Abstract
Peanut yield is severely affected by exchangeable calcium ion (Ca2+) deficiency in the soil. Arbuscular mycorrhizal (AM) symbiosis increases the absorption of Ca2+ for host plants. Here, we analyzed the physiological and transcriptional changes in the roots of Arachis hypogaea L. colonized by Funneliformis mosseae under Ca2+-deficient and -sufficient conditions. The results showed that exogenous Ca2+ application increased arbuscular mycorrhizal fungi (AMF) colonization, plant dry weight, and Ca content of AM plants. Simultaneously, transcriptome analysis showed that Ca2+ application further induced 74.5% of differentially expressed gene transcripts in roots of AM peanut seedlings. These genes are involved in AM symbiosis development, hormone biosynthesis and signal transduction, and carotenoid and flavonoid biosynthesis. The transcripts of AM-specific marker genes in AM plants with Ca2+ deprivation were further up-regulated by Ca2+ application. Gibberellic acid (GA3) and flavonoid contents were higher in roots of AM- and Ca2+-treated plants, but salicylic acid (SA) and carotenoid contents specifically increased in roots of the AM plants. Thus, these results suggest that the synergy of AM symbiosis and Ca2+ improves plant growth due to the shared GA- and flavonoid-mediated pathway, whereas SA and carotenoid biosynthesis in peanut roots are specific to AM symbiosis.
Subject terms: Gibberellins, Plant molecular biology, Plant signalling, Arbuscular mycorrhiza
Introduction
Peanut (Arachis hypogaea L.) is an important oil crop and protein source for humans that contributes 20% to oil production and 11% of the human protein supply per year. The yield is often limited by exchangeable Ca2+ deficiency in soil, which causes early embryo abortion in peanut1,2. Therefore, Ca2+ plays a crucial role in the growth and development of peanut. Calcium is an important macronutrient required for plant growth and development and represents 0.1 to 5% of all plant dry biomass3. Additionally, as a second messenger, Ca2+ has been shown to mediate various aspects of cell and plant development, such as cell division, cell polarity, cell elongation, photomorphogenesis, and biotic and abiotic stress responses4–6.
In peanut, Ca2+ partly regulates turnover of the PSII reaction center components to reduce the stress of photoinhibition to PSII7, and is involved in hormone-induced peanut pod formation by increasing gibberellic acid (GA) and auxin contents1. However, Ca2+ is largely confined to uptake via the young root tips and can only be taken up by the young root system from the soil and delivered to the shoot via the xylem, and it is not remobilized from old to young tissues8. Thus, Ca2+ deficiency commonly affects plant growth and development if the soil cannot be supplemented with exogenous Ca2+. Fortunately, most plants have coped with limited Ca2+ availability via the establishment of symbiotic associations with microbes, more specifically known as AM association. The fungi forming AM symbiosis belong to the subphylum Glomeromycotina9.
This symbiosis plays a significant role in the uptake of nutrients and the carbon cycle, and consequently impacts ecosystem sustainability10. To establish the symbiosis, plant roots recognize chemical signals from AMF, e.g. lipochitooligosaccharides and chitooligosaccharides, which trigger coordinated differentiation and form the symbiotic state11. In turn, AMF require signal communication from the plants that produce strigolactones (which are derived from the carotenoid synthesis pathway), flavonoids, and other diffusible signals exuded by plant roots that induce the germination of AMF spores and branched fungal hyphae12. Then, the AM symbiosis is established by the common symbiosis signaling pathway induced by calcium oscillation after perception of diffusible signals from the symbionts13. In the process of establishing the AM symbiosis, many AM-specific marker genes must be initiated by Ca2+ concentration change14, such as RAM1 (REDUCED ARBUSCULAR MYCORRHIZA 1), RAM2 (glycerol-3-phosphate acetyltransferase), CCD1 (carotenoid cleavage dioxygenase), PT1 (phosphate transporter), and DELLA15–17. These findings suggest that Ca2+ plays an important role in AM development.
From recognition of the fungi to establishment of the symbiosis, complicated transcriptional reprogramming occurs in plant roots, and many specifically expressed genes involved in development of the symbiosis have been identified in legumes18–20. Some of the changes associated with plant hormones were considered to play important roles in this symbiosis21, such as auxin, cytokinins (CKs), gibberellins (GAs), and strigolactones, and were also altered in the roots of AM plants16,22. In addition, increases in flavonoid and anthocyanin were considered to be indispensable in regulating the establishment of AM symbiosis22,23.
Even though the molecular basis of the improvement of plant nutrient acquisitions have been well characterized for phosphorus, nitrogen, sulfur, and potassium18,24,25, the role of plant uptake of Ca2+ needs further study. Several reports showed that a moderate level of Ca2+ supply enhanced the colonization of AMF26,27, and Ca2+ benefited the maintenance of a functioning mycorrhiza28. However, the transcriptional changes in plant roots colonized by an AMF accompanied with sufficient Ca2+ are still unknown.
Cui et al. (2019) demonstrated that AM symbiosis increased the Ca2+ content in peanut seedlings, and Ca2+ application can also promote the development of AM symbiosis29. However, the molecular mechanism of how AMF and Ca2+ application synergistically promote the growth of peanut seedlings is unclear. In this study, we investigated a combination of transcriptional changes, hormone and metabolomic analyses in roots of peanut seedlings inoculated by AMF and Ca2+ application and compared the observed changes with those in AM plants or Ca2+-treated plants. We observed that changes in secondary metabolites in roots of AM and Ca2+-treated plants coincide with the transcriptional regulation of related biosynthesis pathways. These alternations, such as the increases in GA3 and flavonoid content, were considered to be involved in the growth enhancement of peanut seedlings by the synergy of AMF with Ca2+ application.
Results
AM symbiosis improves the dry biomass of peanut
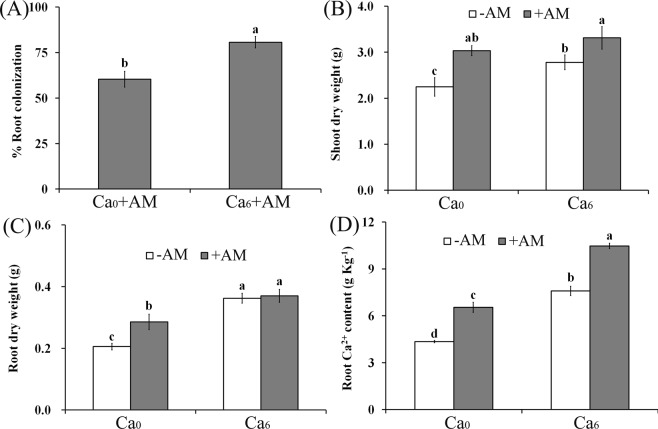
The quantification of AMF colonization showed that 60.33% and 80.67% of plant roots were inoculated by F. mosseae under both Ca2+-deficient and Ca2+-sufficient conditions, respectively (Fig. 1A), indicating that Ca2+ application could significantly improve the number of fungal colonizers.
Figure 1.
Impact of AM symbiosis on plant growth under Ca2+ deficient and sufficient conditions. (A) The rate of AMF colonization in peanut roots was assayed under Ca2+ deprivation and sufficiency. Shoot (B) and root (C) dry weight were determined in AM and NM plants under different Ca2+ treatments. (D) Ca2+ content was measured in roots of AM and NM plants under Ca2+ deficient and Ca2+ sufficient conditions. Letters represent significant differences between treatments and the control (one-way ANOVA, P < 0.05). Bars indicate means ± SD from six plants. DW, dry weight.
Shoot dry weight significantly increased in the AM plants compared with the nonmycorrhized (NM) plants and Ca2+ application further increased the shoot dry weight (Fig. 1B). Moreover, root dry weight was significantly increased in AM plants under Ca0 conditions and Ca2+ further improved the root dry weight; AM symbiosis did not increase the root dry weight (Fig. 1C). Additionally, the Ca2+ content was significantly higher in Ca2+-sufficient seedlings compared with Ca2+-deficient ones, and AM association improved Ca2+ level in roots (Fig. 1D).
Comparative analysis of differentially expressed genes (DEGs) in the AM and Ca2+-treated plants
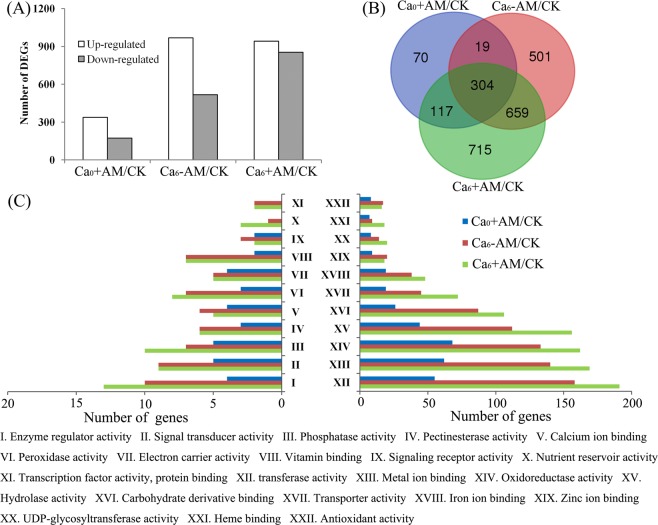
Using Ca0-AM as the control, there were 510, 1483, and 1795 significantly differentially expressed genes (DEGs) from roots of the Ca0 + AM, Ca6 − AM, and Ca6 + AM plants, respectively (Fig. 2A). In all, 304 DEGs were shared by Ca0 + AM, Ca6 − AM, and Ca6 + AM plants and the number of DEGs gradually increased in the plants (Fig. 2B), indicating that AM symbiosis combined with exogenous Ca2+ induced more transcriptional changes. In total, 421 DEGs were shared by Ca0 + AM and Ca6 + AM treatments, representing 82.55% and 23.45% of total DEGs in Ca0 + AM plants (510) and Ca6 + AM plants (1795), respectively. The expression levels of 380 DEGs in Ca0 + AM plants could be further regulated by Ca2+ application (Supplementary Fig. S1); only 40 DEGs were conversely regulated (Supplementary Table S1). This result implied that Ca2+ application could further strengthen the effects of AM on plant growth. In addition, 22 categories involved in molecular functions of GO enrichment analyses were identified, and the number of DEGs involved in transferase activity was the highest, followed by metal ion binding and oxidoreductase activity. Four categories, including calcium ion binding, signaling receptor activity, zinc ion binding, and antioxidant activity were the highest in Ca6 − AM plants; among the other 18 categories, the number of DEGs involved in each molecular function of GO was the highest in the Ca6 + AM plants, followed by the Ca6 − AM plants, and the Ca0 + AM plants (Fig. 2C).
Figure 2.
Transcriptional profiling of peanut roots with or without colonization by AMF under Ca2+ deficient and sufficient conditions. (A) The number of DEGs up-regulated and down-regulated in roots of Ca0 − AM, Ca6 − AM, and Ca6 + AM plants compared with Ca0-AM plants (the control). (B) Venn diagram showing the number of DEGs shared and specifically up- or down-regulated in roots of Ca0 − AM, Ca6 − AM, and Ca6 + AM plants. (C) Significantly enriched GO molecular function terms for the number of DEGs analyzed in different treatments.
To confirm RNA-Seq results, 15 genes were selected randomly from various functional categories and qRT-PCR analysis was conducted using RNA samples from the RNA-Seq experiments. The results were consistent with the expression levels of genes from RNA-Seq data (Fig. 3).
Figure 3.
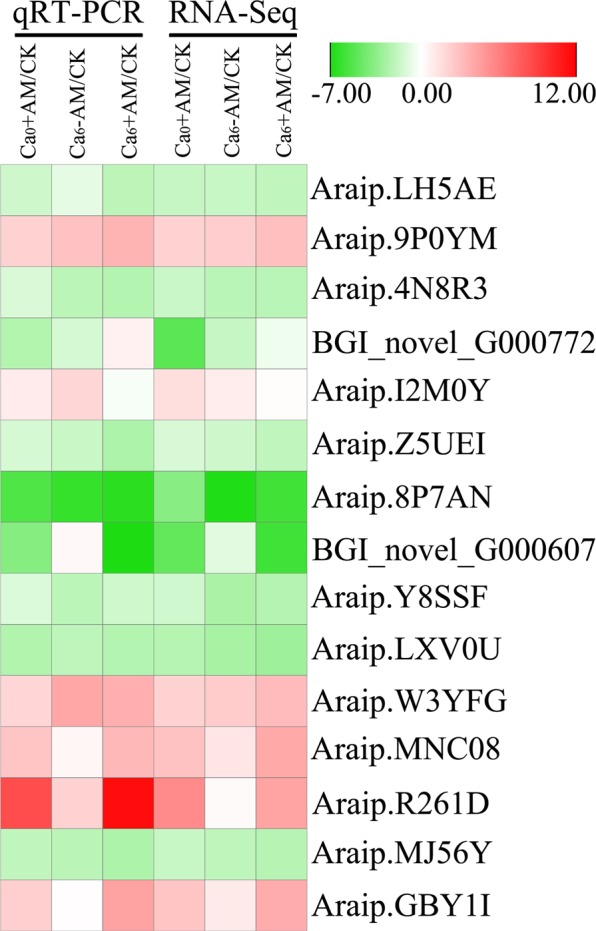
qRT-PCR verification of selected genes. Comparison of gene expression level from transcriptome analyses and qRT-PCR experiments.
Effects of Ca2+ combined with AM symbiosis on the transcripts of AM-specific markers and Ca-related genes
We analyzed the roles of Ca2+ application on the establishment of AM symbiosis. In total, 25 AM-specific marker genes were identified in Ca6 + AM plants, with 12 GRAS family transcription factors (TFs) ten of which were up-regulated and two down-regulated. However, only 12 AM-specific marker genes were induced in Ca0 + AM plants and the transcripts of ten up-regulated genes were further increased by Ca2+ application (Table 1), including MYB, AP2, CCD, DELLA1, RAM1, RAM2, and DELLA. In addition, some AM-specific marker genes were specifically expressed in AM plants by Ca2+ application, e.g. DXS2, DIM2, SbtM1, and PUB1.
Table 1.
Differentially expressed genes of AM-specific markers in roots of Ca0 + AM and Ca6 + AM treated plants compared with controls.
| Gene Name | Gene ID | Annotation | Ca0 + AM/CK | Ca6 + AM/CK |
|---|---|---|---|---|
| DXS2 | Araip.581AC | 1-Deoxy-D-xylulose-5-phosphate synthase | — | −1.37 |
| DIM2 | Araip.7E8G5 | receptor-like kinase | — | 4.14 |
| SbtM1 | Araip.2Y3EX | subtilisin-like protease | — | 2.23 |
| IPD3 | Araip.02MA2 | cyclops protein | — | 1.51 |
| PUB1 | Araip.658mf | E3 ubiquitin ligase | — | 2.24 |
| MYB | Araip.62YF9 | MYB transcription factor | 2.87 | 4.39 |
| AP2 | BGI_novel_G002001 | AP2 transcription factor | 1.93 | 2.98 |
| CCD1 | Araip.S2QC7 | carotenoid cleavage dioxygenase | 2.99 | 5.63 |
| CCD7 | Araip.RJ87T | carotenoid cleavage dioxygenase 7 | 1.13 | 2.65 |
| CCD8 | Araip.MNC08 | carotenoid cleavage dioxygenase 8 | 2.86 | 4.03 |
| PT1 | Araip.QVW26 | phosphate transporter | — | 4.23 |
| PT4 | Araip.WR1Z1 | inorganic phosphate transporter | 5.33 | 5.22 |
| RAM2 | Araip.1QC5L | glycerol-3-phosphate acyltransferase | 3.43 | 5.32 |
| RAM1 | Araip.N9QES | GRAS family transcription factor | 4.15 | 6.30 |
| DELLA1 | BGI_novel_G000145 | GRAS family transcription factor | 2.89 | 5.05 |
| DELLA | Araip.LT9MF | GRAS family transcription factor | 1.80 | 2.47 |
| DELLA | BGI_novel_G000391 | GRAS family transcription factor | 1.81 | 2.84 |
| DELLA | Araip.DNQ5K | GRAS family transcription factor | −2.49 | −3.62 |
| DELLA | Araip.RWP2N | GRAS family transcription factor | — | 4.47 |
| DELLA | Araip.TD6FV | GRAS family transcription factor | — | 3.40 |
| DELLA | BGI_novel_G001778 | GRAS family transcription factor | — | 1.15 |
| DELLA | Araip.W23GC | GRAS family transcription factor | — | −1.51 |
| DELLA | Araip.KB0T7 | GRAS family transcription factor | — | 1.46 |
| DELLA | Araip.KK7TK | GRAS family transcription factor | — | 1.18 |
| DELLA | BGI_novel_G001435 | GRAS family transcription factor | — | 1.08 |
Values represent significant changes in roots of AM plants under Ca2+ deficient and sufficient conditions compared with the control (NM-Ca). Positive and negative ratios indicate up- and down-regulated genes. − Represents no significant alterations at log2FoldChange ≥ 1 and P value ≤ 0.05 level.
We further investigated the effects of AM symbiosis on Ca and Ca2+ signal-related genes. The number of DEGs involved in Ca signals in the Ca6 − AM and Ca6 + AM plants was 29 and 32, respectively (Supplementary Table S2). However, there were 14 DEGs shared by Ca6 − AM and Ca6 + AM plants and the transcript levels of nine of these DEGs were further regulated by AM symbiosis. Additionally, AM symbiosis specifically up-regulated the transcripts of Araip.IZ5U3 and Araip.R6YEY genes, which code the potassium channel KAT3 and AKT2/3, respectively. These results suggest that the Ca2+ signal pathway induced by exogenous Ca2+ is partially different from AM symbiosis.
Effects of AM symbiosis and Ca2+ on genes involved in hormone biosynthesis
DEGs involved in plant hormone biosynthesis were screened, including auxin, CKs, GA, and SA (Table 2). One gene encoding auxin responsive protein indoleacetic acid (IAA) was specifically up-regulated in AM plants without Ca2+ application. Two genes belonging to the auxin responsive GH3 family were down-regulated in AM plants, and Ca2+ application further down-regulated its transcripts. The genes encoding cytokinin dehydrogenase, which catalyze the irreversible degradation of CK, were either up- or down-regulated. In addition, we observed an increase in transcripts of genes involved in the biosynthesis of GA. Compared with the control, all DEGs encoding gibberellin 20-oxidase were up-regulated in AM plants, and more transcripts were observed in Ca6 + AM plants. Two selected DEGs, namely, gibberellins 2-oxidase and gibberellin receptor GID1, were only up-regulated in Ca6 + AM plants. Meanwhile, one TF TGA (Araip.FKG2G) involved in the biosynthesis of SA was specifically up-regulated by Ca2+ application, and was further up-regulated by AM symbiosis.
Table 2.
List of selected altered genes involved in hormone signal transduction in roots of Ca0+AM, Ca6—AM, and Ca6+AMtreated plants compared with controls.
| GeneID | Gene Description | Ca0 + AM/CK | Ca6 − AM/CK | Ca6 + AM/CK |
|---|---|---|---|---|
| Auxin | ||||
| Araip.I2M0Y | auxin responsive protein IAA | 1.54 | — | — |
| Araip.PP5S8 | auxin responsive GH3 gene family | −2.20 | −2.60 | −3.42 |
| Araip.V8NJN | auxin responsive GH3 gene family | −2.31 | −5.41 | −5.42 |
| Cytokinin | ||||
| Araip.DKI8Z | cytokinin dehydrogenase | — | 2.00 | 2.22 |
| Araip.ZXC56 | cytokinin dehydrogenase | −1.44 | −1.97 | −2.48 |
| Araip.2I0VZ | histidine-containing phosphotransfer protein | — | −2.32 | −3.17 |
| Araip.W2KBF | cytokinin dehydrogenase | — | — | −2.05 |
| Gibberellin | ||||
| Araip.9GU4E | gibberellin 20-oxidase | 2.39 | 1.88 | 3.03 |
| Araip.UXP0Y | gibberellin 20-oxidase | 2.03 | 1.62 | 2.48 |
| Araip.X2IEW | gibberellin 20-oxidase | 1.84 | 1.39 | 2.73 |
| Araip.B4LS2 | gibberellin-regulated protein | — | 1.85 | 2.07 |
| Araip.HQ99N | gibberellin 20-oxidase | — | 1.26 | 1.57 |
| Araip.L4RII | gibberellin 20-oxidase | — | 1.45 | 1.60 |
| Araip.E8TE0 | gibberellin 20-oxidase | — | — | 4.62 |
| Araip.4FI3B | gibberellin 20-oxidase | — | — | 1.77 |
| Araip.78FT4 | gibberellin 20-oxidase | — | — | 2.15 |
| Araip.50IUR | gibberellin 20-oxidase | — | — | 1.73 |
| Araip.6PA6C | gibberellin 2-oxidase | — | — | 1.87 |
| Araip.99KY6 | gibberellin receptor GID1 | — | — | 1.58 |
| Salicylic acid | ||||
| Araip.FKG2G | transcription factor TGA | — | 2.80 | 4.08 |
Values represent significant alterations in AM or Ca2+-treated plants compared with the control. Positive and negative ratios indicate up- and down-regulated genes. − Represents no significant alterations at log2FoldChange ≥ 1 and P ≤ 0.05 level.
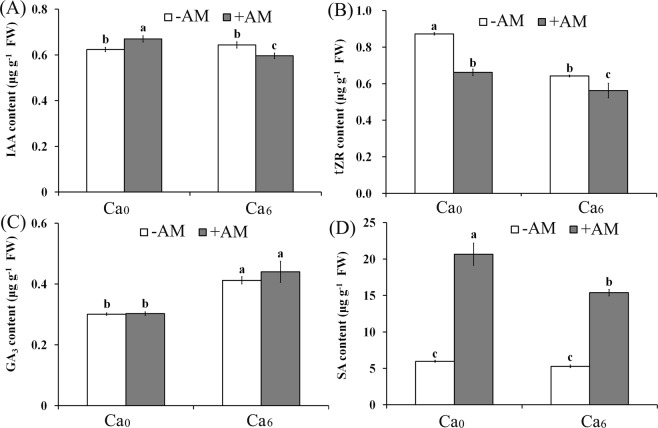
In order to verify whether the hormone level is consistent with the transcriptional changes in DEGs, we tested the content of IAA, trans-zeatin riboside (tZR), GA3, and SA. IAA content significantly increased in AM plants with Ca0 treatment, but decreased in Ca6 treatment (Fig. 4A). Changes in tZR content were consistent with the transcript changes of cytokinin dehydrogenase genes: it significantly decreased in Ca0 + AM and Ca6 − AM plants, and further decreased in Ca6 + AM plants (Fig. 4B). Additionally, the GA3 content only significantly increased in Ca6 treatment (Fig. 4C), which was consistent with the transcriptional changes of GAs biosynthesis. SA content significantly increased only in the roots of AM plants (Fig. 4D).
Figure 4.
Determination of hormone levels in peanut roots. The IAA (A), tZR (B), GA3 (C), and SA (D) content were quantified in roots of the AM plants and NM plants under Ca2+ deficient and sufficient conditions. Bars indicate means ± SD from six plants. Letters represent significant differences between treatments and the control (one-way ANOVA, P < 0.05). FW: fresh weight.
Effects of AM symbiosis and Ca2+ on genes involved in carotenoid and flavonoid biosynthesis
We found an increase in transcripts of DEGs involved in carotenoid biosynthesis. The genes encoding 3-oxoacyl-[acyl-carrier protein] reductase (BGI_novel_G000088), 15-cis-phytoene/all-trans-phytoene synthase (Araip.40 × 13), and 9-cis-beta-carotene 9′,10′-cleaving dioxygenase (CCD7, Araip.RJ87T) involved in carotenoid biosynthesis were only up-regulated in AM plants, and were further up-regulated by Ca6 treatment (Table 3). In addition, the genes encoding unknown protein (BGI_novel_G003217), capsanthin/capsorubin synthase (Araip.3B5FU), and beta-carotene isomerase (Araip.FA949, DWARF27) were specifically up-regulated in Ca6 + AM plants.
Table 3.
Differentially expressed genes involved in carotenoid biosynthesis in roots of AMF and Ca2+ treated plants compared with controls.
| Gene ID | Annotation | Ca0 + AM/CK | Ca6 + AM/CK | Ca6 − AM/CK |
|---|---|---|---|---|
| BGI_novel_G000088 | 3-oxoacyl-[acyl-carrier protein] reductase | 2.49 | 3.80 | — |
| Araip.Y8SSF | abscisate beta-glucosyltransferase | −1.45 | −2.31 | −2.58 |
| BGI_novel_G001960 | momilactone-A synthase | 2.46 | 4.22 | 2.10 |
| Araip.40X13 | 15-cis-phytoene/all-trans-phytoene synthase | 2.23 | 5.38 | — |
| Araip.MNC08 | carotenoid cleavage dioxygenase 8 | 2.86 | 4.03 | 1.20 |
| Araip.D2DUM | xanthoxin dehydrogenase | −1.83 | −1.97 | −2.15 |
| Araip.RJ87T | 9-cis-beta-carotene 9′,10′-cleaving dioxygenase | 1.13 | 2.65 | — |
| Araip.AB0RD | prolycopene isomerase | — | −1.46 | −1.18 |
| Araip.D5CVZ | momilactone-A synthase | — | — | −1.45 |
| BGI_novel_G003217 | unknown protein | — | 2.96 | — |
| Araip.3B5FU | capsanthin/capsorubin synthase | — | 2.18 | — |
| Araip.FA949 | beta-carotene isomerase | — | 2.79 | — |
Values represent significant alterations in AM or Ca2+-treated plants compared with the control. Positive and negative ratios indicate up- and down-regulated genes. − Represents no significant alterations at log2FoldChange ≥ 1 and P ≤ 0.05 level.
Transcriptional changes involved in flavonoid biosynthesis were also observed. The genes encoding chalcone synthase involved in early steps of flavonoid biosynthesis were all down-regulated in Ca0 and Ca6 treatments. In contrast, one gene (BGI_novel_G001027) encoding shikimate O-hydroxycinnamoyltransferase was up-regulated in the Ca0 and Ca6 treatments, and the expression level was the highest in AM plants treated with Ca6. However, the other gene encoding shikimate O-hydroxycinnamoyltransferase was specifically up-regulated in Ca6 + AM plants. In addition, the gene (Araip.6PA6C) encoding flavonol synthase responsible for the biosynthesis of flavanol, was specifically up-regulated in AM plants with Ca6 treatment (Supplementary Table S3).
Next, we verified whether transcriptional changes of carotenoid- and flavonoid-related genes impacted their respective content. As expected, total carotenoid content was higher in Ca0 + AM plants than the control and was the highest in Ca6 + AM plants, but was unchanged in Ca6 − AM plants (Fig. 5A). However, total flavonoid content was the highest in Ca6 treatments, and was higher than the control in Ca0 treatment (Fig. 5B); both increases were significant.
Figure 5.
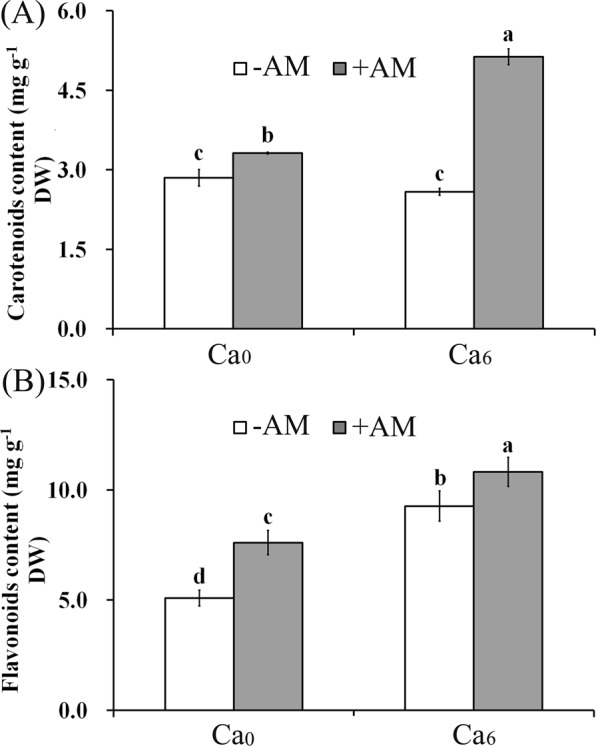
Quantification of carotenoids and flavonoids in roots of peanut seedlings. Carotenoids content (A) and total flavonoids content (B) were determined in roots of the AM plants and NM plants under Ca2+ deficient and sufficient conditions. Bars indicate means ± SD from six plants. Letters represent significant differences between treatments and the control (one-way ANOVA, P < 0.05). DW, dry weight.
Discussion
Calcium is an essential macronutrient for plant growth and development, and also plays various important roles as a secondary messenger. Prolonged Ca2+ deficiency limits root development30. In this study, AM symbiosis increased the Ca2+ content in peanut seedlings (Fig. 1D), because AMF increased the root surface and root projections, which promote plant uptake of nutrients31. Conversely, the increase in Ca2+ content enhanced potassium level in plants by enhancing the transcripts of genes encoding the potassium channel32, and together with AM symbiosis improved plant nutrient uptake9, thus increasing the shoot and root dry weight. This indicated that the interaction between AM symbiosis and exogenous Ca2+ benefited the growth of peanut seedlings.
Our previous study reported that AM symbiosis combined with exogenous Ca2+ was better than AM symbiosis or Ca2+ application alone at improving the growth of peanut seedlings29. This, together with our observations on plant dry weight, could explain why Ca2+ application strengthens the role of AM symbiosis in plant growth by further regulating a major overlap of transcriptional changes in roots of AM plants (380 out of 510 genes, approximately 74%). In addition, the establishment of AM symbiosis requires the expression of AM-specific marker genes33. In this study, Ca2+ further up-regulated and specifically induced the transcripts of AM-specific marker genes in AM plants. It is possible that the Ca2+-calmodulin association with CCaMK induces the phosphorylation of CYCLOPS and forms a complex in the presence of calcium, which acts in concert with GRAS TFs such as DELLA proteins to initiate the expression of AM-specific marker genes that are necessary to establish the AM symbiosis14. These results suggest that Ca2+ plays a vital role in the formation of AM symbiosis.
GRAS family TF encoding DELLA protein is a positive regulator in the formation of AM associations17,34, and is also involved in GA biosynthesis as a negative regulator of GA signaling35. AM symbiosis up-regulation of GA-related genes and GA content in roots has been reported in M. truncatula and tomato36,37. Hence, the observed increase of GA3 content may be the factor involved in Ca2+ further up-regulating the transcripts of DELLAs and the genes encoding gibberellins 20-oxidase, which is a key enzyme that catalyzes the penultimate steps in GA biosynthesis. This result implied that AM symbiosis positively regulated the transcriptional changes involved in GA biosynthesis, and that Ca2+ strengthens this effect.
tZR is the major transport form of CKs from root to shoot in plants22,38. However, it has been reported that CKs act as a negative regulator in lateral root initiation, because overproduction of CKs inhibited lateral root initiation39,40. In this study, the lower tZR in roots of AM plants suggested that the genes encoding cytokinin dehydrogenase, which catalyze the irreversible degradation of cytokinin, were down-regulated by Ca2+ application. Reduced tZR content may be beneficial to the initiation of AM symbiotic roots. This result supports the finding in some studies that CKs might not be involved in the regulation of AM symbiosis development16. In addition, SA and carotenoid have been demonstrated to be activated by AM colonization16,41, and these activations were specific to AM symbiosis but not Ca2+ (Fig. 4), suggesting that increases in SA and carotenoid content can serve as AM-specific marker metabolites.
Flavonoid is involved in hyphal growth and branching23, and in turn, AMF benefit flavonoid biosynthesis and accumulation in roots of M. truncatula42,43. This is in line with our observation that more flavonoids were accumulated in roots of AM plants treated by Ca2+ application, which is attributed to Ca2+ inducing more transcripts of DELLA genes. DELLA-mediated signaling participates in regulating the accumulation of anthocyanin, one of the derivatives of flavonoids44. Additionally, the accumulation of SA can increase the flavonoid content in AM plants45. Thus, more flavonoids were observed in AM plants treated by Ca2+. These results suggested that Ca2+ and AM symbiosis might share the flavonoid biosynthetic pathway for improving plant growth.
Based on our data, we propose a model of interactive pathways that modulate hormone levels, secondary metabolism, and ultimately the growth of AM and Ca2+ plants (Fig. 6). In this model, AM symbiosis promotes the growth of peanut seedlings by increasing contents of GAs, IAA, SA, carotenoids, and flavonoids. However, exogenous Ca2+ application only enhances the GA level and flavonoid content for improving plant growth. The increase in flavonoid content in AM symbiosis or Ca2+-treated plants may be a reason for the regulated DELLA that may enhance flavonoid accumulation. The proposed model reveals that synergy of AM symbiosis with Ca2+ promotes peanut growth by regulating GAs and flavonoid biosynthesis, but carotenoid and SA biosynthesis are specifically regulated by AM symbiosis. These findings should be validated in future research.
Figure 6.
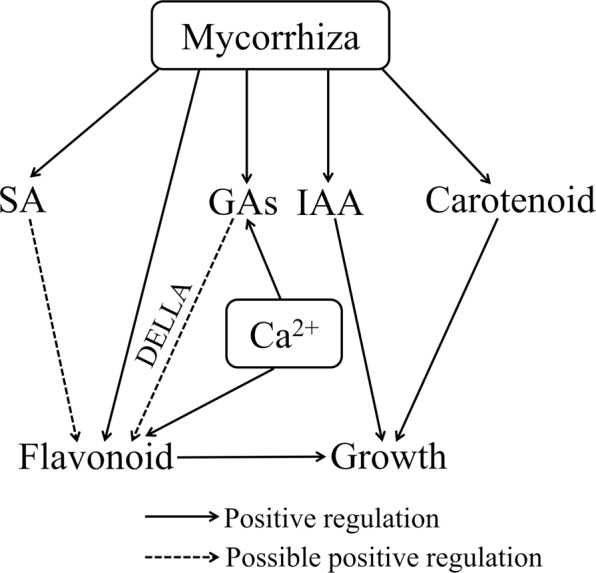
Proposed model of AM- and Ca2+- regulated pathways in peanut roots. AM symbiosis increases the content of IAA, GAs, SA, carotenoids, and flavonoids. Total flavonoids were also accumulated by regulating the transcripts of DELLA genes and the increase of SA in AM plants. Ca2+ application only increases the GA and flavonoid contents.
Methods
Plant material and growth conditions
Peanut cultivar ‘Huayu 22’ seeds were surface sterilized with 70% alcohol for 3 min and rinsed six times with sterile water. They were then germinated in the dark at 25 °C for 3 days. The germinated seeds were transferred to pots filled with quartz sand which was rinsed with deionized water 10 times to remove as much Ca2+ as possible, and then seeds were sterilized at 121 °C for 30 min. Half of the young seedlings were inoculated with about 300 F. mosseae spores (BEG HEB02); the other half were not colonized by F. mosseae. The peanut seedlings were grown in a greenhouse at 24 °C/18 °C with a 16/8 h photoperiod, at a photosynthetic photo flux density of 700 µmol·m−2·s−1, and 60% relative humidity. Each seedling was watered regularly with 80 ml of modified Hoagland’s solution (5 mM KNO3, 2 mM MgSO4·7H2O, 1 mM KH2PO4, 0.1 mM EDTA-Na2, 0.1 mM FeSO4·7H2O, 46 µM H3BO4, 0.32 µM CuSO4·5H2O, 0.77 µM ZnSO4·7H2O and 0.11 µM H2MoO4) supplemented with 6 mM Ca(NO3)2·4H2O (Ca2+ sufficient) or 6 mM NH4·NO3 (Ca2+ deficient, used for balancing nitrogen in Ca(NO3)2). There were four treatments: Ca0-AM, Ca0 + AM, Ca6 − AM, and Ca6 + AM, where 0 and 6 represent the Ca2+ concentrations (mM), + and − represent with or without inoculation of F. mosseae spores. In this study, 6 mM of Ca(NO3)2 was chosen according to our previous report6. Six weeks later, the shoots and roots of AM and NM plants were harvested and further analyzed.
Mycorrhizal quantification and determination of dry weight and Ca2+ content
After six weeks, shoots and roots of the AM and NM plants under Ca2+-deficient or Ca2+-sufficient conditions were harvested separately. Young roots of AM plants were examined by light microscopy (OLYMPUS, CX41, Japan) to estimate the extent to which the roots had been colonized by hyphae and arbuscules46. The fresh shoots and roots were dried at 105 °C for 30 min, and then dried at 80 °C until a constant weight. The Ca2+ contents in the roots from the different treatments were determined according to Yang et al.6.
RNA extraction and sequencing
Total RNA was isolated from roots of AM and NM plants, and then enrichment of mRNA and synthesis of cDNA were conducted1. The cDNA from three biological replicates composed of four plants in each treatment were sequenced using an Illumina HiSeq. 2000 Platform. After filtering, high quality clean reads were aligned with a reference genome (https://peanutbase.org/organism/Arachis/ipaensis) using HISAT47; on average 70.61% reads were mapped, indicating that the samples were comparable.
RNA-Seq analysis and data deposition
After genome mapping, we used StringTie software to reconstruct transcripts with genome annotation information47, then identified novel transcripts using Cuffcompare and predicted the coding ability of those new transcripts using CPC software48,49. After novel transcript detection, the gene expression level was calculated for each sample with RSEM50. Based on the gene expression level, we used DEseq. 2 algorithms to detect differentially expressed genes (DEGs). A threshold of 1 for transcript ratio (log2FoldChange) in treatments versus control (Ca0-AM), and Padj (statistic of adjusted P value) ≤0.05 were set as criteria for the selection of DEGs in NM plants and AM plants under Ca2+-deficient and Ca2+-sufficient conditions. With DEGs, Gene Ontology (GO) classification and functional enrichment were performed using WEGO software51, and the pathway analyses were obtained using the KEGG database (https://www.genome.jp/kegg/pathway.html).
Quantitative real-time PCR
To verify the RNA-Seq results, the expression levels of 15 selected genes were determined by quantitative RT-PCR. mRNA was isolated from the same samples sequenced by RNA-Seq, and the first-strand cDNAs were synthesized for qRT-PCR analyses using SYBR Premix Ex Taq polymerase (Takara) according to the manufacturer’s protocol. The designed primers are shown in Supplementary Table S4. The control reactions were conducted using primers Tua5-F and Tua5-R52. At least three replicates were tested per sample. Relative mRNA (fold) differences were assessed with the 2− ΔΔCt formula53, the values were subsequently transformed to the log2 scale.
Determination of plant hormones
The roots (fresh weight) were ground into a powder in liquid nitrogen, and 1.0 g of powder was used to determine the concentration of endogenous hormones by high performance liquid chromatography (HPLC)54, including IAA, tZR, and GA3. The SA content was measured according to a previous method55. Three independent replicates per sample were statistically analyzed.
Carotenoid and flavonoid content analyses
Carotenoids were extracted from the roots of AM and NM plants56. Total carotenoid content in roots was calculated using absorbance at 450 nm. Flavonoids in roots were measured by chloride colorimetric assay57, and total flavonoid content was determined according to the standard curve of quercetin at an absorbance of 510 nm.
Statistical analysis
Analysis of variance was performed using SSPS software version 16.0 for Windows. One-way analysis of variance (ANOVA) was used, followed by Duncan's test for multiple comparisons. The values obtained are the mean ± SE for the three replicates in each treatment. A P value ≤ 0.05 was considered to be significant.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31601261), the National Key R&D Program of China (2018YFD0201000), Major Scientific and Technological Innovation Projects of Shandong Province (2018YFJH0601), the Innovation Project of Shandong Academy of Agricultural Sciences (CXGC2018D04), the China Postdoctoral Science Foundation (2016M592236), and the Earmarked Fund for Modern Agroindustry Technology Research System (CARS-14).
Author contributions
S.W. and X.L. designed the experiment and drafted the manuscript. L.C. and F.G. carried out most of the experiments and analyzed the transcriptome data. J.Z. and S.Y. determined the hormone levels. J.M. and Y.G. performed the validation of qRT-PCR experiment. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Li Cui and Feng Guo.
Change history
1/15/2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Contributor Information
Xinguo Li, Email: xinguol@163.com.
Shubo Wan, Email: wanshubo2016@163.com.
Supplementary information
is available for this paper at 10.1038/s41598-019-52630-7.
References
- 1.Yang S, et al. Transcriptome and differential expression profiling analysis of the mechanism of Ca2+ regulation in peanut (Arachis hypogaea) pod development. Front Plant Sci. 2017;8:1609. doi: 10.3389/fpls.2017.01609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain M, et al. Calcium dependent protein kinase (CDPK) expression during fruit development in cultivated peanut (Arachis hypogaea) under Ca2+-sufficient and -deficient growth regimens. J. Plant Physiol. 2011;168:2272–2277. doi: 10.1016/j.jplph.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Jaffe LA, Weisenseel MH, Jaffe LF. Calcium accumulations within the growing tips of pollen tubes. J. Cell Biol. 1975;67:488–492. doi: 10.1083/jcb.67.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding F, Chen M, Sui N, Wang BS. Ca2+ significantly enhanced development and salt-secretion rate of salt glands of Limonium bicolor under NaCl treatment. S Afr J Bot. 2010;76:95–101. [Google Scholar]
- 5.Gilroy S, et al. ROS, Calcium, and Electric Signals: Key Mediators of Rapid Systemic Signaling in Plants. Plant Physiol. 2016;171:1606–1615. doi: 10.1104/pp.16.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang S, et al. Exogenous calcium alleviates photoinhibition of PSII by improving the xanthophyll cycle in peanut (Arachis hypogaea) leaves during heat stress under high irradiance. PLoS One. 2013;8:e71214. doi: 10.1371/journal.pone.0071214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang S, et al. Calcium contributes to photoprotection and repair of photosystem II in peanut leaves during heat and high irradiance. J. Integ Plant Biol. 2015;57:486–495. doi: 10.1111/jipb.12249. [DOI] [PubMed] [Google Scholar]
- 8.Sarkar D, Sud KC. The role of calcium nutrition in potato (Solanum tuberosum) microplants in relation to minimal growth over prolonged storage in vitro. Plant Cell Tiss Org. 2005;81:221–227. [Google Scholar]
- 9.Spatafora JW, et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia. 2016;108:1028–1046. doi: 10.3852/16-042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawers R. Progress and challenges in agricultural applications of arbuscular mycorrhizal fungi. Crit Rev Plant Sci. 2011;30:459–470. [Google Scholar]
- 11.Sun J, et al. Activation of symbiosis signaling by arbuscular mycorrhizal fungi in legumes and rice. Plant Cell. 2015;27:1–16. doi: 10.1105/tpc.114.131326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge O. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol. 2013;11:252–263. doi: 10.1038/nrmicro2990. [DOI] [PubMed] [Google Scholar]
- 13.Capoen W, et al. Nuclear membranes control symbiotic calcium signaling of legumes. PANS. 2011;108:14348–14353. doi: 10.1073/pnas.1107912108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maclean AM, Bravo A, Harrison MJ. Plant signaling and metabolic pathways enabling arbuscular mycorrhizal symbiosis. Plant Cell. 2017;29:2319–2335. doi: 10.1105/tpc.17.00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniela SF, et al. DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. PANS. 2013;110:5025–5034. doi: 10.1073/pnas.1308973110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foo E, Ross JJ, Jones WT, Reid JB. Plant hormones in arbuscular mycorrhizal symbioses: an emergingrole for gibberellins. Ann Bot. 2013;111:769–779. doi: 10.1093/aob/mct041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu N, et al. A DELLA protein complex controls the arbuscular mycorrhizal symbiosis in plants. Cell Res. 2014;24:130–133. doi: 10.1038/cr.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia K, Chasman D, Roy S, Ane JM. Physiological responses and gene co-expression network of mycorrhizal roots under K+ deprivation. Plant Physiol. 2017;173:1811–1823. doi: 10.1104/pp.16.01959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pühler A, Becker A. Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol. 2005;137:1283–1301. doi: 10.1104/pp.104.056572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siciliano V, Bonfante P. Transcriptome analysis of arbuscular mycorrhizal roots during development of the prepenetration apparatus. Plant Physiol. 2007;144:1455–1466. doi: 10.1104/pp.107.097980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hause B, Mrosk C, Isayenkov S, Strack D. Jasmonates in arbuscular mycorrhizal interactions. Phytochemistry. 2007;68:101–110. doi: 10.1016/j.phytochem.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 22.Adolfsson L, et al. Enhanced secondary- and hormone metabolism in leaves of arbuscular mycorrhizal medicago truncatula. Plant Physiol. 2017;175:392–411. doi: 10.1104/pp.16.01509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jm S, et al. Flavonoids exhibit fungal species and genus specific effects on the presymbiotic growth of Gigaspora and Glomus. Mycol Res. 2005;109:789–794. doi: 10.1017/s0953756205002881. [DOI] [PubMed] [Google Scholar]
- 24.Garcia K, et al. Take a Trip Through the Plant and Fungal Transportome of Mycorrhiza. Trends plant sci. 2016;21:937–950. doi: 10.1016/j.tplants.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Anderson RC, Liberta AE. Influence of supplemental inorganic nutrients on growth, survivorship, and mycorrhizal relationships of schizachyrium scoparium (Poaceae) grown in fumigated and unfumigated soil. Am J Bot. 1992;79:406–414. [Google Scholar]
- 26.Sawers RJ, et al. Phosphorus acquisition efficiency in arbuscular mycorrhizal maize is correlated with the abundance of root‐external hyphae and the accumulation of transcripts encoding PHT1 phosphate transporters. New Phytol. 2017;214:632–643. doi: 10.1111/nph.14403. [DOI] [PubMed] [Google Scholar]
- 27.Habte M, Soedarjo M. Limitation of vesicular-arbuscular mycorrhizal activity in Leucaena leucocephala by Ca insufficiency in an acid Mn-rich oxisol. Mycorrhiza. 1995;5:387–394. [Google Scholar]
- 28.Jarstfer AG, Farmerkoppenol P, Sylvia DM. Tissue magnesium and calcium affect arbuscular mycorrhiza development and fungal reproduction. Mycorrhiza. 1998;7:237–242. doi: 10.1007/s005720050186. [DOI] [PubMed] [Google Scholar]
- 29.Cui L, et al. Arbuscular mycorrhizal fungi combined with exogenous calcium improves the growth of peanut (Arachis hypogeaea L.) seedlings under continuous cropping. J Integr Agr. 2018;7:60345–7. doi: 10.1016/S2095-3119(18)61982-3. [DOI] [Google Scholar]
- 30.Kirkby EA, Pilbeam DJ. Calcium as a plant nutrient. Plant Cell Environ. 1984;7:397–405. [Google Scholar]
- 31.Wu QS, Zou YN, He XH. Contributions of arbuscular mycorrhizal fungi to growth, photosynthesis, root morphology and ionic balance of citrus seedlings under salt stress. Acta Physiol Plant. 2010;32:297–304. [Google Scholar]
- 32.Lei B, et al. Increased cucumber salt tolerance by grafting on pumpkin rootstoch and after application of calcium. Biol Plantarum. 2014;58:179–184. [Google Scholar]
- 33.Lévy J, et al. A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science. 2004;303:1361–1364. doi: 10.1126/science.1093038. [DOI] [PubMed] [Google Scholar]
- 34.Pimprikar P, et al. A CCaMK-CYCLOPS-DELLA complex activates transcription of RAM1 to regulate arbuscule branching. Curr Biol. 2016;26:987–998. doi: 10.1016/j.cub.2016.01.069. [DOI] [PubMed] [Google Scholar]
- 35.Takeda N, et al. Gibberellins interfere with symbiosis signaling and gene expression and alter colonization by arbuscular mycorrhizal fungi in Lotus japonicus. Plant Physiol. 2015;167:545–557. doi: 10.1104/pp.114.247700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.García Garrido JM, León Morcillo RJ, Martín Rodríguez JÁ, Ocampo Bote JA. Variations in the mycorrhization characteristics in roots of wild-type and ABA-deficient tomato are accompanied by specific transcriptomic alterations. Mol Plant Microbe In. 2010;23:651–664. doi: 10.1094/MPMI-23-5-0651. [DOI] [PubMed] [Google Scholar]
- 37.Martín-Rodríguez JA, et al. Gibberellin–abscisic acid balances during arbuscular mycorrhiza formation in tomato. Front Plant Sci. 2016;7:1273. doi: 10.3389/fpls.2016.01273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kudo T, Kiba T, Sakakibara H. Metabolism and long-distance translocation of cytokinins. J Integr Plant Biol. 2010;52:53–60. doi: 10.1111/j.1744-7909.2010.00898.x. [DOI] [PubMed] [Google Scholar]
- 39.Fukaki H, Tasaka M. Hormone interactions during lateral root formation. Plant Mol Biol. 2009;69:437–449. doi: 10.1007/s11103-008-9417-2. [DOI] [PubMed] [Google Scholar]
- 40.Laplaze L, et al. Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell. 2007;19:3889–3900. doi: 10.1105/tpc.107.055863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baslam M, Esteban R, García-Plazaola JI, Goicoechea N. Effectiveness of arbuscular mycorrhizal fungi (AMF) for inducing the accumulation of major carotenoids, chlorophylls and tocopherol in green and red leaf lettuces. Appl Microbiol Biot. 2013;97:3119–3128. doi: 10.1007/s00253-012-4526-x. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, et al. Arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. Plant J. 2007;50:529–544. doi: 10.1111/j.1365-313X.2007.03069.x. [DOI] [PubMed] [Google Scholar]
- 43.Wulf A, et al. Transcriptional changes in response to arbuscular mycorrhiza development in the model plant Medicago truncatula. Mol Plant Microbe Interact. 2003;16:306–314. doi: 10.1094/MPMI.2003.16.4.306. [DOI] [PubMed] [Google Scholar]
- 44.Jiang C, Fu X. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 2007;145:1460–1470. doi: 10.1104/pp.107.103788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gondor OK, et al. Salicylic Acid Induction of Flavonoid Biosynthesis Pathways in Wheat Varies by Treatment. Front Plant Sci. 2016;7:1447. doi: 10.3389/fpls.2016.01447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGonigle TP, et al. A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol. 1990;115:495–501. doi: 10.1111/j.1469-8137.1990.tb00476.x. [DOI] [PubMed] [Google Scholar]
- 47.Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2016;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kong L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:345–349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye J, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:293–297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chi XY, et al. Validation of reference genes for expression studies in peanut by quantitative real-time RT-PCR. Mol Genet Genomics. 2012;287:167–176. doi: 10.1007/s00438-011-0665-5. [DOI] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔ CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 54.Yun Z, et al. Comparative transcriptomics and proteomics analysis of citrus fruit, to improve understanding of the effect of low temperature on maintaining fruit quality during lengthy post-harvest storage. J. Exp Bot. 2012;63:2873–2893. doi: 10.1093/jxb/err390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herrera-Medina M, et al. Root colonization by arbuscular mycorrhizal fungi is affected by the salicylic acid content of the plant. Plant Sci. 2003;164:993–998. [Google Scholar]
- 56.Sanchez T, et al. Reduction or delay of post-harvest physiological deterioration in cassava roots with higher carotenoid content. J. Sci Food Agr. 2006;86:634–639. [Google Scholar]
- 57.Jia Z, Tang M, Wu J. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem. 1999;64:555–559. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.