Abstract
Progression of fatty liver to non-alcoholic steatohepatitis (NASH) is a rapidly growing health problem. Presence of inflammatory infiltrates in the liver and hepatocyte damage distinguish NASH from simple steatosis. However, the underlying molecular mechanisms involved in the development of NASH remain to be fully understood. Here we perform transcriptional and immune profiling of NASH patients before and after lifestyle intervention (LSI). Analysis of liver microarray data from a cohort of patients with histologically assessed NAFLD reveals a hepatic gene signature, which is associated with NASH and is sensitive to regression of NASH activity upon LSI independently of body weight loss. Enrichment analysis reveals the presence of immune-associated genes linked to inflammatory responses, antigen presentation and cytotoxic cells in the NASH-linked gene signature. In an independent cohort, NASH is also associated with alterations in blood immune cell populations, including conventional dendritic cells (cDC) type 1 and 2, and cytotoxic CD8 T cells. Lobular inflammation and ballooning are associated with the accumulation of CD8 T cells in the liver. Progression from simple steatosis to NASH in a mouse model of diet-driven NASH results in a comparable immune-related hepatic expression signature and the accumulation of intra-hepatic cDC and CD8 T cells. These results show that NASH, compared to normal liver or simple steatosis, is associated with a distinct hepatic immune-related gene signature, elevated hepatic CD8 T cells, and altered antigen-presenting and cytotoxic cells in blood. These findings expand our understanding of NASH and may identify potential targets for NASH therapy.
Non-alcoholic fatty liver disease (NAFLD) is the most common chronic liver disease. Its increasing prevalence is driven by high-calorie diets and sedentary lifestyles. Central obesity, insulin resistance (IR) and type 2 diabetes (T2D) are strong independent risk factors of NAFLD1,2. NAFLD is a histological continuum encompassing stages ranging from isolated steatosis (NAFL) to non-alcoholic steatohepatitis (NASH), characterized by lobular inflammatory infiltrates, hepatocyte ballooning and cell death, to fibrosis and ultimately cirrhosis3. NASH development results from complex interactions of metabolic and stress pathways in hepatocytes, initiated by chronic excessive lipid accumulation, with inflammatory processes driven by various immune cell populations, collectively inducing the histological picture of an active steatohepatitis1. Several lesions can be present, but lobular inflammation and ballooning are the most relevant histological markers of NASH and their combination is referred to as NASH activity, clearly distinguishing the activity of the steatohepatitis from the feature of steatosis4,5.
Ballooned hepatocytes are thought to be stressed and damaged cells (that lose their rectangular shape and swell due to cytoskeleton degeneration), possibly responding inadequately to pro-apoptotic and danger signals6.
Inflammatory infiltrates within the liver lobules are a hallmark of active steatohepatitis, and specialized immune populations, both resident and infiltrating, are linked with NASH7. Although certain circulating and hepatic immune populations have been associated with NASH8–10, a systematic and in-depth analysis of the cellular immune system in NASH is missing. Although current strategies to treat NASH target to reduce both lobular inflammation and ballooning11,12, the molecular mechanisms underlining these components of NASH are poorly understood. Previous studies mainly aimed to identify molecular pathways correlated with NASH versus no NASH12,13, without distinguishing steatosis from disease activity. Functional signatures associated with NASH and focusing on the activity that distinguishes NASH from steatosis as such remain unexplored. Transcriptional signatures of NASH and its activity can be identified by comparing liver transcriptomics from patients with histologically proven lobular inflammation and ballooning versus patients with simple steatosis as well as by longitudinally assessing regression of NASH upon lifestyle intervention (LSI) or bariatric surgery (BS), which can lead to NASH resolution14,15.
Using systems biology and experimental approaches, we set out to identify gene sets associated with NASH presence and activity at baseline and reversible in patients displaying reduced ballooning and lobular inflammation upon LSI. We identified a NASH transcriptomic signature strongly enriched in genes controlling immune inflammatory processes, antigen presentation and cytotoxic cells. We further show that NASH activity is associated with altered blood immune cell populations, including conventional dendritic cell (cDC) subsets and cytotoxic CD8 T cells. Moreover, in an obesity-driven mouse model of NASH exhibiting profound liver inflammation and hepatic damage, we also found increased hepatic expression of genes from the NASH transcriptomic signature, and altered hepatic populations of cDC and CD8 T cells. These results show that distinct immune cell populations play an important role in NASH activity and therefore constitute targets for NASH therapy.
Results
NASH associates with a hepatic immune-related gene module
Using transcriptome data16, we first searched for groups of genes with hepatic expression patterns linked with NASH in a large cohort of obese patients with or without histologically proven NASH (Supplementary Table 1). To this end, weighted gene co-expression network analysis (WGCNA), a method allowing identification of clusters (modules) of co-expressed genes with similar expression patterns in different experimental conditions, was used17. WGCNA of the approximately 30% most variable hepatic transcripts across patients identified 9 co-expressed gene modules (Fig. 1a,b and Supplementary Table 2). To assess whether these gene modules linked with NASH, we first analyzed alterations in the modules' expression in response to improvement in NASH activity in patients with NASH at baseline and in whom a liver biopsy was available 1 year after Roux-en-Y gastric bypass (RYGB) (n=21, Supplementary Table 3). In agreement with the reported effects of RYGB on NAFLD15, steatosis, lobular inflammation, and ballooning were significantly reduced in these patients at 1 year after RYGB (Supplementary Table 3). Interestingly, RYGB resulted in a significant alteration in transcriptional activity of 4 gene modules at follow-up compared to baseline (Fig. 1c). To decipher which gene modules are associated with common mechanisms of NASH regression, rather than with transcriptional alterations in the liver due to body weight loss, we tested these modules in LSI responders (patients with NASH at baseline who decreased lobular inflammation and/or ballooning at 1 year follow-up, for details see methods) and LSI non-responders in terms of NASH activity reduction, but who showed similar body weight loss at 1 year follow-up (Supplementary Table 4). Among the 4 gene modules affected by RYGB, only module “blue” displayed significant down-regulation at follow-up versus baseline in LSI responders, whereas its expression pattern hardly changed in non-responders (Fig. 1c-d). As overall gene expression levels in module “blue” were similarly decreased in RYGB patients and LSI responders, but not in LSI non-responders (Fig. 1c-d), this module is associated with reduction of NASH activity, independent of body weight changes. Indeed, the change in module blue expression was not correlated with % body weight (BW) change in either LSI or BS groups (Supplementary Fig 1b).
Figure 1.
Identification of hepatic transcriptomic signature of NASH. a. WGCNA performed with 11784 transcripts in the liver in patients with or without histologically proven NASH (n=155 patients, see Supplementary Table 1). Clustering of co-expressed genes in 9 gene modules. b. Number of transcripts in each gene module. c. Overall transcriptional regulation of gene modules upon RYGB (n=21 patients), LSI in responders (n=10 patients), and LSI in non-responders (n=10 patients) at one year follow-up compared to baseline (see Supplementary Table 3 and 4). P-values are calculated by mean-rank gene set test using geneSetTest function as described in detail in methods. d. Volcano plots of average log2 fold-changes versus P-values (paired moderated t-test using limma package) of all transcripts (gray dots) and transcripts from gene module “blue” (red dots) in RYGB patients (n=21 patients), LSI responders (n=10 patients), and LSI non-responders at one year follow-up compared to baseline. e. Top hallmark pathways enriched in gene module “blue” (n=786 transcripts), calculated using GSEA software as described in detail in methods. f. Venn diagrams with transcripts in gene module “blue” down-regulated (P<0.05 by moderated paired t-test using limma package) in three groups of patients at follow-up versus baseline, immune-related genes are shown. g. Spearman correlations between NASH activity index and hepatic expression levels of genes in gene module “blue” in the 155 patients at baseline, top genes with maximal positive correlation coefficients are shown. h. CXCL9, CXCL10, and LYZ expression (by microarray) in NAFL (n=22 patients) and NASH (n=106 patients) patients at baseline. Data are presented as median with 1st and 3rd quartiles as the box edges. *P<0.05, **P<0.01 by unpaired two-sided Mann–Whitney U test.
Module “blue” included 786 transcripts (Supplementary Table 2) and was significantly enriched for inflammation-related pathways, such as complement, TNFα and interleukin (IL)-6 signaling, as well as KRAS signaling, coagulation and apoptosis (Fig. 1e). Moreover, fasting plasma insulin and CRP levels were also associated with module “blue” expression, highlighting the close link between insulin resistance, systemic inflammation and NASH (Supplementary Fig 1c). Among the 786 transcripts in module “blue”, 507 were significantly down-regulated at 1 year follow-up, including 195 transcripts significantly down-regulated in both RYGB patients and LSI responders, but not in LSI non-responders, with other transcripts being preferentially decreased in RYGB patients (93 genes) or LSI responders (187 genes) (Fig. 1f). Most of these genes were related to immune system functions, including pro-inflammatory cytokines and chemokines, cytotoxic cells, infiltration of immune cells into tissues, and major histocompatibility complex (MHC) I and II antigen presentation (Fig. 1f). Interestingly, among key IFNα-responsive genes (IFIT1, OAS1, MIX1 and ISG15), only MX1 showed a modest, non-significant association with NAFLD severity at baseline and IFIT1 was significantly increased and ISG15 decreased only after RYGB (Supplementary Fig 1d-e).
To determine whether module “blue” is associated with NASH and its severity in terms of activity at baseline, we correlated the expression levels of its transcripts with the NASH activity index (AI, sum of lobular inflammation and ballooning score) in the 155 patients. Importantly, many genes from module “blue”, including immune-related genes, associated positively with AI (Fig. 1g), suggesting links with NASH disease activity. Moreover, some of these genes were also decreased in RYGB patients or LSI responders at 1 year follow-up (Fig. 1f). In addition, hepatic expression levels of some of these immune-related genes, for instance chemokine (C-X-C motif) ligand (CXCL) 9 and 10, and lysozyme (LYZ), were significantly higher in NASH versus NAFL patients at baseline (Fig. 1h). Taken together, these results indicate that NASH regression associates with the response of a specific gene module, containing multiple co-regulated genes involved in inflammation, antigen presentation, cytotoxic response, and activation of T cells.
Blood immune cells signatures of NASH
As the gene module “blue” identified the immune system as a key player in NASH regression upon LSI, we next assessed whether blood immune cells correlate with the presence of NASH and with its activity by performing a deep immunophenotyping analysis in patients without (n=17) or with NASH (n=21), stratified for T2D, a major risk factor for NASH (Supplementary Table 5). All patients were obese, with the highest BMI in the no-NASH T2D group. Patients with T2D were also older than patients without T2D. Although lobular inflammation was absent, the level of steatosis was already higher in the no-NASH T2D group compared to no-NASH no-T2D group and a few patients featured some ballooning (Supplementary Table 5), indicating that early stages of hepatocyte damage are already present in these patients.
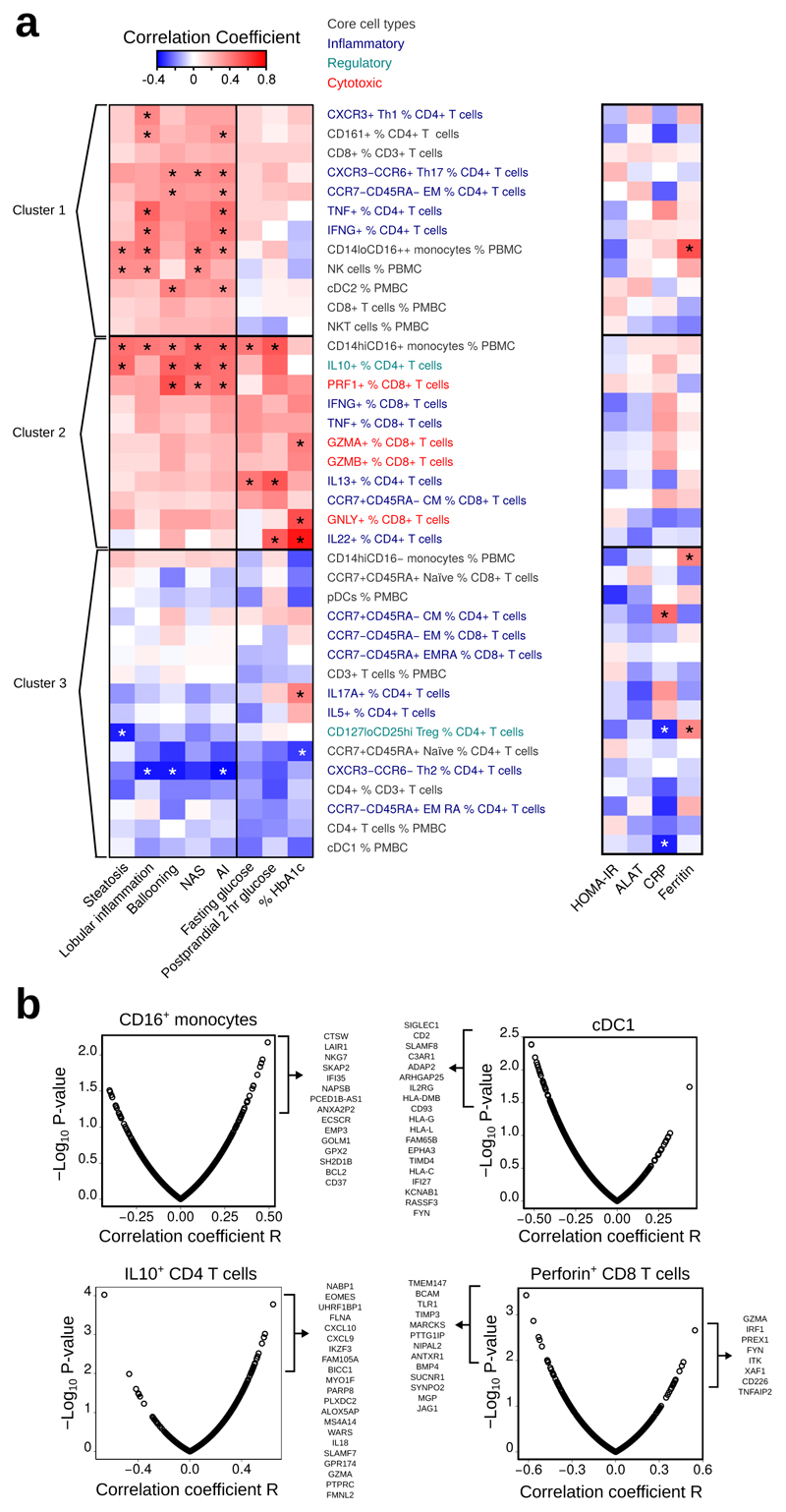
Flow cytometry analysis of blood immune populations was performed in these 38 patients and correlations assessed between the 39 measured immune cell populations and clinical parameters (Fig. 2a, Supplementary Fig. 2). Unsupervised hierarchical clustering grouped immune cell populations based on similarity of their correlation patterns with clinical parameters for NASH and glucose metabolism and yielded three main clusters. Cluster 1 associated with NASH but showed weak or absent correlations with T2D; Cluster 2 positively associated with NASH and T2D and Cluster 3 negatively correlated with most parameters specific for NASH and T2D (Fig. 2a). Within Cluster 1, NASH activity positively correlated with NK cells, atypical CD16++ monocytes and HLA-DR+CD123-CD11c+CD141- cells, similar to classical dendritic cells type 2 (cDC2). Likewise, Th1 lymphocytes and NKT cells were positively associated with lobular inflammation, ballooning and NASH AI. Immune cell populations within Cluster 2 were positively associated with lobular inflammation, ballooning, and with glucose or HbA1c levels, thus linking NASH activity and T2D (Fig. 2a). Cluster 2 included pro-inflammatory CD16+ monocytes and IL-10+ CD4 T lymphocytes, the latter likely reflecting a compensatory anti-inflammatory response to increased disease activity, as previously described in mice18. Interestingly, within Cluster 2 we also found activated and cytotoxic CD8 T lymphocytes correlating with NASH activity (Fig. 2a). CD8 T cells appear to be functionally linked to hepatic damage and inflammation in mouse models of NASH10,19,20, but have not been well studied in human NASH. Cluster 3 immune cell populations were mostly negatively associated with NASH and glucose parameters. Among them, are Th2 lymphocytes, including IL-5+ cells, as well as regulatory T cells (Treg). While cDC2 were positively associated with NASH (Cluster 1), both HLA-DR+CD123-CD11c+CD141+ cDC1 and HLA-DR+CD123+ plasmacytoid DC (pDC) were inversely correlated with NASH and glucose levels (Cluster 3). This indicates that pDC, cDC1, and cDC2 may play opposing roles in NASH. These results show that both NASH and T2D are associated with pronounced changes in blood immune cell populations, which may contribute to the high incidence of NASH in T2D.
Figure 2.
Correlations between blood immune cell populations, disease activity in NASH, and genes in module “blue”. a. Hierarchical clustering of correlation coefficients in 38 patients (see Supplementary Table 5) between proportions of blood immune cell populations and histological liver parameters (Spearman's correlation), T2D-associated parameters (Pearson correlation) and systemic inflammation markers (Pearson correlation). Asterisks indicate P< 0.05 for the given correlation. b. Pearson correlations between selected immune cell populations in blood from a subset of 29 patients (see Supplementary Table 5) and hepatic expression levels of genes from module “blue”. NAS: NAFLD Activity Score. AI: Activity Index
Blood immune populations correlate with liver gene signature
Analysis of hepatic transcriptome and blood immune cell populations suggest that interplay between antigen-presenting cells (likely cDC), CD4 T cells and cytotoxic lymphocytes is linked with the presence and activity of NASH. We thus probed for correlations between blood immune cell populations and expression levels of genes from the NASH transcriptomic signature (module “blue”) in the 28 patients with both liver microarray and blood flow cytometry data. CD16+ monocytes correlated with elevated expression of immune and stress genes from module “blue” (Fig. 2b). cDC1 and cDC2 oppositely linked with NASH activity (Fig. 2a) and cDC1, but not cDC2, displayed negative associations with hepatic expression of genes involved in immune regulation and antigen presentation (Fig. 2b and data not shown). Interestingly, elevated IL-10+ CD4 T cells were linked with increased hepatic expression of genes related to inflammation, chemotaxis, and cytotoxic responses (Fig.2b), again indicative of a possible compensatory increase of immunoregulatory IL-10 expression in CD4 T cells in NASH. Finally, we tested correlations between genes from module “blue” and cytotoxic CD8 T cells expressing perforin (strongly associated with NASH activity and T2D) (Fig. 2a,b). This immune cell population showed associations with hepatic genes related to cytotoxic and IFNγ responses (GZMA, CD226, IRF1), T helper differentiation (ITK), and TNFα signaling (TNFAIP2) (Fig. 2b). Taken together, these results show a remarkable similarity and associations between hepatic immune pathways predicted by gene module “blue” and the repertoire of NASH-associated immune populations in the blood. These data suggest a cross-talk between immune cells and hepatocytes, likely via cytokines, and direct infiltration of immune cells into the liver.
Diet-induced NASH drives increased liver cDC2 and CD8 T cells
Next, we assessed whether lifestyle-induced NASH leads to hepatic accumulation of the immune cell populations identified in the clinical study, such as cDC and cytotoxic cells. Since disease-inducing protocols are unethical and flow cytometric analysis of hepatic immune populations in humans is challenging due to the limited tissue access, we studied liver immune cells in a diet-induced model of murine NASH. Most published NASH models used to analyze hepatic immune cells do not develop obesity2 and hepatic immune populations have not yet been characterized in two published mouse models of obesity-associated NASH21,22. The diet-induced NASH model recapitulated the main known dietary drivers of this disease in humans23,24 and displayed hallmarks of well-established NASH, such as steatosis, hepatocyte damage, and lobular inflammatory infiltrates (Supplementary Fig. 3 and Supplementary Information). Flow cytometric analysis of mouse liver showed unaltered neutrophil population and decreased proportions of B lymphocytes and CLEC4F+ Kupffer cells, whereas inflammatory macrophages and monocytes were increased in livers of mice with NASH (Supplementary Fig. 4a-f). Hepatic NK, NKT, and CD4 T cells were not affected upon NASH induction, whereas hepatic FOXP3+ Treg cells tended to be elevated (Supplementary Fig. 4g-j).
Systems biology analysis in humans predicted an important role of hepatic antigen-presenting cDC and cytotoxic cells in NASH regression upon LSI (Fig. 1f). Interestingly, mouse CD172a+ cDC, phenotypically similar to human cDC2, were significantly increased in mouse livers upon NASH induction (Fig. 3a,b). In contrast, NASH induction in mice led to a decrease of hepatic XCR1+ cDC (analogous to human cDC1) (Fig. 3a,c), driving a marked decrease in the cDC1/cDC2 ratio (Fig. 3d). This shift in cDC populations was specific to the liver, as no changes were found in these cDC subsets in the spleen (data not shown). Moreover, the proportion of CD8 T cells was significantly increased in livers of mice with NASH (Fig. 3e,f), suggesting that hepatic accumulation of CD8 T cells may be linked with antigen presentation by CD172a+ cDC2. These results in a lifestyle-induced NASH model corroborate the hepatic NASH gene expression and blood immune signatures in humans and indicate that hepatic accumulation of CD8 T cells and disturbance in hepatic cDC are main immune hallmarks of NASH activity and progression.
Figure 3.
A diet-induced NASH alters cDC and CD8 T cells and inflammation in the liver. Male C57BL/6J mice were fed conventional diet (CD) or NASH-diet (ND) during 24 weeks (see Supplementary Information). a. Representative flow cytometry plots of cDC in the liver: proportions of XCR1+ and CD172a+ of total cDC are shown (n=8 mice CD; n=6 mice ND). b. CD172a+ cDC2 cells as proportion of CD45+ cells (n=8 mice CD; n=6 mice ND). c. XCR1+ cDC1 cells as proportion of CD45+ cells (n=8 mice CD; n=6 mice ND). d. Ratio of cDC1/cDC2 cells (n=8 mice CD; n=6 mice ND). e. Representative flow cytometry plots of TCRβ+ T cells in the liver: proportions of CD4+ and CD8+ of total TCRβ+ T cells are shown. f. Proportion of CD8+ T cells of CD45+ immune cells in the liver (n=9 mice CD; n=20 mice ND). g. qPCR analysis of inflammatory gene expression in mouse livers. (n=9 mice CD; n=20 mice ND). Data are shown as mean ± SEM. Statistical significance of differences between groups are analyzed by unpaired two-sided t-test (*P < 0.05, **P < 0.01, ***P < 0.001, NS – not significant).
Finally, we studied hepatic expression of inflammatory cytokines and chemokines in the diet-induced NASH model. NASH induction increased expression of TNFα, macrophage colony-stimulating factor (CSF)1, IL23A, IL33, C-C motif chemokine ligand (CCL)2, and CXCL10 (Fig. 3g), the latter four genes present in the gene expression signature of NASH responsive to intervention (Fig. 1f). IFNα-responsive genes Ifit1 and Oas1a were also induced in ND compared to CD fed mice (Supplementary Fig 3g). These data indicate that this diet-induced NASH model in mice affects the same hepatic immune populations and inflammatory pathways that are modified upon regression of NASH activity induced by LSI in humans.
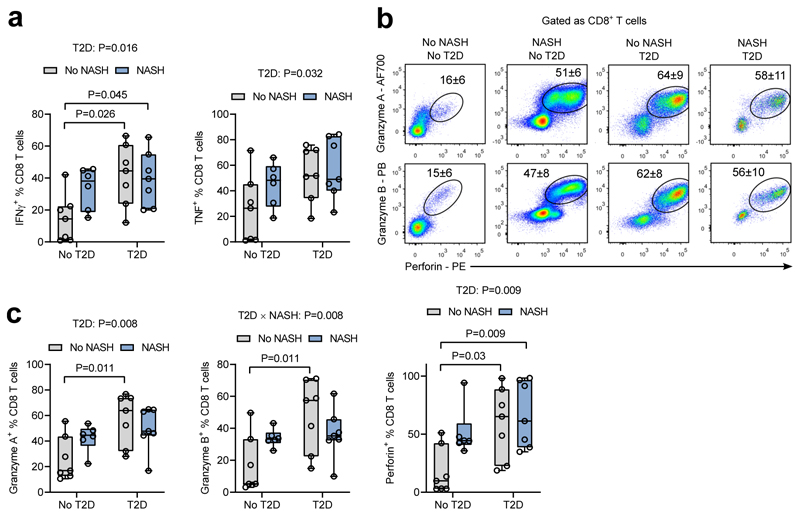
Activated CD8 T cells are elevated in NASH and T2D in humans
Given that both gene expression analysis in human livers and flow cytometric analysis in mouse livers revealed an association of cytotoxic CD8 T cells with obesity-related NASH, we next focused on these cells to better understand their potential role in NASH in humans. Indeed, both NASH and T2D enhanced the proportions of IFNγ+ and TNFα+ circulating CD8 T cells (Fig. 4a). Cytotoxic functions of CD8 T cells were also increased both in NASH and T2D as evidenced by increased expression of granzyme A and B, and perforin (Fig. 4b,c). Because T2D is a major risk factor for NASH, its effects towards CD8 T cells may link these diseases. Indeed, in the absence of NASH (no-NASH T2D group), T2D was associated with elevated expression of IFNγ, TNFα and cytotoxic molecules in CD8 T cells and some of these patients already displayed steatosis and ballooning. These results show that both NASH and T2D patients have a blood signature of increased populations of activated and cytotoxic CD8 T cells, which may link these diseases.
Figure 4.
NASH and T2D alter activity of cytotoxic CD8 T cells. a. Proportion of IFNγ+ and TNFα+ CD8 T lymphocytes in blood from patients with/without NASH and/or T2D (see Supplementary Table 5). Groups: n=7 patients no NASH no T2D, n=6 NASH no T2D, n=7 patients no NASH T2D, n=7 patients NASH T2D. Data are shown as median with 1st and 3rd quartiles. Statistical significance of differences between groups were analyzed by unpaired two-way ANOVA for effects of NASH and T2D followed by Tukey’s post-hoc test. b. Representative flow cytometric plots and c. proportions of perforin, granzyme A and B expression in blood CD8 T lymphocytes from patients. Groups: n=7 patients no NASH no T2D, n=6 NASH no T2D, n=7 patients no NASH T2D, n=7 patients NASH T2D. Data are shown as median with 1st and 3rd quartiles. Statistical significance of differences between groups were analyzed by moderated paired t-test or two-way ANOVA (for effects of NASH and T2D) followed by Tukey's post-hoc test.
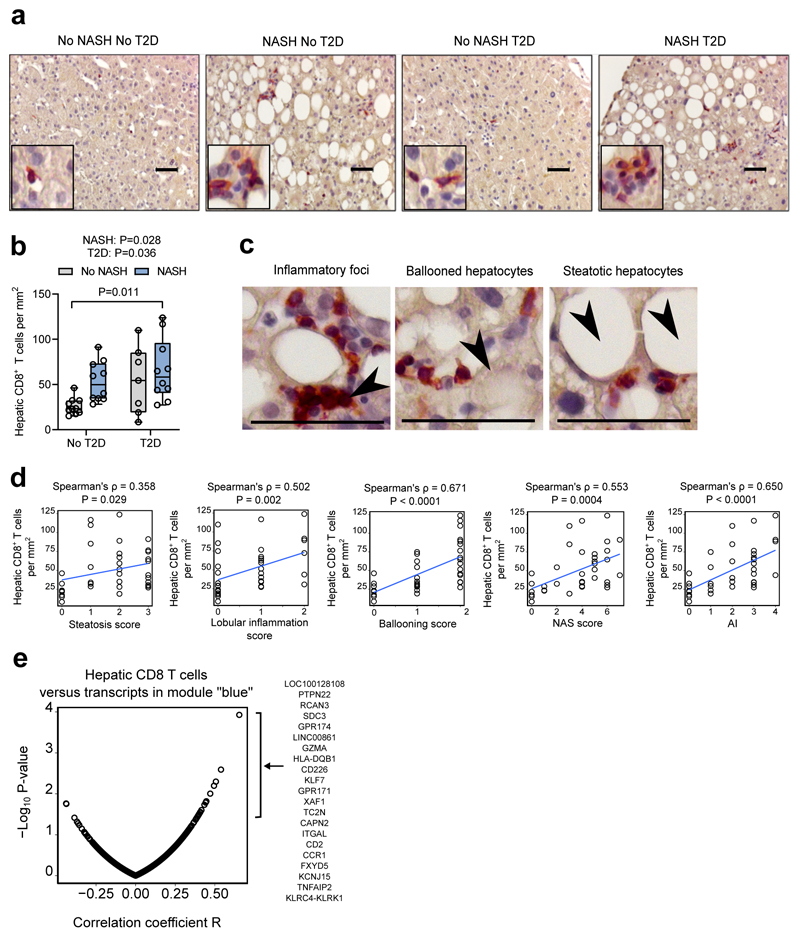
Hepatic CD8 T cells correlate with NASH in human liver
Finally, we investigated whether the increase of circulating cytotoxic CD8 T lymphocytes in obese NASH patients is accompanied by an accumulation of these cells in the liver. CD8-positive cells with lymphoid morphology were detected on sections from all groups of patients (Fig. 5a). While both NASH and T2D were both associated with significantly increased levels of CD8 T lymphocytes in the liver, no further increase was observed in NASH T2D patients (Fig. 5b). CD8 T lymphocytes localised within inflammatory foci in the parenchymal compartment in close proximity to steatotic and ballooned hepatocytes (Fig. 5c). Liver CD8 T lymphocyte numbers showed relatively little association with steatosis, but significantly correlated with lobular inflammation, ballooning, and AI (Fig. 5d). No significant correlation was found between liver CD8 T lymphocytes and glucose levels or HbA1c (data not shown), suggesting that the T2D-associated accumulation of CD8 T lymphocytes is not driven by dysfunctional glucose metabolism. Notably, liver CD8 T lymphocytes significantly correlated with blood CD8 T lymphocytes and with the subset of perforin-expressing CD8 T cells (Supplementary Fig. 5a). Hepatic CD8 T cells also significantly correlated with expression of perforin (PRF1) and granzyme A (GZMA) in the liver, two cytotoxic genes from the module “blue”, but not granzyme B (GZMB) and granulysin (GNLY) (Supplementary Fig, 5b). Finally, we found that hepatic CD8 T cells positively correlated with expression levels of multiple genes from module “blue”, including genes related to T cells (CD2, CD226), cytotoxic responses (KLRC4-KLRK1), and MHCII-mediated antigen presentation (HLA-DQB1) (Fig. 5e). Interestingly, the strongest correlation with hepatic CD8 T cells was observed for PTPN22 (protein tyrosine phosphatase, non-receptor type 22), which controls T cell receptor responsiveness and is associated with inflammatory and autoimmune diseases in humans25. Taken together, these results indicate that disease activity (lobular inflammation and ballooning) in NASH is associated with the accumulation of CD8 T lymphocytes in the liver. Moreover, hepatic CD8 T cells are linked to an increased expression of genes from the signature of active NASH. Thus, hepatic gene expression and immune signatures of NASH activity regression reveals CD8 T cells as a potential target to control hepatic inflammation, cytotoxic responses in the liver and NASH activity.
Figure 5.
Hepatic CD8 T lymphocytes correlate with lobular inflammation, ballooning, and transcriptomic signature of NASH. a. Representative immunostaining for CD8 (red) with haematoxylin counterstaining on liver biopsies from patients with/without NASH and/or T2D. b. Quantification of CD8-positive cells per mm2. No-NASH No-T2D n = 10, NASH No-T2D n = 10, No-NASH T2D n = 7, NASH T2D n = 9. Data are shown as median with 1st and 3rd quartiles. c. Localization of CD8 T lymphocytes (red) near immune infiltrates, steatosis, and ballooned hepatocytes (indicated by arrows) in the liver from NASH patient. Scale bar is 50μm. d. Correlations between hepatic CD8 T lymphocyte number and histological features in the liver (n=36). e. Pearson correlations and −log10 P-values between hepatic CD8 T lymphocyte and expression levels of gene from module “blue” (n=29, Supplementary Table 5). Statistical significance of differences between groups were analysed by unpaired two-way ANOVA (for effects of NASH and T2D) followed by Tukey's post-hoc test (*P < 0.05).
Discussion
The global epidemic of NASH is an important health problem, as effective pharmacological approaches to treat NASH are not yet available. LSI and BS are the best available strategies to reduce NASH activity in some patients14,15,26. Here, we identify a hepatic transcriptomic signature of NASH in humans distinct from NAFL and responsive to RYGB and LSI, suggesting a shared mechanism of NASH regression by these different weight loss methods. Interestingly, weight loss alone was apparently not sufficient to improve NASH in all patients as weight loss was similar in LSI responders and non-responders. The reversible transcriptomic signature of NASH is highly enriched by immune genes such as cytokines and chemokines and their receptors, genes involved in activation of lymphocytes by antigen-presenting cells, and genes linked to cytotoxic cells. Some of these genes have already been described as associated with NASH. For example, CD44, a molecule mediating leukocyte recruitment into the liver, plays a role in methionine-choline deficient dietary (MCD)-induced NASH in preclinical models27. Though the MCD model has obvious limitations in representing human NASH within the framework of the metabolic syndrome, it reproduces NASH in its hepatic phenotype and is therefore an interesting model to study intrahepatic changes in NASH. Furthermore, the same study also found elevated CD44 in human NASH biopsies. The pro-inflammatory cytokine CXCL10 also drives NASH in the MCD NASH model and is increased in blood from NASH patients28. Here we show that down-regulation of hepatic pro-inflammatory pathways accompanies regression of NASH activity upon intervention in patients. Moreover, these pathways increase upon lifestyle induction of NASH in a mouse model. Thus, the reversible transcriptomic signature of NASH reveals new biological targets for specific therapy of NASH focusing on its activity. While the present study focuses on weight-loss or lifestyle modification driven NASH improvement, effective pharmacological treatments could target this immune signature and act in synergy with other, metabolism-focused pathways. Because the transcriptomic analysis here was performed on total RNA from liver biopsies, it is conceivable that single-cell RNA analysis may reveal more subtle differences in hepatocyte vs non-parenchymal cell responses in NASH.
Increasing evidence demonstrates a close interaction between the immune system and metabolism in the development of NASH29. Our data are in line with recent publications on associations of different immune cells with NAFLD in humans8,9,30,31. However, this is the first study in which a global analysis of circulating immune cell populations in relation to the presence of NASH and its severity was performed. Integrative analysis revealed a species-conserved phenotype of NASH implicating antigen-presenting cDC sub-types and cytotoxic CD8 T cells. Whereas cDC1 are thought to present antigens to CD8+ T cells and cDC2 present antigens to CD4+ T cells32,33, we surprisingly found blood cDC1 correlate inversely with NASH activity and blood cDC2. Similarly, CD172a+ cDC2 replace XCR1+ cDC1 in mouse livers upon NASH induction. Moreover, elevated hepatic expression of multiple genes involved in antigen presentation in NASH is reversible upon intervention. In the MCD model, total cDC have been shown to protect against NASH34. However, whether these effects are mediated by cDC1 and/or cDC2 sub-populations is unclear. As cDC subtypes control pro-inflammatory and tolerogenic immune responses depending on the tissue environment35, a shift between cDC1 and cDC2 in mice and humans might be associated with elevated hepatic inflammation and hepatocyte damage in NASH. Interestingly, depletion of CD141+ cDC1 has been shown to occur in human liver upon inflammation36, suggesting an inflammation-mediated mechanism of suppression of cDC1 in relation to NASH activity. Among the hepatic myeloid populations identified in mice (i.e. Kupffer cells, monocyte-derived macrophages, DC, etc.), the present gating strategy was unable to exhaustively phenotype these clearly important immune populations. Further studies are thus necessary to better understand the evolution of the hepatic immune milieu and confirm our findings in human NASH biopsies.
Previous studies demonstrated associations of CD8 T cells with liver damage in mouse models of NASH10,19,20. We show that, in patients, circulating and hepatic cytotoxic CD8 T lymphocytes are significantly linked to histological hallmarks of NASH, such as lobular inflammation and ballooning. Moreover, CD8 T lymphocytes correlate with hepatic markers of inflammation and antigen presentation in NASH. Polymorphisms in one of these genes, HLA-DQB1, a MHCII haplotype, is associated with NAFLD37,38. Another gene, PTPN22, is an important regulator of T-cell receptor (TCR) signaling and linked with several autoimmune diseases25. Furthermore, in the lifestyle-induced NASH model, elevated hepatic CD8 T cells associated with CD172a+ cDC2, suggesting a possible role of antigen-presentation and TCR activation of CD8 T cells in NASH. CD8 T lymphocytes appear to contribute to insulin resistance in a mouse model of diet-induced obesity38. Similarly, hepatic CD8 T lymphocytes and type I IFN signaling promote glucose intolerance and insulin resistance in mice39,40. Interestingly, the reversible transcriptomic signature of NASH that we identified is enriched by cytokines and cytotoxic molecules in circulating CD8 T cells is linked to NASH activity and T2D in humans. Curiously, dysregulated glucose metabolism in T2D patients does not explain the increased numbers of CD8 T cells in the liver, suggesting that other factors contribute to the T2D-mediated accumulation of hepatic CD8 T cells. Moreover, in addition to elevated hepatic CD8 T cells, some T2D patients, without an unequivocal NASH diagnosis, already display ballooning. Together, these results suggest a role of CD8 T cells in the interplay between T2D and NASH. We found that the reversible transcriptomic signature of NASH is enriched by apoptosis pathway and NF-κB target genes (data not shown). These hepatic NF-κB target genes are down-regulated upon NASH activity reduction accompanied by decreasing ballooning. Ballooning is a poorly understood but essential hallmark of NASH, representing a specific form of “undead cells” with features of initiated but not resolved apoptosis6. Because the apoptotic machinery is controlled by the NF-κB signaling pathway41, down-regulation of the pathway upon LSI or BS might resolve apoptosis and thereby eliminate ballooned cells from the liver. Moreover, associations of the blood immune signature of NASH, particularly cytotoxic CD8 T cells, with ballooning suggest a cross-talk between the immune system and stressed hepatocytes in NASH. Although our results identify a pathway contributing to NASH resolution in a manner independent of body weight, it is clear that metabolic control through direct action on the liver or via extra-hepatic organs also contributes to NAFLD. Indeed, adipose tissue may directly contribute to NAFLD progression through systemic cytokine and immune signaling and drive hepatic injury in the context of obesity39,42,43.
Taken together, our study provides insights in the molecular mechanisms implicated in NASH and its regression upon intervention. In patients and a mouse model, we comprehensively and consistently show surprisingly pronounced associations of immune pathways and cell populations with NASH activity. These pathways, as well as the exact molecular targets revealed by this study, underline an important role of innate and acquired immunity in the development and severity of NASH, which may be targeted for treatment.
Materials and Methods
Patients
All patients were consecutively recruited at the Liver Clinic and Obesity Clinic of the Antwerp University Hospital and underwent hepatologic and metabolic work-ups. Blood analysis included blood cell count and white blood cell formula. Exclusion criteria were alcohol consumption >2 U/day for women and >3 U/day for men, liver diseases other than NAFLD, age <18 years, liver cirrhosis. For the baseline gene expression analysis in the liver, patients (n=155, including no liver disease n=27, simple steatosis n=22, NASH n=106) were selected from a cohort recruited since 200616. Selected obese NASH patients with paired biopsies at 1 year follow-up (LSI n=20, Roux-en-Y Gastric Bypass (RYGB) n=21) were included for gene expression analysis) (Figure S1). For immunophenotyping analysis, patients (n=38) were consecutively recruited between 2014 and 2016. Gene expression analysis was performed on a subset of these patients (n=29) who were included in the baseline gene expression analysis (Figure S1). The study protocol is part of the Hepadip protocol (Belgian registration number B30020071389) and was approved by the Ethical Committee of the Antwerp University Hospital (file 6/25/125). Written informed consent was obtained from all patients.
Selection of patients
Among patients visiting the Obesity Clinic at the Antwerp University Hospital recruited from 2006 to 2016, 155 patients with hepatic RNA microarray and clinical data available at baseline were enrolled16,44. Among the 155 patients, 41 obese non-diabetic patients with NASH at baseline were selected for comparative baseline and 1 year follow-up hepatic RNA microarray and clinical data analysis. Among these 41 patients with NASH at baseline, 31 patients displayed an improvement in disease activity, with decreases of lobular inflammation and/or ballooning, one year after RYGB (n=21) or LSI (n=10), whereas 10 patients did not improve one year after LSI. We defined the LSI patients with NASH at baseline who improved ballooning and/or lobular inflammation at 1 year follow-up as responders to and the LSI patients with NASH at baseline who did not improve ballooning and/or lobular inflammation at 1 year follow-up were classified as non-responders to intervention. In a second independent cohort, immune cell populations were analysed in consecutive patients with 4 clearly distinct phenotypes enrolled based on their metabolic (T2D) and histological (NASH) phenotype (total n=38): 17 patients without NASH in which some degree of simple steatosis was allowed (without (n=7) and with (n=10) T2D) and 21 patients with unequivocal NASH based on histological parameters (without (n=11) and with (n=10) T2D). The group without T2D and NASH consisted of a slightly lower number of patients, as liver biopsies were only performed on clinical indication of potential presence of NASH. High quality RNA appropriate for microarray analysis was obtained from liver biopsies in 29 out of 38 patients (13 patients without NASH and 16 patients with NASH).
Clinical assessment and biological measurements
Fasting blood glycaemia was analysed in the morning. A 2 hour oral glucose tolerance test (OGTT) (75 g of glucose), including insulin quantification, was performed. HOMA-IR was calculated45. BMI, glucose, and %HbA1c were measured46. Liver biopsies were performed for suspected NAFLD as indicated by elevated serum transaminase levels or a steatotic liver on ultrasound as described previously44.
Histological assessment of the liver biopsies
All liver biopsies were stained (haematoxylin-eosin, Sirius red, reticulin and Perls' iron stain) and scored by two expert pathologists blinded to all clinical information. Histological features of NAFLD (steatosis, ballooning, lobular inflammation and fibrosis) were assessed using the NASH Clinical Research Network Scoring System criteria47. The NAFLD Activity Score (NAS) was calculated as the sum of steatosis, lobular inflammation and ballooning scores. An activity index (AI) was also calculated as the sum of ballooning (range 0-2) and lobular inflammation (range 0-3) in line with recent observations of the distinct roles of steatosis versus activity of disease5. NASH was defined by the simultaneous presence of steatosis ≥1 AND ballooning ≥1 AND lobular inflammation ≥147. As outlined, the sum of ballooning and lobular inflammation was calculated and indicated as AI.
Mouse model of diet-induced NASH
Wild-type male C57BL/6J mice (8 weeks of age) were purchased from Charles River Laboratories (France). Mice were maintained in pathogen-free environment (12:12 hr light/dark cycle, 21°C-24°C) with ad libitum access to water and food. Alternatively, Foxp3-YFP reporter mice48 maintained on a C57BL/6J background were used to monitor hepatic Treg populations. Littermate animals were randomized by body weight prior to the start of the diet. No power calculations were performed to determine sample size. Mice were fed either a control diet (CD, standard rodent chow, 5% kcal fat) or a “NASH” diet (ND, 45% kcal fat, 40% kcal carbohydrate, 15% kcal protein with 1% (by weight) cholesterol; SAFE diets, Augy, France) for 24 weeks. All experiments were performed following approval by the Ethics Committee for Animal Experimentation from Nord-Pas de Calais Region (APAFIS#5746-2016040109244171 and APAFIS#7160-2017040313471173).
Histology and immunohistochemistry
Human liver biopsy sections were incubated for 1hr at 22°C with rabbit monoclonal anti-CD8a (SP16) (Thermo Fisher Scientific), followed by staining with a mouse anti-rabbit HRP-conjugated antibody, and revealed with Vector ImmPRESS HRP Reagent Kit and Vector NovaRED Substrate Kit. Sections were counterstained by haematoxylin. Mouse liver samples were fixed with 4% PFA, embedded in paraffin and stained with haematoxylin-eosin. Images were obtained by conventional microscopy using a Nikon Ti-U microscope.
Flow cytometry
EDTA blood samples from patients were collected and peripheral blood mononuclear cells (PBMC) isolated using Percoll. For cytokine staining, cells were incubated with 20ng/ml PMA, 1μg/ml ionomycin and 1 μg/ml brefeldin A in RPMI medium for 4hr at 37°C, 5% CO2. 1×106 PBMC were pre-incubated with Fc-block to minimize non-specific binding and labeled with conjugated antibodies CD3 (UCHT1, PE-CF594), CD4 (OKT4, APC-CY7), CD45RA (HI100, V500), CD56 (HCD56, BV605), CD8 (HIT8a, PE-CY7), CCR10 (6588-5, PE), CD11b (ICRF44, AF700), CD11c (3.9, PB), CD123 (6H6, PE-CY7), CD127 (A7R34, BV605), CD14 (HCD14, PerCP-CY5.5), CD141 (B-A35, FITC), CD16 (3G8, V500), CD161 (HP-3G10, PB), CD172a (15-414, APC), CXCR3 (G025H7, APC), CD19 (HIB19, PE-CF594), CCR6 (R6H1, PE), CD197/CCR7 (3D12, FITC), HLA-DR (G46-6, AF700 or APC-CY7), Perforin (dG9, PE), Granulysin (DH2, AF647), Granzyme A (CB9, AF700), Granzyme B (GB11, PB), TNF (MAb11, AF700), IFNγ (4S.B3, BV421), IL-10 (JES3-9D7, eFluor660), IL-17A (BL168, AF700), IL-22 (22URTI, eFluor660), IL-5 (TRFK5, APC). Intracellular staining was performed using the Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer’s instructions.
Immune cells were isolated from mouse liver, digested 45 min with collagenase D, using centrifugation with 30% Percoll. Cells were treated with Zombie UV to discriminate live and dead cells, incubated with Fc-block and labelled with conjugated antibodies: CD45 (BUV737, clone 104), CD11b (BUV395, clone M1/70), CCR2 (BV421, clone SA203G11), Ly6C (BV785, clone HK1.4), F4/80 (BV711, clone BM8), NK1.1 (AF700, clone PK136), CD4 (BV605, clone RM4-5), CD8a (BV510, clone 53-6.7), Ly6G (PE-Cy7, clone 1A8), IA/IE (BV650, clone M5/114.15.2), CD11c (APC-Cy7, clone N418), CD19 (PE-CF594, clone 1D3), TCRb (APC, clone H57-597). For myeloid cell staining, an additional panel was used with the following antibodies: CD45 (PE-CF594, clone 30-F11), CD172a (BUV395, clone P84), CCR2 (BV421, clone SA203G11), B220 (BV510, RA3-6B2), CD26 (BV605, clone H194-112), XCR1 (BV650, clone ZET), F4/80 (BV711, clone BM8), Ly6C (BV786, clone HK1.4), MHCII (FITC, clone M5/114.15.2), CD64 (PE, clone x54-5/7.1), CLEC4F (PE-Cy7, clone #370901), CD19 (APC, clone eBio1D3), CD3 (AF700, clone 500A2), NK1.1 (AF700, clone PK136), CD11c (APC-Cy7, clone N418). The CLEC4F antibody was coupled to PE-Cy7 using the Abcam Antibody Coupling Kit (ref: ab102903) and used at a final dilution of 1:100.
Flow cytometry analysis was performed on a BD LSRFortessa X-20 (Becton Dickinson). Results were acquired with the Diva software (Becton Dickinson) and analysed using FlowJo software (Tree Star). Additional details are provided in the Reporting Summary.
RNA extraction
For human samples, the entire biopsy was homogenized for RNA extraction, purification, and processing as described previously44. Total RNA was isolated from mouse liver using Trizol reagent and used for reverse transcription and real-time PCR. RNA from human liver biopsies was isolated using RNeasy Mini Kit (Qiagen) and used for microarray analysis. Quantification and RNA integrity number (RIN) was tested using the Agilent 2100 Bioanalyzer System. Only RNA samples with RIN≥6 were used for microarray analysis.
Reverse transcription and real-time PCR
Total RNA (500 ng) isolated from mouse liver was treated with DNAse I and used to generate cDNA with High-capacity cDNA reverse transcription kit. Gene expression was measured by SybrGreen based real-time PCR. Results were normalized to the normalization factor calculated as average expression of housekeeping genes Ppia and Tbp, and the ΔΔCt method was used for all real-time PCR analyses. Primers sequences are provided in Supplementary Table 6.
Microarray analysis
Transcriptome analysis was performed with the Affymetrix GeneChip HuGene 2.0 ST arrays16,49. All liquid handling procedures were performed on a GeneChip Fluidic Station 450. GeneChips were scanned with a GeneChip Scanner 3000-7G (Affymetrix) using Command Console v4.1.2. Quality controls were performed using the Affymetrix expression console.
Microarray data processing and WGCNA
Microarray data were normalized by the robust multi-average (RMA) method50 using oligo/Bioconductor and corrected for batch effects using SVA/Bioconductor R packages51. 38598 annotated transcripts were selected for analysis. 11784 transcripts with maximal variability across all patients at baseline (n=155) based on median absolute deviation were selected for WGCNA and tested using the WGCNA R package17. Biweight midcorrelations and weighted adjacency matrix were calculated using the power 11 selected based on the scale-free topology fit model. Gene modules were identified using the "hybrid" method, and parameters deepSplit=4 and mergeCutHeight=0.15. For creation of volcano plots, log10 P-values and log2 fold changes were calculated using the limma R package52. Selected sets of transcripts of gene modules were tested for differential expression with geneSetTest from the limma R package53. Gene Expression Omnibus (GEO) repository accession number for microarray data is GSE106737. Gene set enrichment analysis of gene modules was performed using GSEA software (http://software.broadinstitute.org/gsea/).
Statistical analysis
No statistical method was used to predetermine the sample size. For animal studies, mice were randomized by body weight prior to dietary challenge and no blinding was performed for subsequent analysis. For comparison in patients at baseline and one year after BS or LSI, the paired moderated t-test or the paired Mann-Whitney U-test was used. For comparison of the four groups of patients, data were analysed by two-way ANOVA, using NASH and T2D as factors, followed by Tukey's post-hoc test for multiple comparisons. For histological quantification of hepatic CD8+ cells, the observer was blinded from the clinical parameters of each patient. Parametric Pearson correlation (continuous data) or non-parametric Spearman's rank-order correlation (categorical data) and hierarchical clustering were performed using R. Gene networks were visualized using Cytoscape. P-values <0.05 were considered significant. P-values were adjusted using Benjamini-Hochberg procedure. Statistical analyses were performed with Prism 6 (GraphPad Software, Inc.) or R version 3.4.4.
Supplementary Material
Acknowledgements and Funding
This work was supported by grants from the ANR and the European Union: EGID ANR-10-LABX-46 & Fondation Leducq LEAN 16CVD01 (to BS, DD, PL), ANR-18 NASHILCCD8 (to BS, DD) FP6 HEPADIP LSHM-CT-2005-018734 (to BS, AV, LVG, and SF) and FP7-HEALTH RESOLVE 305707 (to BS, AV, LVG and SF). SF is recipient of Flanders Fund for Scientific Research (FWO klinisch mandaat 1802154N). BS is a recipient of an Advanced European Research Council (ERC) grant (number 694717). JTH was supported by an EMBO Long Term Fellowship (ALTF277-2014).
Footnotes
Data and materials availability: Microarray data used in this study – GSE106737. Requests for other data should be made to the corresponding author.
Author contributions: LV and SF collected human biopsies, histological and biochemical data. LV, SF, LVG and AV supervised human biopsies collection and analysis. AnD performed the histology of the liver biopsies. JTH and DAM performed mouse experiments, flow cytometry, immunological and transcriptomic analysis, and WGCNA. SebF performed immunohistochemistry. BD, HD, CG, PL performed microarray analysis. AuD, AN, EW, LDG and SP performed mouse experiments and flow cytometry. DAM, LV, JTH, SF, BS and DD, conceived the study, interpreted data and wrote the manuscript. DAM, LV and JTH are joint first authors who equally contributed to the work. BS, SF, and DD are senior authors who equally contributed to the work. LV and DD are joint corresponding authors.
Competing interests: BS and SF are consultants for Genfit S.A. SF and LV are consultants for Inventiva. All other authors have nothing to declare.
References
- 1.Haas JT, Francque S, Staels B. Pathophysiology and Mechanisms of Nonalcoholic Fatty Liver Disease. Annu Rev Physiol. 2016;78:181–205. doi: 10.1146/annurev-physiol-021115-105331. [DOI] [PubMed] [Google Scholar]
- 2.Luyckx FH, Lefebvre PJ, Scheen AJ. Non-alcoholic steatohepatitis: association with obesity and insulin resistance, and influence of weight loss. Diabetes Metab. 2000;26:98–106. [PubMed] [Google Scholar]
- 3.Brunt EM. Pathology of nonalcoholic fatty liver disease. Nat Rev Gastroenterol Hepatol. 2010;7:195–203. doi: 10.1038/nrgastro.2010.21. [DOI] [PubMed] [Google Scholar]
- 4.Brunt EM, et al. Nonalcoholic fatty liver disease (NAFLD) activity score and the histopathologic diagnosis in NAFLD: distinct clinicopathologic meanings. Hepatology. 2011;53:810–820. doi: 10.1002/hep.24127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bedossa P, et al. Histopathological algorithm and scoring system for evaluation of liver lesions in morbidly obese patients. Hepatology. 2012;56:1751–1759. doi: 10.1002/hep.25889. [DOI] [PubMed] [Google Scholar]
- 6.Hirsova P, Gores GJ. Ballooned hepatocytes, undead cells, sonic hedgehog, and vitamin E: therapeutic implications for nonalcoholic steatohepatitis. Hepatology. 2015;61:15–17. doi: 10.1002/hep.27279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vonghia L, Michielsen P, Francque S. Immunological mechanisms in the pathophysiology of non-alcoholic steatohepatitis. Int J Mol Sci. 2013;14:19867–19890. doi: 10.3390/ijms141019867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liaskou E, et al. Monocyte subsets in human liver disease show distinct phenotypic and functional characteristics. Hepatology. 2013;57:385–398. doi: 10.1002/hep.26016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rau M, et al. Progression from Nonalcoholic Fatty Liver to Nonalcoholic Steatohepatitis Is Marked by a Higher Frequency of Th17 Cells in the Liver and an Increased Th17/Resting Regulatory T Cell Ratio in Peripheral Blood and in the Liver. J Immunol. 2016;196:97–105. doi: 10.4049/jimmunol.1501175. [DOI] [PubMed] [Google Scholar]
- 10.Wolf MJ, et al. Metabolic activation of intrahepatic CD8+ T cells and NKT cells causes nonalcoholic steatohepatitis and liver cancer via cross-talk with hepatocytes. Cancer Cell. 2014;26:549–564. doi: 10.1016/j.ccell.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Kleiner DE, Bedossa P. Liver histology and clinical trials for nonalcoholic steatohepatitis-perspectives from 2 pathologists. Gastroenterology. 2015;149:1305–1308. doi: 10.1053/j.gastro.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Ryaboshapkina M, Hammar M. Human hepatic gene expression signature of non-alcoholic fatty liver disease progression, a meta-analysis. Sci Rep. 2017;7 doi: 10.1038/s41598-017-10930-w. 12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moylan CA, et al. Hepatic gene expression profiles differentiate presymptomatic patients with mild versus severe nonalcoholic fatty liver disease. Hepatology. 2014;59:471–482. doi: 10.1002/hep.26661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vilar-Gomez E, et al. Weight Loss Through Lifestyle Modification Significantly Reduces Features of Nonalcoholic Steatohepatitis. Gastroenterology. 2015;149:367–378 e365. doi: 10.1053/j.gastro.2015.04.005. quiz e314-365. [DOI] [PubMed] [Google Scholar]
- 15.Lassailly G, et al. Bariatric Surgery Reduces Features of Nonalcoholic Steatohepatitis in Morbidly Obese Patients. Gastroenterology. 2015;149:379–388; quiz e315-376. doi: 10.1053/j.gastro.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 16.Lefebvre P, et al. Interspecies NASH disease activity whole-genome profiling identifies a fibrogenic role of PPARalpha-regulated dermatopontin. JCI Insight. 2017;2 doi: 10.1172/jci.insight.92264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vonghia L, et al. CD4+ROR gamma t++ and Tregs in a Mouse Model of Diet-Induced Nonalcoholic Steatohepatitis. Mediators Inflamm. 2015 doi: 10.1155/2015/239623(2015). 239623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattacharjee J, et al. Hepatic Natural Killer T-cell and CD8+ T-cell Signatures in Mice with Nonalcoholic Steatohepatitis. Hepatol Commun. 2017;1:299–310. doi: 10.1002/hep4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sutti S, et al. Adaptive immune responses triggered by oxidative stress contribute to hepatic inflammation in NASH. Hepatology. 2014;59:886–897. doi: 10.1002/hep.26749. [DOI] [PubMed] [Google Scholar]
- 21.Clapper JR, et al. Diet-induced mouse model of fatty liver disease and nonalcoholic steatohepatitis reflecting clinical disease progression and methods of assessment. Am J Physiol Gastrointest Liver Physiol. 2013;305:G483–495. doi: 10.1152/ajpgi.00079.2013. [DOI] [PubMed] [Google Scholar]
- 22.Asgharpour A, et al. A diet-induced animal model of non-alcoholic fatty liver disease and hepatocellular cancer. J Hepatol. 2016;65:579–588. doi: 10.1016/j.jhep.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen T, et al. Fructose and sugar: A major mediator of non-alcoholic fatty liver disease. J Hepatol. 2018;68:1063–1075. doi: 10.1016/j.jhep.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ioannou GN. The Role of Cholesterol in the Pathogenesis of NASH. Trends Endocrinol Metab. 2016;27:84–95. doi: 10.1016/j.tem.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 25.Bottini N, Peterson EJ. Tyrosine phosphatase PTPN22: multifunctional regulator of immune signaling, development, and disease. Annu Rev Immunol. 2014;32:83–119. doi: 10.1146/annurev-immunol-032713-120249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Froylich D, et al. Effect of Roux-en-Y gastric bypass and sleeve gastrectomy on nonalcoholic fatty liver disease: a comparative study. Surg Obes Relat Dis. 2016;12:127–131. doi: 10.1016/j.soard.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Patouraux S, et al. CD44 is a key player in non-alcoholic steatohepatitis. J Hepatol. 2017;67:328–338. doi: 10.1016/j.jhep.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, et al. CXCL10 plays a key role as an inflammatory mediator and a non-invasive biomarker of non-alcoholic steatohepatitis. J Hepatol. 2014;61:1365–1375. doi: 10.1016/j.jhep.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 29.Tilg H, Moschen AR, Roden M. NAFLD and diabetes mellitus. Nat Rev Gastroenterol Hepatol. 2017;14:32–42. doi: 10.1038/nrgastro.2016.147. [DOI] [PubMed] [Google Scholar]
- 30.Syn WK, et al. Accumulation of natural killer T cells in progressive nonalcoholic fatty liver disease. Hepatology. 2010;51:1998–2007. doi: 10.1002/hep.23599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miura K, Yang L, van Rooijen N, Ohnishi H, Seki E. Hepatic recruitment of macrophages promotes nonalcoholic steatohepatitis through CCR2. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1310–1321. doi: 10.1152/ajpgi.00365.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durai V, Murphy KM. Functions of Murine Dendritic Cells. Immunity. 2016;45:719–736. doi: 10.1016/j.immuni.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vu Manh TP, Bertho N, Hosmalin A, Schwartz-Cornil I, Dalod M. Investigating Evolutionary Conservation of Dendritic Cell Subset Identity and Functions. Front Immunol. 2015;6:260. doi: 10.3389/fimmu.2015.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henning JR, et al. Dendritic cells limit fibroinflammatory injury in nonalcoholic steatohepatitis in mice. Hepatology. 2013;58:589–602. doi: 10.1002/hep.26267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Worbs T, Hammerschmidt SI, Forster R. Dendritic cell migration in health and disease. Nat Rev Immunol. 2017;17:30–48. doi: 10.1038/nri.2016.116. [DOI] [PubMed] [Google Scholar]
- 36.Kelly A, et al. CD141(+) myeloid dendritic cells are enriched in healthy human liver. J Hepatol. 2014;60:135–142. doi: 10.1016/j.jhep.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Doganay L, et al. HLA DQB1 alleles are related with nonalcoholic fatty liver disease. Mol Biol Rep. 2014;41:7937–7943. doi: 10.1007/s11033-014-3688-2. [DOI] [PubMed] [Google Scholar]
- 38.Nishimura S, et al. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med. 2009;15:914–920. doi: 10.1038/nm.1964. [DOI] [PubMed] [Google Scholar]
- 39.Wieser V, et al. Adipose type I interferon signalling protects against metabolic dysfunction. Gut. 2018;67:157–165. doi: 10.1136/gutjnl-2016-313155. [DOI] [PubMed] [Google Scholar]
- 40.Ghazarian M, et al. Type I Interferon Responses Drive Intrahepatic T cells to Promote Metabolic Syndrome. Sci Immunol. 2017;2 doi: 10.1126/sciimmunol.aai7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo JL, Kamata H, Karin M. IKK/NF-kappaB signaling: balancing life and death--a new approach to cancer therapy. J Clin Invest. 2005;115:2625–2632. doi: 10.1172/JCI26322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.du Plessis J, et al. Association of Adipose Tissue Inflammation With Histologic Severity of Nonalcoholic Fatty Liver Disease. Gastroenterology. 2015;149:635–648 e614. doi: 10.1053/j.gastro.2015.05.044. [DOI] [PubMed] [Google Scholar]
- 43.Bijnen M, et al. Adipose tissue macrophages induce hepatic neutrophil recruitment and macrophage accumulation in mice. Gut. 2018;67:1317–1327. doi: 10.1136/gutjnl-2016-313654. [DOI] [PubMed] [Google Scholar]
- 44.Francque S, et al. PPARalpha gene expression correlates with severity and histological treatment response in patients with non-alcoholic steatohepatitis. J Hepatol. 2015;63:164–173. doi: 10.1016/j.jhep.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 45.Matthews DR, et al. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 46.Verrijken A, et al. A gene variant of PNPLA3, but not of APOC3, is associated with histological parameters of NAFLD in an obese population. Obesity (Silver Spring) 2013;21:2138–2145. doi: 10.1002/oby.20366. [DOI] [PubMed] [Google Scholar]
- 47.Kleiner DE, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 48.Rubtsov YP, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 49.Pawlak M, et al. The transrepressive activity of Pparalpha is necessary and sufficient to prevent liver fibrosis. Hepatology. 2014 doi: 10.1002/hep.27297. [DOI] [PubMed] [Google Scholar]
- 50.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 51.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 53.Goeman JJ, Buhlmann P. Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics. 2007;23:980–987. doi: 10.1093/bioinformatics/btm051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.