Abstract
Background
Faecal microbiota transplantation (FMT) is a novel potential therapy for inflammatory bowel diseases, but it is poorly characterised.
Methods
We evaluated the performance of the mouse and rat as a pre-clinical model for human microbiota engraftment. We then characterised the effect of a single human stool transfer (HST) on a humanised model of DSS-induced colitis. Colonic and faecal microbial communities were analysed using the 16S rRNA approach and clinical manifestations were assessed in a longitudinal setting.
Findings
The microbial community of rats showed greater similarity to that of humans, while the microbiome of mice showed less similarity to that of humans. Moreover, rats captured more human microbial species than mice after a single HST. Using the rat model, we showed that HST compensated faecal dysbiosis by restoring alpha-diversity and by increasing the relative abundance of health-related microbial genera. To some extent, HST also modulated the microbial composition of colonic tissue. These faecal and colonic microbial communities alterations led to a relative restoration of colon length, and a significant decrease in both epithelium damage and disease severity. Remarkably, stopping inflammation by removing DSS before HST caused a faster and greater recovery of both microbiome and clinical manifestation features.
Interpretation
Our results indicate that the rat outperforms the mouse as a model for human microbiota engraftment and show that the efficacy of HST can be enhanced when inflammation stimulation is withdrawn. Finally, our findings support a new therapeutic strategy based on the use FMT combined with anti-inflammatory drugs.
Keywords: Inflammatory bowel disease, Faecal microbiota transplantation, Rat model of colitis
Research in context
Evidence before this study
Inflammatory bowel disease (IBD) is a chronic incurable disease affecting approximately 0.5% of the general population in Western countries. Its aetiology remains unknown and its prevalence is expected to increase exponentially over the next decade making it a growing healthcare burden. Faecal microbiota transplantation (FMT), an efficacious treatment for Clostridium difficile infection, has emerged as a potential therapy for IBD. However, the mid/long-term effect of this treatment on the evolution of the host microbiome, immune response and clinical outcomes has not been fully investigated through clinical trials nor through proper animal models. As a result, the efficiency of current FMT protocols is low in IBD.
Added value of this study
In the present study, we first show that a conventional rat model, more reflective of human microbiota, could be an alternative to a mouse model to study microbe-host relationships in human disease. Then, using the humanised rat model of IBD (colitis induced by inflammatory agents), we demonstrate that a single faecal transplantation from a healthy human donor could compensate dysbiosis by restoring alpha-diversity and by increasing the relative abundance of health-related microbial genera. Furthermore, we show that the faecal transplantation also corrected, to some extent, colonic tissue alterations, leading to a relative restoration of colon length, and a significant decrease in epithelium damage and disease severity. Finally, we show that stopping the inflammation by withdrawing the inflammatory agent before faecal transplant results into a faster and greater recovery of both microbiome and clinical manifestation features.
Implications of all the available evidence
Our findings lead to promote the use of a rat model, instead of mouse, as the more appropriate pre-clinical animal model for FMT and IBD. Our first longitudinal study, using a humanised rat model of colitis, proves the efficacy of a single faecal microbiota transplantation and encourages the use of anti-inflammatory drugs during FMT to maintain a remission state in future clinical trials.
1. Introduction
Inflammatory bowel disease (IBD) is a complex and chronic intestinal disorder associated with an exacerbation of the host immune response, a disruption of the intestinal barrier, and alterations in the luminal and mucosal microbial communities [1], [2]. IBD includes two chronic intestinal inflammatory forms, ulcerative colitis (UC), which affects the rectum and colon, and Crohn's disease, which may involve the entire gastrointestinal tract, but is most common in the colon and terminal ileum [3]. Patients with IBD experience episodes of relapse alternating with remission.
Dysbiosis in IBD patients is often characterised by a significant and global alteration of the composition and structure of the microbial community and is associated with a lower microbial alpha-diversity compared to healthy controls [4], [5], [6], [7].
Faecal microbiota transplantation (FMT) has been used for several years to treat recurrent Clostridium difficile infection (CDI) and has recently proven effective in randomised clinical trials [8], [9]. Interest in using FMT to restore a healthy gut microbiota and thereby attenuate inflammatory responses in IBD patients is growing. However, many aspects of this therapeutic approach remain unanswered, including questions regarding patient stratification, donor selection and methods to administer the FMT [10], [11], [12], [13], [14]. While several meta-analyses suggest that FMT is safe, this approach shows variable efficacy in IBD patients [12], [15], [16]. In this regard, more studies are required in order to establish optimal and standardised FMT protocols [17]. Concurrently, many experimental models, including in vivo animal models such as mouse TNBS or DSS-induced colitis [18], [19] and T cell transfer colitis in severe combined immunodeficient (SCID) mice, have been widely used to study IBD and the effect of FMT in this disease [20]. For instance, using a mouse model of colitis and a short-term follow-up study of 8 days, Zhou et al. [20] demonstrated that FMT decreases the severity of colitis. This effect was associated with significantly decreased myeloperoxidase activity, reduced levels of TNF-α and IL-1β, and an increased level of IL-10 in colon tissues. However, these authors did not address the impact of FMT on the microbiome community. In this context, to the best of our knowledge, the present study is the first to evaluate the performance of mouse and rat models for human microbiota transplantation.
The aims of this study were to: (a) Identify which animal species (rat or mouse) was the most appropriate to receive and maintain a single human stool transfer; and (b) to examine the long-term effect of a single human stool transfer on a humanised and chemically-induced colitis murine model at both microbiome and clinical manifestation level.
We demonstrated that the microbiome of rats has a similar composition and structure to that of humans, while the microbiome of mice shows less similarity to that of humans. Moreover, rats captured more human bacterial species than mice after a single human stool transfer. We then showed that a single stool transfer, obtained from a previously selected healthy human donor, modulated the dysbiotic microbiota, which was associated with improvements in clinical and biological manifestations.
2. Materials and methods
2.1. Ethical statement
The animal study was approved by the Ethical Committee of Animal Experimentation at the Vall d'Hebron Research Institute (Barcelona, Spain) and the Department of Agriculture, Livestock and Fisheries of the Generalitat de Catalunya (Departament d'Agricultura, Ramaderia i Pesca). Animal care complied with the criteria outlined in the Guide for the Care and Use of Laboratory Animals Institute for Laboratory Animal Research, Division on Earth and Life Studies, Washington, D.C., USA. All experiments were conducted in the animal facilities of the Vall d'Hebron Research Institute (Barcelona, Spain).
For human stool donors, approval was provided by the University Hospital of Vall d'Hebron Ethics Committee and informed consent was given in all cases.
2.2. Animals
Male Sprague-Dawley rats and BALB/c mice were purchased from Charles River (France). Rats weighed about 200 g and mice about 25 g and were 6 weeks of age. After 7 days of quarantine, each animal was isolated in a conventional cage in order to prevent microbial transmission. They were maintained at a constant room temperature of 21 °C and were exposed to a 12:12 h light-dark cycle. All rats drank sterilised water and were fed an autoclaved standard chow diet. No contact was allowed between rats either through water, chow or faeces.
2.3. Bowel preparation for stool transfer
Omeprazole was diluted in water, filtered, and administered via oral gavage at a dosage of 50 mg/kg/day. All animal groups, except group A-CTL, received the omeprazole solution on day 2 (9 a.m.), day 3 (12 a.m.) and day 4 (8 a.m.). To empty the gastrointestinal tract, on day 3, rats underwent a 24-h fast and mice a 12-h fast. Animals were placed individually in a cage to prevent coprophagy and they have access to water. Then, 0.2 ml of Citrafleet®, prepared following the manufacturer's instructions, was administered twice (9 a.m. and 9 p.m. of day 3) via oral gavage. Citrafleet® (Casen-Fleet, Zaragoza, Spain) was administered two times via oral gavage: 1 ml, 24 h before the stool transfer, and 2 ml for rats (1 ml for mice), 12 h before the transfer [21]. A laparotomy was performed to evaluate the efficiency of Citrafleet® and to collect colon tissues for microbiome analysis. The use of cages with grids prevented coprophagy during the cleansing procedure, which lasted about 24 h. The rats and mice were then given via oral gavage a dose of omeprazole (at the concentration of 50 mg/kg/day).
2.4. Human stool transfer (HST) in mice and in rats
The healthy stool donor was selected in a previous project (Sarrabayrouse et al., unpublished). Briefly, ten healthy volunteers (≥ 18 years of age) provided self-collected stool samples. Two stools with one month interval were collected and immediately frozen at home and brought to the laboratory without breaking the cold chain, where they were stored at −80 °C. Stool and serology screenings were performed for bacterial, parasitic and viral pathogens (Clostridium difficile, Salmonella, Shigella, Yersinia, Aeromonas, Campylobacter, Vibrio, E. coli O157, Rotavirus, Adenovirus, Atrovirus, Norovirus, and Giardia intestinalis). Potential donors did not take antibiotics in the eight weeks preceding the screening. One of the donors was selected on the basis of the stability and high diversity of the stool microbial community and the lack of genera potentially involved in inflammation, such as Escherichia or Fusobacteria, and the presence of Faecalibacterium. The transfer of human stool to rodents was performed once through an oral gavage using 100 mg of a fresh stool sample diluted in 2 ml of PBS for rats (in 1 ml for mice). A second HST containing 100 mg of faeces diluted in 2 ml of PBS was performed 22 days after the induction of colitis. Faecal samples were collected at various time points.
2.5. Induction and assessment of colitis by administration of DSS in rats
Rats were provided with drinking water containing 5% (wt/vol) dextran sodium sulphate (DSS) (mol wt 40,000; TdB Consultancy, Uppsala, Sweden) ad libitum for 5 days in order to induce acute experimental colitis. Chronic colitis was then induced by reducing the concentration of DSS to 2% for 17 (group Dw-DSS) or 57 (group D) days depending on the study group, as previously described by Oishi et al. [22]. Colitis was assessed daily on the basis of the combined scores of body weight, stool consistency and presence of macroscopic blood in stools to calculate disease activity index (DAI), as shown in Supplementary Table S1. The DAI score was graded on a scale of 0–4, as previously described by Murthy et al. [23].
2.6. Laparotomy and euthanasia
On the last day of the study, laparotomy was performed by injecting 10 mg/kg xylazine (Xilagesic®; Calier, Barcelona, Spain) and 75 mg/kg ketamine (Ketolar®; Parke-Dawis, Madrid, Spain). The whole intestine was removed and the colon was washed with PBS and cut into small pieces. Some of the colon fragments were sampled for histological evaluation while others were stored directly at −80 °C for microbiome analyses.
2.7. Histological analysis
For histology, tissues were fixed in Methanol-Carnoy's solution (60% methanol, 30% chloroform, 10% glacial acetic acid) and maintained at room temperature for at least 3 h. Small pieces of colon tissue were placed into cassettes and dehydrated in ethanol baths. They were then immersed in xylene, a clearing agent, to remove ethanol. Next, colon tissues were embedded in molten paraffin and sectioned (5 µm). Slices were stained with hematoxylin and eosin. Finally, the degree of intestinal inflammation was determined following the guide described by Erben et al. [24].
2.8. TNF-α measurement
TNF-α was determined in tissue samples by enzyme-linked immunosorbent assays (ELISAs), following the manufacturer's protocols (eBiosciences). Results are expressed as the ratio of picograms of TNF-α per milligram of total protein. Limits of detection were 30 pg/ml.
2.9. Lymphocyte isolation and staining from spleen
Single cell suspensions were generated from spleens, and erythrocytes were removed from splenic samples by Ficoll gradient centrifugation. 2.5 × 10^5 splenocytes were stained with FITC-conjugated anti-CD3 (Miltenyi Biotec), PB-conjugated anti-CD4 and PE-conjugated anti-CD8 antibodies. Stained cells were analysed by flow cytometry using an LSRFortessa cytometer (BD) and then analysed with FlowJo software (FlowJo LLC).
2.10. Microbiome analysis
2.10.1. 16S rRNA gene amplification
Genomic DNA was extracted using the protocols previously described by Pascal et al. [7] for stool samples and by Santiago et al. [25] for tissue samples. An equivalent of 1 mg of each sample was used for DNA quantification using a Nanodrop ND-1000 Spectrophotometer (Nucliber). DNA integrity was examined by micro-capillary electrophoresis using an Agilent 2100 Bioanalyzer with the DNA 12,000 kit.
For profiling microbiome composition, the hyper-variable region (V4) of the bacterial and archaeal 16S rRNA gene was amplified by PCR using universal primers, as previously described by Pascal et al. Standard PCR using 0.75 units of Taq polymerase (Roche) and 20 pmol/µL of the forward and reverse primers was run in a Mastercycler gradient (Eppendorf) at 94 °C for 3 min, followed by 35 cycles of 94 °C for 45 sec, 56 °C for 60 sec, 72 °C for 90 s, and a final cycle of 72 °C for 10 min. Amplicons were first purified using the QIAquick PCR Purification Kit (Qiagen, Barcelona, Spain), quantified using a Nanodrop ND-1000 Spectrophotometer (Nucliber) and then pooled in equal concentration. For colon samples, a second purification step was applied to the pool of amplicons, as described in Santiago et al. [25]. The pooled amplicons (2 nM) were then subjected to sequencing using Illumina MiSeq technology at the technical support unit of the Autonomous University of Barcelona (UAB, Spain), following standard Illumina platform protocols. In order to identify possible contamination in colonic tissues, considered as low-biomass samples, and subtract the sequences of the potentially contaminated DNA generated, negative controls (blanks) were introduced during two technical steps: genomic DNA extraction and 16S rRNA gene PCR amplifications.
2.10.2. Upstream 16S rRNA sequence analysis
The upstream sequence analysis was performed with QIIME (v.1.9.0) using the guidelines proposed by Navas-Molina [26]. First, we generated a mapping file containing sample identifiers, barcodes, primer sequences, time points, symptoms, treatments and any other additional information of the samples needed for the analyses and validated its compatibility with the QIIME tool. Then, we demultiplexed and performed sequence quality filtering of the obtained fastq files with a minimum acceptable Phred score of 20. From a total of 531 rat faecal samples, we obtained a total of 23.8 millions of high-quality sequences with a number of reads ranging from 1 to 85,679 per sample with an average of 44,201 reads. From a total of 36 rat colon samples, we obtained a total of 4.6 millions of high-quality sequences with a number of reads ranging from 3711 to 460,161 per sample with an average of 127,934 reads. From a total of 60 mice faecal samples, we obtained a total of 2.8 millions of high-quality sequences with a number of reads ranging from 22,663 to 69,572 per sample with an average of 46,230 reads.
We clustered similar filtered sequences into Operational Taxonomic Units (OTUs) with the USEARCH algorithm (v8.1.1861) [27] using a 97% similarity threshold. Although the QIIME developers recommend the open-reference method that first groups sequences using a reference database and then by sequence identity, we preferred, in terms of time consumption and discovery of novel diversity, to use the de novo approach, based only on sequence similarity that focuses on finding new bacteria. In this step, we also identified and removed chimeric sequences using UCHIME [27].
We then performed a taxonomical assignment step using the Basic Local Alignment Search Tool (BLAST) to map each representative sequence against a combined database of Greengenes (gg_13_8 release) and PATRIC databases. Since each OTU could comprise many related sequences, we picked a representative sequence from each OTU and these representative sequences were aligned using PyNAST against Greengenes template alignment. After this step, we did a phylogenetic construction with FastTree. Finally, we generated a table containing the predicted taxonomy and abundance of all the OTUs for each sample.
2.10.3. Removal of unknown sequences not corresponding to the 16S rRNA gene
In the case of colon (low biomass) samples, many of the sequences could correspond to unknown taxa, which could be due to the presence of Eukaryotic host sequences. We identified those sequences assigned as unknown by mapping them against GenBank using BLAST and we removed from the OTU table all sequences that did not correspond to the 16S rRNA gene.
2.10.4. Removal of bacterial contaminants
After removal of non-bacterial DNA, we identified bacterial contamination in colon samples and removed it. We used the 'prevalence' method of the open-source R package decontam [28] with both extraction and PCR negative controls and with the default threshold (P = 0.1). We decontaminated each sequencing run separately.
2.10.5. Downstream sequence analysis
We performed downstream analyses, including diversity analyses and statistical tests with QIIME (v1.9.1) and R software (v3.4.4). We used the biom program to convert tables between biom and txt formats. We assigned arbitrary numbers to unknown species to avoid collapsing possible novel species into one. Finally, we summarised the OTU table by distinct taxonomic levels from phylum (L2) to species (L7).
2.10.6. Diversity
We performed rarefaction (random selection of the same number of reads per sample) in the OTU table in order to account for uneven library sizes. For rat faecal samples, we rarefied the OTU table at 10,912 sequences per sample, which allowed us to keep 521 samples and 16.42 million reads for further analyses. For the analyses involving human and rat faecal samples, we rarefied the OTU table at 7512 sequences per sample (in order to keep most of the samples) and retained 524 samples and 16.46 million reads for further analyses. For rat colon samples, we rarefied the OTU table at 2538 sequences per sample, which maintained all 36 samples and 2.81 million reads for further analyses. Regarding mice faecal samples, we rarefied the OTU table at 7512 sequences per sample, which retained all 60 samples and 2.8 million reads for further analyses.
To estimate the alpha-diversity (within samples), we calculated the Chao1 and Shannon indexes [29], [30]. Beta diversity (between samples) was computed using weighted and unweighted UniFrac [31] to produce distance matrices, which were later used for grouping samples into hierarchical cluster trees with unweighted Pair Group Method with Arithmetic mean (UPGMA) and Principal Coordinate Analyses representations (PCoA).
2.10.7. Differential abundance and statistics
To assess differences in the microbiota composition between groups based on treatment and/or time point, we used the non-parametric statistical Kruskal-Wallis test (more than two groups, unpaired), Wilcoxon signed rank test (pairwise paired comparisons) and Mann-Whitney U test (pairwise unpaired comparisons). To account for multiple comparisons of all taxa, we used FDR correction and considered the results significant when P < 0.1. Then, to compute correlations between the microbiota and clinical or experimental variables, we used non-parametric Spearman correlations. For the statistical analysis of clinical longitudinal data, we used a non-parametric method of the R package nparLD (https://CRAN.R-project.org/package=nparLD).
2.10.8. Microbial source tracking
Microbial source tracking was performed using SourceTracker (v1.0.1) [32], a Bayesian approach that estimates the proportion of OTUs in a given community that come from possible source environments. We used the default parameters (burn-in = 100, restart = 10, alpha and beta Dirichlet parameters = 0.001 and 0.01, respectively), except for the rarefaction depth, which was set to 7512, as in the analyses involving human and rat faecal samples. To analyse the effect of HST in the two rodent models, mouse and rat faecal samples from the first day of the trial and HST donor samples were defined as mouse, rat and human sources, respectively. To determine the proportions of healthy rat microbiota versus microbiota associated with the administration of DSS, rat faecal samples from the day after DSS administration and from after 3 weeks of DSS treatment were defined as healthy and DSS-associated sources and used to train the model. The rest of samples were then assigned some percentages either from these sources or an unknown source.
2.10.9. Data deposition and accession numbers for raw data
Sequence data and metadata have been deposited in the GenBank database with the following access number: PRJNA549436.
3. Results
3.1. Choice of rodent model
3.1.1. Rat microbiome compared to mouse and human microbiome
Conventional mice are often used to study human diseases and, in particular, to examine the effect of alterations in the microbial community in human diseases. To study whether conventional rats outperform mice as recipients for human microbiota, we first compared the faecal microbiome of rats (n = 52), mice (n = 16) and healthy human individuals (n = 72). Human sequence data were recovered from our previous study [7] and data for rats and mice were newly generated.
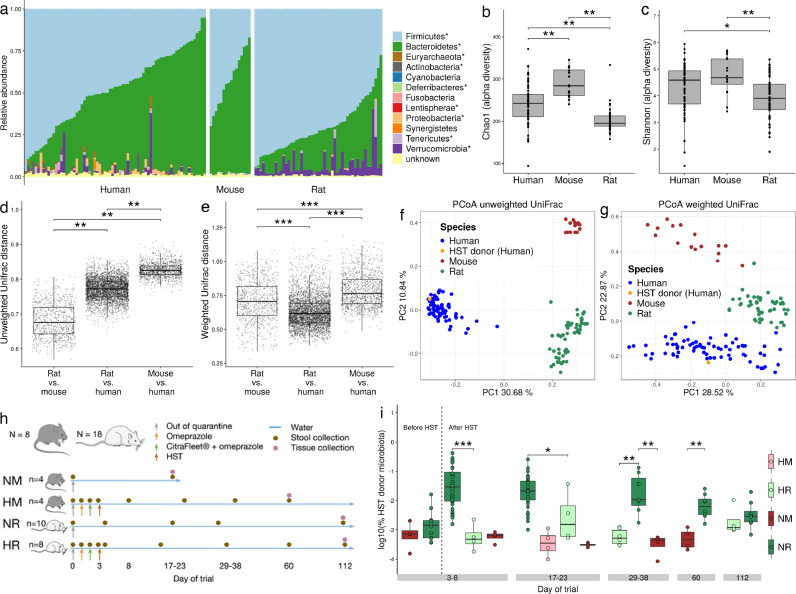
Firmicutes and Bacteroidetes accounted for 93%, 98% and 93% of the sequence data in the human, mouse and rat microbiome, respectively (Fig. 1(a)). The microbial faecal community of the three species was significantly different for the most predominant phyla and genera (Mann Whitney pairwise test, FDR < 0.05). Alpha-diversity analysis using Chao1 estimates showed significant pairwise differences between the three groups (Fig. 1(b)). Shannon indexes revealed significant pairwise differences between all groups, except between mice and humans (Fig. 1(c)). Altogether, mice presented a higher alpha-diversity than rats and humans. Surprisingly, beta-diversity analysis, based on weighted and unweighted UniFrac indexes, revealed that the distance between rats and humans was smaller than that between mice and humans (Fig. 1(d)–(g)). These findings confirmed that the faecal microbial community of rats is more similar to that of humans than that of mice to humans.
Fig. 1.
The microbiome of the rat showed greater similarity to that of humans than the mouse microbiome to humans. (a) Taxonomic profile at the phylum level for human (n = 72), rat (n = 52) and mouse (n = 16) faecal samples. Alpha-diversity analysis using the Chao1 (b) and the Shannon (c) indexes showed differences between humans, mice and rats (FDR < 0.05). Pairwise distances between rats and humans were lower than between mice and humans in both unweighted (d) and weighted (e) UniFrac (FDR < 0.05). Unweighted (f) and weighted (g) UniFrac Principal Coordinate Analysis representation of the three species, highlighting the HST donor sample. (h) Experimental design to assess the engraftment of the human microbiota in mouse and rat. (i) Proportion of predicted microbiota from a human source in rats and mice allocated to different groups as a function of receiving a HST. Significant differences were found between rats receiving a HST or not, and also between rats and mice receiving a HST (Mann-Whitney U test; p < 0.05). NM = No HST mouse; HM = HST mouse; NR = No HST rat; HR = HST rat.
3.1.2. Human stool transfer (HST) in rats and mice
Human microbiota-associated (HMA) animal models are becoming a standard tool through which to study the relation between gut microorganisms and human diseases. We performed a HST into conventional rat and mouse by gavage to evaluate how each rodent shaped the human microbiota over time (Fig. 1(h)). Omeprazole, a proton pump inhibitor, was used to suppress stomach acid secretion and therefore to increase bacterial survival. CitraFleet® (sodium picosulfate), a stimulant laxative, was used to empty the gastrointestinal tract before the HST. The healthy human donor stool was previously selected from ten volunteers on the basis of its high microbial diversity and stability, and tested for the absence of potential common pathogens (see Methods section). Additionally, the healthy donor stool was selected for having the lowest relative abundance of Escherichia and Fusobacteria and a high abundance of Faecalibacterium, characteristics of a healthy stool as compared with an IBD stool [7] (see Methods section). To monitor the evolution of the transferred human microbiome in the rodents, we used SourceTracker, a Bayesian, OTU-based algorithm, to estimate the proportion of the human microbial community remaining in the animals over time, up to 8 weeks (60 days) for mice and 16 weeks (112 days) for rats. To train the model, we used rat and mouse faecal samples collected immediately after quarantine, and the healthy human donor sample. Surprisingly, the proportion of the human microbiome present in the rats was significantly higher than in mice over the 8-week follow-up (Fig. 1(i), day 60), thereby suggesting that rats captured human microbiota more efficiently than mice. The engraftment effect lasted for at least 38 days and was lost at 60 days.
Given that the rat microbiome was more similar to the human microbiome, in terms of composition and structure, than the mouse microbiome to humans and that it showed a better and long-lasting engraftment of this human microbiome, the rat emerges as a better model for studying the relationship between the microbiome and human intestinal diseases.
3.2. Modulation of the rat gut microbiota
3.2.1. Experimental design
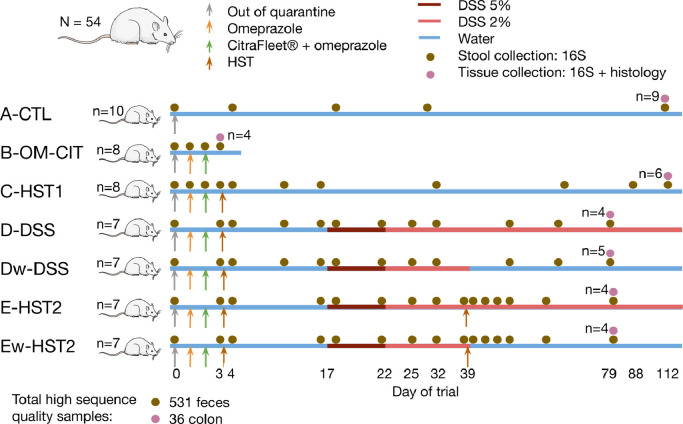
To assess the effect of a HST in a rat model of chronic intestinal inflammation, a total of 50 animals were randomly placed into seven experimental groups (Fig. 2) as follows: a control group receiving a standard diet and water without treatment (group A-CTL, same group as that described in Fig. 1(h) as NR); a group receiving omeprazole and Citrafleet® (group B-OME-CIT), to evaluate the cleansing effect; a group receiving omeprazole, Citrafleet® and a human stool (group C—HST1, same group as that described in Fig. 1(h) as HR), to evaluate the humanization protocol; a group receiving omeprazole, Citrafleet®, human stool and then dextran sodium sulfate (DSS) until the end of the study (group D), to evaluate dysbiosis after inflammation induction; a group receiving omeprazole, Citrafleet®, human stool, DSS until the end of the study and a second human stool (HST2) on day 39 (group E-HST2), to assess the effect of microbiome modulation in response to the transfer of a healthy human stool.
Fig. 2.
Experimental design to evaluate the effect of a human stool transfer on a rat model of chronic intestinal inflammation. Seven groups were distributed as follows: a control group receiving a standard diet and water without treatment (group A-CTL, n = 10); a group receiving omeprazole and Citrafleet® (group B-OME-CIT, n = 8); a group receiving omeprazole, Citrafleet® and a human stool (group C—HST1, n = 8); a group receiving omeprazole, Citrafleet®, human stool, and DSS until day 39, which was replaced by water (w) until the end of the study (group Dw-DSS, n = 7); a group receiving omeprazole, Citrafleet®, human stool and then DSS until the end of the study (group d-DSS, n = 7); a group receiving omeprazole, Citrafleet®, human stool, and DSS until day 39, which was replaced by water (w) until the end of the study, and a second human stool (group Ew-HST2, n = 7); a group receiving omeprazole, Citrafleet®, human stool, DSS until the end of the study, and a second human stool (group E-HST2, n = 7). HST: Human stool transfer; DSS: dextran sodium sulfate. The “n” in the right side of the figure represents the number of animals for which colonic tissues were collected and analysed for microbiome composition.
Two additional groups, Dw-DSS and Ew-HST2, were added. These were similar to groups d-DSS and E-HST2, except that DSS was replaced by water at day 39 before HST2, in order to study the resilience of the mucosal layer and the microbiota after injury to the tissue was stopped. As specified above, omeprazole and CitraFleet® were used to increase microbial survival and to empty the gastrointestinal tract, respectively, before stool transfer. DSS was used to induce chronic intestinal inflammation, which occurred after 5 days of DSS treatment at 5% and then at 2% (group d-DSS vs. Dw-DSS and group E-HST2 vs. Ew-HST2).
The first HST was performed to generate a human microbiota-associated animal model and the second to assess the correction of disease severity (group E-HST2 vs. d-DSS and group Ew-HST2vs. Dw-DSS). Given that human microbiota differs significantly from that of rodents, the transfer of human microbiota was performed to ensure a human microbiota context before inducing inflammation.
In order to calculate the disease activity index (DAI), animals were monitored daily for body weight and for the presence of blood in stool. Histological, immunological and colonic microbial community analyses were performed on colonic samples obtained on the day of sacrifice. Faecal microbial community and DAI scores were evaluated at various time points.
3.2.2. Rat intestinal microbial community viewed across time
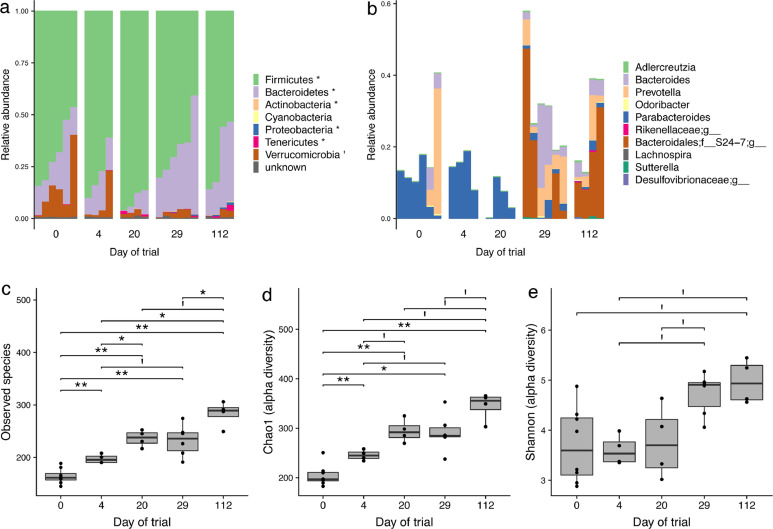
To study the effect of time on the faecal microbial composition, we analysed changes in the taxonomic profile over 16 weeks (112 days, animal group A-CTL) of follow-up (Fig. 3(a)). The relative abundance of six phyla (Fig. 3(b)) and ten genera changed significantly over 16 weeks, with an increase in Adlercreutzia, Sutterella, Prevotella, Bacteroides, Odoribacter, and three unknown genera from Rikenellaceae, Desulfvibrionaceae and S24-7 families, and a decrease or disappearance of Lachnospira and Parabacteroides (Fig. 3(c)). Changes in the taxonomic profile were associated with a significant increase in alpha-diversity, as shown by the Chao1 and Shannon indexes and the number of observed species from D0 to D112, which increased from 161 to 289 (Fig. 3(d)–(f)).
Fig. 3.
Evolution of the rat microbiome over time (112 days). (a) Taxonomic profile at the phylum level. Significant differences (marked with asterisks) were found in most of the phyla over time (Kruskal-Wallis test, FDR < 0.05). Each bar represents an individual faecal sample. (b) Taxonomic profile at the genus level, only significant changes are shown (Kruskal-Wallis test, FDR < 0.05). Each bar represents an individual faecal sample. Alpha-diversity was calculated on the basis of the species observed (c), the Chao1 index (d), and the Shannon index (e). Significant differences were found in all three indexes, indicating a progressive increase in microbial diversity over time (FDR < 0.05; **: FDR < 0.01; I: FDR < 0.1). A total of 24 faecal samples were analysed, with n = 4 or n = 6 at each time point.
3.2.3. Short-term effect of bowel cleansing
The short-term effect of omeprazole alone on the gut microbial community was not significant (animal group B-OME-CIT). However, omeprazole combined with CitraFleet® led to an increase in Verrucomicrobia and Proteobacteria but a decrease in Firmicutes (Supplementary Fig. S1a). At the genus level, omeprazole and CitraFleet® led to increases in several genera, such as Parabacteroides, Morganella and Citrobacter and deceases in others, including Anaerostipes, unknown Clostridiales, and Lactobacillus, among others (Supplementary Fig. S1b). To the best of our knowledge, this is the first study to report this combined effect of omeprazole and CitraFleet® on the composition of the gut microbial community.
3.2.4. Inflammation and dysbiosis induction
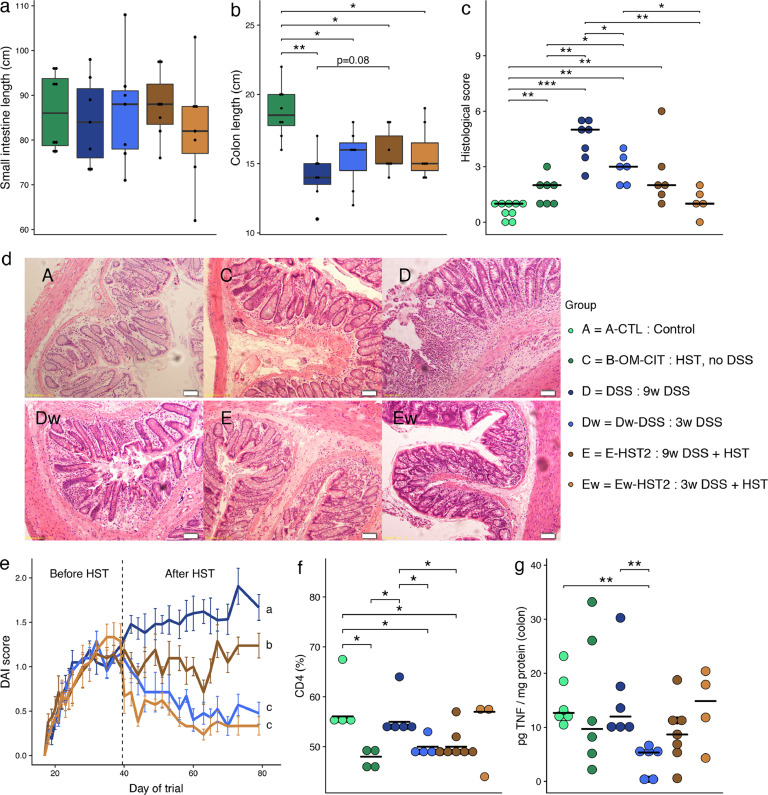
To simulate human IBD, we induced inflammation using dextran sodium sulfate (DSS) in two additional animal groups, d-DSS and Dw-DSS. DSS, a negatively charged sulphated polysaccharide, was given in drinking water at 5% (from day 17) and then at 2% (from day 22) in order to achieve a model of chronic intestinal inflammation [22]. The Dw-DSS group was used to assess resilience after withdrawal of DSS treatment on day 39 (Fig. 3). DSS results in the death of epithelial cells, leading to the disruption of the epithelial monolayer lining. This disruption allows the entry of luminal bacteria and associated antigens into the mucosa and causes an altered mucosal immune response [19]. Small intestine and colon lengths, like the histological scores, were measured on the day each animal was sacrificed. A blinded histological evaluation of colonic tissues was performed by two researchers on the day of animal sacrifice.
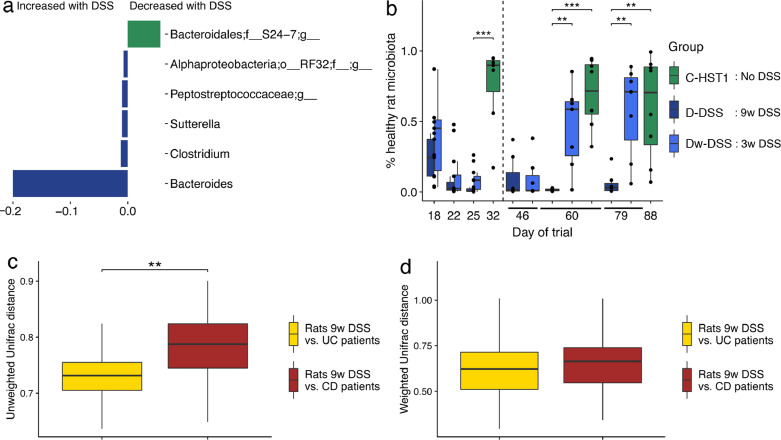
Significant alterations in the faecal microbial communities were observed at the genus level when comparing the two groups of rats with (D-DSS) and without (C—HST1) DSS treatment at day 79. As shown in Fig. 4(a), changes in the microbial community induced by DSS included genera that were significantly affected over time (Fig. 3(b)), such as Bacteroides, Suterella and an unknown member of the S24-7 family. Bacteroides and Suterella increased both over time and upon DSS treatment. Conversely, the unknown S24-7 group increased over time but decreased in response to DSS. These findings suggest that the S24-7 group is involved in maintaining homeostasis in the gut microbiota. No significant correlations were found in colonic tissues—an observation that could be attributed to the low number of samples involved. Notably, the microbiome returned to a baseline and healthier composition when DSS treatment was withdrawn, as shown by SourceTracker (Fig. 4(b)). Indeed, no significant differences were found at the genus level between groups C—HST1 and Dw-DSS.
Fig. 4.
DSS administration causes dysbiosis in rats. (a) Significant differences in the relative abundance of several microbial genera between group C—HST1 and D (no DSS vs. 9 weeks DSS) at the end of the trial (at day 79). (b) Proportion of predicted microbiota from a healthy rat source in rat groups C, D and Dw-DSS, using the SourceTracker software. The vertical, dotted line separates the time points of the 3 weeks in which rats in group Dw-DSS received DSS (left) from the rest of time points in which DSS treatment was withdrawn (right). Significant differences were found between DSS-treated rats and untreated rats (group C—HST1 vs. D) but not between control rats and rats after DSS treatment was withdrawn (group C—HST1 vs. Dw-DSS, at day 60) (Mann-Whitney U test; p < 0.05). (c) Unweighted UniFrac distances between rats in group d-DSS (9 weeks DSS) and UC and CD patients. Significant differences were found, UC patients being closer to rats with DSS-induced colitis than CD patients to this rat model (non-parametric test, p < 0.01). (d) Weighted UniFrac distances between rats with DSS-induced colitis in group d-DSS (9 weeks DSS) and human UC and CD patients. No significant differences were found.
To determine whether the dysbiosis in a rat model of DSS-induced colitis was similar to ulcerative colitis or to Crohn's disease in humans, we compared the microbiome data of rats from group d-DSS collected at day 116 with published sequence data [7] of adult patients with ulcerative colitis (UC, n = 28) and adult patients with Crohn's disease (CD, n = 32), using the UniFrac distances. Our results showed that dysbiosis in the rat model of DSS-induced colitis was closer to dysbiosis in patients with UC than in those with CD, when taking into account only microbial composition (unweighted UniFrac distances) (non-parametric test, p < 0.01) (Fig. 4(c) and (d)). This result validates the description of several clinical characteristics of the rat model, such as colon localization and the lack of transmural inflammation, as reported in this study and others [33].
As shown in Fig. 5(a), small intestine length was not affected by DSS treatment. In contrast, colon length was significantly reduced in all animal groups compared to the control group C—HST1, as reported by others (Fig. 5(b)) [19], [23], [34]. The mucosal layer of group d-DSS was highly damaged compared to group C—HST1, as shown by an average histological score of 4.4 (SD = 1.1) vs. 1.7 (SD = 0.8), respectively (Fig. 5(c) and (d)). When DSS treatment was withdrawn from group Dw-DSS, which implied that mucosal tissues were not exposed to DSS for 40 days, we observed a significant recovery from tissue damage, as shown by an average histological score of 2.9 (SD = 0.8). Furthermore, disease activity indexes (DAI), which were calculated on the basis of animal weight, presence of blood in stool and stool consistency scores for each animal at different time points, significantly increased in group d-DSS compared to Dw-DSS (mixed ANOVA, P < 0.05) when DSS administration was stopped on day 39 (Fig. 5(e)).
Fig. 5.
HST ameliorated clinical manifestations during DSS-induced colitis. Small intestine (a) and colon length (b) measured on the day of sacrifice. (c) Histological scores. (d) Haematoxylin and eosin staining of colonic tissue. (e) Disease activity indexes (DAI) were calculated on the basis of animal weight, presence of blood in stool and stool consistency scores for each animal. Significant differences were found using a non-parametric test equivalent to a mixed ANOVA (P < 0.05, the different letters (a, b and c) indicate which groups were different). (f) Lymphocyte isolation from spleen. (g) TNF-α measurement in colonic tissues. Mann Whitney test, *: P < 0.05; **: P < 0.01.
Moreover, to evaluate the immune response associated with DSS treatment, we studied the frequency of CD4 splenic T lymphocytes in spleen and measured TNF-α in colonic tissues. To this end, splenocytes were isolated and analysed by flow cytometry, as described in the Methods section. On the one hand, we observed that humanization of the rats decreased the frequency of these cells, thereby suggesting that the human microbial community affected the peripheral immune response of the animals (Fig. 5(f)). On the other hand, DSS significantly increased the frequency of these cells, which is in agreement with an increase in the inflammation induced by this treatment. When DSS was maintained until the end of the study, no effect on the production of TNF-α in colonic tissues was observed; however, a significant decrease in this parameter was detected when DSS was stopped on day 39 (Fig. 5(g)).
All together, these results demonstrate that, as expected, DSS induced a significant reduction in colon length and mucosal damage. Moreover, it exacerbated the immune response and caused an increase in disease severity. Furthermore, cessation of DSS after a daily treatment of 22 days revealed an improvement of all these clinical and biological manifestations.
3.2.5. Correction of dysbiosis and disease severity by HST
To evaluate the therapeutic potential of HST to correct DSS-induced inflammation, we added two animal groups, E-HST2 and Ew-HST2, in which we performed a second HST (HST2) on day 39 (22 days after the start of DSS-induced inflammation). The Ew-HST2 group was added to assess the resilience of rats after withdrawal of the DSS treatment (see experimental design in Fig. 2).
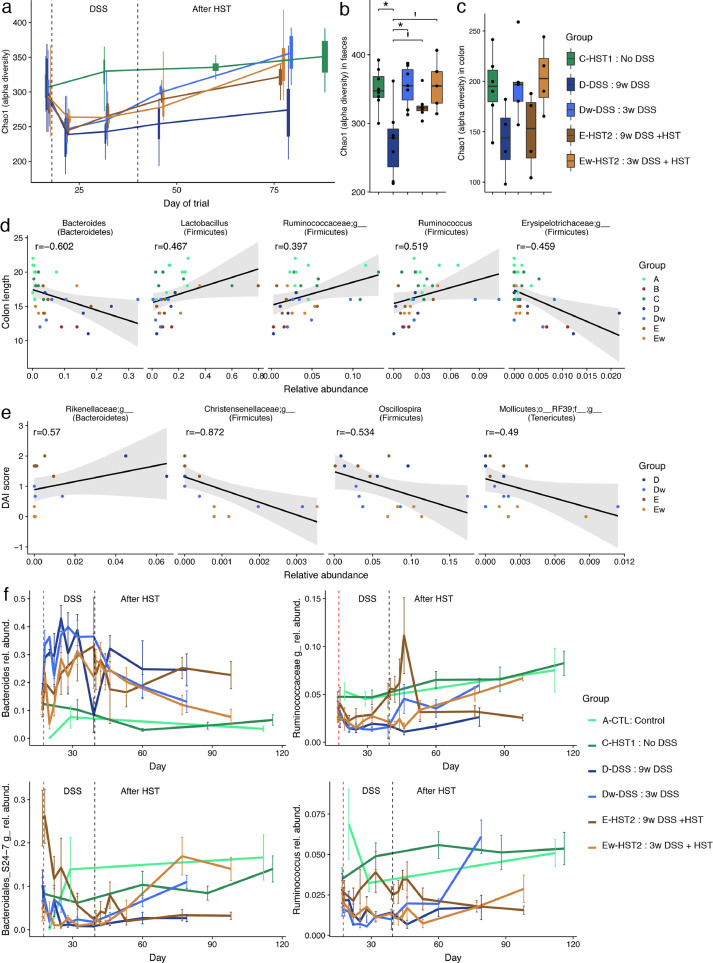
Analysis of group E-HST2 revealed a significant reversal of epithelial damage as compared with group d-DSS, as shown by a relative reduction of the histological score (P = 0.07; Mann-Whitney U test) and a significant reduction of the DAI score (P = 0.003; non-parametric mixed ANOVA-like test). Rats did not present differences in small intestine length with (E-HST2) or without (Ew-HST2) the maintenance of colitis induction. However, colon length tended to increase in group E-HST2 compared to D (Mann-Whitney U test, P = 0.08) (Fig. 5(a) and (b)). Of note, HST2 significantly decreased the frequency of CD4 splenic T lymphocytes (group d-DSS vs. E-HST2), reducing it to basal level (group C—HST1). This finding therefore indicates the efficacy of this treatment on one of the markers of inflammation. At the faecal microbiome level, HST performed in group E-HST2 increased alpha-diversity (Chao1 indexes) as compared with group d-DSS (FDR = 0.057, on the day of sacrifice (Fig. 6(a) and (b)).
Fig. 6.
Effect of HST on DSS-induced dysbiosis. (a) Alpha-diversity in stool samples as measured by the Chao1 indexes in a longitudinal setting. Alpha-diversity in stool (b) and colon (c) samples as measured by the Chao1 indexes on the day of animal sacrifice. There was a significant reduction in faecal microbiome diversity only in group d-DSS compared to control group C—HST1 (n = 5–8 per group, non-parametric test, FDR < 0.1). No significant differences were found in the colonic microbiome (n = 4–6 per group). Correlations between microbiome composition in colonic tissues and clinical manifestations as measured by the Spearman correlation test (P < 0.05). In colon samples, genera were significantly correlated with colon length (d) and with DAI score (e). (f) In stool samples, the evolution of several bacterial genera selected based on their correlation with clinical manifestations are plotted according to the longitudinal setting.
HST2 in group Ew-HST2 caused a significant decrease in disease severity through a reduction in both the DAI scores (P = 1e-13; non-parametric mixed ANOVA-like test) and histological scores (P = 0.005; Mann-Whitney U test) compared to the group that received DSS (D) (Fig. 5(c) and (d)). Simultaneously, at the microbiome level, we observed a recovery of alpha-diversity (Fig. 6(a) and (b)) and a greater proportion of healthier gut microbiota, using the SourceTracker method, in both faecal samples and colonic tissues (Supplementary Fig. S2).
Furthermore, the Spearman correlation test revealed correlations between colonic microbiome composition and clinical manifestations such as DAI scores, histological scores and colon length. Indeed, several microbial phyla and genera in the colonic tissues showed a correlation with colon length and DAI scores (Fig. 6(d) and (e)). Ruminococcus, Lactobacillus and an unknown Ruminococcaceae were positively correlated with colon length, and an unknown Erysipelotrichaceae and Bacteroides were negatively correlated with this parameter. In contrast, an unknown Rikenellaceae was positively correlated with DAI scores, and Oscillospira, an unknown Christensenellaceae and an unknown genus of Mollicutes class were negatively correlated with these scores.
Finally, we also found several correlations between faecal microbiome and clinical manifestation. Among them, an unknown Ruminococcaceae and Ruminococcus were positively correlated with colon length and negatively with histological score. The S24-7 group was positively correlated only with colon length. Conversely, Bacteroides was negatively correlated with colon length but positively correlated with histological score. The longitudinal setting allowed us to assess the effect of the different treatments on the relative abundance of various microbial genera, which evolved as expected from their correlations with clinical manifestations (Fig. 6(f)).
4. Discussion
Here we first studied the performance of the rat and mouse for HST. We then evaluated the therapeutic potential of a stool transfer to treat chronic inflammation induced by a colitogenic chemical. To this end, we analysed clinical manifestations and immunological markers, as well as faecal and colonic microbiota in a longitudinal setting (up to about 100 days) using a conventional animal model. Our experimental design, which included several animal groups, also allowed us to gather information on the natural evolution of the animal gut microbiome over a period of 112 days, the long-term effect of colitis induction, and the resilience of the microbiome when this induction was stopped.
Most studies on IBD using DSS-induced colitis models have been performed on mice [20], [33], [34], [35]. Comparison of rat, mouse and human genomes has revealed that the rat genome contains about the same number of genes as that of humans and mice [36]. A recent study has provided a gene catalogue of the Sprague-Dawley rat gut metagenome, and comparison of rat with human or mouse has revealed a higher pairwise overlap between rats and humans (2.47%) than between mice and humans (1.19%) at the gene level [37]. Our study, using 16S rRNA sequence data, has complemented this previous work, showing that the overall composition and structure of the faecal microbiome of rat has a greater similarity to that of the human than the microbiome of the mouse to human. Furthermore, the rat appeared to capture the human stool microbiome more efficiently than the mouse. Overall, on the basis of these findings, the rat emerges as a potential alternative to the mouse for evaluating a human microbiota-associated rodent, thus highlighting the potential use of rats to study the effect of FMT in treating IBD and other human disorders. Future studies should test the effects of transplanting stools with diverse microbial compositions from various human donors and evaluate a range of conditions to improve the engraftment of human microbiota. For instance, using a mouse model, a recent paper by Staley et al. [38] has shown that an appropriate antibiotic treatment protocol followed by a single gavage of human microbiota provides a useful, complimentary human microbiota-associated model to that established in germ-free facilities. A similar protocol could be applied to the rat model, although in the case of a colitis model, the use of an antibiotic cocktail could cause a bloom of Escherichia (Proteobacteria), which may sustain inflammation. This protocol would therefore need further evaluation on a model of colitis.
Our study presented several limitations. We used a 16S rRNA gene approach to characterise changes in the microbial community. Indeed, to evaluate the effect of treatments, more comprehensive approaches could have been applied. These include DNA and RNA shotgun sequencing, proteomic or metabolomic analysis. However, these approaches are also much more expensive and may not be worth this project using an animal model. Another limitation of the present work is the use of only one colitis model, which may not reproduce all the characteristics of human chronic UC. Another in vivo model such as TNBS-induced colitis might have provided different outcomes. However, DSS is used in many studies for its simplicity (given in drinking water and not by enema administration as for TNBS) and a relative short time to induce chronic colitis. Moreover, the evaluation of the combined effect of Citrafleet® and omeprazole unexpectedly uncovered a dramatic alteration of the microbiome composition. However, we did not assess whether this treatment could influence donor microbiome engraftment or dysbiosis caused by DSS. Future studies should provide details of the long-term effect of these drugs on engraftment and include another control group without this treatment. Finally, we performed HST using a single faecal sample collected from only one healthy donor. This approach did not therefore allow testing of whether a specific microbial community composition affects the efficiency of colonization and recipient immune response. Future studies could also test the effects of stool transfers at several time points on the efficiency of this procedure.
Our longitudinal study has provided valuable information regarding the natural evolution of the rat faecal microbiome over 112 days, demonstrating a continuous increase in alpha-diversity and changes in composition. These results suggest that, although rats reach sexual maturity at six weeks of age, their gut microbial community may still be developing until at least five months of age, the age that these rats reached after the 112-day follow-up. We propose that future studies determine the age at which rats should be tested in order to achieve greater stool transfer efficiency. Moreover, a longitudinal setting has allowed us to evaluate the resilience of the rat microbiome after HST and to monitor the disease severity index after withdrawal of DSS treatment.
Our study not only confirmed previous studies regarding the increase in DAI and histological scores and the decrease in colon length upon DSS treatment but also revealed a lower microbial diversity in stools, as well as in colonic tissues. These characteristics are also observed in both UC and CD patients [6], [39] and are more marked in the latter [7]. Many studies have shown that rat models of DSS-induced colitis share more features with UC than CD patients [19], [40], from a structural, clinical and ultrastructural point of view. Our comparison at microbiome level also confirmed that the rat model of DSS-induced colitis was relatively closer to UC than to CD patients.
In an attempt to understand the mechanism underlying a faecal microbiota transplantation as a potential treatment for IBD, we transferred a single stool collected from a healthy human donor to a humanised DSS-induced colitis rat model. Our findings revealed an improvement in several clinical and microbial variables. The relative recovery of colon length in response to HST confirmed an effect (direct or indirect) of an unaltered stool microbiota on modifying the structure of the colon. This effect overpowered the action of DSS, as its treatment in group E-HST2 was maintained until the end of the study. However, a complete restoration of colon length was not seen even after withdrawal of DSS treatment 40 days before the end of the study. This result is not consistent with the finding of a previous study by Bento et al. using a mouse model of DSS-induced colitis [34]. Those authors demonstrated complete restoration of colon length after withdrawal of DSS treatment. The discrepancy between studies could be attributable to the dose of DSS used (administration of 5% DSS for 5 days before a treatment of 2% for 17 days) in our work. Instead, in Bento et al., DSS was used at only 2% for 20 days. Furthermore, the transfer of a healthy human stool allowed the amelioration of clinical manifestations, leading to a significant decrease in the DAI scores, which were correlated with the relative abundance of several microbial genera in colonic samples. Interestingly, an unknown Christensenellaceae genus presented one of the highest negative correlations with DAI scores, and thus with disease severity. This genus was previously detected in a higher relative abundance in healthy individuals compared to IBD patients [7] and in normal weight individuals compared to overweight subjects [41], thereby validating its role in maintaining a healthy status. The S24-7 family, another remarkable group of bacteria, was depleted upon DSS treatment but increased after DSS withdrawal. Moreover, it was found to be positively correlated with colon length. This result is consistent with a previous study showing that S24-7 was depleted in a model of type 1 diabetes [42], thereby suggesting that this bacterial group may be a good candidate for maintaining homeostasis.
Moreover, HST led to a significant restoration of microbiome alpha-diversity in stool samples but not in colonic tissues unless DSS treatment was withdrawn. This observation indicates that HST alone was not sufficient to correct dysbiosis in mucosal tissues when inflammation was maintained. To the best of our knowledge, there are currently only four randomised published clinical trials on FMT in patients with UC [10], [43], [44], [45]. All of them showed low efficacy of the treatment (between 20 and 30% of success rate), without a clear understanding of the clinical events because of the obvious limitations of working with patients. Our study has pointed out possible reasons why FMT alone was not sufficient to treat IBD and recommends evaluation of the effect of anti-inflammatory drugs on FMT treatment efficacy. Moreover, pre-clinical models, such as the present one, allow the analysis of a wide range of clinical features, including intestinal length, and also mucosal tissue harvesting and a long-term follow-up, which would not be easily performed with patients.
Our study shows for the first time that a conventional rat model, more reflective of human microbiota, is a potential alternative to a mouse model to study microbe-host relationships in human disease. We demonstrate that a healthy stool transfer compensates faecal dysbiosis, with a restoration of alpha-diversity and an increase in the relative abundance of health-related genera. In addition, we show that a single faecal transplantation also corrects, to some extent, colonic tissue alterations, leading to a relative restoration of colon length, and a decrease in epithelium damage and disease severity. Finally, our data suggest that new strategies based on the combination of anti-inflammatory drugs with FMT may allow a better clinical outcome in IBD patients.
5. Authors' contributions
CM: Study concept and design; AS, GS and JW performed the animal experiments and acquired biological samples; AS, GS and JW processed samples for microbiome and immune response analyses; ML and MP analysed all the data; GS, ML and CM interpreted the data; CM drafted the manuscript; all authors provided critical revision of the manuscript for intellectual content; CM: obtained funding. ML and GS contributed equally.
6. Funding
Study funded by the Instituto de Salud Carlos III/FEDER (PI17/00614), a government agency. The funder had no role in study design, data collection, data analysis, interpretation or writing of the report.
Declaration of Competing Interest
None declared.
Acknowledgments
We thank Dr. Anna Barceló for her technical support at the Illumina sequencing platform (Servei de Genòmica i Bioinformàtica, UAB, Barcelona).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2019.10.002.
Appendix. Supplementary materials
References
- 1.Fiocchi C. Inflammatory bowel disease: etiology and pathogenesis. Gastroenterology. 1998;115:182–205. doi: 10.1016/s0016-5085(98)70381-6. [DOI] [PubMed] [Google Scholar]
- 2.Guarner F., Malagelada J.R. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 3.Baumgart D.C., Sandborn W.J. Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet. 2007;369:1641–1657. doi: 10.1016/S0140-6736(07)60751-X. [DOI] [PubMed] [Google Scholar]
- 4.Frank D.N., St Amand A.L., Feldman R.A., Boedeker E.C., Harpaz N., Pace N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ott S.J., Musfeldt M., Wenderoth D.F., Hampe J., Brant O., Folsch U.R. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pascal V., Pozuelo M., Borruel N., Casellas F., Campos D., Santiago A. A microbial signature for Crohn's disease. Gut. 2017;66:813–822. doi: 10.1136/gutjnl-2016-313235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Nood E., Vrieze A., Nieuwdorp M., Fuentes S., Zoetendal E.G., de Vos W.M. Duodenal infusion of donor feces for recurrent clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 9.Ianiro G., Masucci L., Quaranta G., Simonelli C., Lopetuso L.R., Sanguinetti M. Randomised clinical trial: faecal microbiota transplantation by colonoscopy plus vancomycin for the treatment of severe refractory clostridium difficile infection-single versus multiple infusions. Aliment Pharmacol Ther. 2018;48:152–159. doi: 10.1111/apt.14816. [DOI] [PubMed] [Google Scholar]
- 10.Moayyedi P., Surette M.G., Kim P.T., Libertucci J., Wolfe M., Onischi C. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology. 2015;149:102–109. doi: 10.1053/j.gastro.2015.04.001. e6. [DOI] [PubMed] [Google Scholar]
- 11.Paramsothy S., Kamm M.A., Kaakoush N.O., Walsh A.J., van den Bogaerde J., Samuel D. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. 2017;389:1218–1228. doi: 10.1016/S0140-6736(17)30182-4. [DOI] [PubMed] [Google Scholar]
- 12.Paramsothy S., Paramsothy R., Rubin D.T., Kamm M.A., Kaakoush N.O., Mitchell H.M. Faecal microbiota transplantation for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis. 2017;11:1180–1199. doi: 10.1093/ecco-jcc/jjx063. [DOI] [PubMed] [Google Scholar]
- 13.Sartor R.B., Wu G.D. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology. 2017;152:327–339. doi: 10.1053/j.gastro.2016.10.012. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vermeire S., Joossens M., Verbeke K., Wang J., Machiels K., Sabino J. Donor species richness determines faecal microbiota transplantation success in inflammatory bowel disease. J Crohns Colitis. 2016;10:387–394. doi: 10.1093/ecco-jcc/jjv203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colman R.J., Rubin D.T. Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis. 2014;8:1569–1581. doi: 10.1016/j.crohns.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang H., Fu L., Wang J. Protocol for fecal microbiota transplantation in inflammatory bowel disease: a systematic review and meta-analysis. Biomed Res Int. 2018;2018 doi: 10.1155/2018/8941340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pigneur B., Sokol H. Fecal microbiota transplantation in inflammatory bowel disease: the quest for the holy grail. Mucosal Immunol. 2016;9:1360–1365. doi: 10.1038/mi.2016.67. [DOI] [PubMed] [Google Scholar]
- 18.Antoniou E., Margonis G.A., Angelou A., Pikouli A., Argiri P., Karavokyros I. The TNBS-induced colitis animal model: an overview. Ann Med Surg (Lond) 2016;11:9–15. doi: 10.1016/j.amsu.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaudio E., Taddei G., Vetuschi A., Sferra R., Frieri G., Ricciardi G. Dextran sulfate sodium (DSS) colitis in rats: clinical, structural, and ultrastructural aspects. Dig Dis Sci. 1999;44:1458–1475. doi: 10.1023/a:1026620322859. [DOI] [PubMed] [Google Scholar]
- 20.Zhou J., Zhou Z., Ji P., Ma M., Guo J., Jiang S. Effect of fecal microbiota transplantation on experimental colitis in mice. Exp Ther Med. 2019;17:2581–2586. doi: 10.3892/etm.2019.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El-Salhy M., Wendelbo I.H., Gundersen D., Hatlebakk J.G., Hausken T. Colonoscopy with mucosal biopsies in young rats: a model for experimental gastroenterology. Mol Med Rep. 2013;7:1757–1760. doi: 10.3892/mmr.2013.1422. [DOI] [PubMed] [Google Scholar]
- 22.Oishi M., Tokuhara K., Miki H., Tanaka Y., Yamaki S., Kaibori M. Temporal and spatial dependence of inflammatory biomarkers and suppression by fluvastatin in dextran sodium sulfate-induced rat colitis model. Dig Dis Sci. 2014;59:2126–2135. doi: 10.1007/s10620-014-3163-x. [DOI] [PubMed] [Google Scholar]
- 23.Murthy S.N., Cooper H.S., Shim H., Shah R.S., Ibrahim S.A., Sedergran D.J. Treatment of dextran sulfate sodium-induced murine colitis by intracolonic cyclosporin. Dig Dis Sci. 1993;38:1722–1734. doi: 10.1007/BF01303184. [DOI] [PubMed] [Google Scholar]
- 24.Erben U., Loddenkemper C., Doerfel K., Spieckermann S., Haller D., Heimesaat M.M. A guide to histomorphological evaluation of intestinal inflammation in mouse models. Int J Clin Exp Pathol. 2014;7:4557–4576. [PMC free article] [PubMed] [Google Scholar]
- 25.Santiago A., Sanchez E., Clark A., Pozuelo M., Calvo M., Yanez F. Sequential changes in the mesenteric lymph node microbiome and immune response during cirrhosis induction in rats. mSystems. 2019;4 doi: 10.1128/mSystems.00278-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navas-Molina J.A., Peralta-Sanchez J.M., Gonzalez A., McMurdie P.J., Vazquez-Baeza Y., Xu Z. Advancing our understanding of the human microbiome using qiime. Methods Enzymol. 2013;531:371–444. doi: 10.1016/B978-0-12-407863-5.00019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar R.C. Search and clustering orders of magnitude faster than blast. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 28.Davis N.M., Proctor D.M., Holmes S.P., Relman D.A., Callahan B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chao A., Chazdon R.L., Colwell R.K., Shen T.-.J. Abundance-based similarity indices and their estimation when there are unseen species in samples. Biometrics. 2006;62:361–371. doi: 10.1111/j.1541-0420.2005.00489.x. [DOI] [PubMed] [Google Scholar]
- 30.Shannon C.E. A mathematical theory of communication. Bell System Technical Journal. 1948;27:379–423. [Google Scholar]
- 31.Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knights D., Costello E.K., Knight R. Supervised classification of human microbiota. FEMS Microbiol Rev. 2011;35:343–359. doi: 10.1111/j.1574-6976.2010.00251.x. [DOI] [PubMed] [Google Scholar]
- 33.Chassaing B., Aitken J.D., Malleshappa M., Vijay-Kumar M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr Protoc Immunol. 2014;104:15–25. doi: 10.1002/0471142735.im1525s104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bento A.F., Leite D.F., Marcon R., Claudino R.F., Dutra R.C., Cola M. Evaluation of chemical mediators and cellular response during acute and chronic gut inflammatory response induced by dextran sodium sulfate in mice. Biochem Pharmacol. 2012;84:1459–1469. doi: 10.1016/j.bcp.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 35.Park Y.H., Kim N., Shim Y.K., Choi Y.J., Nam R.H., Choi Y.J. Adequate dextran sodium sulfate-induced colitis model in mice and effective outcome measurement method. J Cancer Prev. 2015;20:260–267. doi: 10.15430/JCP.2015.20.4.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibbs R.A., Weinstock G.M., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S. Genome sequence of the brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 37.Pan H., Guo R., Zhu J., Wang Q., Ju Y., Xie Y. A gene catalogue of the Sprague-Dawley rat gut metagenome. Gigascience. 2018;7 doi: 10.1093/gigascience/giy055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staley C., Kaiser T., Beura L.K., Hamilton M.J., Weingarden A.R., Bobr A. Stable engraftment of human microbiota into mice with a single oral gavage following antibiotic conditioning. Microbiome. 2017;5:87. doi: 10.1186/s40168-017-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sokol H., Leducq V., Aschard H., Pham H.P., Jegou S., Landman C. Fungal microbiota dysbiosis in ibd. Gut. 2017;66:1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okayasu I., Hatakeyama S., Yamada M., Ohkusa T., Inagaki Y., Nakaya R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990;98:694–702. doi: 10.1016/0016-5085(90)90290-h. [DOI] [PubMed] [Google Scholar]
- 41.Garcia-Mantrana I., Selma-Royo M., Alcantara C., Collado M.C. Shifts on gut microbiota associated to mediterranean diet adherence and specific dietary intakes on general adult population. Front Microbiol. 2018;9:890. doi: 10.3389/fmicb.2018.00890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krych L., Nielsen D.S., Hansen A.K., Hansen C.H. Gut microbial markers are associated with diabetes onset, regulatory imbalance, and IFN-gamma level in nod mice. Gut Microbes. 2015;6:101–109. doi: 10.1080/19490976.2015.1011876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costello S.P., Hughes P.A., Waters O., Bryant R.V., Vincent A.D., Blatchford P. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA. 2019;321:156–164. doi: 10.1001/jama.2018.20046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paramsothy S., Nielsen S., Kamm M.A., Deshpande N.P., Faith J.J., Clemente J.C. Specific bacteria and metabolites associated with response to fecal microbiota transplantation in patients with ulcerative colitis. Gastroenterology. 2019;156:1440–1454. doi: 10.1053/j.gastro.2018.12.001. e2. [DOI] [PubMed] [Google Scholar]
- 45.Rossen N.G., Fuentes S., van der Spek M.J., Tijssen J.G., Hartman J.H., Duflou A. Findings from a randomized controlled trial of fecal transplantation for patients with ulcerative colitis. Gastroenterology. 2015;149:110–118. doi: 10.1053/j.gastro.2015.03.045. e4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.