Abstract
Background
Non-ischemic dilated cardiomyopathy (NIDCM) responds variably to intramyocardial injection of mesenchymal stem cells (MSCs). We hypothesized that NIDCM genotype may influence responsiveness to MSC therapy and performed genotyping on all patients in the POSEIDON-DCM trial.
Methods
POSEIDON-DCM patients (n = 34) underwent genetic sequence analysis and deletion/duplication testing. The results were classified as positive for pathological variants (PV+; n = 8), negative for any variants (V−; n = 6), or as variants of uncertain significance (VUS; n = 20). All outcomes of therapy were analysed for each category of genetic results.
Findings
The 3 groups were indistinguishable at baseline with regard to ejection fraction (EF), demographics, medication use, or functional parameters. V− patients had an increase in EF at 12 months: +13.6% (IQR = +7.8%; +20.5%; p = 0.002), compared with VUS (+6.5%; IQR = +0.9%, +11.1%; p = 0.005) and PV+(−5.9%; IQR = −12.7%, +1.0; p = 0.2; p = 0.01 between groups). Six-minute walk distance improved in V- patients, but not in VUS and PV+. V− patients improved MLHFQ, compared to the other 2 groups, which did not improve over time. EPC—CFUs increased by 9.7 ± 1.9 in V− (p = 0.009) compared to VUS and PV+ patients. V− patients had one-year survival (100%) compared with VUS (85%) and PV+ (40%; p = 0.015 log-rank). Similarly, MACE rates were lower in V− (0%) than PV+ (61.9%) or VUS (42.2%; p = 0.021 log-rank).
Interpretation
Our findings support the concept that the genetic profile of NIDCM patients plays a role in responsiveness to MSC therapy, with V− patients more likely to benefit and the converse for PV+. This observation emphasizes the need for further genetic studies, because of important implications for the management of NIDCM syndromes.
Keywords: Precision medicine, Variants of uncertain significance, Positive for pathologic/likely pathologic variant, Dilated cardiomyopathy
Abbreviations and acronyms
- 6MWT
six-minute walk test
- DCM
dilated cardiomyopathy
- EDM
end-diastolic mass
- EDV
end-diastolic volume
- EF
ejection fraction
- ESV
end-systolic volume
- HF
heart failure
- HFrEF
heart failure with reduced ejection fraction
- HFrecEF
hear failure with recovered ejection fraction
- LV
left ventricular
- LVEDD
LV end-diastolic diameter
- MI
myocardial infarction
- MLHFQ
Minnesota Living with Heart Failure Questionnaire
- MSCs
mesenchymal stem cells
- NIDCM
non-ischemic dilated cardiomyopathy
- NYHA
New York Heart Failure Association
- PV+
positive for pathologic/likely pathologic variant
- QOL
quality of life
- SI
sphericity index
- SV
stroke-volume
- TESI
transendocardial stem cell injection
- V-
negative for any pathologic variants
- VUS
variants of uncertain significance
Research in context
Evidence before this study
When the main POSEIDON DCM clinical trial was planned in 2010, TOPCARE-DCM trial (2009) and ABCD trial (2010) had reported the benefit of intracoronary delivery of autologous bone marrow derived cells by improving cardiac function and quality of life. During the time of development of our trial, some trials had controversial results; some supporting the use of other cells including CD 34+ (Vrtovec, 2013) cells. However, other data did not fully support the effect of allogeneic stromal cells (Perin, 2015) and multicellular therapy (ixmyelocel-T Henry (2014) in the NIDCM population. POSEIDON DCM demonstrated evidence for the superiority of allogeneic compared with autologous bone marrow derived MSCs in a subpopulation of patients that transitioned from HFrEF to HFrecEF. A meta-analysis of over 8000 patients (Kayvanpour, 2017) demonstrated that despite optimal standard of care in NIDCM patients, efficacy was affected by the genotype of the patient.
Added value of this study
To our knowledge, this post-hoc analysis of the POSEIDON DCM trial is the first one to evaluate the role of genetic variants in determining responsiveness to cell delivery, comparing genetically-associated NIDCM to those cases that do not appear to have genetic associations. Patients negative for any variants in genes associated with cardiovascular conditions are more likely to respond to cell delivery by improving cardiac function, quality of life, major adverse cardiovascular events, and survival, and the converse for patients positive for pathological variations.
Implications of all the available evidence
Future clinical trials evaluating cell therapy in NIDCM should take these results into account. The findings emphasize the need for further studies on genetic influences of cell therapy in NIDCM population and the major implications for personalized management of advanced cardiomyopathic syndromes.
Introduction
The PercutaneOus StEm Cell Injection Delivery Effects On Neomyogenesis in Dilated CardioMyopathy (The POSEIDON-DCM) trial identified a meaningful increase in ejection fraction (EF) in a cohort of patients with non-ischemic dilated cardiomyopathy (NIDCM) who received mesenchymal stem cells (MSCs) [1]. One-third of the patients transitioned from heart failure with reduced ejection fraction (HFrEF) to heart failure with recovered EF (HFrecEF). However, a variability in responses to intramyocardial injection of MSCs raised the question of patient-specific factors underlying this response dichotomy. Since a significant proportion of the NIDCM population burden appears to be familial, and are associated with variants in specific genes [2,3], we sought to test the hypothesis that genetic factors predict clinical responses and correlate with patient recovery.
Improvement of left ventricle function (LV) and/or restoration of LV geometry, referred to as reverse remodeling, is associated with improved quality of life (QOL) [4], and with reductions in mortality [5], left ventricular assist device (LVAD) implantation, heart transplants, and hospitalization [6]. The therapeutic response of HFrecEF is a recognized outcome with improved prognosis compared to HF with a persistently reduced EF [7]. Thus recovery of LV function in patients with cardiomyopathy is a meaningful goal of therapy in HFrEF.
Among patients with NIDCM, a substantial proportion have familial clustering and identifiable genetic variants [8], including mutations in cytoskeletal, nuclear membrane, sarcomere, mitochondria, desmosome, and RNA binding proteins. Several secondary modifiers, such as environmental factors, comorbidities, or other factors that modulate the phenotype and outcome, can alter these primary pathogenic variants. Our hypothesis was focused on the question whether a subgroup of NIDCM patients with an apparent genetic basis for the disease responded differently to MSC therapy than did those without associated variants.
The interpretation of variants identified by genetic testing is complex, based upon a gradient ranging from disease-causing variants, to variants of uncertain significance (VUS), and likely-benign or benign single nucleotide polymorphisms (SNPs). A consensus statement by the American College of Medical Genetics and Genomics/Association for Molecular Pathology (ACMG/AMP) provides a methodology for estimating probabilities of pathogenicity and the potential for a pathophysiological association between variants and phenotypes. This provides a system for classifying genetic variants as pathologic, likely pathogenic, variant of uncertain significance, likely benign, and benign [9]. This classification is intended to provide a uniform method for evaluating the likelihood of relevance of variants, especially those that are unique to individual families, and to assist medical decision-making by fulfilling the clinical diagnostic and predictive significance in the perspective of a genetic sequence change [10]. In this study, we sought to investigate the role of genetic variants in determining responsiveness to intramyocardial delivery of MSCs, comparing genetically-associated NIDCM to those cases that do not appear to have genetic associations.
Materials and methods
The prospectively designed Percutaneous Stem Cell Injection Delivery Effects On Neomyogenesis in Dilated Cardiomyopathy trial (POSEIDON-DCM; NCT01392625)(1), randomized 37 patients and tested 100 million MSCs cells of autologous versus allogeneic origin for patient outcomes.
Patients consented to genetic testing for genetic variants associated with NIDCM. Cells were delivered by 10 transendocardial stem cells injection (TESI), into 10 LV sites, distributed in a uniform pattern, using the Biosense Webster MyoStar NOGA Catheter System (Johnson & Johnson). Complete details are available in the full text manuscript [1] and the trial protocol can be found in the Supplementary Appendix.
Cardiac function and anatomy were evaluated by cardiac computed tomography (CT) or magnetic resonance imaging (MRI), depending on implantable cardiac devices. Functional capacity was assessed by six-minute walk distance test (6MWT) and forced expiratory volume at one second (FEV1). Quality of life (QOL) assessment included Minnesota Living with Heart Failure Questionnaire (MLHFQ) total score and New York Heart Failure Association (NYHA) functional class. All subjects provided written informed consent. This study was approved by the institutional review board (IRB) of the University Of Miami Miller School Of Medicine.
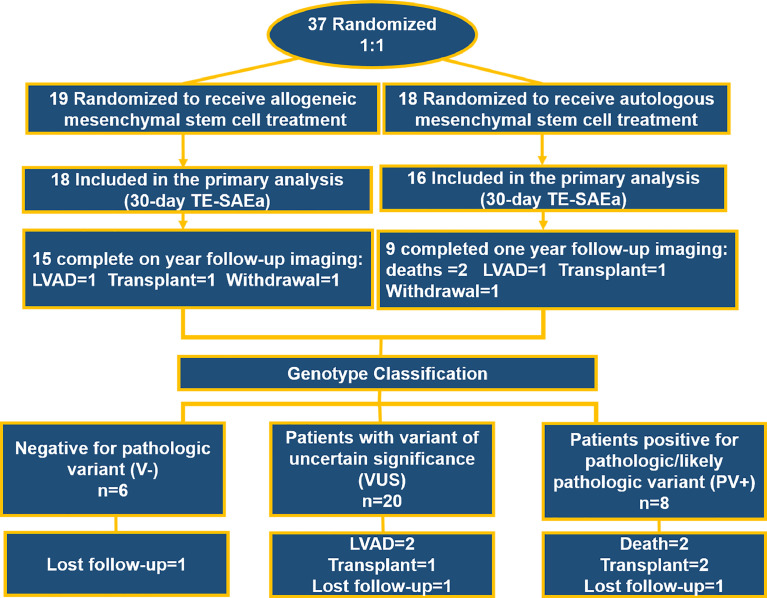
Imaging parameters were measured at baseline and 12 months after MSC injection, including EF and end diastolic (EDV) and systolic (ESV) volume, sphericity index (SI), as measured by cardiac CT or MRI. Functional parameters such as MLHFQ score, 6MWT, and NYHA classification were also evaluated at 3 and 6 months after cell delivery. Endothelial function parameters, including endothelial progenitor cell (EPC) colony formation and flow-mediated dilatation (FMD), were assessed at three months after cell delivery. Ten patients did not receive one-year imaging parameters and eight patients did not receive functional parameter assessment due to: Death (n = 2, not related to treatment), heart transplant (n = 3), automated implantable cardioverter-defibrillator placement (AICD) (n = 2) and withdrawal from the study (n = 3) (Fig. 1). Data measurements were performed as described and collected using a central electronic data system [1].
Fig. 1.
Consort diagram including initial randomization of original trial and genetic analysis. Thirty-four patients were included into the genetic analysis and divided into three groups; positive (PV+) or negative (V−) for pathologic variant and uncertain (VUS).
2.1. Genetic testing
POSEIDON-DCM patients (n = 34) underwent genetic analysis and were classified into groups according to the ACMG guidelines, including positive for pathologic/likely pathologic variant (PV+; n = 8), negative for any potentially-relevant variants (V-; n = 6), or those identified as variants of uncertain significance (VUS; n = 20). (Fig. 1) The analysis was based upon a comprehensive cardiomyopathy panel that included 105 genes, composed of a primary panel of 50 genes associated with inherited cardiomyopathies, 30 genes with preliminary evidence for association with cardiomyopathy, eight genes related to autosomal recessive syndromic paediatric cardiomyopathy genes and 17 genes associated to RASopathy, syndromes affected by mutations on genes of the Ras-MAPK pathway. The analysis included both gene sequencing and testing for deletions/duplications. [11,12]
2.2. Next generation sequencing and bioinformatics
Genetic testing was performed at Invitae (San Francisco, CA) as previously described [13]. Briefly, genomic DNA obtained from blood samples was subjected to target enrichment using hybridization capture with a custom bait pool, and sequenced using Illumina sequencing chemistry. A validated bioinformatics pipeline incorporating community standard and custom algorithms was used to identify sequence changes and exonic deletions/duplications simultaneously. Clinically significant observations were confirmed by orthogonal technologies, except individually validated variants and variants previously confirmed in a first-degree relative. Depending on the variant type, confirmation technologies may include any of the following: Sanger sequencing, Pacific Biosciences SMRT sequencing, MLPA, MLPA-seq, Array CGH.
2.3. Statistical analysis
Data distribution was evaluated using Shapiro-Wilk-Pearson normality test, for continuous measures. Continuous variables normally distributed were assessed by one-way ANOVA and presented as Mean ± SE. Non-normally distributed variables were assessed by Mann-Whitney test and reported by median and interquartile range [IQR]. Within data normally distributed were analysed by paired t-test, otherwise by Wilcoxon matched-pairs. Categorical variables were analysed by the Pearson chi-squared and Fisher's exact test as corresponding. Kaplan-Meier curve was used in the evaluation of survival distribution for the composite terminal event of all-cause death, cardiac transplantation, or LVAD placement. Imputation was not performed for missing data. All statistics were tested using two-sided at alpha=0.05. Analyses were done using GraphPad Prism7 (GraphPad Software, Inc. La Jolla, CA).
3. Results
3.1. Patient characteristics
Thirty-four patients with NIDCM were randomized and treated with autologous versus allogeneic MSCs. The patient population was 70.6% male. The mean age at the time of cell delivery was 55.12 ± 1.92 years and the duration of the disease prior to cell delivery was 4.68 (1.93, 9.23) years. Pathogenic variants were identified in 8 subjects (23.5%), among whom 5 had a positive family history of heart failure, 1 had an unknown family history and 2 had an identified DCM phenotype in the absence of a documented or reported family history of DCM. One-half of the V- patients had a positive family history of HF and in the VUS group, eight patients reported a positive family history of HF and two unknown family history for HF. Baseline characteristics of the genetic subgroups negative for any pathologic variants (V-), variants of uncertain significance (VUS), and patients harbouring known pathologic/likely pathologic variants (PV+), are shown in Table 1.
Table 1.
Baseline characteristics.
| V- | VUS | PV+ | Total | p-value | |
|---|---|---|---|---|---|
| Baseline Characteristics | n = 6 | n = 20 | n = 8 | n = 34 | |
| Age at cell delivery (years) | 57.83 ± 3.16 | 55.05 ± 2.79 | 53.25 ± 3.77 | 55.12 ± 1.9 | 0.76 |
| Years of Diagnosis | 2.1(2.09, 3.14) | 5.88(2.14, 10.97) | 6.2(1.84, 10.69) | 4.68(1.93–9.23) | 0.18 |
| Sex | 0.31 | ||||
| Male | 3(50%) | 16 (80%) | 5(62•5%) | 24(70.58%) | |
| Female | 3(50%) | 4 (20%) | 3(37.5%) | 10(29.41%) | |
| Cell delivery | 0.70 | ||||
| Allogeneic | 4 (66%) | 9 (45%) | 5(62.5%) | 18(52.94%) | |
| Autologous | 2 (33%) | 11(55%) | 3(37.5%) | 16(47.05%) | |
| History of Hypertension | 2(33.3) | 8 (40%) | 0(0%) | 10(29.41%) | 0.12 |
| History of Smoking | 3(50%) | 10(50%) | 4(50%) | 17(50%) | >0.9 |
| History of Hyperlipidemia | 1(16.6) | 7(35%) | 1(12.5%) | 9(26.47%) | 0.39 |
| History of Diabetes | 0(0%) | 1(5%) | 0(0%) | 1(2.94%) | 0.69 |
| History of TIA or CVA | 0(0%) | 2(2%) | 1(12.5%) | 3(8.82%) | 0.69 |
| Atrial Ventricular Arrhythmia | 1(16.6%) | 3(15%) | 2(25%) | 6(17.64%) | 0.82 |
| AICD | 4(66.6%) | 17(85%) | 8(100%) | 29(85.29%) | 0.22 |
| Medications | |||||
| Statins | 2(33.3%) | 8(40%) | 3(37.5%) | 13(38.2%) | 0.95 |
| ASA | 4(66.6%) | 10(50%) | 3(37.5%) | 17(50%) | 0.56 |
| Angiotensin 2 Blocker | 2(33.3%) | 4(20%) | 4(40%) | 10(29.4%) | 0.29 |
| B Blockers | 5(83.3%) | 19(95%) | 7(87.5%) | 31(91.2%) | 0.61 |
| ACE inhibitors | 2(33.3%) | 13(65%) | 3(37.5%) | 18(52.9%) | 0.23 |
| Diuretics | 4(66.6%) | 18(90%) | 8(100%) | 30(88.2%) | 0.14 |
| Other Anti hypertensives | 0 | 1(5%) | 0 | 1(2.9%) | 0.69 |
| Anti-arrhythmic | 1(16.6%) | 8(40%) | 3(37.5%) | 12(35.3%) | 0.57 |
| Ca+ channel inhibitors | 1(16.6%) | 0 | 0 | 1(2.9%) | 0.09 |
| Aldosterone inhibitors | 0 | 2(10%) | 1(12.5%) | 3(8.8%) | 0.69 |
| Pro-BNP | 806(278.9, 4506) | 822(246.8, 2313) | 1640(645.3, 2955) | 892(413.4, 2107) | 0.54 |
| NYHA | |||||
| Class I - No Limitation | 2 (33.3%) | 7 (35%) | 1 (12.5%) | 10(29.41%) | |
| Class II - Slight Limitation of Physical Activity | 3(50%) | 8 (40%) | 6 (75%) | 17(50%) | |
| Class III - Marked Limitation of Physical Activity | 1 (16.6%) | 5 (25%) | 1 (12.5) | 7(20.58%) | |
| Class IV - Marked Limitation at rest | 0(0%) | 0(0%) | 0(0%) | 0(0%) | |
| Peak VO2 (mL/kg/min) Median | 16.37 ± 1.82 | 18.34 ± 1.26 | 14.41 ± 1.42 | 17.03 ± 0.89 | 0.19 |
| Six Minute Walk Test (meters) | 437.3 ± 20.94 | 430.4 ± 24.13 | 390.5 ± 18.05 | 422 ± 15.12 | 0.51 |
| Forced Expiratory Volume in one second (%) | 2.07 ± 0.18 | 2.69 ± 0.16 | 2.57 ± 0.25 | 2.55 ± 0.12 | 0.16 |
| MLHFQ | 44.5 ± 12.13 | 33.75 ± 4.99 | 49.13 ± 7.32 | 36(17.75,64) | 0.26 |
| LV Size and Function | |||||
| Ejection Fraction (%) | 31.21(18.74, 31.95) | 24(17.37, 33.9) | 28.39 (21.87, 38.33) | 26.53(18.74, 32.6) | 0.84 |
| Left Ventricular End Diastolic Volume (ml): | 262.9(217.9, 285.4) | 345.9(281.8, 462.2) | 246.4(197.3, 396.6) | 296.7(246.4–429.3) | 0.07 |
| Left Ventricular Systolic Volume (ml): | 190.3(151.4222.6) | 267.2(182.3, 358.4) | 178.3(123.6, 306.2) | 233.7(168.5–325.7) | 0.16 |
| End Diastolic Sphericity Index | 0.519 ± 0.041 | 0.544 ± 0.024 | 0.576 ± 0.053 | 0.55 ± 0.02 | 0.69 |
| End Systolic Sphericity Index | 0.381 ± 0.033 | 0.411 ± 0.029 | 0.414 ± 0.045 | 0.407 ± 0.021 | 0.13 |
| Long Axis Diameter (mm) | 97.9 ± 3.19 | 111.6 ± 3.80 | 102.4 ± 7.57 | 107.3 ± 3.062 | 0.20 |
| End Diastolic Diameter (mm) | 66.13(59.49, 69.65) | 77.27(65.68, 85.03) | 67.75(58.65, 78.8) | 70.4(63.75, 80.1) | 0.10 |
| End Systolic Diameter (mm) | 60.5 42.91, 61.34) | 69.05(56.75, 76.93) | 61.8(46.1, 69.3) | 63.5(54.55,73.95) | 0.09 |
Values are n (%), mean ± SEM, or median (interquartile range). AICD indicates the automated cardioverter-defibrillator; MLHFQ, Minnesota Living with Heart Failure Questionnaire; NYHA, New York Heart Association (NYHA) Functional Classification.Table 2. Genetic profiles of individual patients. Colored cells represent V- (green), VUS (orange), or PV+ (red), Variants not relevant for cardiomyopathy (blue). Patient 10 and 28 have duplicated box because they are negative for PV's potentially related to DCM, but reported as present in the genes for completeness of genetic data.
Blood chemistries and biomarkers prior to cell delivery, including TNF-α, pro-BNP, blood urea, CRP, renal function, electrolytes, haemoglobin, red blood cells, platelets and cholesterol levels, were similar between the groups. Medications prior to cell delivery, including aldosterone blocker, angiotensin receptor blocker, beta-blocker and statins, were similar in all groups. Likewise, the presence of implantable cardioverter-defibrillators (ICD) were similar among the groups.
3.2. Sequencing summary and genetic basis
Variants were detected mainly in the structural myocardial coding genes, including sarcomere and z disk, and nuclear envelope. Ion channel and mitochondrial coding genes were not detected in this patient population. We observed multiple variants in 11 genes associated with DCM, including Ankyrin Repeat Domain (ANKRD), BCL2 Associated Athanogene 3 (BAG3), Dystrophin (DMD), GATA Zinc Finger Domain Containing 1 (GATAD1), LIM Domain Binding 3 (LDB3), Lamin A/C (LMNA), Myosin Binding Protein C, Cardiac (MYBPC3), Myosin Heavy Chain 6 (MYH6), RNA Binding Motif Protein 20 (RBM20), Troponin T2, Cardiac Type (TNNT2) and Titin (TTN). Eight positive pathological variants associated with DCM were identified, including 3 variants in TTN, 2 in LMNA, 2 in Desmoplakin (DSP) and 1 in MYBPC3. In addition, 9 variants of uncertain significance, that were possibly significant for pathological associations, were identified in ANKRD, BAG3, MYH6, RBM20, TNNT2 and TTN (Table 2 and Online Table 1).
Table 2.
Genetic profiles of individual patients. Colored cells represent V- (green), VUS (orange), or PV+ (red), Variants not relevant for cardiomyopathy (blue). Patient 10 and 28 have duplicated box because they are negative for PV's potentially related to DCM, but reported as present in the genes for completeness of genetic data. (For interpretation of the references to color in this Table legend, the reader is referred to the web version of this article.)
 |
Among the 34 patients, a total of 105 genes were sequenced, 39 positive pathological variants or variants of uncertain significance were identified in 28 subjects according to the ACMG guidelines. There were 8 positive pathological variants in 3 genes among 8 patients (100%) and 31 variants of uncertain significance in 23 genes among 20 patients. In the entire cohort, 21 patients (61.8%) had a single positive pathological variants or variant of uncertain significance, 5 had two positive pathological variants (15%), and 2 had more than three (6%) positive pathological variants or variants of uncertain significance in the same subject.
The most commonly affected gene was TTN, with variants observed in 6 patients, among which 3 (50%) were PV+. Importantly, one unique variant of uncertain significance in TTN Intron 248 had not been reported previously in normal or DCM patients, while other TTN VUSs are reported in both DCM and normal patients. The TTN subpopulation, which affected mostly male patients, was associated with severely impaired cardiac function, with EFs below 30%. TNNT2 was the second most frequent variant in our study and was found in 4 patients. Importantly, one unique variant of uncertain significance in TNNT2 [p.Arg151Cys], co-expressed in two unrelated patients, which gives greater likelihood to its pathological relevance. LMNA was the third most frequent gene, with variants found in three male patients, none of whom had a 12-month follow-up because all underwent heart transplantation owing to progression of disease in the absence of a response to MSC therapy.
3.3. Patient outcomes
Of the total of 34 NIDCM patients, 55% of those who were V- improved to HFrecEF, 55% of the VUS patients remained in the non-HFrecEF group, and 62.5% of the PV+ patients had an adverse outcome, defined as no significant benefit or worsening of cardiovascular status, requirement of LVAD or heart transplant, or death.
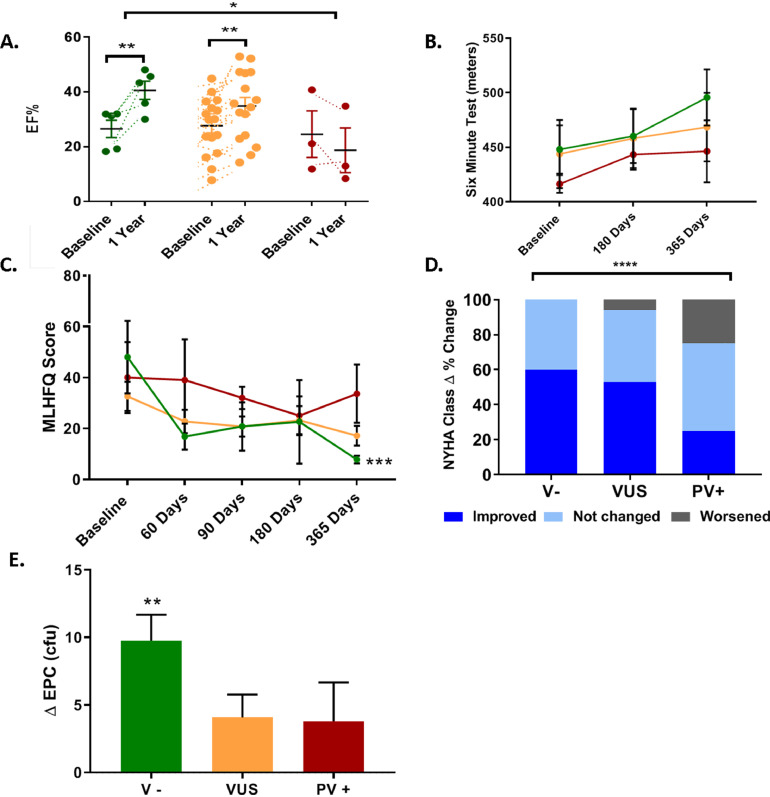
Among the specific measures of these clinical outcomes, V− patients had a significant increase in EF at 12 months: median change +13.6% (IQR = 7.8, 20.5; p = 0.002; V− vs VUS p = 0.24; V− vs. PV+ p = 0.009). This is compared to +6.5% (0.9, 11.1; p = 0.005; VUS vs PV+ p = 0.13) in the VUS category, and a trend toward a decline in PV+ patients, with a median change of −5.9% (IQR = −12.7, +1.0; p = 0.2; p = 0.01 between groups) (Fig. 2A). Cardiac volumes, including EDV and ESV, showed no significant difference between the groups. ESV was similar between VUS −24 mL (−54.5, 14.1; p = 0.04) and V−: −38.5 mL (−97.1, 0.8; p = 0.31) compared to PV+: +27.9 mL (17.0, 40.9; p = 0.25; p = 0.26 between groups). Similarly, EDV in the V− was −42.8 mL (−81.7, 48.2; p = 0.6) and VUS −10.3 mL (−38.6, 24.7; p = 0.5) vs. +43.3 mL (−37.7, 0.5) in the PV+ group (p = 0.74 between groups). Reverse cardiac remodelling was not different over time between groups.
Fig. 2.
Genotype modify responsiveness in cardiac function and functional capacity. A) V- patients had the greatest increase in EF at 12 months compared with VUS. EF decreased in PV+ patients. B) Six-minute walk distance increased in the V− patients vs. VUS and PV+. C) V− patients improved to a greater extent in MLHFQ contrasted with VUS and PV+. D) Percentage change in NYHA shows significant difference between groups. V− improved by 60% in contrast to 53% in VUS and 25% in PV+. E) EPC-cfu significantly increased over time only in V− group. V− = negative for any pathologic variants group (green), VUS = variants of uncertain significance group (orange), PV+ = positive for pathologic/likely pathologic variant group (red). *p = <0.05, **p = 0.01 ***p = 0.001 ****p<0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Functional capacity and QOL improved to a greater extent in the V− patients. Six-minute walk distance (6MWT), improved notably in V−: +33 m (30, 72.5; p = 0.06), but not in VUS: 32.5 m (0, 53; p = 0.24) and PV+: 39 m (−17, 68; p = 0.5; p = 0.9 between groups) (Fig. 2B). V− patients had the greatest improvement in MLHFQ by: −40.2 ± 14 (p = 0.0005) contrasted with VUS: −15.4 ± 6.1 (p = 0.07) and PV+: −6.3 ± 11.6 (p=>0.9; p = 0.3 between groups) (Fig. 2C). Interestingly, NYHA class improved by 60% in V- patients, compared to 53% in VUS patients and 25% in PV+ patients (between group delta% change p = <0.0001) (Fig. 2D). Endothelial function evaluated by EPC—CFUs was increased by 9.7 ± 1.9 ucf in V− (p = 0.009) in contrast to VUS: 4.1 ± 1.7 ucf (p = 0.07) and PV+: 3.8 ± 2.9 ucf (p = 0.4) (Fig. 2E). However, FMD from V− patients had the greatest increase over time, 3.1(2.16, 5.41, p = 0.12), it didn't reach any significant increase over time or compared to VUS = 0.27 (−1.52, 2.42, p = 0.64) and V+ = 0.88 (−0.065, 7, p = 0.18). Importantly, TNFα levels decreased significantly in all the groups from baseline to one-year follow-up. V− patients decreased by −6.93 ± 2•59pg/mL (p = 0.044), VUS by −8.68 ± 1.47pg/mL (<0.0001) and PV+ by-10.77 ± 1.97pg/mL (p = 0.0016).
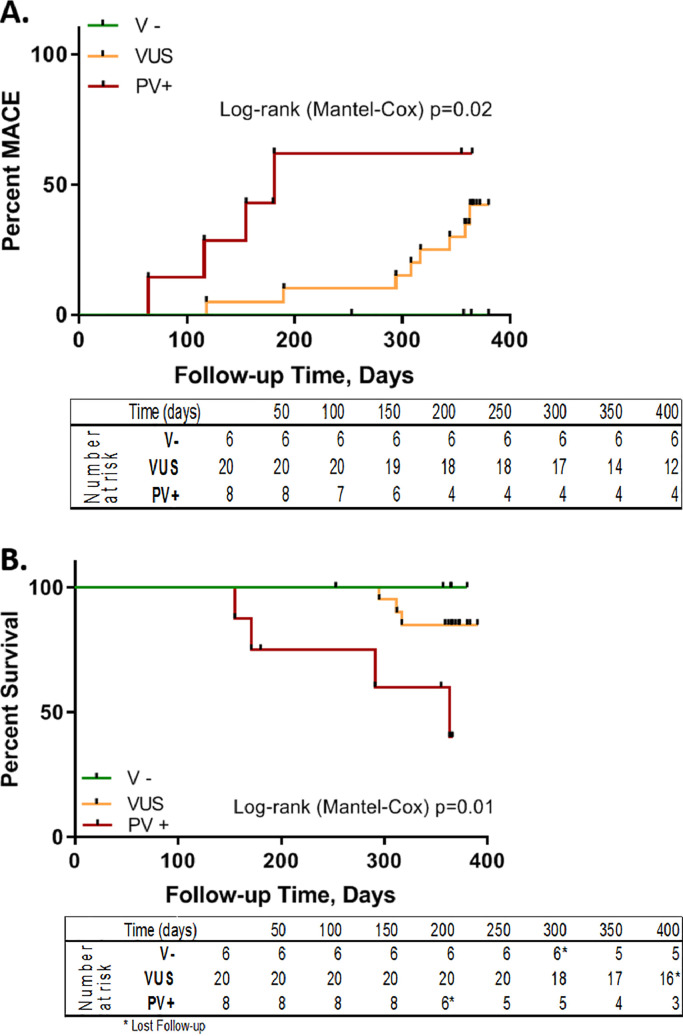
After a follow-up of up to one year, seven subjects had a terminal event, including death (n = 2), transplant (n = 3), or LVAD placement (n = 2). Three patients withdrew from the study, one in each group. Patients negative for any variants had greater one-year survival (100%) compared with VUS (85%) and PV+ (40%) (Log-rank (Mantel-Cox) test, p = 0.015; V− vs PV+ (P = 0.048); VUS vs VP+ (p = 0.018); V− vs VUS (p = 0.373)) (Fig. 3A). Interestingly, all patients who received a transplant had a mutation in LMNA. PV+ patients had a substantial increase of risk for death, transplant or LVAD by one-year follow-up. Similarly, MACE events differed between V- patients, who did not have any events, compared to VUS with seven events (42.2%) and PV+ patients who had fourth events (61.9%; Log-rank (Mantel-Cox) test, p = 0.021) (Fig. 3B).
Fig. 3.
Genetic variation affect MACE and Survival in response to MSC delivery. A) V− patients had 100% survival, VUS had 85% survival, and PV+ had 40% survival (Log-rank (Mantel-Cox) test p = 0.015). Overall, PV+ patient's had a substantial increase in death, transplant, or LVAD risk by 1 year follow-up. B) MACE events differed between groups; V- patients had 0 events, VUS had 7 events, and PV+ had 6 events in 4 patients (Log-rank (Mantel-Cox) test= 0.021). V- = negative for any pathologic variants group (green), VUS = variants of uncertain significance group (orange) PV+ = positive for pathologic/likely pathologic variant group (red). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Despite multiple phase I and II clinical trials, there is a continuing variability in responses to intramyocardial injection of MSCs and controversy as to the ideal patient population for cell treatment. In this study, we explored the genotypes of a NIDCM population to test the hypothesis of an association between genotype and effectiveness of MSC therapy. The results support the concept that these subpopulations are not discernible at baseline by phenotypic or demographic characteristics. However, the V- group demonstrated a significantly greater clinical benefit compared to VUS and PV+ groups, including improved cardiac function and functional capacity. Moreover, patients with pathogenic or likely-pathogenic variants in cardiomyopathy-associated genes had a decreased survival and increase in MACE events at one year. Overall, these findings strongly support for the idea that NIDCM genotype plays a major role in determining individual patient responsiveness to intramyocardial MSC delivery. Accordingly, these findings highlight the potential value of incorporating genetic testing to prospectively identify the potential for beneficial responses to cell delivery in patients with a similar disease phenotype.
Genotype–phenotype correlations and prognosis in genetic DCM must be cautiously analysed, as the majority of available data emerges from small, single institution studies, and single-family information. Although DCM has multifactorial pathogenesis that can lead to a final common phenotype, there are major challenges in identifying genetic bases or contributions to the disease and its response to therapy. These include incomplete knowledge of the genes associated with DCM, heterogeneity of phenotype due to incomplete penetrance and the potential for influences of modifier genes, and environmental, demographic[14], age[15], sex[16], and epigenetic modifications. In this study, we used the ACMG/AMP guidelines classification which includes variant pathogenicity and its effect with phenotype interpretation. [9] The World Heart Federation classification scheme for cardiomyopathies, using the evaluation of: Morphofunctional, Organ involvement, Genetic or familial, Etiology, Stage (MOGES) [17] are designed to integrate more parameters that can give a more complete landscape into the genetic DCM evaluation.
In our study, patients negative for pathogenic variants had a greater improvement in EF, with 55% of the V- patients transitioning to HFrecEF. This provides support for the concept that a subpopulation of NIDCM might have a preferential benefit from MSC therapy. Several hypotheses about the potential mechanisms of EF improvement induced by cell therapy are postulated. Etiologies of NIDCM, such as infectious, autoimmune, and toxicity[18,19], are characterized by persistent inflammation[20,21], cytokine and reactive oxygen species accumulation, nitroso-redox imbalance, fibrosis, apoptosis and impaired angiogenesis which consequently lead to left ventricular remodelling. MSC therapy has robust immunomodulatory effects, evidenced by the significant decrease TNF-α in all groups, and improved restoration of endothelial function [22], which could play a primordial mechanism in patients with negative mutations and favour the concept of independent contribution of the genotype. This is supported by the fact that the greatest increase in EPCs was in the V− patients, while conversely raising the possibility that genetically-based DCM may be resistant to these responses on an inherent cellular basis.
The genetic data provides evidence that VUS and PV+ patients had mild-to-absent responses and maintain as HFrEF, expressed by a higher mortality, LVAD requirements, transplant, and MACE events compared to HFrecEF [6]. Independent factors associated with blunted responses include long HF duration, greater EDV, NYHA classes III–IV, and lower systolic blood pressure [23]. Although we did not observe any significant differences in baseline characteristics between the groups, including medical therapy and cardiac resynchronization therapy, further studies are needed with larger cohorts to clarify these potential mechanisms. Our data did not shown associations between specific genetic variants and cell therapy response. This is not surprising since there is substantial evidence for a spectrum of genes and specific variants associated with inherited DCM.
Several single centre studies and meta-analyses have evaluated the relationship of phenotype at presentation and outcomes associated with variants in specific genes. Most showed clinical presentations in the fourth and fifth decades of life with prevalence in males. The more common associations include variants in LMNA, RBM20 and sarcomeric gene mutations including TTN, MYH6, MYBPC3 and TNNT [24], similar to those described previously in our population. Interestingly, patients with LMNA, PLN and RBM20 mutations were associated with a higher transplant rate and poor outcome, similar to our transplanted population. Tobita et al. evaluated left ventricular function and the presence or absence of reverse remodelling associated with genetic variants in patients with DCM. TTN truncating variants were the most common [15] and were associated with lower baseline EF. While patient with TTN truncating variants in our study (N = 6) also exhibit lower baseline ejection fraction, in contrast to Tobita's data, our patients who carried these variants did not improve left ventricle function over time. Nevertheless, better prognosis evidenced by lower transplant and death rate was observed in patients with TTN truncating variant after treatment compared to patients with LMNA truncating variant [25]. Similarly to previous studies, patients harbouring LMNA PV+ truncating variant had a worse outcome and higher (100%) heart transplantation [16,26]. Another factor evaluated in the responsiveness to cell delivery was the contribution of multiple positive pathological variants into worse outcome. Similar to previous studies, our study did not show evidence that multiple VUSs were associated with a worse prognosis. Hereditary defects in cardiac structural proteins might influence the response of cell delivery compared to favourable genetic background. Thus, the evaluation of the genetic burden should be considered in the contribution of each variant to the disease severity, as predicted by the natural history and thus to therapy indication.
Prognosis in patients with DCM has improved during the last decade, due to the earlier diagnosis and introduction of effective neurohormonal treatments [27,28] and device therapy. However, DCM is still the leading condition leading to heart transplant [29]. MACE events occur early after diagnosis and remain unpredictable. Merlo et al. found that 37% of DCM patients receiving tailored medical therapy, demonstrate an increase in EF or reverse remodelling of cardiac chambers. Moreover this improvement was associated to increase in survival [29]. Similarly, V− patients had a significantly increase in EF and survival was greatly increased, compared to PV+ patients. Interestingly, V− patients not only had a higher survival rate, but also had a significant decrease in MACE events after cell delivery after one year follow-up. Relevant genetic variants were identified in patients that had higher mortality and MACE events. This finding may refine the clarity of identification of patients who can potentially benefit from stem cell delivery and the meaningful impact in the treatment of NIDCM.
This study is limited by a small sample size and absence of a placebo group. The relatively limited power due to the sample size reduces the detection of small differences in the baseline factors that could influence the outcome. A subsequent larger study is currently being planned based upon the hypothesis generated in the current that will provide further insights into the genetic determinants of the therapeutic response and will advance precision medicine insights. Previous analysis in the original trial evaluated the difference between allogeneic vs autologous MSC treatment. The power to examine outcomes based on subgroups by genetic testing and treatment is limited because of the sample size. However, the distribution of the patients, based upon the treatment received, did not show any significant differences in baseline characteristics. We acknowledge the limitation of over interpreting these data due to the multiple comparisons analysis. Because of the natural history of DCM, longer follow-up to evaluate the recurrence of heart failure symptoms and clinical events will be preferable in trials. The cut-off used to differentiate patients with or without HFrecEF was 40%, based on the AHA guidelines [30]. However, this cut-off is controversial as 45 and 50% are used by other authors. Patients were distributed into 3 main groups depending the presence or absence of pathological variants, plus the spectrum of VUSs which are difficult to interpret, based upon factors that challenge their interpretation and lead to a range of likelihoods from likely benign to likely pathogenic. However, many studies use this classification, ideally ACMG/AMP classification including all the subcategories should be used. Further larger, double blinded, controlled studies need to validate the use of genotype-phenotype association testing as a prognostic factor of cell delivery effectiveness in patients with negative cardiac mutations. Finally, we do not have molecular information to evaluate whether remodelling features, such as cell hypertrophy, changes in excitation-contraction coupling of the myocyte, progressive loss of myofilaments, β-adrenergic desensitization, abnormal myocardial energetics, or progressive loss and/or disarray of the cytoskeleton, were corrected after cell delivery.
5. Conclusions
This hypothesis generating study has potential major implications for the management of advanced cardiomyopathy syndromes and supports the need of larger placebo-controlled studies that will provide further insights into the concept that genetic variability determines responsiveness to stem cell therapy in patients with NIDCM and will provide further precision medicine insights.
Clinical trial registration
POSEIDON-DCM: NCT01392625 https://clinicaltrials.gov/ct2/show/NCT01392625
Funding sources
This work was supported by NIH grant RO1 HL RO110737 and Miami Heart Research Institute (MHRI). Dr. Myerburg is supported in part by the American Heart Association Chair in Cardiovascular Research at the University of Miami Miller School of Medicine.
Author contributions
JH served as PI of POSEIDON-DCM study and oversaw its conduct AR, RM, JH conceptualizated and designed the study. MVC and DD data . AR, VF, BT, MN and CP did formal analysis: anonymize, preparation and analysis of CT and endothelial function. RM and AH intramyocardial injection of the cells, AK manufactured the cells, GMP facility director. AR and RM; DNA preparation, extraction, and analysis of genetic variants. AR primary statistical analysis. AR, RM, JH wrote the initial draft. IS reviewed & edited. All authors read and approved the final report
Declaration of Competing Interest
Aisha Khan reports holds an equity in AssureImmune Cord Blood Bank. Courtney Premer has a patent 20170360837 pending. Alan W. Heldman reports holds equity in Vestion Inc. and maintains a professional relationship as Chief Executive and member of the Board of Directors. Joshua M. Hare reports having a patent for cardiac cell-based therapy and holds equity in Vestion Inc. and maintains a professional relationship with Vestion Inc. as a consultant and member of the Board of Directors and Scientific Advisory Board. Vestion Inc. did not play a role in the design, conduct, or funding of the study. Dr. Joshua Hare is the Chief Scientific Officer, a compensated consultant and advisory board member for Longeveron and holds equity in Longeveron. Dr. Hare is also the co-inventor of intellectual property licensed to Longeveron. Longeveron did not play a role in the design, conduct, or funding of the study. Angela C. Rieger has no disclosures, Robert J. Myerburg has no disclosures, Victoria Florea has no disclosures, Bryon A. Tompkins has no disclosures, Makoto Natsumeda has no disclosures, Ivonne H. Schulman has no disclosures, Mayra Vidro-Casiano has no disclosures, Darcy L DiFede has no disclosures, Raul Mitrani, has no disclosures.
Acknowledgements
The authors thank to John P. Hussman Institute for Human Genomics Biorepository Core at the University of Miami, Miller School of Medicine for DNA extraction assistance and INVITAE for DNA analysis.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2019.09.043.
Appendix. Supplementary materials
References
- 1.Hare J.M., DiFede D.L., Rieger A.C. Randomized comparison of allogeneic versus autologous mesenchymal stem cells for nonischemic dilated cardiomyopathy: POSEIDON-DCM trial. J Am Coll Cardiol. 2017;69(5):526–537. doi: 10.1016/j.jacc.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haas J., Frese K.S., Peil B. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J. 2015;36(18):1123–1135a. doi: 10.1093/eurheartj/ehu301. [DOI] [PubMed] [Google Scholar]
- 3.Hershberger R.E., Givertz M.M., Ho C.Y. Genetic evaluation of cardiomyopathy: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG) Genet Med. 2018 doi: 10.1038/s41436-018-0039-z. [DOI] [PubMed] [Google Scholar]
- 4.Konstam M.A. Reliability of ventricular remodeling as a surrogate for use in conjunction with clinical outcomes in heart failure. Am J Cardiol. 2005;96(6):867–871. doi: 10.1016/j.amjcard.2005.05.037. [DOI] [PubMed] [Google Scholar]
- 5.Kalogeropoulos A.P., Fonarow G.C., Georgiopoulou V. Characteristics and outcomes of adult outpatients with heart failure and improved or recovered ejection fraction. JAMA Cardiol. 2016;1(5):510–518. doi: 10.1001/jamacardio.2016.1325. [DOI] [PubMed] [Google Scholar]
- 6.Basuray A., French B., Ky B. Heart failure with recovered ejection fraction: clinical description, biomarkers, and outcomes. Circulation. 2014;129(23):2380–2387. doi: 10.1161/CIRCULATIONAHA.113.006855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yancy C.W., Jessup M., Bozkurt B. 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on practice guidelines. J Am Coll Cardiol. 2013;62(16):e147–e239. doi: 10.1016/j.jacc.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 8.Kayvanpour E., Sedaghat-Hamedani F., Amr A. Genotype-phenotype associations in dilated cardiomyopathy: meta-analysis on more than 8000 individuals. Clin Res Cardiol. 2017;106(2):127–139. doi: 10.1007/s00392-016-1033-6. [DOI] [PubMed] [Google Scholar]
- 9.Richards S., Aziz N., Bale S. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards S., Aziz N., Bale S. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hershberger R.E., Givertz M.M., Ho C.Y. Genetic evaluation of cardiomyopathy - a Heart Failure Society of America practice guideline. J Card Fail. 2018;24(5):281–302. doi: 10.1016/j.cardfail.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly M.A., Caleshu C., Morales A. Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: recommendations by ClinGen's Inherited Cardiomyopathy Expert Panel. Genet Med. 2018;20(3):351–359. doi: 10.1038/gim.2017.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Truty R., Paul J., Kennemer M. Prevalence and properties of intragenic copy-number variation in Mendelian disease genes. Genet Med. 2018 doi: 10.1038/s41436-018-0033-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hershberger R.E., Siegfried J.D. Update 2011: clinical and genetic issues in familial dilated cardiomyopathy. J Am Coll Cardiol. 2011;57(16):1641–1649. doi: 10.1016/j.jacc.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herman D.S., Lam L., Taylor M.R. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366(7):619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Rijsingen I.A., Nannenberg E.A., Arbustini E. Gender-specific differences in major cardiac events and mortality in lamin A/C mutation carriers. Eur J Heart Fail. 2013;15(4):376–384. doi: 10.1093/eurjhf/hfs191. [DOI] [PubMed] [Google Scholar]
- 17.Hazebroek M.R., Moors S., Dennert R. Prognostic relevance of gene-environment interactions in patients with dilated cardiomyopathy: applying the MOGE(S) classification. J Am Coll Cardiol. 2015;66(12):1313–1323. doi: 10.1016/j.jacc.2015.07.023. [DOI] [PubMed] [Google Scholar]
- 18.Hare J.M., Walford G.D., Hruban R.H., Hutchins G.M., Deckers J.W., Baughman K.L. Ischemic cardiomyopathy: endomyocardial biopsy and ventriculographic evaluation of patients with congestive heart failure, dilated cardiomyopathy and coronary artery disease. J Am Coll Cardiol. 1992;20(6):1318–1325. doi: 10.1016/0735-1097(92)90243-g. [DOI] [PubMed] [Google Scholar]
- 19.Felker G.M., Thompson R.E., Hare J.M. Underlying causes and long-term survival in patients with initially unexplained cardiomyopathy. N Engl J Med. 2000;342(15):1077–1084. doi: 10.1056/NEJM200004133421502. [DOI] [PubMed] [Google Scholar]
- 20.Aukrust P., Ueland T., Lien E. Cytokine network in congestive heart failure secondary to ischemic or idiopathic dilated cardiomyopathy. Am J Cardiol. 1999;83(3):376–382. doi: 10.1016/s0002-9149(98)00872-8. [DOI] [PubMed] [Google Scholar]
- 21.Kittleson M.M., Minhas K.M., Irizarry R.A. Gene expression analysis of ischemic and nonischemic cardiomyopathy: shared and distinct genes in the development of heart failure. Physiol Genomics. 2005;21(3):299–307. doi: 10.1152/physiolgenomics.00255.2004. [DOI] [PubMed] [Google Scholar]
- 22.Premer C., Blum A., Bellio M.A. Allogeneic mesenchymal stem cells restore endothelial function in heart failure by stimulating endothelial progenitor cells. EBioMedicine. 2015;2(5):467–475. doi: 10.1016/j.ebiom.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merlo M., Pivetta A., Pinamonti B. Long-term prognostic impact of therapeutic strategies in patients with idiopathic dilated cardiomyopathy: changing mortality over the last 30 years. Eur J Heart Fail. 2014;16(3):317–324. doi: 10.1002/ejhf.16. [DOI] [PubMed] [Google Scholar]
- 24.Merlo M., Sinagra G., Carniel E. Poor prognosis of rare sarcomeric gene variants in patients with dilated cardiomyopathy. Clin Transl Sci. 2013;6(6):424–428. doi: 10.1111/cts.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tobita T., Nomura S., Fujita T. Genetic basis of cardiomyopathy and the genotypes involved in prognosis and left ventricular reverse remodeling. Sci Rep. 2018;8(1):1998. doi: 10.1038/s41598-018-20114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hasselberg N.E., Haland T.F., Saberniak J. Lamin A/C cardiomyopathy: young onset, high penetrance, and frequent need for heart transplantation. Eur Heart J. 2018;39(10):853–860. doi: 10.1093/eurheartj/ehx596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Groenning B.A., Nilsson J.C., Sondergaard L., Fritz-Hansen T., Larsson H.B., Hildebrandt P.R. Antiremodeling effects on the left ventricle during beta-blockade with metoprolol in the treatment of chronic heart failure. J Am Coll Cardiol. 2000;36(7):2072–2080. doi: 10.1016/s0735-1097(00)01006-8. [DOI] [PubMed] [Google Scholar]
- 28.Di Lenarda A., Secoli G., Perkan A. Changing mortality in dilated cardiomyopathy. Br Heart J. 1994;72(6 Suppl):S46–S51. doi: 10.1136/hrt.72.6_suppl.s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merlo M., Pyxaras S.A., Pinamonti B., Barbati G., Di Lenarda A., Sinagra G. Prevalence and prognostic significance of left ventricular reverse remodeling in dilated cardiomyopathy receiving tailored medical treatment. J Am Coll Cardiol. 2011;57(13):1468–1476. doi: 10.1016/j.jacc.2010.11.030. [DOI] [PubMed] [Google Scholar]
- 30.Yancy Clyde W., Jessup M., Bozkurt B. 2013 ACCF/AHA guideline for the management of heart failure. Circulation. 2013;128(16):e240–e327. doi: 10.1161/CIR.0b013e31829e8776. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.