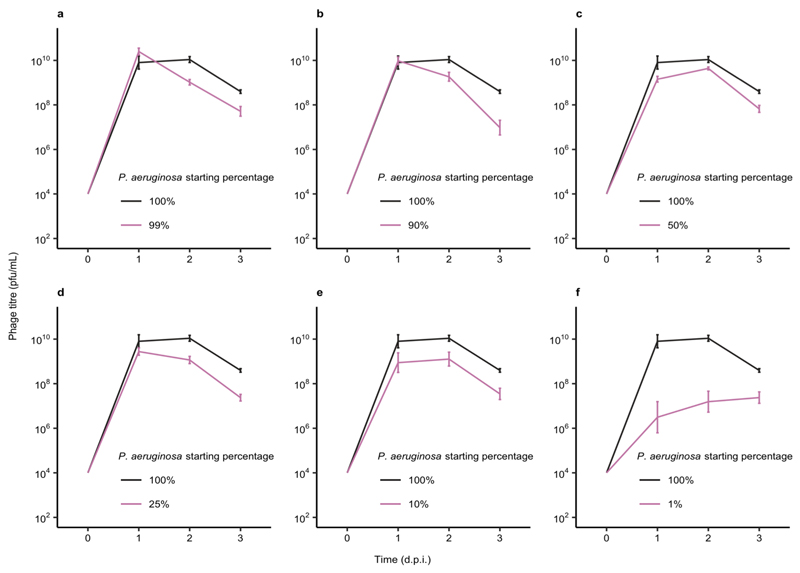
Extended Data Figure 3. Increased CRISPR-based resistance evolution across a range of microbial community compositions over time.
Proportion of P. aeruginosa that acquired surface modification (SM) or CRISPR-based immunity (or remained sensitive) at up to 3 days post infection (d.p.i.) with phage DMS3vir when grown either in monoculture (100%), or in polyculture mixtures consisting of the mixed microbial community but with varying starting percentages of P. aeruginosa based on volume. (6 replicates for most samples, with 24 colonies per replicate, n = 2784 biologically independent replicates). (a) Resistance evolution at 1 d.p.i. Error bars correspond to ± one standard error, with the mean as the measure of centre. Deviance test: Relationship between CRISPR and P. aeruginosa starting percentage at timepoint 1; Residual deviance(34, n = 41) = 4.42, p = 0.004; 1%; z = -3.27, p = 0.002; 10%; z = 1.21, p = 0.23; 25%; z = 1.62, p = 0.11; 50%; z = 2.20, p = 0.034; 90%; z = 2.07, p = 0.046; 99%; z = 0.47, p = 0.65; 100%; z = 1.47, p = 0.15. (b) Resistance evolution at 2 d.p.i. Error bars correspond to ± one standard error, with the mean as the measure of centre. Deviance test: Relationship between CRISPR and P. aeruginosa starting percentage at timepoint 2; Residual deviance(25, n = 32) = 3.86, p = 2.51 × 10-6; 1%; z = -2.14, p = 0.04; 10%; z = 1.19, p = 0.25; 25%; z = 2.07, p = 0.049; 50%; z = 1.89, p = 0.07; 90%; z = 1.12, p = 0.27; 99%; z = 1.21, p = 0.24; 100%; z = 1.11, p = 0.28. (c) Resistance evolution at 3 d.p.i. Error bars correspond to ± one standard error, with the mean as the measure of centre. Deviance test: Relationship between CRISPR and P. aeruginosa starting percentage at Timepoint 3; Residual deviance(35, n = 42) = 8.24, p = 0.0004; 1%; z = -3.38, p = 0.002; 10%; z = 2.12, p = 0.04; 25%; z = 2.77, p = 0.009; 50%; z = 3.07, p = 0.004; 90%; z = 2.46, p = 0.019; 99%; z = 1.55, p = 0.13; 100%; z = 0.87, p = 0.39.