Abstract
Antimicrobial agents are crucial for the treatment of many bacterial diseases in pigs, however, the massive use of critically important antibiotics such as colistin, fluoroquinolones and 3rd–4th-generation cephalosporins often selects for co-resistance. Based on a comprehensive characterization of 35 colistin-resistant Escherichia coli from swine enteric colibacillosis, belonging to prevalent Spanish lineages, the aims of the present study were to investigate the characteristics of E. coli clones successfully spread in swine and to assess the correlation of the in vitro results with in silico predictions from WGS data. The resistome analysis showed six different mcr variants: mcr-1.1; mcr-1.10; mcr-4.1; mcr-4.2; mcr-4.5; and mcr-5.1. Additionally, blaCTX–M–14, blaCTX–M–32 and blaSHV–12 genes were present in seven genomes. PlasmidFinder revealed that mcr-1.1 genes located mainly on IncHI2 and IncX4 types, and mcr-4 on ColE10-like plasmids. Twenty-eight genomes showed a gyrA S83L substitution, and 12 of those 28 harbored double-serine mutations gyrA S83L and parC S80I, correlating with in vitro quinolone-resistances. Notably, 16 of the 35 mcr-bearing genomes showed mutations in the PmrA (S39I) and PmrB (V161G) proteins. The summative presence of mechanisms, associated with high-level of resistance to quinolones/fluoroquinolones and colistin, could be conferring adaptive advantages to prevalent pig E. coli lineages, such as the ST10-A (CH11-24), as presumed for ST131. SerotypeFinder allowed the H-antigen identification of in vitro non-mobile (HNM) isolates, revealing that 15 of the 21 HNM E. coli analyzed were H39. Since the H39 is associated with the most prevalent O antigens worldwide within swine colibacillosis, such as O108 and O157, it would be probably playing a role in porcine colibacillosis to be considered as a valuable subunit antigen in the formulation of a broadly protective Enterotoxigenic E. coli (ETEC) vaccine. Our data show common features with other European countries in relation to a prevalent clonal group (CC10), serotypes (O108:H39, O138:H10, O139:H1, O141:H4), high plasmid content within the isolates and mcr location, which would support global alternatives to the use of antibiotics in pigs. Here, we report for first time a rare finding so far, which is the co-occurrence of double colistin-resistance mechanisms in a significant number of E. coli isolates.
Keywords: Escherichia coli, colistin, mcr, ESBL, fluoroquinolones, ST10, colibacillosis, swine
Introduction
Multidrug-resistant Enterobacteriaceae, such as Escherichia coli, represent a threat to both human and veterinary health. E. coli has a great capacity to accumulate resistance genes, mostly through horizontal gene transfer. The major problematic mechanisms correspond to the acquisition of genes coding for extended-spectrum beta-lactamases (ESBL), carbapenemases, 16S rRNA methylases, plasmid-mediated quinolone resistance (PMQR) and mcr genes conferring resistance to polymyxins (Poirel et al., 2018).
Colistin has been widely used in Spain for the control of neonatal and post-weaning diarrhoea (PWD) in pigs caused by certain E. coli pathotypes: Enterotoxigenic E. coli (ETEC), defined by the presence of genes encoding enterotoxins (eltA, and/or estA, and/or estB); atypical Enteropathogenic E. coli (aEPEC), carriers of eae but negative for bfpA (aEPEC); Shiga toxin–producing E. coli (STEC), positive for stx2e; STEC/ETEC, positive for both shiga toxin type 2e and enterotoxin-encoding genes (stx2e and estB and/or estA) (García-Meniño et al., 2018). PWD results in significant economic losses for the pig industry due to costs derived of treatment and handling, decreased weight gain, and mortality. These circumstances have promoted the use and abuse of antibiotics in intensive farming (Luppi, 2017; Rhouma et al., 2017). However, specific regulations have been set up in Europe due to the concern that colistin resistance could be transmitted from food-production animals to humans which makes necessary the investigation of sustainable alternatives to antimicrobials (EUROPEAN COMMISSION, 2018).
In Spain, the rates of antibiotic resistance in pig farming were recently analyzed in a collection of 499 E. coli isolates from 179 outbreaks of enteric colibacillosis occurred during a period of 10 years (2006–2016) (García et al., 2018; García-Meniño et al., 2018). The results revealed a prevalence of colistin-resistant E. coli implicated in PWD in Spanish farms as high as 76.9% within 186 ETEC, STEC and STEC/ETEC isolates. Besides, PCR and sequencing identified the presence of mcr-4 in 102 isolates, mcr-1 in 37 isolates and mcr-5 in five isolates. Interestingly, almost all mcr-4 isolates belonged to the clonal group ST10-A (CH11-24) (García et al., 2018), which was shown to be highly present (more than 50%) within the mcr-1 diarrheagenic isolates of a second study (García-Meniño et al., 2018). Both studies reinforced other countries’ findings that the pig industry is an important reservoir of colistin-resistant E. coli, as well as being carriers of other additional risk genes such as blaESBL genes (García et al., 2018; García-Meniño et al., 2018; Magistrali et al., 2018; Manageiro et al., 2019). Based on reported evidences (Beyrouthy et al., 2017; Gilrane et al., 2017), there is great concern about the in vivo acquisition of mcr- and blaESBL-bearing plasmids by human E. coli isolates following treatment with colistin, or via animal transmission through direct contact or via food chain. Particular attention is given to those named as high-risk clones of (ESBL)-producing bacteria, worldwide spread within humans and animals, including Escherichia coli sequence types ST10, ST131, ST405, and ST648 (Mathers et al., 2015; Sellera and Lincopan, 2019).
The aims of this study were (i) the characterization of resistances and plasmid profiles of successfully spread mcr-1, mcr-4, and mcr-5 E. coli in Spanish pig farming; (ii) the assessment of WGS-based approaches for the characterization of pathogenic E. coli, through the correlation of the in vitro results with in silico predictions using the bioinformatics tools of the Center for Genomic Epidemiology (CGE).
Materials and Methods
E. coli Collection
Thirty-five swine E. coli, positive by PCR for mcr-genes, were fully sequenced. Specifically, the 35 E. coli were selected from 499 diarrheagenic isolates of different geographic areas of Spain (2006–2016) (García et al., 2018; García-Meniño et al., 2018), taking into account the results of prevalence and significant association observed between pathotypes, presence of mcr and certain serogroups. In brief, the serogroups O108, O138, O141, O149, O157 were found significantly associated with ETEC; serogroups O26, O49, O80, O111 with aEPEC; serogroups O138 and O141 with STEC/ETEC; serogroup O139 with STEC; and serogroups O2, O15, O26, O45, O111, O138, O141, O157 with mcr-positive isolates (García-Meniño et al., 2018). Therefore, the collection analyzed here included 27 ETEC isolates (of serogroups O7, O8, O15, O45, O108, O138, O141, O149, O157, ONT); four STEC (O2, O139); three STEC/ETEC (O138 and O141) and one aEPEC (O111). The 35 representative isolates were carriers of the three mcr-types (mcr-1, mcr-4, and mcr-5) detected so far in our E. coli collection of porcine origin. Conventional pheno- and geno-typing was performed to complete classical characterization of serotypes, phylogroups, pathotypes and resistance profiles.
Conventional Typing
The H antigen was established for motile isolates by serotyping using H1 to H56 antisera, while non-motile isolates (HNM) were analyzed by PCR to determine their flagellar genes as described elsewhere (García-Meniño et al., 2018). The phylogroup was assigned by means of the quadruplex PCR of Clermont et al. (2013). Antimicrobial susceptibility was determined by minimal inhibitory concentrations (MICs) using the MicroScan WalkAway®-automated system (Siemens Healthcare Diagnostics, Berkeley, CA, United States) according to the manufacturer’s instructions for: amikacin, ampicillin-sulbactam, aztreonam, cefepime, ceftazidime, ciprofloxacin, colistin, fosfomycin, gentamicin, imipenem, levofloxacin, meropenem, minocycline, nitrofurantoin, piperacillin-tazobactam, ticarcillin, tigecycline, and tobramycin. Additionally, resistance to ampicillin, amoxicillin/clavulanate, cefazolin, cefotaxime, cefoxitin, cefuroxime, chloramphenicol, doxycycline, nalidixic acid and trimethoprim-sulfamethoxazole was determined by disk (Becton Dickinson, Sparks, MD, United States) diffusion assays. All results were interpreted according to the CLSI break points (Clinical and Laboratory Standards Institute, 2019). Genetic identification of the ESBLs was performed by PCR using the TEM, SHV, CTX-M-1 and CTX-M-9 group-specific primers followed by amplicon sequencing (García-Meniño et al., 2018).
Whole Genome Sequencing (WGS) and Sequence Analysis
The libraries for sequencing were prepared following the instructions provided by the TruSeq Illumina PCR-Free protocol. Mechanical DNA fragmentation was performed with Covaris E220, and the final quality of the libraries assessed with Fragment Analyzer (Std. Sens. NGS Fragment Analysis kit 1-6000 bp). Lastly, the libraries were sequenced in an Illumina HiSeq1500, obtaining 100–150 bp paired-end reads which were trimmed (Trim Galore 0.5.0) and filtered according to quality criteria (FastQC 0.11.7). The reconstruction of the genomes and plasmids in the genomes was carried out using the methodology PLAsmid Constellation NETwork (PLACNETw)1 (Lanza et al., 2014). The assembled contigs, with genomic size ranging between 4.9 and 5.9 Mbp (mean size 5.5 Mbp), were analyzed using the bioinformatics tools of the Center for Genomic Epidemiology (CGE)2 for the presence of antibiotic resistance (ResFinder V2.1.), virulence genes (VirulenceFinder v1.5.), plasmid replicon types (PlasmidFinder 1.3./PMLST 1.4.), and identification of clonotypes (CHTyper 1.0), sequence types (MLST 2.0) and serotypes (SerotypeFinder 2.0). All the CGE predictions were called applying a select threshold for identification and a minimum length of 95 and 80%, respectively. Phylogroups were predicted using the ClermonTyping tool at the iame-research center web3. The mcr gene location was determined using PlasmidFinder/ResFinder prediction, together with PLACNETw references, and automatic annotation with Prokka v1.13 (Seemann, 2014).
Results and Discussion
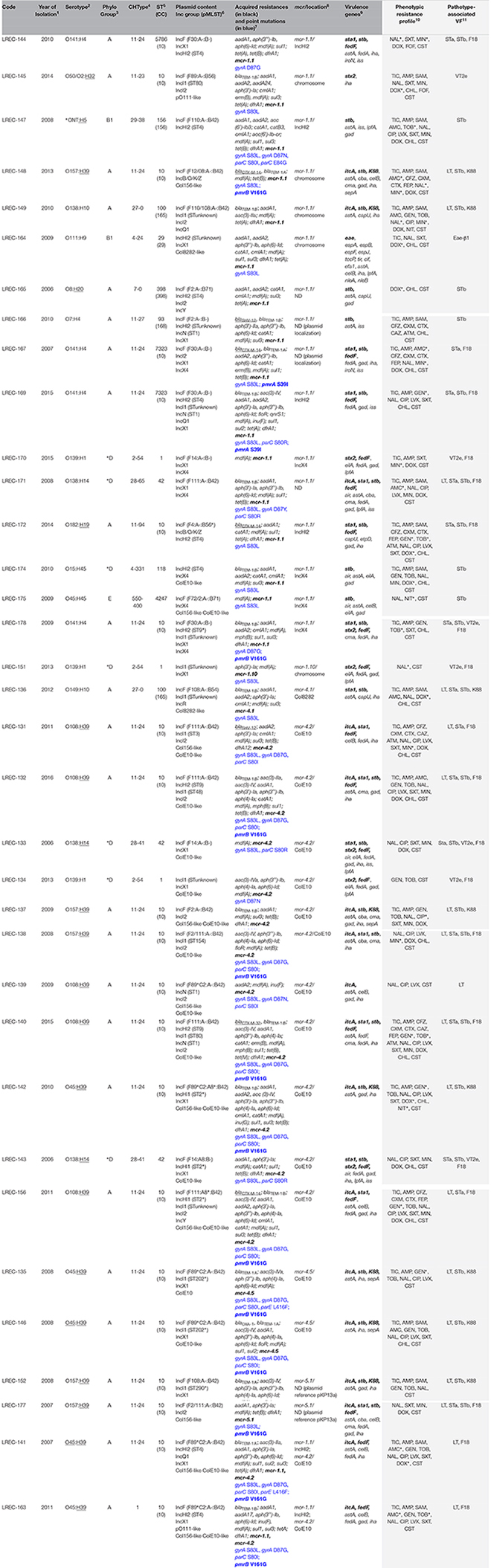
The phenotypic and genotypic traits of the 35 mcr-positive E. coli of swine origin, as well as their resistome and mobilome are summarized in Table 1. ResFinder confirmed that all genomes were mcr carriers. Likewise, VirulenceFinder predicted the acquired virulence genes encoding for the enterotoxins (sta1, stb, itcA), for fimbriae (fedF, k88), verotoxin (stx2) and intimin (eae), correlating in all cases with the pathotype assignation previously determined by PCR (García et al., 2018; García-Meniño et al., 2018).
TABLE 1.
Features of the 35 colistin-resistant E. coli genomes of swine origin based on in silico characterization (light columns) and on conventional typing (gray columns).
 |
Serotype Identification
In most studies, there is lack of information on E. coli serotypes since serotyping is performed by very few laboratories worldwide, hindering epidemiological comparisons. Here, we not only proved that there is a very good correlation between serotyping and SerotypeFinder predictions, but also the advantage of in silico H-antigen identification for those non-mobile (HNM) isolates. It is of note that 15 of the 21 HNM isolates were predicted as H39 (Table 1), namely O108:H39, O157:H39 and O45:H39 (five genomes, each). Given that the H39 is associated with the most prevalent O antigens within swine colibacillosis, such as O108 and O157, as well as ONT (García-Meniño et al., 2018), it would be probably playing a role in porcine colibacillosis to be considered as a valuable subunit antigen in the formulation of a broadly protective ETEC vaccine (Roy et al., 2009). The remaining six HNM isolates showed different O:H combinations: O138:H14, ONT:H5, O8:H20, O50/O2:H32, and O182:H19. SerotypeFinder also allowed the O45-antigen determination of two non-typeable (ONT) isolates (LREC-141 and LREC-146) and O182 of LREC-172; while LREC-147, belonging to O157 serogroup (Table 1), was predicted as ONT, probably due to the limitation of the assembly based on Illumina short reads (100–150 bp paired-end reads here) (Wick et al., 2017).
Phylogroups, Sequence Types and Clonotypes
The phylogroups established for the 35 genomes were the common ones reported for porcine E. coli isolates (A, B1, D-E) (Shepard et al., 2012; Bosak et al., 2019). However, we found discrepancies in the assignation obtained with the quadruplex PCR of Clermont et al. (2013) in comparison with that predicted by ClermonTyping for seven isolates: phylogroup E by PCR, while phylogroup D in silico (Table 1).
MLST and CHTyper tools determined 12 different STs, but mostly belonging to CC10 (21 genomes) and clonotype CH11-24 (18 genomes) (Table 1). The predominance of CC10, and specifically ST10, is in accordance with published data on E. coli isolates of swine origin, independently of the pathogenicity or antibiotic-resistance/susceptibility status (Shepard et al., 2012; Kidsley et al., 2018; Magistrali et al., 2018).
Resistome, Plasmidome and Phenotypic Expression of Resistances
The resistome analysis revealed that 34 of the 35 genomes encoded mechanisms of antibiotic resistance for ≥three different antimicrobial categories (Table 1). Seven E. coli were carriers of blaESBL, namely blaCTX–M–14 (four genomes), blaCTX–M–32 (one) and blaSHV–12 (two). Besides, six different mcr variants were identified within the 35 E. coli: mcr-1.1 (in 18 genomes, including two mcr-4.2 carriers); mcr-1.10 (one); mcr-4.1 (one); mcr-4.2 (13 genomes, including the two mcr-1.1 carriers); mcr-4.5 (two) and mcr-5.1 (two).
PlasmidFinder revealed a high plasmid diversity based on the identified replicons, with two to seven different plasmid types per genome (Table 1). Within this heterogeneity, mcr-1.1 genes were found mainly on plasmids of the IncHI2 and IncX4 types (six and four of the 12 mcr-1.1 plasmid-located genes, respectively); however, mcr-1.1 was also found integrated in the chromosome of LREC-145, LREC-148, LREC-149 and LREC-164 genomes. The mcr-1.10 gene of LREC-151 was located on the chromosome, while mcr-4 and mcr-5 variants were on Col8282-like (mcr-4.1), ColE10-like (for all 13 mcr-4.2 and two mcr-4.5 carriers) and pKP13a-like (mcr-5.1) plasmids. Furthermore, we found that there was no mcr plasmid co-occurrence in LREC-141 and LREC-163, but rather the mcr-1.1 and mcr-4.2 genes were located in independent plasmids (IncHI2 and ColE10-like types, respectively). The mcr location remained undetermined for four isolates.
Since the mcr-1 plasmid gene was first described (Liu et al., 2016), it has been identified in members of the Enterobacteriaceae family encoded in different plasmid types, including IncI2, IncX4, IncHI1, IncHI2, IncFI, IncFII, IncP, IncK (Sun et al., 2018). Different authors corroborate that large conjugative plasmids of types IncHI2, IncX4 and IncI2 would be the maximum responsible for the dissemination of the mcr-1 gene among E. coli isolates from different sources and geographical locations (Doumith et al., 2016; Li et al., 2017; Manageiro et al., 2019). To date, other mcr genes (2–9) have been described (Carroll et al., 2019); among them, the mcr-4 and mcr-5 genes appear mostly encoded in small and non-conjugative ColE-like type plasmids (Sun et al., 2018). Here we found similar results, since mcr-1.1 genes were located mainly on IncHI2 and IncX4 types, and mcr-4 on ColE10-like plasmids. It is of note that the mcr-5.1 gene, predicted in LREC-152 and LREC-177, was linked to a Kp13-like plasmid (CP003996.1), location previously described by Hammerl et al. (2018) for one mcr-5 isolate recovered from a fecal pig sample at farm. Chromosomally-encoded mcr-1 location remains rare, however, it was described soon after the discovery of this plasmid-borne gene (Falgenhauer et al., 2016; Veldman et al., 2016). Here, we determined chromosomal location in five genomes by means of PLACNETw, and according to the predictive annotation of the mcr-contigs, the only common element flanking the mcr-1 was a putative ORF, pap2, which is part of the Tn6360 and encodes a Pap2 superfamily protein. Thus, Pap2 was detected in LREC-145, LREC-148, LREC-149, and LREC-164, while the ISApI1 element typically associated with the initial mobilization of mcr-1, was missing within the five contigs (Snesrud et al., 2018).
Overall, our findings are in accordance with those reported by Magistrali et al. (2018) on 13 mcr-positive E. coli isolated from swine colibacillosis in Belgium, Italy and Spain. Both studies show common features in relation to a prevalent clonal group (CC10), serotypes (O108:H39, O138:H10, O139:H1, O141:H4), and mcr-plasmid types. The confirmation of these similarities are of interest for the global design of alternatives to antibiotics that would curb the dissemination of specific clones in the pig farming.
The in vitro analysis of resistances showed that 30 of the 35 E. coli were multidrug-resistant (MDR) according to Magiorakos et al. (2012) definition (Table 1). Phenotypic results corresponded broadly to those predicted by ResFinder (Supplementary Table S1) as detailed below.
The quinolones/fluoroquinolones (FQ), together with polymyxins and 3rd–4th-generation cephalosporins, all are included in Category B of restricted antimicrobials in the EMA categorization, considering that the risk to public health resulting from its veterinary use needs to be mitigated by specific restriction (EMA/CVMP/CHMP, 2019). Two major mechanisms are implicated in the resistance to FQ, namely, mutations in the genes for DNA gyrase and topoisomerase IV, and decreased intracellular drug accumulation. In addition, plasmid-mediated quinolone resistances also play a role but usually conferring low-level FQ resistance (van Duijkeren et al., 2018). Phenotypically, 17 of the 35 isolates showed resistance to both nalidixic acid and ciprofloxacin, and other eight resistance to nalidixic acid only (Supplementary Table S1). In the majority of cases, phenotypic results correlated with those predicted by ResFinder. Particularly, 28 of the 35 genomes carried the gyrA S83L substitution, with 12 of those 28 showing double-serine mutations (gyrA S83L and parC S80I). An additional substitution (gyrA D87N) was detected in two of the 12 gyrA S83L/parC S80I genomes. Thus, nalidixic acid resistance in vitro corresponded to one single substitution (gyrA S83L), and FQ resistance to double or triple substitutions (gyrA S83L/parC S80I/gyrA D87N). Plasmid-mediated quinolone resistant genes acc(6′)-Ib-cr and qnrS1 were also present together with chromosomal mutations in LREC-147 and LREC-169, respectively. Double-serine mutations in specific positions of the gyrA and parC genes have been reported as a dominant feature of MDR lineages within E. coli, S. aureus and K. pneumoniae, with favorable fitness balance linked to high levels of resistance to FQ (Fuzi et al., 2017). This finding, in 12 out of the 28 in silico predicted FQ-resistant could be conferring adaptive advantages to certain widely spread pig pathogenic clonal groups of E. coli, such as the ST10-A (CH11-24) (García et al., 2018). This hypothesis is presently assumed for ST131 and other risk clones linked to high FQ-resistance (Johnson et al., 2015; Fuzi et al., 2017).
On the other hand, colistin has been widely used for the control of enteric diseases, mainly in swine and poultry (Rhouma et al., 2016; Hammerl et al., 2018). Several mechanisms of resistance due to chromosomal mutations or acquired resistance genes have been described so far (Olaitan et al., 2014; Poirel et al., 2018). The 35 colistin-resistant E. coli of this study showed MIC values > 4 mg/L. As detailed above, ResFinder confirmed that all the analyzed genomes were mcr-carriers. In addition to the plasmid mechanism (mcr) of resistance, polymyxin resistance in E. coli can be due to genes encoding LPS-modifying enzymes, particularly to mutations in the two-component systems PmrAB and PhoPQ, or in the MgrB regulator. Quesada et al. (2015) detected two colistin-resistant E. coli recovered in 2011 and 2013 from the stools of two pigs, which showed mutations in PmrB V161G and PmrA S39I, reporting the finding as a rare event. Subsequently, Delannoy et al. (2017) analyzed 90 strains of E. coli isolated from diseased pigs: 81 were phenotypically resistant to colistin and 72 mcr-1 carriers (including two colistin-susceptible). Although different mutations were found in the amino acid sequences of the MgrB, PhoP, PhoQ, and PmrB proteins of eight isolates, only two of them were mcr-1 positive (but colistin-susceptible). Surprisingly, we found here the double mechanism of colistin resistance in 16 E. coli, harboring mcr-genes together with one amino acid substitution: PmrB V161G (14 genomes) or PmrA S39I (two genomes). In a recent study on Parisian inpatient fecal E. coli (Bourrel et al., 2019), the authors found 12.5% of colistin-resistant E. coli carriers among 1,217 patients; however, mcr-1 gene was identified in only seven of 153 isolates, while 72.6% harbored mutations in the PmrA and PmrB proteins. According to the authors, their findings indicate two evolutionary paths leading to colistin resistance in human fecal E. coli, one corresponding to a minority of plasmid-encoded mcr-1 isolates of animal origin, and a second corresponding to a vast majority of human isolates exhibiting chromosomally encoded mechanisms (Bourrel et al., 2019). Thus, and given the limited data regarding the co-occurrence of double resistance mechanism, it is of note that 16 of the 35 mcr-bearing genomes of our study showed mutations in the PmrA and PmrB proteins. Furthermore, two E. coli (LREC-141 and LREC-163) shown to be carriers of two different mcr-bearing plasmids together with PmrB V161G mutation. An explanation for this rare finding is that these isolates would be reflecting a cumulative evolution to antibiotic pressure and, as a consequence, enhancing the transmission (vertical and horizontal) of colistin resistance. In any case, further investigation is needed to evaluate the implication of chromosomal mutations and mcr co-occurrence regarding colistin resistance phenotype.
In this study, 22 out of the 25 isolates showing phenotypic resistance to beta-lactams (Supplementary Table S1), were positive in the analysis in silico for the presence of blaTEM–1 genes, alone (14 genomes), or in combination with other bla genes (blaCTX–M–14, blaSHV–12, blaCTX–M–32 and blaOXA–1); additionally, two genomes showed the presence of blaCTX–M–14 and blaSHV–12, respectively. With the exception of LREC-147, LREC-164, and LREC-170, which were phenotypically resistant to narrow-spectrum beta-lactamases but negative for the presence of genes, a good correlation was observed between genes predicted and resistance shown in vitro. It is of note that blaTEM–135, determined in LREC-156 by conventional typing, was not identified in silico. Beta-lactams are the most widely used family in current clinical practice. Numerous genes in E. coli confer resistance to this group, being some of them, such as blaTEM–1 widespread in E. coli from animals coding for narrow-spectrum beta-lactamases that can inactivate penicillins and aminopenicillins. However, genes encoding ESBLs/AmpCs have increasingly emerged in E. coli from humans and animals, including food-producing animals (Cortes et al., 2010).
Thirty out of the 35 genomes showed high frequency of resistance genes to aminoglycosides, specifically encoding AAC(3)-II/IV and AAC(6)-Ib, which are the most frequently encountered acetyltransferases among E. coli of human and animal origins. The subclass AAC(3)-II, which is characterized by resistance to gentamicin, netilmicin, tobramycin, sisomicin, 2′-N-ethylnetilmicin, 6′-N-ethylnetilmicin and dibekacin (Shaw et al., 1993), and AAC(6′) enzymes that specify resistance to several aminoglycosides and differ in their activity against amikacin and gentamicin C1 (Ramirez and Tolmasky, 2010) seemed to correlate with the phenotypic detection of resistance to gentamicin and/or tobramycin (12 of the 17 resistant isolates) (Supplementary Table S1). We also detected high prevalence of genes encoding nucleotidyltransferases (aadA), which specify resistance to spectinomycin and streptomycin, alone or together with phosphotransferases (APHs) (Ramirez and Tolmasky, 2010), but they were not tested in the phenotypic antimicrobial susceptibility tests.
It is noteworthy that the 35 genomes of our study were carriers of mdf(A). Edgar and Bibi (1997) described that cells expressing MdfA from a multicopy plasmid are substantially more resistant to a diverse group of cationic or zwitterionic lipophilic compounds. Besides, the authors found that MdfA also confers resistance to chemically unrelated, clinically important antibiotics such as chloramphenicol, erythromycin, and certain aminoglycosides and fluoroquinolones. This capability could correlate with the in vitro resistance observed for some isolates to tetracyclines and aminoglycosides, in absence of other specific genes. In our collection, of the 24 isolates showing phenotypic resistance to minocycline and, or doxycycline (Supplementary Table S1), 20 showed carriage of tet genes: 12 tet(B), six tet(A), one tet(A) + tet(B) and one tet(B) + tet(M). However, two tet(A) isolates were susceptible to those antibiotics (LREC-163, LREC-169). Additionally, tet genes were not detected in silico in four phenotypically resistant isolates. In general, tet(A) and tet(B) are the most prevalent tetracycline resistance genes in E. coli of animal origin, and specifically in isolates from pigs (Tang et al., 2011; Holzel et al., 2012; Jurado-Rabadan et al., 2014).
Although the use of chloramphenicol was banned in the European Union in food-producing animals in 1994, fluorinated derivative florfenicol is allowed for the treatment of bacterial infection in these animals (Schwarz et al., 2004; OIE, 2019). In the present study, all 19 chloramphenicol-resistant isolates (Supplementary Table S1) correlated with the presence of genes catA1 (12 genomes), catB3 (one genomes), cmlA (ten genomes) or floR (three genomes) detected in silico. Travis et al. (2006) showed that chloramphenicol resistant genes are frequently linked to other antibioresistance genes. Thus, through transformation experiments conducted with E. coli from pigs demonstrated that aadA and sul1 were located with catA1 on a large ETEC plasmid, and plasmids carrying cmlA also carried sul3 and aadA. According to the authors, this linkage might partly explain the long-term persistence of chloramphenicol resistance in ETEC despite its withdrawal years ago. In our study, ResFinder also showed an association of genes cmlA, sul3 and aadA present in the same contig (7 of the 10 genomes positive for cmlA), and cmlA/aadA in all cases. Additionally, aadA and sul1 were located with floR in LREC-146.
In E. coli from food-producing animals, sulfonamide resistance is mediated by sul genes (sul1, sul2, sul3), widely disseminated, and frequently found together with other antimicrobial resistance genes, while dfr genes confer trimethoprim resistance in E. coli and other gram-negative bacteria (van Duijkeren et al., 2018). Within our collection, 20 of the 35 isolates were in vitro resistant to trimethoprim/sulfamethoxazole (Supplementary Table S1), and most of them correlated with the presence of sul + dfrA genes in their genomes, with the exception of LREC-133 and LREC-170 (negative for the in silico detection of sul, dfrA genes) and LREC-146 (in which only sul1 and sul2 genes were predicted). Besides, ResFinder showed that sul1 (present in 16 genomes), sul3 (14 genomes) and sul2 (three genomes) were located together with dfrA, and other resistance genes, as mentioned previously.
The fosfomycin resistance showed in vitro by two isolates of the study collection, was not predicted for LREC-144 and LREC-145 (Supplementary Table S1) by ResFinder, which analyzes the presence of fos genes encoding for fosfomycin-modifying enzymes. The use of this antibiotic has been limited to the treatment of infections by Gram-positive and negative pathogens, included E. coli, mainly in pig and poultry farming (Poirel et al., 2018). However, phosphonic acid derivates such as fosfomycin, have been recently categorized by the EMA (EMA/CVMP/CHMP, 2019) as Category A (antimicrobial classes not currently authorized in veterinary medicine in EU).
Conclusion
Swine colibacillosis control has been traditionally managed through the extensive use of antibiotics. Our results are a reflection of the situation within the industrial pig farming, where global hygiene procedures and vaccinations are essential for improvement in antimicrobial stewardship. The summative presence of antibioresistances could be conferring adaptive advantages to prevalent pig E. coli lineages, such as the ST10-A (CH11-24). Based on the different replicons identified by PlasmidFinder (up to seven), it is of note the high plasmid diversity found within these isolates; further research is needed to know mechanisms of maintenance and advantages conferred to them.
Here, we report for first time a rare finding so far, which is the co-occurrence of double colistin-resistance mechanisms (mcr-genes and chromosomal mutations in the PmrA and PmrB proteins) in a significant number of E. coli isolates. This fact could be increasing the risk of colistin resistance-acquisition by means of food transmission. Globally, we found a very good correlation between resistances determined in vitro and genes predicted using CGE tools, and the same observation applies to the E. coli pathotype determination.
Data Availability Statement
The nucleotide sequence of the 35 LREC genomes have been deposited in the NCBI sequence databases with accession codes SAMN11523829 to SAMN11523863. These sequences are part of BioProject ID PRJNA540146.
Author Contributions
IG-M, DD-J, and SF-S undertook the laboratory work. AM and JB conceived and designed the study. IG-M, DD-J, VG, MT, and AM performed the data analysis. IG-M, DD-J, VG, MT, JB, and AM drafted the manuscript. All authors provided critical input and approved the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported by projects PI16/01477 from Plan Estatal de I+D+I 2013–2016, Instituto de Salud Carlos III (ISCIII), Subdirección General de Evaluación y Fomento de la Investigación, and FEDER; AGL2016-79343-R from the Agencia Estatal de Investigación (AEI, Spain) and FEDER; ED431C 2017/57 from the Consellería de Cultura, Educación e Ordenación Universitaria (Xunta de Galicia) and FEDER. IG-M and VG acknowledge the Consellería de Cultura, Educación e Ordenación Universitaria, Xunta de Galicia for their pre-doctoral and post-doctoral grants (Grant Numbers ED481A-2015/149 and ED481B-2018/018, respectively). SF-S acknowledges the FPU programme from the Secretaría General de Universidades, Ministerio de Educación, Cultura y Deporte, Gobierno de España (Grant Number FPU15/02644).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02469/full#supplementary-material
References
- Beyrouthy R., Robin F., Lessene A., Lacombat I., Dortet L., Naas T., et al. (2017). MCR-1 and OXA-48 in vivo acquisition in KPC-producing Escherichia coli after colistin treatment. Antimicrob. Agents Chemother. 61:e2540-16. 10.1128/AAC.02540-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosak J., Hrala M., Pirkova V., Micenkova L., Cizek A., Smola J., et al. (2019). Porcine pathogenic Escherichia coli strains differ from human fecal strains in occurrence of bacteriocin types. Vet. Microbiol. 232 121–127. 10.1016/j.vetmic.2019.04.003 [DOI] [PubMed] [Google Scholar]
- Bourrel A. S., Poirel L., Royer G., Darty M., Vuillemin X., Kieffer N., et al. (2019). Colistin resistance in parisian inpatient faecal Escherichia coli as the result of two distinct evolutionary pathways. J. Antimicrob. Chemother. 74 1521–1530. 10.1093/jac/dkz090 [DOI] [PubMed] [Google Scholar]
- Carroll L. M., Gaballa A., Guldimann C., Sullivan G., Henderson L. O., Wiedmann M. (2019). Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium Isolate. MBio 10:e853-19. 10.1128/mBio.00853-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont O., Christenson J. K., Denamur E., Gordon D. M. (2013). The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5 58–65. 10.1111/1758-2229.12019 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute, (2019). Performance Standars for Antimicrobial Susceptibility Testing, 29th Edn Wayne, PA: CLSI. [Google Scholar]
- Cortes P., Blanc V., Mora A., Dahbi G., Blanco J. E., Blanco M., et al. (2010). Isolation and characterization of potentially pathogenic antimicrobial-resistant Escherichia coli strains from chicken and pig farms in Spain. Appl. Environ. Microbiol. 76 2799–2805. 10.1128/AEM.02421-2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delannoy S., Le Devendec L., Jouy E., Fach P., Drider D., Kempf I. (2017). Characterization of colistin-resistant Escherichia coli isolated from diseased pigs in France. Front. Microbiol. 8:2278. 10.3389/fmicb.2017.02278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumith M., Godbole G., Ashton P., Larkin L., Dallman T., Day M., et al. (2016). Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J. Antimicrob. Chemother. 71 2300–2305. 10.1093/jac/dkw093 [DOI] [PubMed] [Google Scholar]
- Edgar R., Bibi E. (1997). MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J. Bacteriol. 179 2274–2280. 10.1128/jb.179.7.2274-2280.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMA/CVMP/CHMP (2019). Answer to the Request From the European Commission for Updating the Scientific Advice on the Impact on Public Health and Animal Health of the Use of Antibiotics in Animals - Categorisation Of Antimicrobials Ema/Cvmp/Chmp/682198/2017. London: European Medicines Agency. [Google Scholar]
- EUROPEAN COMMISSION (2018). Overview Report on Measures to Tackle Antimicrobial Resistance (Amr) Through the Prudent Use of Antimicrobials in Animals. Brussels: European Commission. [Google Scholar]
- Falgenhauer L., Waezsada S.-E., Gwozdzinski K., Ghosh H., Doijad S., Bunk B., et al. (2016). Chromosomal locations of mcr-1 and blaCTX−M−15 in fluoroquinolone resistant Escherichia coli ST410. Emerg. Infect. Dis. 22 1689–1691. 10.3201/eid2209.160692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuzi M., Szabo D., Csercsik R. (2017). Double-serine fluoroquinolone resistance mutations advance major international clones and lineages of various multi-drug resistant bacteria. Front. Microbiol. 8:2261. 10.3389/fmicb.2017.02261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García V., García-Meniño I., Mora A., Flament-Simon S. C., Díaz-Jiménez D., Blanco J. E., et al. (2018). Co-occurrence of mcr-1, mcr-4 and mcr-5 genes in multidrug-resistant ST10 enterotoxigenic and shiga toxin-producing Escherichia coli in Spain (2006-2017). Int. J. Antimicrob. Agents. 52 104–108. 10.1016/j.ijantimicag.2018.03.022 [DOI] [PubMed] [Google Scholar]
- García-Meniño I., García V., Mora A., Díaz-Jiménez D., Flament-Simon S. C., Alonso M. P., et al. (2018). Swine enteric colibacillosis in Spain: pathogenic potential of mcr-1 ST10 and ST131 E. coli isolates. Front. Microbiol. 9:2659. 10.3389/fmicb.2018.02659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilrane V. L., Lobo S., Huang W., Zhuge J., Yin C., Chen D., et al. (2017). Complete genome sequence of a colistin-resistant Escherichia coli strain harboring mcr-1 on an IncHI2 plasmid in the United States. Genome Announc. 5:e1095-17. 10.1128/genomeA.01095-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerl J. A., Borowiak M., Schmoger S., Shamoun D., Grobbel M., Malorny B., et al. (2018). mcr-5 and a novel mcr-5.2 variant in Escherichia coli isolates from food and food-producing animals, Germany, 2010 to 2017. J. Antimicrob. Chemother. 73 1433–1435. 10.1093/jac/dky020 [DOI] [PubMed] [Google Scholar]
- Holzel C. S., Harms K. S., Bauer J., Bauer-Unkauf I., Hormansdorfer S., Kampf P., et al. (2012). Diversity of antimicrobial resistance genes and class-1-integrons in phylogenetically related porcine and human Escherichia coli. Vet. Microbiol. 160 403–412. 10.1016/j.vetmic.2012.06.010 [DOI] [PubMed] [Google Scholar]
- Johnson J. R., Johnston B., Kuskowski M. A., Sokurenko E. V., Tchesnokova V. (2015). Intensity and mechanisms of fluoroquinolone resistance within the H30 and H30Rx subclones of Escherichia coli sequence type 131 compared with other fluoroquinolone-resistant E. coli. Antimicrob. Agents. Chemother. 59 4471–4480. 10.1128/AAC.00673-615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado-Rabadan S., de la Fuente R., Ruiz-Santa-Quiteria J. A., Orden J. A., de Vries L. E., Agerso Y. (2014). Detection and linkage to mobile genetic elements of tetracycline resistance gene tet(M) in Escherichia coli isolates from pigs. BMC Vet. Res. 10:155. 10.1186/1746-6148-10-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidsley A. K., Abraham S., Bell J. M., ÓDea M., Laird T. J., Jordan D., et al. (2018). Antimicrobial susceptibility of Escherichia coli and Salmonella spp. isolates from healthy pigs in Australia: results of a pilot national survey. Front. Microbiol. 9:1207. 10.3389/fmicb.2018.01207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza V. F., de Toro M., Pilar Garcillan-Barcia M., Mora A., Blanco J., Coque T. M., et al. (2014). Plasmid flux in Escherichia coli ST131 sublineages, analyzed by plasmid constellation network (PLACNET), a new method for plasmid reconstruction from whole genome sequences. PLoS Genet. 10:e1004766. 10.1371/journal.pgen.1004766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Xie M., Zhang J., Yang Z., Liu L., Liu X., et al. (2017). Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob. Chemother. 72 393–401. 10.1093/jac/dkw411 [DOI] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet. Infect. Dis. 16 161–168. 10.1016/S1473-3099(15)00424-427 [DOI] [PubMed] [Google Scholar]
- Luppi A. (2017). Swine enteric colibacillosis: diagnosis, therapy and antimicrobial resistance. Porcine Health Manag. 3:16 10.1186/s40813-017-0063-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos A. P., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- Magistrali C. F., Curcio L., Luppi A., Pezzotti G., Orsini S., Tofani S., et al. (2018). Mobile colistin resistance genes in Escherichia coli from pigs affected by colibacillosis. Int. J. Antimicrob. Agents. 52 744–746. 10.1016/j.ijantimicag.2018.08.008 [DOI] [PubMed] [Google Scholar]
- Manageiro V., Clemente L., Romao R., Silva C., Vieira L., Ferreira E., et al. (2019). IncX4 plasmid carrying the new mcr-1.9 gene variant in a CTX-M-8-producing Escherichia coli isolate recovered from swine. Front. Microbiol. 10:367. 10.3389/fmicb.2019.00367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers A. J., Peirano G., Pitout J. D. (2015). The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 28 565–591. 10.1128/CMR.00116-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE (2019). List of Antimicrobial Agents of Veterinary Importance. Available at: https://www.oie.int/fileadmin/Home/eng/Our_scientific_expertise/docs/pdf/AMR/A_OIE_List_antimicrobials_July2019.pdf (accessed October 23, 2019). [Google Scholar]
- Olaitan A. O., Morand S., Rolain J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. 10.3389/fmicb.2014.00643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Madec J. Y., Lupo A., Schink A. K., Kieffer N., Nordmann P., et al. (2018). Antimicrobial resistance in Escherichia coli. Microbiol. Spectr. 6:ARBA-0026-2017. 10.1128/microbiolspec.ARBA-0026-2017 [DOI] [PubMed] [Google Scholar]
- Quesada A., Porrero M. C., Tellez S., Palomo G., Garcia M., Dominguez L. (2015). Polymorphism of genes encoding PmrAB in colistin-resistant strains of Escherichia coli and Salmonella enterica isolated from poultry and swine. J. Antimicrob. Chemother. 70 71–74. 10.1093/jac/dku320 [DOI] [PubMed] [Google Scholar]
- Ramirez M. S., Tolmasky M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Updat. 13 151–171. 10.1016/j.drup.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhouma M., Beaudry F., Theriault W., Letellier A. (2016). Colistin in pig production: chemistry, mechanism of antibacterial action, microbial resistance emergence, and one health perspectives. Front. Microbiol. 7:1789. 10.3389/fmicb.2016.01789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhouma M., Fairbrother J. M., Beaudry F., Letellier A. (2017). Post weaning diarrhea in pigs: risk factors and non-colistin-based control strategies. Acta Vet. Scand. 59:31. 10.1186/s13028-017-0299-297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K., Hamilton D., Ostmann M. M., Fleckenstein J. M. (2009). Vaccination with EtpA glycoprotein or flagellin protects against colonization with enterotoxigenic Escherichia coli in a murine model. Vaccine 27 4601–4608. 10.1016/j.vaccine.2009.05.076 [DOI] [PubMed] [Google Scholar]
- Schwarz S., Kehrenberg C., Doublet B., Cloeckaert A. (2004). Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28 519–542. 10.1016/j.femsre.2004.04.001 [DOI] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Sellera F. P., Lincopan N. (2019). Zooanthroponotic transmission of high-risk multidrug-resistant pathogens: a neglected public health issue. J. Infect. Public Health. 12 294–295. 10.1016/j.jiph.2018.12.013 [DOI] [PubMed] [Google Scholar]
- Shaw K. J., Rather P. N., Hare R. S., Miller G. H. (1993). Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57 138–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard S. M., Danzeisen J. L., Isaacson R. E., Seemann T., Achtman M., Johnson T. J. (2012). Genome sequences and phylogenetic analysis of K88- and F18-positive porcine enterotoxigenic Escherichia coli. J. Bacteriol. 194 395–405. 10.1128/JB.06225-6211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snesrud E., McGann P., Chandler M. (2018). The birth and demise of the ISApl1-mcr-1-ISApl1 composite transposon: the vehicle for transferable colistin resistance. MBio 9 e2381–17. 10.1128/mBio.02381-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhang H., Liu Y. H., Feng Y. (2018). Towards understanding MCR-like colistin resistance. Trends Microbiol. 26 794–808. 10.1016/j.tim.2018.02.006 [DOI] [PubMed] [Google Scholar]
- Tang X., Tan C., Zhang X., Zhao Z., Xia X., Wu B., et al. (2011). Antimicrobial resistances of extraintestinal pathogenic Escherichia coli isolates from swine in China. Microb. Pathog. 50 207–212. 10.1016/j.micpath.2011.01.004 [DOI] [PubMed] [Google Scholar]
- Travis R. M., Gyles C. L., Reid-Smith R., Poppe C., McEwen S. A., Friendship R., et al. (2006). Chloramphenicol and kanamycin resistance among porcine Escherichia coli in Ontario. J. Antimicrob. Chemother. 58 173–177. 10.1093/jac/dkl207 [DOI] [PubMed] [Google Scholar]
- van Duijkeren E., Schink A. K., Roberts M. C., Wang Y., Schwarz S. (2018). Mechanisms of bacterial resistance to antimicrobial agents. Microbiol. Spectr. 6 51–82. 10.1128/microbiolspec.ARBA-0019-2017 [DOI] [PubMed] [Google Scholar]
- Veldman K., van Essen-Zandbergen A., Rapallini M., Wit B., Heymans R., van Pelt W., et al. (2016). Location of colistin resistance gene mcr-1 in Enterobacteriaceae from livestock and meat. J. Antimicrob. Chemother. 71 2340–2342. 10.1093/jac/dkw181 [DOI] [PubMed] [Google Scholar]
- Wick R. R., Judd L. M., Gorrie C. L., Holt K. E. (2017). Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genom. 3:e000132. 10.1099/mgen.0.000132 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The nucleotide sequence of the 35 LREC genomes have been deposited in the NCBI sequence databases with accession codes SAMN11523829 to SAMN11523863. These sequences are part of BioProject ID PRJNA540146.


