Abstract
Background
Gene expression analysis of breast cancer largely relies on homogenized tissue samples. Due to the high degree of cellular and molecular heterogeneity of tumor tissues, bulk tissue-based analytical approaches can only provide very limited system-level information about different signaling mechanisms and cellular interactions within the complex tissue context.
Methods
We describe an analytical approach using in situ sequencing (ISS), enabling highly multiplexed, spatially and morphologically resolved gene expression profiling. Ninety-one genes including prognostic and predictive marker profiles, as well as genes involved in specific cellular pathways were mapped within whole breast cancer tissue sections, covering luminal A/B-like, HER2-positive and triple negative tumors. Finally, all these features were combined and assembled into a molecular-morphological OncoMap for each tumor tissue.
Findings
Our in situ approach spatially revealed intratumoral heterogeneity with regard to tumor subtype as well as to the OncotypeDX recurrence score and even uncovered areas of minor cellular subpopulations. Since ISS-resolved molecular profiles are linked to their histological context, a deeper analysis of the core and periphery of tumor foci enabled identification of specific gene expression patterns associated with these morphologically relevant regions.
Interpretation
ISS generated OncoMaps represent useful tools to extend our general understanding of the biological processes behind tumor progression and can further support the identification of novel therapeutical targets as well as refine tumor diagnostics.
Fund
Swedish Cancerfonden, UCAN, Vetenskapsrådet, Cancer Genomics Netherlands, Iris, Stig och Gerry Castenbäcks Stiftelse, BRECT, PCM Program, King Gustaf V Jubilee Fund, BRO, KI and Stockholm County Council, Alice Wallenberg Foundation.
Keywords: Breast cancer, Tumor heterogeneity, Gene expression analysis, Diagnostic, Prognostic
Research in Context.
Evidence before this study
Given the high degree of cellular and molecular heterogeneity of tumor tissues, bulk tissue-based analytical approaches can only provide very limited system-level information and dismiss information about the spatial distribution and interaction of different cell types and their signaling mechanisms within the complex tissue context.
Review of most recent literature on precision medicine notes, that the clinical benefits derived till date based on this bulk approaches remain relatively limited and are therefore in need to be more refined. Such refinement could be the addition of spatial information connecting and relating highly multiplexed transcriptomic data points to each other as well as to the histological and cellular tissue context.
Added value of this study
In this study, we demonstrate the development of an analytical approach using our established in situ sequencing (ISS) assay to generate highly multiplexed, spatially and morphologically resolved gene expression profiles of breast cancer tissue. All these features are combined and assembled into a molecular-morphological OncoMap for each tumor tissue, which allows refined diagnostics through the extended molecular and morphological context of the data. But the utility of OncoMaps goes further beyond the diagnostic aspect as they can concurrently help to gain better understanding of regulatory signaling mechanisms in different tumor niches.
Implications of all available evidence
Taken together ISS-based OncoMaps represent a useful tool for refined tumor diagnostics but also to extend our general understanding of the biological processes behind tumor progression and therewith guide the development of future therapies.
Alt-text: Unlabelled Box
1. Introduction
The concept of precision medicine within oncology has emerged over the last decade. Precision oncology addresses the need for molecular characterization of individual tumors to enable tailored treatment for each patient. However, increasing knowledge about intratumoral heterogeneity is challenging this concept, since multiple subclones with varied therapeutic sensitivity and/or phenotypic characteristics can exist within the same tumor [1], thus making the decision towards selection of a specific therapy more difficult. Furthermore, this highlights the need for spatially resolving techniques, which allow an in depth characterization of the different cellular niches and their signaling pathways within the tumor tissue in order to reveal information about the biology of their regulation.
Current breast cancer diagnostics relies on the combined evaluation of histopathology including tumor grade and immunohistochemical staining of ER, PR, HER2 (to be combined with ISH/FISH) and KI67. Additionally, complementary molecular analyses such as next generation sequencing, Mammaprint [2], OncotypeDX recurrence score [3] and PAM50 [4] are done on bulk cell lysates from homogenized tissues. Breast cancer displays both inter- and intratumoral genetic heterogeneity with thousands of different mutations and several copy number variations uniquely combined in each tumor [[5], [6], [7]], as well as subclonal variation with diverse genomic alterations [8]. Moreover, intratumoral heterogeneity of ER [9] and KI67 [10] expression is frequently observed in breast tumors, also heterogeneous ERBB2 (HER2) amplification in HER2-positive breast cancer [[11], [12], [13], [14]]. However, molecular analysis on bulk tissue only captures the average of all subclones within the tumor and patient stratification will be based on the largest clone present. Moreover, great genetic similarity has been observed between primary tumors and their metastasis, suggesting a late dissemination of metastatic cells from the primary lesion [[15], [16], [17], [18], [19]]. By identifying subclones with different transcriptomic profiles (although genetically similar) that could be associated with metastasis already in the primary tumor, we could gain novel insight in the biological mechanisms of invasion and metastasis. This knowledge could assist the development of future therapeutical strategies to prevent disease recurrence [20,21].
Thus, there is a clear need for spatially resolved tissue analytics in breast cancer to resolve both inter- and intratumoral heterogeneity and to address the significance and regulation of subclonal variations at system level. During the past few years, a number of methods has been developed for spatially resolved transcriptomics [22]. Some are untargeted transcriptome wide methods, where RNA in tissue is transferred to spatially barcoded slides, where spatial barcode sequences are incorporated during the cDNA synthesis step of next-generation sequencing library preparation [23,24]. The spatial resolution is limited by the feature size of the spatially barcoded slides, and the sensitivity is currently low (<0.1% capture efficiency). The strength is the hypothesis free generation of transcriptome-wide spatially resolved expression data. Other methods are targeted heavily multiplexed in situ methods that can be divided into unamplified single-molecule FISH (sm-FISH) based methods, and rolling-circle amplification (RCA) based methods. Multiplexing in sm-FISH is achieved by non-combinatorial labelling [25], combinatorial labelling [26,27], or a combination thereof [28]. The in situ based methods have sub-cellular resolution and expression data can directly be linked to morphology. The main advantage of sm-FISH methods is the high sensitivity, where close to 100% detection efficiency can be achieved. The main disadvantage is the low signal intensity of the fluorescence signals, which requires high-NA, high-magnification (60-100×) objectives, and thus slow imaging across tissues. Moreover, the fluorescence signals are typically lower than the autofluorescence level of human tumor tissues, which makes tumor samples less tractable for this method [29]. The main advantage of the RCA based methods is the strength of the amplified detection signal, which permits fast imaging using 20× magnification, and the signals stand out over the autofluorescence background even in tumor tissue [30]. Multiplexing is achieved by carrying out next-generation sequencing chemistry within the preserved tissue context [31]. This in situ sequencing (ISS) method has been demonstrated for multiplexed expression profiling in breast and prostate tumor tissues [31,32], for immune profiling of heavily crosslinked tuberculosis infected mouse lung sections [33], and for lineage tracing in mouse brain by reading virus encoded cellular barcodes in situ [34].
Here we describe an analytical approach based on ISS [31] that allows highly multiplexed and spatially resolved gene expression analysis. For each sequencing read, x and y coordinates are generated that correspond to its location within the tissue, thereby coupling molecular features directly to tumor morphology.
2. Materials and methods
2.1. Tissue specimens
Fresh frozen tissue sections (5 μm) from nine breast cancer tumors, collected at the Erasmus MC in Rotterdam were used for gene expression profiling. A protocol for studying biological markers associated with disease outcome was approved by the medical ethics committee of the Erasmus Medical Center Rotterdam, The Netherlands (MEC 02.953). The present study, in which coded tumor tissues were used, was performed in accordance with the Code of Conduct of the Federation of Medical Scientific Societies in the Netherlands (https://www.federa.org/codes-conduct). Tumors were from lymph node negative patients which did not receive systemic adjuvant treatment (clinical data is summarized in Table S1).
2.2. In situ sequencing (ISS)
ISS was performed as described previously [31] using a padlock-probe panel described below. Briefly, tissue sections were mounted onto Superfrost Plus slides (ThermoFisher Scientific), fixed in 4% PFA for 45 min and permeabilized with 0.1 mg/ml pepsin (Sigma) in 0.1 M HCl 37 °C for 90 s. SecureSeal™ reaction chambers were mounted on top of the tissues or cells (Grace Biolabs, Bend, United States). cDNA was synthesized in situ using random decamer primers (IDT, Leuven Belgium). Single-stranded cDNA was created through Rnase H cleavage and padlock probes were hybridized and ligated, followed by rolling circle amplification (RCA). For ISS, the padlock probes were equipped with short unique barcode sequences that become clonally amplified in the RCA products. The RCA products were then identified and sequenced by ligation (according to [35]) in several cycles using fluorophore-labelled interrogation probes (sequences are listed in Table S2). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). There were some changes compared to the published protocol [31], in addition to the specific base stain, every sequencing cycle was also stained with a general anchor stain (coupled with AF750) that was used for alignment of the sequencing cycles. Images were acquired with an automated Zeiss Axioplan II epifluorescence microscope (Zeiss, Oberkochen, Germany) using a z-stack of 0.49 μm × 5 and a tile overlap of 15%. Images were scanned with a 20× objective and the exposure times DAPI: 1 ms, FITC: 272.5 ms, Cy3: 178.6 ms, Cy5: 385.6 ms, Texas Red: 103.1 ms and AF750: 700 ms. Orthogonal projections and stitching of tiles were done with the ZEN software (Zeiss). Images from the respective sequencing cycles were aligned and decoded using Cellprofiler v.2.1.1 (Broad Institute, MA, United States) and an in-house Matlab script (Mathworks, Sweden). Quality scores ranged from 0.33–0.4, unexpected and homomer reads ranged from 3.7–9.1 and 0.7–6.6% of total reads, respectively. After the ISS analysis, the tissue sections where stained with hematoxylin (Sigma) or PanCK antibody.
2.3. Gene expression profiling
In total 91 genes were selected for expression profiling in situ, including the OncotypeDX scoring [3], molecular subtyping [36,37] and genes involved in proliferation, DNA repair, EMT, invasiveness, stemness and angiogenesis. Also targets for stromal and immunological features were included (Table S3). One padlock probe per target was designed, sequences are listed in Table S3 (IDT, Leuven Belgium).
2.4. Microarray and SNP data
Microarray and SNP data were previously generated from the nine tumors collected at Erasmus MC, Rotterdam. Microarray data was from the Affymetrix Human Genome U133A 2.0 Array [2,38] (GSE2034, GSE5327) and SNP data from the Affymetrix GeneChip Human Mapping 100 K Array [39] (GSE10099). GEO numbers are listed in Table S4. The data was generated before sections were prepared for our ISS analysis, thus the tissue parts used for microarray and SNP calling could vary.
2.5. Correlation of consecutive sections and ISS data to microarray data
ISS data and microarray data were log2-transformed and correlations between the data sets were tested using Pearson's correlation. In order to enable log2-transformation of the dataset in the presence of zero values for certain genes, “1” was added as a constant to all values of the dataset. Missing genes in the corresponding microarray data for the tumors were NANOG, POU5F1 and ZNF703 and these were excluded from the analysis.
2.6. Immunohistochemistry of PR, KI67 and PanCK
Immunohistochemistry (IHC) of PR and KI67 were performed on 5 μm sections consecutive to the ones used for ISS. PanCK was performed on the same section subjected to ISS. Slides were fixed for 10 min with 3.7% Formaldehyde. Peroxidase-blocking solution (DAKO REAL) was applied for 15 min, followed by incubation with serum-free protein blocking solution (DAKO) for 30 min. Sections were incubated with mouse monoclonal KI67 (clone MIB1) or PR (clone PgR 636) antibody (both DAKO) or PanCK antibody (clone AE1/AE3, DAKO) diluted 1:100 in DAKO REAL antibody diluent overnight at 4 °C. Thereafter secondary ImmPRESS HRP Anti-mouse IgG (VectorLaboratories) was applied for 30 min and chromogenic visualization was performed with DAB Peroxidase substrate kit (Vector laboratories) according to the manufacturer's instructions. Slides were counterstained with Mayer's HTX Hematoxylin (Histolab, Goteborg, Sweden) and mounted. PR and KI67 stained IHC images were further aligned to the H&E stained images from the ISS experiments.
2.7. ISS-based molecular subtyping
For subtyping of tumors we considered the gene expression of ER (ESR1), PR (PGR), HER2 (ERBB2), KRT5/6/8, KI67 (MKI67) and EGFR. These genes were used to approximate the breast cancer molecular subgroups according to the following criteria, luminal A-like (ER+ and/or PR+, HER2-, KI67 low and KRT8+), luminal B-like (ER+ and or PR+, HER2-, KI67 high and KRT8+), HER2-positive (ER−/+, PR−/+ and HER2+) and triple negative breast cancer (TNBC) (ER-, PR-, HER2-, EGFR+ and/or KRT5/6+). Subtyping was performed on whole tissue sections or on binned data (tissue scans were divided into hexagon bins, 500 μm in diameter). ISS data, from whole tissue sections or bins, were normalized to the sum of all reads for each tumor/bin. Based on the microarray data, thresholds were set to distinguish between ER/PR/HER2-positive or negative tumors as well as KI67, KRT5/6/8 and EGFR high or low tumors (Fig. S1). Bins with <100 total ISS reads and < 10 EPCAM/CDH1 reads were excluded from the analysis.
2.8. OncotypeDX recurrence scoring in situ
The 21 genes included in the OncotypeDX recurrence scoring were reference genes (ACTB, GAPDH, RPLP0, GUSB and TFRC), proliferation markers (MKI67, AURKA, BIRC5, CCNB1 and MYBL2), genes associated with invasiveness (MMP11 and CTSL2), ER hormone receptor status (ESR1, PGR, BCL2 and SCUBE2), HER2 status (ERBB2 and GRB7) and ungrouped (GSTM1, CD68 and BAG1) [3,40]. OncotypeDX recurrence scoring in situ was based on the published algorithm [3] with some modifications. Count data was first log-transformed using the rlog function of the DESeq2 package from within the R statistical environment. Logged expression values for each gene were then normalized to the mean value of the five reference genes. Normalized data was further used to calculate scores for each of the four gene groups “HER2”, “ER”, “proliferation” and “invasion” and combined with the normalized expression values of the remaining three ungrouped genes. The final unscaled recurrence score was scaled to a value between 0 and 100 and divided into three groups (low risk; RS < 18, intermediate risk; RS ≥ 18 and < 31, high risk; RS ≥ 31). OncotypeDX recurrence scoring was performed on whole tissue scans and scans divided into hexagonal bins (500 μm in diameter). Bins with <100 total ISS reads and < 10 EPCAM/CDH1 reads were excluded from the analysis.
2.9. Gene expression analysis of selected regions
For regions selected based on gene markers or morphology, each read count was normalized to the total read count for that region. Genes in the whole tissue with a read count of <0.02% of total read counts were excluded from the analysis. Normalized region-specific expression was compared to the normalized expression of another region (markerlow for all of the marker-based regions and tumor core for the two morphology-based regions). The most differently expressed genes were the genes with highest fold change in the respective comparison. Specific gene expression patterns were illustrated with Kernel density estimation using a bandwidth of 200–300 μm. ISS expression fold changes of MKI67 and PGR in the morphology-based regions of sample 880356 were further validated by IHC (Fig. S2).
2.10. Trendsceek
Trendsceek identifies spatial gene expression patterns through a nonparametric approach that uses marked point processes. The method tests for significant dependency between the spatial distribution of cells (points) and their gene expression levels (marks) [41]. The tissue scans were divided into hexagon bins (100 μm in diameter) and analysed using Trendsceek. Zero values for certain genes were corrected through adding “1” as a constant to all values of the dataset. Genes having at least 3 bins with a minimum read count of 5 were included in the analysis. The Benjamini-Hochberg correction for multiple comparisons was performed and alpha value was set to 0.01. Genes tested for spatial gene expression trends were CXCL12 and CXCL14 for tumor section 870556 and 900196_2, CTSL2 and MMP11 for 880356 and 900196_1 as well as VAV1 and XBP1 for 880842 and 930492.
2.11. Dimensionality reduction of ISS data by tSNE analysis
The tissue scans were divided into hexagonal bins (100 μm in diameter) and read counts were extracted from each bin. The genes ACTB, GAPDH, GUS1, TFRC, RPLP0 (reference genes) and COL3A1 (highly expressed in all tumors) were excluded from the analysis, also genes with a read count <0.02% of total read count, as well as bins with <3 total ISS reads were omitted. tSNE was used for dimensionality reduction of the ISS binned gene expression data into three dimensions and translated into RGB colors.
3. Results
3.1. Spatially resolved gene expression profiling by in situ sequencing (ISS)
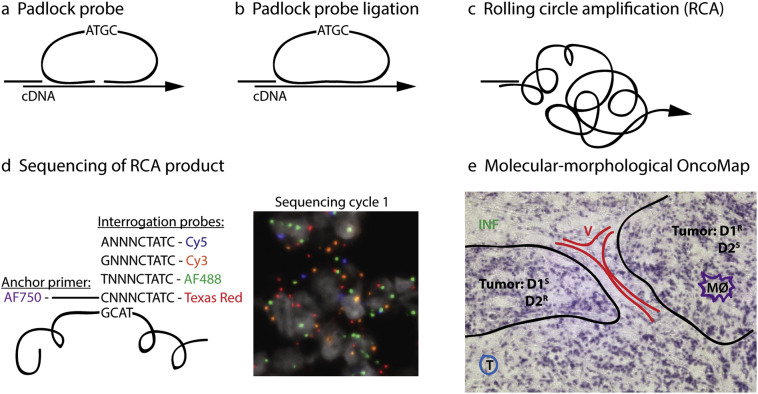
We used the ISS method to spatially resolve the expression of 91 genes simultaneously in breast cancer tumors, thereby generating molecular-morphological maps of the tumor tissues. The method employs target-specific DNA barcoded padlock probes to detect mRNA in fixed tissue and has been described previously [31]. In brief, after a reverse transcription of mRNA in situ, the padlock probes are hybridized to their specific targets and circularized through ligation. Circularized padlock probes are amplified by rolling circle amplification (RCA). Target identification is achieved through sequencing of the barcodes in the RCA product using sequencing by ligation chemistry and fluorophore-labelled interrogation probes. Stepwise, an anchor primer is hybridized next to the barcode sequence and extended by ligation with one of four random nonamer interrogation probes, labelled with a fluorophore according to the base of the barcode position that will be sequenced. Sequencing by ligation is performed over four cycles until all bases of the barcodes are sequenced and the barcode can be decoded and matched to its target (Fig. 1).
Fig. 1.
Gene expression profiling by in situ sequencing. Tissue sections were fixed and RNA was reversely transcribed in situ. (a) Padlock probes carrying target-specific barcode sequences were hybridized to the cDNA and (b) circularized by ligation. (c) Amplification of the circularized padlock probe was done using rolling circle amplification (RCA). The generated RCA products were subjected to (d) sequencing by ligation to retrieve the barcode information. Therefore, an anchor primer is hybridized next to the barcode sequence and extended by ligation with one of four random (N) nonamer interrogation probes, labelled with a fluorophore according to the base of the barcode position to be sequenced. Sequencing by ligation is performed over four cycles until all four bases of the barcode are sequenced and the barcode can be matched to its target (e) A molecular-morphological map (OncoMap) is generated through combination of the molecular ISS data with the histological context. Morphological tumor features are outlined based on hematoxylin staining of the same tissue section. D1/2 = drug 1/2, S = sensitive clones, R = resistant clones, V = vessel, T = T-cells, INF = inflammation and MØ = macrophages.
Selected gene targets included prognostic and predictive markers for breast cancer such as molecular subtyping, OncotypeDX recurrence scoring, and genes involved in proliferation, DNA repair, EMT, invasiveness, stemness and angiogenesis (Table S3). Also targets for stromal and immunological features were included. Here we describe different approaches for analysing ISS data, such as spatially resolved recurrence scoring/subtyping, molecular marker- and morphology-driven characterization of specific regions, as well as tSNE dimensionality reduction [42] for visualization of tumor heterogeneity. Hematoxylin stains of all tumor sections subjected to ISS can be visualized in Fig. S3.
3.2. Validation of the ISS gene expression panel
In order to validate the performance of the ISS gene expression panel, we tested the panel for its reproducibility and correlation with microarray data. For two tumors (860502 and 900196), the reproducibility of ISS was tested by analysis of two non-consecutive sections from the same tumor (referred to as _1 and _2, respectively), showing high agreement (Pearson's R = 0.93 and 0.96 respectively, Fig. S4). For all genes in the gene expression panel, total ISS read counts (similar to bulk measurements) were compared to microarray data from the same tumors. Pearson's R values ranged from 0.46 to 0.69, only one sample displayed values R < 0.50 (Table S5).
3.3. Establishment of ISS-based molecular subtyping and OncotypeDX recurrence scoring
Intrinsic subtyping (according to [36]) of microarray data was previously done on the tumors subjected to ISS [43]. In total, four of the tumors were classified as luminal A-like, one as luminal B-like, two as HER2-positive and two as TNBC (Table 1). In an initial step, ISS-based molecular subtyping using the expression of ER (ESR1), PR (PGR), HER2 (ERBB2), KRT5/6/8, KI67 (MKI67) and EGFR was done with total read counts of ISS data (similar to bulk measurements) through comparison with the available microarray data. The thresholds for each of the molecular subtyping targets were set guided through correlation scatter plots with microarray and ISS data from each tumor, applying the intrinsic subtype information as a guide to define thresholds for negative/positive target expression to the ISS data (Fig. S1).
Table 1.
Comparison of protein, microarray and ISS spatial data with regards to molecular subtype.
Comparison between ISS (blue) and previously acquired microarray (MA, green), and histological protein stain (purple). Discrepancies between the data sets are highlighted in grey. For the ISS subtype column, / indicates the various subtypes detected. TNBC = triple negative breast cancer.
Of note, gene expression based on total ISS read counts correlated very well with the microarray data for all subtyping genes (Pearson's R > 0.84) except for PR, that did not correlate (Pearson's R = 0.47) (Fig. S1). However, when comparing the microarray data with the spatially resolved ISS data (described in detail in the next section) and the histological protein stain, three tumors that were negative for PR with microarray data were positive with ISS (positive in <30% of the tumor) and in the protein staining (860502, 880842 and 930492, Table 1). Discrepancies were seen for two tumors (870556 and 880356) that were positive for PR with ISS (in 81% and 31% of tumors, respectively) but negative with both microarray and histological protein stain. Therefore, PR expression was additionally tested by IHC on tissue sections consecutive to the ones used for ISS. The staining was in agreement with the ISS data and indicated that the two differentiating tumor tissues were highly heterogeneous in their distribution of PR positive cells (Fig. S5), which could explain the discrepancies to the microarray and protein data. Additionally, one tumor (870556) was ER positive (8%) only with ISS and two tumors (900196 and 900317) were positive for ER with microarray and ISS data but not in the histological protein stain (Table 1).
OncotypeDX recurrence scoring includes expression of 21 genes divided into four functional groups, the HER2, ER, proliferation and invasion gene groups. Based on the published algorithm [3] with some minor adjustments for ISS data (see Materials and Methods for detailed description), each gene group was scored and the recurrence score was then calculated. The OncotypeDX score based on total ISS read counts (similar to a bulk measurement) agreed with RT-qPCR based scoring previously done on the same tumors (Table S6). Six of the tumors displayed a high RS, one tumor showed an intermediate score and two tumors low RS (Table S6).
Overall, the results indicate that ISS was well suited for tumor subtyping and OncotypeDX recurrence scoring.
3.4. Diagnostic and prognostic heterogeneity observed within tumors
As a next step, we aimed to investigate the diagnostic and prognostic heterogeneity within tumor tissues with regard to molecular subtype and RS score by ISS. We therefore divided the tumor scans into large hexagonal spatial bins (500 μm in diameter) to ensure a sufficient number of ISS reads of subtyping and OncotypeDX recurrence scoring related genes within each bin. The same thresholds as determined based on total ISS read counts were applied to the subtyping genes in each bin (Fig. S1 and S6).
Four tumors displayed intratumoral heterogeneity, where different areas of the same tumor were classified into different subtypes (Fig. 2a and Fig. S6) due to regionally high expression of KI67 (MKI67, tumor 880356 and 860502), HER2 (ERBB2, tumor 880842) or the hormone receptor PR (PGR, tumor 930492) (Fig. 2b). The two tumors showing heterogeneity of luminal A and B regions, also displayed a heterogeneous distribution of KI67 positive cells by immunohistochemical staining (Fig. 5 and Fig. S7). Interestingly, one tumor classified as TNBC in the microarray bulk subtyping approach showed PR positive bins that correlated with the PR positive histological protein staining (tumor 930492, Table 1 and Fig. S6). A summary of the comparison between ISS, microarray and histological protein data is given in Table 1. Of note, for one tumor only (900196), the subtyping intratumoral heterogeneity did not fully relate with the observed ISS KI67 expression levels and the IHC stain done on a consecutive section (Fig. S8).
Fig. 2.
Intratumoral heterogeneity with regards to subtype within breast cancer tumors. Tumors were divided into hexagonal bins (500 μm in diameter) and subjected to molecular subtyping. a) Images illustrate the four tumors displaying subtype heterogeneity. Yellow = luminal A bins, magenta = luminal B, cyan = HER2-positive and grey = triple-negative breast cancer (TNBC). The colored bins are overlaid on the hematoxylin stain. b) Kernel density maps showing expression patterns of four subtyping genes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
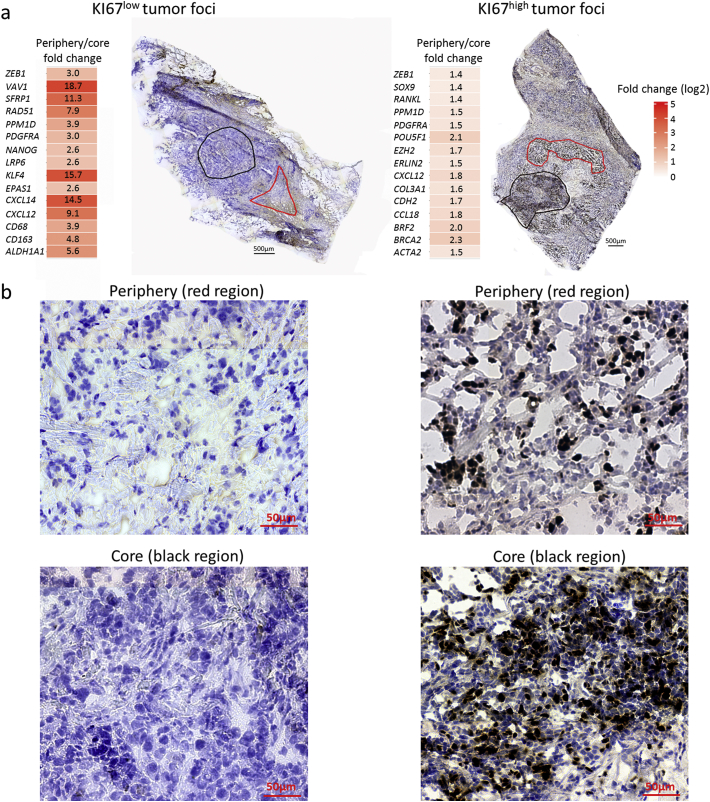
Fig. 5.
Characterization of morphologically relevant regions. (a) Regions selected for molecular profiling are outlined in red on top of the KI67 immunohistochemistry stain. Log2-transformed fold changes of the 15 most upregulated genes in each region are displayed with heat maps, non-transformed fold changes are indicated. Left, comparison of the peripheral region (red outline) and core (black outline) of a KI67low tumor foci (tumor 880842). Right, comparison of the peripheral region (red outline) and core (black outline) of a KI67high tumor foci (tumor 880356). (b) Enlarged images of the morphology in each region. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
With regard to intratumoral heterogeneity, four tumors showed different RS scores within the tumor (Fig. 3a). In three cases, high RS scores were associated with less ER gene group expression (t-test p < .05) and in one case with increased proliferative (t-test p < .05) or increased invasive (t-test p < .05) gene groups (Fig. 3b and c).
Fig. 3.
OncotypeDX recurrence score heterogeneity within breast cancer tumors. Tumors were divided into hexagonal bins (500 μm in diameter) and subjected to recurrence scoring. a) Images illustrate four tumors displaying OncotypeDX recurrence scoring heterogeneity. Colour-coded bins are overlaid on the hematoxylin stain. b) Mean of OncotypeDX gene group scores (HER2-, ER-, proliferation- and invasive group) in bins with low, medium or high recurrence scores, error bars represent standard deviation (t-test). c) Distribution of ISS reads for the OncotypeDX gene groups, plotted as dots in their spatial location within the tissues. Colour of dots represents: black = HER2 group, green = ER group, blue = proliferation group and red = invasive group. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.5. Specific gene expression patterns with inter- and intratumoral heterogeneity
Since we observed intratumoral heterogeneity of diagnostic and prognostic gene profiles, we next examined if this was also reflected for other molecular markers such as genes involved in immune signaling, invasiveness and chemokine signaling (Table S3). Indeed, we observed both inter- and intratumoral heterogeneity in the spatial expression of specific genes, e.g. VAV1, XBP1, CTSL2, MMP11, CXCL14 and CXCL12 (Fig. 4a, verified by Trendsceek p < .01, that identifies spatial gene expression trends [41] and Methods). For instance, specific genes were expressed within the tumor compartment for some samples, whereas other samples displayed expression of the same gene mainly in the stroma or at the tumor/stroma boundary. This is illustrated in Fig. 4a, showing spatial expression patterns of the six genes in relation to the tumor compartment, demonstrated by EPCAM/CDH1 spatial expression. For three of the genes (XBP1, MMP11 and CXCL14), regions with high gene expression were selected and their respective expression patterns and morphological features were evaluated. Gene expression analysis of the selected regions revealed increased expression of BRCA1 and decreased expression of KRT8, VEGFA, MET, CLDN3 and three genes involved in EMT (CTSL2, KLF4 and MMP11) for the XBP1high region. The MMP11high region showed enrichment of CCL18 and two genes involved in DNA repair (EMSY and RAD51) as well as less expression of PDGFRB, BCL2 and KRT5. The CXCL14high regions showed gene expression characteristic of SFRP1, GRB7, CDH2 and TNF (Fig. 4b). Morphologically, the XBP1high region contained mostly immune cells, the MMP11high region fibroblast and immune cells, and the CXCL14high region a mixture of epithelial, fibroblast and immune cells (Fig. 4c).
Fig. 4.
Heterogeneous expression of specific genes across tumor samples. (a) Presentation of selected genes that showed distinct expression patterns across tumor sections. Top; Kernel density maps of the selected genetic marker presented in red or magenta. Bottom; Kernel density maps of EPCAM/CDH1 expression displayed as grey to indicate the epithelial region within the section. (b) For XBP1, MMP11 and CXCL14 specific regions were selected based on the Kerneldensity map displayed in (a). The outlined regions (markerhigh) are shown on top of the PanCK IHC stain. ISS data was retrieved from these regions and compared to the rest of the tissue (markerlow). Log2-transformed fold changes of the 10 most differently expressed genes for each selected region are displayed with heat maps, non-transformed fold changes are indicated. Red squares indicate regions shown in (c). (c) High power views of the morphology in each region. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.6. Molecular profiling of morphologically relevant regions within tumor samples
Then, we proceeded from a histological perspective by identifying morphological features, and characterized their molecular profiles. Current breast cancer diagnostics largely relies on histological evaluation of tumors [37]. Hence, we investigated if morphological phenotypes could be correlated to their molecular characteristics. For this, two tumor regions selected from the core and the periphery of a tumor foci were characterized. This analysis was performed on two samples (samples 880842 and 880356) which differed strongly in their number of KI67 positive cells as measured by immunohistochemistry (Fig. 5a and b). Molecular analysis revealed, that within the KI67low sample (880842), the periphery of a tumor foci showed increased expression of PPM1D, VAV1, SFRP1, LRP6 and genes involved in stemness (ALDH1A1 and NANOG), EMT (KLF4 and ZEB1), macrophages (CD163 and CD68) and chemokine signaling (CXCL12 and CXCL14) as compared to the core region. In contrast, within the KI67high sample (880356) the periphery of a tumor foci displayed an increase in expression of POU5F1, BRF2 and BRCA2 (Fig. 5a) as compared to its core region. Morphologically, the peripheral areas of both tumor foci was enriched in stromal cells and extracellular matrix with clusters of epithelial cells migrating from the core out into the surrounding tissue, whereas the core regions for both tumors showed very dense epithelial cells (Fig. 5b).
3.7. Tumor heterogeneity visualized using tSNE analysis
We continued to explore the ISS data with an unsupervised approach using tSNE cluster analysis [42]. With this approach, heterogeneity within the tissue could be visualized through clustering into distinct colour schemes. Tumor scans were divided into small hexagonal spatial bins (100 μm in diameter; required size to obtain sufficient read number for subsequent tSNE) and tSNE dimensionality reduction was performed on the binned ISS gene expression data for each tumor. The data were reduced into three dimensions and translated into RGB colors where bins with similar gene expression profiles get similar colors (Fig. S9). Examples for four tumors are displayed in Fig. 6, showing the tSNE clustered and colored bins plotted in their original location within the tissue section. In general, two main shades for each tumor were identified, one shade corresponded to the epithelial part of the tumor tissue section whereas the other corresponded to the stromal part. Many tSNE clusters were defined by the expression of one specific gene, although most genes were not expressed in a single cluster (Fig. S10, S11 and S12). Expectedly, the tumors showed similar heterogeneous spatial patterns as observed in the previous analyses, where tumors 880356 and 880842 had indicated distinct prognostic intratumoral heterogeneity (Fig. 3) and tumor 930492 a distinct XBP1high region (Fig. 4), which is seen as a dark blue/green region in the tSNE analysis.
Fig. 6.
Intratumoral heterogeneity visualized by tSNE analysis. The tumors were divided into small hexagonal bins (100 μm in diameter). tSNE dimensionality reduction into three dimensions was performed on each bin and translated into RGB colors, bins with similar colors have similar gene expression patterns. For visualization of tumor heterogeneity and gene expression patterns, the tSNE clustered and colored bins were plotted in their original location within the tissue.
3.8. Tumor-specific OncoMaps generated by ISS
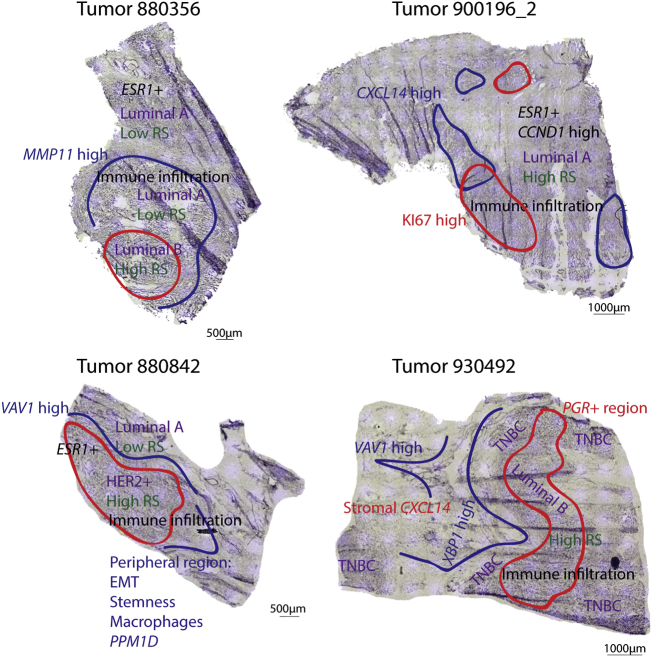
To summarize the information acquired, findings from the different analysis approaches were combined to generate a molecular-morphological map of the tumor tissue (OncoMap). OncoMaps were generated for four tumors, displaying the spatial intratumoral heterogeneity with regards to subtype, RS and specific expression patterns, thereby enabling a detailed map of each tumors' molecular characteristics and possible variation in therapeutic sensitivity. For example, tumor 900196, classified as an ER+ luminal A-like tumor with bulk RNA analysis, displayed KI67 and CXCL14 intratumoral heterogeneity as well as generally high expression and amplification of CCND1 (Fig. S13), associated with poor prognosis in ER+ tumors [44]. This patient showed metastases 37 month after surgical treatment for the primary tumor (Table S1). Tumor 930492, a medullary carcinoma, displayed an immunosupressive/ER stress response region (XBP1high [45,46]) at the tumor/stroma boundary, stromal CXCL14 (Fig. S14, associated with shorter survival in breast cancer [47]) and subtype intratumoral heterogeneity due to regionally high expression of PR (PGR). This tumor developed metastases after 9 months (Table S1). For the tumors, 880356 and 880842, specific regions with higher RS and a more aggressive subtype were identified (Fig. 7). Tumor 880842 presented with metastasis 7 months after primary surgery (Table S1).
Fig. 7.
Molecular-morphological OncoMaps. Findings from the different analysis approaches were summarized for each tumor and assigned to the histological context. Spatial intratumoral heterogeneity with regards to OncotypeDX recurrence score (RS; green), tumor subtype (magenta) and specific gene expression patterns (red and blue) as well as histological observations (black) are displayed on top of the hematoxylin stain. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Of note, immune cell related targets were commonly enriched in peritumoral stroma areas.
4. Discussion
Current applications of precision medicine in cancer are mainly informed by the results of bulk molecular profiling. However, the clinical benefits derived till date based on this bulk approach remain relatively limited [48,49]. This could possibly be explained by the lack of morphologically and subclonally resolved molecular profiling approaches. Extended information that correlates both the molecular and morphological information at various levels would be practically helpful to bring these studies closer to clinical relevance.
We have developed an analytical method that couples highly multiplexed gene expression profiling to the morphological features of the tumor tissue. Here we present different approaches for analysing this type of data and how to spatially assemble all information together within a so-called OncoMap of each tumor. These OncoMaps reveal intratumoral heterogeneity in the expression of molecular markers with regard to subregions of different tumor subtype, recurrence score or gene expression profile directly within the tissue context. Thereby, OncoMaps offer the unique feature that they can complement cancer diagnostics and at the same time deliver important information about the underlying biological processes within the different tumor niches.
From a diagnostic point of view, heterogeneity with regards to molecular tumorsubtype could have great impact on patient outcome, due to possible variations in therapeutic sensitivity within the tumor. For instance, we identified a tumor with a region positive for PR (PGR) resulting in a different subtype, which is in accordance with the histological protein stain but missed out with microarray bulk RNA analysis approach. Furthermore, we observed intratumoral subtyping heterogeneity due to regionally high expression of KI67 (MKI67). Recently, heterogeneity with regards to presence of both ER positive and negative regions within a tissue, was associated with worse long-time survival in breast cancer, also for patients with luminal A tumors that are commonly considered to be the least aggressive of the invasive breast tumors [50]. This finding demonstrated the importance of diagnostic approaches that take heterogeneity into account. Of note, most clinical routine approaches already score prognostic KI67 as percentage of positive tumor cells within multiple hot spot areas of the tumor tissue [51,52].
The intratumoral prognostic heterogeneity we observed by spatial mapping of the OncotypeDX RS, indicates that the scoring could differ depending on the tumor part analysed, and that smaller regions with potentially aggressive features could be missed in bulk tissue processing.
Besides the diagnostic aspects, ISS-based OncoMaps also deliver relevant links between histological and molecular tumor features and can thereby help to gain better understanding of regulatory signaling mechanisms in different tumor niches. For example, we demonstrate molecular profiling of clinically-relevant regions exemplified by in depth analysis of the core and periphery of tumor foci, thus enabling a more distinct characterization of potentially metastatic molecular profiles. This approach can extend our general understanding of the biology behind tumor invasion and metastasis, as well as potentially guide development and selection of targeted therapies to prevent disease recurrence.
Our OncoMaps assign molecular profiles to different stromal, immune or tumor compartments of the tissue, enabling identification of differences in specific gene expression patterns both within and between tumor samples. Indeed, we observed both inter- and intratumoral heterogeneity with regards to the spatial expression of specific genes, as exemplified for three genes generally associated with poor prognosis (Fig. 4). Being able to spatially characterize and define the expression of biomarkers should be advantageous for improving both the prognostic capability of molecular analysis as well as therapeutic strategies. For example, it has been shown that increased stromal, but not epithelial, CXCL14 expression is associated with shorter survival in breast cancer [47]. And indeed, we observed stromal CXCL14 expression in the tumor stroma of one patient with poor outcome (tumor 930492, Fig. S14 and Table S1).
A limitation of the presented proof-of-concept study is though the relative low number of analysed breast cancer patients. In order to correlate features of ISS-based OncoMaps to clinical outcome or treatment response, follow-up studies need to be performed on larger clinically well-annotated patient cohorts. Such studies will also offer the opportunity to develop novel algorithms needed to connect multi-layer biomarker signatures to clinical endpoints. Nevertheless, for future high-throughput analyses with minimal technical variation as well as for integration into clinical pathology routines, the ISS assay needs to be fully automatized combining ideally both fluidics and optics systems. Current efforts are heading in this direction and will be interesting to follow [22].
We further demonstrate the use of an unsupervised approach, tSNE dimensionality reduction, for analysing spatial gene expression data that allows visualization of heterogeneous gene expression patterns. In a large sample cohort, such an approach could possibly be useful for identifying tissue areas of common prognostic and predictive gene signatures or markers of biological processes within different patients. Generally, the spatial intratumoral heterogeneity observed with other analysis approaches could also be visualized using tSNE analysis. However, some findings were not picked up by the unsupervised approach, for example the CXCL14 intratumoral heterogeneity observed in tumor 900196 (Fig. 4, Fig. 6), indicating that the use of several different analysis approaches might be advantageous.
In conclusion, we provide the first proof of principle application of spatially resolved molecular profiling by ISS-based OncoMaps. Through spatial mapping of heterogeneity in the expression of therapeutic sensitivity and prognostic markers, our approach can complement diagnostic information and potentially improve patient stratification. Simultaneously, OncoMaps can deliver corresponding gene expression information on system level to clinically relevant tumor regions and thereby offer new opportunities to gain a better insight of the biological processes regulating tumor progression.
Funding sources
JS, CS, XQ, KJCZ and MN were funded by Swedish Cancerfonden, (150838), the strategic research area UCAN, and Vetenskapsrådet (2014-3380). Cancer Genomics Netherlands financially sponsored co-author AMS. NPT was funded by Iris, Stig och Gerry Castenbäcks Stiftelse for cancer research and JB by BRECT, the Swedish Cancer Society, the Cancer Society in Stockholm-Personalised Cancer Medicine (PCM), the King Gustaf V Jubilee Foundation, the Swedish Breast Cancer Association (BRO), Karolinska Institutet and Stockholm County Council, and Alice Wallenberg Foundation.
Author contributions
JS designed the study, acquired, analysed and interpreted data, and drafted the article. CS acquired, analysed and interpreted data, and participated in drafting the article. XQ helped with analysis, interpretation and illustrations of data and wrote scripts for this purpose. KJCZ acquired, analysed and interpreted data. NPT developed the ISS-based OncotypeDX RS scoring. JB contributed with clinically relevant input on the study. AMS helped out with the initial study design and contributed with samples for ISS including matching microarray, RT-qPCRand SNP data. MN designed the study, interpreted the data and participated in drafting the article. All authors critically revised the article.
Declaration of Competing Interest
MN, ZK, JS and CS report grants from Swedish Cancerfonden, grants from strategic research area UCAN, grants from Vetenskapsrådet, during the conduct of the study. MN and XQ hold shares in Cartana AB, a company commercializing in situ sequencing reagents. JB's research group receives research funding from Merck paid to Karolinska Institutet and from Amgen, Bayer, Pfizer, Roche and Sanofi-Aventis paid to Karolinska University Hospital. No personal payments. Payment from UpToDate for a chapter in breast cancer prediction paid to Asklepios Medicine HB.
Acknowledgements
Dr. Nicola Crosetto for valuable comments on the manuscript. Members of the MN and AMS group including Prof. Dr. John A. Foekens and Dr. John WM Martens are acknowledged for all their support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.09.009.
Appendix A. Supplementary data
Supplementary material
References
- 1.McGranahan N., Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168(4):613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y., Klijn J.G., Zhang Y. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet (London, England) 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 3.Paik S., Shak S., Tang G. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 4.Parker J.S., Mullins M., Cheang M.C. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtis C., Shah S.P., Chin S.F. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Network C.G.A. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nik-Zainal S., Davies H., Staaf J. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534(7605):47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yates L.R., Gerstung M., Knappskog S. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med. 2015;21(7):751–759. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung G.G., Zerkowski M.P., Ghosh S., Camp R.L., Rimm D.L. Quantitative analysis of estrogen receptor heterogeneity in breast cancer. Lab Investig. 2007;87(7):662–669. doi: 10.1038/labinvest.3700543. [DOI] [PubMed] [Google Scholar]
- 10.Dowsett M., Nielsen TO, A'Hern R. Assessment of Ki67 in breast cancer: recommendations from the international Ki67 in breast cancer working group. J Natl Cancer Inst. 2011;103(22):1656–1664. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee H.J., Kim J.Y., Park S.Y. Clinicopathologic significance of the intratumoral heterogeneity of HER2 gene amplification in HER2-positive breast cancer patients treated with adjuvant trastuzumab. Am J Clin Pathol. 2015;144(4):570–578. doi: 10.1309/AJCP51HCGPOPWSCY. [DOI] [PubMed] [Google Scholar]
- 12.Lee H.J., Seo A.N., Kim E.J. HER2 heterogeneity affects trastuzumab responses and survival in patients with HER2-positive metastatic breast cancer. Am J Clin Pathol. 2014;142(6):755–766. doi: 10.1309/AJCPIRL4GUVGK3YX. [DOI] [PubMed] [Google Scholar]
- 13.Ng C.K., Martelotto L.G., Gauthier A. Intra-tumor genetic heterogeneity and alternative driver genetic alterations in breast cancers with heterogeneous HER2 gene amplification. Genome Biol. 2015;16:107. doi: 10.1186/s13059-015-0657-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seol H., Lee H.J., Choi Y. Intratumoral heterogeneity of HER2 gene amplification in breast cancer: its clinicopathological significance. Modern Pathol. 2012;25(7):938–948. doi: 10.1038/modpathol.2012.36. [DOI] [PubMed] [Google Scholar]
- 15.Brown D., Smeets D., Szekely B. Phylogenetic analysis of metastatic progression in breast cancer using somatic mutations and copy number aberrations. Nat Commun. 2017;8 doi: 10.1038/ncomms14944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demeulemeester J., Kumar P., Moller E.K. Tracing the origin of disseminated tumor cells in breast cancer using single-cell sequencing. Genome Biol. 2016;17(1):250. doi: 10.1186/s13059-016-1109-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meric-Bernstam F., Frampton G.M., Ferrer-Lozano J. Concordance of genomic alterations between primary and recurrent breast cancer. Mol Cancer Ther. 2014;13(5):1382–1389. doi: 10.1158/1535-7163.MCT-13-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel M.B., He X., Hoadley K.A. Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J Clin Invest. 2018;128(4):1371–1383. doi: 10.1172/JCI96153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ullah I., Karthik G.M., Alkodsi A. Evolutionary history of metastatic breast cancer reveals minimal seeding from axillary lymph nodes. J Clin Invest. 2018;128(4):1355–1370. doi: 10.1172/JCI96149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yates L.R. Intratumoral heterogeneity and subclonal diversification of early breast cancer. Breast (Edinburgh, Scotland) 2017;34(Suppl. 1):S36–s42. doi: 10.1016/j.breast.2017.06.025. [DOI] [PubMed] [Google Scholar]
- 21.Yates L.R., Desmedt C. Translational genomics: practical applications of the genomic revolution in breast cancer. Clin Cancer Res. 2017;23(11):2630–2639. doi: 10.1158/1078-0432.CCR-16-2548. [DOI] [PubMed] [Google Scholar]
- 22.Strell C., Hilscher M.M., Laxman N. Placing RNA in context and space - methods for spatially resolved transcriptomics. FEBS J. 2019;286(8):1468–1481. doi: 10.1111/febs.14435. [DOI] [PubMed] [Google Scholar]
- 23.Rodriques S.G., Stickels R.R., Goeva A. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science (New York, NY) 2019;363(6434):1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stahl P.L., Salmen F., Vickovic S. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science (New York, NY) 2016;353(6294):78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 25.Codeluppi S., Borm L.E., Zeisel A. Spatial organization of the somatosensory cortex revealed by osmFISH. Nat Methods. 2018;15(11):932–935. doi: 10.1038/s41592-018-0175-z. [DOI] [PubMed] [Google Scholar]
- 26.Chen K.H., Boettiger A.N., Moffitt J.R., Wang S., Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science (New York, NY) 2015;348(6233):aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lubeck E., Coskun A.F., Zhiyentayev T., Ahmad M., Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014;11(4):360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moffitt J.R., Bambah-Mukku D., Eichhorn S.W. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science (New York, NY) 2018;362(6416) doi: 10.1126/science.aau5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu C., Simonetti M., Rossell C. RollFISH achieves robust quantification of single-molecule RNA biomarkers in paraffin-embedded tumor tissue samples. Commun Biol. 2018;1:209. doi: 10.1038/s42003-018-0218-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grundberg I., Kiflemariam S., Mignardi M. In situ mutation detection and visualization of intratumor heterogeneity for cancer research and diagnostics. Oncotarget. 2013;4(12):2407–2418. doi: 10.18632/oncotarget.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ke R., Mignardi M., Pacureanu A. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods. 2013;10(9):857–860. doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- 32.Kiflemariam S., Mignardi M., Ali M.A., Bergh A., Nilsson M., Sjoblom T. In situ sequencing identifies TMPRSS2-ERG fusion transcripts, somatic point mutations and gene expression levels in prostate cancers. J Pathol. 2014;234(2):253–261. doi: 10.1002/path.4392. [DOI] [PubMed] [Google Scholar]
- 33.Carow B., Hauling T., Qian X., Kramnik I., Nilsson M., Rottenberg M.E. Spatial and temporal localization of immune transcripts defines hallmarks and diversity in the tuberculosis granuloma. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-09816-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X., Sun Y.C., Church G.M., Lee J.H., Zador A.M. Efficient in situ barcode sequencing using padlock probe-based BaristaSeq. Nucleic Acids Res. 2018;46(4):e22. doi: 10.1093/nar/gkx1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drmanac R., Sparks A.B., Callow M.J. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science (New York, NY) 2010;327(5961):78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 36.Perou C.M., Sorlie T., Eisen M.B. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 37.Senkus E., Kyriakides S., Ohno S. Primary breast cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26(Suppl. 5):v8–30. doi: 10.1093/annonc/mdv298. [DOI] [PubMed] [Google Scholar]
- 38.Yu J.X., Sieuwerts A.M., Zhang Y. Pathway analysis of gene signatures predicting metastasis of node-negative primary breast cancer. BMC Cancer. 2007;7:182. doi: 10.1186/1471-2407-7-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., Martens J.W., Yu J.X. Copy number alterations that predict metastatic capability of human breast cancer. Cancer Res. 2009;69(9):3795–3801. doi: 10.1158/0008-5472.CAN-08-4596. [DOI] [PubMed] [Google Scholar]
- 40.Cronin M., Sangli C., Liu M.L. Analytical validation of the Oncotype DX genomic diagnostic test for recurrence prognosis and therapeutic response prediction in node-negative, estrogen receptor-positive breast cancer. Clin Chem. 2007;53(6):1084–1091. doi: 10.1373/clinchem.2006.076497. [DOI] [PubMed] [Google Scholar]
- 41.Edsgard D., Johnsson P., Sandberg R. Identification of spatial expression trends in single-cell gene expression data. Nat Methods. 2018;15(5):339–342. doi: 10.1038/nmeth.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maaten L.J.P.V.D., Hinton G.E. 2008. Visualizing high-dimensional data using t-SNE; pp. 2579–2605. (9(Nov) [Google Scholar]
- 43.Smid M., Wang Y., Zhang Y. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68(9):3108–3114. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- 44.Roy P.G., Pratt N., Purdie C.A. High CCND1 amplification identifies a group of poor prognosis women with estrogen receptor positive breast cancer. Int J Cancer. 2010;127(2):355–360. doi: 10.1002/ijc.25034. [DOI] [PubMed] [Google Scholar]
- 45.Cubillos-Ruiz J.R., Bettigole S.E., Glimcher L.H. Molecular pathways: immunosuppressive roles of IRE1alpha-XBP1 signaling in dendritic cells of the tumor microenvironment. Clin Cancer Res. 2016;22(9):2121–2126. doi: 10.1158/1078-0432.CCR-15-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 47.Sjoberg E., Augsten M., Bergh J., Jirstrom K., Ostman A. Expression of the chemokine CXCL14 in the tumour stroma is an independent marker of survival in breast cancer. Br J Cancer. 2016;114(10):1117–1124. doi: 10.1038/bjc.2016.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Letai A. Functional precision cancer medicine-moving beyond pure genomics. Nat Med. 2017;23(9):1028–1035. doi: 10.1038/nm.4389. [DOI] [PubMed] [Google Scholar]
- 49.Pauli C., Hopkins B.D., Prandi D. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017;7(5):462–477. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindstrom L.S., Yau C., Czene K. Intratumor heterogeneity of the estrogen receptor and the long-term risk of fatal breast cancer. J Natl Cancer Inst. 2018;110(7):726–733. doi: 10.1093/jnci/djx270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leung S.C.Y., Nielsen TO, Zabaglo L. Analytical validation of a standardized scoring protocol for Ki67: phase 3 of an international multicenter collaboration. NPJ Breast Cancer. 2016;2 doi: 10.1038/npjbcancer.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yerushalmi R., Woods R., Ravdin P.M., Hayes M.M., Gelmon K.A. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol. 2010;11(2):174–183. doi: 10.1016/S1470-2045(09)70262-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material