Abstract Abstract
Rhus chinensis represents a commercially and ecologically important tree species in China, but suffers from canker diseases in Jiangxi Province. Synnemata, pycnidia and ascomata were discovered on cankered tissues. Strains were obtained from single ascospore or conidium within the fruiting bodies and identified based on morphological comparison and the phylogenetic analyses of partial ITS, LSU, tef1 and rpb2 gene sequences. As a result, two species were confirmed to represent two kinds of synnemata. One of these species is described herein as Flavignomonia rhoigenagen. et sp. nov.; and Synnemasporella aculeans is illustrated showing ascomata, pycnidia and synnemata. Flavignomonia is distinguished from Synnemasporella by the colour of the synnematal tips. Additionally, Flavignomonia can be distinguished from the other gnomoniaceous genera by the formation of synnemata.
Keywords: Diaporthales , Gnomoniaceae , systematics, taxonomy
Introduction
Many Diaporthales species are important branch canker pathogens, forming acervuli or pycnidia on diseased tissues (Rossman et al. 2007, Senanayake et al. 2017, Jiang et al. 2018, 2019, Wijayawardene et al. 2018, Yang et al. 2018, Voglmayr et al. 2019). However, two diaporthalean species with synnemata were reported to cause cankers, namely Synnemasporella aculeans (syn. Cryptodiaporthe aculeans) and S. toxicodendri (Fan et al. 2018). These two species differ from the other diaporthalean taxa in conidiomata and form a distinct clade phylogenetically, which was named Synnemasporellaceae and distinguished by Fan et al. (2018).
Gnomoniaceae was initially introduced with Gnomonia as the type (Winter 1886). Species in Gnomoniaceae formed upright perithecia, with or without long or short necks and presence or absence of stromatic tissues (Barr 1978, Sogonov et al. 2008, Walker et al. 2012). In the recent monograph of Diaporthales, 34 genera were accepted in the family Gnomoniaceae (Senanayake et al. 2018). Subsequently, Neognomoniopsis and Tenuignomonia were added based on both molecular and morphological evidence (Crous et al. 2019, Minoshima et al. 2019).
Chinese gall (Rhus chinensis Mill.) has a range of uses as source of medicine, dye and oil, and has a wide distribution in China (Wang et al. 2014). However, cankers were found to be associated with different ascomata during our fungal collection trips in Jiangxi Province, China. The objectives of the present study were to identify these fungi based on morphological and phylogenetic evidence.
Materials and methods
Sample collections and isolation
We conducted our fungal collection surveys from April to October in China, and found Rhus chinensis to be one of the major tree species in Jiangxi Province. Twigs, branches and stems were carefully checked, and diseased tissues were cut into small pieces and packed in paper bags. Isolates were obtained by transferring the ascospores or conidial masses from ascomata to sterile PDA plates, incubating at 25 °C until spores germinated. Single germinating spores were transferred onto new PDA plates, which were kept at 25 °C in darkness. Specimens were deposited in the Museum of the Beijing Forestry University (BJFC) and axenic cultures maintained in the China Forestry Culture Collection Centre (CFCC).
Morphological analysis
Recognition and identification of the fungal species on Rhus chinensis was based on fruiting bodies formed on the bark. Ascomata and conidiomata were sectioned by hand using a double-edged blade, and microscopic structures were observed under a dissecting microscope. At least 10 conidiomata/ascomata, 10 asci, and 50 conidia/ascospores were measured to calculate mean and standard deviation. Measurements are reported as maxima and minima in parentheses and the range representing the mean plus and minus the standard deviation of the number of measurements given in parentheses (Voglmayr et al. 2017). Microscopy photographs were captured with a Nikon Eclipse 80i compound microscope equipped with a Nikon digital sight DS-Ri2 high definition colour camera, using differential interference contrast illumination. Nomenclatural novelties and descriptions were deposited in MycoBank (Crous et al. 2004).
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted from colonies grown on cellophane-covered PDA plates using a modified CTAB method (Doyle and Doyle 1990). PCR amplifications were performed in a DNA Engine Peltier Thermal Cycler (PTC-200; Bio-Rad Laboratories, Hercules, CA, USA). The primer sets ITS1/ITS4 (White et al. 1990) were used to amplify the ITS region. The primer pair LR0R/LR5 (Vilgalys and Hester 1990) was used to amplify the LSU region. The primer pairs EF1-688F/EF1-986R or EF1-728F/TEF1-LLErev (Carbone and Kohn 1999; Jaklitsch et al. 2005; Alves et al. 2008) were used to amplify tef1 gene. The primer pair dRPB2-5f/dRPB2-7r (Voglmayr et al. 2016) was used to amplify the rpb2 gene. The polymerase chain reaction (PCR) assay was conducted as described by Fan et al. (2018). PCR amplification products were assayed via electrophoresis in 2 % agarose gels. DNA sequencing was performed using an ABI PRISM 3730XL DNA Analyzer with a BigDye Terminater Kit v.3.1 (Invitrogen, USA) at the Shanghai Invitrogen Biological Technology Company Limited (Beijing, China).
Phylogenetic analyses
The preliminary identities of the isolates sequenced in this study were obtained by conducting a standard nucleotide BLAST search using the sequences generated from the above primers of the different genomic regions (ITS, LSU, tef1 and rpb2). The BLAST results showed that three isolates were grouped in the family Gnomoniaceae, and five isolates in the genus Synnemasporella. The phylogenetic analyses for the three gnomoniaceous isolates were conducted based on Senanayake et al. (2018), supplemented by sequences of Tenuignomonia styracis and Neognomoniopsis quercina from Crous et al. (2019) and Minoshima et al. (2019). Melanconis marginalis (CBS 109744) in Melanconidaceae was selected as the out-group taxon. All sequences were aligned using MAFFT v. 6 (Katoh and Toh 2010) and edited manually using MEGA v. 6 (Tamura et al. 2013). Phylogenetic analyses were performed using PAUP v. 4.0b10 for maximum parsimony (MP) analysis (Swofford 2003), and PhyML v. 3.0 for Maximum Likelihood (ML) analysis (Guindon et al. 2010).
MP analysis was run using a heuristic search option of 1000 search replicates with random additions of sequences with a tree bisection and reconnection algorithm. Other calculated parsimony scores were tree length (TL), consistency index (CI), retention index (RI), and rescaled consistency (RC). ML analysis was performed using a GTR site substitution model including a gamma-distributed rate heterogeneity and a proportion of invariant sites (Guindon et al. 2010). The branch support was evaluated using a bootstrapping method of 1000 replicates (Hillis and Bull 1993). The matrix was partitioned for the different gene regions. Phylograms were shown using FigTree v. 1.4.3 (Rambaut 2016). Novel sequences generated in the current study were deposited in GenBank (Table 1) and the aligned matrices used for phylogenetic analyses in TreeBASE (accession number: S25047).
Table 1.
Strains used in the phylogenetic tree and their culture accession and GenBank numbers. Strains from this study are in bold.
| Species | Strains | GenBank numbers | |||
| ITS | LSU | tef1 | rpb2 | ||
| Alnecium auctum | CBS 124263 | KF570154 | KF570154 | KF570200 | KF570170 |
| Ambarignomonia petiolorum | CBS 116866 | EU199193 | AY818963 | NA | EU199151 |
| CBS 121227 | EU254748 | EU255070 | EU221898 | EU219307 | |
| Amphiporthe tiliae | CBS 119289 | EU199178 | EU199122 | NA | EU199137 |
| Anisogramma anomala | 529478 | EU683064 | EU683066 | NA | NA |
| Anisogramma virgultorum | 529479 | EU683062 | EU683065 | NA | NA |
| Apiognomonia errabunda | AR 2813 | DQ313525 | NG027592 | DQ313565 | DQ862014 |
| Apiognomonia veneta | MFLUCC 16-1193 | MF190114 | MF190056 | NA | NA |
| Apioplagiostoma populi | 858501 | KP637024 | NA | NA | NA |
| Asteroma alneum | CBS 109840 | EU167609 | EU167609 | NA | NA |
| Asteroma sp. | Masuya 8Ah9-1 | NA | AB669035 | NA | NA |
| Cryptosporella hypodermia | CBS 116866 | EU199181 | AF408346 | NA | EU199140 |
| Discula destructiva | MD 254 | AF429741 | AF429721 | AF429732 | NA |
| Ditopella biseptata | MFLU 15-2661 | MF190147 | MF190091 | NA | MF377616 |
| Ditopella ditopa | CBS 109748 | DQ323526 | EU199126 | NA | EU199145 |
| Ditopellopsis sp. | CBS 121471 | EU254763 | EU255088 | EU221936 | EU219254 |
| Flavignomonia rhoigena | CFCC 53118 | MK432674 | MK429917 | NA | MK578102 |
| CFCC 53119 | MK432675 | MK429918 | NA | MK578103 | |
| CFCC 53120 | MK432676 | MK429919 | NA | MK578104 | |
| Gnomonia gnomon | CBS 199.53 | DQ491518 | AF408361 | EU221885 | EU219295 |
| CBS 829.79 | AY818957 | AY818964 | EU221905 | NA | |
| Gnomoniopsis alderdunensis | CBS 125680 | GU320825 | NA | NA | NA |
| Gnomoniopsis chamaemori | CBS 803.79 | EU254808 | EU255107 | NA | NA |
| Gnomoniopsis racemula | AR 3892 | EU254841 | EU255122 | EU221889 | EU219241 |
| Mamianiella coryli | BPI 877578 | EU254862 | NA | NA | NA |
| Marsupiomyces quercina | MFLUCC 13-0664 | MF190116 | MF190061 | NA | NA |
| Marsupiomyces epidermoidea | MFLU 15-2921 | NA | MF190058 | NA | NA |
| Melanconis marginalis | CBS 109744 | EU199197 | AF408373 | EU221991 | EU219301 |
| Neognomoniopsis quercina | CBS 145575 | MK876399 | MK876440 | NA | NA |
| Occultocarpon ailaoshanense | LCM 524.01 | JF779849 | JF779853 | NA | JF779856 |
| LCM 522.01 | JF779848 | JF779852 | JF779862 | JF779857 | |
| Ophiognomonia melanostyla | LCM 389.01 | JF779850 | JF779854 | NA | JF779858 |
| Ophiognomonia vasiljevae | AR 4298 | EU254977 | EU255162 | EU221999 | EU219331 |
| Plagiostoma aesculi | AR 3640 | EU254994 | EU255164 | NA | EU219269 |
| Linospora capreae | CBS 372.69 | NA | AF277143 | NA | NA |
| Pleuroceras oregonense | AR 4333 | EU255060 | EU255196 | EU221931 | EU219313 |
| Pleuroceras pleurostylum | CBS 906.79 | EU255061 | EU255197 | EU221962 | EU219311 |
| Phragmoporthe conformis | AR 3632 | NA | AF408377 | NA | NA |
| Valsalnicola oxystoma | AR 5137 | JX519561 | NA | NA | NA |
| AR 4833 | JX519559 | JX519563 | NA | NA | |
| Sirococcus tsugae | AR 4010 | EF512478 | EU255207 | EU221928 | EU219289 |
| CBS 119626 | EU199203 | EU199136 | EF512534 | EU199159 | |
| Synnemasporella aculeans | CFCC 52094 | MG682086 | MG682026 | MG682066 | MG682046 |
| Synnemasporella aculeans | CFCC 53123 | MK432679 | MK429920 | MK578148 | MK578105 |
| CFCC 53124 | MK432680 | MK429921 | MK578149 | MK578106 | |
| CFCC 53125 | MK432681 | MK429922 | MK578150 | MK578107 | |
| CFCC 53126 | MK432682 | MK429923 | MK578151 | MK578108 | |
| CFCC 53127 | MK432683 | MK429924 | MK578152 | MK578109 | |
| Synnemasporella toxicodendri | CFCC 52097 | MG682089 | MG682029 | MG682069 | MG682049 |
| Tenuignomonia styracis | BPI 89278 | NA | LC379288 | LC379282 | LC379294 |
Results
Phylogenetic analyses
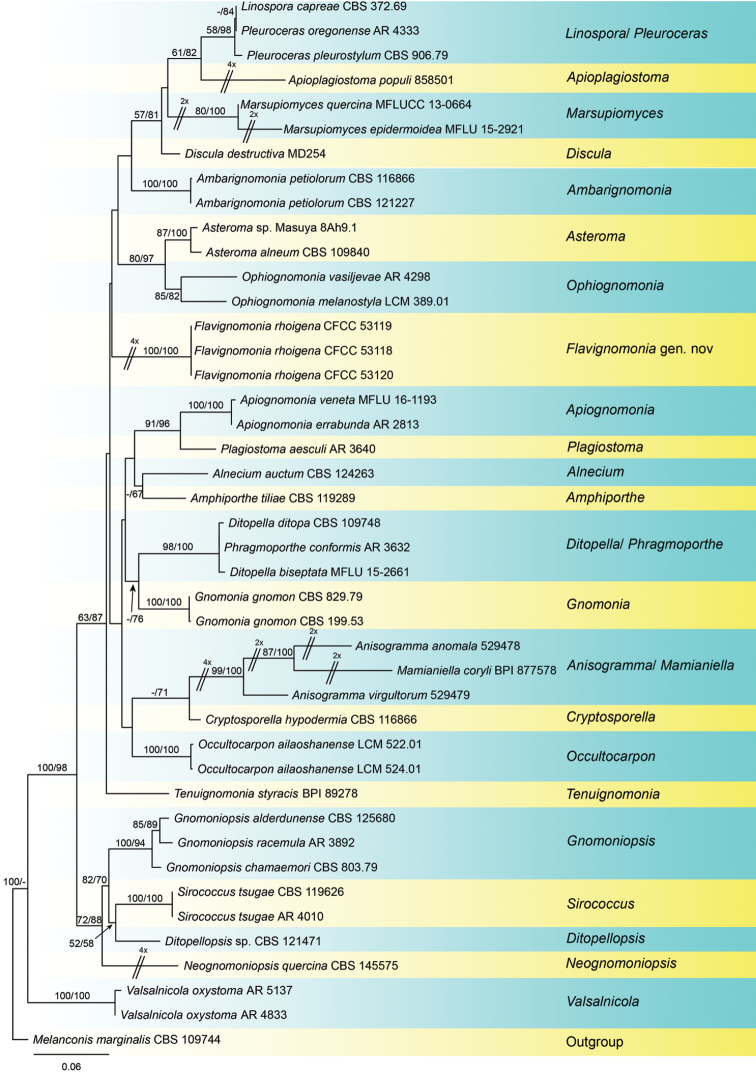
The alignment based on the combined sequence dataset (ITS, LSU, tef1, and rpb2) included 42 in-group taxa and one out-group taxon, comprising 3368 characters in the aligned matrix. Of these, 2201 characters were constant, 224 variable characters were parsimony-uninformative and 943 characters were parsimony informative (282 from the ITS-LSU, 280 from tef1, 381 from rpb2). The MP analysis resulted in nine equally most parsimonious trees with identical tree backbone. The best ML tree (lnL = −20604.0384) was compatible with the MP strict consensus tree, except for unsupported clades in Fig. 1. As the trees obtained from different analytical methods were similar, only the ML tree was present in Fig. 1. The phylogram based on the four gene sequence matrix indicated that the three strains from the present study represent a novel genus in Gnomoniaceae.
Figure 1.
Phylogenetic tree based on an ML analysis of a combined DNA dataset of ITS, LSU, tef1 and rpb2 gene sequences for all genera with DNA data and some species of Gnomoniaceae. Bootstrap values ≥ 50 % for MP and ML analyses are presented at the branches. The scale bar represents the number of changes per site.
Taxonomy
Flavignomonia
C.M. Tian, Q. Yang & N. Jiang gen. nov.
DAD63D30-357D-53BE-8FEA-5EBACB4FB8AA
829530
Diagnosis.
Flavignomonia is distinguished from Synnemasporella by the orange tips of its synnemata.
Type species.
Flavignomonia rhoigena C.M. Tian & Q. Yang
Etymology.
The generic name is derived from the colour of synnemata (flavus = yellow) and the genus name Gnomonia.
Description.
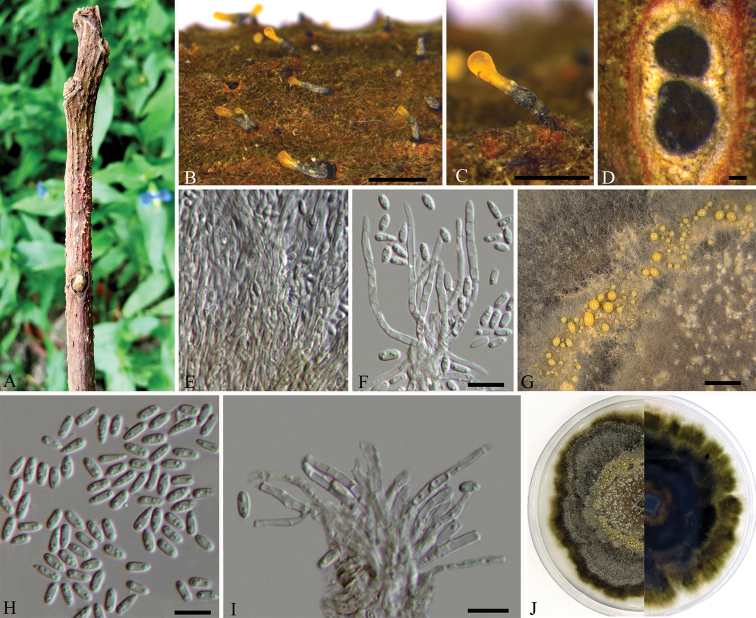
Sexual morph: not observed. Asexual morph: Conidiomata synnematal. Synnemata long and determinate, growing from host tissue, with brown base and orange tip, straight to curved, parallel, with flat to slightly concave and dark zone of conidiogenous cells and host tissue at their bases. Conidiophores reduced to conidiogenous cells. Conidiogenous cells phialidic, aggregated, hyaline, straight to curved, cylindrical, arranged adjacent to one another at the end of the synnema, producing a single conidium. Conidia cylindrical to oblong, smooth, multiguttulate, hyaline.
Notes.
Flavignomonia is included in Gnomoniaceae based on DNA sequences data. Flavignomonia is morphologically similar to Synnemasporella in forming synnemata (Wehmeyer 1933, Fan et al. 2018). However, Flavignomonia, typified with Flavignomonia rhoigena, is distinguished from Synnemasporella species by its orange synnematal tips and hyaline conidia (Fan et al. 2018).
Flavignomonia rhoigena
C.M. Tian & Q. Yang sp. nov.
7F7D0EAD-2C04-5A91-B125-2DF50F71C843
829531
Figure 2.
Flavignomonia rhoigena on Rhus chinensis (BJFC-S1766, holotype) A–C habit of conidiomata on twigs D transverse section through synnema E longitudinal section through synnema F, I conidiogenous cells attached with conidia G conidiomata on PDA H condia J the colony on PDA. Scale bars: 1 mm (B); 500 μm (C); 100 μm (D); 10 μm (F, H–I); 200 μm (G).
Diagnosis.
Flavignomonia rhoigena can be distinguished from other gnomoniaceous species by the formation of synnemata.
Etymology.
Named after the host genus, Rhus.
Description.
Sexual morph: not observed. Asexual morph: Conidiomata synnematal. Synnemata (650–)750–1100 µm high, 150–300 µm diam, determinate, growing from host tissue, with brown base and orange tip, straight to curved, parallel, with flat to slightly concave and dark zone of conidiogenous cells and host tissue at their bases. Conidiophores reduced to conidiogenous cells. Conidiogenous cells (12.5–)16–22(–25) × 2 μm, phialidic, aggregated, hyaline, straight to curved, cylindrical, arranged adjacent to one another at the end of the synnema, producing a single conidium. Conidia cylindrical to oblong, smooth, multiguttulate, hyaline, (5–)5.5–7(–8) × 1.5–2 µm.
Culture characters.
On PDA at 25 °C in darkness, initially white, becoming olive-green to black after 3 wk, zonate with 3–4 well defined zones. Conidiomata distributed concentrically over agar surface.
Specimen examined.
CHINA, Jiangxi Province, Ganzhou City, Xunwu County, 24°52'31.34"N, 115°35'39.53"E, on branches of Rhus chinensis, 14 May 2018, Q. Yang, Y. Liu & Y.M. Liang (holotype BJFC-S1766, ex-type living cultures CFCC 53118, CFCC 53119 and CFCC 53120).
Notes.
Flavignomonia rhoigena is the type species of Flavignomonia in the family Gnomoniaceae. It can be easily distinguished from the other gnomoniaceous genera by its unique conidiomata (Walker et al. 2004, Senanayake et al. 2018, Crous et al. 2019, Minoshima et al. 2019).
Synnemasporella aculeans
(Schwein.) X.L. Fan & J.D.P. Bezerra, Persoonia 40: 130. 2018.
C8503EB3-6F7E-529D-BB7A-97E1F130DB8E
Figure 3.
Asexual morphology of Synnemasporella aculeans on Rhus chinensis (BJFC-S1740) A, B habit of pycnidia on twigs C transverse section of pycnidium D longitudinal section through pycnidium E conidia F, G conidiogenous cells and conidia H, I habit of synnemata on twigs J longitudinal section through synnema K the colony on PDA L, N conidiogenous cells bearing conidia M conidia. Scale bars: 500 μm (B–D, I, J); 10 μm (E–G, L–N).
Figure 4.
Sexual morphology of Synnemasporella aculeans on Rhus chinensis (BJFC-S1745) A, B habit of ascomata on twigs C transverse section of ascomata D longitudinal section through ascomata E, F asci G ascospores. Scale bars: 500 μm (B–D); 10 μm (E–G).
Description.
Sexual morph: See Wehmeyer (1933) and Fan et al. (2018). Asexual morph: See Fan et al. (2018).
Specimens examined.
CHINA, Jiangxi Province, Ganzhou City, Xunwu County, 24°52'31.34"N, 115°35'39.53"E, on branches of Rhus chinensis, 14 May 2018, Q. Yang, Y. Liu & Y.M. Liang (BJFC-S1740, living culture CFCC 53123); Ganzhou City, Fengshan forest park, 25°44'32.14"N, 114°59'25.54"E, on branches of Rhus chinensis, 15 May 2018, Q. Yang, Y. Liu & Y.M. Liang (BJFC-S1753, living culture CFCC 53124 and CFCC 53125). 24°38'38.18"N, 115°33'58.45"E, on branches of Rhus chinensis, 16 May 2018, Q. Yang, Y. Liu & Y.M. Liang (BJFC-S1745, living culture CFCC 53126 and CFCC 53127).
Notes.
Synnemasporella aculeans was proposed as a new combination in the new genus Synnemasporella based on the description of Cryptodiaporthe aculeans (Fan et al. 2018), which was introduced producing perithecial ascomata, and an asexual morph producing sporodochial and/or pycnidial conidiomata (Wehmeyer 1933). In the present study, five isolates from canker tissues on Rhus chinensis were congruent with S. aculeans based on morphology and DNA sequences data. This was the first time that the sexual morph of Synnemasporella aculeans in China had been collected.
Discussion
In this study, two diaporthalean species forming synnemata on Rhus chinensis were identified based on morphology and ITS, LSU, tef1, and rpb2 sequence datasets. As a result, Flavignomonia typified with F. rhoigena is proposed as a new genus in Gnomoniaceae for its distinct phylogenic position and distinctive asexual fruiting body, Also, Synnemasporella aculeans strains were successfully isolated from perithecia, pycnidia and synnemata, which was confirmed by molecular data.
Nineteen fungal species have been recorded from the commercially and ecologically important tree species in China, including Cladosporium cladosporioides, Cronartium quercuum, Mycosphaerella fushinoki, Pestalotiopsis diospyri, P. guepinii, P. mangiferae, P. sorbi, Phaeoramularia rhois, Phyllactinia corylea, Ph. rhoina, Pileolaria klugkistiana, Pi. shiraiana, Pseudocercospora rhoina, Ps. toxicodendri, Septoria sp., Tubercularia phyllophila, Uncinula verniciferae, and two synnematal species from branch cankers in this study (Farr and Rossman 2019). Flavignomonia rhoigena and Synnemasporella aculeans, described and illustrated in the present study can be easily recognized by the asexual fruiting bodies, and they differ from each other in the colour of the synnematal tips.
Gnomoniaceae is a globally distributed fungal family on diverse plant hosts (Mejía et al. 2008, 2011a, 2011b, 2012, Sogonov et al. 2008, Walker et al. 2012, Senanayake et al. 2017, 2018). Host specificity of this family has been confirmed to be important in the evolution (Walker et al. 2014). Our newly discovered genus Flavignomonia was only found on Rhus chinensis, and more Flavignomonia species might be collected from the plant family Anacardiaceae in the future.
Supplementary Material
Acknowledgements
This study is financed by National Natural Science Foundation of China (Project No.: 31670647). We are grateful to Chungen Piao, Minwei Guo (China Forestry Culture Collection Center (CFCC), Chinese Academy of Forestry, Beijing.
Citation
Jiang N, Yang Q, Liang Y-M, Tian C-M (2019) Taxonomy of two synnematal fungal species from Rhus chinensis, with Flavignomonia gen. nov. described. MycoKeys 60: 17–29. https://doi.org/10.3897/mycokeys.60.46395
References
- Alves A, Crous PW, Correia A, Phillips AJL. (2008) Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Diversity 28: 1–13. [Google Scholar]
- Barr ME. (1978) The Diaporthales in North America with emphasis on Gnomonia and its segregates. Mycologia Memoir 7: 1–232. [Google Scholar]
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 3: 553–556. 10.2307/3761358 [DOI] [Google Scholar]
- Crous PW, Carnegie AJ, Wingfield MJ, et al. (2019) Fungal Planet description sheets: 868–950. Persoonia 42: 291–473. 10.3767/persoonia.2019.42.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. (2004) MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Doyle JJ, Doyle JL. (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15. 10.2307/2419362 [DOI] [Google Scholar]
- Fan XL, Bezerra JD, Tian CM, Crous PW. (2018) Families and genera of diaporthalean fungi associated with canker and dieback of tree hosts. Persoonia 40: 119–134. 10.3767/persoonia.2018.40.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr DF, Rossman AY. (2019) Fungal Databases. US National Fungus Collections, ARS, USDA. https://nt.ars-grin.gov/fungaldatabases/ [Retrieved September 1, 2019]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hillis DM, Bull JJ. (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192. 10.1093/sysbio/42.2.182 [DOI] [Google Scholar]
- Jaklitsch WM, Komon M, Kubicek CP, Druzhinina IS. (2005) Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia 97: 1365–1378. 10.1080/15572536.2006.11832743 [DOI] [PubMed] [Google Scholar]
- Jiang N, Fan XL, Tian CM. (2019) Identification and pathogenicity of Cryphonectriaceae species associated with chestnut canker in China. Plant Pathology 68(6): 1132–1145. 10.1111/ppa.13033 [DOI] [Google Scholar]
- Jiang N, Voglmayr H, Tian CM. (2018) New species and records of Coryneum from China. Mycologia 110: 1172–1188. 10.1080/00275514.2018.1516969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. (2010) Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26: 1899–1900. 10.1093/bioinformatics/btq224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejía LC, Castlebury LA, Rossman AY, Sogonov MV, White JF. (2008) Phylogenetic placement and taxonomic review of the genus Cryptosporella and its synonyms Ophiovalsa and Winterella (Gnomoniaceae, Diaporthales). Mycological Research 112(1): 23–35. 10.1016/j.mycres.2007.03.021 [DOI] [PubMed] [Google Scholar]
- Mejía LC, Castlebury LA, Rossman AY, Sogonov MV, White JF. (2011a) A systematic account of the genus Plagiostoma (Gnomoniaceae, Diaporthales) based on morphology, host-associations, and a four-gene phylogeny. Studies in Mycology 68: 211–235. 10.3114/sim.2011.68.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejía LC, Rossman AY, Castlebury LA, White JF. (2011b) New species, phylogeny, host-associations and geographic distribution of genus Cryptosporella (Gnomoniaceae, Diaporthales). Mycologia 103(2): 379–399. 10.3852/10-134 [DOI] [PubMed] [Google Scholar]
- Mejía LC, Rossman AY, Castlebury LA, Yang ZL, White JF. (2012) Occultocarpon, a new monotypic genus of Gnomoniaceae on Alnus nepalensis from China. Fungal Diversity 52(1): 99–105. 10.1007/s13225-011-0108-y [DOI] [Google Scholar]
- Minoshima A, Walker DM, Takemoto S, Hosoya T, Walker AK, Ishikawa S, Hirooka Y. (2019) Pathogenicity and taxonomy of Tenuignomonia styracis gen. et sp. nov., a new monotypic genus of Gnomoniaceae on Styrax obassia in Japan. Mycoscience 60(1): 31–39. 10.1016/j.myc.2018.08.001 [DOI] [Google Scholar]
- Rambaut A. (2016) FigTree, version 1.4.3. University of Edinburgh, Edinburgh.
- Rossman AY, Farr DF, Castlebury LA. (2007) A review of the phylogeny and biology of the Diaporthales. Mycoscience 48: 135–144. 10.1007/S10267-007-0347-7 [DOI] [Google Scholar]
- Senanayake IC, Crous PW, Groenewald JZ, Maharachchikumbura SSN, Jeewon R, Phillips AJL, Bhat JD, Perera RH, Li QR, Li WJ, Tangthirasunun N, Norphanphoun C, Karunarathna SC, Camporesi E, Manawasighe IS, Al-Sadi AM, Hyde KD. (2017) Families of Diaporthales based on morphological and phylogenetic evidence. Studies in Mycology 86: 217–296. 10.1016/j.simyco.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake IC, Jeewon R, Chomnunti P, Wanasinghe DN, Norphanphoun C, Karunarathna A, Pem D, Perera RH, Camporesi E, McKenzie EHC, Hyde KD, Karunarathna SC. (2018) Taxonomic circumscription of Diaporthales based on multigene phylogeny and morphology. Fungal Diversity 93: 241–443. 10.1007/s13225-018-0410-z [DOI] [Google Scholar]
- Sogonov MV, Castlebury LA, Rossman AY, Mejía LC, White JF. (2008) Leaf-inhabiting genera of the Gnomoniaceae, Diaporthales. Studies in Mycology 62: 1–77. 10.3114/sim.2008.62.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. (2003) PAUP*: Phylogenetic Analyses Using Parsimony, * and Other Methods, Version 4.0b10. Sinauer Associates, Sunderland.
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Akulov OY, Jaklitsch WM. (2016) Reassessment of Allantonectria, phylogenetic position of Thyronectroidea, and Thyronectria caraganae sp. nov. Mycological Progress 15: 921–937. 10.1007/s11557-016-1218-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Castlebury LA, Jaklitsch WM. (2017) Juglanconis gen. nov. on Juglandaceae, and the new family Juglanconidaceae (Diaporthales). Persoonia 38: 136–155. 10.3767/003158517X694768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Jaklitsch WM, Mohammadi H, Chakusary MK. (2019) The genus Juglanconis (Diaporthales) on Pterocarya. Mycological Progress 18: 425–437. 10.1007/s11557-018-01464-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang N, Li T, Chen HZ. (2014) Sumac (Rhus chinensis Mill) biomass refinery engineering. Chinese Journal of Biotechnology 30(5): 695–706. 10.13345/j.cjb.140058 [DOI] [PubMed] [Google Scholar]
- Walker DM, Castlebury LA, Rossman AY, Mejía LC, White JF. (2012) Phylogeny and taxonomy of Ophiognomonia (Gnomoniaceae, Diaporthales), including twenty-five new species in this highly diverse genus. Fungal Diversity 57(1): 85–147. 10.1007/s13225-012-0200-y [DOI] [Google Scholar]
- Walker DM, Castlebury LA, Rossman AY, Struwe L. (2014) Host conservatism or host specialization? Patterns of fungal diversification are influenced by host plant specificity in Ophiognomonia (Gnomoniaceae: Diaporthales). Biological Journal of the Linnean Society 111(1): 1–6. 10.1111/bij.12189 [DOI] [Google Scholar]
- Wehmeyer LE. (1933) The genus Diaporthe Nitschke and its segregates. University of Michigan, USA. 10.1016/S0007-1536(33)80010-6 [DOI]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications 18: 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Wijayawardene NN, Hyde KD, Lumbsch HT, Liu JK, Maharachchikumbura SSN, Ekanayaka AH, Tian Q, Phookamsak R. (2018) Outline of Ascomycota: 2017. Fungal Diversity 88: 167–263. 10.1007/s13225-018-0394-8 [DOI] [Google Scholar]
- Winter G. (1886) Fungi Australienses. Revue Mycologique Toulouse 8: 207–213. [Google Scholar]
- Yang Q, Fan XL, Guarnaccia V, Tian CM. (2018) High diversity of Diaporthe species associated with dieback diseases in China, with twelve new species described. Mycokeys 39: 97–149. 10.3897/mycokeys.39.26914 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.