Abstract
The endoplasmic reticulum (ER) produces about 40% of the nucleated cell’s proteome. ER size and content in molecular chaperones increase upon physiologic and pathologic stresses on activation of unfolded protein responses (UPR). On stress resolution, the mammalian ER is remodeled to pre-stress, physiologic size and function on activation of the LC3-binding activity of the translocon component SEC62. This elicits recov-ER-phagy, i.e., the delivery of the excess ER generated during the phase of stress to endolysosomes (EL) for clearance. Here, ultrastructural and genetic analyses reveal that recov-ER-phagy entails the LC3 lipidation machinery and proceeds via piecemeal micro-ER-phagy, where RAB7/LAMP1-positive EL directly engulf excess ER in processes that rely on the Endosomal Sorting Complex Required for Transport (ESCRT)-III component CHMP4B and the accessory AAA+ ATPase VPS4A. Thus, ESCRT-III-driven micro-ER-phagy emerges as a key catabolic pathway activated to remodel the mammalian ER on recovery from ER stress.
Subject terms: Autophagy, Stress signalling, Endoplasmic reticulum
The ER increases in size upon stress in response to the UPR and is remodelled after the stressor is removed by a process called recov-ER-phagy. Here, the authors show remodelling to pre-stress ER size occurs by micro-ER-phagy requiring ESCRT-III components CHMP4B and VPS4A.
Introduction
The endoplasmic reticulum (ER) is the site of protein, lipid, and oligosaccharide synthesis, calcium storage, and drugs detoxification. Its size (and functions) is maintained at steady state and is adapted to environmental and developmental conditions by a homeostatic equation that comprises the anabolic transcriptional and translational programs of the unfolded protein response (UPR) and the catabolic programs relying on receptor-mediated, lysosomal clearance of select ER subdomains. The UPR increases the size of the ER and its content in resident proteins1,2. In contrast, the lysosomal-regulated ER turnover maintains ER size at steady state, prevents excessive ER expansion during ER stress, and, as we recently discovered, regulates ER return at physiologic size during recovery from ER stresses3,4. Selective clearance of the ER by endolysosomes (ELs) has originally been observed, in the mid-1960s, during butterfly pupation5 and remained confined to morphological analyses for over 50 years. It is only with the discovery that lysosomal turnover of the ER is controlled by dedicated ER-resident proteins (FAM134B6, SEC627, RTN38, ATL39, CCPG110, TEX26411,12 in mammalian cells, Atg39 and Atg40 in yeast13) that the studies entered a phase of mechanistic dissection (reviewed in refs. 4,14–18). All these proteins engage components of the autophagic machinery via their cytosolic domains, which display FIP200-10 and/or LC3-6–8,10–12 and/or GABARAP-interacting9 regions in mammals, and Atg11- and/or Atg8-interacting motifs in yeast13. Nutrient deprivation indiscriminately enhances several autophagic pathways to rapidly mobilize amino acids and other cellular building blocks and it has extensively been used to activate and investigate the mechanisms of receptor-mediated ER clearance by mammalian and yeast macro-ER-phagy pathways6,8–13. Starvation-induced mammalian macro-ER-phagy relies on engulfment of ER subdomains decorated with FAM134B6, RTN38, ATL39, CCPG110, and TEX26411,12 by double-membrane autophagosomes. These eventually fuse with EL for cargo clearance. However, ER-centric signals do exist that trigger clearance of select ER subdomains on activation of individual receptors as reported for CCPG1-mediated control of ER expansion during ER stress10, for SEC62-controlled reduction of the ER volume to physiologic state after conclusions of acute ER stresses7, and for ER-to-lysosome-associated degradation (ERLAD) pathways activated to deliver proteasome-resistant misfolded proteins from the ER to EL for destruction19–21 (and reviewed in refs. 15,17). Mechanistic dissection of all these pathways is in its infancy and characterization of signal-specific (ER-centric) activation of individual LC3 receptors at the ER membrane awaits further studies and is assessed here in the case of SEC62-regulated recov-ER-phagy. SEC62 is an essential component of the SEC61 protein translocation machinery, where it acts in a functional complex with SEC63 to promote the post-translational entrance of newly synthesized polypeptides in the ER22,23. Notably, the function of SEC62 in selective delivery of ER subdomains to EL for clearance is not activated by nutrient deprivation12, nor at steady state or during ER stress7. Our studies revealed that SEC62 controls delivery of excess ER to RAB7/LAMP1-positive EL for clearance during the recovery phase that follows the conclusion of acute ER stresses. In our experiments, acute ER stresses were triggered on transient perturbation of calcium or redox homeostasis to mimic original observation in liver cells showing lysosomal removal of excess ER after cessation of treatments with antiepileptic drugs such as phenobarbital24,25 (please refer to the detailed description of the protocols for reversible and non-toxic induction of ER stress in ref. 7 and in the Methods section). SEC62-controlled ER turnover during recovery from ER stress, recov-ER-phagy, can also be induced on SEC62 overexpression or on silencing of SEC63, which participates in SEC62-containing heterodimers7,26. Here we report that in contrast to starvation-induced, receptor-mediated ER-clearance6,8–13, SEC62-driven ER turnover, which is activated in response to an ER-centric signal, that is, the conclusion of an acute ER stress, does not rely on engagement of the macro-autophagy pathway. Rather, resolution of ER stress activates catabolic processes where RAB7/LAMP1-positive EL directly engulf excess ER subdomains via ESCRT-III-mediated piecemeal micro-ER-phagy.
Results
ER subdomains delivery within EL on ER stress resolution
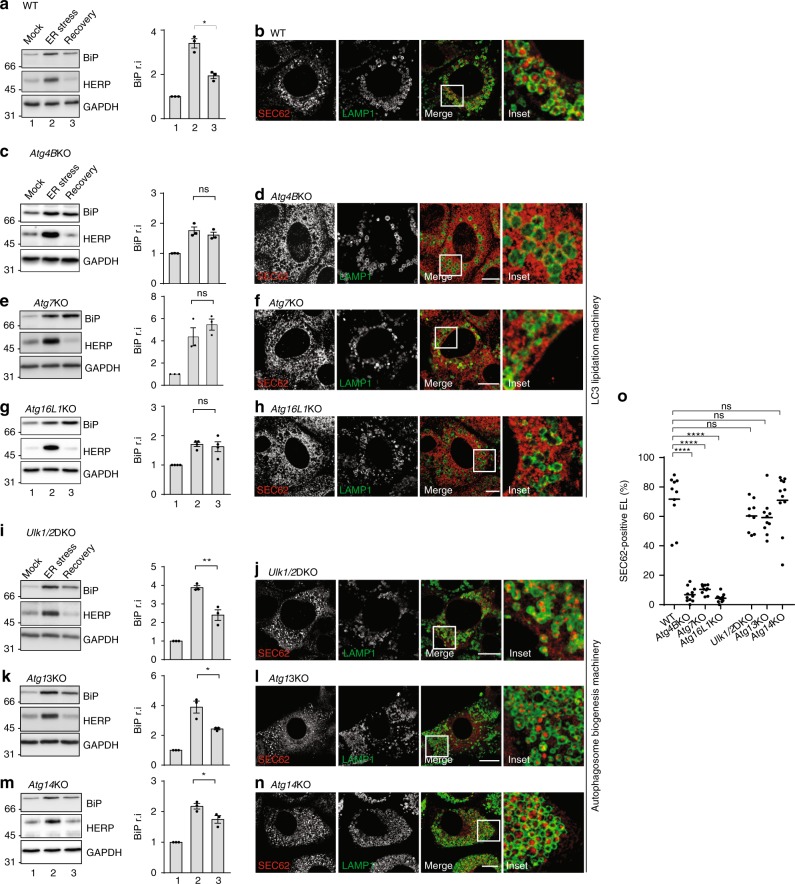
To characterize the mechanisms of mammalian ER remodeling that re-establish physiologic (pre-stress) condition on resolution of ER stresses, we made use of a previously established protocol for acute induction of ER stress on transient exposure of mouse embryonic fibroblasts (MEFs) to cyclopiazonic acid (CPA)7, a reversible inhibitor of the sarco/ER calcium ATPase27. Western blot analyses show that the level of ER stress marker proteins increases in wild-type (WT) MEFs exposed to CPA (Fig. 1a, lane 1 vs. 2, upper and middle panels for BiP and HERP, respectively) and decreases after interruption of the pharmacologic treatment (lane 3)7. For ERAD factors like HERP, return at the pre-stress level relies on the activity of cytosolic proteasomes and other ERAD tuning mechanisms7,28–30. For conventional ER-resident chaperones and members of the protein disulfide isomerase superfamily, it relies on SEC62-controlled delivery of excess ER within EL for clearance in catabolic processes collectively defined as recov-ER-phagy7. Inactivation of lysosomal hydrolases with bafilomycin A1 (BafA1) delays return of these ER-resident chaperones to the pre-stress level7 and causes their accumulation within EL displaying RAB7 and LAMP1 at the limiting membrane7 (Fig. 1b, o, Supplementary Fig. 1a). Recov-ER-phagy is faithfully recapitulated on SEC62 overexpression (Supplementary Fig. 1b) and on release of orphan endogenous SEC62 upon silencing of SEC63 expression7.
Fig. 1.
Delivery of endogenous SEC62-labeled EV within EL during recovery from ER stress. a Upper panel, WB analysis showing steady-state level of BiP in WT MEF (Mock), BiP induction on cell exposure to CPA (ER stress, lane 2), and return of BiP to the pre-stress level after CPA wash-out (Recovery, lane 3); middle panel, same for HERP; lower panel, GAPDH as a loading control. Quantification of BiP levels in WB, n = 3 independent experiments, mean ± SEM, unpaired, two-tailed t test, *P = 0.0037. b Delivery of SEC62-decorated EV within LAMP1-positive EL in WT MEF during 12 h recovery from an ER stress in the presence of 50 nM BafA1. c Same as a in Atg4BKO MEF; n = 3 independent experiments, mean ± SEM, unpaired, two-tailed t test, P = 0.4001. d Same as b in Atg4BKO MEF. e Same as a in Atg7KO MEF, n = 3 independent experiments, mean ± SEM; unpaired, two-tailed t test, P = 0.3159. f Same as b in Atg7KO MEF. g Same as a in Atg16L1KO MEF; n = 4 independent experiments, mean ± SEM; unpaired, two-tailed t test, P = 0.6909. h Same as b in Atg16L1KO MEF. i Same as a in Ulk1/2 double-KO MEF; n = 3 independent experiments, mean ± SEM; unpaired, two-tailed t test, **P = 0.0083. j Same as b in Ulk1/2 double-KO MEF. k Same as a in Atg13KO MEF; n = 3 independent experiments, mean ± SEM; unpaired, two-tailed t test, *P = 0.0232. l Same as b in Atg13KO MEF. m Same as a in Atg14KO MEF; n = 3 independent experiments, mean ± SEM; unpaired, two-tailed t test, *P = 0.0497. n Same as b in Atg14KO MEF. See Supplementary Fig. 1 for recov-ER-phagy recapitulated on overexpression of SEC62-HA. o Quantification of SEC62-positive EV delivery within LAMP1-positive EL (n = 10, 11, 10, 10, 9, 10, 11 cells, respectively). One-way analysis of variance (ANOVA) and Dunnett’s multiple comparisons test, n.s.P > 0.05, ****P < 0.0001. Molecular weight markers in WB are in kDa. Scale bars for CLSM: 10 μm. WB and IF panels are representative of at least three independent experiments
LC3 lipidation in resolution of ER stress
LC3 lipidation is required for FAM134B-, RTN3-, CCPG1-, ATL3-, and TEX264-dependent macro-ER-phagy3,6,8–12,14,18. Not surprisingly, individual ablation of Atg4B, Atg7, or Atg16L1, three components of the LC3 lipidation machinery (Supplementary Fig. 2)31, inhibits return of ER stress-induced marker proteins to the pre-stress level (BiP in Fig. 1c, e, g, upper panels, lanes 2 vs. 3). Return of HERP at the pre-stress level, which relies on cytosolic proteasomes7,28, remains unaffected (Fig. 1c, e, g, middle panels). Consistently, ablation of LC3 lipidation abolishes delivery of ER subdomains within EL during recovery from ER stress (Fig. 1d, f, h, o) and when recov-ER-phagy is recapitulated by overexpression of HA-tagged SEC62 (Supplementary Fig. 1d).
Autophagosome dispensability to resolve ER stress
To corroborate the notion that SEC62-driven clearance of excess ER on UPR resolution occurs via macro-ER-phagy, we verified the involvement of the autophagosome biogenesis machinery in recov-ER-phagy. Surprisingly, individual ablation of Ulk1, Ulk2, Atg13, and Atg14, which are dispensable for LC3 lipidation (Supplementary Fig. 2) but are required for biogenesis of double-membrane autophagosomes31–34, does not prevent return of BiP to pre-stress level on ER stress resolution (Fig. 1i, k, m, upper panels, lanes 1 vs. 3). Consistently, inactivation of autophagosome biogenesis does not affect delivery of excess ER within EL during recovery from stress (Fig. 1j, l, n, o) and when recov-ER-phagy is recapitulated by SEC62 overexpression (Supplementary Fig. 1e–g).
SNAREs dispensability to resolve ER stress
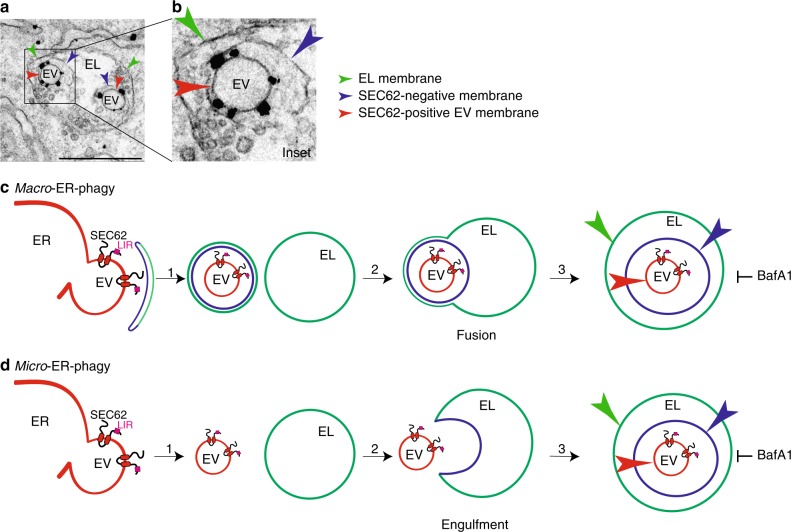
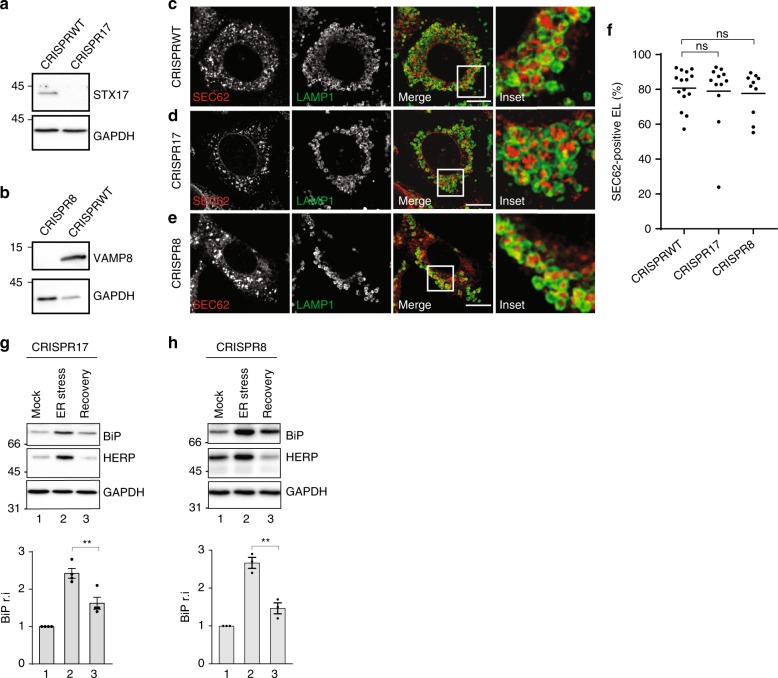
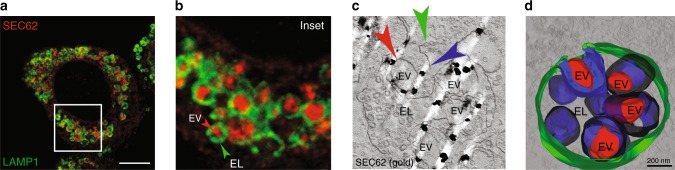
To investigate lysosomal delivery of ER subdomains with ultrastructural resolution, we turned to immuno electron microscopy (IEM) and invariably found that ER-derived vesicles (EVs) displaying SEC62 at their limiting membrane (red arrows, Fig. 2a, b) are sequestered by a SEC62-negative membrane (blue arrows, Fig. 2a, b), within the EL (green arrows, Fig. 2a, b). This topology is consistent with macro-ER-phagy, where ER subdomains are captured by double-membrane autophagosomes that eventually fuse with EL to clear their cargo (Fig. 2c), or with micro-ER-phagy, where ER subdomains are directly engulfed by EL (Fig. 2d). A major difference between macro- and micro-autophagy is the requirement for the former of an heterotypic membrane fusion event (arrow 2, Fig. 2c, fusion vs. Fig. 2d, engulfment), which is substantially impaired on ablation of the SNARE proteins STX17 and VAMP835,36. Ablation of STX17 (Fig. 3a, d, f) or of VAMP8 (Fig. 3b, e, f) does not affect delivery of excess ER within EL and does not delay return of chaperones at their pre-stress level during recovery from ER stress (Fig. 3g, h). In agreement with our genetic analyses showing dispensability of autophagosomes and macro-ER-phagy (Figs. 1i–o, 3a–h), IEM analyses do not reveal SEC62-labeled ER fragments within double-membrane autophagosomes when cells are recovering from ER stress. Rather, they show EL caught in the act of capturing SEC62-positive EV by inward invagination of their membranes (Fig. 4a–d and Supplementary Movie 1). Thus, recov-ER-phagy is topologically equivalent to micro-autophagy, a poorly characterized type of autophagy involved in clearance of organelles including large ER whorls in yeast37–40.
Fig. 2.
Distribution of gold-labeled, endogenous SEC62 by IEM. a WT MEFs are exposed 12 h to 10 μM CPA. Recovery is initiated on CPA wash-out and progresses for 12 h in the presence of 50 nM BafA1 to inhibit clearance of EV delivered within EL. b Inset of a. IEM images are representative of two independent experiments with similar results. c Macro-ER-phagy pathway. d Micro-ER-phagy pathway. EV, ER-derived vesicle; EL, endolysosome; LIR, LC3-interacting region. Color code of arrows is given in the panels. Scale bar for IEM: 1 μm
Fig. 3.
SNAREs dispensability for delivery of endogenous SEC62-labeled EV within EL. a WB analysis showing the efficiency of STX17 knockout in CRISPR17 MEF (please also refer to ref. 19). b Same as a for VAMP8 in CRISPR8 MEF (please also refer to ref. 19). c Delivery of endogenous SEC62-labeled EV within LAMP1-positive EL during recovery from CPA-induced ER stress in WT MEF exposed to 50 nM BafA1 for 12 h. d Same as c in MEF lacking STX17. e Same as c in MEF lacking VAMP8. Scale bars: 10 μm. f Quantification of EV delivery within EL in c–e (n = 15, 12, 10 cells, respectively). One-way ANOVA and Dunnett’s multiple comparisons test, n.s.P > 0.05. IF panels are representative of three independent experiments. g Same as Fig. 1a in cells lacking STX17, n = 4 independent experiments, mean ± SEM, unpaired, two-tailed t test, **P = 0.0083. h Same as Fig. 1a in cells lacking VAMP8; n = 3 independent experiments, mean ± SEM, unpaired, two-tailed t test, **P = 0.0043. Molecular weight markers in WB are in kDa
Fig. 4.
Engulfment of endogenous SEC62-labeled EV by EL. a WT MEF after 12 h recovery from an ER stress in the presence of 50 nM BafA1. IF panel is representative of >10 independent experiments. b Inset from a. c Single slice of an electron tomogram showing the distribution of gold-labeled endogenous SEC62 during recovery from ER stress. IEM image is representative of two independent experiments with similar results. Color code of arrows in b, c as in Fig. 2a–d. d 3D visualization by electron tomography of EL containing EV that display gold-labeled, endogenous SEC62 at the limiting membrane. See also Supplementary Movie 1. Scale bar for CLSM: 10 μm; scale bar for IEM: 200 nm
CHMP4B and VPS4A intervention during ER stress resolution
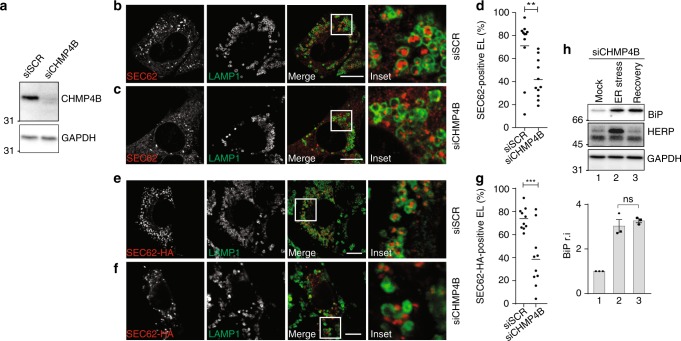
Inward membrane invagination (i.e., reverse-topology membrane remodeling and scission) is driven by ESCRT-III41–44. Consistently, silencing of the charged multivesicular body protein 4B (CHMP4B), an essential ESCRT-III subunit, prevents capture of cytosolic proteins by inward endolysosomal membrane budding45. In our experimental setup, silencing of CHMP4B expression (Fig. 5a) substantially inhibits delivery of EV within LAMP1-positive EL during recovery from ER stress (Fig. 5b–d) and when recov-ER-phagy is recapitulated by SEC62 induction (Fig. 5e–g). Consistently, CHMP4B silencing delays return of ER stress-induced chaperones at their pre-stress level (Fig. 5h, upper panel), without affecting return of HERP (middle panel).
Fig. 5.
A role of CHMP4B in recov-ER-phagy. a WB showing knockdown efficiency for siCHMP4B. b Same as Fig. 1b in MEF transfected with a scrambled siRNA. c Same as b in cells transfected with a siRNA silencing CHMP4B expression. d Quantification of EV delivery within EL in b, c (n = 11, 11 cells, respectively). Unpaired, two-tailed t test, **P = 0.0052. e Same as b in cells where recov-ER-phagy is recapitulated on SEC62 overexpression. f Same as c in cells where recov-ER-phagy is recapitulated on SEC62 overexpression. g Quantification of EV delivery within EL in e, f (n = 10, 10 cells, respectively). Unpaired t test, ***P = 0.0006. Data are representative of at least two independent experiments. Scale bars: 10 μm. h Same as Fig. 1a in cells transfected with a siRNA silencing CHMP4B expression. Quantification of BiP levels in WB, n = 3 independent experiments, mean ± SEM; unpaired, two-tailed t test, P = 0.4776. Molecular weight markers in WB are in kDa; n.s. not significant
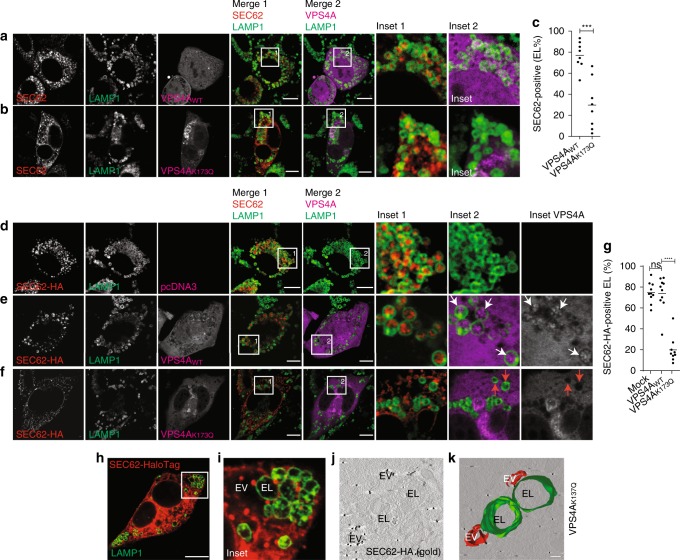
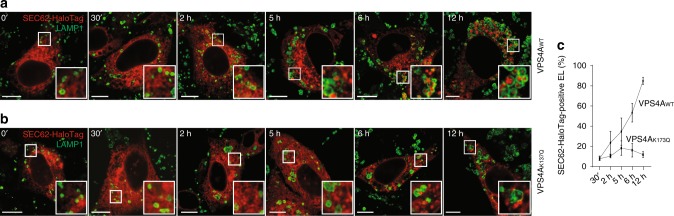
ESCRT-III-driven membrane remodeling and scission relies on the energy delivered on ATP hydrolysis by the auxiliary AAA+ ATPase VPS4A41–44. In our experiments, engulfment of SEC62-positive EV by EL during recovery from ER stress was normal in cells expressing fluorescently labeled VPS4AWT (Fig. 6a, merge 1, inset 1, Fig. 6c), but it was substantially inhibited in cells expressing VPS4AK173Q, a dominant-negative mutant that cannot bind and hydrolyze ATP46 (Fig. 6b, merge 1, inset 1, Fig. 6c). The role of VPS4A in EV engulfment by LAMP1-positive EL was confirmed on overexpression of SEC62-HA to faithfully recapitulate recov-ER-phagy (Fig. 6d–g). Ultrastructural analyses of cells where the engulfment of excess ER by LAMP1-positive EL is inhibited on inactivation of the ESCRT-III machinery show that SEC62-positive EV remain in close proximity of the EL and are not delivered within the degradative organelles (Fig. 6h–k, Supplementary Movie 2). Notably, VPS4AWT, which drives EV engulfment, accumulates within LAMP1-positive EL on inactivation of proteolytic enzymes with BafA1 (Fig. 6a, merge 2, inset 2, Fig. 6e, merge 2, white arrows in inset 2 and in inset VPS4A). The inactive VPS4AK173Q is not found within EL (Fig. 6b, merge 2, inset 2, Fig. 6f, merge 2, red arrows in inset 2 and in inset VPS4A). This is consistent with partitioning of VPS4A within the inward budding structure after the ESCRT-III mediated scission event44. Finally, we confirm VPS4A-dependent engulfment of excess ER by LAMP1-positive EL by HaloTag pulse chase (Fig. 7), a protocol for time-resolved analyses of EV segregation recently developed in our lab19. Briefly, to monitor by time-resolved fluorescence microscopy the sequential steps of ER subdomains delivery within EL for clearance, WT MEF were transfected with SEC62-HaloTag (Supplementary Fig. 3) and with VPS4AWT (Fig. 7a, c) or VPS4AK173Q (Fig. 7b, c). The fate of newly synthesized SEC62-HaloTag is followed by pulsing cells, for 15 min, with the fluorescent HaloTag ligand PBI 5030 (ref. 19 legend of Fig. 7 and Materials and methods). Initially (0–2 h chase), SEC62-HaloTag is not visible within LAMP1-positive EL. Only in cells expressing VPS4AWT, it is eventually delivered within the EL, where it progressively accumulates on EL inactivation with BafA1 (Fig. 7a, c, 5–12 h chase). In cells expressing inactive VPS4AK173Q, SEC62-HaloTag remains virtually excluded from the EL throughout the chase (Fig. 7b, c).
Fig. 6.
A role of VPS4A in recov-ER-phagy. a Same as Fig. 1b in MEF transfected with a plasmid for expression of GFP-VPS4A (VPS4AWT). b Same as a in cells transfected with a plasmid for expression of GFP-VPS4AK137Q (VPS4AK137Q). c Quantification of EV delivery within EL in a, b (n = 8, 8 cells, respectively). Unpaired, two-tailed t test, ***P = 0.0002. Data are representative of three independent experiments with similar results. d Delivery of SEC62-HA within LAMP1-positive EL in MEF transfected with an empty plasmid exposed to BafA1 for 12 h. e Same as d in cells transfected with a plasmid for expression of GFP-VPS4AWT. f Same as d in cells transfected with a plasmid for expression of GFP-VPS4AK137Q. g Quantification of EV delivery within EL in d–f (n = 10, 10, 9 cells, respectively); ANOVA and Dunnett’s multiple comparisons test, n.s.P > 0.05, ****P < 0.0001. Data are representative of at least three independent experiments. h SEC62-HaloTag-positive EV outside LAMP1-positive EL in cells expressing VPS4AK137Q. See also Supplementary Fig. 3. i inset of h. Scale bars: 10 μm. j Single slice of an electron tomogram. k 3D visualization by electron tomography of SEC62-positive EV adjacent to EL. For j, k, IEM images are representative of two independent experiments with similar results. Scale bar: 250 nm. See Supplementary Movie 2
Fig. 7.
Time-resolved analyses of VPS4A-dependent delivery of EV within EL. a Cells expressing SEC62-HaloTag and VPS4AWT are incubated for 30 min with 6-chlorohexanol, a non-fluorescent HaloTag ligand that quenches fluorescent detection of the SEC62-HaloTag chimera. Cells are subsequently incubated with a cell-permeable fluorescent HaloTag ligand (PBI 5030) to label, for 15 min, newly synthesized SEC62-HaloTag (fluorescent pulse). Cells are fixed at different times after interruption of the fluorescent pulse on PBI 5030 replacement with 6-chlorohexanol (0 to 12 h19) to monitor the progressive delivery of SEC62-HaloTag-labeled EV within LAMP1-positive EL. b Same as a in MEF expressing SEC62-HaloTag and VPS4AK137Q, where EV delivery within EL is virtually abolished. c Quantification of SEC62-HaloTag positive EL (VPS4AWT n = 3, 3, 3, 4, 4; VPS4AK173Q n = 3, 3, 4, 4, 4 cells for the corresponding time points). Mean ± SEM. IF panels are representative of two independent experiments. Scale bar: 10 μm
Endogenous LC3B decorates EV
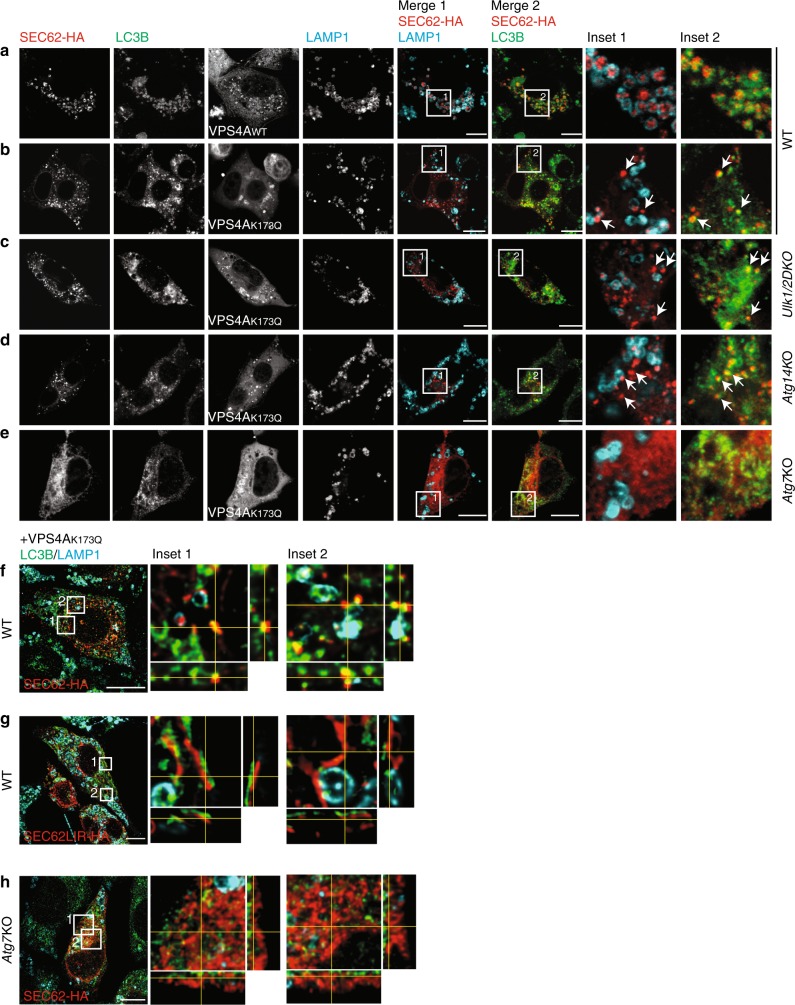
In macro-autophagy, LC3 is lipidated on the phagophore membrane and eventually recruits cargo within double-membrane autophagosomes31. Since autophagosome biogenesis is dispensable for ER turnover at the end of acute ER stresses, we assessed LC3 localization during recov-ER-phagy. Our analyses reveal that endogenous LC3 co-localizes with SEC62-labeled ER accumulating within LAMP1-positive EL on inhibition of lysosomal activity with BafA1 (Fig. 8a). The inhibition of EV engulfment by LAMP1-positive EL on expression of the inactive variant of VPS4A reveals the presence of endogenous LC3B on SEC62-labeled structures both in WT MEF (Fig. 8b) and in MEF with defective autophagosome biogenesis (Fig. 8c, d). The vesicular nature of the SEC62/LC3-positive ER-derived structures that deliver excess ER within EL during recovery from ER stress is confirmed by orthogonal sections of deconvoluted images (Fig. 8f). The formation of EV is abolished in cells expressing a SEC62LIR variant that cannot bind LC37 (Fig. 8g) and in Atg7KO MEF with defective LC3 lipidation (Fig. 8e, h). All in all, these results support the notion that the ER-resident LC3-binding protein SEC62, LC3 lipidation and the VPS4A-powered ESCRT-III machinery, but not autophagosome biogenesis, regulate ER fragmentation and piecemeal micro-ER-phagy characterizing the ER turnover on ER stress resolution.
Fig. 8.
LC3B localization on SEC62-positive EV. a SEC62-HA co-localizes with endogenous LC3B inside LAMP1-positive EL in WT MEF-expressing VPS4AWT exposed to BafA1 for 12 h. b SEC62-HA/LC3B-positive EV (white arrows) fails to be delivered inside the EL in WT MEF-expressing VPS4AK173Q. c Same as b in Ulk1/2 double-KO MEF. d Same as b in Atg14KO MEF. e Same as b in Atg7KO MEF. f Max projection of a WT cell expressing SEC62-HA and VPS4AK173Q after deconvolution. Insets show orthogonal sections of selected region. g Same as in f with SEC62LIR-HA. h Same as f in Atg7KO MEF cells. Scale bars: 10 μm. IF panels are representative of at least two independent experiments
Discussion
The ER is a plastic organelle of eukaryotic cells, whose volume and activities are adapted in response to intra- and extracellular signals. The anabolic pathways that enlarge the ER and increase its content in resident proteins and enzymes are collectively defined as UPR and have been characterized in molecular details1,2. The catabolic pathways that reduce ER volume and content by delivering ER subdomains to acidic compartment for destruction are much less understood. The identification of several ER-resident proteins that may engage, upon appropriate stimuli, cytosolic factors such as LC3s/Atg8, GABARAPs, and FIP200/Atg11 to promote delivery of ER fragments within EL/vacuole for clearance has opened the run to dissect receptor-mediated, lysosomal-controlled, ER turnover pathways. At the end of an ER stress, the reduction of the ER volume and of the ER content in resident proteins to pre-stress, physiologic status is crucial to re-establish ER homeostasis. In this phase, cells must, among other things, also re-gain the capacity to produce the secretOME (i.e., the 40% of the proteOME destined to the organelles of the secretory and endocytic compartments, the plasma membrane, and the extracellular space), whose production is temporarily halted during the ER stress phase47. The secretOME enters the ER co-translationally via the SEC61 protein translocation machinery that is endowed with a SEC62:SEC63 functional complex to facilitate access of those polypeptides that are synthesized in the cytosol and imported only after completion of the polypeptide chain via post-translational translocation22,23. We recently reported that conclusion of ER stresses activates the LC3-binding function of SEC62 and triggers lysosomal turnover of ER subdomains containing folding factors, but lacking ERAD factors via catabolic pathways collectively defined as recov-ER-phagy7. The finding that selective ER delivery to EL for clearance is recapitulated by SEC62 overexpression and by SEC63 silencing7 hint at possible mutually exclusive role of SEC62 in ER protein import vs. receptor-mediated, selective ER turnover. Notably, the LC3-binding region is dispensable for the former and required for the latter function of SEC62 (ref. 7 and Supplementary Fig. 3c). It will be of interest to verify whether the changes in newly synthesized proteins getting access to the ER during steady state (presumably more cargo) vs. ER stress (presumably more ER-resident proteins) vs. recovery from ER stress (back to more cargo proteins import?) phases results in variations in the SEC61:SEC62:SEC63 oligomeric composition22,23 and if this in turn generates orphan SEC62 to promote ER turnover. Alternatively, the triggering signal for a switch in SEC62 function from protein translocation to ER turnover could be a change in the luminal fraction of free BiP (which is regulated in complex manner48–50). In fact, SEC63 is a BiP-binding protein and BiP association could set SEC62 free for its role in recov-ER-phagy. All these issues will be assessed in future work.
Here, we report that lipidation of LC3 is required, but the autophagosome biogenesis machinery and macro-ER-phagy are dispensable for clearance of excess ER characterizing the recovery phase from acute ER stresses. As such, recov-ER-phagy, which is triggered by an ER-centric signal, is mechanistically different from starvation-induced turnover of the mammalian ER, which activates (unselective?) macro-autophagic clearance of ER subdomains decorated with FAM134B6, RTN38, ATL39, CCPG110, TEX26411,12. Rather, SEC62-driven return of ER size and content at pre-stress, physiologic status, entails the LC3 lipidation machinery, the ESCRT-III component CHMP4B and the accessory AAA+ ATPase VPS4A. During recovery from ER stress, CHMP4B and VPS4A, whose involvement in physiologic and pathogen-induced membrane repair, remodeling and fission events has been reported43,44,51, ensure the inward budding of the EL membrane required for the engulfment and clearance of pre-formed ER vesicles that display SEC62 and LC3 at the limiting membrane and contain excess ER chaperones and membranes generated during the phase of ER stress. The function of LC3 in recov-ER-phagy (and in other types of receptor-mediated ER turnover by lysosomes/vacuole) as well as the mode of recruitment of the ESCRT-III machinery that welcomes the incoming ER-derived vesicle containing material to be cleared from cells remains a matter for further studies. However, these data highlight the variety of pathways that cells can activate in response to pleiotropic and to ER-centric signals to ensure lysosomal turnover of the ER. For the mechanistic dissection of catabolic regulation of ER function via lysosomal-controlled ER turnover, it seems crucial to examine in detail cellular responses to ER-centric signals that may activate individual ER-resident LC3-binding proteins. ER-centric signals activate client-specific autophagic and not autophagic pathways that deliver fragmented ER subdomains containing defective material (certainly misfolded proteins15,17, possibly aberrant lipids or ER-resident proteins, whose activity is determined post-translationally by regulated turnover29) to the lysosomal/vacuolar compartments. Significant on this line, is the characterization of ERLAD pathways that ensure disposal of ER or of ER-exit site subdomains containing misfolded proteins that cannot be dislocated across the membrane for proteasomal degradation15,17. Emerging evidence reveal the involvement in ERLAD of mammalian ER-resident LC3-binding proteins such as FAM134B19,20 and CCPG110 and the yeast ER-resident Atg8-binding protein Atg4052. These receptors operate conventionally to engage autophagosomes in macro-ER-phagy-like processes20,52, or unconventionally by ensuring direct delivery of the ER subdomain to be cleared from cells to EL via vesicular trafficking19 or via poorly characterized micro-autophagy-like processes21.
Back to recov-ER-phagy, the broader, possible implications of our findings relate to the fact that amplification of the SEC62 gene and the consequent enhanced ER turnover confers a stress tolerance that correlates with resistance to anti-cancer therapies to a number of carcinomas, including breast, prostate, thyroid, lung adenocarcinoma, and head and neck squamous cell carcinoma53–55. Our characterization of the catabolic pathways activated by cells recovering from ER stress to remodel the ER paves the way for identification of therapeutic targets to treat diseases caused by impaired proteostatic control and to unravel in more detail the novel function of the ESCRT-III/VPS4A machinery in membrane remodeling to better define molecular aspects of organelle and membrane dynamics.
Materials and methods
Antibodies, expression plasmids, and chemicals
Antibodies against CNX (Western blot (WB) 1:3000, immunofluorescence (IF) 1:100), CHMP4B (1:1000), SEC62 (WB 1:1000, IF 1:100), and ERj3 (1:250) are kind gifts from A. Helenius, H. Stenmark, and R. Zimmermann. Plasmid encoding GFP-RAB7 is a kind gift from T. Johansen. Commercial antibodies used in this study are from Stressgen (BiP, 1:1000), Chondrex (HERP, 1:1500), Developmental Studies Hybridoma Bank (LAMP1, 1:50), Sigma and Santa Cruz (HA, WB 1:3000, IF 1:100), Merck Millipore (GAPDH, 1:30,000), Sigma (STX17, 1:1000), Abcam (VAMP8, 1:1000), Santa Cruz (Actin 1:500), Novus and Sigma (LC3B, WB 1:1000, IF 1:50), Promega (HaloTag, 1:1000), and MBL (p62, 1:1500). The secondary horse radish peroxidase (HRP)-conjugated antibodies were from Jackson Immunoresearch (rabbit, 1:10,000), Santa Cruz (goat, 1:20,000), and SouthernBiotech (mouse, 1:20,000). Alexa Fluor-conjugated secondary antibodies (1:300) from Invitrogen, Jackson Immunoresearch, Thermo Fisher, and BioLegend. Plasmids encoding GFP-VPS4AWT and GFP-VPS4AK173Q were a kind gift from John McCullough. SEC62 was subcloned in a pcDNA3.1 expression plasmid with the addition of a C-terminal HA-tag or HaloTag7. CPA (Sigma) and BafA1 (Sigma) were used for 12 h, if not otherwise specified, at final concentrations of 10 μM and 50 nM, respectively. PS341 (Millennium Pharmaceuticals) was used for 8 h at a final concentration of 10 μM.
Induction of transient ER stress
To induce a mild and reversible ER stress, cultured cells were exposed for up to 12 h to CPA, a reversible inhibitor of the sarco/ER calcium ATPase7,27. An alternative protocol implies cell exposure to dithiothreitol7 (DTT, a reversible perturbator of redox homeostasis56), whereas other ER stress-inducing drugs such as tunicamycin and thapsigargin, which are irreversible inhibitors of GlcNAc phosphotransferase57 and of the sarco/ER calcium ATPase58, respectively, are toxic7. Consistent with UPR induction, CPA treatment caused splicing of XBP1 transcripts, 25% attenuation of global protein synthesis, and induction of ER stress marker transcripts and proteins7. The catabolic events characterizing recovery from ER stress (i.e., the delivery of ER subdomains within EL for clearance) were investigated after CPA wash-out as specified in the figure legends and in ref. 7.
Cell culture, transient transfection, and RNA interference
MEF and HEK cells were grown at 37 °C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) medium supplemented with 10% fetal calf serum (FCS). WT and Atg7KO MEF cells were gifts from M. Komatsu. Atg4BKO, Atg13KO, Atg14 and Atg16L1KO, and Ulk1/2DKO MEF cells were kindly provided by G. Marino, F. Reggiori, T. Saitoh, and S. Tooze, respectively, and have been characterized in ref. 19 and in Supplementary Fig. 2. To induce autophagy, cells were washed three times with Earle’s balanced salt solution (EBSS, Thermo Fisher) and then incubated in EBSS for 2 h in the presence or absence of 100 nM BafA1.
Transient transfections were performed using JetPrime transfection reagent (PolyPlus) according to the manufacturer’s instructions. RNA interferences were performed in MEF cells plated at 50–60% confluence. Cells were transfected with scrambled small interfering RNA or small interfering RNA (siRNA) against CHMP4B (5′-AAACAGUCCCUCUACCAAAtt-3′, 50 nM per dish, Silencer Select Pre-designed, Ambion). Cells were processed for immunofluorescence or for biochemical analyses 48 h after transfection (see below).
Preparation of KO cells
STX17- and VAMP8-KO MEF and SEC62-KO HEK cells were generated by CRISPR/Cas9 genome editing. For the generation of the guideRNA‐Cas9 plasmids, lentiCRISPRv2‐puro system (Addgene52961) was obtained from Addgene (http://www.addgene.org). Guide sequences were obtained from the Cas9 target design tools (crispr.mit.edu:8079 and/or www.addgene.org/pooled-library). All protocols and information can be found at the website https://www.addgene.org/crispr. The target sequences for guide RNA were synthesized by Microsynth. Two annealed oligonucleotides (5′-CACCGCTGTGGTTGACTACTGCAAC-3′, 5′-AAACGTTGCAGTAGTCAACCACAGC-3′ for human SEC62; 5′‐GCGCTCCAATATCCGAGAAA‐3′, 5′‐TTTCTCGGATATTGGAGCGC‐3′ for murine STX17; 5′‐CCACCTCCGAAACAAGACAG‐3′, 5′‐CTGTCTTGTTTCGGAGGTGG‐3′ for murine VAMP8) were inserted into the lentiCRISPRv2‐puro vector using the BsmBI restriction site. Vectors were transfected in HEK and MEF cells with JetPrime (Polyplus) according to the manufacturer’s instructions7,19. Cells were cultured in DMEM supplemented with 10% FBS. Two days after transfection, the medium was supplemented with 2 μg/ml puromycin. After 10 days, puromycin‐resistant clones were picked and gene KO was verified by WB (ref. 7 for SEC62; Fig. 3a, b and ref. 19 for STX17 and VAMP8) and with a translocation assay for SEC62 (ref. 7 and Supplementary Fig. 3c).
Cell lysis, Western blot
Cells were washed in ice cold phosphate-buffered saline (PBS) containing 20 mM N-ethylmaleimide (NEM) and lysed in 1% sodium dodecyl sulfate (SDS) in HEPES-buffered saline, pH 6.8, or with RIPA buffer (1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate in HBS, pH 7.4) containing 20 mM NEM and protease inhibitor cocktail (1 mM PMSF, chymostatin, leupeptin, antipain, and pepstatin, 10 μg/ml each). Post-nuclear supernatants were collected by centrifugation at 10,000 × g for 10 min at 4 °C. Samples were denatured and reduced in DTT-containing sample buffer for 5 min at 95 °C and separated by SDS-PAGE (polyacrylamide gel electrophoresis). Proteins were transferred to polyvinylidene fluoride membranes with the Trans-Blot Turbo Transfer System (Bio-Rad). Membranes were blocked 10 min with 10% (w/v) non-fat dry milk (Bio-Rad) and stained with the above-mentioned primary antibodies for 90 min and for 45 min with HRP-conjugated secondary antibodies. Membranes were developed using the Luminata Forte ECL detection system (Millipore) and signals were acquired with the ImageQuant LAS 4000 system (GE Healthcare Life Sciences) or with the Amersham Imager 680 system. Image quantifications were performed with the Multi Gauge Analysis tool (Fujifilm). Membrane stripping was performed using Re-Blot Plus Strong Solution (Millipore) following the manufacturer’s instructions. Uncropped blots can be found as Supplementary Fig. 4.
Confocal laser scanning microscopy
MEF cells plated on alcian blue-coated glass coverslips were treated according to the experimental setup. Cells were washed twice in PBS and fixed at room temperature for 20 min with 3.7% formaldehyde in PBS or 5 min in 100% methanol at −20 °C for endogenous LC3B detection. Cells were then incubated for 15 min with permeabilization solution (PS, 0.05% saponin, 10% goat serum, 10 mM HEPES, 15 mM glycine, pH 7.4) to improve antigen accessibility. Cells were incubated with the primary antibodies diluted 1:50–1:200 in PS for 90 min, washed for 15 min in PS, and then incubated with Alexa Fluor-conjugated secondary antibodies diluted 1:300 in PS for 30 min. Cells were rinsed with PS and water and mounted with Vectashield (Vector Laboratories) supplemented with 4′,6-diamidino-2-phenylindole. Confocal pictures were acquired using a Leica TCS SP5 microscope with a 63.0 × 1.40 Oil UV objective. FIJI was used for image analysis and processing. Figure 8f–h were acquired with LEICA HCX PL APO CS 100.0 × 1.44 Oil UV objective with a XY pixel size of 50 nm and Z step of 125 nm and pinhole 0.8 AU. Images were deconvoluted with Autoquant 3.1.1 (Media Cybernetics) with a spherical aberration correction.
Immunogold electron microscopy
Cells were plated on alcian blue-coated glass coverslips and fixed 10 min with 0.05% glutaraldehyde in 4% paraformaldehyde (PFA) EM grade and 0.2 M HEPES buffer and 50 min in 4% PFA EM grade in 0.2 M HEPES buffer. After three washes in PBS, cells were incubated 10 min with 50 mM glycine and blocked 1 h in blocking buffer (0.2% bovine serum albumin, 5% goat serum, 50 mM NH4Cl, 0.1% saponin, 20 mM PO4 buffer, 150 mM NaCl). Staining with primary antibodies and nanogold-labeled secondary antibodies (Nanoprobes) were performed in blocking buffer at room temperature. Cells were fixed 30 min in 1% glutaraldehyde and nanogold was enlarged with gold enhancement solution (Nanoprobes) according to the manufacturer’s instructions. Cells were post fixed with osmium tetroxide, embedded in epon, and processed into ultrathin slices. After contrasting with uranyl acetate and lead citrate, the sections were analyzed with Zeiss LEO 512 electron microscope. Images were acquired by 2k × 2k bottom-mounted slow-scan Proscan camera controlled by the EsivisionPro 3.2 software.
HaloTag pulse-chase analyses
MEF cells were plated on alcian blue-coated glass coverslip and transfected with SEC62-HaloTag and VPS4WT or VPS4K173Q. Seventeen hours after transfection, cells were incubated with 15 μM 6-chlorohexanol (Sigma) in DMEM 10% FCS for 30 min. 6-Chlorohexanol is cell-permeable black ligand that irreversibly binds the SEC62-HaloTag-binding pocket. After three washes in DMEM 10% FCS, cells are pulsed 15 min with 1 μM of the fluorescent ligand PBI 5030 (Promega), which exclusively enters the HaloTag ligand binding pocket of newly synthetized SEC62-HaloTag, and 100 nM BafA1 in DMEM 10% FCS. The fluorescent ligand is removed and after three washes in DMEM 10% FCS, cells are incubated with 15 μM of the black ligand to block incorporation of the fluorescent ligand in the newly synthetized SEC62-HaloTag, and 100 nM BafA1 in DMEM 10% FCS. Cells were fixed at increasing time points (0, 30 min, 2, 5, 6, and 12 h) and processed for confocal laser scanning microscopy as described above. HaloTag pulse chase has been described in ref. 19.
Statistical analyses and reproducibility
In panels showing WB or IF, unless stated otherwise, images are representative of three independent experiments with similar results. The number of independent experiments or cell numbers is given in the figure’s legend. Statistical analyses were performed only if sample size was ≥3 using GraphPad Prism 7 software. One-way analysis of variance with Dunnett’s multiple comparisons test or unpaired, two-tailed t test were used to asses statistical significance. P values are given in the figure legends; n.sP < 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank G. Brambilla Pisoni, F. Fumagalli, A. Helenius, M. Komatsu, G. Marino, N. Mizushima, J. Noack, P. Paganetti, F. Reggiori, T. Saitoh, C. Settembre, S. Tooze, X. Wang, and the members of Molinari’s lab for gift of reagents, discussions, and critical reading of the manuscript. We are grateful to the ALEMBIC facility at San Raffaele Scientific Institute, Milan, Italy for the help in electron microscopy analyses. M.M. is supported by Signora Alessandra, AlphaONE Foundation, Foundation for Research on Neurodegenerative Diseases, Swiss National Science Foundation, and Comel and Gelu Foundations.
Author contributions
Conceptualization: M.M. together with M.L.; methodology: M.L., A.R., D.M., and M.M.; investigation: M.L., A.R., D.M., and M.M.; writing original draft: M.M.; supervision: M.M.
Data availability
The data that support the findings of this study are available from the authors on reasonable request. The source data underlying the figures can be found in the Source Data file.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communication thanks Simon Wilkinson and other, anonymous, reviewers for their contributions to the peer review of this work. Peer review reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-019-12991-z.
References
- 1.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 2.Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529:326–335. doi: 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- 3.Smith M, Wilkinson S. ER homeostasis and autophagy. Essays Biochem. 2017;61:625–635. doi: 10.1042/EBC20170092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fregno I, Molinari M. Endoplasmic reticulum turnover: ER-phagy and other flavors in selective and non-selective ER clearance. F1000Res. 2018;7:454. doi: 10.12688/f1000research.13968.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Locke M, Collins JV. The structure and formation of protein granules in the fat body of an insect. J. Cell Biol. 1965;26:857–884. doi: 10.1083/jcb.26.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khaminets A, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 7.Fumagalli F, et al. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat. Cell Biol. 2016;18:1173–1184. doi: 10.1038/ncb3423. [DOI] [PubMed] [Google Scholar]
- 8.Grumati P, et al. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife. 2017;6:e25555. doi: 10.7554/eLife.25555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q, et al. ATL3 is a tubular ER-phagy receptor for GABARAP-mediated selective autophagy. Curr. Biol. 2019;29:846–855. doi: 10.1016/j.cub.2019.01.041. [DOI] [PubMed] [Google Scholar]
- 10.Smith MD, et al. CCPG1 is a non-canonical autophagy cargo receptor essential for ER-phagy and pancreatic ER proteostasis. Dev. Cell. 2018;44:217–232. doi: 10.1016/j.devcel.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An H, et al. Tex264 is an endoplasmic reticulum-resident ATG8-interacting protein critical for ER remodeling during nutrient stress. Mol. Cell. 2019;74:891–908. doi: 10.1016/j.molcel.2019.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chino H, Hatta T, Natsume T, Mizushima N. Intrinsically disordered protein TEX264 mediates ER-phagy. Mol. Cell. 2019;74:909–921. doi: 10.1016/j.molcel.2019.03.033. [DOI] [PubMed] [Google Scholar]
- 13.Mochida K, et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522:359–362. doi: 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- 14.Loi M, Fregno I, Guerra C, Molinari M. Eat it right: ER-phagy and RecovER-phagy. Biochem. Soc. Trans. 2018;46:699–706. doi: 10.1042/BST20170354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilkinson S. ER-phagy: shaping up and destressing the endoplasmic reticulum. FEBS J. 2019;286:2645–2663. doi: 10.1111/febs.14932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkinson S. Emerging principles of selective ER autophagy. J. Mol. Biol. 2019;S0022-2836:30279–7. doi: 10.1016/j.jmb.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fregno I, Molinari M. Proteasomal and lysosomal clearance of faulty secretory proteins: ER-associated degradation (ERAD) and ER-to-lysosome-associated degradation (ERLAD) pathways. Crit. Rev. Biochem. Mol. Biol. 2019;54:153–163. doi: 10.1080/10409238.2019.1610351. [DOI] [PubMed] [Google Scholar]
- 18.Grumati P, Dikic I, Stolz A. ER-phagy at a glance. J. Cell Sci. 2018;131:1–6. doi: 10.1242/jcs.217364. [DOI] [PubMed] [Google Scholar]
- 19.Fregno I, et al. ER-to-lysosome-associated degradation of proteasome-resistant ATZ polymers occurs via receptor-mediated vesicular transport. EMBO J. 2018;37:e99259. doi: 10.15252/embj.201899259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forrester A, et al. A selective ER-phagy exerts procollagen quality control via a Calnexin-FAM134B complex. EMBO J. 2019;38:e99847. doi: 10.15252/embj.201899847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omari S, et al. Noncanonical autophagy at ER exit sites regulates procollagen turnover. Proc. Natl. Acad. Sci. USA. 2018;115:E10099–E10108. doi: 10.1073/pnas.1814552115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conti BJ, Devaraneni PK, Yang ZY, David LL, Skach WR. Cotranslational stabilization of Sec62/63 within the ER Sec61 translocon is controlled by distinct substrate-driven translocation events. Mol. Cell. 2015;58:269–283. doi: 10.1016/j.molcel.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jadhav B, et al. Mammalian SRP receptor switches the Sec61 translocase from Sec62 to SRP-dependent translocation. Nat. Commun. 2015;6:1–11. doi: 10.1038/ncomms10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feldman D, Swarm RL, Becker J. Elimination of excess smooth endoplasmic reticulum after phenobarbital administration. J. Histochem. Cytochem. 1980;28:997–1006. doi: 10.1177/28.9.7410819. [DOI] [PubMed] [Google Scholar]
- 25.Bolender RP, Weibel ER. A morphometric study of the removal of phenobarbital-induced membranes from hepatocytes after cessation of treatment. J. Cell Biol. 1973;56:746–761. doi: 10.1083/jcb.56.3.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer HA, et al. Mammalian Sec61 is associated with Sec62 and Sec63. J. Biol. Chem. 2000;275:14550–14557. doi: 10.1074/jbc.275.19.14550. [DOI] [PubMed] [Google Scholar]
- 27.Pirot P, et al. Global profiling of genes modified by endoplasmic reticulum stress in pancreatic beta cells reveals the early degradation of insulin mRNAs. Diabetologia. 2007;50:1006–1014. doi: 10.1007/s00125-007-0609-0. [DOI] [PubMed] [Google Scholar]
- 28.Yan L, et al. Ube2g2-gp78-mediated HERP polyubiquitylation is involved in ER stress recovery. J. Cell Sci. 2014;127:1417–1427. doi: 10.1242/jcs.135293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernasconi R, Molinari M. ERAD and ERAD tuning: disposal of cargo and of ERAD regulators from the mammalian ER. Curr. Opin. Cell Biol. 2011;23:176–183. doi: 10.1016/j.ceb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagiwara M, Ling JJ, Koenig PA, Ploegh HL. Posttranscriptional regulation of glycoprotein quality control in the endoplasmic reticulum is controlled by the E2 Ub-conjugating enzyme UBC6e. Mol. Cell. 2016;63:753–767. doi: 10.1016/j.molcel.2016.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bento CF, et al. Mammalian autophagy: how does it work? Annu. Rev. Biochem. 2016;85:685–713. doi: 10.1146/annurev-biochem-060815-014556. [DOI] [PubMed] [Google Scholar]
- 32.McAlpine F, Williamson LE, Tooze SA, Chan EYW. Regulation of nutrient-sensitive autophagy by uncoordinated 51-like kinases 1 and 2. Autophagy. 2013;9:361–373. doi: 10.4161/auto.23066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaizuka T, Mizushima N. Atg13 is essential for autophagy and cardiac development in mice. Mol. Cell. Biol. 2016;36:585–595. doi: 10.1128/MCB.01005-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kageyama S, et al. The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol. Biol. Cell. 2011;22:2290–2300. doi: 10.1091/mbc.e10-11-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Matsui T, et al. Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J. Cell Biol. 2018;217:2633–2645. doi: 10.1083/jcb.201712058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tekirdag K, Cuervo AM. Chaperone-mediated autophagy and endosomal microautophagy: jointed by a chaperone. J. Biol. Chem. 2018;293:5414–5424. doi: 10.1074/jbc.R117.818237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lefebvre C, Legouis R, Culetto E. ESCRT and autophagies: endosomal functions and beyond. Semin. Cell Dev. Biol. 2018;74:21–28. doi: 10.1016/j.semcdb.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 39.Schuck S, Gallagher CM, Walter P. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J. Cell Sci. 2014;127:4078–4088. doi: 10.1242/jcs.154716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lipatova, Z. & Segev, N. in Autophagy: Cancer, Other Pathologies, Inflammation, Immunity, Infection, and Aging (ed. Hayat, M. A.) (Elsevier, 2018).
- 41.Schoneberg J, et al. ATP-dependent force generation and membrane scission by ESCRT-III and Vps4. Science. 2018;362:1423–1428. doi: 10.1126/science.aat1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 43.McCullough J, Frost A, Sundquist WI. Structures, functions, and dynamics of ESCRT-III/Vps4 membrane remodeling and fission complexes. Annu. Rev. Cell Dev. Biol. 2018;34:85–109. doi: 10.1146/annurev-cellbio-100616-060600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoneberg J, Lee IH, Iwasa JH, Hurley JH. Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell. Biol. 2017;18:5–17. doi: 10.1038/nrm.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mejlvang J, et al. Starvation induces rapid degradation of selective autophagy receptors by endosomal microautophagy. J. Cell Biol. 2018;217:3640–3655. doi: 10.1083/jcb.201711002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bishop N, Woodman P. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol. Biol. Cell. 2000;11:227–239. doi: 10.1091/mbc.11.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uhlen M, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 48.Preissler S, et al. AMPylation matches BiP activity to client protein load in the endoplasmic reticulum. Elife. 2015;4:e12621. doi: 10.7554/eLife.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Preissler S, Ron D. Early events in the endoplasmic reticulum unfolded protein response. Cold Spring Harb. Perspect. Biol. 2019;11:1–18. doi: 10.1101/cshperspect.a033894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chambers JE, Petrova K, Tomba G, Vendruscolo M, Ron D. ADP ribosylation adapts an ER chaperone response to short-term fluctuations in unfolded protein load. J. Cell Biol. 2012;198:371–385. doi: 10.1083/jcb.201202005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Radulovic M, Stenmark H. ESCRTs in membrane sealing. Biochem. Soc. Trans. 2018;46:773–778. doi: 10.1042/BST20170435. [DOI] [PubMed] [Google Scholar]
- 52.Cui Y, et al. A COPII subunit acts with an autophagy receptor to target endoplasmic reticulum for degradation. Science. 2019;365:53–60. doi: 10.1126/science.aau9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Linxweiler M, Schick B, Zimmermann R. Let’s talk about Secs: Sec61, Sec62 and Sec63 in signal transduction, oncology and personalized medicine. Signal Transduct. Target Ther. 2017;2:1–10. doi: 10.1038/sigtrans.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bergmann TJ, Fumagalli F, Loi M, Molinari M. Role of SEC62 in ER maintenance: a link with ER stress tolerance in SEC62-overexpressing tumors? Mol. Cell Oncol. 2017;4:e1264351. doi: 10.1080/23723556.2016.1264351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takacs FZ, et al. Identification of 3q oncogene SEC62 as a marker for distant metastasis and poor clinical outcome in invasive ductal breast cancer. Arch. Gynecol. Obstet. 2019;299:1405–1413. doi: 10.1007/s00404-019-05081-4. [DOI] [PubMed] [Google Scholar]
- 56.Braakman I, Helenius J, Helenius A. Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. EMBO J. 1992;11:1717–1722. doi: 10.1002/j.1460-2075.1992.tb05223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takatsuki A, Tamura G. Effect of tunicamycin on synthesis of macromolecules in cultures of chick embryo fibroblasts infected with Newcastle Disease Virus. J. Antibiot. 1971;24:785–794. doi: 10.7164/antibiotics.24.785. [DOI] [PubMed] [Google Scholar]
- 58.Lytton J, Westlin M, Hanley MR. Thapsigargin inhibits the sarcoplasmic or endoplasmic reticulum Ca-ATPase family of calcium pumps. J. Biol. Chem. 1991;266:17067–17071. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the authors on reasonable request. The source data underlying the figures can be found in the Source Data file.