested whether dysregulation of amino acids (AA) was a pervasive phenomenon in individuals with ASD. This is the first paper to report results from the Children’s Autism Metabolome Project (CAMP, ClinicalTrials.gov Identifier: NCT02548442), a large-scale effort to define autism biomarkers based on metabolomic analyses of blood samples from young children.
Methods
Dysregulation of AA metabolism was identified by comparing plasma metabolites from 516 children with ASD with those from 164 age-matched typically-developing (TYP) children recruited into CAMP. ASD subjects were stratified into subpopulations based on shared metabolic phenotypes associated with BCAA dysregulation.
Results
We identified groups of AAs with positive correlations that were, as a group, negatively correlated with BCAA levels in ASD. Imbalances between these two groups of AAs identified three ASD associated Amino Acid Dysregulation Metabotypes (AADM). The combination of glutamine, glycine, and ornithine AADMs identified a dysregulation in AA/BCAA metabolism that is present in 16.7% of the CAMP ASD subjects and is detectable with a specificity of 96.3% and a PPV of 93.5%.
Conclusions
Identification and utilization of metabotypes of ASD can lead to actionable metabolic tests that support early diagnosis and stratification for targeted therapeutic interventions.
Keywords: autism, biomarker, amino acids, metabotype, metabolomics, diagnosis
INTRODUCTION
Autism Spectrum Disorder (ASD) is characterized by core symptoms of altered social communication and repetitive behaviors or circumscribed interests (1) and has a prevalence of 1:59 children in the United States. Affected individuals vary enormously in the severity of their autistic characteristics as well as in the occurrence of many co-morbid conditions. Co-morbid conditions include intellectual disability which affects at least 40% of individuals with autism (24); anxiety in approximately 50% (5); epilepsy in approximately 25% (3, 4); and gastrointestinal disorders in approximately 25% of autistic individuals (6). Twin studies (7, 8) have indicated that genetic factors play a prominent role in the etiology of ASD although the genetics of autism appears to be extremely complex. There has been enormous progress in establishing the genetic architecture of ASD and there are at least 100 genes known to confer risk of ASD (9, 10). There is also increasingly strong evidence that environmental factors, alone or in conjunction with genotype, can contribute to the risk for ASD (11). These findings have led to a widespread consensus that there are different biological forms of ASD that may necessitate different diagnostic, preventative and treatment strategies.
ASD is currently diagnosed based on behavioral characteristics exhibited by an affected child (12). While specialist clinicians are able to confidently diagnose children as young as 24 months (13), the average age of diagnosis in the United States is over 4 years (2, 14). Families often experience long waits to receive a definitive diagnosis due to the paucity of trained clinicians able to perform diagnostic assessment. Early diagnosis is important because intensive behavioral therapies are not only effective in reducing disability in many children with autism (15–17), but, the benefit of early intervention is greater the earlier the intervention is started.
Unfortunately, there is currently no reliable biomarker that can be used to identify children at risk for ASD (18). Because of the genetic complexity of ASD, there is currently no clinically meaningful genotyping carried out to detect ASD. There have been recent intriguing neuroimaging studies indicating that alterations of brain function or structure as early as 6 months may be valuable indicators of a higher risk for autism (19, 20). However, it is unlikely that comprehensive structural and functional magnetic resonance imaging is a practical approach to detecting ASD in young children. Other, more cost effective and widely applicable biomarker strategies must be discovered.
We previously demonstrated that a metabolomics approach for the detection of autism risk holds substantial promise (21). In our preliminary study, we identified a subset of 179 features that classified ASD and TYP children with 81% accuracy. Metabolism-based analysis has the merit of being sensitive to interactions between the genome, gut microbiome, diet, and environmental factors that contribute to the unique metabolic signature of an individual (22). Metabolic testing can provide important biomarkers toward identifying the perturbations of biological processes underlying an individual’s ASD. Past studies have been underpowered to identify metabolic perturbations that lead to actionable metabolic subtypes (23).
To test for metabolic imbalances that can reveal subtypes of ASD subjects, we conducted the Children’s Autism Metabolome Project (CAMP, ClinicalTrials.gov Identifier NCT02548442). CAMP recruited 1,100 young children (18 to 48 months) with ASD, intellectual disability or typical development. Research reliable clinicians confirmed the child’s diagnosis and blood samples were collected under protocols designed specifically for metabolomics analyses. The CAMP study is the largest metabolomics study of ASD to date.
The current study was motivated by observations of AA dysregulation in West et al. (21) and in preliminary analysis of the CAMP samples. The relevance of AA dysregulation to ASD is reinforced by Novarino (24) who demonstrated loss of function mutations in the gene BCKDK (Branched Chain Ketoacid Dehydrogenase Kinase) resulting in reductions of BCKDK messenger RNA and protein, E1a phosphorylation, and plasma branched-chain AAs in consanguineous families with autism, epilepsy, and intellectual disability. Follow on studies by Tarlungeanu (25) demonstrated that altered transport of BCAAs across the blood brain barrier led to dysregulation of AA levels and neurological impairments. We sought to determine whether dysregulation of AAs was a more pervasive phenomenon in individuals with ASD.Thus, we set out to identify metabotypes indicating the dysregulation of AAs in individuals with ASD and to determine whether these metabotypes might be diagnostic of subsets of individuals. A metabotype is a subpopulation defined by a common metabolic signature that can be differentiated from other members of the study population (26). Metabotypes of ASD can be useful in stratifying the broad autism spectrum into more biochemically homogeneous and clinically significant subtypes. Stratification of ASD based on distinct metabolism can inform pharmacological and dietary interventions that prevent or ameliorate clinical symptoms within a metabotype.
METHODS AND MATERIALS
CAMP Participants
The CAMP study recruited children, ages 18 to 48 months, from 8 centers across the United States (Supplemental Table S1). Informed consent of a parent or legal guardian was obtained for each participant. The study protocol was approved and monitored by local IRBs at each of the sites. Enrollment was limited to one child per household to minimize genetic or family environmental effects. Children participating in other clinical studies could not have used any investigational agent within 30 days of participation. Children were excluded from the study if they were previously diagnosed with a genetic condition such as fragile × syndrome, Rett syndrome, Down syndrome, tuberous sclerosis, or inborn errors of metabolism. Subjects that had fetal alcohol syndrome, or other serious neurological, metabolic, psychiatric, cardiovascular, or endocrine system disorders were also excluded. In addition, children exhibiting signs of illness within 2 weeks of enrollment, including vomiting, diarrhea, fever, cough, or ear infection were rescheduled. Each participant underwent physical, neurological and behavioral examinations. Metadata was obtained about the children’s birth, developmental, medical and immunization histories, dietary supplements and medications. Parents’ medical histories were also obtained.
The Autism Diagnostic Observations Schedule-Second Version (ADOS-2) was performed by research reliable clinicians to confirm ASD diagnoses. The Mullen Scales of Early Learning (MSEL) was administered to establish a developmental quotient (DQ) for all children in the study. A prior ADOS-2 or MSEL was accepted if performed within 90 days of enrollment by qualified personnel. Children without ASD receiving a clinical diagnosis of developmental delay were not included in the current study. Children entering the study as TYP were not routinely administered the ADOS-2. The Social Communications Questionnaire (SCQ) was administered for a subset of 65 TYP children as a screen for ASD. Of these, 9 were referred for subsequent ADOS-2 evaluation. Four received a diagnosis of ASD (and were included in this study) and 5 received a diagnosis of TYP.
Training and Test Sets
A training set was used to identify metabotypes associated with ASD and a test set was used to evaluate the reproducibility of the metabotypes. The sample size of the training set was designed to detect metabotypes with a sensitivity (metabotype prevalence) > 3% and specificity > 85% with a power of 0.90 (Supplemental Table S2). The training set (N=338, ASD=253, TYP=85) was established and analyzed, then as recruitment continued, the test set (N=342, ASD=263, TYP=79) was established when sufficient subjects were available to match the training set demographics (Supplemental Table S3).
Phlebotomy Procedures
Blood was collected from subjects after at least a 12 hour fast by venipuncture into 6ml sodium heparin tubes on wet ice. A minimum of a 2 ml blood draw was required for sample inclusion in the computational analyses. The plasma was obtained by centrifugation (1200 × G for 10 minutes) and frozen to −70°C within 1 hour.
Triple Quadrupole LC-MS/MS Method for Quantitative Analysis of Biological Amines
The Waters AccQTag™ Ultra kit (Waters Corporation, Milford, MA), which employs derivatization of amine moieties with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate was employed for all samples prior to multiple reaction monitoring (MRM) on a liquid chromatography (LC) mass spectrometry (MS) system consisting of an Agilent 1290 ultra-high performance liquid chromatograph (UHPLC) coupled to an Agilent G6490 Triple Quadrupole Mass Spectrometer (Agilent Technologies Santa Clara, CA) (Supplemental Methods, Supplemental Table S4 and S5).
Bioinformatic Analysis
The concentration values of each metabolite were log base 2 transformed and Z-score normalized prior to analyses. Analysis of covariance (ANCOVA) and pairwise Pearson correlation analysis were performed on each amine compound. False discovery rates were controlled for multiple testing using the Benjamini and Hochberg (27) method of p-value correction. A comparison was considered significant if the corrected p-value was less than 0.05. Dissimilarity measurements of 1 – the absolute value of the Pearson correlation coefficients (ρ) was used to calculate distances for clustering. Wards’ method was utilized for hierarchical clustering. Bootstrap analysis of the clustering result was performed using the pvclust package in R (28). Clusters were considered significant when the unbiased p-value was ≥ 0.95. The non-linear iterative partial least squares (NIPALS) algorithm was used for principal component analysis (PCA) and confidence intervals drawn at 95th percentile of the PCA scores using Hotelling’s T2 statistic using the package PCAmethods (29). Welch T-tests were used to test for differences in study populations. The independence of subject metadata relative to the metabotypes was tested using the Fisher Exact test statistic with an alpha of 0.05 to reject the null hypothesis. These analyses were conducted with R (version 3.4.3).
Establishing and Assessing Diagnostic Thresholds
A heuristic algorithm was developed to identify individual biomarkers able to discriminate ASD subpopulations, indicative of a metabotype, using a threshold (Supplemental Figure S1) for metabolite abundance or ratios. Diagnostic thresholds were established in the training set to generate a subpopulation with at least 10% of the ASD population while minimizing the number of TYP subjects in the subpopulation. A subject exceeding the diagnostic threshold was scored as a metabotype-positive ASD subject and the remaining subjects as metabotype-negative. Diagnostic performance metrics of specificity (detection of TYP), sensitivity (detection of ASD) and positive predictive value (PPV, percent of metabotype-positives that are ASD) were calculated based on metabotype status (positive or negative) and ADOS-2 diagnosis (ASD or TYP).
Permutation analysis was performed to test the probability that the observed diagnostic performance values from threshold setting and subpopulation prediction could be due to chance. Chance was assessed using 1000 permutations of subject diagnoses in the training set for threshold setting and subpopulation prediction or test set following subpopulation prediction. In both permutation procedures, the probability that observed biomarker performance metrics were due to chance was calculated based on the frequency that the observed sensitivity, specificity, and PPV were met or exceeded in the random permutation set.
When the diagnostic ratios were combined into panels of ratios to test for ASD associated metabotypes, the minimum performance required to consider a metabotype as reproducible were a sensitivity ≥ 5%, a specificity ≥ 95%, and a PPV ≥ 90% in both training and test sets.
RESULTS
Children’s Autism Metabolome Project (CAMP) Study Demographics
The training and test sets of subjects were chosen with appropriate power and randomization (Supplemental Table S3). The ASD prevalence, DQ, and gender composition between the training and test sets are equivalent (p value > 0.05). However, the ASD population contains 16.5 % more males than the TYP population (p value < 0.01). The ASD population is slightly older than the TYP by 3.3 months (p value < 0.01) and the ASD subjects within the training set are 1.4 months older than ASD subjects in the test set (p value < 0.01).
Analysis of Amine-Containing Metabolites between ASD and TYP Study Populations
Analysis of covariance (ANCOVA) was performed on 31 amine containing metabolites in the training set of subjects to test the effect of gender or diagnoses controlling for subject age on metabolite means. No significant differences were identified in metabolite abundance values for diagnosis, age, sex, gender or their interactions (Supplemental Table S6). These results suggest that within the demographic ranges in this study, the differences in subject age or sex have little impact on metabolite abundance. Therefore, the differences in the composition of ASD and TYP study populations are unlikely to have significant impact on study results.
Metabolite Correlations within ASD Reveal Distinct Clusters of Amine Metabolites
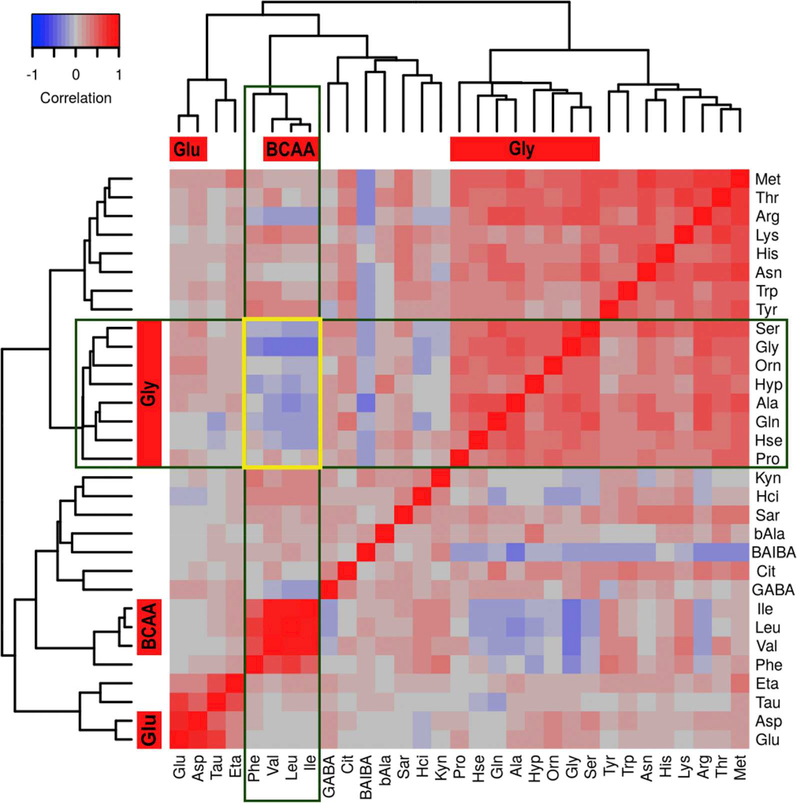
We then examined the relationship among the amine metabolites in the training set of ASD subjects by pairwise Pearson correlation analysis and hierarchical clustering to identify metabolites with co-regulated metabolism (Figure 1). Three clusters of metabolites with positive correlations were identified. Cluster 1 contains the metabolites serine, glycine, ornithine, 4-hydroxyproline, alanine, glutamine, homoserine, and proline (i.e., the glycine cluster - mean ρ 0.45 ± 0.02). Cluster 2 contains the BCAAs (leucine, isoleucine, and valine) and phenylalanine where the BCAAs are more highly correlated with each other (mean pairwise ρ of 0.86 ± 0.02) than the BCAAs are with phenylalanine (mean pairwise ρ of 0.56 ± 0.02) (i.e., the BCAA cluster red boxes, Figure 1). Cluster 3 contains glutamate and aspartate (i.e., the glutamate cluster - ρ of 0.78, Figure 1). The intersection of the glycine and BCAA clusters yielded a block of negative correlations (Figure 1, intersection of boxes). We decided to focus our analysis on the glycine cluster metabolites that are negatively correlated with BCAA metabolites. Proline was removed from further analysis because it was not negatively correlated with the BCAAs. Phenylalanine was removed because it is not a BCAA metabolite.
Figure 1.
Heat map with hierarchical clustering dendrograms from pairwise Pearson correlations of metabolite abundances for the training set ASD subjects. Red filled boxes associated with the dendrograms identify statistically significant clusters following bootstrap resampling. The names of these clusters appear within the red boxes. The green open boxes highlight the BCAA cluster in the columns and the glycine cluster in the rows. The intersection of the two green boxes, marked by a yellow open rectangle, identifies the block of negative correlations shared by the glycine and BCAA clusters. Abbreviations: BCAA, branched chain amino acids; BAIBA, β-Aminoisobutyric Acid; GABA, γ-Aminobutyric acid, bAla, β-alanine; Hci, Homocitrulline; Hse, Homoserine; ETA, Ethanolamine; Sar, Sarcosine; Tau, Taruine; Hyp, 4-Hydroxyproline; Cit, Citrulline
Identification of Amino Acid:BCAA Imbalance Metabotypes Associated with ASD
The negative correlation between the BCAA and glycine cluster led us to evaluate ratios of these AA as predictors of ASD diagnosis. Ratios can uncover biological properties not evident with individual metabolites and increase the signal when two metabolites with a negative correlation are evaluated. This strategy, for example, formed the basis of the standard phenylketonuria (PKU) diagnostic using a ratio of phenylalanine and tyrosine (30). Based on analysis of boxplots (Supplemental Figure S2), we created ratios with one of the BCAAs in the denominator and one of the glycine cluster metabolites in the numerator. Thresholds for the ratios were set in the training set and evaluated in the test set of subjects (Table 1). The BCAA ratios of glutamine, glycine, ornithine and serine identified subpopulations of subjects associated with an ASD diagnosis at a rate higher than chance in both training and test sets (Supplemental Tables S7 and S8).
Table 1:
Diagnostic performance metrics of amine ratios to discriminate subpopulations of ASD subjects in the training and test sets of subjects. The ratios all include branched chain amino acid (BCAA) values in the denominators and negatively correlated Gly-cluster metabolites in the numerator. Abbreviations: Pos., positive, Pred., predictive; AADM, amino acid dysregulation metabotype; ASD, autism spectrum disorder; Train, training set; Test, test set.
| Ratio | Sensitivity | Specificity | Pos. Pred. Value | Permutation Test | ||||
|---|---|---|---|---|---|---|---|---|
| Train | Test | Train | Test | Train | Test | Train | Test | |
| Ratios used to create AADMAlanine | ||||||||
| Ala:Ile | 0.202 | 0.175 | 0.894 | 0.937 | 0.850 | 0.902 | 3.40E-02 | 9.00E-03 |
| Ala:Leu | 0.245 | 0.133 | 0.894 | 0.937 | 0.873 | 0.875 | 2.00E-03 | 5.50E-02 |
| Ala:Val | 0.178 | 0.114 | 0.918 | 0.924 | 0.865 | 0.833 | 2.20E-02 | 2.22E-01 |
| Ratios used to create AADMGlutamine | ||||||||
| Gln:Ilea | 0.126 | 0.163 | 0.976 | 0.937 | 0.941 | 0.896 | 3.00E-03 | 1.30E-02 |
| Gln:Leu | 0.103 | 0.217 | 0.976 | 0.886 | 0.929 | 0.864 | 9.00E-03 | 2.40E-02 |
| Gln:Val | 0.130 | 0.186 | 0.976 | 0.886 | 0.943 | 0.845 | 1.00E-03 | 9.90E-02 |
| Ratios used to create AADMGlycine | ||||||||
| Gly:Ilea | 0.174 | 0.152 | 0.953 | 0.962 | 0.917 | 0.930 | 0.00E+00 | 2.00E-03 |
| Gly:Leua | 0.146 | 0.148 | 0.953 | 0.987 | 0.902 | 0.975 | 8.00E-03 | 0.00E+00 |
| Gly:Val | 0.126 | 0.160 | 0.976 | 0.937 | 0.941 | 0.894 | 3.00E-03 | 1.50E-02 |
| Ratios used to create AADMHomoserine | ||||||||
| Hse:Ile | 0.063 | 0.137 | 0.976 | 0.949 | 0.889 | 0.900 | 1.16E-01 | 2.50E-02 |
| Hse:Leu | 0.107 | 0.122 | 0.953 | 0.975 | 0.871 | 0.941 | 8.20E-02 | 3.00E-03 |
| Hse:Val | 0.067 | 0.160 | 0.965 | 0.962 | 0.850 | 0.933 | 2.09E-01 | 4.00E-03 |
| Ratios used to create AADMOrnithine | ||||||||
| Orn:Ilea | 0.115 | 0.103 | 0.965 | 0.987 | 0.906 | 0.964 | 2.10E-02 | 9.00E-03 |
| Orn:Leua | 0.103 | 0.202 | 0.965 | 0.949 | 0.897 | 0.930 | 3.70E-02 | 0.00E+00 |
| Orn:Val | 0.119 | 0.141 | 0.953 | 0.937 | 0.882 | 0.881 | 3.70E-02 | 4.30E-02 |
| Ratios used to create AADMSerine | ||||||||
| Ser:Ile | 0.130 | 0.129 | 0.953 | 0.949 | 0.892 | 0.895 | 2.00E-02 | 2.20E-02 |
| Ser:Leua | 0.138 | 0.167 | 0.941 | 0.924 | 0.875 | 0.880 | 3.00E-02 | 3.80E-02 |
| Ser:Val | 0.190 | 0.228 | 0.941 | 0.873 | 0.906 | 0.857 | 1.00E-03 | 3.90E-02 |
| Ratios used to create AADMHydroxyproline | ||||||||
| Hyp:Ile | 0.111 | 0.087 | 0.941 | 0.911 | 0.848 | 0.767 | 1.14E-01 | 6.03E-01 |
| Hyp:Leu | 0.115 | 0.095 | 0.918 | 0.899 | 0.806 | 0.758 | 2.69E-01 | 6.64E-01 |
| Hyp:Val | 0.241 | 0.213 | 0.871 | 0.772 | 0.847 | 0.757 | 1.70E-02 | 6.64E-01 |
The observed diagnostic performance occurred in less than 5% of 1000 permutations of subject diagnosis in both training and test sets.
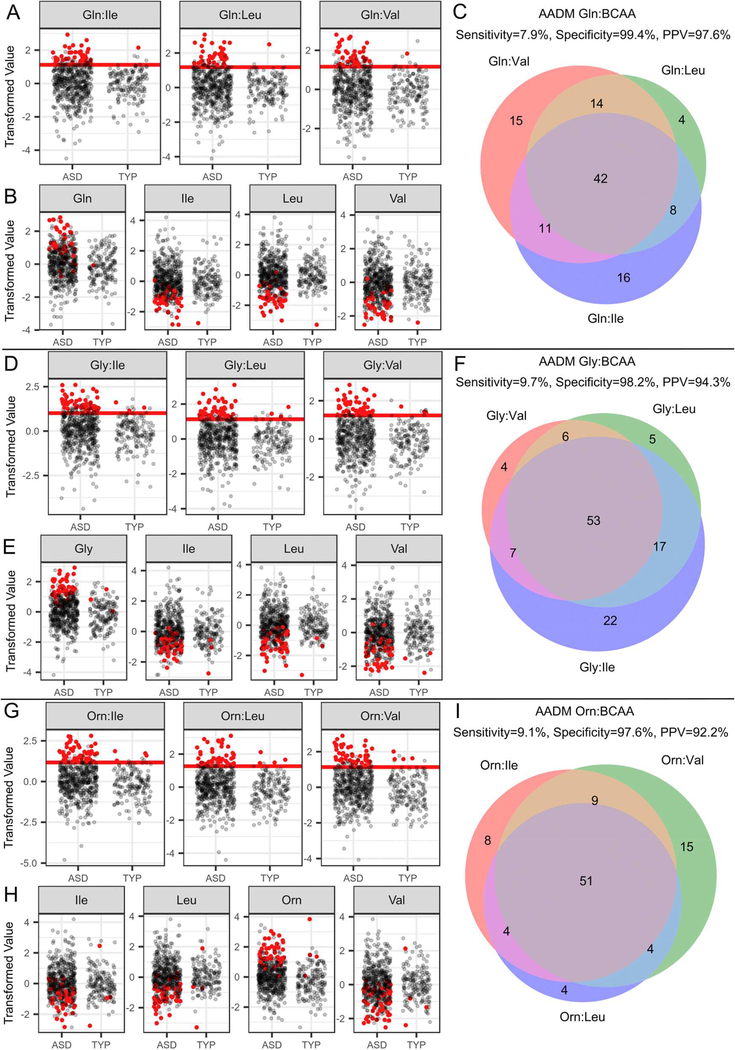
The correlation of the BCAAs with each other (ρ = 0.86 ± 0.02) and the overlap of affected-subjects (Figure 2, Venn diagrams) identified by the AA:BCAA ratios suggested that a combination of ratios containing a single numerator and each of the three BCAAs as denominators could uncover BCAA metabolic dysregulation. Exploiting the positive correlation among the BCAAs in this way improves the specificity and PPV. For example, each of the Glycine:BCAA ratios (i.e. glycine:leucine or glycine:valine or glycine:isoleucine) results in a specificity of 94.1% and PPV of 91.1% (Supplemental Table S9). However, requiring that the subject be positive for all three Glycine:BCAA ratios, results in a specificity of 98.8% and PPV of 96.0%. Through this process, we identified groups of subjects that exhibited an Amino Acid Dysregulation Metabotype (AADM). Subjects were identified by AADM when they exceeded an established threshold for all three AA:BCAA ratios. Since the nomenclature for these biomarkers can quickly become confusing, we have designated different AADMs using the numerator metabolite e.g. AADMglutamine (Figure 2). Not all ratios of AAs to BCAA resulted in diagnostic differences between the ASD and TYP groups (Table 2, Supplemental Figures S3-S6). We focused, therefore, on those AA:BCAA ratios that had the greatest predictive power including glutamine AADM (AADMglutamine; Figure 2, A-C), glycine AADM (AADMglycine; Figure 2, D-F) and ornithine AADM (AADMornithine; Figure 2, G-I).
Figure 2.
Levels of amino acid ratios and of individual amino acids; diagnostic threshold set in the training set (red lines). Venn diagrams of the metabotype-positive subjects identified by each of the AA:BCAA diagnostics for AADMglutamine (A-C), AADMglycine (D-F), and AADMornithine (G-I) in the training and test sets. A, D, and G) Scatter plots of the AA:BCAA ratios used to create an AADM diagnostic test. Red points represent AADM positive subjects and black points represent AADM negative subjects. The red horizontal line is the diagnostic threshold set in the training set. B, E, and H) Scatter plots of individual amino acids used in the creation of the ratios. Red dots indicate AADM positive subjects and black points represent those that are AADM negative. C, F, and I) Venn diagram of metabotype-positive subjects identified by the three ratios used to create each AA:BCAA diagnostic. Each circle represents the subjects identified by the diagnostic threshold for a given ratio. The intersection of the Venn diagram indicates the subjects called AADM positive (red dots in scatter plots). Performance metrics above the Venn diagram represent entire study population (training and test sets).Abbreviations: AA, amino acid; BCAA, branched chain amino acid; ASD, autism spectrum disorder; TYP, typically developing; AADM, amino acid dysregulation metabotype.
Table 2:
Diagnostic performance metrics of Amino Acid Dysregulation Metabotypes (AADM). Each AADM consists of three ratios with a different branched chain amino acid (BCAA) in the denominator. Abbreviations: Pos., positive, Pred., predictive; Train, training set; Test, test set; Hyp, 4-Hydroxyproline; Hse, Homoserine; Orn, Ornithine.
| AADM Diagnostic |
Sensitivity | Specificity | Pos. Pred. Value | |||
|---|---|---|---|---|---|---|
| Train | Test | Train | Test | Train | Test | |
| Ala:BCAA | 0.150 | 0.141 | 0.929 | 0.937 | 0.864 | 0.881 |
| Gln:BCAAa | 0.079 | 0.080 | 0.988 | 1.000 | 0.952 | 1.000 |
| Gly:BCAAa | 0.095 | 0.099 | 0.988 | 0.975 | 0.960 | 0.929 |
| Hse:BCAA | 0.036 | 0.080 | 0.988 | 1.000 | 0.900 | 1.000 |
| Orn:BCAAa | 0.079 | 0.103 | 0.976 | 0.975 | 0.909 | 0.931 |
| Ser:BCAA | 0.091 | 0.106 | 0.965 | 0.949 | 0.885 | 0.875 |
| Hyp:BCAA | 0.087 | 0.080 | 0.965 | 0.924 | 0.880 | 0.778 |
AADMs are a reproducible metabotype that is identified across training and test populations with a sensitivity greater than 5% and a positive predictive value greater than 90%.
AADMs Define a Diagnostic for BCAA Dysregulation Associated with ASD
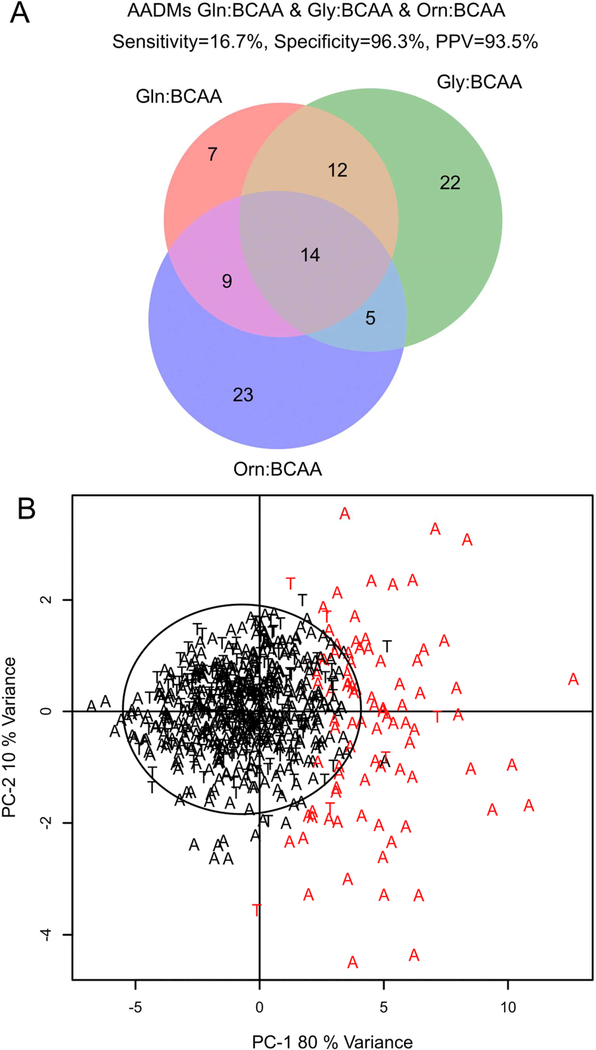
The ASD subjects identified by each AADM were evaluated to assess the extent of overlap. We found that there is substantial overlap of the subjects identified by each of the metabotypes (Figure 3). However, each of the metabotypes also identifies a unique group of subjects. The AADMglutamine identified 7.9% of the ASD subjects in the total CAMP population, AADMglycine 9.7%, and the AADMornithine 9.1%, with PPVs of 97.6%, 94.3% and 92.2% respectively. Combining all three AADM subtypes together (AADMtotal), identified 16.7% of ASD subjects in the CAMP population with a specificity of 96.3% and a PPV of 93.5% (Figure 3 A). Principal component analysis (PCA) of the metabolite ratios utilized in AADMglycine, AADMglutamine, and AADMornithine was performed to test if an unsupervised method could identify subjects with AA dysregulation. A majority (80%, 74/92) of the AADM-positive subjects were separated from the unaffected subjects (Figure 3 B).
Figure 3.
A) Venn diagram of the 92 AADMtotal subjects identified by each of the AADMs. At least 50% of the subjects identified by one AADM were identified by the other 2 AADMs. The AADMtotal population is comprised of 86 ASD and 6 TYP subjects. The overall prevalence of metabolic dysregulation in the CAMP ASD population is 16.7% (86 AADMtotal ASD / 516 CAMP ASD), specificity 96.3 % (158 AADM-negative TYP / 164 CAMP TYP), PPV 93.5% (86 AADMtotal ASD / 92 AADMtotal). B) PCA analysis of the metabolite ratios used in the metabolic signature of the reproducible AADMs creating the AADMtotal estimates in the CAMP study population. Black circle is the 95% confidence interval from the Hoetellings T2. Red letters are AADMtotal positive (N=92), Black letters are AADMtotal negative (N=588). A=ASD and T=TYP. Abbreviations: BCAA, branched chain amino acid; Orn, Ornithine; ASD, autism spectrum disorder; TYP, typically developing; AADM, amino acid dysregulation metabotype; CAMP, Children’s Autism Metabolome Project.
AADMOrnithine and AADMGlutamine are More Sensitive at Detecting Females with ASD.
Since the composition of subject sex and age differed between the ASD and TYP populations, the impact of these variables was evaluated in the AADM positive and negative populations. Differential analysis of reproducible AADM positive and negative subjects’ metabolite levels with respect to age or sex did not identify statistically significant changes in abundance (Supplemental Results, Supplemental Tables S11 and S12). Females with ASD were 2.1 fold (odds ratio 2.8, p value 0.002) more likely to be identified by AADMornithine and AADMglutamine than would be expected by chance (Supplemental Table S13). The AADMglycine did not demonstrate a predictive sex bias.
DISSCUSSION
CAMP is the largest study of the metabolism of children with autism spectrum disorder and age-matched typically developing children carried out to date. Metabolomics offers the opportunity to examine associations between small molecule abundance levels and the presence of a disorder such as ASD as well as influences such as sex, severity of the disorder, comorbid conditions, diet, supplements and other environmental factors. Given the known heterogeneity of ASD, the size of CAMP offers the prospect of identifying metabolically defined subtypes (or metabotypes) that can identify groups with a prevalence as low as 5%. Diagnostic tests for metabotypes of ASD create an opportunity for earlier diagnosis and the potential to inform more targeted treatment.
Our goal is to analyze data from the CAMP population to identify metabotypes associated with ASD that could enable stratification of the disorder based on shared metabolic characteristics. Based on our own observations and growing literature (23–25, 31, 32) reporting a dysregulation of amino acid metabolism associated with ASD, we began our analysis by studying free plasma amine levels. A simple analysis of the mean concentrations of free plasma amines did not reveal meaningful differences between the ASD and TYP populations of children. However, scatterplots of amine levels indicated that there were subsets of children with ASD with amine levels at the extreme upper or lower end of the abundance distribution. Moreover, correlation analyses revealed two negatively correlated clusters of related metabolites. We tested if ratios of these metabolites could identify subpopulations that exhibit dysregulation of AA metabolism associated with ASD. Diagnostic thresholds established in the training set of subjects using ratios of glutamine, glycine, ornithine and serine with leucine, isoleucine and valine (BCAAs) reproducibly detected subpopulations in an independent test set. Three AADMs based on an imbalance of glutamine, glycine, or ornithine with the BCAAs were reproduced across training and test sets of subjects. Separately, each AADM identified ASD subjects with 7–10% sensitivity and 92–98% PPVs. Taken together, all AADMs identified an altered metabolic phenotype of imbalanced BCAA metabolism in 16.7% of CAMP ASD subjects with a specificity of 96.3% and PPV of 93.5%.
Identification of ASD children with altered AADMs represents an important step toward understanding the etiology of one form of ASD. Imbalances in BCAAs in plasma have been shown to alter not only brain levels of BCAAs, but also other amino acids important for key metabolic processes including intermediary metabolism, protein synthesis, and neurotransmission. For example, when plasma BCAA levels are reduced due to a rare genetic defect in branched chain ketoacid dehydrogenase kinase (BCKDK) (24) leading to accelerated BCAA degradation, the transporters that are normally responsible for their import into the brain transport an excess of other amino acids instead. And, this condition is associated with ASD (24). Similarly, Tarlungeanu (25) demonstrated that rare disruption of amino acid transport associated with defects in the LAT1 transporter reduced the uptake of BCAAs into the brain; again this was associated with ASD-like symptoms. Interestingly, neither study reported elevated plasma levels of glycine, ornithine, or glutamine. The imbalance of amino acid levels in CAMP strongly suggests that other perturbations in BCAA metabolism may be a risk factor for the development of ASD. Importantly, the metabolomic results reported here provide a mechanism for stratifying the larger group of children with ASD into an AADM positive subgroup to enable a more targeted approach to understanding the etiology of this form of ASD. For example, the AADMs we identified may reveal a disruption of the mTORC1 system which could be an underlying reason for lower free plasma BCAA levels. Cellular levels of BCAA as well as other amino acids are maintained through signaling associated mTORC1 and the transcription factor ATF4 (33). Dysregulation of the mTOR pathway is an underlying cause of amino acid dysregulation that is associated with ASD and tuberous sclerosis (34).
The AADMs provide one pathway to much earlier diagnosis of a substantial subset of children with ASD. Earlier diagnosis may also provide the opportunity for earlier biological intervention. BCAA supplementation or high protein diet has been used in mouse models (24) and human patients (31) with BCKDK deficiency to successfully reduce ASD symptoms and improve cognitive function. Defining a group of AADM positive children may enable stratification of the autistic population as a precursor to targeted intervention through dietary supplementation or specialized diet. Currently, clinical trials of common therapies such as vitamin and mineral supplements, carnitine and gluten-free casein-free diets, apply these therapies to all participants. Metabotyping subjects prior to treatment and monitoring metabolite levels provides the opportunity to assess patient compliance and response, and to make adjustments to treatment based on objective measurement of the metabolic profile of the individual subject. It is likely that this strategy would substantially improve positive treatment outcomes.
This study does have some limitations. The levels of blood plasma amine metabolites are not directly relatable to brain levels (35) making direct association of changes in plasma levels to changes in brain levels difficult. The CAMP study focused on recruitment of a large sample of children with ASD and age-matched typically developing controls. Logistical and financial constraints precluded our ability to recruit a large enough sample of children with developmental delays without ASD. Thus, the specificity of ADDM for ASD relative to other neurodevelopmental disorders is currently unclear. This is an important issue that will need to be resolved in future studies. In addition, longitudinal samples are not available to analyze whether AADMs are stable over time. Finally, this study lacks animal models or tissue samples that could be used to dissect enzymatic and expression analysis to identify the molecular mechanisms underlying AADM. While we cannot explain the alterations in metabolism, we have demonstrated that our approach provides stratification of subjects for which future studies and perhaps targeted treatments could be carried out.
This study demonstrates one approach to analyzing the metabolism of ASD to successfully identify reproducible metabotypes. Analysis of the CAMP study samples is ongoing and there will be additional metabotypes which will be diagnostic for subsets of children with ASD. Stratifying ASD based on metabotypes offers an opportunity to identify efficacious interventions within metabotypes that can lead to more precise and individualized treatment. The hope is that by combining the established metabotypes into a more comprehensive diagnostic system, that a substantial percentage of children at risk for ASD will be identifiable at a very early age.
Supplementary Material
Figure S1. Outline of computational procedures utilized to set diagnostic thresholds and to evaluate diagnostic performance. Diagnostic thresholds were set for each metabolite or metabolite ratio in the training set and the threshold was applied to the training or test set to identify the affected subpopulation and determine the observed diagnostic performance. Permutation analysis was performed to evaluate the frequency at which the observed diagnostic performance occurred by chance. A diagnostic test was considered to identify a relevant metabolic subpopulation if the observed performance metrics occurred in less than 5% of 1000 iterations of random permutations of the subjects’ diagnoses.
Figure S2. Scatter plot of the training set’s transformed amine concentration values. Blue boxes indicate groups that are comprised of greater than 90% ASD subjects (90% PPV). These groups include at least 5% of the training set of ASD subjects (5% sensitivity). Glycine, alanine, asparagine, aspartic acid, GABA, glutamic acid, homoserine, ethanolamine, sarcosine, serine and taurine exhibited elevated metabolite levels in ASD subjects, while leucine exhibited decreased metabolite levels in ASD subjects. Red = ASD, Black = TYP; TYP, Typically Developing; ASD, Autism Spectrum Disorder. BAIBA, β-Aminoisobutyric Acid; GABA, γ-Aminobutyric acid, bAla, β-alanine; Hci, Homocitrulline; Hse, Homoserine; ETA, Ethanolamine; Sar, Sarcosine; Tau, Taurine; Hyp, 4-Hydroxyproline; Cit, Citrulline.
Figure S3. Alanine:BCAA diagnostic for AADMalanine. A) Scatter plots of the ratios used to create AADMalanine diagnostic test. Red points represent AADMalanine positive subjects and black points represent AADMalanine negative subjects. The red horizontal line is the diagnostic threshold set in the training set. B) Scatter plots of individual metabolites used in the creation of the ratios. Red dots indicate AADMalanine positive subjects and black points represent those that are AADMalanine negative. C) Venn diagram of subjects identified by the three ratios. Each circle represents the subjects identified by the diagnostic threshold for a given ratio. The intersection of the Venn diagram indicates the subjects called AADMalanine positive (red dots). BCAA, branched chain amino acid; ASD, autism spectrum disorder; TYP, typically developing; AADM, amino acid dysregulation metabotype.
Figure S4. Homoserine:BCAA diagnostic for AADMhomoserine. A) Scatter plots of the ratios used to create AADMhomoserine diagnostic test. Red points represent AADMhomoserine positive subjects and black points represent AADMhomoserine negative subjects. The red horizontal line is the diagnostic threshold set in the training set. B) Scatter plots of individual metabolites used in the creation of the ratios. Red dots indicate AADMhomoserine positive subjects and black points represent those that are AADMhomoserine negative. C) Venn diagram of subjects identified by the three ratios. Each circle represents the subjects identified by the diagnostic threshold for a given ratio. The intersection of the Venn diagram indicates the subjects called AADMhomoserine positive (red dots). Hse, homoserine; BCAA, branched chain amino acid; ASD, autism spectrum disorder; TYP, typically developing; AADM, amino acid dysregulation metabotype.
Figure S5. Serine:BCAA diagnostic for AADMserine. A) Scatter plots of the ratios used to create AADMserine diagnostic test. Red points represent AADMserine positive subjects and black points represent AADMserine negative subjects. The red horizontal line is the diagnostic threshold set in the training set. B) Scatter plots of individual metabolites used in the creation of the ratios. Red dots indicate AADMserine positive subjects and black points represent those that are AADMserine negative. C) Venn diagram of subjects identified by the three ratios. Each circle represents the subjects identified by the diagnostic threshold for a given ratio. The intersection of the Venn diagram indicates the subjects called AADMserine positive (red dots). BCAA, branched chain amino acid; ASD, autism spectrum disorder; TYP, typically developing; AADM, amino acid dysregulation metabotype.
Figure S6. 4-Hydroxyproline:BCAA diagnostic for AADMhydroxyproline. A) Scatter plots of the ratios used to create AADMhydroxyproline diagnostic test. Red points represent AADMhydroxyproline positive subjects and black points represent AADMhydroxyproline negative subjects. The red horizontal line is the diagnostic threshold set in the training set. B) Scatter plots of individual metabolites used in the creation of the ratios. Red dots indicate AADMhydroxyproline positive subjects and black points represent those that are AADMhydroxyproline negative. C) Venn diagram of subjects identified by the three ratios. Each circle represents the subjects identified by the diagnostic threshold for a given ratio. The intersection of the Venn diagram indicates the subjects called AADMhydroxyproline positive (red dots). BCAA, branched chain amino acid; ASD, autism spectrum disorder; TYP, typically developing; AADM, amino acid dysregulation metabotype.
Figure S7. Scatter plots of the ratios and individual metabolites utilized in identification of AADMs. Red points are AADMtotal positive subjects. Black points AADMtotal negative subjects. ASD, autism spectrum disorder; TYP, typically developing; AADM, amino acid dysregulation metabotype.
Table S1. Children’s Autism Metabolome Project (CAMP) clinical sites and locations.
ACKNOWLEDGEMENTS
Funding sources:
Grants: NIH 5 R44 MH107124–03 to B. Burrier; 1R01MH103371 to D. G. Amaral
Other financial support: Nancy Lurie Marks Family Foundation; The Robert E. and Donna Landreth Family Fund
The Children’s Autism Metabolome Project (CAMP) investigators group includes: David G. Amaral from the MIND Institute, University of California, Davis, Amanda E. Bennett from Children’s Hospital of Philadelphia, Daniel L. Coury from Nationwide Children’s Hospital, Richard E. Frye from The University of Arkansas for Medical Sciences, Raun D. Melmed from The Melmed Center, Ann Neumeyer from Massachusetts General Hospital, Kevin B. Sanders from Vanderbilt University Medical Center and Logan K. Wink and Craig Erickson from Cincinnati Children’s Hospital. We appreciate the CAMP investigators and their staff’s contributions to making CAMP a successful study.
We are grateful to Dana Kelly and Gina Wentling for their administrative and social media support of subject recruitment, DelRay Sugden for subject sample intake, handling and integrity quality control, and Lindsay Feuling for development and maintenance of study subject and sample databases.
We are particularly indebted to the families and children who participated in the CAMP Study; without their collaboration, this study would not have been possible.
Footnotes
DISCLOSURES
Alan M. Smith is an employee of Stemina Biomarker Discovery and is an inventor on provisional patent application 62/623,153 entitled “Amino Acid Analysis and Autism Subsets” filed on January 29, 2018.
Joseph J. King was an employee of Stemina Biomarker Discovery and is an inventor on provisional patent application 62/623,153 entitled “Amino Acid Analysis and Autism Subsets” filed on January 29, 2018.
Paul R. West was an employee of Stemina Biomarker Discovery and is an inventor on provisional patent application 62/623,153 entitled “Amino Acid Analysis and Autism Subsets” filed on January 29, 2018.
Michael A. Ludwig is an employee of Stemina Biomarker Discovery and is an inventor on provisional patent application 62/623,153 entitled “Amino Acid Analysis and Autism Subsets” filed on January 29, 2018.
Elizabeth L.R. Donley is an equity owner in Stemina Biomarker Discovery and is an inventor on provisional patent application 62/623,153 entitled “Amino Acid Analysis and Autism Subsets” filed on January 29, 2018.
Robert E. Burrier is an employee of Stemina Biomarker Discovery and is an inventor on provisional patent application 62/623,153 entitled “Amino Acid Analysis and Autism Subsets” filed on January 29, 2018.
David G. Amaral receives research funding from the NIH, the Simons Foundation and Stemina Biomarker Discovery, Inc. He is on the Scientific Advisory Boards of Stemina Biomarker Discovery, Inc. and Axial Therapeutics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Association AP (2013): Diagnostic and statistical manual of mental disorders (5th ed.). Arlington, VA: American Psychiatric Publishing. [Google Scholar]
- 2.Autism, Developmental Disabilities Monitoring Network Surveillance Year Principal I, Centers for Disease C, Prevention (2012): Prevalence of autism spectrum disorders--Autism and Developmental Disabilities Monitoring Network, 14 sites, United States, 2008. MMWR Surveill Summ. 61:1–19. [PubMed] [Google Scholar]
- 3.El Achkar CM, Spence SJ (2015): Clinical characteristics of children and young adults with co-occurring autism spectrum disorder and epilepsy. Epilepsy Behav. 47:183–190. [DOI] [PubMed] [Google Scholar]
- 4.Spence SJ, Schneider MT (2009): The role of epilepsy and epileptiform EEGs in autism spectrum disorders. Pediatr Res. 65:599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerns CM, Kendall PC, Berry L, Souders MC, Franklin ME, Schultz RT, et al. (2014): Traditional and atypical presentations of anxiety in youth with autism spectrum disorder. J Autism Dev Disord. 44:2851–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buie T, Campbell DB, Fuchs GJ 3rd, Furuta GT, Levy J, Vandewater J, et al. Evaluation, diagnosis, and treatment of gastrointestinal disorders in individuals with ASDs: a consensus report. Pediatrics. 125 Suppl 1:S1–18. [DOI] [PubMed] [Google Scholar]
- 7.Bourgeron T (2016): Current knowledge on the genetics of autism and propositions for future research. C R Biol. 339:300–307. [DOI] [PubMed] [Google Scholar]
- 8.Sandin S, Lichtenstein P, Kuja-Halkola R, Hultman C, Larsson H, Reichenberg A (2017): The Heritability of Autism Spectrum Disorder. JAMA. 318:1182–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geschwind DH, State MW (2015): Gene hunting in autism spectrum disorder: on the path to precision medicine. Lancet Neurol. 14:1109–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, et al. (2015): Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 87:1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandy W, Lai MC (2016): Annual Research Review: The role of the environment in the developmental psychopathology of autism spectrum condition. J Child Psychol Psychiatry. 57:271–292. [DOI] [PubMed] [Google Scholar]
- 12.Lord C, Risi S, Lambrecht L, Cook EH Jr., Leventhal BL, DiLavore PC, et al. (2000): The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord. 30:205–223. [PubMed] [Google Scholar]
- 13.Lord C, Risi S, DiLavore PS, Shulman C, Thurm A, Pickles A (2006): Autism from 2 to 9 years of age. Arch Gen Psychiatry. 63:694–701. [DOI] [PubMed] [Google Scholar]
- 14.Autism, Developmental Disabilities Monitoring Network Surveillance Year Principal I, Centers for Disease C, Prevention (2009): Prevalence of autism spectrum disorders - Autism and Developmental Disabilities Monitoring Network, United States, 2006. MMWR Surveill Summ. 58:1–20. [PubMed] [Google Scholar]
- 15.Dawson G (2010): Recent advances in research on early detection, causes, biology, and treatment of autism spectrum disorders. Curr Opin Neurol. 23:95–96. [DOI] [PubMed] [Google Scholar]
- 16.Dawson G, Rogers S, Munson J, Smith M, Winter J, Greenson J, et al. (2010): Randomized, controlled trial of an intervention for toddlers with autism: the Early Start Denver Model. Pediatrics. 125:e17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganz ML (2007): The lifetime distribution of the incremental societal costs of autism. Arch Pediatr Adolesc Med. 161:343–349. [DOI] [PubMed] [Google Scholar]
- 18.McPartland JC (2017): Developing Clinically Practicable Biomarkers for Autism Spectrum Disorder. J Autism Dev Disord. 47:2935–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hazlett HC, Gu H, Munsell BC, Kim SH, Styner M, Wolff JJ, et al. (2017): Early brain development in infants at high risk for autism spectrum disorder. Nature. 542:348–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen MD, Kim SH, McKinstry RC, Gu H, Hazlett HC, Nordahl CW, et al. (2017): Increased Extra-axial Cerebrospinal Fluid in High-Risk Infants Who Later Develop Autism. Biol Psychiatry. 82:186–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West PR, Amaral DG, Bais P, Smith AM, Egnash LA, Ross ME, et al. (2014): Metabolomics as a tool for discovery of biomarkers of autism spectrum disorder in the blood plasma of children. PLoS One. 9:e112445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beger RD DW, Schmidt MA, Gross SS, Kirwan JA, Cascante M, Brennan L,, Wishart DSOM, Hankemeier T, Broadhurst DI, Lane AN, Suhre K, Kastenmüller GSS, Thiele I, Fiehn O, Kaddurah-Daouk R (2016): Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metabolomics. 12:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Żurawicz E, Kałużna-Czaplińska J (2015): Analysis of amino acids in autism spectrum disorders. TrAC Trends in Analytical Chemistry. 73:91–118. [Google Scholar]
- 24.Novarino G, El-Fishawy P, Kayserili H, Meguid NA, Scott EM, Schroth J, et al. (2012): Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science. 338:394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarlungeanu DC, Deliu E, Dotter CP, Kara M, Janiesch PC, Scalise M, et al. (2016): Impaired Amino Acid Transport at the Blood Brain Barrier Is a Cause of Autism Spectrum Disorder. Cell. 167:1481–1494 e1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riedl A, Gieger C, Hauner H, Daniel H, Linseisen J (2017): Metabotyping and its application in targeted nutrition: an overview. Br J Nutr. 117:1631–1644. [DOI] [PubMed] [Google Scholar]
- 27.Benjamini Y, and Hochberg Y (1995): Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing Journal of the Royal Statistical Society Series B. 57:289–300. [Google Scholar]
- 28.Suzuki R, Shimodaira H (2006): Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 22:1540–1542. [DOI] [PubMed] [Google Scholar]
- 29.Stacklies W RH, Scholz M, Walther D, Selbig J. (2007): pcaMethods--a bioconductor package providing PCA methods for incomplete data. Bioinformatics. 23:1164–1167. [DOI] [PubMed] [Google Scholar]
- 30.Eastman JW, Sherwin JE, Wong R, Liao CL, Currier RJ, Lorey F, et al. (2000): Use of the phenylalanine:tyrosine ratio to test newborns for phenylketonuria in a large public health screening programme. J Med Screen. 7:131–135. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Cazorla A, Oyarzabal A, Fort J, Robles C, Castejon E, Ruiz-Sala P, et al. (2014): Two novel mutations in the BCKDK (branched-chain keto-acid dehydrogenase kinase) gene are responsible for a neurobehavioral deficit in two pediatric unrelated patients. Hum Mutat. 35:470–477. [DOI] [PubMed] [Google Scholar]
- 32.Zheng HF, Wang WQ, Li XM, Rauw G, Baker GB (2017): Body fluid levels of neuroactive amino acids in autism spectrum disorders: a review of the literature. Amino Acids. 49:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park Y, Reyna-Neyra A, Philippe L, Thoreen CC (2017): mTORC1 Balances Cellular Amino Acid Supply with Demand for Protein Synthesis through Post-transcriptional Control of ATF4. Cell Rep. 19:1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maynard TM, Manzini MC (2017): Balancing Act: Maintaining Amino Acid Levels in the Autistic Brain. Neuron. 93:476–479. [DOI] [PubMed] [Google Scholar]
- 35.Scholl-Burgi S, Haberlandt E, Heinz-Erian P, Deisenhammer F, Albrecht U, Sigl SB, et al. (2008): Amino acid cerebrospinal fluid/plasma ratios in children: influence of age, gender, and antiepileptic medication. Pediatrics. 121:e920–926. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Outline of computational procedures utilized to set diagnostic thresholds and to evaluate diagnostic performance. Diagnostic thresholds were set for each metabolite or metabolite ratio in the training set and the threshold was applied to the training or test set to identify the affected subpopulation and determine the observed diagnostic performance. Permutation analysis was performed to evaluate the frequency at which the observed diagnostic performance occurred by chance. A diagnostic test was considered to identify a relevant metabolic subpopulation if the observed performance metrics occurred in less than 5% of 1000 iterations of random permutations of the subjects’ diagnoses.
Figure S2. Scatter plot of the training set’s transformed amine concentration values. Blue boxes indicate groups that are comprised of greater than 90% ASD subjects (90% PPV). These groups include at least 5% of the training set of ASD subjects (5% sensitivity). Glycine, alanine, asparagine, aspartic acid, GABA, glutamic acid, homoserine, ethanolamine, sarcosine, serine and taurine exhibited elevated metabolite levels in ASD subjects, while leucine exhibited decreased metabolite levels in ASD subjects. Red = ASD, Black = TYP; TYP, Typically Developing; ASD, Autism Spectrum Disorder. BAIBA, β-Aminoisobutyric Acid; GABA, γ-Aminobutyric acid, bAla, β-alanine; Hci, Homocitrulline; Hse, Homoserine; ETA, Ethanolamine; Sar, Sarcosine; Tau, Taurine; Hyp, 4-Hydroxyproline; Cit, Citrulline.
Figure S3. Alanine:BCAA diagnostic for AADMalanine. A) Scatter plots of the ratios used to create AADMalanine diagnostic test. Red points represent AADMalanine positive subjects and black points represent AADMalanine negative subjects. The red horizontal line is the diagnostic threshold set in the training set. B) Scatter plots of individual metabolites used in the creation of the ratios. Red dots indicate AADMalanine positive subjects and black points represent those that are AADMalanine negative. C) Venn diagram of subjects identified by the three ratios. Each circle represents the subjects identified by the diagnostic threshold for a given ratio. The intersection of the Venn diagram indicates the subjects called AADMalanine positive (red dots). BCAA, branched chain amino acid; ASD, autism spectrum disorder; TYP, typically developing; AADM, amino acid dysregulation metabotype.
Figure S4. Homoserine:BCAA diagnostic for AADMhomoserine. A) Scatter plots of the ratios used to create AADMhomoserine diagnostic test. Red points represent AADMhomoserine positive subjects and black points represent AADMhomoserine negative subjects. The red horizontal line is the diagnostic threshold set in the training set. B) Scatter plots of individual metabolites used in the creation of the ratios. Red dots indicate AADMhomoserine positive subjects and black points represent those that are AADMhomoserine negative. C) Venn diagram of subjects identified by the three ratios. Each circle represents the subjects identified by the diagnostic threshold for a given ratio. The intersection of the Venn diagram indicates the subjects called AADMhomoserine positive (red dots). Hse, homoserine; BCAA, branched chain amino acid; ASD, autism spectrum disorder; TYP, typically developing; AADM, amino acid dysregulation metabotype.
Figure S5. Serine:BCAA diagnostic for AADMserine. A) Scatter plots of the ratios used to create AADMserine diagnostic test. Red points represent AADMserine positive subjects and black points represent AADMserine negative subjects. The red horizontal line is the diagnostic threshold set in the training set. B) Scatter plots of individual metabolites used in the creation of the ratios. Red dots indicate AADMserine positive subjects and black points represent those that are AADMserine negative. C) Venn diagram of subjects identified by the three ratios. Each circle represents the subjects identified by the diagnostic threshold for a given ratio. The intersection of the Venn diagram indicates the subjects called AADMserine positive (red dots). BCAA, branched chain amino acid; ASD, autism spectrum disorder; TYP, typically developing; AADM, amino acid dysregulation metabotype.
Figure S6. 4-Hydroxyproline:BCAA diagnostic for AADMhydroxyproline. A) Scatter plots of the ratios used to create AADMhydroxyproline diagnostic test. Red points represent AADMhydroxyproline positive subjects and black points represent AADMhydroxyproline negative subjects. The red horizontal line is the diagnostic threshold set in the training set. B) Scatter plots of individual metabolites used in the creation of the ratios. Red dots indicate AADMhydroxyproline positive subjects and black points represent those that are AADMhydroxyproline negative. C) Venn diagram of subjects identified by the three ratios. Each circle represents the subjects identified by the diagnostic threshold for a given ratio. The intersection of the Venn diagram indicates the subjects called AADMhydroxyproline positive (red dots). BCAA, branched chain amino acid; ASD, autism spectrum disorder; TYP, typically developing; AADM, amino acid dysregulation metabotype.
Figure S7. Scatter plots of the ratios and individual metabolites utilized in identification of AADMs. Red points are AADMtotal positive subjects. Black points AADMtotal negative subjects. ASD, autism spectrum disorder; TYP, typically developing; AADM, amino acid dysregulation metabotype.
Table S1. Children’s Autism Metabolome Project (CAMP) clinical sites and locations.