Abstract
Cellular behavior is controlled by the interplay of diverse biomolecules. Most experimental methods, however, can monitor only a single molecule class or reaction type at a time. We developed an in vitro Nuclear Magnetic Resonance spectroscopy (NMR) approach, which permitted dynamic quantification of an entire “heterotypic” network – simultaneously monitoring three distinct molecule classes (metabolites, proteins, RNA) and all elementary reaction types (bimolecular interactions, catalysis, unimolecular changes). Focusing on an 8-reaction co-transcriptional RNA folding network, in a single sample we recorded over 35 time-points with over 170 observables each, and accurately determined 5 core reaction constants in multiplex. This reconstruction revealed unexpected cross-talk between the different reactions. We further observed dynamic phase-separation in a system of five distinct RNA binding domains in the course of the RNA transcription reaction. Our Systems NMR approach provides a deeper understanding of biological network dynamics by combining the dynamic resolution of biochemical assays and the multiplexing ability of “omics”.
Introduction
The regulation of cellular behavior is complex, and emerges from the dynamic interplay of diverse biomolecules, including proteins, RNA and metabolites. Most experimental methods, however, can monitor only a single molecule class or reaction type at a time (catalysis, bimolecular interactions, unimolecular state changes), limiting our ability to measure complex cellular dynamics.
Furthermore, studies of networks often face a choice between biochemical methods – measuring dynamic (e.g. time-resolved) data only for a few network components, and "omics" methods – measuring a large number of components, but usually with little dynamic information in a single sample 1. This lack of dynamic data covering multiple network components is among the main limitations 2–4 in developing validated mechanistic, mathematical models for cellular networks, which are key to understand the underlying logic of these networks 5.
To address the above challenges, we sought to devise a Nuclear Magnetic Resonance spectroscopy (NMR)-based approach which would (i) allow to monitor “heterotypic” networks and pathways – involving different molecule or reaction types – entirely, in a single in vitro sample; and (ii) provide quantitative dynamic data for modeling of the network mechanisms. With certain limitations on molecule size and concentration (≲ 50-100 kDa, ≳ 10-50 µM) 6,7, solution NMR can monitor any reaction type or molecular class in a wide range of conditions, including unfractionated cell extracts and living cells 8. The use of NMR to monitor reactions is common in chemistry 9, and in recent years, NMR has also been used to follow the dynamics of small-scale reaction networks in biology. However, those studies focused on individual molecule classes, i.e. metabolites 10–14, proteins 13,15,16, or RNA 17–19.
We sought to monitor a more complex network by NMR – that comprises a wide range of different molecule and reaction types. Co-transcriptional RNA folding is an important cellular process which simultaneously involves RNA, proteins and metabolites, and is still poorly understood. RNA molecules of the same sequence may form distinct folded structures, with distinct functions and fates, depending on the effectors present during RNA transcription 20,21. Insights are still limited on how the final RNA structures are influenced by co-transcriptional interactions of the transcribing RNA. The core reactions of the underlying network are RNA synthesis from metabolites, RNA folding and protein-RNA interactions (Fig. 1a). Our aims were (i) to design an assay to monitor all main components of the network simultaneously by NMR spectroscopy, using specific signatures of different molecules in NMR spectra; (ii) to establish a mathematical model explaining our observations; and (iii) to perturb the network with proteins and drug molecules to gain system-level insight into its dynamics. This assay revealed competitive weakening of specific hnRNP A1 protein-RNA interactions by unspecific nucleotide-bearing molecules, and exposed the dynamic phase-separation of proteins in the course of the RNA transcription. We termed this methodological approach "Systems NMR" – a potential generic name for NMR-driven reconstruction of biomolecular reaction networks.
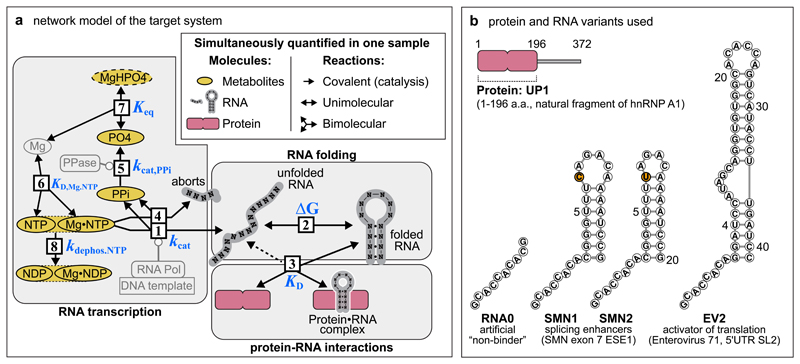
Figure 1. Molecules and reactions of the target co-transcriptional network.
(a) Organization of reactions in the reconstructed network: [1] RNA transcription, [2] RNA folding, [3] binding of regulator protein to RNA (dashed line indicates that protein can also bind to the unfolded RNA), [4] formation of RNA aborts, [5] hydrolysis of pyrophosphate (PPi), [6] formation of Mg NTP (nucleotide triphosphate) complexes, [7] formation of MgHPO4 salt aggregates, [8] dephosphorylation of NTPs to NDPs (nucleotide diphosphates. merged ovals indicate that NMR observables of free and Mg-bound nucleotide-phosphates, e.g. NTP and MgNTP, cannot be separately discriminated). PO4 refers ambiguously to multiple states of phosphate ion, including PO43– and its protonated forms. Indirectly observed MgHPO4 species is shown with dashed contour. Non-observable species – Mg, pyrophosphatase (PPase), RNA Polymerase (abbreviated with RNA Pol), DNA template – are shaded. Metabolites, proteins and RNA are shown in ochre, rose and grey respectively.
(b) Protein and RNA variants used in Systems NMR experiments. To make the products of the abortive RNA transcription uniform in all constructs (panel (a), reaction 4), the hairpin RNAs include a non-native single-stranded 5’ overhang matching the sequence of the control RNA0. The hairpins include two non-native closing GC pairs to offset the instability caused by the 5’-single-stranded overhang. The single C>U mutation between SMN1 and SMN2 RNAs is highlighted in orange. Abbreviations, ESE1 – exonic splicing enhancer 1, UTR - untranslated region, SL2 – stem-loop 2.
Monitoring a co-transcriptional RNA folding network
We first sought to investigate if the RNA binding protein UP1 (a natural fragment of hnRNP A1) would perturb the co-transcriptional folding of three cognate RNA hairpins of this protein: SMN1, SMN2 – two hairpins present in exon 7 of human SMN1 and SMN2 genes respectively 22, and the stem loop II (EV2) of the IRES of Enterovirus 71 23 (Fig. 1b). To distinguish the RNA-specific UP1 perturbations from other changes in the network dynamics (pH, nucleotide tri-phosphate concentrations), a fourth, “non-binding” RNA0 (RNA zero) was designed and tested as a control (Fig. 1b and Methods).
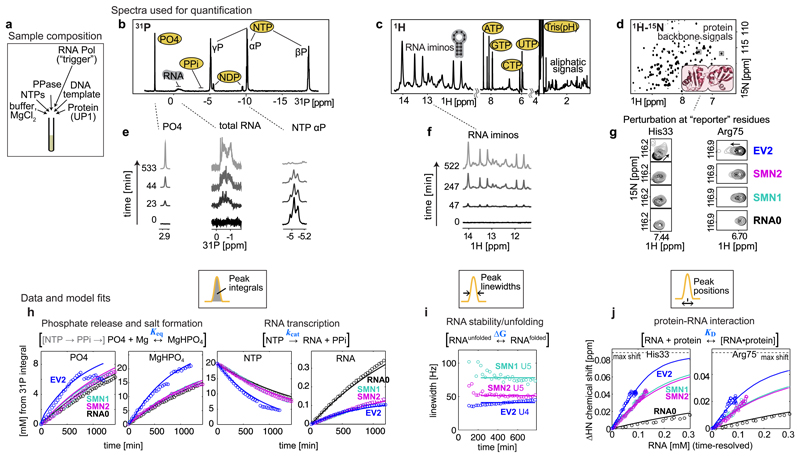
In our experiments, the DNA template, the nucleotide tri-phosphates (NTPs), MgCl2, the pyrophosphatase and the RNA-binding protein are initially mixed in an NMR tube (Fig. 2a), and then transcription is triggered by addition of the T7 RNA Polymerase. The reaction network (Fig. 1a) is subsequently monitored for ~20 hours by repeating several NMR experiments (Fig. 2b,c,d and Supplementary Video 1): 1D (one-dimensional) 31P – to monitor the levels of metabolites and RNA, 1D 1H – to monitor RNA folding, and 2D (two-dimensional) 1H-15N – to monitor protein interactions. Each set of measurements takes ~30 minutes to record, yielding an overall dataset of ~120-160 NMR spectra with ~40 time-points for each individual spectrum type. The combined number of resolved quantifiable NMR signals at each time point exceeded 170: 8 in the 31P spectrum (Fig. 2b), more than 20 in the 1H spectrum (Fig. 2c), and over 150 protein backbone amide signals in the 2D 1H-15N spectrum (Fig. 2d). For quantitative modelling of the target network 10 signals were used (Supplementary Table 1): the 31P signals of inorganic phosphate (PO4 – referring to PO43– and its protonated forms), RNA phosphate, α, β and γ-phosphate of the NTP, α and β-phosphate of NDP (Fig. 2b,e); the 1H imino signals of RNA uracils U5 (SMN1 and SMN2) or U4 (EV2) (Fig. 2f); and the 1H-15N signals of the selected UP1 residues reporting on RNA binding – His33 and Arg75 (Fig. 2g). In this study, these 10 signals were sufficient to quantify the key parameters of the target 8-reaction network. The data from remaining signals can still be used in subsequent studies to investigate the system in more detail. E.g. to analyse individual conversion rates of four NTPs, or to analyse RNA and protein perturbations not just via overall reaction constants, but with residue-level resolution.
Figure 2. NMR observables, data and mathematical model fits used for network parameterization.
(a) Sample composition, 450 µL NMR tube with 150 µM UP1 protein, 33 nM T7 RNA Polymerase, and 20 mM starting NTPs, from which 0.1-0.4 mM RNA is synthesized by the end of transcription. (b,c,d) Full spectra recorded in the NMR assay. (e,f,g) Selected, time-resolved, spectra regions used for network quantification. From 1D 31P spectra (b,e) the PO4, RNA, PPi, NDPs and NTPs species are quantified using integrals of the corresponding signals. From 1D 1H spectra (c,f) the folded RNA is quantified using linewidths of imino proton signals. 2D 1H-15N spectra (d,g) show a “fingerprint” of all amino acid 1H-15N moieties in the protein, and the positions of selected reporter residues at the RNA binding interface are used to quantify protein-RNA interaction. Experiments were repeated at least three times independently, using different batches of protein and/or DNA template, with similar results (n = 3 (RNA0, SMN1), n = 4 (SMN2, EV2).
(h,i,j) Time-resolved quantified observables used for network parameterization (circles), and the resulting model fits (solid lines). (h) Integrals from 31P spectra converted into mM concentrations using the starting 20 mM NTPs as calibration reference. Abrupt intensity jumps in free PO4 and MgHPO4 are discussed in main text. (i) Linewidths of U5 imino signals in SMN RNAs and U4 in EV2. (j) Chemical shift perturbations of His33 and Arg75 residues of UP1 protein plotted against time-resolved RNA concentration. Time-resolved animation of exemplary data is shown in Supplementary Video 1.
Among the key features of NMR is the intrinsically quantitative nature of the observed signals, which permits direct determination of certain physico-chemical molecular properties with little or no calibrations. To quantify the metabolite and RNA concentrations, we measured the integrals of corresponding signals in 31P spectra (Fig. 2h). Linewidths of NMR signals combine information about the molecule size (tumbling rate) and dynamics (lifetime) of molecular states, we therefore measured the linewidths of the well-separated imino signals of the folded RNA in 1H spectra to quantify the RNA stability (Ura5, Ura4, Fig. 2i). The positions of NMR signals report on the chemical environment of the corresponding atoms. Therefore, to quantify the RNA binding to the protein, we measured the shifts in the positions of selected protein “reporter” signals, which shifted systematically in the 1H-15N spectra between the free and bound protein states as the more RNA was bound (His33, Arg75, Fig. 2j).
In summary, a quantitative NMR assay was established with dedicated reporter signals (Supplementary Table 1 and Video 1) to monitor metabolite, RNA and protein dynamics in one sample.
Network model from NMR data
To integrate the measured data and evaluate our understanding of the network dynamics, a mathematical model combining ordinary differential equations (ODE) was formulated (Methods). The initial model consisted of 3 reactions: RNA synthesis, RNA folding and protein-RNA binding (Fig. 1a, reactions 1-3). Unexpectedly, a reduction in the total integral of 31P-containing species was observed over time (T1-relaxation-corrected), sometimes followed by sharp drops in the concentration of free PO4 at the end of the transcription (Fig. 2h, first panel, blue trace). Further analysis revealed that the designed assay could also sense the formation of soluble MgHPO4 aggregates, which are not directly visible in solution NMR. Extension of the network model with the relevant reactions (Fig. 1a, reactions 4-8) allowed to quantify the MgHPO4 solubility, which matched the literature data (see below).
Correlating time-resolved concentrations of the synthesized RNA with the shifts of protein reporter signals within the same mathematical model, we could see that the established assay can sense the differences in UP1 protein affinity to the four tested RNAs. In particular, the smallest UP1 perturbations were observed in presence of the “non-binder” RNA0, intermediate perturbations observed with the “moderate”-affinity SMN RNAs, and strongest perturbations – with “high”-affinity EV2 RNA (Fig. 2g,j).
Validation of Systems NMR derived reaction constants
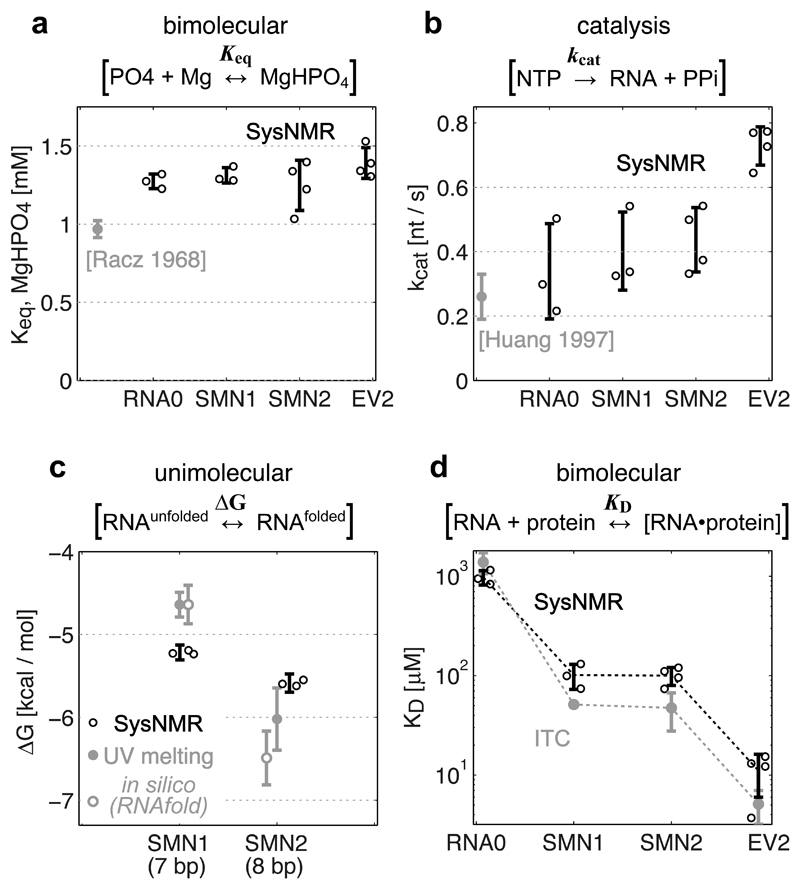
The behavior of a reaction network can be predicted at any concentrations of reactants if the constants – fundamental parameters – of all reactions are known. Deriving these fundamental constants from experimental data is one of the main goals of mathematical modeling of reaction networks. From a single NMR assay, we could determine the constants of 5 out of the 8 network reactions (# 1-3, 7-8, Fig. 1a), while the constants of the other reactions (# 4,5,6) were fixed (Methods and Supplementary Table 2). The five unconstrained constants were determined by fitting the mathematical model to the time-resolved NMR observables. For each RNA (RNA0, SMN1, SMN2, EV2) at least three NMR assay replicates were recorded and fitted (Supplementary Fig. 1). For validation, four out of five multiplex-derived Systems NMR constants – kcat, ΔG, KD, Keq,MgHPO4 – were compared with the constants derived by classical approaches, when a single reaction is perturbed at a time. Remarkably, all tested constants were in agreement with classical methods.
The equilibrium constant for the formation of soluble MgHPO4 aggregates (Keq,MgHPO4) showed an average value of 1.31 ± 0.06 mM, closely matching the 0.97 ± 0.05 mM value reported in the literature 24 (Fig. 3a).
Figure 3. Validation of Systems-NMR-derived constants.
Systems NMR data shown in black, validation data in grey. (a) Equilibrium formation of MgHPO4 salt aggregate constant (from the decrease of total 31P integral), and (b) RNA synthesis rate constant (from the decrease of NTP signals), compared to literature data. (c) Unimolecular free energy of RNA folding (from the linewidths of 1H imino signals) compared to experimental UV-melting data (filled grey circles) and theoretical predictions by RNAfold (empty grey circles). (d) Dissociation constant for protein-RNA complex formation (from the chemical shifts of protein 1H-15N signals) compared to ITC measurements.
In Systems NMR data, the experimental parameter values (Keq, kcat, ΔG, KD) and error bars correspond to the means ± s.d. from independent network model fits using independent experimental dataset replicates (n = 3 (RNA0, SMN1), n = 4 (SMN2, EV2), with different batches of DNA template and/or protein). In the validation experiments, the constants (UV-ΔG, ITC-KD) correspond to the optimized parameter ± s.d. uncertainty of data fits for the melting curve (UV-ΔG) and binding isotherm (ITC-KD). Individual data points, where available, are shown as circles next to the error bars.
The expected catalytic rate constant kcat = 0.26 ± 0.07 nt s–1 of T7 RNA Polymerase 25 closely matched the average of 0.4 ± 0.12 nt s–1 for the three short RNAs in Systems NMR datasets (Fig. 3b). Both the literature reference and the NMR kcat constants are averaging the initiation and elongation phases of transcription. This is manifested in the roughly 2-fold increase in the overall kcat = 0.73 ± 0.06 nt s–1 for the longer EV2 RNA (Fig. 3b), when the polymerase spends more time in the faster elongation phase.
Based on the measured UV-melting experiments (Supplementary Note 1), the free energy (ΔG) of folding of the two RNA hairpins (SMN1 and SMN2, differing by single base-pair) are expected to be –4.64 ± 0.15 and –6.02 ± 0.37 kcal/mol (Fig. 3c). Systems NMR measured –5.2 ± 0.1 and –5.6 ± 0.1 kcal/mol for the respective constants (Fig. 3c). SMN2 stabilities thus matched within the standard deviation limits, while SMN1 stability was overestimated by ~0.3 kcal/mol in NMR compared to UV data. Analysis of the SMN1 U5 imino signal peak shapes revealed partial peak doubling (Supplementary Fig. 2), suggesting that a more complex than a two-state model would be required for accurate analysis of SMN1 stability. RNA0 does not form hairpins, and the EV2 hairpin is too stable for the accurate UV-melting ΔG determination, therefore their stabilities were not evaluated.
Isothermal Titration Calorimetry (ITC) was used to validate the affinity constants (KD) of UP1 with the four different RNAs: one control (RNA0), the two hairpins of moderate-affinity (SMN1, SMN2) and the "high"-affinity EV2 RNA. The ITC affinity constants were 1,391 ± 331 µM for RNA0, 51.3 ± 2.5 µM for SMN1, 47.4 ± 19.7 µM for SMN2 and 5.1 ± 1.9 µM for EV2 (Fig. 3d and Supplementary Fig. 3). The corresponding constants from Systems NMR for the same four RNAs were 978 ± 162, 101 ± 29, 100 ± 20 and 11.1 ± 5.1 µM, respectively, based on 3-4 replicate measurements for each RNA (Fig. 3d). Systems NMR appeared therefore accurate for the KD measurement of the unspecific RNA0 control, but showed systematically weaker binding for the three specific RNA targets. This level of weakening could originate from unspecific UP1 interactions with RNA aborts (2-8 nt in length) and with free NTPs. The ability of UP1 to bind RNA aborts is evident from its 1,391 µM ITC-derived affinity to the “non-binding” RNA0, whose sequence matches the sequence of RNA aborts in all four RNAs. Affinity of UP1 to free NTPs was also experimentally confirmed by standard NMR titrations, measuring an overall KD,UP1-NTPs of 16,200 ± 2,100 µM (Supplementary Fig. 4). RNA aborts increase from 0 to ~2,000 µM, and NTPs decrease from 20,000 to 5,000 µM during the transcription reaction (Supplementary Fig. 1), thus both of these can weaken the affinity of UP1 to specific RNAs under these conditions.
In summary, Systems NMR accurately quantified all core reaction constants of the target network in multiplex. All validated reaction constants matched the reference values with < 2.5-fold difference.
RNA perturbations by proteins and small molecules
The reconstructed network was then perturbed by proteins and drug candidate molecules to gain insight into the network dynamics.
To probe the effect of protein on the folded RNA, the assays were performed under two conditions: “co-transcriptionally” – when the UP1 protein was added from the start and present during the entire period of RNA synthesis, and “post-transcriptionally” – when UP1 protein was added only near the end of transcription. The experiments showed that UP1 appears to (1) at least partially unwind the SMN2 hairpin; and (2) forms a 2:1 complex with the EV2 RNA when UP1 is added post-transcriptionally, and only a 1:1 complex with EV2 when UP1 is present co-transcriptionally (Supplementary Fig. 2).
To probe the effect of small molecules, the reactions with SMN2 RNA were performed in presence of drug candidates molecules, recently developed to correct the aberrant splicing of exon 7 from SMN2 gene 26. The experiments with SMN2 ESE1 suggested that under given co-transcriptional conditions one of the molecules may influence RNA folding, and another one – reduces RNA transcription rate (Supplementary Fig. 5).
Multiplexed monitoring of protein perturbations during RNA transcription
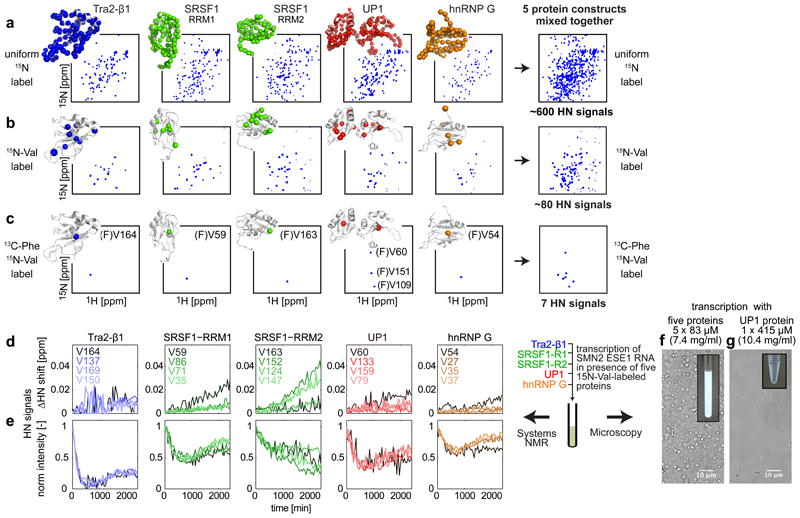
RNA binding proteins often synergize or compete for binding to the same RNA. For example the splicing of the SMN2 exon 7 is regulated by hnRNP A1, SRSF1, hnRNP G and Tra2-β1 27. To facilitate the multiplexed analysis of interactions in this system of several RNA-binding proteins, we devised two labeling schemes that visualized the protein-RNA interaction interfaces in the RNA Recognition Motifs (RRM) of these proteins in one sample at the same time (Fig. 4a-c and Supplementary Note 2). RNA transcription was then performed in the presence of five 15N-valine-labeled protein constructs mixed together (two independent RRMs in case of SRSF1), monitoring all valines and quantifying their perturbations in real time (Fig. 4d,e).
Figure 4. Monitoring multiple proteins in one NMR sample employing selective labeling.
NMR spectra of five protein constructs (in columns) with three different labeling schemes – uniform 15N (a), selective 15N-Val (b) and double-selective [13C-Phe, 15N-Val] (c). Protein structures with observable NMR signals as colored spheres are shown next to all the spectra. (a, b) show 2D HN spectra, (c) shows 2D HN(CO) spectra. Uniform 15N labeling reveals HN signals for all residues except prolines in the 2D HN experiment; 15N-Val labeling – only HN signals of valines, 13C-Phe,15N-Val labeling – HN signals of valines preceded by phenylalanines. Last column shows the spectral overlap upon combining five proteins in one sample.
Chemical Shift Perturbation (CSP) (d) and intensity (e) changes of HN signals in a mixture of five proteins during transcription of SMN2 ESE1 RNA. Traces of four valine residues, including the most strongly perturbed ones, for each protein are shown.
Microscope images of the RNA transcription performed with (f) 5 protein constructs and (g) only UP1 protein at identical 415 µM total protein concentration (in mass units 7.4 mg/ml for 5-protein sample, and 10.4 mg/ml for pure UP1). Five-protein transcription reactions were repeated twice (n = 2 independent experiments) with similar results.
During the reaction, all observed constructs except SRSF1-RRM2 showed an unexpected bi-modal response (Fig. 4e) – with valine signals first decreasing and then increasing their intensity, many without significant change in the signal positions. The samples showed evidence of Liquid-Liquid-Phase-Separation 28, which was confirmed by microscopy (Fig. 4f and Supplementary Fig. 6). Notably, none of the protein constructs included disordered regions. The number of phase-separated droplets decreased more than 10-fold when the transcription was performed in presence of individual proteins at the same total concentration as in the mixture of five (Fig. 4g and Supplementary Fig. 6). This suggests that this RNA-dependent phase-separation is driven here not simply by high protein concentrations, but involves interactions between specific proteins.
Decrease followed by increase of protein NMR signal intensities in the absence of substantial change of the NMR signal positions suggests that the proteins phase-separate into larger assemblies at the start of transcription reaction, when RNA/protein ratio is low, and are partially re-dissolved later, when the RNA concentration increases. This matches the recently reported RNA-dependent phase-separation of hnRNP A1, TDP43 and FUS proteins in vitro and in vivo 29.
To evaluate whether the presented NMR setup could be implemented under physiological conditions, we measured nuclear concentrations of the UP1/hnRNP A1, SRSF1, hnRNP G and Tra2-β1 proteins in HEK293 cells, and found those to be 30, 7.6, 3.3 and 5.2 µM respectively (Supplementary Fig. 7). This is close to the current NMR sensitivity limits (≥ 10-50 µM), suggesting that our setup, at least for hnRNP A1, can be tuned for observations under near-physiological conditions.
Discussion
The derivation of individual catalytic 30, unimolecular 31 and bimolecular 32 reaction constants by NMR is not uncommon, but Systems NMR approach enables us to quantify a network with all elementary reaction types and main biomolecule classes in a single sample. Due to the non-destructive nature of NMR, each sample yields not just a snapshot of the network, but reveals its dynamics over time or another variable condition, thereby giving deeper insight into the network logic.
The different reaction constants determined from individual multiplexed NMR assays in our study appear accurate, showing < 2.5-fold difference with validation values (Fig. 3). The differences between network-based and single-reaction-based assays can reveal unaccounted cross-talk reactions, such as the unspecific interactions of UP1 protein with the abortive RNAs and free NTPs detected here. Our results correlate with the recent UP1 specificity screens 33 and suggest that in vivo UP1/hnRNP A1 protein affinity to specific RNA targets will likely be ~1000-fold weaker than the nM-range affinities anticipated from single-reaction in vitro assays 23.
Another emergent behavior we detected was the RNA-driven in vitro phase-separation in a system of five protein domains (Fig. 4d), which was largely absent for individual domains under the same conditions. This observation suggests that Systems NMR could be used to probe structural perturbations of proteins in phase-separated droplets and membraneless organelles 29, an emerging research area with connections to various age-related disorders 28. The method can resolve residue-level signals of multiple proteins at once, does not require chemical modifications of proteins and allows monitoring of enzymatic activities within the same assay.
NMR assay limits
One specific requirement of the assay developed here is the need to design a ~8-10 nucleotide-long 5’ overhang RNA sequence which minimizes interference of short abortive RNAs with specific protein-RNA interactions and RNA folding. This sequence is designed algorithmically and can be used as a separate control to identify specific RNA effects from the other network perturbations.
More broadly, for a generic reaction network, present-day solution NMR permits the direct observation of rigid molecules below ~50-100 kDa in size 6,34 and at minimal concentration of ~10-50 µM 7,35. Under certain conditions, observation of 1 MDa complexes can be achieved 36, and in combination with hyperpolarization methods, molecules at sub-µM concentrations can be transiently observed 37,38.
For catalytic reactions, NMR permits quantification of kinetic (non-equilibrium) processes on the time-scales going from seconds to hours and days 39. For unimolecular reactions, many NMR techniques are available 31,40, potentially allowing quantification of low-populated molecular states down to fractions of percent from the main species (~5-10 kcal/mol in free energy difference). For bimolecular interactions, NMR currently permits direct quantification of dissociation constants in the low-µM to medium-mM range 32. And by monitoring competitive displacement of weak-affinity ligands, also low-nM dissociation constants can be quantified 41.
As suggested by the selective labeling experiments shown here (Fig. 4) and the recent multiplexed NMR kinase assays 42, at least a few dozen of protein-focused reactions should be observable by NMR in one sample in parallel. The same multiplexing is also feasible for metabolites 43,44, but may be challenging for RNAs due to the higher degeneracy of their NMR signals 45.
While small molecule NMR signals can mostly be interpreted ab initio, the assignment of observed signals to specific molecular epitopes in macromolecules requires time. Nevertheless, the NMR signal assignments from ~7000 unique protein and ~600 unique RNA NMR structures are available in the Protein Data Bank (pdb.org), providing an already vast starting ground for NMR network reconstructions.
Mathematical ODE models of reaction networks can be easily formulated using Rule-Based-Modeling 46, and computational methods exist to efficiently estimate network parameters and perform model selection 47–49, with virtually no limitations for moderately-large networks expected in NMR assays.
Applications
The generalized workflow in Systems Biology consists of four steps: experiment, modeling, prediction, and testing of predictions – often repeated iteratively 50. By uniquely providing both multiplexed and dynamic data from single samples at the first experimental stage, Systems NMR can accelerate the downstream development of accurate mathematical models, the understanding of network dynamics and the resulting predictions. Because NMR can dynamically monitor molecules in complex environments including living cells 10,12,13, the determination of true rates and constants for cellular networks in their natural context can generate reusable data for modeling and prediction of network dynamics.
Another advantage is that in vitro Systems NMR reconstructions provide experimental ground of intermediate complexity – between simplified single-reaction in vitro assays, and often very complex in vivo networks. Such moderate complexity may already reveal emergent network properties, like phase-separation of RNA binding domains observed here.
Considering specific applications, Systems NMR can give strong advance to the studies of "heterotypic" networks – involving different molecule and/or reaction types. For example, concurrent quantification of perturbations in different parts of a biochemical network like RNA transcription, folding and protein interactions observed here; or simultaneous quantification of catalysis and allosteric interactions in synthetic biology networks 51, or monitoring cross-talk between metabolic and signaling pathways 52,53.
In conclusion, combining the dynamic resolution of biochemical assays and the multiplexing ability of “omics”, we expect Systems NMR to pave the way to a deeper systems-level understanding of biological network dynamics both in fundamental and applied contexts.
Online Methods
A Supplementary Protocol describing how to set up and analyze data using Systems NMR for the presented here network is available at Protocol Exchange (doi: 10.21203/rs.2.9160/v1), and the most recent version at github.com/systemsnmr/ivtnmr.
RNA construct design
The sequence of the control RNA0 was designed algorithmically, using custom-built MATLAB scripts (github.com/systemsnmr/ivtnmr), from all possible sequences using four requirements: starts with G; contains no purine pairs – which are recognized specifically by UP1 protein; contains ≥ 30% purines – to reduce RNA Polymerase dissociation/abortion at initiation stage; does not form stable dimers or hairpins with itself or target SMN and EV2 RNA sequences. This resulted in 7 variants, of which (5’-GCACCACACG-3’) was chosen, as it showed fewest unspecific signals in the NMR imino region during transcription. The hairpin RNAs included non-native single-stranded 5’ overhang matching the sequence of the control RNA0 – to make the abortive RNA products uniform in all constructs, and contained two non-native closing GC pairs to offset the instability caused by the 5’-single-stranded overhang.
DNA templates
For RNA transcription corresponding sequences were cloned into pTX1 vector 54 at SapI sites, using dsDNAs from commercial (Microsynth AG) single-stranded oligos: RNA0 (ATAGCACCACACG, TCACGTGTGGTGC), SMN1 (ATAGCACCACACGGGTTTCAGACAAAATCCG, TCACGGATTTTGTCTGAAACCCGTGTGGTGC), SMN2 (ATAGCACCACACGGGTTTTAGACAAAATCCG, TCACGGATTTTGTCTAAAACCCGTGTGGTGC), EV2 (ATAGCACCACAGGATCAATAGCAGGTGTGGCACACCAGTCATACCTTGATCC, TCAGGATCAAGGTATGACTGGTGTGCCACACCTGCTATTGATCCTGTGGTGC). Plasmids were purified using Nucleobond Xtra Midi kit (Macherey Nagel), final pellets washed three times with 70% ethanol, dried and linearized by BsaI (NEB) enzyme for 15 hours at 50ºC in NEB3.1 buffer.
Proteins
All constructs and purification procedures were described earlier: UP1 55, SRSF1-RRM1 and RRM2 56, Tra2-β1 57, hnRNP G 58. After purification proteins were transferred into transcription-NMR buffer (40 mM Tris-HCl, 0.01% Triton-X100, 5 mM Dithiothreitol (DTT), pH 7.7) by dialysis, flash-frozen and stored at –20ºC.
In vitro transcription in NMR tube
Reactions were performed at 30ºC, 40 mM Tris-HCl, 0.01% Triton-X100, 5 mM DTT, pH 7.7 supplemented with 5 mM of each nucleotide-triphosphate (AppliChem), 24 mM MgCl2, 1 U/ml Inorganic Pyrophosphatase from baker’s yeast (Sigma), 5% D2O, 50 µM 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS), 280 nM T7 RNA polymerase and 33 nM DNA template. Proteins were 150 µM – in single-protein reactions, and 83 µM of each protein – for multi-protein reactions. In multi-protein experiments 50 mM L-Arg and 50 mM L-Glu (AppliChem) were added to reduce protein aggregation, which have likely also reduced the systems’ propensity for phase-separation. T7 RNA Polymerase was purified at 4ºC using Ni–NTA HisTrap chromatography (GE Healthcare), and stored at 70 µM concentration in 25 mM Tris-HCl pH 8, 50 mM NaCl, 0.5% β-mercaptoethanol (β-ME), 50% w/v glycerol at –20ºC.
NMR experiments
Experiments were measured on Bruker AVIII-600 MHz with CPQCI cryoprobe, and consisted of repeating series of 1D 1H-watergate (spectral width (SW) 22 ppm; acquisition time (AQ) 0.62 s; D1 (interscan delay) 1 s; number of scans (NS) 128), 2D 1H-TOCSY (SW 10 / 9 ppm; AQ 0.17 / 0.018 s; D1 0.5 s; NS 4), 1D 31P (SW 50 ppm; AQ 0.66 s; NS 256; carrier –8.22 ppm; D1 0.8 s), 1D 1H-SOFAST (SW 24 ppm; AQ 0.053 s; D1 0.1 s; NS 1536; 1H excitation with Pc9 pulse, 5 ppm wide, centered at 12.9 ppm) and 2D 1H15N-SOFAST-HMQC (H/N: SW 16 / 23.5 ppm; AQ 0.106 / 0.035 s; D1 0.2 s; NS 24; 15N carrier 117.8; 1H excitation with Pc9 pulse, 4 ppm wide, centered at 7.95 ppm). BEST-TROSY 2D HN(CO) for analysis of selectively labeled proteins was provided by Frank Lohr (BMRZ, Goethe Universitat Frankfurt) and measured with (H/N: SW 12 / 16 ppm; AQ 0.107 / 0.090 s; D1 0.2 s; NS 16; 15N carrier 117.8; 1H excitation with Pc9 pulse, 4.2 ppm wide, centered at 8.5 ppm). NMR spectra were sorted, processed and analyzed using TopSpin 3.x (Bruker), custom-built Python and MATLAB scripts and the rbnmr routine (Nyberg N., RBNMR, MATLAB Central File Exchange #40332, (2013)). Chemical shifts of protein residues in 2D HN spectra were traced in CARA (cara.nmr.ch).
NMR observables
For final network modelling 10 signals were used: 31P spectra – 1) PO4, 2) RNA, 3-5) αNTP, βNTP, γNTP, 6-7) αNDP, βNDP; 8) 1H spectra – U5 (SMN1/2) or U4 (EV2) imino signals; 9-10) 2D HN spectra – His33 and Arg75 residues of UP1. Populations of phosphate-containing species were calculated stoichiometrically from 31P integrals using αNTP integral at time=0 as 20 mM internal calibration. Each 31P integral was T1-relaxation-weighted using the ratios of corresponding integrals measured in the reference 31P spectra with 30 s and 0.8 s interscan delay at the end of the transcription reaction, i.e. [I31P, corrected = I31P × (I31P ref, d1=30s / I31P ref, d1=0.8s)]. NTP and NDP populations were quantified from αP integrals, MgHPO4 was calculated from decrease of the total T1-weighted 31P integral of all species. For long RNAs (>≈ 20 nt), the 1D 31P signals became too broad, preventing accurate quantification at a reasonable time-resolution, so RNA concentrations were calculated from the decay of NTP signals. Due to the NMR signal degeneracy, the fractions of RNA and aborts could not be quantified within the NMR assay, and were fixed to 30 and 70%, by nucleotide mass, based on quantitative UV(260 nm)-HPLC analysis of reaction end-products.
The fraction of bound UP1 protein was derived from the chemical shift perturbation (CSP) of HN signals of His33 and Arg75. These residues were chosen as reporters for two reasons. Firstly, they appeared to sense the same molecular epitope in all four protein:RNA complexes, as they all displayed signals moving in the same direction during transcription (Fig. 2g). Based on the existing UP1-RNA/DNA structures (PDB: 4YOE, 2UP1), these two residues are located near the RNA binding pocket, but are not directly interacting with the RNA, which explains how they could sense the same epitope changes independent of the RNA sequence. Secondly, these signals were sensitive to the differences in affinity of the RNA binding, as the magnitude of the 1H-15N chemical shift perturbation varied for four different protein:RNA complexes (Fig. 2g,j). In assays with 150 µM UP1 concentration, both signals appeared predominantly in fast exchange with respect to the NMR time scale, and so the fast exchange assumption 59 was used during modeling. The experimental HN CSPs were calculated using The effect of pH change on histidine signal position was assumed negligible because the transcription buffer pH = 7.7 is far from the histidine pKa ≈ 6, and the chemical shifts of other surface-exposed UP1 histidines did not exhibit the same perturbation effects. Calculating the fraction of the bound protein under the fast exchange regime requires information on protein signal positions in the free and fully bound states. The shifts of the bound state are usually estimated as one of the parameters of the KD fitting procedure, as the asymptote of the protein saturation curve. The 150 µM UP1 protein data did not approach this saturation under the assay conditions, because of the low protein:RNA ratio, since the final concentrations of specific RNAs reached only ~120 µM, giving only ~0.8:1 RNA:protein ratio. Addition of pure RNA to saturate the protein under these conditions consistently led to protein precipitation, which correlates with UP1’s ability to phase-separate and aggregate in presence of RNA. Therefore, to estimate the HN signal positions of His33 and Arg75 in the saturated protein, an additional set of transcription experiments was recorded using 20 and 30 µM UP1 and the “high-affinity” EV2 RNA. This allowed to increase protein saturation by reaching ~6:1 and 4:1 RNA:protein ratios (~120:20 and 120:30 µM respectively) at the end of the reaction. Due to the poor NMR sensitivity at 20-30 µM protein concentrations, the 2D HN spectra in these experiments required ~10 hour acquisition time, and could only be recorded as non time-resolved spectra at end of transcription reaction, when the system reached equilibrium. The combined CSP data from the datasets with high (150 µM) and low (20-30 µM) protein concentrations gave an imperfect fit with the single-site binding model, suggesting that UP1 is already close to saturation in the assays with 150 µM protein concentration (Supplementary Fig. 8). This is likely due to additional weak UP1 binding sites in the EV2 RNA, as suggested by Arg75 peak splitting in assays with 20-30 µM UP1 concentration. The chemical shifts of His33 and Arg75 residues in the fully saturated protein state for the final ODE modeling of the four main datasets (RNA0, SMN1, SMN2, EV2 at 150 µM UP1) were taken as the shifts giving best fit when simultaneously fitting the data from EV2 RNA datasets with 150, 30 and 20 µM UP1 protein concentration (Supplementary Fig. 8).
All above 31P and HN data was used for global parameterization of the ODE model, and 1H-imino signals were used for lineshape analysis and RNA ΔG derivation.
Mathematical modeling
The model was built using the BioNetGen language 60 and resulted in 8 rate equations and 9 differential equations:
Rate equations
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
Differential equations
| (9) |
| (10) |
| (11) |
| (12) |
| (13) |
| (14) |
| (15) |
| (16) |
| (17) |
At the given hairpin stability (≈ –5.5 kcal/mol) the SMN and EV2 RNAs are predominantly in a single state (99,99% folded). The expected rate of hairpin folding (v2F = 8-51 * 103 s–1 for ~30 nt hairpin) 61 is three orders of magnitude faster than protein-RNA encounters at the unbiased diffusion rate (~105 M–1 s–1) 62. For these reasons the final ODE model treats the folded and unfolded RNA as a single species, and the stability of RNA is derived independently from imino signal lineshape analysis at each reaction time point (see below). Because of the sequence identity with abortive products, RNA0 length is used as a weighted sum of 10 nt full length RNA0 and the 4 nt average length of corresponding abortive products: 0.3 * 10 + (1–0.3) * 4 = 5.8 nt. The final ODE model was deposited in BioModels 63 (MODEL1812270001).
Model fitting
The ODE model was fitted to the seven NMR-based observables employing the gradient based Trust-Region method, using a custom-built set of MATLAB routines (github.com/systemsnmr/ivtnmr) based on earlier code 47. Model parameters, their boundaries and initial optimization conditions are given in Supplementary Table 2. The experimental errors were assumed to follow a Gaussian distribution and the standard deviations of individual data points were calculated from spectral noise for 31P integrals, and from the variance of protein HN chemical shifts at the end of the reaction (when the network is approaching equilibrium state). The values and standard deviations of the derived network parameters were obtained with two alternative approaches. In the first method, we calculated the standard deviation between estimates that were obtained in independent ODE model fits of several NMR replicate datasets, varying batches of the protein and/or DNA template preparations (Fig. 3 and Supplementary Fig. 1). In the second method, parameter uncertainties were estimated with bootstrap analysis. Here, we generated 200 model fits using resampling of the full data vector with replacement. The confidence intervals obtained by bootstrapping (Supplementary Fig. 9) were narrower than the variability between the 3-4 experimental replicates, and therefore the values from the replicate analysis were chosen as the ones more realistically reflecting the parameter variance.
Derivation of final constants
The enzyme (280 nM T7 RNAP) is assumed to be saturated by the substrate (4-20 mM NTPs) during the reaction, so the KM can be ignored and the concentration of the active enzyme can be defined by the limiting concentration of the DNA template (33 nM).
| (18) |
Because reactions # 3 and #7 (Fig. 1a, protein binding and aggregation of MgHPO4) equilibrate faster than the time-resolution of the current setup, only the equilibrium constants (Keq, KD – ratios of kinetic rates) for these reactions could be parameterized.
| (19) |
Pyrophosphate hydrolysis can be quantified from the specific PPi observable, but this rate was too rapid for meaningful quantification. Therefore, this constant was fixed based on the enzyme activity provided by the manufacturer (Sigma).
| (20) |
The formation of soluble Mg-PO4 aggregates depends on the concentration of the free Mg. Apart from Mg-PO4, the free Mg can also participate in Mg-NTP and Mg-RNA complexes. The fraction of the free Mg was estimated from the published KD,Mg.NTP = 0.3 mM 64, using the general bimolecular binding isotherm 67 denoted with 𝑓 in the equation below. For the calculation the concentration of free Mg available for Mg-PO4 salt formation was assumed constant during the reaction.
| (21) |
| (22) |
RNA folding ΔG from imino signal lineshape analysis
For fitting, 1D 1H-SOFAST spectra were Fourier-transformed with no apodization function. Imino signals were fit to a single-lorentzian function using the lorentzfit routine (Wells J., Lorentzfit, MATLAB Central File Exchange #33775, (2015)). The fits used 0.2 ppm fitting window, and assumed a baseline fixed at zero signal intensity. In the single-lorentzian fit, the intrinsic broadening of a signal cannot be distinguished from broadening due to overlapping signals, thus accurate quantification requires well-resolved signals. The fitted linewidth parameter (full width at half maximum) was used to derive the unfolding-driven imino exchange rate (kex,unfolding).
| (23) |
| (24) |
The contribution of B0 field inhomogeneity (R2(B0)) is considered negligible. The combined contribution of base-flipping (kex,base-flipping) 31 and transverse relaxation rate (R2) was determined from linewidths of imino signals in purified, SMN2 hairpin additionally stabilized by terminal GCs (13-bp, GGCGGGUUUUGGC-AGAC-GCCAAAAUCCGCC). In this stabilized RNA the exchange by global unfolding is suppressed (ΔG = –23 kcal/mol), which was confirmed by the negligible dependence of its imino integrals on temperature (not shown). The combined (kex,base-flipping+R2) value for imino signals in an U-A pair flanked by GU and UA pairs, under transcription buffer conditions, was 61.4 s–1.
Imino linewidths depended on pH (concentration of imino-exchange catalyst), indicating that the system is under the bimolecular exchange regime (“EX2”) 65, and hence the measured kex reports on the equilibrium constant of RNA unfolding/opening (Keq,unfolding = Kop). The intrinsic exchange rate kex,intrinsic (same as the exchange from the open state, kex,open) in the transcription buffer was measured to be ~106 s–1 for both UTP and GTP, using a protocol described elsewhere 66. The final free energy of folding was determined using:
| (25) |
| (26) |
Where R is the gas constant and T is the absolute temperature.
RNA purification
RNAs for ITC and UV-melting experiments were purified by anion-exchange HPLC under denaturing 6 M Urea, 80ºC conditions, followed by n-butanol extraction, snap-cooling and lyophilization.
UV temperature-melting
For the melting experiments the RNA hairpins were produced without the single-stranded 5’ overhang to eliminate UV baseline distortions caused by this single-stranded region. The experiments used 2 µM RNAs in 10 mM Sodium-Cacodylate, pH 7.35, 5 mM MgCl2, 25 mM L-Arg/L-Glu buffer. Details of the analysis shown in Supplementary Note 1.
ITC
Experiments used conditions approximating those at the end of transcription-NMR reaction: 40 mM Tris-HCl, 0.01% Triton-X100, 2.5 mM β-ME, pH 7.5, 37 mM NaPO4, 2.6 mM NTPs, 24 mM MgCl2, 303K. DTT was replaced with β-ME due to its instability and background heat changes. For each RNA an RNA-to-buffer titration was performed and subtracted from the RNA-to-protein data. Data from the first injection point was discarded. To better represent the pool of unspecific RNAs in the transcription reaction, RNA0 was purified as a combined pool of 2-10 nt RNAs from transcription reaction. The concentration of this RNA0+aborts pool was normalized by the average size of 5.8 nts based on HPLC-UV-weighted composition of the corresponding transcription mixture (30% full length RNA * 10 nt + 70% aborts * 4 nt average). For RNA0, SMN1 and SMN2 RNAs the ITC data was fitted with the one-site interaction model using MicroCal Origin (Supplementary Fig. 3a-c). A one-site KD was needed for all RNAs to compare it with the one-site binding model used in the ODE analysis. The standard fitting protocol of MicroCal Origin encountered local minima when fitting the one-site model to the EV2-UP1 ITC data (likely due to the known bi-modal binding of UP1 to this RNA 23). Therefore for EV2-UP1, the apparent one-site KD constant was derived as a mean of three different fitting protocols (Supplementary Fig. 3d-g): (1) high-affinity KD from two-site model in MicroCal Origin, (2) one-site KD fitted using stoichiometric 1:1 equilibrium model in Affinimeter software (affinimeter.com), and (3) one-site KD fitted using general ligand:target equilibrium binding isotherm 67, which parameterizes only KD constant (without considering ΔH) and assumes N=1. All three fitting procedures yielded comparable KD for EV2-UP1 binding (4.8, 7.1 and 3.3 µM).
Perturbation experiments
Post-transcriptional perturbations by UP1 protein were performed by adding, 13 hours after transcription start, 67.5 nmol of 15N-UP1. Protein was ~1500 µM concentration, to achieve ≤ 10% dilution of the mixture. In small-molecule perturbation experiments, the SMN2 ESE1 transcription mixture was spiked co-transcriptionally with 200 µM of one of three molecules (NVS-SM1, smn-C5 and smn-C7 26 in 1% DMSO), or 1% DMSO alone. In these experiments SMN2 DNA template used an earlier version of the 5’-overhang sequence (5’-GCGCCGUA-3’), before it was optimized at three positions (3,7,8) to reduce its self-complementarity.
Imino signal broadening upon UP1 protein binding
Imino signal linewidths could be primarily influenced by the (1) changes in kex exchange rate of iminos due to unfolding of the stem, (2) kex changes due to base-flipping, and (3) line broadening due to enhanced transverse relaxation (R2) and B0 field inhomogeneity (R2(B0)). Thus, overall linewidth equals Δν1/2 = (kex, unfolding + kex, base-flipping + R2 + R2(B0))/π. Contribution of B0 field inhomogeneity is considered negligibly small. The R2 relaxation increase upon formation of a 1:1 RNA:UP1 protein complex was estimated to be 41/π Hz, given 22.25 kDa UP1 mass, at 303K, in phosphate buffer saline (0.001 kg m−1 s−1), assuming spherical shape of the protein with rw=2.4 Å hydration layer, and R2 = 5*τc (correlation time) 69.
Cell culture and nuclear extracts
Extracts were prepared using published procedures 68 from HEK293 cells grown to confluence.
Mass Spectrometry for quantification of specific protein concentrations in cells
Pure recombinant uniformly 15N-labeled proteins were spiked into nuclear extracts at 0.15-0.2 µM concentration. Resulting extracts were reduced, alkylated and digested using trypsin prior to peptide desalting and purification as previously described 70. Selected reaction monitoring (SRM) on a triple-quadrupole mass spectrometer was used for targeted proteomic measurements. SRM assays were generated as previously described 71 by selecting the 4-5 most intense transitions from samples with pure 15N-labeled recombinant proteins digested with the same protocol. Sum peak areas of transitions for each peptide were used to calculate the intensity ratio between 15N reference and 14N endogenous peptide signals. The mean and standard deviation of all peptides for each protein were used to find the concentration of endogenous proteins in nuclear extracts.
Selective labeling
Proteins were expressed in minimal M9 medium supplemented with 15N-Val and 13C-Phe, and with all amino acids and nucleosides in unlabeled form (Supplementary Table 3). A limiting amount of unlabeled Phe/Val was added for transaminase suppression. 13C/15N-labeled amino acids were added only 10 minutes before induction. Cells were harvested 3-5 hours post-induction.
Statistics
Statistical analyses and experiment replicate numbers, where applicable, are described in the corresponding figure legends and method sections. Unless otherwise indicated the derived values and error bars correspond to the mean ± s.d.
Supplementary Material
Acknowledgements
We thank J. Vollmer and G. Fengos for the help with network modeling. We acknowledge G. Wider and all members of the ETH BNSP platform for excellent maintenance of the NMR infrastructure, S. Heinrich for the help with microscopy, and E. Lehmann for providing T7 polymerase. We thank all members of the Allain Lab, in particular F. Damberger, and the Parpan retreat participants for helpful discussions. This work was supported by the Promedica Stiftung, Chur (Grant 1300/M to Y.N.), Novartis Foundation and Krebsliga Zurich (Y.N.), NCCR RNA and Disease by the Swiss National Science Foundation (F.A.).
Footnotes
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability
Main code with examples and a protocol for the Systems NMR setup is available at github.com/systemsnmr/ivtnmr.
Data availability
Raw NMR data from the main experiments was deposited in Zenodo (10.5281/zenodo.2554066). Source data for the main figures are included in the online article. SRM-MS-Proteomics data was deposited in PeptideAtlas (PASS01365). The ODE network model was deposited in BioModels 63 (MODEL1812270001). RNA-encoding plasmids were deposited in AddGene (ID #126040, #126041, #126042, #126043). Further data and code are available from the corresponding authors upon request.
Author Contributions
Y.N. conceived the idea, obtained initial funding, performed experiments, data analysis and network modeling. F.A. supervised the project, provided infrastructure and financial support. D.I. supervised the network modeling part of the project. N.R. expressed and purified selectively labeled proteins, and contributed to the transcription-NMR experiments. P.P. and M.S. supervised and provided technical and analytical support on mass spectrometry analyses. Y.N. wrote the manuscript with input from all of the authors.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Szallasi Z. System Modeling in Cellular Biology Ch. 10. The MIT Press; Cambridge: 2006. [DOI] [Google Scholar]
- 2.Barabási A-L, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 3.Howsmon DP, Hahn J. Regularization Techniques to Overcome Over-Parameterization of Complex Biochemical Reaction Networks. IEEE Life Sci Lett. 2016;2:31–34. doi: 10.1109/LLS.2016.2646498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yugi K, Kuroda S. Metabolism as a signal generator across trans-omic networks at distinct time scales. Curr Opin Syst Biol. 2017;8:59–66. [Google Scholar]
- 5.Hart Y, Alon U. Review The Utility of Paradoxical Components in Biological Circuits. Mol Cell. 2013;49:213–221. doi: 10.1016/j.molcel.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Frueh DP, Goodrich AC, Mishra SH, Nichols SR. NMR methods for structural studies of large monomeric and multimeric proteins. Curr Opin Struct Biol. 2013;23:734–739. doi: 10.1016/j.sbi.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwan AH, Mobli M, Gooley PR, King GF, Mackay JP. Macromolecular NMR spectroscopy for the non-spectroscopist. FEBS J. 2011;278:687–703. doi: 10.1111/j.1742-4658.2011.08004.x. [DOI] [PubMed] [Google Scholar]
- 8.Freedberg DI, Selenko P. Live cell NMR. Annu Rev Biophys. 2014;43:171–192. doi: 10.1146/annurev-biophys-051013-023136. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein MA. Reaction monitoring using NMR. Magn Reson Chem. 2016;54:422. doi: 10.1002/mrc.4436. [DOI] [PubMed] [Google Scholar]
- 10.Wolak J, et al. Chapter 11 Noninvasive Fluxomics in Mammals by Nuclear Magnetic Resonance Spectroscopy. Handb Metabolomics. 2012:321–392. doi: 10.1007/978-1-61779-618-0. [DOI] [Google Scholar]
- 11.Eicher JJ, Snoep JL, Rohwer JM. Determining Enzyme Kinetics for Systems Biology with Nuclear Magnetic Resonance Spectroscopy. Metabolites. 2012;2:818–843. doi: 10.3390/metabo2040818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lerche MH, Jensen PR, Karlsson M, Meier S. NMR Insights into the Inner Workings of Living Cells. Anal Chem. 2015;87:119–132. doi: 10.1021/ac501467x. [DOI] [PubMed] [Google Scholar]
- 13.Smith MJ, et al. Real-time NMR monitoring of biological activities in complex physiological environments. Curr Opin Struct Biol. 2015;32:39–47. doi: 10.1016/j.sbi.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Fan TW, Lane AN. Applications of NMR spectroscopy to systems biochemistry. Prog Nucl Magn Reson Spectrosc. 2016;92–93:18–53. doi: 10.1016/j.pnmrs.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordier F, et al. Ordered Phosphorylation Events in Two Independent Cascades of the PTEN C-tail Revealed by NMR. J Am Chem Soc. 2012;134:20533–20543. doi: 10.1021/ja310214g. [DOI] [PubMed] [Google Scholar]
- 16.Theillet F-X, et al. Cell signaling, post-translational protein modifications and NMR spectroscopy. J Biomol NMR. 2012;54:217–236. doi: 10.1007/s10858-012-9674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee M-K, Gal M, Frydman L, Varani G. Real-time multidimensional NMR follows RNA folding with second resolution. Proc Natl Acad Sci USA. 2010;107:9192–9197. doi: 10.1073/pnas.1001195107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wacker A, Buck J, Richter C, Schwalbe H, Wöhnert J. Mechanisms for differentiation between cognate and near-cognate ligands by purine riboswitches. RNA Biol. 2012;9:672–680. doi: 10.4161/rna.20106. [DOI] [PubMed] [Google Scholar]
- 19.Steinert H, et al. Pausing guides RNA folding to populate transiently stable RNA structures for riboswitch-based transcription regulation. Elife. 2017;6:1–18. doi: 10.7554/eLife.21297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breaker RR. Prospects for Riboswitch Discovery and Analysis. Mol Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Landick RA. Two-Way Street : Regulatory Interplay between RNA Polymerase and Nascent RNA Structure. Trends Biochem Sci. 2016;41:293–310. doi: 10.1016/j.tibs.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh RN. Evolving concepts on human SMN pre-mRNA splicing. RNA Biol. 2007;4:7–10. doi: 10.4161/rna.4.1.4535. [DOI] [PubMed] [Google Scholar]
- 23.Levengood JD, Tolbert M, Li M, Tolbert B. High-affinity interaction of hnRNP A1 with conserved RNA structural elements is required for translation and replication of enterovirus 71. RNA Biol. 2013;10:1136–1145. doi: 10.4161/rna.25107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Racz GJ, Soper RJ. Solubility of Dimagnesium Phosphate Trihydrate and Trimagnesium Phosphate. Can J Soil Sci. 1968;48:265–269. [Google Scholar]
- 25.Huang Y, Beaudry A, Mcswiggen J, Sousa R. Determinants of Ribose Specificity in RNA Polymerization : Effects of Mn 2+ and Deoxynucleoside Monophosphate Incorporation into Transcripts. Biochemistry. 1997;36:13718–13728. doi: 10.1021/bi971609o. [DOI] [PubMed] [Google Scholar]
- 26.Sivaramakrishnan M, et al. Binding to SMN2 pre-mRNA-protein complex elicits specificity for small molecule splicing modifiers. Nat Commun. 2017;8:1476. doi: 10.1038/s41467-017-01559-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh NN, et al. An intronic structure enabled by a long-distance interaction serves as a novel target for splicing correction in spinal muscular atrophy. Nucleic Acids Res. 2013;41:8144–8165. doi: 10.1093/nar/gkt609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boeynaems S, et al. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maharana S, et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science (80-.) 2018;360:918–921. doi: 10.1126/science.aar7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowen S, Hilty C. Time-resolved dynamic nuclear polarization enhanced NMR spectroscopy. Angew Chem Int Ed Engl. 2008;47:5235–5237. doi: 10.1002/anie.200801492. [DOI] [PubMed] [Google Scholar]
- 31.Bothe JR, et al. Characterizing RNA dynamics at atomic resolution using solution-state NMR spectroscopy. Nat Methods. 2011;8:919–931. doi: 10.1038/nmeth.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fielding L. NMR methods for the determination of protein-ligand dissociation constants. Prog Nucl Magn Reson Spectrosc. 2007;51:219–242. [Google Scholar]
- 33.Jain N, Lin H-C, Morgan CE, Harris ME, Tolbert BS. Rules of RNA specificity of hnRNP A1 revealed by global and quantitative analysis of its affinity distribution. Proc Natl Acad Sci. 2017;114:2206–2211. doi: 10.1073/pnas.1616371114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeuchi K, Wagner G. NMR studies of protein interactions. Curr Opin Struct Biol. 2006;16:109–117. doi: 10.1016/j.sbi.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Bieri M, et al. Macromolecular NMR spectroscopy for the non-spectroscopist: beyond macromolecular solution structure determination. FEBS J. 2011;278:704–715. doi: 10.1111/j.1742-4658.2011.08005.x. [DOI] [PubMed] [Google Scholar]
- 36.Wiesner S, Sprangers R. Methyl groups as NMR probes for biomolecular interactions. Curr Opin Struct Biol. 2015;35:60–67. doi: 10.1016/j.sbi.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Frydman L, Blazina D. Ultrafast two-dimensional nuclear magnetic resonance spectroscopy of hyperpolarized solutions. Nat Phys. 2007;3:415–419. [Google Scholar]
- 38.Kim Y, Hilty C. Affinity Screening Using Competitive Binding with Fluorine-19 Hyperpolarized Ligands. Angew Chemie Int Ed. 2015;54:1–5. doi: 10.1002/anie.201411424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gal M, Schanda P, Brutscher B, Frydman L. UltraSOFAST HMQC NMR and the repetitive acquisition of 2D protein spectra at Hz rates. J Am Chem Soc. 2007;129:1372–1377. doi: 10.1021/ja066915g. [DOI] [PubMed] [Google Scholar]
- 40.Baldwin AJ, Kay LE. NMR spectroscopy brings invisible protein states into focus. Nat Chem Biol. 2009;5:808–814. doi: 10.1038/nchembio.238. [DOI] [PubMed] [Google Scholar]
- 41.Dalvit C, Fagerness PE, Hadden DTa, Sarver RW, Stockman BJ. Fluorine-NMR experiments for high-throughput screening: Theoretical aspects, practical considerations, and range of applicability. J Am Chem Soc. 2003;125:7696–7703. doi: 10.1021/ja034646d. [DOI] [PubMed] [Google Scholar]
- 42.Thongwichian R, et al. A Multiplexed NMR-Reporter Approach to Measure Cellular Kinase and Phosphatase Activities in Real-Time. J Am Chem Soc. 2015;137:6468–6471. doi: 10.1021/jacs.5b02987. [DOI] [PubMed] [Google Scholar]
- 43.Soininen P, et al. High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst. 2009;134:1781–1785. doi: 10.1039/b910205a. [DOI] [PubMed] [Google Scholar]
- 44.Lindon JC, London IC, Kensington S. Biofluids Studied by NMR Spectroscopy. Encycl Spectrosc Spectrom. 2010;1:128–141. [Google Scholar]
- 45.Lu K, Miyazaki Y, Summers MF. Isotope labeling strategies for NMR studies of RNA. J Biomol NMR. 2010;46:113–125. doi: 10.1007/s10858-009-9375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chylek La, et al. Rule-based modeling: a computational approach for studying biomolecular site dynamics in cell signaling systems. Wiley Interdiscip Rev Syst Biol Med. 2014;6:13–36. doi: 10.1002/wsbm.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geier F, Fengos G, Felizzi F, Iber D. In: Computational Modeling of Signaling Networks. Liu X, Betterton MD, editors. Humana Press; 2012. pp. 23–39. [DOI] [Google Scholar]
- 48.Maiwald T, Timmer J. Dynamical modeling and multi-experiment fitting with PottersWheel. Bioinformatics. 2008;24:2037–2043. doi: 10.1093/bioinformatics/btn350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raue A, et al. Data2Dynamics: A modeling environment tailored to parameter estimation in dynamical systems. Bioinformatics. 2015;31:3558–3560. doi: 10.1093/bioinformatics/btv405. [DOI] [PubMed] [Google Scholar]
- 50.Vanlier J, Tiemann CA, Hilbers PAJ, van Riel NAW. Parameter uncertainty in biochemical models described by ordinary differential equations. Math Biosci. 2013;246:305–314. doi: 10.1016/j.mbs.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 51.Chen Z, Rappert S, Zeng A. Rational Design of Allosteric Regulation of Homoserine Dehydrogenase by a Nonnatural Inhibitor. ACS Synth Biol. 2015;4:126–131. doi: 10.1021/sb400133g. [DOI] [PubMed] [Google Scholar]
- 52.Smith MJ, Ikura M. Integrated RAS signaling defined by parallel NMR detection of effectors and regulators. Nat Chem Biol. 2014;10:223–230. doi: 10.1038/nchembio.1435. [DOI] [PubMed] [Google Scholar]
- 53.Tripodi F, Nicastro R, Reghellin V, Coccetti P. Post-translational modifications on yeast carbon metabolism: Regulatory mechanisms beyond transcriptional control. Biochim Biophys Acta - Gen Subj. 2015;1850:620–627. doi: 10.1016/j.bbagen.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 54.Michel E, Duss O, Allain FH-T. In: Bacterial Regulatory RNA: Methods and Protocols. Arluison V, Valverde C, editors. Springer; New York: 2018. pp. 177–195. [DOI] [Google Scholar]
- 55.Barraud P, Allain FH-T. Solution structure of the two RNA recognition motifs of hnRNP A1 using segmental isotope labeling: how the relative orientation between RRMs influences the nucleic acid binding topology. J Biomol NMR. 2012;55:119–138. doi: 10.1007/s10858-012-9696-4. [DOI] [PubMed] [Google Scholar]
- 56.Cléry A, et al. Isolated pseudo-RNA-recognition motifs of SR proteins can regulate splicing using a noncanonical mode of RNA recognition. Proc Natl Acad Sci U S A. 2013;110:E2802–11. doi: 10.1073/pnas.1303445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cléry A, et al. Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-β1. Nat Struct & Mol Biol. 2011;18:443. doi: 10.1038/nsmb.2001. [DOI] [PubMed] [Google Scholar]
- 58.Moursy A, Allain FH-T, Cléry A. Characterization of the RNA recognition mode of hnRNP G extends its role in SMN2 splicing regulation. Nucleic Acids Res. 2014;42:6659–6672. doi: 10.1093/nar/gku244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Williamson MP. Progress in Nuclear Magnetic Resonance Spectroscopy Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 60.Harris LA, et al. BioNetGen 2.2: advances in rule-based modeling. Bioinformatics. 2016;32:3366–3368. doi: 10.1093/bioinformatics/btw469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hyeon C, Thirumalai D. Chain length determines the folding rates of RNA. Biophys J. 2012;102:L11–L13. doi: 10.1016/j.bpj.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qin S, Pang X, Zhou H-X. Automated prediction of protein association rate constants. Structure. 2011;19:1744–1751. doi: 10.1016/j.str.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chelliah V, et al. BioModels: Ten-year anniversary. Nucleic Acids Res. 2015;43:D542–D548. doi: 10.1093/nar/gku1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Donghi D, Pechlaner M, Finazzo C, Knobloch B, Sigel RKO. The structural stabilization of the κ three-way junction by Mg(II) represents the first step in the folding of a group II intron. Nucleic Acids Res. 2013;41:2489–2504. doi: 10.1093/nar/gks1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Russu BIM. Probing Site-Specific Energetics in Proteins and Nucleic Acids by Hydrogen Exchange and Nuclear Magnetic Resonance Spectroscopy. Methods. 2004;379:152–175. doi: 10.1016/S0076-6879(04)79009-6. [DOI] [PubMed] [Google Scholar]
- 66.Rinnenthal J, Klinkert B, Narberhaus F, Schwalbe H. Direct observation of the temperature-induced melting process of the Salmonella fourU RNA thermometer at base-pair resolution. Nucleic Acids Res. 2010;38:3834–3847. doi: 10.1093/nar/gkq124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pollard TD. A Guide to Simple and Informative Binding Assays. Mol Biol Cell. 2010;21:4061–4067. doi: 10.1091/mbc.E10-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kislinger T, et al. PRISM, a generic large scale proteomic investigation strategy for mammals. Mol Cell Proteomics. 2003;2:96–106. doi: 10.1074/mcp.M200074-MCP200. [DOI] [PubMed] [Google Scholar]
- 69.Cavanagh, et al. Protein NMR Spectroscopy: Principles and Practice. Ch 1.4. New York: Academic Press; 1996. [Google Scholar]
- 70.Soste M, et al. A sentinel protein assay for simultaneously quantifying cellular processes. Nat Methods. 2014;11:1045–1048. doi: 10.1038/nmeth.3101. [DOI] [PubMed] [Google Scholar]
- 71.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.