Abstract
Background
The transcriptome of Plasmodium falciparum clinical isolates varies according to strain, mosquito bites, disease severity and clinical history. Therefore, it remains a challenge to directly interpret the parasite's transcriptomic information into a more general biological signature in a natural human malaria infection. These confounding variations can be potentially overcome with parasites derived from controlled-human malaria infection (CHMI) studies.
Methods
We performed CHMI studies in healthy and immunologically naïve volunteers receiving the same P. falciparum strain ((Sanaria® PfSPZ Challenge (NF54)), but with different sporozoite dosage and route of infection. Parasites isolated from these volunteers at the day of patency were subjected to in vitro culture for several generations and synchronized ring-stage parasites were subjected to transcriptome profiling.
Findings
We observed clear deviations between CHMI-derived parasites from volunteer groups receiving different PfSPZ dose and route. CHMI-derived parasites and the pre-mosquito strain used for PfSPZ generation showed significant transcriptional variability for gene clusters associated with malaria pathogenesis, immune evasion and transmission. These transcriptional variation signature clusters were also observed in the transcriptome of P. falciparum isolates from acute clinical infections.
Interpretation
Our work identifies a previously unrecognized transcriptional pattern in malaria infections in a non-immune background. Significant transcriptome heterogeneity exits between parasites derived from human infections and the pre-mosquito strain, implying that the malaria parasites undergo a change in functional state to adapt to its host environment. Our work also highlights the potential use of transcriptomics data from CHMI study advance our understanding of malaria parasite adaptation and transmission in humans.
Fund
This work is supported by German Israeli Foundation, German ministry for education and research, MOE Tier 1 from the Singapore Ministry of Education Academic Research Fund, Singapore Ministry of Health's National Medical Research Council, National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA and the German Centre for Infection Research (Deutsches Zentrum für Infektionsforschung-DZIF).
Keywords: Malaria, CHMI, Sporozoite, Transcriptome
Research in context.
Evidence before this study
The compendium of transcriptome datasets either from laboratory or clinical isolates showed divergent functional responses associated with variation in strain genotype, host pre-existing immunity and in vitro culture conditions. The functional importance of transcriptome variation during infection is unclear due to the underlying complexity in a natural human malaria infection, but is central to our understanding of parasite adaptation in the human hosts. Thus, the the establishment of controlled human malaria infection (CHMI) trials provide a unique experimental model to study parasite adaptation upon transmission and infection.
Added value of this study
Here, we identified a unique transcriptional signature of a single strain malaria infection in a non-immune background. Transcriptional changes between sporozoite route of infection (intradermal versus intravenous), sporozoite dose (high versus low) and before-after mosquito passage (CHMI-derived strain versus pre-mosquito strain) reveal intriguing functional gene clusters associated with host immune evasion and parasite transmission. We applied and showed significant association of the variable transcriptome component from this study into the transcriptome component of parasites obtained from acute human infections from Africa, Bangladesh, Mekong, Myanmar and Cambodia.
Implications of all the available evidence
Parasites derived from CHMI studies can yield novel insights into the adaptation process in different host environments through transcriptome profiling studies. We have identified the functional gene clusters with high transcriptional divergence in genetically identical parasites from non-immune background, likely conferring a fitness advantage to the parasites during infection. In this context, this study will also bring new concept into new CHMI-trials in semi-immune individual, where stronger selective pressures and more profound changes in parasite biology are likely to be observed.
Alt-text: Unlabelled Box
1. Introduction
Malaria parasites have a complex life cycle alternating between the intermediate vertebrate (human) and the definitive insect (mosquito) hosts. The asexual life cycle within the red blood cell (RBC) is responsible for the pathology within the host mainly due to development of anemia and the sequestration of infected RBC in the deep tissues leading to decreased blood flow, tissue anoxia and ultimately resulting in organ failure. Development of different severe malaria syndromes has been associated with the transcription and expression of parasite multigene families essential for binding to the vasculature endothelium and aggregate formation between RBCs in the vasculature [[1], [2], [3], [4], [5], [6]]. Stress and reproductive constraints can trigger sexual commitment and differentiation of late-stage asexual parasites into gametocytes that are transmissible to the mosquito vector [7,8]. Transmission fate is essentially governed by an Apicomplexan transcriptional factor; the ApiAP2-G, through the coordinated transcriptional activation of several sexually-commitment genes [9,10]. Once in the mosquito host, the parasites undergo another complex developmental cycle, leading to the production of motile sporozoites (SPZ) that are able to travel from the mosquito salivary gland into the human host dermis and capillaries during the ingestion of a blood meal. The SPZ undergo asexual reproduction in the liver resulting in the release of merozoites into the blood circulation which then initiates the asexual, symptomatic blood stage infection. All the changes the parasite undergoes during its complex life cycle are controlled by about 5000 protein-coding genes within the malaria parasite genome [11]. Transcriptional profiling of the malaria parasites has led to an enormous understanding of the malaria parasites biology showing that throughout the blood stage life cycle, it activates and represses each gene in a just-in-time fashion under in vitro culture condition [[12], [13], [14], [15]]. The transcriptional portrait of in vitro-adapted parasites however provides only a fraction of information on the overall parasite biology. Genome-wide transcriptomic studies on P. falciparum clinical isolates in fact suggested notable transcriptional differences between parasite samples from human infection and those obtained from long term culture adapted parasites [16,17]. However, the majority of these clinical isolates were derived from patients with pre-existing immunity, with variation in the number of mosquito bites and sporozoite disposition as well as mixed infection with multiple strains [[18], [19], [20], [21]]. Therefore, it remains a challenge to directly interpret the parasite's transcriptomic information into a more general biological signature due to the underlying complexity in a natural human malaria infection.
One approach to address this challenge is the establishment of controlled human malaria infection (CHMI) trials, where immunologically naïve human volunteers were inoculated either by intradermal (ID), intramuscular (IM) or direct venous inoculation (DVI) injection with purified PfSPZ generated from a defined P. falciparum strain. Such trials not only offer a new approach to test new clinical intervention approaches like vaccine and drug efficacy [22,23], but also provides a unique opportunity to gain at the same time a better understanding of parasite biology in a controlled and standardized environment. PfSPZ infections in non-immune populations using a defined P. falciparum strain can also overcome the inherent variation amongst field isolates, hosts and the impact of variations in PfPSZ inoculum injected from mosquito salivary glands [[24], [25], [26]]. Thus far, transcriptional profiling of parasites derived from CHMI individuals, either immediately after patency period [27,28] or subjected to in vitro cultures [25,29,30] were limited to the var multigene repertoires using quantitative PCR analysis. More recently, transcriptome profiling analysis of whole blood from 10 adults following CHMI with PfSPZ Challenge suggested early key transcriptional changes during pre-erythrocytic stage [31]. Here, we report the global transcriptome of the erythrocytic stage malaria parasites obtained from the dose escalation CHMI of Sanaria® PfSPZ Challenge (TÜCHMI-001-NCT01624961) derived from a single P. falciparum NF54 (PfNF54) working cell bank [25]. Our work provides information on the transcriptome dynamics of the human malaria parasites, and exemplifies the selective transcriptome variation in a controlled-human infection setting. We have explored the variable transcriptome of the CHMI-derived samples and show that it is influenced by the duration of intrahost replication and the duration of in vitro culture progression. We also further distinguish the transcriptional differences between parasites from CHMI and pre-mosquito PfNF54 transcriptome, suggesting that the human malaria parasites reprogram expression pattern for a subset of genes in the blood stage ensuing mosquito passage and human infection. Importantly, we recapitulated the transcriptionally variable genes from CHMI-derived parasites in field isolates transcriptome from patients with acute malaria infection, which circumvent the issue related in vitro culturing phase. Our work here identifies a previously unrecognized transcriptional pattern of a single strain malaria infection in a non-immune background and deconvolutes the transcriptome complexity of clinical malaria infection.
2. Materials and methods
2.1. Ethics statement and study participants
This study is designated as TÜCHMI-001; an open label, single centre, randomized and controlled human malaria infection study in healthy human volunteers, where the volunteers and samples were identical to previous study by Bachmann et al. and Dimonte et al. [25,27]. Written informed consent was obtained from all participating volunteers from this study. Ethics approval was obtained from the ethics committee of the University Clinic of the University of Tubingen, Germany. This study was conducted in accordance to the Declaration of Helsinki (6th revision) and the International Conference on Harmonization with Good Clinical Practice guidelines. The registration code for this study is under ClinicalTrials.gov identifier numeric NCT01624961.
2.2. PfSPZ challenge and parasite sample collection
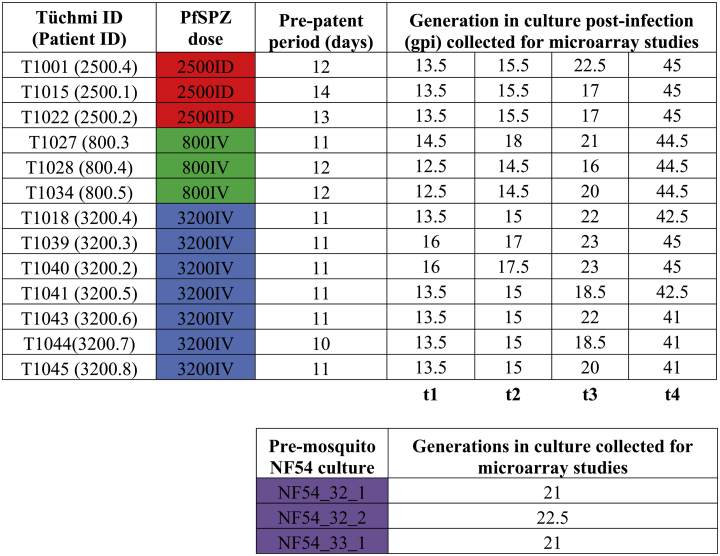
Thirteen participating volunteers received either intradermal or intravenous inoculation of PfSPZ Challenge at designated dosage; 3 with PfSPZ 2500 intradermally, 3 with PfSPZ 800 intravenously and 7 with PfSPZ 3200 intravenously (Table 1). All PfSPZ Challenge were provided by Sanaria Inc. In brief, the NF54 WCB SAN02-073009 was propagated in vitro for 3 generations prior the induction of gametocytogenesis and 1 generation in the mosquito stage for the production of PfSPZ. A positive thick blood smear with a least 1 to 2 parasites per μl of blood indicates successful blood stage infection or day of positivity in the PfSPZ challenge volunteers [25].
Table 1.
Summary of cryopreserved PfSPZ challenge in malaria naïve healthy volunteers (top) and pre-mosquito NF54 culture generation (bottom). Corresponding PfSPZ dose 2500ID (red); represents 2500 intradermal PfPSZ injection, 800IV (green); represents 800 intravenous PfSPZ injection, 3200IV (blue); represents 3200 intravenous PfSPZ injection. t1, t2, t3 and t4 represent the approximate generation stage during in vitro culture.
2.3. Parasite cultures
On the day of positivity, 0·25 ml of patient blood was used to establish 10 ml in vitro cultures. As soon as the parasitemia reached 3–4%, 4 ml of the culture were expanded into a 20 ml culture. 5 ml cultures with 5% haematocrit were maintained continuously for eight weeks. Expansion into 20 ml cultures was subsequently performed twice per week for a period of 8 weeks. In addition, two cryopreserved vials of the original NF54 strain WCB SAN02–073009 used to generate PfSPZ Challenge were provided by Sanaria Inc. Both vials were thawed and taken into in vitro culture under the same conditions as the volunteers' isolates. All P. falciparum isolates were cultivated at 5% haematocrit of O+ erythrocytes. RPMI 1640 medium was completed with 10% Albumax (Gibco), 25 mM Hepes Buffer, 2 mM l-Glutamine and 0·05 mg/ml gentamicin (all PAA Laboratories). Parasites were incubated at 37 °C in 90% nitrogen, 5% oxygen and 5% carbon dioxide.
2.4. Sorbitol synchronization, RNA extraction and cDNA synthesis
RNA was harvested as soon as an individual 20 ml parasite culture reached a parasitemia of 3–5%, and then every 2–3 generations thereafter. 20 ml parasite cultures were synchronized twice per day with 5% (v/v) sorbitol in water to obtain ring-stage parasites. The double-synchronized parasite culture was pelleted, washed two times with 1× phosphate buffered saline (PBS) and the erythrocytes were lysed with 0.02% (v/v) saponin. The pellet was washed three times with 1× PBS and resolved in 750 μl of Trizol® LS Reagent (Invitrogen). The samples were stored at −20 °C until further processing. The samples were shipped to Nanyang Technological University, Singapore for RNA isolation and microarray processing. Frozen samples were immediately thawed at 65 °C for 3 min and total RNA was isolated as previously described [32]. The quality of RNA was assessed by nanodrop spectrophotometer ND-1000 (Supplementary Table 6).
2.5. cDNA synthesis and microarray hybridization
Details on the materials used for cDNA synthesis and microarray hybridization are as described [33] Briefly, first strand cDNA was synthesized from 500 ng of total RNA using SuperScriptII Reverse Transcriptase kit (Thermo Fisher Scientific). The cDNA was then subjected to Switch Mechanism at the 5′ end of Reverse Transcription (SMART)-PCR amplification for 19 cycles to generate at least 4 μg of aminoallyl-coupled cDNA materials, which was subsequently purified using MiniElute DNA purification kit (Qiagen). 4 μg of SMART-PCR amplified products were labelled with fluorescent dye Cy5 (Amersham) and mixed with equal amount of Cy3-labelled P. falciparum 3D7 reference pool comprising of equal mass of total RNA samples representing all developmental stages of the parasite [34]. Hybridization was carried at 65 °C for 20 h on our customized microarray chip [32] using the Agilent hybridization system. Scanning and image acquisition of the hybridized microarray chips was performed using the PowerScanner™ (Tecan) at 10 μM resolution under automated photomultiplier (PMT) balancing. Hybridized spots or features were analysed and pre-processed using GenePix® ProMicroarray Image Analysis Software v6.0 (Axon Instruments).
2.6. Microarray data processing
Raw microarray data are further subjected to local feature background correction to remove background fluorescence signal that would result in the spurious variations in the gene expression datasets. All features were normalized using normexp function followed by Lowess normalization within each array using the ‘limma’ package [35] from R statistical software v3.5. Normalization using these methods minimize the intensity-dependent variation in two colour dye between batches. Outlier features with foreground intensity lesser or equal to 1·5-fold median of background intensity will be removed from subsequent analysis. Overall, 5061 genes display specific expression profile with normalized log2-transformed expression ratio of the RNA sample/3D7 reference pool, which was calculated by averaging the ratio for all oligo probes map to a gene open reading frame. These gene expression datasets were used for downstream analyses. The raw and processed microarray data has been deposited into GEO repository with accession number GSE136076.
2.7. Estimation of parasite age and gametocyte density
Mixture model and maximum likelihood estimates [36] was used to estimate the projected asexual parasite age in hour post-invasion (hpi) and gametocyte densities from the transcriptome datasets of CHMI-derived samples and the parent PfNF54 samples. Previously published P. falciparum Dd2 asexual time course transcriptome [13] and 3D7 sexual time course transcriptome [37] was used as reference.
2.8. Data analysis
All statistical and clustering analysis was conducted using R statistical software version 3·5. Unpaired Wilcoxon test was used to test the significance difference between any two groups of samples. For comparing the transcriptome datasets with previously published in vivo datasets under the same NCT01624961 clinical trials with matched volunteers' identification [27], expression values from each sample was separately normalized into z-score using the scale function. Pearson correlation coefficient and significance level were applied to measure the similarity of expression distribution. Unsupervised hierarchical clustering was done with the hcclust function using the complete linkage method and Euclidean distance metric. Principal component analysis was performed using prcomp function. Variation index was measured for each time-point (t1, t2 t3 or t4) as the expression standard deviation differences between individual for each PfSPZ group and throughout culturing phase, relative to the overall expression standard deviation throughout culturing phase.
where σ is the standard deviation expression levels of i-th gene between volunteer samples; p and throughout the gpi culturing phase from t1 to t4; g. Differential gene expression analysis for each gene was calculated as the average log2 expression ratio differences between CHMI and PfNF54 samples. Significance of differential expression was assessed with t-tests analysis corrected for multiple testing using Benjamini and Hochberg, controlling for the false discovery rate (FDR). Functional enrichment analysis was analysed by hypergeometric testing, by comparing to annotated functional pathways extracted from the Malaria Parasite Metabolic Pathway (MPM) database version 2016 [38].
3. Results
3.1. Early culture passage of freshly isolated infected erythrocytes has minimal effect on the transcriptional variation between CMHI-derived parasites from volunteers receiving the same PfSPZ dose
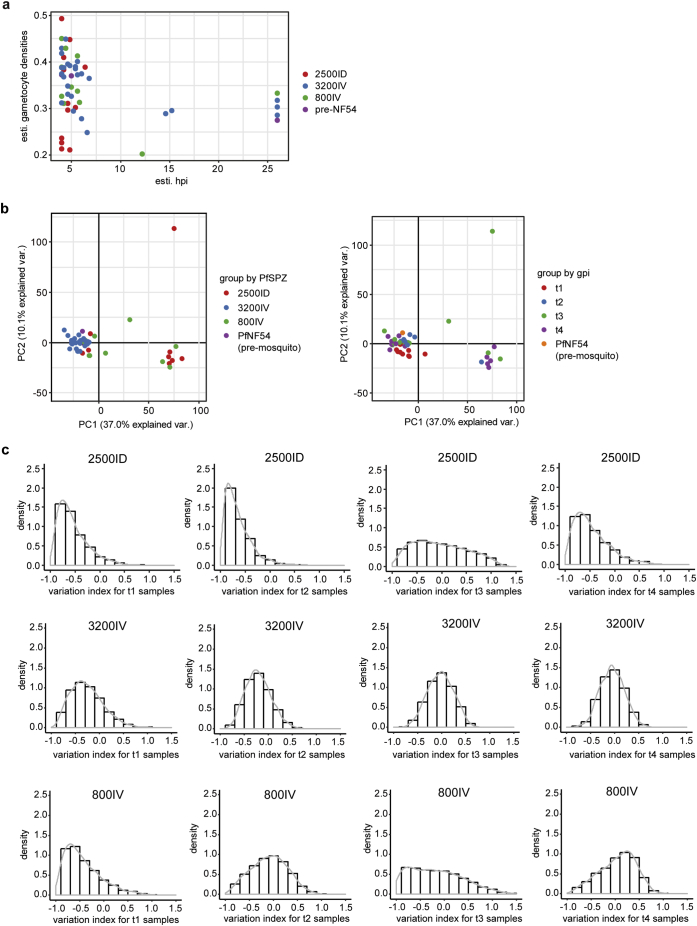
Infectivity of PfSPZ administration at 2500 PfSPZ injected intradermally and 3200 or 800 PfSPZ injected intravenously was assessed by thick film microscopy and PCR every 12 h from day 5 until day 21 post-infection (Table 1). Patient inclusion and exclusion criteria were deposited to ClinicalTrials.gov with the following identifier; NCT01624961. Due to limited amount of parasite RNA material that can be isolated directly from the freshly drawn blood of CHMI volunteers, as opposed to field samples often with higher parasitemia count, the CHMI-derived blood samples obtained from 13 volunteers were immediately subjected to in vitro culture prior to RNA extraction (Table 1). The parental PfNF54 asexual stage cultures initiated from two separate vials of the working cell bank (WCB) (vial 32 and vial 33) used for the production of PfSPZ Challenge [25,39] was also subjected to in vitro culture and RNA extraction (Table 1). A total of 52 CHMI-derived and 3 parental PfNF54 culture-adapted samples were synchronized and collected progressively throughout the in vitro culturing phase (from t1 to t4) at increasing generation post infection (gpi) from early to mid-ring stage to emulate the major developmental stages of infection observed in the human peripheral blood (Table 1). A total of 5061 genes were detected by microarray hybridization in at least one of the 55 samples (Supplementary Table 1). Samples with a poor synchronicity window as determined by maximum likelihood estimates (MLE) of the core transcriptomes against the asexual P. falciparum Dd2 references (13, 36, 37) were removed from subsequent analysis (Fig. 1a). This includes samples with an estimated hpi score of >16, which enters the late ring stage (Fig. 1a). To our surprise, MLE analysis also estimated high gametocyte proportion for some of the CHMI-derived samples including the parental PfNF54 using the day 5 to 7 sexual life-cycle reference transcriptome of P. falciparum 3D7, with values ranging from 20% to 50% (Fig. 1a). Recent analysis on the parental PfNF54 strain used for CHMI study from Sanaria Inc. showed significantly higher gametocyte production rate as compared to 3D7 strain [40]. This suggests that the inherent expression of gametocyte-specific genes, which was used for MLE on the gametocyte proportion in our study, were higher in our parental PfNF54 and this effect was subsequently seen in the CMHI-derived samples during asexual progression. We then showed that duration of culture passage of freshly isolated infected erythrocytes has an effect on the transcriptional variation between CMHI-derived parasites from volunteers receiving the same group of PfSPZ dose (2500ID, 3200IV and 800IV) using principal component and transcriptome variation index analysis (see Methods) (Fig. 1b, c). Principal component analysis (PCA) analysis showed a characteristic association between samples with different PfSPZ dosage and route of infection, where samples from 3200IV-derived dosage are more similar than 800IV or 2500ID (Fig. 1b). Samples from gpi t1 and t2 are also more closely clustered than samples from later gpi (Fig. 1b). Furthermore, the differences in gametocyte proportion estimation from the above MLE analysis does not showed significant clustering pattern in PCA analysis (Supplementary Fig. 1). Transcriptome variation index analysis allows us to visualize between individual or interindividual transcriptome differences at the same culturing stage (gpi). This is necessary to determine if culture progression have any effect on transcriptome variation between individuals receiving the same PfSPZ doses. For instance, the transcriptome derived from the three patients receiving PfSPZ 2500ID (2500.4, 2500.1 and 2500.2) has lesser interindividual variability when at t1 culturing phase compared with t3 culturing phase (Fig. 1c). Transcriptional variation between volunteers receiving the same PfSPZ doses was more evident with increasing culture passages, suggesting that a larger part of the parasite's transcriptome begins to fluctuate significantly as the in vitro culture progresses. The transcriptional variation observed in part of culture progression also resonates with the overall transcriptional profile of the var multigene family expression. We matched and analysed the Z-scores normalized var transcriptional profiles from t1 to t4 CHMI-derived cultures with the in vivo var gene transcriptional data for the same 9 volunteers from Bachmann et al [27]. Z-score normalization was used due to the differences in the var gene expression profile derived from this study (microarray) and Bachmann et al. (quantitative Real-time PCR). As a general trend, the distribution for var expression from the in vivo samples showed positive and significant correlation (P < .05) with the overall var expression profiles from CHMI-derived t1, t2, t3 and t4 cultured samples, with several group B var dominating the overall var genes expression hierarchy (Supplementary Fig. 2a). Notably, a trend to towards preferred expression and variation of expression for group B/C and C var genes from in vitro samples was apparent across parasites from all CHMI parasite cultures as the in vitro culture generation progresses from t1 to t4 (Supplementary Fig. 2b). Specifically, pairwise comparison between all the in vivo and culture-derived CHMI showed overall stronger and significant correlation for t1 and t2 samples (Supplementary Fig. 3), suggesting that the var genes expression profile are relatively conserved from the point of patency to at least 15 gpi in culture.
Fig. 1.
Transcriptional variation throughout the in vitro CHMI-derived samples. (A) Estimation of parasite age in hours post-invasion (hpi) and gametocyte densities based on the overall transcriptome dataset for 2500ID (red), 3200IV (blue) and 800IV (green) and pre-mosquito PfNF54 (purple) group. (B) Transcriptome principal component analysis of 2500ID, 3200IV and 800IV from t1 to t4 culturing phase and as well as the parental PfNF54 samples for a total of 3479 genes. Coloured scatterplot represents grouping by PfSPZ dose (left panel) and gpi (right panel). (C) Density plot shows the distribution of variation index for 3479 genes during t1, t2, t3 and t4 culturing phase for 2500ID, 3200IV and 800IV PfSPZ group. Variation index is defined as the standard deviation of expression levels of a gene between the volunteer samples and across culturing phase from t1 to t4. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Transcriptome variation associated with differences in intrahost replication time and distinct PfSPZ dose
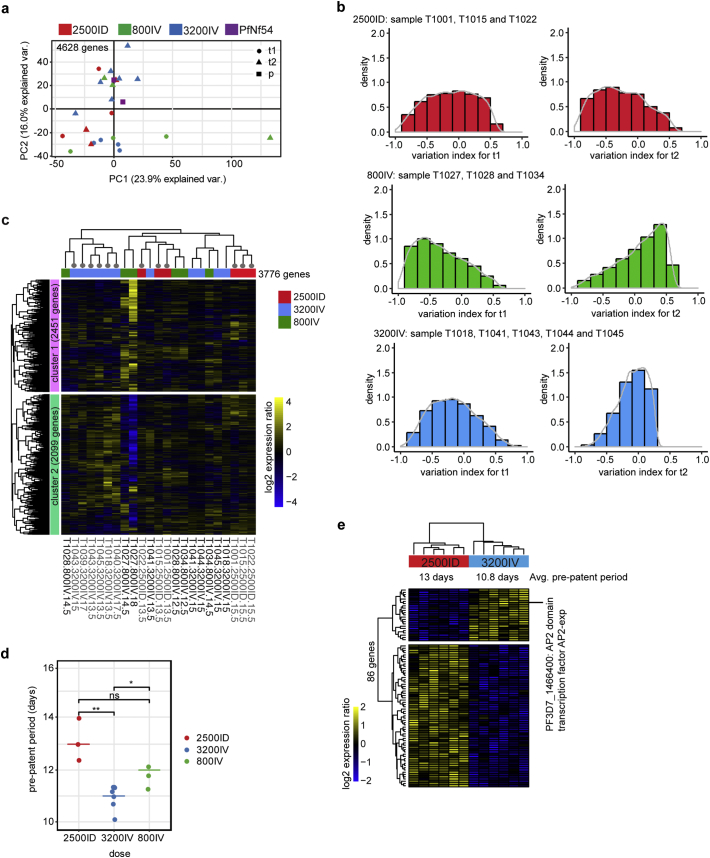
A total of 4628 genes were expressed across all CHMI-derived samples derived after at least 15 gpi in culture (t1 and t2 phase). Transcriptome principal component analysis (PCA) and variation index analysis of these t1 and t2 cultures showed dynamic transcriptome variation between the CHMI-derived parasites originating from the same PfSPZ dose, in particular the 800IV and 2500ID-derived parasites (Fig. 2a, b). A small fraction of genes (18%) exhibited significant transcriptional changes (P < 0·05, FDR < 0·05) as the culture progresses from t1 to t2, largely involved in parasite metabolic processes, post-translational modification and transport in the malaria parasites (Supplementary Fig. 4a, b; Supplementary Table 2). These set of genes were removed from further analysis to further reduce the transcriptional variation dimensionality during in vitro culturing. Next, expression profile for the remaining 3776 genes was subjected to unsupervised hierarchical clustering analysis. We identified two distinct sample clusters; six of the 3200IV-derived samples were clustered together while the rest of the 3200IV, 2500ID and 800IV-derived samples dominated the second cluster suggesting that transcriptome variation indeed exists between the interindividual samples (Fig. 2c). Notably, when we compared six of 3200IV and 2500ID sample clusters that showed significant differences in intrahost replication time of an average of 10·8 days vs 13 days respectively (Fig. 2d), we observed a small fraction of 86 differentially expressed genes between the two groups (Fig. 2e) (P < 0·05, FDR < 0·05). A large majority of the genes were up-regulated in the 2500ID cluster and involved in functions such membrane organization and vesicle-mediated transport (Supplementary Table 3). One of the genes with striking transcriptional differences between the 2500ID and 3200IV cluster is PF3D7_1466400 which encodes for the ApiAP2 transcriptional regulator family member, PfAP2-exp. PfAP2-exp have been shown to positively and/or negatively regulates the expression of a subset of clonally variant genes in P. falciparum, leading to the morphological alteration on the surface of infected RBC [41]. We next determine whether the changes in the transcriptional profile of the transcriptional regulator PfAP2-exp between PfSPZ 2500ID and 3200IV samples have any effect on the putative PfAP2-exp gene targets. Comparison for the expression of putative PfAP2-exp gene targets obtained from a previous study by Martins et al [41] between the 2500ID and 3200IV cluster revealed distinct transcriptional pattern, suggesting that divergence in Pf-AP2-exp PF3D7_1466400 between the two clusters can alter the expression of its putative gene targets as well (Supplementary Fig. 5). Collectively, the PfSPZ dose and consequently the duration of intrahost replication time (2500ID vs 3200IV) were associated with differences in the core transcriptome profile.
Fig. 2.
Transcriptome variation associated with distinct PfSPZ dose and intrahost replication rate. (A) Transcriptome principal component analysis of 2500ID (red), 3200IV (blue), 800IV (green) and parental PfNF54 group (purple) for 4628 genes that are expressed in t1 (circles), t2 (triangles) and pre-mosquito, p (squares) culturing phase. (B) Density plot shows the distribution of variation index for 4628 genes during t1 and t2, culturing phase for 2500ID (red), 800IV (green) and 3200IV (blue) PfSPZ group. (C) Heatmap shows hierarchical clustering on row scaled log2 expression ratio for 3776 genes for all PfSPZ samples at t1 and t2 culturing phase. PfSPZ samples with labelled dendrogram nodes (circle, grey) were selected for further comparative analysis. (D) Differences in pre-patent period represent by days between infection and positive thick blood smear for 2500ID (red), 3200IV (blue) and 800IV (green) group. Asterisks represent *P < 0·05, **P < 0·01. (E) Heatmap shows hierarchical clustering on row scaled log2 expression ratio for 86 genes between 2500ID and 3200IV PfSPZ clusters with significant expression differences at P < 0·05, FDR < 0·05. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Transcriptome diversity between parasites recovered from CHMI volunteers is also reflected in parasites recovered from clinical infection
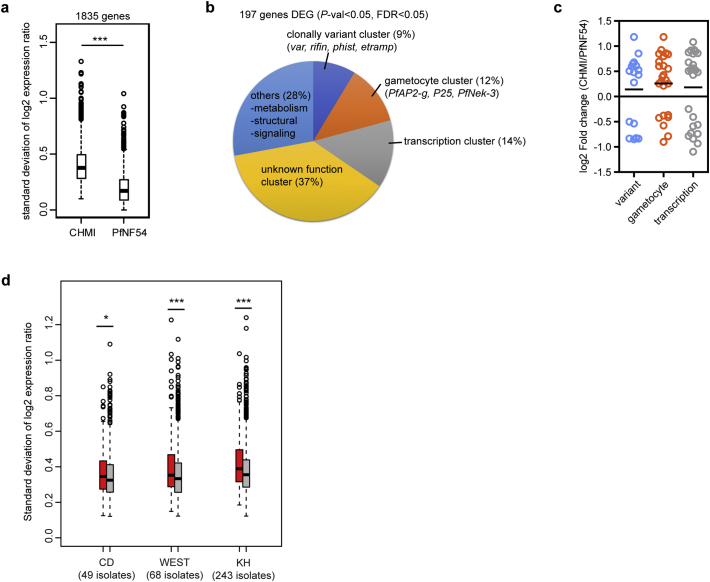
The diversity in gene expression profile may also reflect a form of response to natural selection that confers an optimal fitness phenotype in the given parasite population [[42], [43], [44]]. To evaluate this we first identified genes with higher transcriptional heterogeneity between the CHMI-derived samples from 2500ID and 3200IV cluster (Fig. 2c) versus the pre-mosquito, parental PfNF54 cluster. 1835 genes from the CHMI-derived 2500ID and 3200 PfSPZ cluster samples showed higher transcriptional variation compared to the parental PfNF54 (Fig. 3a, Supplementary Table 4), suggesting these genes are more likely to differ in parasites from different individual volunteers in response to infection after mosquito passage. It is important to note that the NF54 strain used for PfSPZ generation was propagated for 3 generations before gametocytes and sporozoites were induced. After PfSPZ Challenge and in vitro adaptation, t1 and t2 CHMI-derived samples were obtained from 13·5 to 17·5 gpi. Thus the total number of mitotic division for the in vitro t1 and t2 samples ranged from 16·5 to 20·5. The total number of mitotic division for the parental NF54 culture was 21 and thus in the same range as the number of mitotic division of the CHMI-derived cultures.
Fig. 3.
Differential expression and transcriptional variation analysis between pre-mosquito parasites and parasites recovered from CHMI volunteers. (A) Boxplots show within sample transcriptional variation of 1835 genes within the CHMI and PfNF54-derived parasite samples. Asterisk represent ***P < 0·001 for between group differences. (B) Pie chart represents proportion of genes associated with specific functional cluster for 197 differentially expressed genes between CHMI and PfNF54-derived parasite samples. (C) Scatter dotplot represents the log2 fold change of genes belonging to clonally variant, gametocyte and transcriptional clusters. (D) Boxplots show within sample transcriptional variation of 197 genes (red) within the clinical isolates group from Congo (CD), Bangladesh and Myanmar (WEST) and Cambodia (KH). For each group, significance in variation was compared with a control gene group by 100 times randomly selecting 197 genes from a list of non-differentially expressed genes. Asterisks represent *P < 0·05, ***P < 0·001. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We then identify genes with significant transcriptional differences between the CHMI-derived and parental PfNF54 samples within this variable transcriptome by calculating the average expression ratio differences between CHMI-derived and PfNF54 samples. 197 genes fulfilled the P < 0·05 and the overall adjusted P value at FDR < 0·05 differential expression criteria (Supplementary Table 4). Genes with unknown function account for the vast majority, as well as genes associated with clonal expression [45], gametocyte commitment [46], transcription, host cell structural organization and signalling events (Fig. 3b, Supplementary Table 5). Genes belonging to the clonally variant, gametocyte and transmission clusters have a higher average differential expression pattern in the CHMI-derived versus PfNF54 parent (Fig. 3c). The differential expression of this subset of genes could indicate that they provide the parasite with an adaptive advantage. If this is indeed the case it would be expected that these genes also show significant variation in natural infections. To assess this, we mapped the transcriptionally variable and differentially expressed 197 genes to the transcriptomes of P. falciparum clinical isolates from Africa, Bangladesh and the Mekong region, represented by Myanmar and Cambodia with early ring developmental stage between 8 and 10 hpi [34]. The transcriptomes of these clinical isolates were previously generated using the same batch of 3D7 reference pool and microarray printing and hybridization protocol, thus enabling us to directly compare both transcriptomes datasets. Clinical samples from Africa were collected from symptomatic children with acute malaria infection, whereas the remaining samples were collected from adult population across the Asia region with acute malaria [34]. For each geographical cluster, we observed significantly higher expression variation differences for the transcriptionally variable 197 genes compared to a control gene group by randomly selecting 197 genes for 100 permutations from a list of non-differentially expressed genes (Fig. 3d). Interestingly, samples from West Asia and South East Asia showed higher interindividual variation (P = 0·0006078, P = 5·333e-06) compared to samples from Africa (P = 0·03794), possibly reflecting the heterogeneity of the parasite transcriptome in patients with acute malaria from different geographical regions (Fig. 3d). Collectively these findings show that the transcriptome diversity observed in CHMI-derived samples, for a subset of genes involved in evasion of host immunity and parasite transmission, is also prevalent in actual clinical samples after mosquito passage and human infection suggesting a potential role in host adaptation.
4. Discussion
There have been renewed efforts and strategies towards malaria eradication, in particular a new drug pipeline, rapid diagnostics and vaccine development [47,48]. More recently, immunization with radiation attenuated, aseptic, purified, cryopreserved PfSPZ (Sanaria® PfSPZ Vaccine) [[49], [50], [51], [52], [53]] and infectious, cryopreserved PfSPZ with chemoprophylaxis (Sanaria® PfSPZ-CVac) [54] showed protection of 10 weeks to 14 months against homologous, heterologous, and heterogeneous P. falciparum infections by CHMI and natural exposure, thus offering the potential of a vaccine that could be used for malaria elimination campaigns. Questions concerning the biology of the malaria parasites during human infection are crucial in understanding the parasites virulence and pathogenesis. A greater part of malaria parasites transcriptomics profiling studies has been largely focused on field isolates directly isolated from patients, with infection phenotypes ranging from uncomplicated to severe malaria [16,17,55,56]. These studies have identified activation of potential genes or functional pathways important for the malaria parasites physiology, pathogenesis and adaptation to the human hosts; some of which are not seen in the long-term laboratory adapted strains. However, major impediments of these transcriptional profiling studies are associated with strain genetic variation [16], variation in disease severity and clinical conditions between patients [55] which could possibly affect the overall transcriptional output. The impact of CHMI studies not only allow for evaluation of malaria vaccine and drug efficacy, but are also useful for comparing parasite biology after mosquito passage and human infection as volunteers are infected with standardized number of aseptic, purified infectious PfSPZ with reproducible and consistent infections [23,26,57,58]. Nonetheless, parasitemia derived from CHMI studies, from non-immune and asymptomatic infection, are significantly lower compared with a natural infection setting due to ethical rationale. This ultimately leads to insufficient RNA material for whole transcriptomics profiling studies [16,59], and therefore in vitro culture expansion is inevitable.
Here, we report a comprehensive overview of genome-wide expression changes of the malaria parasites isolated from 13 immunologically naïve volunteers from the TÜCHMI-001 CHMI clinical trial. CMHI-derived parasites were subjected to in vitro culture under homogenous culturing conditions and RNA was collected at successive gpi for transcriptomics profiling. Previously, it was reported that in vitro culture adaptation of clinical isolates [16] and even isogenic, genetically identical parasites [60] can result in extensive transcriptional variation. Moreover, variation was also seen at the genome level compared with the parental source [61,62]. Such variation affecting the transcriptomic and genetic portrait are presumably due to the loss in immune selection pressure, differences in life-cycle stage and variation in donor blood batch, reagents, haematocrit and incubations conditions [63]. Here, we integrate the global transcriptomic data of the parental PfNF54 and CHMI-derived samples and performed a comparative analysis between these two groups. All CHMI-derived parasites were immediately transferred into in vitro culture on the day of smear positivity, thus avoiding potential a potential bottleneck effect due to cryopreservation of parasites. Pre-mosquito NF54 and CHMI derived parasites were cultured in parallel in a homogenous batch of culturing materials and culture conditions to avoid possible confounding factors due to batch variation.
Three important variables could shape the transcriptional profile observed in the CHMI-derived parasites; [1] culture synchrony, [2] environmental variation associated with blood donor batch, culture media and temperature and [3] parasite population heterogeneity resulting from genetic drift as the culture-adaptation progresses [63]. We addressed each of the variables by comparing the transcriptional output of CHMI-derived parasites with the laboratory-adapted P. falciparum reference strain and the previously isolated in vivo CHMI-derived used for var gene expression profiling [27]. Life-cycle estimation for each of the CHMI-derived parasites transcriptome profile corresponds to young mid ring stages, thus eliminating potential bias resulting from culture asynchrony. Comparisons with the limited transcriptional profile of PfEMP1 multigene families from matched volunteer ID and parasites obtained immediately post-CHMI [27] implied that in vitro expansion of these culture are still able to preserve moderate similarities with the in vivo generation. Interindividual sample variation, also evidenced by the var gene expression profile, became more apparent when culture progresses up to 45 gpi. Likewise, significant transcriptional variation was also seen beyond the t2 culturing phase when we mapped the interindividual variation for the entire transcriptome reflecting the transcriptome stochasticity in culture environment. Independent of this universal culture adaptation phenomenon, marked transcriptional differences were observed between CHMI-derived parasites from intravenously and intradermally infected individuals throughout the entire period of in vitro growth. Together, these results suggest that the above-mentioned factor variables may result in the apparent transcriptome fluctuation after each proceeding in vitro life cycle.
In order to best delineate transcriptional changes associated with the in vivo transcriptome, our study design takes several filtering approaches to mitigate problems arising from several variable effects; first, by removing samples which do not match to early-mid ring stage window and second, by removing samples with high interindividual transcriptome variation after in vitro culture expansion i.e. stage t3 and t4 samples for further analysis. We then only proceed to identify the true transcriptional response changes between groups of individuals infected with distinct PfSPZ dose, different intrahost replication time and between the parental references prior to mosquito infection. The choice of early gpi phase was according to the correlation analysis with Bachmann et al.'s samples which best represent the in vivo parasites. Our findings suggest that expression of transcripts associated with membrane organization, variant surface antigen, vesicle-mediated transport and transcriptional regulator of the clonal variant genes were significantly modulated between parasites samples originating from high (3200IV) and low (2500ID) PfSPZ dose, both which showed significantly different intrahost replication time. Evidence from previous CHMI studies suggested that variation in intrahost replication time reflects the differences in parasites load in the liver, which in turn is affected by the number of inoculated sporozoites through deposition in the dermis or direct into the vasculature [22,57,64]. Furthermore, transcriptional profiling of whole blood following a high dose of PfSPZ infection by intradermal versus intravenous route revealed differential gene expression cluster corresponding to the differences of pre-patency between individual volunteers, supporting that variation in parasite load in the blood can alter the host response [31] and that the duration of host-parasite interaction influences parasite transcription.
Variation in the parasite liver load in turn can also affect the antibody and cellular immune responses; high IV PfSPZ dosing can elicit strong and sustain T cell immunity in the liver whereas ID administration showed limited efficacy [49]. However, in view of the fact that all study participants were immunologically naïve and hence represent a potentially more homogenous environment, it appears unlikely that liver immunity differed amongst the study participants though variations in the innate immune response could make some contribution. As the transcriptional differences predominantly associated with transcripts that encodes for structural or variant proteins on the infected RBC membrane, it is tempting to speculate that the observed transcriptional differences between high and low PfSPZ dosing reflect host factors that favour expansion of parasite populations with the most beneficial transcriptional pattern, and that the observed transcriptional differences would be even greater in parasites obtained from semi-immune individuals. Indeed, a previous study investigating parasites from a natural chronic asymptomatic infection showed marked differences in expression of variant surface antigens compared to NF54 parasites [65] that persisted for the entire period of in vitro growth. In line with this observation Bachmann et al. recently reported a strong effect of host semi-immunity on in vivo var gene expression [66]. Transcriptome analyses of parasites from CHMI of semi-immune versus non-immune individuals are necessary to further delineate the importance of host factors for dynamic changes in the P.falciparum transcriptome.
A large proportion of the CHMI-derived transcriptome resembled the transcriptional profile of pre-mosquito parental PfNF54 in vitro cultures, suggesting conservation of >90% of the transcriptional profile after mosquito and human infection from a genetically identical pool of parasites. Nevertheless, we also observed transcriptional variation for a group of genes in response to mosquito passage and human infection. In addition to a small proportion of the major variant gene families such as vars, rifin and phist, it was noticeable that transcripts involved in transcriptional control, protein degradation, signalling machinery and invasion were up-regulated in CHMI-derived parasites. Up-regulation of genes involved in these pathways may enable the parasites to cope with the switch in host environment and the early host-parasite interaction mechanism after mosquito passage and human infection. Interestingly, this effect was most apparent in CHMI-derived parasites from intradermal infections as opposed to intravenous infection. In animal infection studies, mosquito passage of P. chabaudi changes the expression profiles of surface variant antigen in asexual blood-stage parasites which led to variation in disease severity [67].
We observed that a small number of 197 genes show significant interindividual expression changes following mosquito passage. To ensure that these changes were indeed a result of adaptation to host differences rather than representing stochastic variations due to the inherently small sample size of a CHMI study we investigated the variability of these genes in >500 transcriptomes obtained from natural infections [34]. In line with our findings using the CHMI samples, the transcriptome variation was also seen under natural infection setting, in individual transcriptome of clinical isolates from the Africa, West Asia and South East Asia region [34]. These 197 genes represent a range of different functionalities including clonally variant expression, signalling, metabolism, transcriptional control and as well transcripts associated with early sexual commitment stages, including the PfAP2-g [10,46]. The prevalence in activation of early gametocyte genes was also seen for some of P. falciparum field isolates either directly analysed from in vivo [17] or from culture-adaptation [16], analogous to our observations here and suggests that certain parasites may have a transcriptional setting that makes them more adapt for gametocyte production. Furthermore, a repertoire of the pre-versus post-mosquito/human infection genes encode proteins of unknown function with no known homologs in other species, suggesting a previously unrecognized role of these proteins. We postulate that transcriptional variation of these genes may be a general strategy of selection for parasites that would best adapt and response to common environmental conditions during infection. Indeed, functional variation in gene expression has shaped the principle of natural selection for the best phenotypic trait at the expense of rapid environmental changes [68]. Variation in gene expression also creates diversity of phenotypes, which confer fitness especially for genetically identical population [42,44,69,70]. In P. falciparum, variation in the transcriptional response consequential of environmental changes primarily occurs for clonally variant gene families, which is necessary for host-parasite interaction and parasite's survival under unfavourable conditions [60].
By eliminating the transcriptome variation and complexity associated with mixed infection, variable mosquito bites, immune response, culture progression and interindividual variation, our work here identifies a previously unrecognized transcriptional pattern of P. falciparum after passage through mosquito and human. We show that differences in PfSPZ dose and duration of intrahost replication resulted in parasite populations that showed differential gene expression profiles throughout the entire period of in vitro growth, whilst a small proportion of genes showed greater transcriptional variation amongst parasites derived from different individual volunteers. Most notably, the transcriptional variation of these genes was also seen in a larger pool of parasite samples from real clinical infection, implying that parasites consequent of CHMI studies can yield novel insights into the adaptation of parasites in different host environments. Adaptation by changing the overall transcriptional profiles may be a key survival and transmission strategy for the malaria parasites. Therefore, identification of transcriptome signature variation in malaria parasites and pathway genes associated with such variation is of potential clinical relevance for instance, identification of new pathway genes that can be selectively targeted by new antimalarials. In this context it is important to keep in mind that the parasites analysed in this work were obtained from a dose finding trial in immunologically naïve volunteers, therefore the observed transcriptional differences simply reflect the effect of a relative short difference in asexual replication time in immunologically naïve hosts. It is therefore likely that CHMI-trials in semi-immune individuals will result in stronger selective pressures and more profound changes in parasite biology. Transcriptome analysis of tissue culture adapted parasites from semi-immune individuals could therefore serve as a basis to understand the biologic processes that enable asymptomatic chronic infections. So far CHMI studies have been largely utilized for immunization strategy, evaluation of malaria vaccine and drug candidates [71]. Our work here supports the use of CHMI derived parasites as an avenue for the analysis of malaria parasite biology.
The following are the supplementary data related to this article.
Transcriptome of ring stage, culture-adapted CHMI and pre-mosquito NF54 parasites expressed as log2 ratio (Cy5/Cy3).
Functional enrichment analysis for 852 genes with significance difference in expression profile between t1 and t2 genes using hypergeometric distribution function on the Malaria Parasite Metabolic Pathway (MPM) database. Pathways testing was performed using hypergeometric test, significance at P < 0·05.
List of differentially expressed genes between 2500ID and 3200IV sample cluster. Significance of differential expression was assessed with t-tests analysis corrected for multiple testing using Benjamini and Hochberg, controlling for FDR at <0·05. Unpaired Wilcoxon test was used to test the significance difference between the two groups of samples.
List of genes with higher transcriptional variation for CHMI-derived 2500ID and 3200 PfSPZ cluster samples versus parental NF54. Significance of differential expression was assessed with t-tests analysis corrected for multiple testing using Benjamini and Hochberg, controlling for FDR at <0·05. Unpaired Wilcoxon test was used to test the significance difference between the PfSPZ and parental PfNF54 samples.
List of differentially expressed genes between the CHMI-derived and parental PfNF54 samples, with corresponding functional groupings. Significance of differential expression was assessed with t-tests analysis corrected for multiple testing using Benjamini and Hochberg, controlling for FDR at <0·05.
Concentration of RNA and absorbance ratio measured by Nanodrop 1000 Spectrophotometer.
Supplementary material
Funding sources
The parasite culture and RNA generation work was supported by the German Israeli Foundation (for S.D.) and the German Ministry for Education and Research (BMBF) (Grant Nr. 01KA110) (for E.B. and M.F.). This work was supported by MOE Tier 1 funding from the Singapore Ministry of Education Academic Research Fund Tier 1 (RG41/16) as well as the Singapore Ministry of Health's National Medical Research Council under its Individual Research Grant (NMRC/1292/2011) (for P.R.P. and R.H). We would like to acknowledge the support of a grant from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA (SBIR award number 2R44AI058375) awarded to S.L.H. The CHMI work was supported by the German Centre for Infection Research (Deutsches Zentrum für Infektionsforschung – DZIF). The funders have no role in this study design, data collection, data analysis, data interpretation and writing of this report.
Author contributions
E.B and S.D. performed and collected the P. falciparum parasite in vitro culture for RNA extraction. R.H performed the RNA extraction, cDNA synthesis, microarray hybridization, data processing and data analysis. L.Z provided bioinformatics support and performed estimation of parasite age. M.F conceived the project and oversaw the in vitro cultivation and experimental analysis of the P. falciparum parasites in Tübingen. R.H, P.R.P and M.F wrote the manuscript with inputs from B.M, B.K.L.S, P.G.K, S.L.H and Z.B. P.R.P and M.F conceived and designed the study.
Declaration of Competing Interest
B. K. L. S. is an employee of and S. L. H. the chief executive and scientific officer of Sanaria Inc. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Acknowledgements
We thank all volunteers and personnel who participated in the TÜCHMI-001 trial. We thank Michal Kucharski for his assistance with printing of the microarray chips. We thank Sachel Mok for providing the pool reference Pf 3D7 RNA for microarray hybridization.
Contributor Information
Matthias Frank, Email: Matthias.frank@praxis-frank-tuebingen.de.
Peter R. Preiser, Email: prpreiser@ntu.edu.sg.
References
- 1.Salanti A., Dahlback M., Turner L., Nielsen M.A., Barfod L., Magistrado P. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J Exp Med. 2004;200(9):1197–1203. doi: 10.1084/jem.20041579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turner L., Lavstsen T., Berger S.S., Wang C.W., Petersen J.E.V., Avril M. Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature. 2013;498(7455):502. doi: 10.1038/nature12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salanti A., Staalsoe T., Lavstsen T., Jensen A.T., Sowa M.P., Arnot D.E. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol. 2003;49(1):179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- 4.Albrecht L., Moll K., Blomqvist K., Normark J., Chen Q., Wahlgren M. var gene transcription and PfEMP1 expression in the rosetting and cytoadhesive Plasmodium falciparum clone FCR3S1.2. Malar J. 2011;10:17. doi: 10.1186/1475-2875-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goel S., Palmkvist M., Moll K., Joannin N., Lara P., Akhouri R.R. RIFINs are adhesins implicated in severe Plasmodium falciparum malaria. Nat Med. 2015;21(4):314–317. doi: 10.1038/nm.3812. [DOI] [PubMed] [Google Scholar]
- 6.Niang M., Bei A.K., Madnani K.G., Pelly S., Dankwa S., Kanjee U. STEVOR is a Plasmodium falciparum erythrocyte binding protein that mediates merozoite invasion and rosetting. Cell Host Microbe. 2014;16(1):81–93. doi: 10.1016/j.chom.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Josling G.A., Llinas M. Sexual development in Plasmodium parasites: knowing when it's time to commit. Nat Rev Microbiol. 2015;13(9):573–587. doi: 10.1038/nrmicro3519. [DOI] [PubMed] [Google Scholar]
- 8.Carter L.M., Kafsack B.F., Llinas M., Mideo N., Pollitt L.C., Reece S.E. Stress and sex in malaria parasites: why does commitment vary? Evol Med Public Health. 2013;2013(1):135–147. doi: 10.1093/emph/eot011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinha A., Hughes K.R., Modrzynska K.K., Otto T.D., Pfander C., Dickens N.J. A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium. Nature. 2014;507(7491):253–257. doi: 10.1038/nature12970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kafsack B.F., Rovira-Graells N., Clark T.G., Bancells C., Crowley V.M., Campino S.G. A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature. 2014;507(7491):248–252. doi: 10.1038/nature12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419(6906):498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bozdech Z., Llinas M., Pulliam B.L., Wong E.D., Zhu J., DeRisi J.L. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1(1) doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foth B.J., Zhang N., Chaal B.K., Sze S.K., Preiser P.R., Bozdech Z. Quantitative time-course profiling of parasite and host cell proteins in the human malaria parasite Plasmodium falciparum. Mol Cell Proteomics. 2011;10(8) doi: 10.1074/mcp.M110.006411. (M110 006411) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Roch K.G., Zhou Y., Blair P.L., Grainger M., Moch J.K., Haynes J.D. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301(5639):1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 15.Young J.A., Fivelman Q.L., Blair P.L., de la Vega P., Le Roch K.G., Zhou Y. The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology-based pattern identification. Mol Biochem Parasitol. 2005;143(1):67–79. doi: 10.1016/j.molbiopara.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Mackinnon M.J., Li J., Mok S., Kortok M.M., Marsh K., Preiser P.R. Comparative transcriptional and genomic analysis of Plasmodium falciparum field isolates. PLoS Pathog. 2009;5(10) doi: 10.1371/journal.ppat.1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daily J.P., Scanfeld D., Pochet N., Le Roch K., Plouffe D., Kamal M. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature. 2007;450(7172):1091–1095. doi: 10.1038/nature06311. [DOI] [PubMed] [Google Scholar]
- 18.Yuan L., Zhao H., Wu L., Li X., Parker D., Xu S. Plasmodium falciparum populations from northeastern Myanmar display high levels of genetic diversity at multiple antigenic loci. Acta Trop. 2013;125(1):53–59. doi: 10.1016/j.actatropica.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Druilhe P., Daubersies P., Patarapotikul J., Gentil C., Chene L., Chongsuphajaisiddhi T. A primary malarial infection is composed of a very wide range of genetically diverse but related parasites. J Clin Invest. 1998;101(9):2008–2016. doi: 10.1172/JCI119890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray L., Mobegi V.A., Duffy C.W., Assefa S.A., Kwiatkowski D.P., Laman E. Microsatellite genotyping and genome-wide single nucleotide polymorphism-based indices of Plasmodium falciparum diversity within clinical infections. Malar J. 2016;15(1):275. doi: 10.1186/s12936-016-1324-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nkhoma S.C., Nair S., Cheeseman I.H., Rohr-Allegrini C., Singlam S., Nosten F. Close kinship within multiple-genotype malaria parasite infections. Proc Biol Sci. 2012;279(1738):2589–2598. doi: 10.1098/rspb.2012.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomez-Perez G.P., Legarda A., Munoz J., Sim B.K., Ballester M.R., Dobano C. Controlled human malaria infection by intramuscular and direct venous inoculation of cryopreserved Plasmodium falciparum sporozoites in malaria-naive volunteers: effect of injection volume and dose on infectivity rates. Malar J. 2015;14:306. doi: 10.1186/s12936-015-0817-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mordmuller B., Supan C., Sim K.L., Gomez-Perez G.P., Ospina Salazar C.L., Held J. Direct venous inoculation of Plasmodium falciparum sporozoites for controlled human malaria infection: a dose-finding trial in two centres. Malar J. 2015;14:117. doi: 10.1186/s12936-015-0628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richie T.L., Billingsley P.F., Sim B.K., James E.R., Chakravarty S., Epstein J.E. Progress with Plasmodium falciparum sporozoite (PfSPZ)-based malaria vaccines. Vaccine. 2015;33(52):7452–7461. doi: 10.1016/j.vaccine.2015.09.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimonte S., Bruske E.I., Hass J., Supan C., Salazar C.L., Held J. Sporozoite route of infection influences in vitro var gene transcription of Plasmodium falciparum parasites from controlled human infections. J Infect Dis. 2016;214(6):884–894. doi: 10.1093/infdis/jiw225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheehy S.H., Spencer A.J., Douglas A.D., Sim B.K., Longley R.J., Edwards N.J. Optimising controlled human malaria infection studies using cryopreserved P. falciparum parasites administered by needle and syringe. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0065960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bachmann A., Petter M., Krumkamp R., Esen M., Held J., Scholz J.A. Mosquito passage dramatically changes var gene expression in controlled human Plasmodium falciparum infections. PLoS Pathog. 2016;12(4) doi: 10.1371/journal.ppat.1005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang C.W., Hermsen C.C., Sauerwein R.W., Arnot D.E., Theander T.G., Lavstsen T. The Plasmodium falciparum var gene transcription strategy at the onset of blood stage infection in a human volunteer. Parasitol Int. 2009;58(4):478–480. doi: 10.1016/j.parint.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 29.Lavstsen T., Magistrado P., Hermsen C.C., Salanti A., Jensen A.T., Sauerwein R. Expression of Plasmodium falciparum erythrocyte membrane protein 1 in experimentally infected humans. Malar J. 2005;4:21. doi: 10.1186/1475-2875-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peters J.M., Fowler E.V., Krause D.R., Cheng Q., Gatton M.L. Differential changes in Plasmodium falciparum var transcription during adaptation to culture. J Infect Dis. 2007;195(5):748–755. doi: 10.1086/511436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothen J., Murie C., Carnes J., Anupama A., Abdulla S., Chemba M. Whole blood transcriptome changes following controlled human malaria infection in malaria pre-exposed volunteers correlate with parasite prepatent period. PLoS One. 2018;13(6) doi: 10.1371/journal.pone.0199392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mok S., Imwong M., Mackinnon M.J., Sim J., Ramadoss R., Yi P. Artemisinin resistance in Plasmodium falciparum is associated with an altered temporal pattern of transcription. BMC Genomics. 2011;12:391. doi: 10.1186/1471-2164-12-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bozdech Z.M.S., Gupta A.P. DNA microarray-based genome-wide analyses of Plasmodium parasites. In: Menard R., editor. Malaria: Methods and protocols, methods in molecular biology. 2nd ed. Vol. 923. Springer Distributor; New York: Humana: 2013. pp. 189–211. [DOI] [PubMed] [Google Scholar]
- 34.Mok S., Ashley E.A., Ferreira P.E., Zhu L., Lin Z., Yeo T. Drug resistance. Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science. 2015;347(6220):431–435. doi: 10.1126/science.1260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 36.Lemieux J.E., Gomez-Escobar N., Feller A., Carret C., Amambua-Ngwa A., Pinches R. Statistical estimation of cell-cycle progression and lineage commitment in Plasmodium falciparum reveals a homogeneous pattern of transcription in ex vivo culture. Proc Natl Acad Sci U S A. 2009;106(18):7559–7564. doi: 10.1073/pnas.0811829106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu L., Tripathi J., Rocamora F.M., Miotto O., van der Pluijm R., Voss T.S. The origins of malaria artemisinin resistance defined by a genetic and transcriptomic background. Nat Commun. 2018;9(1):5158. doi: 10.1038/s41467-018-07588-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ginsburg H. Progress in in silico functional genomics: the malaria metabolic pathways database. Trends Parasitol. 2006;22(6):238–240. doi: 10.1016/j.pt.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 39.Hoffman S.L., Billingsley P.F., James E., Richman A., Loyevsky M., Li T. Development of a metabolically active, non-replicating sporozoite vaccine to prevent Plasmodium falciparum malaria. Hum Vaccin. 2010;6(1):97–106. doi: 10.4161/hv.6.1.10396. [DOI] [PubMed] [Google Scholar]
- 40.Gebru T., Lalremruata A., Kremsner P.G., Mordmuller B., Held J. Life-span of in vitro differentiated Plasmodium falciparum gametocytes. Malar J. 2017;16(1):330. doi: 10.1186/s12936-017-1986-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martins R.M., Macpherson C.R., Claes A., Scheidig-Benatar C., Sakamoto H., Yam X.Y. An ApiAP2 member regulates expression of clonally variant genes of the human malaria parasite Plasmodium falciparum. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-12578-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kussell E., Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309(5743):2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- 43.Viney M., Reece S.E. Adaptive noise. Proc Biol Sci. 2013;280(1767) doi: 10.1098/rspb.2013.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elowitz M.B., Levine A.J., Siggia E.D., Swain P.S. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 45.Lopez-Rubio J.J., Mancio-Silva L., Scherf A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe. 2009;5(2):179–190. doi: 10.1016/j.chom.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 46.Pelle K.G., Oh K., Buchholz K., Narasimhan V., Joice R., Milner D.A. Transcriptional profiling defines dynamics of parasite tissue sequestration during malaria infection. Genome Med. 2015;7(1):19. doi: 10.1186/s13073-015-0133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J., Modrek S., Gosling R.D., Feachem R.G. Malaria eradication: is it possible? Is it worth it? Should we do it? Lancet Glob Health. 2013;1(1):e2–e3. doi: 10.1016/S2214-109X(13)70002-0. [DOI] [PubMed] [Google Scholar]
- 48.Tanner M., Greenwood B., Whitty C.J., Ansah E.K., Price R.N., Dondorp A.M. Malaria eradication and elimination: views on how to translate a vision into reality. BMC Med. 2015;13:167. doi: 10.1186/s12916-015-0384-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seder R.A., Chang L.J., Enama M.E., Zephir K.L., Sarwar U.N., Gordon I.J. Protection against malaria by intravenous immunization with a nonreplicating sporozoite vaccine. Science. 2013;341(6152):1359–1365. doi: 10.1126/science.1241800. [DOI] [PubMed] [Google Scholar]
- 50.Ishizuka A.S., Lyke K.E., DeZure A., Berry A.A., Richie T.L., Mendoza F.H. Protection against malaria at 1 year and immune correlates following PfSPZ vaccination. Nat Med. 2016;22(6):614–+. doi: 10.1038/nm.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lyke K.E., Ishizuka A.S., Berry A.A., Chakravarty S., DeZure A., Enama M.E. Attenuated PfSPZ vaccine induces strain-transcending T cells and durable protection against heterologous controlled human malaria infection. Proc Natl Acad Sci U S A. 2017;114(10):2711–2716. doi: 10.1073/pnas.1615324114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sissoko M.S., Healy S.A., Katile A., Omaswa F., Zaidi I., Gabriel E.E. Safety and efficacy of PfSPZ vaccine against Plasmodium falciparum via direct venous inoculation in healthy malaria-exposed adults in Mali: a randomised, double-blind phase 1 trial. Lancet Infect Dis. 2017;17(5):498–509. doi: 10.1016/S1473-3099(17)30104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Epstein J.E., Paolino K.M., Richie T.L., Sedegah M., Singer A., Ruben A.J. Protection against Plasmodium falciparum malaria by PfSPZ vaccine. JCI Insight. 2017;2(1) doi: 10.1172/jci.insight.89154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mordmuller B., Surat G., Lagler H., Chakravarty S., Ishizuka A.S., Lalremruata A. Sterile protection against human malaria by chemoattenuated PfSPZ vaccine. Nature. 2017;542(7642):445–449. doi: 10.1038/nature21060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Almelli T., Nuel G., Bischoff E., Aubouy A., Elati M., Wang C.W. Differences in gene transcriptomic pattern of Plasmodium falciparum in children with cerebral malaria and asymptomatic carriers. PLoS One. 2014;9(12) doi: 10.1371/journal.pone.0114401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daily J.P., Le Roch K.G., Sarr O., Fang X., Zhou Y., Ndir O. In vivo transcriptional profiling of Plasmodium falciparum. Malar J. 2004;3:30. doi: 10.1186/1475-2875-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roestenberg M., O'Hara G.A., Duncan C.J., Epstein J.E., Edwards N.J., Scholzen A. Comparison of clinical and parasitological data from controlled human malaria infection trials. PLoS One. 2012;7(6) doi: 10.1371/journal.pone.0038434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roestenberg M., Bijker E.M., Sim B.K., Billingsley P.F., James E.R., Bastiaens G.J. Controlled human malaria infections by intradermal injection of cryopreserved Plasmodium falciparum sporozoites. Am J Trop Med Hyg. 2013;88(1):5–13. doi: 10.4269/ajtmh.2012.12-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vignali M., Armour C.D., Chen J., Morrison R., Castle J.C., Biery M.C. NSR-seq transcriptional profiling enables identification of a gene signature of Plasmodium falciparum parasites infecting children. J Clin Invest. 2011;121(3):1119–1129. doi: 10.1172/JCI43457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rovira-Graells N., Gupta A.P., Planet E., Crowley V.M., Mok S., Ribas de Pouplana L. Transcriptional variation in the malaria parasite Plasmodium falciparum. Genome Res. 2012;22(5):925–938. doi: 10.1101/gr.129692.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Claessens A., Affara M., Assefa S.A., Kwiatkowski D.P., Conway D.J. Culture adaptation of malaria parasites selects for convergent loss-of-function mutants. Sci Rep. 2017;7 doi: 10.1038/srep41303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Biggs B.A., Kemp D.J., Brown G.V. Subtelomeric chromosome deletions in field isolates of Plasmodium falciparum and their relationship to loss of cytoadherence in vitro. Proc Natl Acad Sci U S A. 1989;86(7):2428–2432. doi: 10.1073/pnas.86.7.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duffy S., Avery V.M. Routine in vitro culture of Plasmodium falciparum: experimental consequences? Trends Parasitol. 2018;34(7):564–575. doi: 10.1016/j.pt.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 64.McCall M.B.B., Wammes L.J., Langenberg M.C.C., van Gemert G.J., Walk J., Hermsen C.C. Infectivity of Plasmodium falciparum sporozoites determines emerging parasitemia in infected volunteers. Sci Transl Med. 2017;9(395) doi: 10.1126/scitranslmed.aag2490. [DOI] [PubMed] [Google Scholar]
- 65.Bruske E.I., Dimonte S., Enderes C., Tschan S., Flotenmeyer M., Koch I. In vitro variant surface antigen expression in Plasmodium falciparum parasites from a semi-immune individual is not correlated with var gene transcription. PLoS One. 2016;11(12) doi: 10.1371/journal.pone.0166135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bachmann A., Bruske E., Krumkamp R., Turner L., Wichers J.S., Petter M. Controlled human malaria infection with Plasmodium falciparum demonstrates impact of naturally acquired immunity on virulence gene expression. PLoS Pathog. 2019;15(7) doi: 10.1371/journal.ppat.1007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spence P.J., Jarra W., Levy P., Reid A.J., Chappell L., Brugat T. Vector transmission regulates immune control of Plasmodium virulence. Nature. 2013;498(7453):228–231. doi: 10.1038/nature12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gilad Y., Oshlack A., Rifkin S.A. Natural selection on gene expression. Trends Genet. 2006;22(8):456–461. doi: 10.1016/j.tig.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 69.Raser J.M., O'Shea E.K. Control of stochasticity in eukaryotic gene expression. Science. 2004;304(5678):1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rifkin S.A., Kim J., White K.P. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat Genet. 2003;33(2):138–144. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- 71.Stanisic D.I., McCarthy J.S., Good M.F. Controlled human malaria infection: applications, advances, and challenges. Infect Immun. 2018;86:1. doi: 10.1128/IAI.00479-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transcriptome of ring stage, culture-adapted CHMI and pre-mosquito NF54 parasites expressed as log2 ratio (Cy5/Cy3).
Functional enrichment analysis for 852 genes with significance difference in expression profile between t1 and t2 genes using hypergeometric distribution function on the Malaria Parasite Metabolic Pathway (MPM) database. Pathways testing was performed using hypergeometric test, significance at P < 0·05.
List of differentially expressed genes between 2500ID and 3200IV sample cluster. Significance of differential expression was assessed with t-tests analysis corrected for multiple testing using Benjamini and Hochberg, controlling for FDR at <0·05. Unpaired Wilcoxon test was used to test the significance difference between the two groups of samples.
List of genes with higher transcriptional variation for CHMI-derived 2500ID and 3200 PfSPZ cluster samples versus parental NF54. Significance of differential expression was assessed with t-tests analysis corrected for multiple testing using Benjamini and Hochberg, controlling for FDR at <0·05. Unpaired Wilcoxon test was used to test the significance difference between the PfSPZ and parental PfNF54 samples.
List of differentially expressed genes between the CHMI-derived and parental PfNF54 samples, with corresponding functional groupings. Significance of differential expression was assessed with t-tests analysis corrected for multiple testing using Benjamini and Hochberg, controlling for FDR at <0·05.
Concentration of RNA and absorbance ratio measured by Nanodrop 1000 Spectrophotometer.
Supplementary material