Abstract
OXA-48 is the most common carbapenemase in Enterobacterales in Germany and one of the most frequent carbapenemases worldwide. Several reports have associated blaOXA–48 with a virulent host phenotype. To challenge this hypothesis, 35 OXA-48-producing clinical isolates of Escherichia coli (n = 15) and Klebsiella pneumoniae (n = 20) were studied in vitro, in vivo employing the Galleria mellonella infection model and by whole-genome sequencing. Clinical isolates belonged to 7 different sequence types (STs) in E. coli and 12 different STs in K. pneumoniae. In 26/35 isolates blaOXA–48 was located on a 63 kb IncL plasmid. Horizontal gene transfer (HGT) to E. coli J53 was high in isolates with the 63 kb IncL plasmid (transconjugation frequency: ∼103/donor) but low in isolates with non-IncL plasmids (<10–6/donor). Several clinical isolates were both highly cytotoxic against human cells and virulent in vivo. However, 63 kb IncL transconjugants generated from these highly virulent isolates were not more cytotoxic or virulent when compared to the recipient strain. Additionally, no genes associated with virulence were detected by in silico analysis of OXA-48 plasmids. The 63 kb plasmid was highly stable and did not impair growth or fitness in E. coli J53. In conclusion, OXA-48 clinical isolates in Germany are diverse but typically harbor the same 63 kb IncL plasmid which has been reported worldwide. We demonstrate that this 63 kb IncL plasmid has a low fitness burden, high plasmid stability and can be transferred by highly efficient HGT which is likely the cause of the rapid dissemination of OXA-48 rather than the expansion of a single clone or gain of virulence.
Keywords: OXA-48, carbapenemases, IncL, β-lactamases, virulence, fitness, plasmid
Introduction
Escherichia coli and Klebsiella pneumoniae are the most common species causing Gram-negative sepsis (Goto et al., 2016). The increasing incidence of multidrug-resistant (MDR) E. coli and K. pneumoniae strains has complicated the treatment of Gram-negative sepsis and other invasive infections. Among MDR bacteria, carbapenemase-producing Enterobacterales (CPE) are a major threat to our health system, because of resistance to most or all β-lactam antibiotics and a lack of therapeutic options. As a consequence, the mortality in patients infected with CPE is 26–44% higher than in patients infected with carbapenem-susceptible isolates (Falagas et al., 2014).
Among the different carbapenemases, OXA-48 has been particularly successful and is the most common carbapenemase in Germany. It has rapidly disseminated from Europe and the Middle East to every continent (Canton et al., 2012). OXA-48 is most frequently detected in K. pneumoniae and E. coli, but can also occur in other Enterobacterales (Khajuria et al., 2013; Manageiro et al., 2014). The β-lactamase gene (bla) encoding OXA-48 (blaOXA–48) is usually located on plasmids (pOXA-48) and bracketed by two identical insertion sequences, IS1999, forming the composite transposon Tn1999 (Giani et al., 2012; Poirel et al., 2012). Different variants of OXA-48, commonly referred to as OXA-48-like (e.g., OXA-181, OXA-232), have been described which differ from OXA-48 by several amino acids, but have not spread as successfully as OXA-48 (Lascols et al., 2013). However, the reasons for the successful dissemination of OXA-48 are currently not fully understood.
Interestingly, virulence has been linked to the presence of blaOXA–48 in clinical E. coli and K. pneumoniae isolates. Several studies reported on clinical isolates with an unusual high lethality in murine infection models as well as the presence of genes associated with virulence or host colonization, but the specific role of OXA-48 had not been addressed (Beyrouthy et al., 2013, 2014; de Toro et al., 2017). Since the dissemination is not driven by a single sequence type (ST), it is possible that OXA-48 itself or blaOXA–48 harboring plasmids contribute to fitness and virulence of clinical isolates which would provide a survival benefit. We therefore investigated if acquisition of blaOXA–48 carrying plasmids affects fitness or virulence of E. coli and K. pneumoniae. For this purpose, OXA-48 producing clinical isolates of E. coli and K. pneumoniae were analyzed by whole genome sequencing (WGS) including single molecule real time sequencing (SMRT). Additionally, factors involved in dissemination and virulence were investigated in silico, in vitro, and in vivo.
Materials and Methods
Clinical Isolates
A total of 35 clinical, non-copy blaOXA–48 carrying E. coli and K. pneumoniae strains were included, which were isolated in 2010–2017 in two large German tertiary care hospitals (University Hospitals of Cologne and Frankfurt). All isolates were phenotypically characterized and tested for blaOXA–48 by PCR and Sanger sequencing as described previously (Göttig et al., 2015; Hamprecht et al., 2016; Koroska et al., 2017). Minimal inhibitory concentrations (MIC) were determined by MIC test strips (Liofilchem, Roseto degli Abruzzi, Italy) and interpreted according to CLSI breakpoints. Multilocus sequence typing (MLST) was performed either by PCR employing the Pasteur scheme1 or by WGS using the CGE Sequence Typer Tool.
Horizontal Gene Transfer and Analysis of blaOXA–48 Plasmids
Horizontal gene transfer (HGT) of blaOXA–48 was evaluated by liquid mating assays using clinical Enterobacteriaceae isolates as donors and sodium azide-resistant strains E. coli J53 (EC-J53) and K. pneumoniae PRZ (KP-PRZ), Serratia marcescens PRaT, Enterobacter cloacae DdL, and Citrobacter freundii ULTN as recipients as described (Hamprecht et al., 2013; Gruber et al., 2015). Transconjugants (Tc) were selected employing chromogenic agar plates containing 20 μg/mL amoxicillin-clavulanate (ACA) and 100 μg/mL sodium azide. Transconjugation frequency was determined by dividing the numbers of Tc by the numbers of donors. Estimation of the blaOXA–48 carrying plasmid size was done by S1 nuclease digestion and pulsed-field gel electrophoresis (PFGE). PCR and Sanger sequencing was employed to determine the incompatibility group of OXA-48 plasmids (Carattoli et al., 2015) and the Tn1999 transposon variants (Supplementary Table 1).
Next Generation Sequencing and Data Analysis
Whole genome sequencing was carried out using the Illumina MiSeq platform applying a v3 reagent kit (Illumina, San Diego, CA, United States) generating 150 or 250 bp paired-end reads with coverage of ≥50. De novo assembly and scaffolding after quality trimming of the reads was conducted using SPAdes v3.12.0 with standard parameters (Bankevich et al., 2012). To generate complete genome sequences, SMRT long-read sequencing (Pacific Biosciences, Menlo Park, CA, United States) in combination with Illumina short-read sequencing was applied in selected isolates as described (Steglich et al., 2018).
Genomes were annotated using Prokka 1.8 software (Seemann, 2014), and annotation was corrected manually. Genome sequences were visualized using the BRIG-Software and SnapGene® Viewer (Alikhan et al., 2011). Sequence comparison of plasmids was performed and visualized using an in-house Perl script utilizing BLAST. Short reads of isolates harboring the 63 kb IncL plasmid were mapped to the plasmid sequence from EC7215 (NZ_LR025098.1) used as a reference employing Bowtie2 and visualized using Geneious 11.1.52 (Langmead and Salzberg, 2012). Fully closed genome sequences were submitted to NCBI GenBank and can be found under accession number LR025099–LR025100 (KP12536) and LR025096–LR025098 (EC7215). Screening for virulence genes was performed using K. pneumoniae BIGS database, VFDB3 and a database which was created based on selected publications (Supplementary Figures 1, 2). Antimicrobial resistance genes were identified using the tool ABRicate4 applying CGE ResFinder database (Zankari et al., 2012).
Determination of Cytotoxicity
A549 human lung epithelial cells (ATCC® CCL-185) were grown in RPMI medium (Biochrom GmbH, Berlin, Germany) with 10% fetal calf serum (FCS) (Sigma, Taufkirchen, Germany) at 37°C until almost confluent and infected with bacteria at a multiplicity of infection (MOI) of 100. After 24 h, the supernatant was filtered (0.45 μm) and the lactate dehydrogenase (LDH) activity was determined by spectrophotometry (Cobas 8000, module 701; Roche, Grenzach-Wyhlen, Germany).
Evaluation of in vivo Virulence Using the Galleria mellonella Infection Model
Larvae of the greater wax moth (G. mellonella) were obtained from UK Waxworms (Sheffield, United Kingdom). Infection of larvae, read-out, and controls were performed as previously described (Göttig et al., 2016). Median lethal doses (LD50) were calculated by non-linear regression analysis using GraphPad Prism 5.0 (La Jolla, CA, United States) (Gruber et al., 2015).
Evaluation of Bacterial Fitness and Plasmid Stability
For analysis of non-competitive growth kinetics, brain heart infusion (BHI) broth (Becton Dickinson, Heidelberg, Germany) were inoculated with an overnight culture and incubated at 37°C and shaking until an optical density of 0.5 at 600 nm (OD600) was reached. Bacteria were adjusted to an OD600 of 1 in phosphate-buffered saline (PBS), serially diluted and inoculated into an aerobe BD Bactec bottle (BD, Heidelberg, Germany), resulting in a final inoculum of 103 colony-forming units (cfu) per bottle. Bacterial growth was recorded for 24 h at 37°C in a Bactec Fx instrument (BD) by measuring the fluorescence intensity every 15 min. The doubling time was calculated using MS Excel and the Solver tool.
To assess the impact of blaOXA–48 carriage on bacterial fitness, pairwise competition assays with Tc carrying the natural plasmid pOXA-48 from EC7215 were employed. EC-J53 Tc7215 and its parental strain EC-J53 were analyzed as described before (Lenski et al., 1994; Göttig et al., 2016). For determination of cfu, serial dilutions of each culture were plated on Mueller Hinton agar (MHA) and MHA containing 12 mg/L ACA (MHA-ACA). Selection rate constant (s) was calculated as described by Lenski et al. (1994) with negative s-values indicating a loss of fitness. To estimate plasmid stability, a pure culture of EC-J53 Tc7215 and KP-PRZ Tc7215 was cultured under the same conditions as for the pairwise competitions experiments in antibiotic-free medium. The ratio of colonies growing on MHA-ACA compared to antibiotic-free MHA was determined after three transfers at 90 h in triplicate.
Statistical Analysis
Categorical variables were compared using the Chi-square or Fisher’s exact test where appropriate. Continuous variables were assessed by Mann–Whitney U test. A P-value of <0.05 was considered significant.
Ethics Statement
All bacterial strains were isolated as part of routine microbiological diagnostic and stored in an anonymized database. According to the ethic committee of the Hospital of Johann Wolfgang Goethe-University, Frankfurt am Main, no informed consent or ethical approval of the study is necessary.
Results
Whole-Genome Sequencing and Molecular Characterization of OXA-48 Producing Clinical Isolates
Thirty-five non-copy clinical isolates producing OXA-48 including 15 E. coli and 20 K. pneumoniae were characterized (Tables 1, 2). For E. coli, ST38 (n = 5) and ST354 (n = 4) were the most frequent STs. E. coli isolates harbored blaOXA–48 on a mobilizable plasmid in 10/15 cases. In 6/15 isolates blaOXA–48 was embedded in a Tn1999 structure which was located on a 63 kb IncL plasmid, whereas one isolate harbored an IncL plasmid with a size of 75 kb. Three isolates carried IncF plasmids of different sizes (80–89 kb). In five isolates, blaOXA–48 could be transferred neither by conjugation nor by electroporation, indicating a chromosomal location, which was confirmed by WGS and PCRs amplifying Tn6237 embedded blaOXA–48 and the adjacent chromosomal genetic environment (Turton et al., 2016).
TABLE 1.
Characteristics of OXA-48 harboring E. coli isolates.
|
Isolate |
Genetic support of OXA-48 |
MIC (mg/L) |
||||||||||
| Isolatea | ST (Warwick) | ST (Pasteur) | Specimen | Transposonb | Plasmid size | IncL | CTX | CAZ | ETP | IMP | MEM | CIP |
| EC1027 | 297 | 666 | Rectal swab | Tn1999.2 | 63 kb | + | 256H | 16 | >32 | >32 | 16 | >32 |
| EC1639 | 393 | 494 | Blood | invTn1999.2 | 63 kb | + | 1 | 0.25 | 4 | 4 | 0.5 | >32 |
| EC2058 | 38 | 8 | Rectal swab | Tn6237 | Chromosomal | – | 64 | 16 | 2 | 0.5 | 0.25 | 0.25 |
| EC2196 | 38 | 8 | Rectal swab | Tn6237 | Chromosomal | – | 64 | 2 | 16 | 8 | 2 | 0.25 |
| EC2269 | 38 | 8 | Rectal swab | ΔTn6237 | Chromosomal | – | 64 | 1 | 2 | 2 | 2 | 0.25 |
| EC2667 | 354 | 39 | Rectal swab | Tn1999 | 63 kb | + | >256 | >256 | 4 | 4 | 2 | >32 |
| EC2700 | 354 | 39 | Rectal swab | Tn1999 | 63 kb | + | 256H | 256H | 16 | 16 | 2 | >32 |
| EC3124 | 38 | 8 | Rectal swab | Tn6237 | Chromosomal | – | 256H | 2 | 4 | 4 | 1 | 0.25 |
| EC3239 | 10 | 262 | Bile fluid | invTn1999.2 | 75 kb | + | 4 | 0.5 | 4 | 4 | 2 | >32 |
| EC3338 | 354 | 39 | Rectal Swab | ΔTn6237 | 87 kb | – | 256H | 4 | 2 | 1 | 0.25 | >32 |
| EC3428 | 38 | 8 | Rectal swab | Tn6237 | Chromosomal | – | 32 | 1 | 2 | 4 | 1 | 0.25 |
| EC3471 | 1598 | 398 | Rectal swab | Tn1999.2 | 63 kb | + | 32 | 64 | 1 | 2 | 0.25 | 0.5 |
| EC7215 | 448 | 58 | Rectal swab | Tn1999.2 | 63 kb | + | 0.5 | 0.25 | 1 | 1 | 0.5 | >32 |
| EC9629 | 354 | 39 | Blood | ΔTn6237 | 80 kb | – | 64 | 2 | 1 | 1 | 0.5 | >32 |
| EC14604 | 1598 | 398 | Rectal swab | ΔTn6237 | 89 kb | – | >256 | 16 | 0.25 | 0.5 | 0.12 | 0.25 |
H, heteroresistance; CTX, cefotaxime; CAZ, ceftazidime; ETP, ertapenem; IMP, imipenem; MEM, meropenem; CIP, ciprofloxacin; aIsolates which were further assessed in virulence assays are highlighted in bold. bTn6237 refers to KT444705 (Genbank); ΔTn6237 of EC2269, EC3338, EC9629, and EC14604 refer to a 19.9 kb fragment of KT444704, KT444705 (Genbank), 3.3 kb fragment of KT444704, and 21.5 kb fragment of KT444705, respectively.
TABLE 2.
Characteristics of OXA-48 harboring K. pneumoniae isolates.
|
Isolate |
Genetic support of OXA-48 |
MIC (mg/L) |
|||||||||
| Isolatea | ST (Pasteur) | Specimen | Transposon | Plasmid size | IncL | CTX | CAZ | ETP | IMP | MEM | CIP |
| KP659 | 15 | Stool | Tn1999.2 | 63 kb | + | >256 | 32 | >32 | 0.5 | 2 | >32 |
| KP920 | 16 | Wound swab | Tn1999.2 | 63 kb | + | >256 | >256 | >32 | >32 | >32 | >32 |
| KP980 | 16 | Rectal swab | Tn1999.2 | 63 kb | + | >256H | 16 | >32 | 2 | 2 | >32 |
| KP1151 | 15 | Rectal swab | Tn1999.2 | 63 kb | + | >256 | >256 | >32 | >32 | >32 | >32 |
| KP1664 | 16 | Throat swab | Tn1999.2 | 63 kb | + | 256H | 64 | 32 | 32 | 32 | >32 |
| KP1673 | 101 | Urine | Tn1999.2 | 63 kb | + | >256 | >256 | >32 | 32 | >32 | >32 |
| KP1696 | 377 | Skin swab | invTn1999.2 | 63 kb | + | >256 | >256 | 3 | 1 | 1 | >32 |
| KP1766 | 1658 | Skin swab | invTn1999.2 | 63 kb | + | >256 | >256 | >32 | 16 | 32 | >32 |
| KP1982 | 377 | Wound swab | Tn1999.2 | 63 kb | + | >256 | >256 | 32 | 8 | 16 | >32 |
| KP2255 | 437 | Rectal swab | invTn1999.2 | 63 kb | + | >256 | 128 | 32 | 32 | 32 | >32 |
| KP2451 | 392 | Rectal swab | Tn1999.2 | 63 kb | + | >256 | >256 | 2 | 16 | 4 | >32 |
| KP2540 | 392 | Rectal swab | invTn1999.2 | 63 kb | + | 64 | 128 | 2 | 4 | 2 | > 32 |
| KP2575 | 3384 | Rectal swab | Tn1999 | 63 kb | + | 4 | 2 | >32 | 8 | 8 | 0.06 |
| KP2670 | 101 | Tracheal secretion | Tn1999 | 63 kb | + | 0.5 | 0.25 | 2 | 2 | 1 | 0.03 |
| KP2746 | 101 | Liver biopsy | invTn1999.2 | 63 kb | + | 2 | 0.5 | 16 | 8 | 4 | 0.03 |
| KP3379 | 307 | Urine | Tn1999.2 | 63 kb | + | 2 | 1 | 4 | 2 | 4 | 16 |
| KP3907 | 307 | Stool | Tn1999.2 | 63 kb | + | >256 | 32 | >32 | 2 | 1 | >32 |
| KP12536 | 3385 | Tracheal secretion | Tn1999.2 | 63 kb | + | 0.5 | 0.25 | 16 | 2 | 4 | 0.03 |
| KP12883 | 323 | Urine | invTn1999.2 | 63 kb | + | 0.5 | 0.25 | 1 | 8 | 1 | >32 |
| KP13815 | 11 | Wound swab | Tn1999.2 | 63 kb | + | >256 | >256 | 2 | 4 | 2 | >32 |
H, heteroresistance; CTX, cefotaxime; CAZ, ceftazidime; ETP, ertapenem; IMP, imipenem; MEM, meropenem; CIP, ciprofloxacin; aIsolates which were further assessed in virulence assays are highlighted in bold.
In K. pneumoniae isolates, STs varied considerably (12 different STs). Strikingly, in all isolates blaOXA–48 was located on an IncL plasmid with a size of ∼63 kb and embedded in a Tn1999 family transposon (Table 2). Tn1999.2 was the most common transposon variant (n = 12), followed by its inverted version invTn1999.2 (n = 6) and two isolates harboring Tn1999.
In 30 of 35 isolates, blaOXA–48 was identified on plasmids of which 26 were 63 kb IncL plasmids. Of these, five plasmids were completely sequenced by both Illumina® short-read and SMRT long-read technology revealing a sequence identity of each ≥99.9%. To analyze the 63 kb IncL plasmids of the 21 isolates, which were solely sequenced by short-read technology, the sequencing reads of each isolate were mapped to the complete sequence of the 63 kb plasmid IncL from EC7215 (Brehony et al., 2019). Thereby, a median coverage of >50 of each isolate to the reference sequence from EC7215 was noticed, suggesting the presence of highly similar plasmids in all these isolates (Supplementary Figure 1). The IncL plasmid from EC3239 had a length of 75,552 bp, a sequence identity of >99% compared to the 63 kb IncL plasmids and carried an additional 8.3 kb Tn3 transposon and a 4.4 kb fragment of a Col440I plasmid (highest sequence homology to CP039328.1 and AF527822.1, respectively).
The three isolates EC14604, EC3338, and EC9629 harbored blaOXA–48 on IncF plasmids of different sizes (80–89 kb). The sequences of the plasmids from EC3338 and EC9629 were highly similar and both plasmids shared the same IncF type F-:A1:B32. In contrast, the blaOXA–48 plasmid from EC14604 had a different IncF type (F2:A-:B-) and displayed a lower sequence similarity compared to the two other EC isolates (Supplementary Figure 2). The transposon structures containing blaOXA–48 on these three IncF plasmids were more diverse compared to the IncL plasmids. The plasmid from EC14604 harbored a 21,571 bp fragment of Tn6237 in which one of the two copies of the flanking IS1R mobile elements was truncated by a Tn3-type and a IS6-like type insertion element. In EC3338, the blaOXA–48 plasmid harbored a 12,192 bp fragment of Tn6237, flanked on one side by a complete 767 bp IS1R-element and a 472 bp fragment of an IS1R-element on the other side. The IncF plasmid from isolate EC9629 harbored only a short fragment of Tn6237 consisting of two copies of IS1R sequences up and downstream of blaOXA–48 and a truncated lysR gene.
Antimicrobial susceptibility testing revealed highly variable MICs for carbapenems with meropenem MICs ranging from 0.125 to 16 mg/L in E. coli and from 1 to >32 mg/L in K. pneumoniae. Ceftazidime remained susceptible in 8/15 E. coli and 4/20 K. pneumoniae isolates. Ciprofloxacin resistance was common in both E. coli (8/15) and K. pneumoniae (16/20). WGS revealed that 8 of 15 E. coli isolates carried genes encoding ESBLs, with CTX-M-24 being the most frequent (7/9), and two isolates expressing CMY-42 AmpC β-lactamases, which is in line with the resistance phenotype (Supplementary Table 2). Interestingly, CTX-M-15 was present in 13/14 ESBL-producing K. pneumoniae isolates but detected in only one strain of E. coli, whereas CTX-M-24 was found only in E. coli.
Taken together, clinical OXA-48 isolates from Germany are diverse, but mostly carry blaOXA–48 on a 63 kb sized IncL plasmid. In K. pneumoniae blaOXA–48 was exclusively linked to a Tn1999 type transposon and the 63 kb sized IncL plasmid. In E. coli, genetic support of blaOXA–48 was more diverse, but the same 63 kb plasmid was detected in half of the isolates. From the 35 clinical isolates, ten representative K. pneumoniae and E. coli each were selected based on different criteria (ST type, transposon type, isolation site, and MIC) to study HGT, virulence, and fitness.
Horizontal Gene Transfer of blaOXA–48
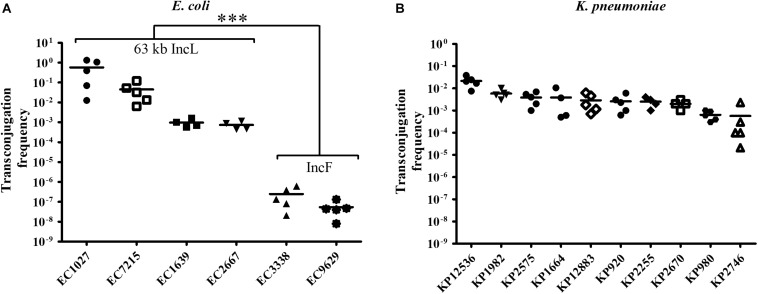
To investigate HGT efficiency of pOXA-48, liquid mating assays were performed using E. coli isolates as donors and sodium azide-resistant EC-J53 as a recipient. Except for the five E. coli isolates with chromosomal localization of blaOXA–48, pOXA-48 could be transferred indicating successful intraspecies transconjugation. The conjugation frequency was high for all donors carrying the 63 kb IncL plasmid (median frequency: 1.7 × 10–1 ± 3.8 × 10–1). In contrast, a significantly lower frequency of 1.5 × 10–7 ± 1.9 × 10–7 was determined for the isolates with IncF plasmids (P < 0.0001, Figure 1A). In K. pneumoniae, pOXA-48 could be transferred to EC-J53 in all cases with a high conjugation frequency of 4.7 × 10–3 ± 7.1 × 10–3, indicating efficient intergenus transconjugation (Figure 1B). The 63 kb IncL plasmid could furthermore be conjugated from E. coli and K. pneumoniae into other sodium azide-resistant Enterobacterales species including K. pneumoniae PRZ, S. marcescens PRaT, E. cloacae DdL and C. freundii ULTN with mean transconjugation frequencies of 6.8 × 10–3–1.3 × 10–6 (Supplementary Figure 3).
FIGURE 1.
Transconjugation frequency during horizontal transfer of plasmids harboring blaOXA–48 from E. coli and K. pneumoniae clinical isolates to EC-J53. Each dot represents the frequency of a single transconjugation experiment, whereas the bars indicate the arithmetic mean of all experiments. (A) Transconjugation frequency from E. coli (EC) donors. The first four EC isolates harbor a 63 kb IncL blaOXA–48 plasmid with a Tn1999 type plasmid (“63 kb IncL”), whereas EC3338 and EC9629 contain an IncF plasmid. ∗∗∗P < 0.0001 (Mann–Whitney U test). (B) Transconjugation frequency from K. pneumoniae (KP) donors.
These results suggest that the 63 kb IncL blaOXA–48 plasmid occurs in different K. pneumoniae and E. coli lineages, has a broad host range in Enterobacterales and can be mobilized by efficient intraspecies and intergenus HGT with a high frequency of up to 5.7 × 10–1.
Virulence of OXA-48 Producing Clinical Isolates
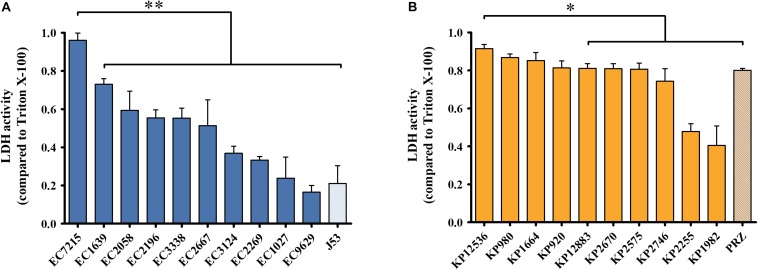
To evaluate virulence traits of clinical OXA-48 producing isolates, cytotoxicity toward human host cells and virulence in an in vivo infection model was investigated. First, human lung epithelial cells were infected with the 20 selected E. coli and K. pneumoniae isolates and the release of the intracellular enzyme LDH was measured as a surrogate marker for cytotoxicity (Figure 2). After infection with E. coli isolates, the mean LDH activity compared to the positive control was 37.8 ± 24.1% (Figure 2A). EC7215 was significantly more cytotoxic than all other strains (P < 0.0001). LDH activity was high after infection with K. pneumoniae (mean: 76.0 ± 18.6%), indicating high species-specific membrane disintegration of K. pneumoniae compared to E. coli (P < 0.0001). KP12536 was the most cytotoxic K. pneumoniae isolate, whereas KP2255 and KP1982 were significantly less cytotoxic compared to all other isolates (P < 0.05).
FIGURE 2.
Cytotoxicity of OXA-48 isolates. (A) E. coli and (B) K. pneumoniae. LDH activity was measured from supernatants of infected A549 lung epithelial cells. Mean values from four independent experiments were compared to that of Triton X-100 which served as the positive control. Error bars represent standard deviations. ∗∗P < 0.0001; ∗P < 0.05 (Mann–Whitney U test).
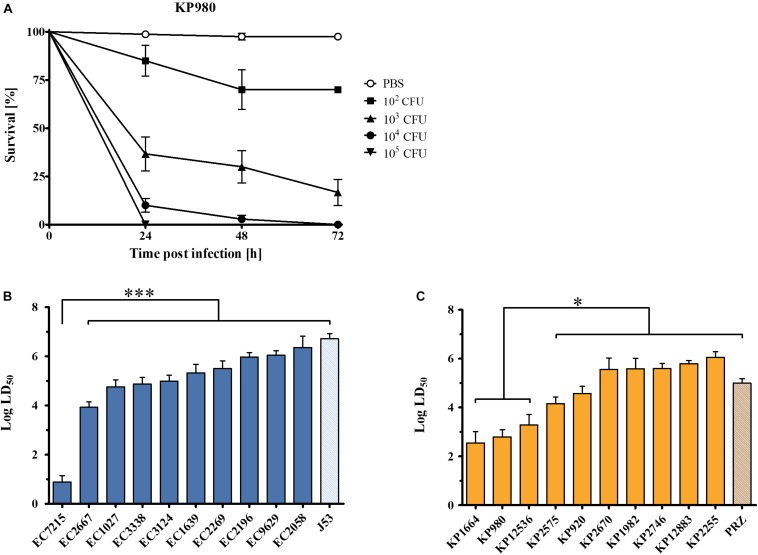
To assess pathogenicity in vivo, the G. mellonella infection model was employed. To compare virulence across the isolates, time-kill curves were compared and median lethal dose (LD50) values were calculated. All E. coli and K. pneumoniae isolates caused a time- and dose-dependent killing of larvae (Figure 3A). The mean LD50 was 4.9 for E. coli (range: 0.9–6.4) and 4.6 for K. pneumoniae (range: 2.5–6.1). Among E. coli isolates, EC7215 was highly virulent [LD50 0.9 (95% CI 0.63–1.14)] compared to all other clinical strains and the reference strain J53, which was the least virulent [LD50 6.7 (95% CI 6.50–6.91)]. In K. pneumoniae, the isolates KP980, KP1664, and KP12536 were significantly more virulent (LD50 2.5–3.3) than the other clinical isolates and KP-PRZ (P < 0.05). In summary, EC7215 and KP12536 were highly virulent in both assays (Figures 2, 3).
FIGURE 3.
Virulence of OXA-48 producing Enterobacteriaceae clinical isolates in the Galleria mellonella in vivo infection model. (A) Dose-dependent mortality of G. mellonella infected with KP980. Different cfu were injected into the larvae and survival was monitored for 72 h. Error bars show standard error of the mean. (B) Log median lethal dose (LD50) of clinical E. coli isolates. (C) LD50 of K. pneumoniae isolates. Mean values from three to four independent experiments with 12 larvae in each group are shown. Error bars represent the 95% confidence interval. ∗∗∗P < 0.0001; ∗P < 0.05 (Mann–Whitney U test).
In silico Analysis of Virulence
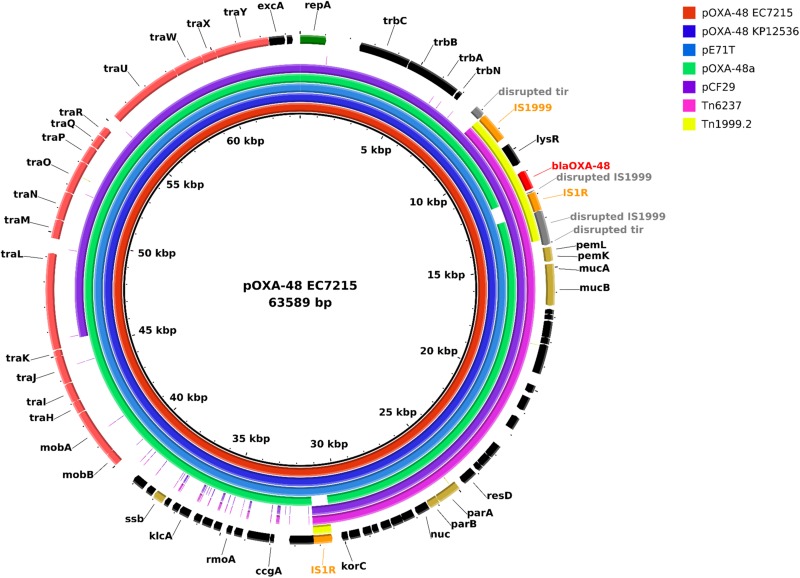
To investigate virulence mediated by the blaOXA–48 carrying plasmids in silico, the two highly virulent isolates EC7215 and KP12536 were sequenced by SMRT to obtain the complete sequence of the plasmids. The chromosome of EC7215 harbored a genome of 4,830,938 bp and a GC content of 50.8%, compared to 5,323,688 bp and 57.5% in KP12536. Both isolates contained an identical blaOXA–48 encoding plasmid of 63,589 bp (Figure 4), which showed a 99.9% similarity to strain KPN-El-Nr.7 (Carattoli et al., 2015). The plasmid contained 41 genes and 28 open reading frames (ORF) encoding hypothetical proteins of unknown function. Among the predicted genes located on the plasmid, no virulence gene could be detected by any of the virulence gene databases (see section “Materials and Methods”).
FIGURE 4.
Plasmid map of pOXA-48 and comparison of plasmids harboring blaOXA–48 and the transposons Tn1999.2 and Tn6237. Each ring represents the nucleotide sequence of a plasmid or the transposon Tn6237 as indicated by the color code. Sequences of pE71T (accession no. KC335143.1), pOXA-48a (JN626286.1), pCF29 (LN864820.1), and Tn6237 (HG977710.1) were retrieved from the NCBI nucleotide database. Gene annotation of the reference plasmid pOXA-48 EC7215 is shown in the outer ring. Tn1999.2 indicates the transposon region on all plasmids in this graph. Colored arrows indicate blaOXA–48 (red), insertion elements (orange), genes for plasmid stabilization (gold), and replication (green), disrupted genes (gray) and genes of unknown function (black).
Whole genome analysis demonstrated that E. coli isolates harbored 70–122 genes associated with virulence, while K. pneumoniae isolates possessed 42–59 virulence genes and were less diverse regarding the presence of virulence genes (Supplementary Figures 1, 2). Analysis of all thirteen isolates sequenced by SMRT showed that the vast majority of virulence genes was located on the chromosome, whereas only five different virulence genes were identified on plasmids which, however, did not harbor blaOXA–48. E. coli isolates harbored genes encoding virulence factors of different cellular function, e.g., adhesion, invasion, effector protein secretion, and the pathogenicity island PAIIICFT073. The virulence factors wabG and uge, which are important for host colonization and virulence and kfu which is involved in ion homeostasis, were present in all K. pneumoniae clinical isolates. Twenty-one virulence genes were detected in all isolates, e.g., the adhesin focA or the toxin astA which is commonly found in enterohemorrhagic E. coli (EHEC) and enteroaggregative E. coli (EAggEC). The mean number of virulence genes was 101 in E. coli and 47 in K. pneumoniae isolates. The isolates with the highest virulence in the Galleria model, E. coli EC7215 (86 virulence genes) and K. pneumoniae KP1664 (42 virulence genes) did not show higher numbers or distinct patterns of virulence genes compared to other isolates of the same species. In contrast, more virulence genes were found in EC1639 (122 virulence genes), which was isolated from a blood stream infection, but which neither showed a higher cytotoxicity nor a lower LD50 compared to the other isolates.
Virulence and Fitness of blaOXA–48 Harboring Transconjugants
The plasmid pOXA-48 EC7215 contained 28 ORFs of unknown function which potentially could encode for proteins mediating virulence or fitness. Therefore, transconjugants carrying blaOXA–48 (Tc) were used to investigate the impact of pOXA-48 on pathogenicity and fitness and compared to their isogenic E. coli and K. pneumoniae parental strains.
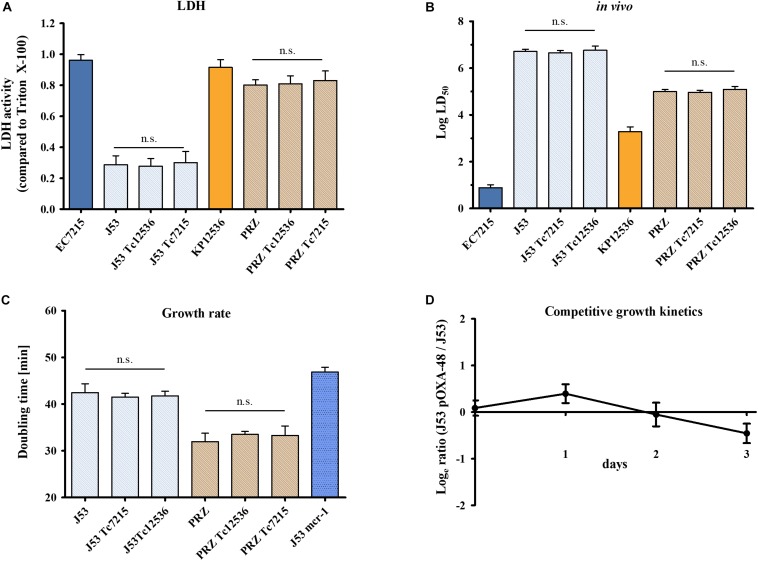
The infection of A549 cells with EC-J53 and J53 transconjugants harboring pOXA-48 from EC7215 (J53 Tc7215) and from KP12536 (J53 Tc12536) caused a LDH activity of 0.29 ± 0.06, 0.28 ± 0.05, and 0.30 ± 0.07, respectively, thereby showing no effect of pOXA-48 carriage on cytotoxicity (Figure 5A). Likewise, carriage of pOXA-48 in K. pneumoniae Tc and parental strains did not alter cytotoxicity toward A549 cells. In comparison, the clinical strains EC7215 (LDH 0.96 ± 0.04) and KP12536 (LDH 0.92 ± 0.05) were significantly more cytotoxic than all transconjugants (P < 0.01).
FIGURE 5.
Impact of pOXA-48 carriage on virulence and fitness. Cytotoxicity, virulence, and growth kinetics of clinical isolates EC7215 and KP12536 harboring pOXA-48, acceptor strains J53 and PRZ, and pOXA-48 transconjugants (Tc) were analyzed. (A) Cytotoxicity was determined by the LDH assay and (B) virulence by G. mellonella infection experiments as described above. (C) Doubling times were calculated from growth kinetics. An EC-J53 control strain (“J53 mcr-1”) harboring the plasmid pKP2442-mcr-1, whose carriage causes a fitness loss, was used as a control (Tietgen et al., 2018). Bars represent the mean (n = 4). Error bars denote standard error (A,C) or the 95% CI (B). n.s., no statistically significant differences as determined by the Mann–Whitney U test. (D) Dynamics of replicate competition experiments for EC-J53 and transconjugants containing the natural plasmid pOXA-48. The ratio of resistant versus susceptible cfu is shown (mean values from four independent experiments). Error bars denote the standard deviation of mean cfu counts.
To further explore virulence mediated by OXA-48, G. mellonella larvae were infected with transconjugants and their isogenic parental strains (Figure 5B). LD50 values were similar for EC-J53 Tc (log LD50 = 6.65 ± 0.10 and 6.76 ± 0.18) and the parental strain (log LD50 = 6.71 ± 0.10), P = 0.74, and P = 0.98, respectively. Likewise, carriage of pOXA-48 in KP-PRZ did not have a significant impact on LD50 values, indicating no substantial effect of blaOXA–48 expression on virulence in G. mellonella (P = 0.85 and P = 0.73). In summary, the acquisition of pOXA-48 did not result in higher cytotoxicity or in vivo virulence.
Impact of pOXA-48 on Bacterial Fitness
To analyze the impact of the acquisition of pOXA-48 on fitness, growth kinetics were compared between EC-J53 Tc7215 and Tc12536 and their parental strains (Figure 5C). Doubling times were 42.4 ± 1.9 min for J53, 41.5 ± 0.8 min for J53 Tc7215, and 41.8 ± 1.0 min for J53 Tc12536 (P > 0.05). In contrast, transconjugants harboring the plasmid pKP2442 containing the colistin resistance gene mcr-1 had a doubling time of 46.9 min (P = 0.0095), indicating a loss of fitness (Tietgen et al., 2018). In KP-PRZ, transconjugants grew as fast as the parental strain (doubling times: 32.0 ± 1.8 min for PRZ, 33.5 ± 0.6 min for PRZ Tc12536, and 33.3 ± 2.0 min for PRZ Tc7215, P > 0.05). In conclusion, acquisition of pOXA-48 did not have an impact on the doubling time when compared to the parental strains.
In addition, pairwise competitive growth experiments using minimal media were performed to further assess the impact of pOXA-48 on bacterial fitness. For this purpose, E. coli transconjugant J53 Tc7215 competed with its isogenic parental strain EC-J53 (Figure 5D) and only a minor impact on fitness was recorded in the absence of antibiotics (selection rate constant s = −0.18 ± 0.02).
To assess plasmid stability, the plasmid loss of transconjugants EC-J53 Tc7215 and KP-PRZ Tc7215 was assessed by serial subcultures in antibiotic-free medium. In E. coli, the ratio of cells with and without plasmid was 1.026 ± 0.035 and 0.999 ± 0.156 in KP-PRZ, indicating a stable insertion of pOXA-48 with no plasmid loss.
Discussion
The successful dissemination of OXA-48 producing isolates is currently not completely understood. In contrast to other successful β-lactamase producing clonal lineages (e.g., K. pneumoniae KPC-2 ST258 or E. coli CTX-M-15 ST131), OXA-48 isolates belong to different STs but frequently harbor similar plasmids (Beyrouthy et al., 2013; Potron et al., 2013). In this study, 35 OXA-48 producing E. coli and K. pneumoniae clinical isolates were molecularly characterized and factors involved in dissemination and virulence were investigated in silico, in vitro, and in vivo.
Antibiotic susceptibility, resistance determinants and STs were highly diverse in the isolates investigated and did not correlate with virulence properties. In contrast, all K. pneumoniae and the majority of E. coli isolates harbored the same blaOXA–48 encoding 63 kb IncL plasmid, which was highly transmissible in both intraspecies and intergenus transconjugation assays. This plasmid was highly similar to the first isolate of OXA-48 in Turkey in 2001 (Poirel et al., 2004) and the isolates KPN-El-Nr.7 from Switzerland (Carattoli et al., 2015). In contrast to the first description with Tn1999 in 2001, most of our clinical isolates harbored a Tn1999.2 transposon. Isolates with Tn1999.2 have been reported to hydrolyze imipenem at higher rates than isolates carrying Tn1999 due to increased expression of blaOXA–48 (Carrer et al., 2008). Genetic support of blaOXA–48 was more diverse in E. coli with blaOXA–48 being located on the chromosome or on different plasmids including non-IncL variants. These non-IncL plasmids could be mobilized with much lower frequencies of only 10–7 compared to 10–3 for IncL plasmids, indicating that dissemination of blaOXA–48 is facilitated by the 63 kb sized IncL plasmid. The OXA-48 transposon structures on these non-IncL plasmids consisted of fragments of the chromosomally located Tn6237. However, IS1R sequences were truncated by additional mobile genetic elements in two of three plasmids, leading to a likely dysfunctional transposon structure. In the third IncF plasmid, a 3,360 bp fragment of Tn6237 was found, forming a presumable intact and mobile transposon structure flanked by two IS1R sequences, which might be mobilizable into other plasmids or the chromosome. The high conjugation frequency of this plasmid has been attributed to the disruption of the tir gene, encoding a transfer inhibition protein, by insertion of the Tn1999 in pOXA-48 (Potron et al., 2014).
Transconjugants showed high plasmid stabilities in the absence of antibiotic pressure. Furthermore, the 63 kb plasmid could also be conjugated to other Enterobacterales species such as E. cloacae, S. marcescens, or C. freundii, indicating a broad host range. This is also evidenced by reports of OXA-48 expressing clinical isolates of different species (Dimou et al., 2012; Chen et al., 2015; Findlay et al., 2017). These properties might lead to persistence of OXA-48 in environmental niches, healthy humans and animals without antibiotic pressure, contributing to further dissemination (Pulss et al., 2018).
Because of the overwhelming epidemiological success of OXA-48, several studies previously suggested that OXA-48 does not only confer resistance to beta-lactams but is also associated with bacterial virulence and fitness (Beyrouthy et al., 2013, 2014; de Toro et al., 2017). To address this hypothesis, the LDH cytotoxicity assay using A549 human lung epithelial cells and the G. mellonella in vivo infection model were chosen. These two infection systems have been successfully used to compare virulence across strains from different isolation sites, to identify virulence factors and to analyze the impact of antibiotic resistance genes on fitness and virulence (Insua et al., 2013; Ciesielczuk et al., 2015; Göttig et al., 2016; Tietgen et al., 2018). Even though different virulence properties are analyzed by the LDH assay and the G. mellonella infection, a positive correlation between these infection models could be observed for both E. coli and K. pneumoniae in our study (R = 0.43 and R = 0.51, respectively).
The clinical isolates were highly diverse in their ability to damage human A549 cells, with LDH release ranging from 16.5 to 96.1% compared to the positive control. K. pneumoniae isolates were on average more cytotoxic compared to E. coli isolates indicating species-specific virulence (Figure 2). However, EC7215 and KP12536 were the most cytotoxic isolates from the two species. Likewise, these two isolates were highly virulent in the Galleria assay. For example, just 8 cfu of EC7215 were able to kill 50% of larvae after 24 h (Figure 3). Beyrouthy et al. (2013, 2014) and de Toro et al. (2017) reported that the presence of virulence genes, e.g., pap, sfa/foc, hly, and afa/dra are correlated with a virulent phenotype when analyzing eight isolates. However, when analyzing 20 isolates neither of these factors correlated with increased virulence or the numbers of virulence genes (Supplementary Figures 4, 5).
Since the dissemination of OXA-48 is not driven by a single clone but rather by the highly prevalent 63 kb-sized IncL plasmid, we investigated the impact of pOXA-48 on virulence and fitness employing isogenic OXA-48-expressing E. coli and K. pneumoniae transconjugants and parental strains lacking OXA-48. We completely sequenced pOXA-48 but could not identify any genes with known association to virulence or fitness. Yet, 28 ORFs of unknown function were found which hypothetically could have an impact on virulence. However, survival of larvae was not different in transconjugants harboring pOXA-48 compared with the plasmid-free parental strains. Likewise, pOXA-48 carriage did not influence cytotoxicity toward human cells. This is in line with the observation that most of the clinical isolates displayed different virulence phenotypes but harbored the same OXA-48 plasmid. Even though no effect of the 63 kb IncL pOXA-48 was observed in the strains J53 and PRZ, it cannot be excluded that pOXA-48 contributes to virulence in other host strains, since the plasmid-encoded hypothetical ORFs may uniquely interact with other bacterial hosts. Hence, future studies should analyze the impact of pOXA-48 on virulence in additional host strains and also plasmid-cured clinical isolates.
This work has some limitations. Clinical isolates from two large tertiary care hospitals were used, which might not be representative for the whole country. Furthermore, it is not possible to transfer the virulence and fitness data from in vitro and in vivo model systems directly to a clinical setting. For example, the bacterial isolates may have organ-specific adaptations during infection in patients which cannot be monitored with our infection models, or the uptake of the IncL plasmid into E. coli J53 may not accurately recapitulate transmissibility in real clinical settings. The in vitro fitness assay allows the comparison of different isolates under standardized conditions. However, the fitness observed is influenced by the choice of media and can therefore not fully comparable to the clinical situation where different nutrient conditions are present and other, more complex factors like the immune system could further influence growth. Nevertheless, to the best of our knowledge this is the most comprehensive study on OXA-48 clinical isolates and plasmids from Germany. Compared to previous studies, we investigated HGT and virulence of OXA-48 producers employing a large number of isolates using in vitro, in vivo, and in silico methods. Furthermore, we could demonstrate for the first time that the 63 kb IncL plasmid which is disseminated worldwide can be transferred more efficiently compared to IncF plasmids, which might be a reason for its successful dissemination. In addition, acquisition of the 63 kb OXA-48 plasmid had only a minor impact on the fitness of EC-J53 in the competitive fitness assay and also did not lead to impaired growth in vitro. This is in contrast to other carbapenemases like NDM-1, which resulted in a highly decreased fitness of EC-J53 (Göttig et al., 2016). The high prevalence and rapid dissemination of OXA-48 can therefore be rather attributed to a low fitness burden and high plasmid stability combined with a highly efficient HGT of the 63 kb IncL plasmid and not increased fitness or virulence.
Data Availability Statement
The datasets generated for this study can be found at NCBI BioProject accession no. PRJEB27471, PRJEB27468, PRJEB27481, and PRJEB27482.
Author Contributions
AH, MW, and SG designed the study. JS, CB, YS, FK, FW, TT, SR-C, and JK performed the experiments. JS, MW, CI, AG, and UN carried out sequencing and bioinformatic analyses. AH, JS, and SG analyzed the data and drafted the manuscript. All authors reviewed the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ahmad Saleh, Emily Huth, and Vera Junker for excellent technical assistance, Linda Falgenhauer, Paul G. Higgins, and Julia Wille for Illumina sequencing of blaOXA–48 harboring isolates, and Simone Severitt, Nicole Heyer, and Cathrin Spröer for continued SMRT sequencing support. We also thank the team of curators of the Institute Pasteur MLST and whole genome MLST databases for curating the data and making them publicly available.
Funding. This work was supported by DZIF (German Center for Infection Research, TTU 08.905; ANGEL study to AH, MW, CI, UN, and SG and the COSIMA study to AH, MW, UN, and SG) and the Frankfurter Promotionsförderung (FPF) to JS and CB. Additional support to UN was provided by the EU Horizon 2020 program, grant agreement number 643476.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02509/full#supplementary-material
References
- Alikhan N. F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyrouthy R., Robin F., Cougnoux A., Dalmasso G., Darfeuille-Michaud A., Mallat H., et al. (2013). Chromosome-mediated OXA-48 carbapenemase in highly virulent Escherichia coli. J. Antimicrob. Chemother. 68 1558–1561. 10.1093/jac/dkt051 [DOI] [PubMed] [Google Scholar]
- Beyrouthy R., Robin F., Dabboussi F., Mallat H., Hamze M., Bonnet R. (2014). Carbapenemase and virulence factors of Enterobacteriaceae in North Lebanon between 2008 and 2012: evolution via endemic spread of OXA-48. J. Antimicrob. Chemother. 69 2699–2705. 10.1093/jac/dku181 [DOI] [PubMed] [Google Scholar]
- Brehony C., Mcgrath E., Brennan W., Tuohy A., Whyte T., Brisse S., et al. (2019). An MLST approach to support tracking of plasmids carrying OXA-48-like carbapenemase. J. Antimicrob. Chemother. 74 1856–1862. 10.1093/jac/dkz136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton R., Akova M., Carmeli Y., Giske C. G., Glupczynski Y., Gniadkowski M., et al. (2012). Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 18 413–431. 10.1111/j.1469-0691.2012.03821.x [DOI] [PubMed] [Google Scholar]
- Carattoli A., Seiffert S. N., Schwendener S., Perreten V., Endimiani A. (2015). Differentiation of IncL and IncM plasmids associated with the spread of clinically relevant antimicrobial resistance. PLoS One 10:e0123063. 10.1371/journal.pone.0123063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrer A., Poirel L., Eraksoy H., Cagatay A. A., Badur S., Nordmann P. (2008). Spread of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Istanbul, Turkey. Antimicrob. Agents Chemother. 52 2950–2954. 10.1128/AAC.01672-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Al Laham N., Chavda K. D., Mediavilla J. R., Jacobs M. R., Bonomo R. A., et al. (2015). First report of an OXA-48-producing multidrug-resistant Proteus mirabilis strain from Gaza, Palestine. Antimicrob. Agents Chemother. 59 4305–4307. 10.1128/AAC.00565-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesielczuk H., Betts J., Phee L., Doumith M., Hope R., Woodford N., et al. (2015). Comparative virulence of urinary and bloodstream isolates of extra-intestinal pathogenic Escherichia coli in a Galleria mellonella model. Virulence 6 145–151. 10.4161/21505594.2014.988095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Toro M., Fernandez J., Garcia V., Mora A., Blanco J., De La Cruz F., et al. (2017). Whole genome sequencing, molecular typing and in vivo virulence of OXA-48-producing Escherichia coli isolates including ST131 H30-Rx, H22 and H41 subclones. Sci. Rep. 7:12103. 10.1038/s41598-017-12015-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimou V., Dhanji H., Pike R., Livermore D. M., Woodford N. (2012). Characterization of Enterobacteriaceae producing OXA-48-like carbapenemases in the UK. J. Antimicrob. Chemother. 67 1660–1665. 10.1093/jac/dks124 [DOI] [PubMed] [Google Scholar]
- Falagas M. E., Tansarli G. S., Karageorgopoulos D. E., Vardakas K. Z. (2014). Deaths attributable to carbapenem-resistant Enterobacteriaceae infections. Emerg. Infect. Dis. 20 1170–1175. 10.3201/eid2007.121004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay J., Hopkins K. L., Loy R., Doumith M., Meunier D., Hill R., et al. (2017). OXA-48-like carbapenemases in the UK: an analysis of isolates and cases from 2007 to 2014. J. Antimicrob. Chemother. 72 1340–1349. 10.1093/jac/dkx012 [DOI] [PubMed] [Google Scholar]
- Giani T., Conte V., Di Pilato V., Aschbacher R., Weber C., Larcher C., et al. (2012). Escherichia coli from Italy producing OXA-48 carbapenemase encoded by a novel Tn1999 transposon derivative. Antimicrob. Agents Chemother. 56 2211–2213. 10.1128/aac.00035-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M., O’shea A. M. J., Livorsi D. J., Mcdanel J. S., Jones M. M., Richardson K. K., et al. (2016). The effect of a nationwide infection control program expansion on hospital-onset gram-negative rod bacteremia in 130 veterans health administration medical centers: an interrupted time-series analysis. Clin. Infect. Dis. 63 642–650. 10.1093/cid/ciw423 [DOI] [PubMed] [Google Scholar]
- Göttig S., Gruber T. M., Stecher B., Wichelhaus T. A., Kempf V. A. (2015). In vivo horizontal gene transfer of the carbapenemase OXA-48 during a nosocomial outbreak. Clin. Infect. Dis. 60 1808–1815. 10.1093/cid/civ191 [DOI] [PubMed] [Google Scholar]
- Göttig S., Riedel-Christ S., Saleh A., Kempf V. A., Hamprecht A. (2016). Impact of blaNDM-1 on fitness and pathogenicity of Escherichia coli and Klebsiella pneumoniae. Int. J. Antimicrob. Agents 47 430–435. 10.1016/j.ijantimicag.2016.02.019 [DOI] [PubMed] [Google Scholar]
- Gruber T. M., Gottig S., Mark L., Christ S., Kempf V. A., Wichelhaus T. A., et al. (2015). Pathogenicity of pan-drug-resistant Serratia marcescens harbouring blaNDM-1. J. Antimicrob. Chemother. 70 1026–1030. 10.1093/jac/dku482 [DOI] [PubMed] [Google Scholar]
- Hamprecht A., Poirel L., Göttig S., Seifert H., Kaase M., Nordmann P. (2013). Detection of the carbapenemase GIM-1 in Enterobacter cloacae in Germany. J. Antimicrob. Chemother. 68 558–561. 10.1093/jac/dks447 [DOI] [PubMed] [Google Scholar]
- Hamprecht A., Rohde A. M., Behnke M., Feihl S., Gastmeier P., Gebhardt F., et al. (2016). Colonization with third-generation cephalosporin-resistant Enterobacteriaceae on hospital admission: prevalence and risk factors. J. Antimicrob. Chemother. 71 2957–2963. 10.1093/jac/dkw216 [DOI] [PubMed] [Google Scholar]
- Insua J. L., Llobet E., Moranta D., Perez-Gutierrez C., Tomas A., Garmendia J., et al. (2013). Modeling Klebsiella pneumoniae pathogenesis by infection of the wax moth Galleria mellonella. Infect. Immun. 81 3552–3565. 10.1128/IAI.00391-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajuria A., Praharaj A. K., Grover N., Kumar M. (2013). First report of an Enterobacter ludwigii isolate coharboring NDM-1 and OXA-48 carbapenemases. Antimicrob. Agents Chemother. 57 5189–5190. 10.1128/aac.00789-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroska F., Göttig S., Kaase M., Steinmann J., Gatermann S., Sommer J., et al. (2017). Comparison of phenotypic tests and an immunochromatographic assay and development of a new algorithm for detection of OXA-48-like carbapenemases. J. Clin. Microbiol. 55 877–883. 10.1128/JCM.01929-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascols C., Peirano G., Hackel M., Laupland K. B., Pitout J. D. (2013). Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-like enzymes in North America. Antimicrob. Agents Chemother. 57 130–136. 10.1128/AAC.01686-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski R. E., Simpson S. C., Nguyen T. T. (1994). Genetic analysis of a plasmid-encoded, host genotype-specific enhancement of bacterial fitness. J. Bacteriol. 176 3140–3147. 10.1128/jb.176.11.3140-3147.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manageiro V., Ferreira E., Pinto M., Canica M. (2014). First description of OXA-48 carbapenemase harbored by Escherichia coli and Enterobacter cloacae from a single patient in Portugal. Antimicrob. Agents Chemother. 58 7613–7614. 10.1128/aac.02961-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Bonnin R. A., Nordmann P. (2012). Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob. Agents Chemother. 56 559–562. 10.1128/AAC.05289-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Heritier C., Tolun V., Nordmann P. (2004). Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48 15–22. 10.1128/aac.48.1.15-22.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potron A., Poirel L., Nordmann P. (2014). Derepressed transfer properties leading to the efficient spread of the plasmid encoding carbapenemase OXA-48. Antimicrob. Agents Chemother. 58 467–471. 10.1128/AAC.01344-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potron A., Poirel L., Rondinaud E., Nordmann P. (2013). Intercontinental spread of OXA-48 beta-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro Surveill. 18:20549. 10.2807/1560-7917.es2013.18.31.20549 [DOI] [PubMed] [Google Scholar]
- Pulss S., Stolle I., Stamm I., Leidner U., Heydel C., Semmler T., et al. (2018). Multispecies and clonal dissemination of OXA-48 carbapenemase in Enterobacteriaceae from companion animals in Germany, 2009-2016. Front. Microbiol. 9:1265. 10.3389/fmicb.2018.01265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Steglich M., Hoffmann J. D., Helmecke J., Sikorski J., Spröer C., Riedel T., et al. (2018). Convergent loss of ABC transporter genes from Clostridioides difficile genomes is associated with impaired tyrosine uptake and p-cresol production. Front. Microbiol. 9:901. 10.3389/fmicb.2018.00901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietgen M., Semmler T., Riedel-Christ S., Kempf V. A. J., Molinaro A., Ewers C., et al. (2018). Impact of the colistin resistance gene mcr-1 on bacterial fitness. Int. J. Antimicrob. Agents 51 554–561. 10.1016/j.ijantimicag.2017.11.011 [DOI] [PubMed] [Google Scholar]
- Turton J. F., Doumith M., Hopkins K. L., Perry C., Meunier D., Woodford N. (2016). Clonal expansion of Escherichia coli ST38 carrying a chromosomally integrated OXA-48 carbapenemase gene. J. Med. Microbiol. 65 538–546. 10.1099/jmm.0.000248 [DOI] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67 2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found at NCBI BioProject accession no. PRJEB27471, PRJEB27468, PRJEB27481, and PRJEB27482.