Abstract
Through experimental information available from antioxidant assays of seventeen anthocyanins, and six common anthocyanidins, quantitative structure–activity relationships (QSAR) have been established in the present work. The antioxidant bioactivity has been predicted in three different lipid environments: emulsified and bulk oil (methyl linoleate) (in vitro tests) at concentrations of 50 and 250 μM, and 50 μM of the inhibitor, respectively, and in human LDL (low-density lipoprotein; “bad cholesterol”) (ex vivo test) at concentrations of 2.5, 10, and 25 μM of the inhibitor. Radical scavenging activity was predicted in the assay with the 1,1-diphenyl-2-picrylhydrazyl radical (DPPH·). The QSAR models developed for each test and concentration used allowed to obtain prospective information on the constitutional and topological molecular characteristics for anthocyanin/anthocyanidin compounds. Therefore, the antioxidant activity was predicted for twenty-one compounds with unknown experimental values, leading for some of them to a favorable predicted bioactivity.
Electronic supplementary material
The online version of this article (10.1007/s13197-019-04024-w) contains supplementary material, which is available to authorized users.
Keywords: Anthocyanins, Antioxidant activity, Quantitative structure–activity relationships, Molecular descriptors
Introduction
Anthocyanins are natural hydrosoluble dyes occurring as O-glycosides in flowers, fruits, leaves, bracts, and stems that belong to the family of flavonoids (Harborne and Williams 2000; He and Giusti 2010; Du et al. 2018; Fujiwara et al. 2018). These compounds are constituted by an aglycone (anthocyanidin) that contains a positive charge on the oxygen atom (oxonium salt), commonly known as a flavylium salt or a 2-phenyl-chromilium ion. Several 3,5,7-trisubstituted hydroxylated anthocyanidins have been isolated and further identified, 5- and 7-hydroxyls being sometimes methylated. Also 6- or 8-hydroxylated anthocyanidins, and 3- and 5-deoxyanthocyanidins have been reported (Sousa et al. 2016).
The structural variability of anthocyanins is due to the hydroxylation and methoxylation patterns of the aglycones, and also to the glycosylation pattern, including sugar acylation (Sakuta 2014). Acylated anthocyanins are of interest due to their restricted taxonomic distribution, bioactivity, and biogenetic relationship with sugar esters of cinnamic acid derivatives (Pomilio and Mercader 2017; Zhang et al. 2019).
Anthocyanins (O-glycosides) and their aglycones (anthocyanidins) show several bioactivities of therapeutic value against chronic diseases, such as anti-inflammatory, antiangiogenic, antimutagenic, antiproliferative, antioxidant, anticarcinogenic, cardioprotective, estrogenic, chondroprotective, and anti-neurodegenerative properties (Bonesi et al. 2019; Li et al. 2019; Tsakiroglou et al. 2019; Ullah et al. 2019; Vishnu et al. 2019; Wongwichai et al. 2019; Zhang et al. 2019). Mechanisms of action have been reported, including effects on gene and protein expression as well as interactions with micro-RNAs (miRNAs) and cell-signaling pathways (Khan et al. 2019; Krga and Milenkovic 2019; Ma and Ning 2019; de Sousa Moraes et al. 2019). Target cells of anthocyanins seem to be adipocytes, endothelial cells, inflammatory cells, hepatocytes, and intestinal cells (Jiang et al. 2019).
In particular, antioxidant activities (Kaurinovic and Vastag 2019) explain the value of some functional foods in human health, and the importance of nutraceuticals to prevent diseases, such as cardiovascular, and inflammatory diseases, including diabetes and metabolic syndrome. Plant-derived antioxidants have been shown to function as singlet and triplet oxygen quenchers, free radical scavengers, peroxide decomposers, metal chelators, enzyme inhibitors and synergists (Jaganath and Crozier 2010).
It is worth noting that free radicals are highly reactive and, therefore, very unstable, with a very short life time [minutes for hydrogen peroxide (H2O2), and organic hydroperoxides; seconds for the peroxyl radical; milliseconds for peroxynitrite; microseconds for superoxide anion, singlet oxygen, alkoxy radicals; and nanoseconds for the hydroxyl radical]. The presence of free radicals in living organisms can only be explained by compartmentalization at cellular level (Thankam Finosh and Jayabalan 2013). Therefore, the main product of cell metabolism is H2O2, whose formation is thermodynamically favorable because of a positive redox potential. Anthocyanins are known to remove H2O2 from cells that mainly occurs by the Fenton reaction. It takes place through the hydroxyalkene group of of the anthocyanin structures via the mechanism proposed for this reaction (Vitale et al. 2016). Hence, a large part of their bioactivities can be explained by this important function.
The potential chemopreventive use of anthocyanins from functional foods or as supplements, together with the possibility of using anthocyanins as natural food colorants led us to perform studies of quantitative structure–activity relationships (QSAR). In this way, mathematical models with predictive power were obtained by associating the molecular structure, encoded within the so-called molecular descriptors, with the biological activity of interest, e.g., the antioxidant activity.
Therefore, the aim of the present work was to obtain simple, novel, and easily interpretable QSAR models to predict the antioxidant properties of twenty-three anthocyanins (aglycones, O-glycosides, and O-acylglycosides) by evaluating the free radical scavenging activity, and percentage inhibition of lipid oxidation.
Structural features such as the extent of hydroxylation and methoxylation of ring B, and the type of glycosylation/acylglycosylation of the anthocyanins have been previously reported to modify the stability, bioavailability, and antioxidant/anti-inflammatory effects of anthocyanins (Kähkönen and Heinonen 2003; Miguel 2011). These retrospective analyses based on experimental values are very useful, but we encouraged the present QSAR study due to the need to deepen into the whole molecular structure. QSAR developments involved prospective information based on the constitutional and topological molecular characteristics of the anthocyanins. Therefore, the antioxidant activity was analyzed and quantified for the predictive design of new antioxidants.
Finally, a practical application of the established QSAR models was performed. Acceptable predicted antioxidant bioactivities were obtained for twenty-one anthocyanins/anthocyanidins with unknown experimental values, most of them being considered as reliable predictions according to their leverage values.
Materials and methods
Anthocyanins selected for this study
Seventeen anthocyanins and six anthocyanidins were selected, from which experimental information on antioxidant tests was available (Kähkönen and Heinonen 2003). The chemical structures of the selected anthocyanins, with increasing complexity, corresponded to:
Anthocyanidins (without glycosidation), such as: pelargonidin (1), cyanidin (2), delphinidin (3), peonidin (4), petunidin (5), and malvidin (6) (Fig. 1A).
Anthocyanins: 3-O-monoglycosides of anthocyanidins, with monosaccharides (glucose, galactose, and arabinose) in the 3-position of the aglycones, such as: pelargonidin 3-O-β-d-glucopyranoside (7), cyanidin 3-O-β-d-glucopyranoside (8), delphinidin 3-O-β-d-glucopyranoside (9),peonidin 3-O-β-d-glucopyranoside (10), petunidin 3-O-β-d-glucopyranoside (11), malvidin 3-O-β-d-glucopyranoside (12); cyanidin 3-O-β-d-galactopyranoside (13), peonidin 3-O-β-d-galactopyranoside (14), malvidin 3-O-β-d-galactopyranoside (15); cyanidin 3-O-α-l-arabinopyranoside (16), peonidin 3-O-α-l-arabinopyranoside (17) (Fig. 1B).
Anthocyanins: 3-O-diglycosides of anthocyanidins, with disaccharide (rutin = disaccharide 6-O-α-l-rhamnopyranosyl-β-d-glucopyranose), in the 3-position of the aglycone, such as: cyanidin 3-O-β-d-rutinoside (18), delphinidin 3-O-β-d-rutinoside (19);
Anthocyanins: 3,5-O-diglycosides of anthocyanidins, with monosaccharides (glucose) in the 3- and 5-positions of the aglycone, such as: cyanidin 3,5-di-O-β-d-glucopyranoside (20), malvidin 3,5-di-O-β-d-glucopyranoside (21) (Fig. 1C).
Anthocyanin: 3-O-diglycoside-5-O-monoglycoside of anthocyanidin, with disaccharide (xylosylglucoside = sambubioside = disaccharide 2-O-β-d-xylopyranosyl-β-d-glucopyranose) in the 3-position, and monosaccharide (galactose) in the 5-position of the anthocyanidin, such as: cyanidin 3-O-β-d-sambubioside-5-O-β-d-galactopyranoside (22);
Monoacylated anthocyanin: 3-O-monoacyldiglycoside-5-O-monoglycoside of anthocyanidin, with monoacylated disaccharide (coumaroyl-xylosylglucoside = E-p-coumaroylsambubioside = 6″-O-E-p-coumaroylsambubioside) in the 3-position, and monosaccharide (galactose) in the 5-position of the anthocyanidin; the acyl corresponds to a derivative of cinnamic acid as p-coumaroyl, such as: cyanidin 3-O-β-d-(6″-O-E-p-cumaroyl)sambubioside-5-O-β-d-galactopyranoside (23) (Fig. 1D).
Fig. 1.
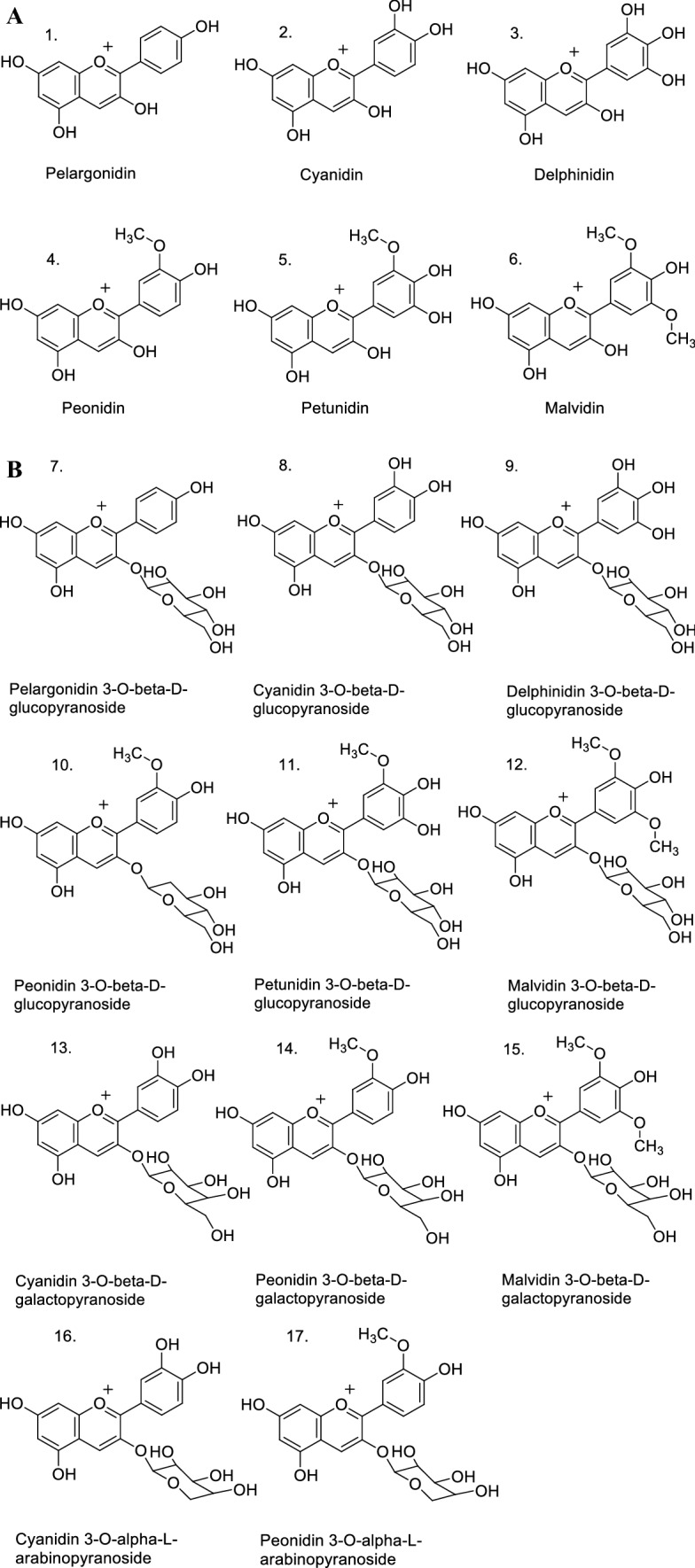
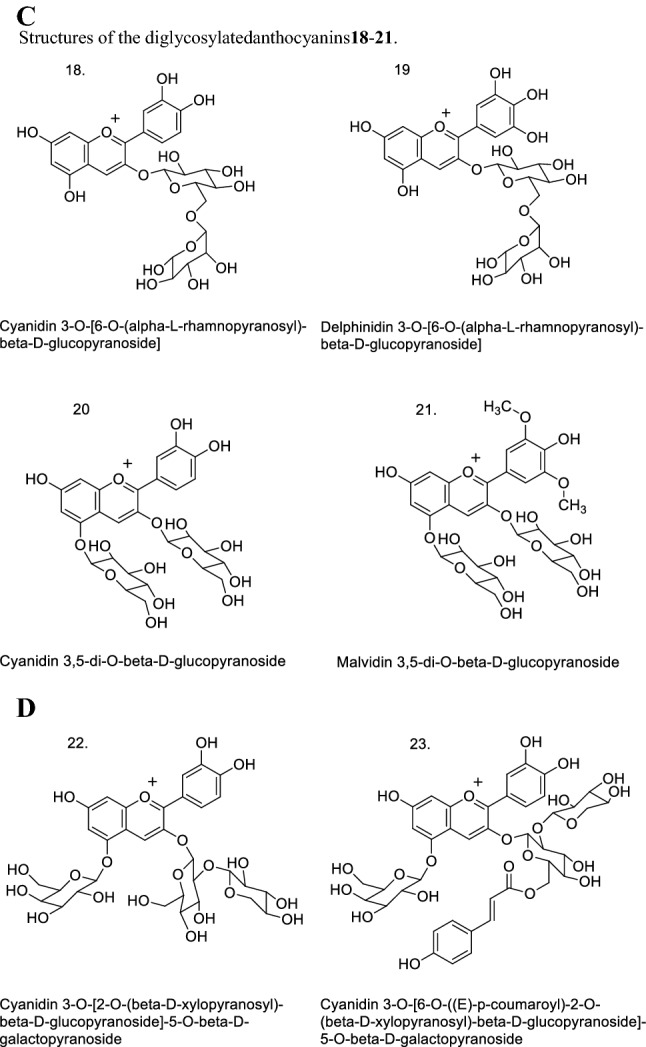
Structures of: A anthocyanidins 1–6; B monoglycosylated anthocyanins7–17; C diglycosylated anthocyanins 18–21; D triglycosylated anthocyanin 22, and acylated triglycosylated anthocyanin 23
Well-known antioxidants were also tested for comparative purposes (Kähkönen and Heinonen 2003): ascorbic acid, α-tocopherol, and Trolox, three phenolic acids: gallic, caffeic, and chlorogenic acids, and three flavonoids: quercetin, rutin, and (+)-catechin (Bentz et al. 2017).
Antioxidant activity evaluation
The antioxidant activity of anthocyanidins and their glycosides was evaluated in three different lipid environments: emulsified and bulk oil (methyl linoleate: MeLo) (in vitro tests) at concentrations of 50 and 250 μM, and 50 μM of the inhibitor, respectively, and in human low-density lipoprotein (LDL; “bad cholesterol”) (ex vivo test) at concentrations of 2.5, 10, and 25 μM of the inhibitor. In addition, the radical scavenging activity of the compounds was studied against the 1,1-diphenyl-2-picrylhydrazyl radical (DPPH·). The evaluation methodologies have been previously reported (Kähkönen and Heinonen 2003).
QSPR studies on the anthocyanins
The structures of the anthocyanins were represented with conformation-independent molecular descriptors in order to consider their constitutional and topological nature. The molecules were drawn in the MDL mol (V2000) format in ACD/ChemSketch (2016). PaDEL (2016), Mold2 (Hong et al. 2008), EPI Suite (2018), RECON (Lavine et al. 2003), and QuBiLS-MAS (Valdes-Martini et al. 2012) softwares were used to calculate 46,994 linearly independent descriptors. All these programs are free and open access.
Data set of anthocyanins
The molecular set was divided into a training set (train) to select the molecular descriptors and adjustable parameters of the multivariable linear regression (MLR), and an external testset (test) to determine the predictive capacity of the obtained model. The test set compounds were selected by approximately spanning the experimental bioactivity interval.
Apart from using a test set, each model was theoretically validated by means of a ‘Leave-One-Out’ (loo) cross-validation. Further validation parameters were calculated as proposed in the literature (Chirico and Gramatica 2012; Roy et al. 2013).
Table S1 includes the experimental antiradical and antioxidant activities in scaled and logarithmic units [] of the selected anthocyanidins and anthocyanins (Kähkönen and Heinonen 2003). The test set compounds are indicated with ^ for each activity analyzed.
The selected data reduction technique will make the QSAR models differ in their accuracy, complexity, and predictive capacity. The most direct strategy is MLR, where the variables of the linear equation are a few descriptors that best predict the activity. As it is known, the selection of descriptors is not easy, that is why the QSAR models were established with the “Replacement Method” (RM) (Duchowicz et al. 2006). This technique consists of a mathematical algorithm designed in our research group, which allows selecting the best descriptors from a large number of them. This selection is performed by minimizing the square root of the mean square error of the training set ().
The approximate solutions achieved with RM sometimes agree with the exact solutions (complete analysis of all possible variable combinations). In other cases they are very close (Duchowicz et al. 2006; Mercader et al. 2010). All Matlab algorithms used were developed in our research group, and are available by request.
Once the best solution was identified, the techniques of the external validation set, cross validation, and applicability domain (Roy et al. 2015) allowed determining whether the model was acceptable.
Results and discussion
Quantitative structure–activity relationships (QSAR) models were established in order to predict the inhibition percentage of oxidation product formation () when the inhibitors were added at different concentrations. The structures of the anthocyanins were represented with conformation-independent molecular descriptors in order to take into account their constitutional and topological nature.
The main advantage of not considering molecular conformations is that the only experimental information required to establish the QSAR models is the biological activity that is analyzed (Duchowicz 2018), in our case the antioxidant activity. No additional experimental information was required, such as that provided, for example, by X-ray structural crystallographic data for some anthocyanin conformation in its molecular environment, data that are often unknown. In this way, a QSAR model was established based on the minimum essential experimental information (experimental biological activity, I%). Therefore, conformation-independent QSAR studies were very useful for application to any chemical system of interest.
The most representative non-conformational molecular descriptors found in this work for the studied bioactivities are briefly described in Table 1, being involved in the different quantitative structure–antioxidant activity relationships of the 23 anthocyanins analyzed. These descriptors were selected from 46,994 linearly independent descriptors available at PaDEL (2016), Mold2 (Hong et al., 2008), EPI Suite (2018), RECON (Lavine et al. 2003), and QuBiLS-MAS (Valdes-Martini et al. 2012) programs. The numerical values of such descriptors are displayed in Table S2.
Table 1.
A brief description for non-conformational descriptors appearing in this QSAR study, obtained from PaDEL (2016), Mold2 (Hong et al. 2008), and QuBiLS-MAS (Valdes-Martini et al. 2012) programs
| Type of descriptor | Symbol | Description |
|---|---|---|
| QuBiLS-MAS | qub1, qub2, qub3, qub4, qub5, qub6, qub7a | Mathematical indexes obtained from quadratic, bilinear, and linear-type algebraic forms, based on matrices of the Graph theory and atomic weights |
| Electrotopological | minHBint4 | Minimum E-state descriptor of strength for potential hydrogen bonds of path length 4 |
| Mold2 | D284 | structural information content index of order-3 |
| D714 | count of CH3R and CH4 groups | |
| 2D Autocorrelation | D458 | Geary autocorrelation—lag 4, weighed by atomic van der Waals volumes |
| GATS6p | Geary autocorrelation—lag 6, weighed by atomic polarizabilities | |
| ATSC8e | Centered Broto-Moreau autocorrelation—lag 8/weighted by atomic Sanderson electronegativities | |
| Baryszmatrix | VR1.Dzm | Randic-type eigenvector-based index from Barysz matrix/weighted by mass |
| VR2.Dzs | Normalized Randic-type eigenvector-based index from Barysz matrix/weighted by I-state |
aqub1 = SD_B_AB_nCi_2_SS8_T_KA_psa-p_MAS;qub2 = SD_B_AB_nCi_2_SS8_T_KA_psa-e_MAS; qub3 = AM_B_AB_nCi_2_DS3_M_KA_v-c_MAS; qub4 = SD_B_AB_nCi_2_SS7_D_KA_psa-v_MAS; qub5 = AM_B_AB_nCi_2_SS15_T_KA_psa-r_MAS; qub6 = SD_B_AB_nCi_2_SS8_A_KA_psa-p_MAS; qub7 = AM_Q_AB_nCi_2_DS5_T_KA_psa_MAS
The best QSAR linear regressions, which predict the inhibition percentage of oxidation products formation (I%) when the inhibitors are added at different concentrations are shown as Eqs. (1)–(7). Each model had an acceptable number of molecules to number of descriptors ratio () in order to avoid data overfitting.
DPPH· (17 μM):
| 1 |
MeLo emulsion (50 μM):
| 2 |
MeLo emulsion (250 μM):
| 3 |
Human LDL (2.5 μM):
| 4 |
Human LDL (10 μM):
| 5 |
Human LDL (25 μM):
| 6 |
MeLo in bulk (50 μM):
| 7 |
In these equations, is the linear correlation coefficient; is the number of calibration molecules that have a residual greater than the value, where is the standard deviation for the training set; and is the maximum intercorrelation between descriptor pairs.
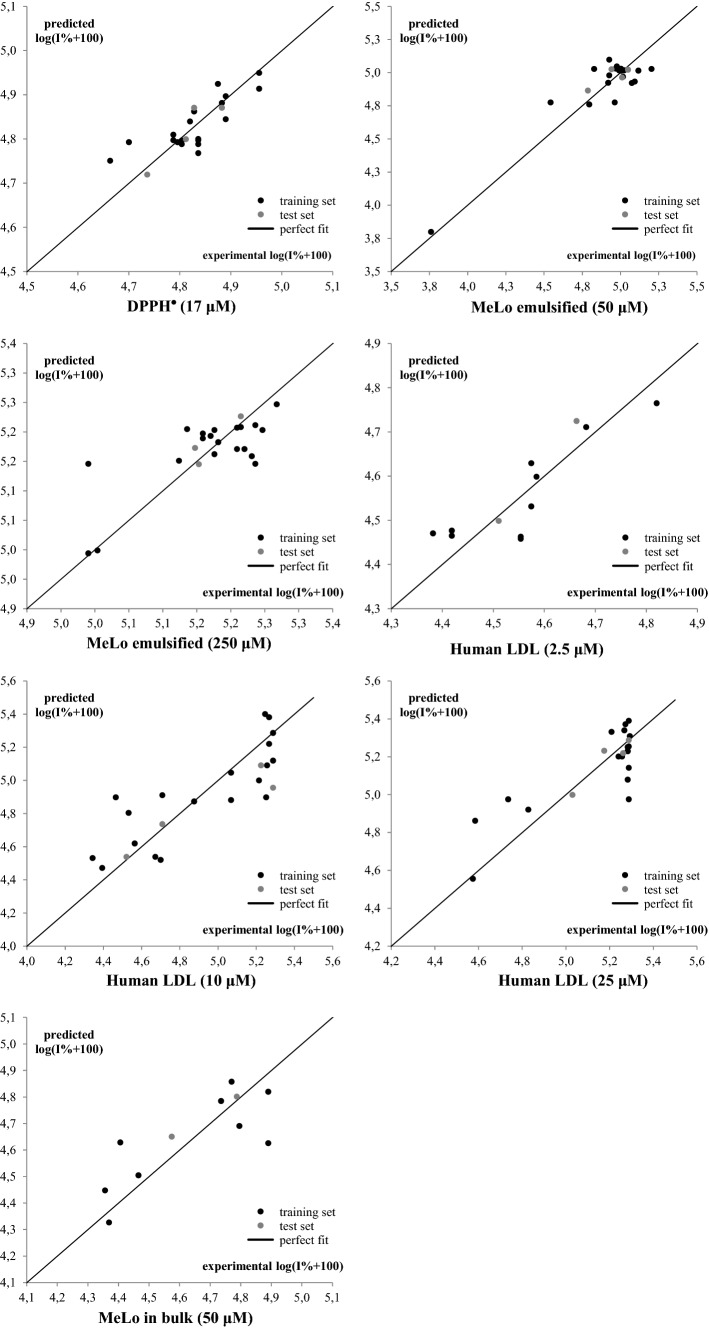
The molecular descriptors involved in Eqs. (1)–(7) tended to accurately predict the antioxidant potential, as shown in Fig. 2. A linear trend was observed when plotting the predicted antioxidant activity as a function of the experimental activity. The maximum intercorrelation between pairs of descriptors () was not a serious problem in Eqs. (1)–(7), suggesting that each descriptor involved in the linear regression included non-overlapping structural information.
Fig. 2.
Predictions of the antioxidant activity according to QSAR models of Eqs. (1)–(7)
The residual scatter plots of the QSAR models showed that the data tended to be randomly distributed around the zero line, thus suggesting the absence of systematic errors during modeling (Fig. S1). It is worth to mention that it was not possible to obtain an absolute conclusion of the pattern shown by such graphs, due to the small number of observations involved (). In addition, although some anthocyanins of these graphs exhibited high residual (difference between the value of the experimental activity, and the predicted one), all structures were included, deciding not to remove any of them of the model in order to be able to predict and analyze them in the present dataset. Finally, none of the established models showed molecules with residues greater than the value, revealing the absence of molecules with anomalous behavior in terms of their structure–activity relationship.
The predictive capacity of these models for antioxidant activity was satisfactory, as suggested by the parameter, which was lower or comparable to in Eqs. (1)–(7). Therefore, the anthocyanins of the external test set were adequately predicted with the QSAR models that were obtained only on the basis of the calibration set. Here, it should be mentioned that was not a significant validation parameter in the present analysis due to the small number of validation molecules used ().
Apart from using a test set, each model was theoretically validated by means of ‘Leave-One-Out’ (loo) cross-validation. The and parameters revealed that the Eqs. (1)–(7) were stable when any molecule was removed from the training set, and was predicted with the recalculated model with the remaining calibration molecules (using the same model descriptors).
Further validation parameters from the literature were provided in Table 2 for each QSAR equation, achieving moderate results. However, it was noted that these parameters (e.g., , , , CCC, and ) would have worked better if a greater number of data points were included in the QSAR study. A better behavior would also have been obtained if these data points uniformly covered the range of experimental activity, which was not the case in the bioactivities of the present work.
Table 2.
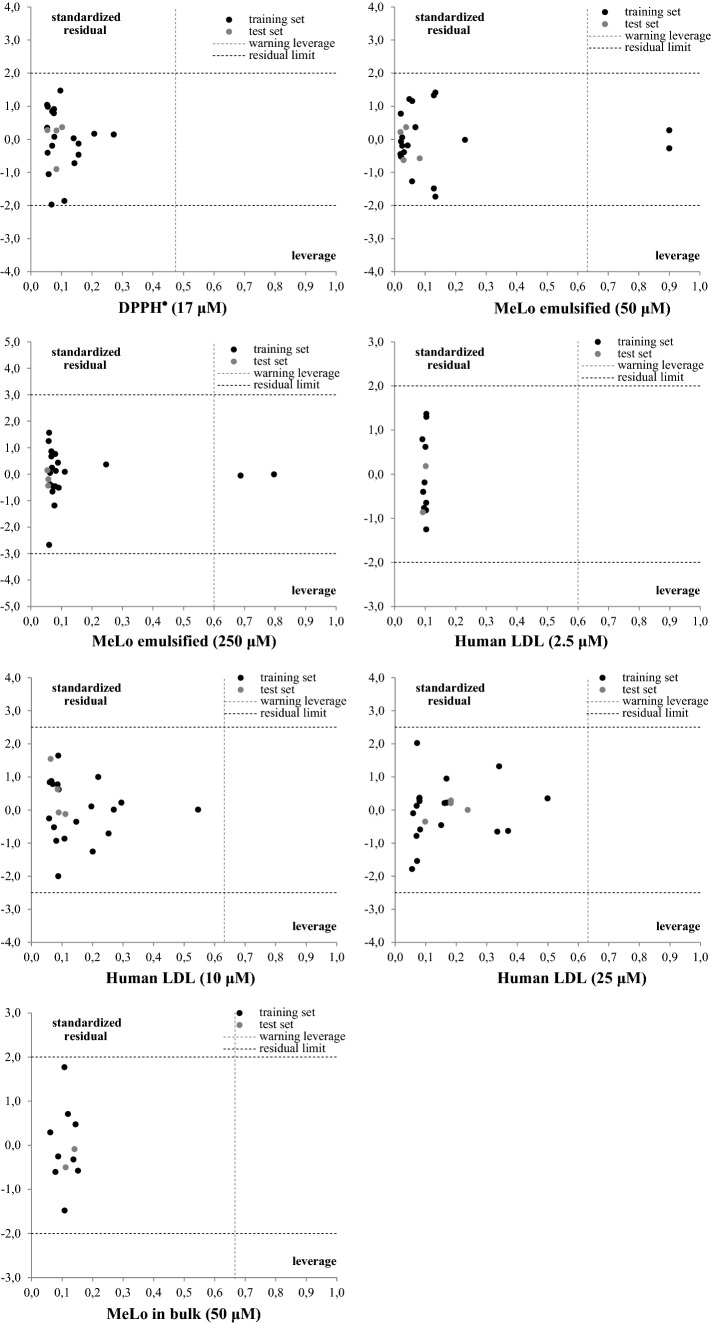
As it is known, a predictive QSAR model is only able to predict molecules that belong to its applicability domain (AD), so that the activity predicted by the model is not the result of extrapolation (unreliable prediction) (Roy et al. 2015). In the present work, the AD was evaluated by the influence parameter (): the prediction of a test set molecule was reliable provided when its influence was less than the limit influence (). As shown in Fig. 3, the validation anthocyanins of Eqs. (1)–(7) were contained within the applicability domain of each model (), so that their predictions could be considered reliable.
Fig. 3.
Standardized residuals as function of the influence parameter
The molecular descriptors involved in the QSAR models of the current study were independent of the conformations. These descriptors only depended on the constitutional and topological features. Table 1 showed that many of these descriptors were abstract, and did not have a direct physical interpretation. Despite this, the proposed models were useful in providing a QSAR guide for the chemical synthesis of new antioxidant structures, provided that the anthocyanins, whose activity was predicted, belonged to the applicability domains of the developed models.
For example, in the case of the QSAR model given by Eq. (1) for DPPH· (17 μM), the molecular structures of inhibitors that simultaneously had lower values of the descriptors GATS6p and qub1, would have exhibited higher predicted values of antioxidant activity . The reason was due to the fact that the descriptors of Eqs. (1)–(7) took positive values, and the degree of contribution of each descriptor to the predicted activity was then given by the sign and magnitude of its regression coefficient. Table 3 summarizes the simultaneous trend that the descriptors of the QSAR Eqs. (1)–(7) should have, so that the predicted inhibition was higher for an anthocyanin structure.
Table 3.
Trend of the molecular descriptors for a greater predicted antioxidant activity
Anthocyanins and anthocyanidins used in this QSAR study have shown antioxidant properties in in vitro and ex vivo tests as previously reported (Kähkönen and Heinonen 2003). Twenty-three compounds were studied. The six more common anthocyanidins were selected for this study, e.g., pelargonidin, cyanidin, delphinidin, peonidin, petunidin, and malvidin; also, their 3-O-monoglycosides, 3-O-diglycosides, 3,5-O-diglycosides, 3-O-diglycoside-5-O-monoglycosides, and a 3-O-acyldiglycoside-5-O-monoglycoside were tested, including different types of hexoses in the pyranose form (see “Anthocyanins selected for this study” section). The radical scavenging activity was in vitro studied against the 1,1-diphenyl-2-picrylhydrazyl radical (DPPH·). The ability to prevent the oxidation of lipids was tested in three different lipid environments, such as emulsified and bulk methyl linoleate at different inhibitor concentrations (50 and 250 μM, and 50 μM, respectively), and human low-density lipoprotein (LDL) (2.5, 10, and 25 μM). Results revealed the structural- and concentration-dependence of this activity.
The effects of structural features on the antioxidant activity have been previously analyzed according to the inhibition experimental values (Kähkönen and Heinonen 2003). As expected, hydroxylation and methoxylation patterns showed remarkable effects on the antioxidant power of the aglycones, and glycosylation/acylglycosylation pattern in the case of anthocyanins (Miguel 2011).
From the in vitro and ex vivo tests, retrospective considerations arose on the influence of substituents of the ring B and of the presence of 3-O-mono- or di-glycosides, 3,5-O-diglycosides and acyglycosides, that is, direct interpretations of the experimental values taking into account the possible mechanisms of action involved, but not always the characteristics of the medium involved. But these evaluations of the assays do not serve to design new antioxidants, nor to establish a possible antioxidant value (unknown) for an anthocyanin that has not been previously tested. To be able to do this, and either to obtain numerical values of antioxidant activity of a given anthocyanin (prediction) or to design a novel antioxidant of anthocyanin nature (drug design) it was necessary to delve into the molecular electronic structure and its properties to establish the relationships with that bioactivity according to bioinformatics and statistics as currently achieved by the QSAR theory. Furthermore, statistically-based QSAR approaches are of value in Medicinal Chemistry, and help to guide lead optimization (Cherkasov et al. 2014; Mercader et al. 2016).
Therefore, we developed QSAR modeling, so that small differences at the molecular level, which are not so obvious, can be explained. For the coding process a large number of independent variables or molecular descriptors were computed and used to develop the QSAR models. Descriptors quantify several constitutional, topological, and structural properties of a molecule, but may lack the capacity for structural interpretation. Each descriptor is a reproducible and invariable value since it depends on the calculation algorithm used, while experimental bioassay values are dependent variables. A set of computational techniques were necessary for a virtual spatial design and visualization of molecules, calculation of the molecular descriptors, bioinformatics, and statistics (see “Materials and Methods” section).
The statistical predictive QSAR models obtained in the present study correlated the antioxidant bioactivity of anthocyanins with representative descriptors of their molecular structure and/or molecular properties. Results showed that different descriptors account for each antioxidant test used, and concentrations of the anthocyanins/anthocyanidins due to the test- and concentration-dependence of the antioxidant activity of these compounds.
The QSAR models of this study were obtained from the two-dimensional descriptors mentioned above. Results show that the molecular skeleton provides enough information for useful relationships, three-dimensional topography not being necessary.
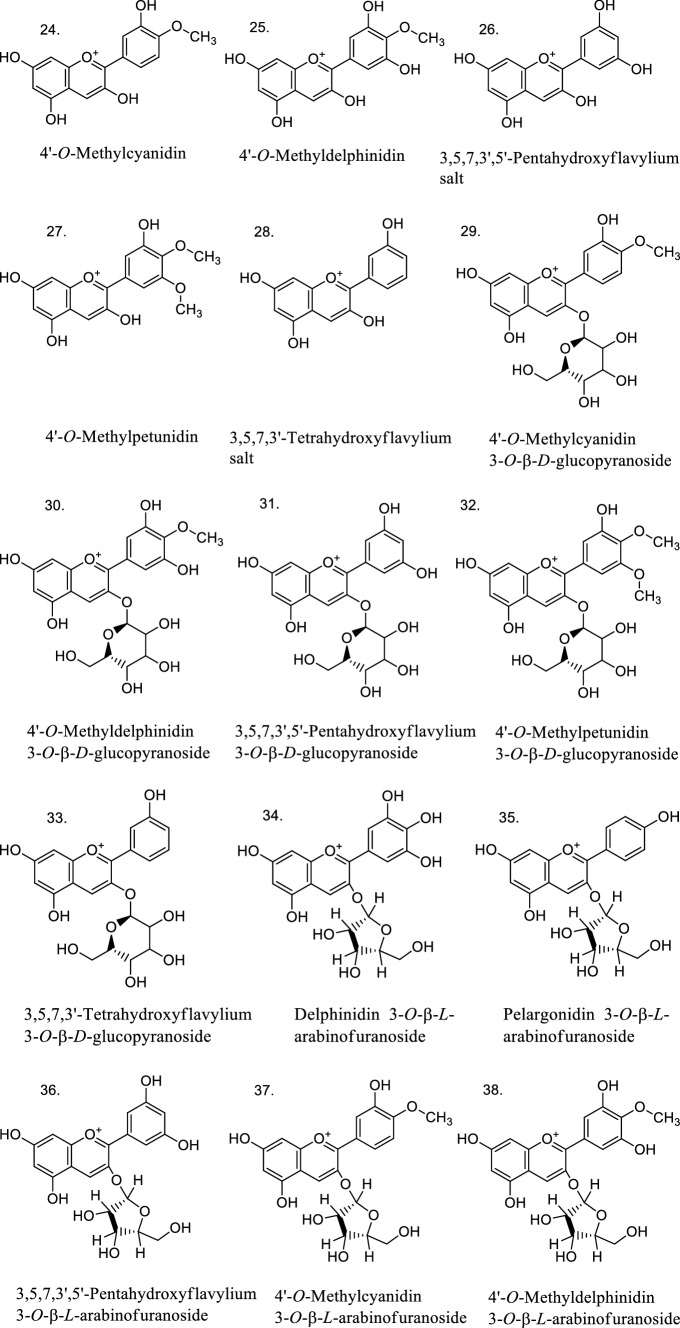
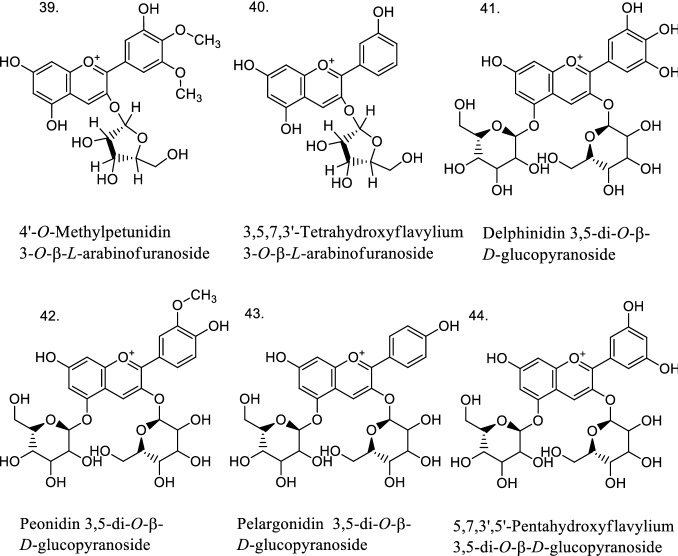
Finally, a practical application of the established QSAR models of Eqs. 1–7 was performed. Antioxidant bioactivities were predicted for 21 anthocyanins/anthocyanidins with unknown experimental values (Fig. 4). As shown in Table 4, most of them can be considered as reliable predictions according to their leverage values, and most importantly, some of the proposed anthocyanins had favorable predicted bioactivities. For instance, the predicted antioxidant activity had a high value for the following compounds in different lipid environments: 26 and 38 in DPPH· (17 μM); 27, 28, and 39 in MeLo emulsion (50 μM); 26, 28, and 31 in Human LDL (10 μM); and 24 and 25 in MeLo in bulk (50 μM). From the results of Table 4, experimental antioxidant tests should be carried out to verify such predicted molecular structures.
Fig. 4.
Structures of 21 anthocyanins with unknown experimental antioxidant activities
Table 4.
Predicted antioxidant activities and leverages for 21 anthocyanins with unknown experimental values
| ID | Equation 1 | Equation 2 | Equation 3 | Equation 4 | Equation 5 | Equation 6 | Equation 7 |
|---|---|---|---|---|---|---|---|
| 24 | 5.15 (0.53) | 5.00 (0.03) | 5.19 (0.11) | 4.71 (0.09) | 4.96 (0.22) | 5.45 (0.38) | 5.01 (0.19) |
| 25 | 5.23 (1.03) | 4.73 (0.16) | 5.05 (0.66) | 4.66 (0.10) | 4.72 (0.25) | 5.36 (0.25) | 5.32 (0.26) |
| 26 | 5.01 (0.38) | 5.00 (0.03) | 5.24 (0.06) | 4.70 (0.09) | 5.75 (0.07) | 5.03 (0.25) | 4.88 (0.16) |
| 27 | 4.79 (0.07) | 5.04 (0.02) | 5.23 (0.06) | 4.74 (0.09) | 4.55 (0.48) | 4.21 (0.21) | 4.59 (0.10) |
| 28 | 4.92 (0.31) | 5.04 (0.02) | 5.25 (0.05) | 4.70 (0.09) | 5.56 (0.19) | 5.35 (0.37) | 4.73 (0.13) |
| 29 | 4.96 (0.28) | 4.84 (0.09) | 5.13 (0.07) | 4.46 (0.10) | 4.41 (0.10) | 5.23 (0.08) | 4.94 (0.17) |
| 30 | 4.80 (0.05) | 4.92 (0.06) | 5.16 (0.06) | 4.47 (0.10) | 4.88 (0.07) | 5.33 (0.07) | 4.63 (0.11) |
| 31 | 4.92 (0.15) | 5.02 (0.02) | 5.17 (0.07) | 4.55 (0.10) | 5.60 (0.25) | 4.99 (0.07) | 4.90 (0.16) |
| 32 | 4.76 (0.06) | 4.48 (0.40) | 5.17 (0.05) | 4.46 (0.10) | 4.36 (0.13) | 5.10 (0.08) | 4.67 (0.12) |
| 33 | 4.84 (0.25) | 5.03 (0.02) | 5.15 (0.07) | 4.48 (0.10) | 4.99 (0.26) | 5.19 (0.07) | 4.85 (0.15) |
| 34 | 4.83 (0.06) | 4.87 (0.08) | 5.20 (0.06) | 5.19 (0.19) | 5.32 (0.09) | ||
| 35 | 4.75 (0.06) | 4.90 (0.06) | 5.18 (0.07) | 4.46 (0.14) | 4.94 (0.09) | ||
| 36 | 4.86 (0.05) | 4.91 (0.06) | 5.20 (0.06) | 5.46 (0.13) | 5.08 (0.06) | ||
| 37 | 4.83 (0.06) | 4.87 (0.08) | 5.20 (0.06) | 5.19 (0.19) | 5.32 (0.09) | ||
| 38 | 5.01 (0.13) | 5.01 (0.06) | 5.10 (0.22) | 4.60 (0.11) | 5.28 (0.08) | ||
| 39 | 4.72 (0.20) | 5.07 (0.04) | 5.20 (0.04) | 4.28 (0.24) | 5.16 (0.15) | ||
| 40 | 4.79 (0.06) | 4.95 (0.04) | 5.19 (0.06) | 4.91 (0.18) | 5.30 (0.13) | ||
| 41 | 4.83 (0.10) | 5.02 (0.03) | 5.17 (0.07) | 5.05 (0.27) | 5.04 (0.31) | ||
| 42 | 4.72 (0.07) | 5.02 (0.03) | 5.13 (0.06) | 4.17 (0.10) | 5.03 (0.09) | ||
| 43 | 4.73 (0.06) | 5.01 (0.04) | 5.15 (0.07) | 4.22 (0.07) | 4.73 (0.12) | ||
| 44 | 4.83 (0.06) | 5.02 (0.03) | 5.16 (0.07) | 5.25 (0.17) | 4.86 (0.17) |
Conclusion
Most anthocyanins and their aglycones behaved as strong antioxidants in emulsion and LDL, showing activities comparable to known antioxidants, such as α-tocopherol, Trolox, catechin, and quercetin. In bulk methyl linoleate, anthocyanins and anthocyanidins showed only weak antioxidant or even oxidation-promoting activities. Mostly, the activities of the glycosides and aglycones did not differ markedly in emulsion. In LDL, the aglycones generally showed higher activities than glycosides. In contrast, in bulk oil, glycosides were more effective than aglycones.
The QSAR models obtained in this work allowed a prospective analysis leading to proper antioxidant prediction and design of new antioxidant structures in the anthocyanin field, depending solely on constitutional and electrotopological features of the molecular structures. Therefore, these results allowed to establish quantitative structure–activity relationships that are satisfactory for predicting properties through molecular descriptors; and to show that QSAR studies are suitable for designing molecules with optimal desired antioxidant activities.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the National Scientific and Technical Research Council of Argentina [Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina] (PIP0311) and Universidad de Buenos Aires (Argentina) for financial support; and Secretaría de Ciencia, Tecnología e Innovación Productiva (formerly Ministerio de Ciencia, Tecnología e Innovación Productiva) for electronic library facilities. N.A.S. thanks the Scientific Research Comission [Comisión de InvestigacionesCientíficas (CIC), La Plata city, Argentina] for a fellowship. A.B.P. and P.R.D. are Research Members of CONICET.
Compliance with ethical standards
Conflict of interest
All authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- ACD/ChemSketch (2016) www.acdlabs.com
- Bentz EN, Pomilio AB, Lobayan RM. Donor–acceptor interactions as descriptors of the free radical scavenging ability of flavans and catechin. Comput Theor Chem. 2017;1110:14–24. doi: 10.1016/j.comptc.2017.03.028. [DOI] [Google Scholar]
- Bonesi M, Leporini M, Tenuta MC, Tundis R. The role of anthocyanins in drug discovery: recent developments. Curr Drug Discov Technol. 2019 doi: 10.2174/1570163816666190125152931. [DOI] [PubMed] [Google Scholar]
- Cherkasov A, Muratov EN, Fourches D, Varnek A, Baskin II, Cronin M, Dearden J, Gramatica P, Martin YC, Todeschini R, Consonni V, Kuz’min VE, Cramer R, Benigni R, Yang C, Rathman J, Terfloth L, Gasteiger J, Richard A, Tropsha A. QSAR modeling: Where have you been? Where are you going to? J Med Chem. 2014;57:4977–5010. doi: 10.1021/jm4004285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirico N, Gramatica P. Real external predictivity of QSAR models. Part 2. New intercomparable thresholds for different validation criteria and the need for scatter plot inspection. J Chem Inf Model. 2012;52:2044–2058. doi: 10.1021/ci300084j. [DOI] [PubMed] [Google Scholar]
- de Sousa Moraes LF, Sun X, Peluzio MDCG, Zhu MJ. Anthocyanins/anthocyanidins and colorectal cancer: What is behind the scenes? Crit Rev Food Sci Nutr. 2019;59:59–71. doi: 10.1080/10408398.2017.1357533. [DOI] [PubMed] [Google Scholar]
- Du H, Lai L, Wang F, Sun W, Zhang L, Li X, Wang L, Jiang L, Zheng Y. Characterisation of flower colouration in 30 Rhododendron species via anthocyanin and flavonol identification and quantitative traits. Plant Biol (Stuttg) 2018;20:121–129. doi: 10.1111/plb.12649. [DOI] [PubMed] [Google Scholar]
- Duchowicz PR. Linear regression QSAR models for Polo-Like Kinase-1 Inhibitors. Cells. 2018;7:1–11. doi: 10.3390/cells7020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchowicz PR, Castro EA, Fernández FM. Alternative algorithm for the search of an optimal set of descriptors in QSAR-QSPR studies. MATCH Commun Math Comput Chem. 2006;55:179–192. [Google Scholar]
- Epi Suite 4.11 (2018) https://www.epa.gov/tsca-screening-tools/epi-suitetm-estimation-program-interface
- Fujiwara Y, Kono M, Ito A, Ito M. Anthocyanins in Perilla plants and dried leaves. Phytochemistry. 2018;147:158–166. doi: 10.1016/j.phytochem.2018.01.003. [DOI] [PubMed] [Google Scholar]
- Harborne JB, Williams CA. Advances in flavonoid research since 1992. Phytochemistry. 2000;55:481–504. doi: 10.1016/S0031-9422(00)00235-1. [DOI] [PubMed] [Google Scholar]
- He J, Giusti MM. Anthocyanins: natural colorants with health-promoting properties. Annu Rev Food Sci Technol. 2010;1:163–187. doi: 10.1146/annurev.food.080708.100754. [DOI] [PubMed] [Google Scholar]
- Hong H, Xie Q, Ge W, Qian F, Fang H, Shi L, Su Z, Perkin R, Tong W. Mold(2), molecular descriptors from 2D structures for chemoinformatics and toxicoinformatics. J Chem Inf Model. 2008;48:1337–1344. doi: 10.1021/ci800038f. [DOI] [PubMed] [Google Scholar]
- Jaganath IB, Crozier A. Dietary flavonoids and phenolic compounds. In: Fraga CG, editor. Plant phenolics and human health: biochemistry, nutrition, and pharmacology. Hoboken: Wiley; 2010. [Google Scholar]
- Jiang X, Li X, Zhu C, Sun J, Tian L, Chen W, Bai W. The target cells of anthocyanins in metabolic syndrome. Crit Rev Food Sci Nutr. 2019;59:921–946. doi: 10.1080/10408398.2018.1491022. [DOI] [PubMed] [Google Scholar]
- Kähkönen MP, Heinonen M. Antioxidant activity of anthocyanins and their aglycons. J Agric Food Chem. 2003;51:628–633. doi: 10.1021/jf025551i. [DOI] [PubMed] [Google Scholar]
- Kaurinovic Biljana, Vastag Djendji. Antioxidants [Working Title] 2019. Flavonoids and Phenolic Acids as Potential Natural Antioxidants. [Google Scholar]
- Khan MS, Ali T, Kim MW, Jo MH, Chung JI, Kim MO. Anthocyanins improve hippocampus-dependent memory function and prevent neurodegeneration via JNK/Akt/GSK3β signaling in LPS-treated adult mice. Mol Neurobiol. 2019;56:671–687. doi: 10.1007/s12035-018-1101-1. [DOI] [PubMed] [Google Scholar]
- Krga I, Milenkovic D. Anthocyanins: from sources and bioavailability to cardiovascular-health benefits and molecular mechanisms of action. J Agric Food Chem. 2019;67:1771–1783. doi: 10.1021/acs.jafc.8b06737. [DOI] [PubMed] [Google Scholar]
- Lavine BK, Davidson CE, Breneman C, Katt W, Sundling CM. Electronic van der Waals surface property descriptors and genetic algorithms for developing structure-activity correlations in olfactory databases. J Chem Inf Comput Sci. 2003;43:1890–1905. doi: 10.1021/ci030016j. [DOI] [PubMed] [Google Scholar]
- Li S, Wu B, Fu W, Reddivari L. The anti-inflammatory effects of dietary anthocyanins against ulcerative colitis. Int J Mol Sci. 2019 doi: 10.3390/ijms20102588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Ning S. Cyanidin-3-glucoside attenuates the angiogenesis of breast cancer via inhibiting STAT3/VEGF pathway. Phytother Res. 2019;33:81–89. doi: 10.1002/ptr.6201. [DOI] [PubMed] [Google Scholar]
- Matlab 7.6 http://www.mathworks.com
- Mercader AG, Duchowicz PR, Fernández FM, Castro EA. Replacement method and enhanced replacement method versus the genetic algorithm approach for the selection of molecular descriptors in QSPR/QSAR Theories. J Chem Inf Model. 2010;50:1542–1548. doi: 10.1021/ci100103r. [DOI] [PubMed] [Google Scholar]
- Mercader AG, Duchowicz PR, Sivakumar PM, editors. Chemometrics applications and research: QSAR in medicinal chemistry. Boca Raton: CRC Press; 2016. [Google Scholar]
- Miguel MG. Anthocyanins: antioxidant and/or anti-inflammatory activities. J Appl Pharm Sci. 2011;1:7–15. [Google Scholar]
- PaDEL (2016). http://www.yapcwsoft.com/dd/padeldescriptor
- Pomilio AB, Mercader AG. Natural acylated anthocyanins and other related flavonoids: structure elucidation of Ipomoea cairica compounds and QSAR studies including multidrug resistance. In: Atta-ur-Rahman D, editor. Studies in natural products chemistry. (Bioactive natural products) The Netherlands: Elsevier; 2017. pp. 291–321. [Google Scholar]
- Roy K, Chakraborty P, Mitra I, Ojha PK, Kar S, Das RN. Some case studies on application of ‘‘r2 m’’ metrics for judging quality of Quantitative Structure-Activity Relationship predictions: emphasis on scaling of response data. J Comput Chem. 2013;34:1071–1082. doi: 10.1002/jcc.23231. [DOI] [PubMed] [Google Scholar]
- Roy K, Kar S, Ambure P. On a simple approach for determining applicability domain of QSAR models. Chemom Intell Lab Syst. 2015;145:22–29. doi: 10.1016/j.chemolab.2015.04.013. [DOI] [Google Scholar]
- Sakuta M. Diversity in plant red pigments: anthocyanins and betacyanins. Plant Biotechnol Rep. 2014;8:37–48. doi: 10.1007/s11816-013-0294-z. [DOI] [Google Scholar]
- Sousa A, Araújo P, Azevedo J, Cruz L, Fernandes I, Mateus N, de Freitas V. Antioxidant and antiproliferative properties of 3-deoxyanthocyanidins. Food Chem. 2016;192:142–148. doi: 10.1016/j.foodchem.2015.06.108. [DOI] [PubMed] [Google Scholar]
- Thankam Finosh G, Jayabalan M. Reactive oxygen species—control and management using amphiphilic biosynthetic hydrogels for cardiac applications. Adv Biosci Biotechnol. 2013;4:1134–1146. doi: 10.4236/abb.2013.412150. [DOI] [Google Scholar]
- Tsakiroglou P, VandenAkker NE, Del Bo’ C, Riso P, Klimis-Zacas D. Role of berry anthocyanins and phenolic acids on cell migration and angiogenesis: an updated overview. Nutrients. 2019 doi: 10.3390/nu11051075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah R, Khan M, Shah SA, Saeed K, Kim MO. Natural antioxidant anthocyanins—a hidden therapeutic candidate in metabolic disorders with major focus in neurodegeneration. Nutrients. 2019 doi: 10.3390/nu11061195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes-Martini JR, García Jacas CR, Marrero-Ponce Y, Silveira Vaz‘d Almeida Y, Morrel C (2012) Versión 1.0. CAMD-BIR Unit, CENDA Number of register: 2373-2012
- Vishnu VR, Renjith RS, Mukherjee A, Anil SR, Sreekumar J, Jyothi AN. Comparative study on the chemical structure and in vitro antiproliferative activity of anthocyanins in purple root tubers and leaves of sweet potato (Ipomoea batatas) J Agric Food Chem. 2019;67:2467–2475. doi: 10.1021/acs.jafc.8b05473. [DOI] [PubMed] [Google Scholar]
- Vitale AA, Bernatene EA, Vitale MG, Pomilio AB. New insights of the Fenton reaction using glycerol as experimental model. Effect of O2, inhibition by Mg2+, and oxidation state of Fe. J Phys Chem A. 2016;120:5435–5445. doi: 10.1021/acs.jpca.6b03805. [DOI] [PubMed] [Google Scholar]
- Wongwichai T, Teeyakasem P, Pruksakorn D, Kongtawelert P, Pothacharoen P. Anthocyanins and metabolites from purple rice inhibit IL-1β-induced matrix metalloproteinases expression in human articular chondrocytes through the NF-κB and ERK/MAPK pathway. Biomed Pharmacother. 2019;112:108610. doi: 10.1016/j.biopha.2019.108610. [DOI] [PubMed] [Google Scholar]
- Zhang ZC, Zhou Q, Yang Y, Wang Y, Zhang JL. Highly acylated anthocyanins from purple sweet potato (Ipomoea batatas L.) alleviate hyperuricemia and kidney inflammation in hyperuricemic mice: possible attenuation effects on allopurinol. J Agric Food Chem. 2019;67:6202–6211. doi: 10.1021/acs.jafc.9b01810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.