Abstract
Background
Aberrant expression of p53 and its downstream gene p21 is closely related to alterations in cell cycle and cell proliferation, and is common among cancer patients. However, the underlying molecular mechanism has not been fully unravelled. ZER6 is a zinc-finger protein with two isoforms possessing different amino termini (N-termini) in their proteins, p52-ZER6 and p71-ZER6. The biological function of ZER6 isoforms, as well as their potential involvement in tumourigenesis and the regulation of p53 remain elusive.
Methods
The effect of ZER6 isoforms on p53 and p21 was determined using specific knockdown and overexpression. p52-ZER6 expression in tumours was analysed using clinical specimens, while gene modulation was used to explore p52-ZER6 roles in regulating cell proliferation and tumourigenesis. The mechanism of p52-ZER6 regulation on the p53/p21 axis was studied using molecular biology and biochemical methods.
Findings
p52-ZER6 was highly expressed in tumour tissues, and was closely related with tumour progression. Mechanistically, p52-ZER6 bound to p53 through a truncated KRAB (tKRAB) domain in its N-terminus and enhanced MDM2/p53 complex integrity, leading to increased p53 ubiquitination and degradation. p52-ZER6-silencing induced G0-G1 phase arrest, and subsequently reduced cell proliferation and tumourigenesis. Intriguingly, this regulation on p53 was specific to p52-ZER6, whereas p71-ZER6 did not affect p53 stability, most likely due to the presence of a HUB-1 domain.
Interpretation
We identified p52-ZER6 as a novel oncogene that enhances MDM2/p53 complex integrity, and might be a potential target for anti-cancer therapy.
Keywords: p53 ubiquitination, Cell cycle, Zinc-finger protein, ZER6, p52-ZER6
Research in context.
Evidence before this study
ZER6 is a zinc-finger protein that encodes two isoforms with different N-termini: p71-ZER6 and p52-ZER6. In our previous study, through a high-throughput screening for factors regulating the transcriptional activity of cell cycle regulator p21 using an shRNA expression vector library, we identified ZER6 as a candidate inhibitor of p21. However, the biological and pathological functions of ZER6 isoforms remain unknown.
Added value of this study
This study provides a first characterisation of the oncogenic functions of p52-ZER6, one of the ZER6 isoforms. p52-ZER6 possesses a truncated KRAB domain at its N-terminus, whose function has not been identified previously. We found that p52-ZER6 is highly expressed in tumour tissues, and is closely related to tumour progression. We revealed that p52-ZER6 is critical for inducing p53 degradation by enhancing MDM2/p53 complex stabilisation; furthermore, its truncated KRAB domain is essential for p53 binding. Concomitantly, p52-ZER6 silencing significantly increases p21 expression, leading to G0-G1 phase arrest, and subsequently reduces cell proliferation and tumour progression. However, p71-ZER6, another splicing isoform of ZER6, does not affect MDM2/p53 axis, most likely due to the presence of a HUB-1 domain.
Implications of all the available evidence
Our study provides new insights on the regulation of the MDM2/p53 axis and is the first report regarding the function of p52-ZER6 in tumourigenesis. Furthermore, our study suggests the potential of targeting p52-ZER6 for anti-cancer therapy.
Alt-text: Unlabelled Box
1. Introduction
p53 is one of the most important tumour suppressor genes and a key determinant of genome integrity [1,2]. p53 regulation occurs mainly at the level of protein stability, enabling its rapid accumulation and activation [3,4]. Its homeostasis is crucial for maintaining cellular and physiological functions, including cell cycle, DNA repair, and cell death [5]. Aberrant p53 expression is closely related to various diseases: over-activated p53 induces premature aging and radiation sickness; whereas its mutation could be found in approximately 50% of cancer patients [[6], [7], [8], [9]]. Furthermore, p53 is frequently down-regulated even in tumour patients with the wild-type gene, indicating that its altered expression is critical in carcinogenesis [10,11]. Despite its importance, the regulatory mechanism of p53 expression has not been fully elucidated.
Aberrant p53 expression is closely related to improper cell cycle regulation, leading to uncontrolled cell proliferation in tumour cells. p21 is a downstream target of p53 that blocks cell cycle progression by binding to cyclins and cyclin-dependent kinases, whose tightly controlled expression serves to fine-tune the cell cycle [[12], [13], [14], [15]]. As with p53, decreased p21 expression is also found in various tumours. In an effort to unravel the p53/p21 regulatory mechanism, we previously performed a high-throughput screening for factors regulating the transcriptional activity of p21 using a small hairpin RNA (shRNA) expression vector library covering 2065 genes [11]. From those candidates, we identified a unique isoform of zinc-finger-oestrogen receptor interaction, clone 6 (ZER6, also called ZNF398), a Krüppel C2H2-type zinc-finger protein family containing six C2H2-type zinc-fingers, as a novel p53 regulator. ZER6 encodes two isoforms with different N-termini: p71-ZER6, whose N-terminus contains a full-length Krüppel-associated box (KRAB) domain and a HTLV-I U5RE-binding protein 1 (HUB-1) domain; and p52-ZER6, whose N-terminus contains only 30 C-terminal amino acids of the KRAB domain (hereafter named truncated KRAB or tKRAB domain) [16]. To date, the biological and pathological functions of ZER6 isoforms remain unknown.
We report herein that p52-ZER6 is up-regulated in tumour tissue, and is crucial for tumourigenesis. p52-ZER6, but not p71-ZER6, is critical for the binding of mouse double minute 2 (MDM2) to p53 through its tKRAB domain; and is crucial for MDM2-induced p53 ubiquitination and proteasomal degradation, a major regulatory pathway for p53 homeostasis [[17], [18], [19], [20]]. Intriguingly, p71-ZER6, another isoform of ZER6, fails to enhance p53 ubiquitination, most plausibly due to the presence of the HUB-1 domain, which suppresses the above effect of p52-ZER6. Together, these findings not only identify p52-ZER6 as a novel oncogene, but also describe, for the first time, the role of a tKRAB domain. Moreover, our study sheds new light on the mechanism regulating p53 ubiquitination and stabilisation.
2. Materials and methods
2.1. Vectors construction
shRNA expression vectors were constructed as described previously [21]. Briefly, target sites were predicted using the algorithm for predicting specific target sites. The oligonucleotides bringing the hairpin structure, as well as overhanging and terminator sequences were synthesized, annealed and inserted into the BspM I sites of a pcPUR+U6i cassette vector containing a U6 promoter [21].
For p52-ZER6 and p71-ZER6 overexpression vectors (pcFLAG-p52 and pcFLAG-p71, respectively), human cDNA was obtained by reverse-transcribing total RNA extracted from HCT116 cells using the PrimeScript Reagent Kit with gDNA Eraser (Takara Bio, Dalian, China). The corresponding regions were then amplified from the cDNA using the Takara Prime STAR Max DNA Polymerase (Takara Bio), and the amplicons were cloned into the Hind III and EcoR I sites of pcFLAG [11].
Vectors expressing MDM2, p53, and ubiquitin, (pcMDM2, pcp53, and pcUbi, respectively), vector bringing FLAG sequence (pcFLAG), as well as wild-type p21 promoter luciferase reporter vector (p21-luc) and p21 luciferase reporter vector lacking p53 binding site (p21mut-luc), were constructed as described previously [11]. For vectors expressing N terminal of p52-ZER6 (pcFLAG-p52-N), C terminal of p52-ZER6 (FLAG-p52-C), p52-ZER6 with full length KRAB domain (pcFLAG-p52K), p52-ZER6 with full length KRAB domain and HUB-1 domain (pcFLAG-p52KH), and p52-ZER6 without tKRAB domain (pcFLAG-p52tKdel), the fragments were amplified from pcFLAG-p71 and cloned into the Hind III and EcoR I sites of pcFLAG. For vector expressing N terminal of p52-ZER6 fragment bringing nuclear localisation signal (NLS) sequence (pcFLAG-p52-N-NLS) and NLS-fused p52-ZER6 N terminal lacking tKRAB domain (pcFLAG-p52-N-NLStKdel), the NLS sequence were inserted at the C terminal of the fragments. For vector expressing His-tagged ubiquitin (His-Ubi), 6 × His sequence was inserted into the Nhe I and Hind III sites of pcUbi.
For vectors expressing p52-ZER6 (pET-22b-His-p52) and N terminal of p52-ZER6 (pET-22b-His-p52-N) bringing 6 × His, the fragments were amplified from pcFLAG-p71 and cloned into the Nde I and Xho I sites of pET-22b (Novagen, Darmstadt, Germany).
2.2. Cell cultures and cell lines
HCT116 cell lines were cultured in McCoy's 5A medium (Gibco, Life Technologies, Grand Island, NY) supplemented with 10% foetal bovine serum (Biological Industries, Beit Haemek, Israel), while HepG2 and U2OS cells were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum. HCT116 cell lines were verified using short-tandem repeat profiling method at Genetic Testing Biotechnology (Suzhou, China), while HepG2 and U2OS cells were verified using short-tandem repeat profiling method at Cell Bank, Type Culture Collection, Chinese Academy of Science (Shanghai, China). All cell lines have been routinely tested and found negative for mycoplasma contamination using Mycoplasma Detection Kit-QuickTest (Biotool, Houston, TX). Transfection was performed using Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer's instruction.
For constructing shp52 stable cell lines, shp52–1 and shp52–2 vectors were transfected separately into HCT116 cells. The transfected cells were selected using puromycin (final concentration: 1.2 μg/ml), and stable cell lines were established from single clones. Control stable cell line was established from single clone using shCon vector.
For gene silencing experiments, cells were seeded in 6-well plate and transfected with 2 μg of indicated shRNA expression vector. Twenty-four hour later, cells were subjected to puromycin selection (final concentration: 1.2 μg/ml) to eliminate untransfected cells. mRNA and protein samples were collected 24 h after puromycin selection.
For overexpression experiments, 5 × 105cells were seeded in 6-well plate and transfected with 2 μg of indicated overexpression vector. Twenty-four hour later, mRNA and protein samples were collected and subjected for further analysis.
For double-knockdown and rescue experiments, 5 × 105cells were seeded in 6-well plate and co-transfected with 1 μg of each indicated vector. Cells were subjected to puromycin selection (final concentration: 1.2 μg/ml) to eliminate untransfected cells. mRNA and protein samples were collected 24 h after puromycin selection.
For actinomycin D and cycloheximide treatment, 1 × 106 cells were seeded in 6-well plate and cultured for 24 h. Proteins were collected 1 h after the cells were treated with actinomycin D (final concentration: 1 μM) or cycloheximide (final concentration: 30 μg/ml) for 1 h.
For treatment with nutlin-3 (Medchem Express, Monmouth Junction, NJ), 5 × 105 cells were seeded in 6-well plate and cultured for 24 h, then the cells were transfected with 2 μg of indicated overexpression vectors. Six hours later, the medium was changed with medium containing nutlin-3 (final concentration: 5 μM). Proteins were collected after the cells being treated for 24 h.
2.3. Animal experiment
For the in vivo tumour study, BALB/c-nu/nu mice (male, body weight: 18–22 g, 6 weeks old) were purchased from the Third Military Medical University (Chongqing, China; Permit Number SYXK-PLA-20120031). Animal studies were approved by the Laboratory Animal Welfare and Ethics Committee of the Third Military Medical University, and carried out in the Third Military Medical University. All animal experiments conformed to the approved guidelines of Animal Care and Use Committee of Third Military Medical University. All efforts were made to minimize suffering.
Xenograft experiment was performed as previously described [22]. Briefly, BALB/c-nu/nu mice were randomly divided into three groups (n = 8), and each group was injected subcutaneously with 5 × 106 HCT116/shCon, HCT116/shp52–1 or HCT116/shp52–2 stable cells. The investigator was blinded to the group allocation and during the assessment.
2.4. Clinical human colon carcinoma specimens
Human colon carcinoma specimens were obtained from colon carcinoma patients undergoing surgery at Chongqing University Cancer Hospital (Chongqing, China), and stored in Biological Specimen Bank of Chongqing University Cancer Hospital. Patients did not receive chemotherapy, radiotherapy or other adjuvant therapies prior to the surgery. The specimens were snap-frozen in liquid nitrogen. Prior patient's written informed consents were obtained, and the experiments were approved by the Institutional Research Ethics Committee of Chongqing University Cancer Hospital, and conducted in accordance with Declaration of Helsinki.
2.5. Dual luciferase assay
For dual luciferase assay, 8 × 104 cells were seeded in 24-well plates. Twenty-four hour later, the cells were co-transfected with the indicated shRNA expression vectors, reporter vector, and the Renilla luciferase expression vector (pRL-SV40, Promega, Madison, WI) as internal control. Twenty-four hour after co-transfection, the luciferase activities were then measured with the Dual Luciferase Assay System (Promega).
2.6. RNA extraction and quantitative RT-PCR (qPCR) analysis
Total RNAs were extracted using Trizol (Invitrogen Life Technologies) according to the manufacturer's instruction. Each total RNA sample (1 μg) was reverse-transcribed into cDNA using the PrimeScript Reagent Kit with gDNA Eraser (Takara Bio), and qPCR was performed to assess the mRNA expression levels with SYBR Premix ExTaq (Takara Bio). The sequences of the primers used for qPCR were shown in Supplementary Table S1. β-Actin was used to normalise sample amplification. The results are shown as relative to the expression level in the corresponding controls, which are assumed as 1.
2.7. Western blotting analysis
For cell culture experiments, cells were collected and lysed with RIPA lysis buffer with protease inhibitor and phosphatase inhibitor cocktail (complete cocktail; Roche Applied Science, Mannheim, Germany). Western blot was performed as described previously [23]. Briefly, equal amounts of the sample proteins were electrophoresed on sodium dodecyl sulphate polyacrylamide gel and transferred to a polyvinylidene fluoride membrane (Millipore, Billerica, MA) with 0.45 μm pore. The antibodies used are listed in Supplementary Table S2, and immunoblotting with anti-β-Actin antibody was conducted to ensure equal protein loading. The signals were measured using Super Signal West Femto Maximum Sensitivity Substrate detection system (Thermo Scientific, Waltham, MA). Quantification was performed using Quantity One, and the result was normalized using β-Actin.
For protein degradation assay, 1 × 106 cells were seeded in 3.5 cm dish. Cycloheximide was added at the final concentration of 200 μg/ml. Protein samples were collected at indicated time points, and were subjected to western blotting analysis as described above.
2.8. Quantification of total cell number
Cells were transfected with indicated vectors, and 24 h after transfection, puromycin selection (final concentration: 1.2 μg/ml) was performed to eliminate the untransfected cells. The transfected cells were seeded into 96-well plates. The cell numbers were measured by Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) at indicated time points.
2.9. 5-ethynyl-2′-deoxyuridine (EdU) incorporation assay and colony formation assay
Cells were transfected with indicated shRNA expression vectors or overexpression vector and selected using puromycin as indicated above. EdU incorporation and staining were performed using Cell-Light™ EdU Apollo®488 In Vitro Imaging Kit (RiboBio, Guangzhou, China) according to the manufacturer's instruction. Nuclei were stained with DAPI. Images were taken with DMI6000B (Leica, Heidelberg, Germany). Quantification of EdU positive and DAPI positive cells was performed using Microsystems LAS AF-TCS MP5 (Leica), and the results are shown as the percentage of EdU positive cells to DAPI positive cells.
For colony formation assay, 500 cells were cultured in a six-well plate for 10 days. Cells were then fixed with 4% paraformaldehyde and stained with methylene blue. The colonies were then counted. The investigator was blinded during the assessment.
2.10. Cell cycle analysis
Cells were transfected with indicated shRNA expression vectors as described above. Selection were performed using puromycin selection (final concentration: 1.2 μg/ml) to eliminate untransfected cells. Cells were subjected for starvation for 24 h before being incubated further for 24 h under normal condition. Cells were then harvested and stained with propidium iodide (KeyGen Biotech, Nanjing, China). The percentages of the cells in each cell cycle phase were determined by flow cytometry.
2.11. Immunoprecipitation assay
Cells were seeded in 10 cm dish (4 × 106 cells/dish), and transfected with total 16 μg of indicated vectors. Total protein samples were collected and lysed with RIPA lysis buffer with protease inhibitor and phosphatase inhibitor cocktail (complete cocktail, Roche Applied Science), and cleared by centrifugation at 12,000 rpm. The supernatants were incubated at 4 °C for 4 h with Protein A+G Agarose beads (Beyotime Biotechnology, Shanghai, China) in the presence of indicated antibodies or IgG as control. Then, the immunoprecipitated proteins were subjected to immunoblotting analysis as described in the western blotting section.
2.12. Sequential immunoprecipitation assay
Sequential immunoprecipitation was performed as described previously [11,24]. Briefly, HCT116 cells seeded in 10 cm dish (4 × 106 cells/dish) were transfected with pcFLAG-p52, pcMDM2 and pcp53 (6 μg each) using Lipofectamine 2000 (Invitrogen Life Technologies). Total protein samples were treated as described in immunoprecipitation section, except that after immunoprecipitation with IgG or anti-FLAG antibody, the beads were washed three times and eluted by FLAG peptide solution (final concentration of 100 μg/ml) twice, and the immunoprecipitants were again immunoprecipitated with IgG or anti-MDM2 antibody. Then, the immunoprecipitants were subjected to immunoblotting analysis as described above in the western blotting section. The antibodies used were listed in Supplementary Table S2.
2.13. His fusion proteins preparations and His pull-down assay
His-p52 and His-p52-N fusion proteins were expressed in E. coli by transforming pET-22b-His-p52 and pET-22b-His-p52-N into E. coli strain BL21. The expression of His-p52 fusion proteins were then induced using isopropyl β-D-thiogalactopyranoside (final concentration: 5 mM), and the cells were then grown for 12 h at 16 °C. The E. coli were lysed and purified using Ni-NTA beads. The purified protein was then resolved using 12% SDS-PAGE for quantification and assessment of their purities.
The His-p52 proteins, GST-MDM2 or p53 were mixed with Ni-NTA beads in the binding buffer (PBS + 0.05% Tween). The mixtures were then washed with wash buffer (20 mM Tris-HCl +500 mM NaCl +5% glycerol +1 mM PMSF +20 mM imidazole) for 4 times, and the protein complexes were eluted using 1% SDS. The pulled-down proteins were subjected to western blotting analysis as described above.
2.14. In vivo ubiquitination assay
For ubiquitination assay of p53, cells were transfected with pcUbi, pcMDM2, pcp53 and indicated shRNA expression vector or overexpression vector (4 μg each). Twenty-four hour later, cells were treated with MG132 (final concentration 20 μM) for 8 h to block proteasomal degradation. Cells were lysed in RIPA buffer and immunoprecipitated using anti-p53 antibody followed by immunoblotting with an anti-ubiquitin antibody. Alternatively, cells were transfected with pcHis-Ubi, pcMDM2, pcp53 and indicated shRNA expression vector or overexpression vector (4 μg each). Cells were lysed in denaturing buffer (0.1 M Na2HPO4/NaH2PO4, 10 mM imidazole, 6 M guanidine-HCl pH 8.0) [25]. Cell lysate was pulled-down by incubation with Ni-NTA beads as described above in the His pull-down assay, followed by immunoblotting with the anti-p53 antibody.
2.15. Immunohistochemical analysis
Paraffin-embedded sections were obtained from fresh colon carcinoma and adjacent tissues or xenografted tumours at 4 μm thickness using a cryostat and subjected to immunohistochemistry. Briefly, the tissue sections were incubated with primary antibodies for 1 h. The specimens were then incubated with corresponding secondary antibodies conjugated with horse-radish peroxidase. Visualization was performed using a DAB Kit (DAKO, Beijing, China) under microscope. The nuclei were then counterstained with hematoxylin, followed by dehydration and coverslip mounting. The antibodies used were listed in Supplementary Table S2. Images were taken using Pannoramic Midi (3DHistech, Budapest, Hungary).
2.16. In situ hybridization
For in situ hybridization, paraffin-embedded tissue sections were rehydrated and digested with protease K for 25 min at 37 °C. After pre-hybridization at 37 °C for 1 h, the hybridization was carried out by overnight incubation at 37 °C with 8 ng/μl digoxin-conjugated probe specific for the p52-ZER6 isoform (91–113 at the 5’ UTR of p52-ZER6 mRNA; TCTCG TCTTC GACCG CATCC CTC). After being washed with SSC washing buffer at 37 °C, blocking was performed with bovine serum albumin (BSA) for 30 min. Specifically bound probes were detected by anti-DIG-HRP antibody (Servicebio, Wuhan, China). Nuclei were stained with hematoxylin. Images were taken using Pannoramic Midi (3DHistech).
2.17. Immunofluorescence staining
Cells were seeded in 3.5 cm culture dishes (2 × 105 cells per dish), fixed for 30 min at room temperature with 4% paraformaldehyde, permeabilized for 5 min with PBS containing 0.1% Triton X-100, blocked with 1% BSA for 1 h and incubated with primary antibodies for 2 h. The cells were then incubated with fluorescent-conjugated secondary antibodies for 1 h. The nuclei were stained with DAPI for 15 min. Images were taken with laser scanning confocal microscopy (Leica Microsystems-TCS SP5). The antibodies used were listed in Supplementary Table S2.
2.18. Statistical analysis
Quantification data were analysed by One-way ANOVA conducted using SPSS Statistics v. 17.0. For bioinformatics analysis, bivariate correlation analysis (Pearson's r test) was used to examine the correlation of two variables in human specimens. Kaplan-Meier method was used to analyse the correlation between relapse-free survival and variables in survival analysis, and the numbers at risk was analysed using GraphPad Prism 5. Survival curves were analysed using the log-rank test.
2.19. Data sharing
Patient data supporting the analyses in Figs. 1a and b, 2f and S3a were accessed from the public colon carcinoma and hepatocarcinoma data sets Gene Expression Omnibus (GEO, accession numbers GSE39582, GSE42284 and GSE25097). All other data and key resources table of this study are available in http://dx.doi.org/10.17632/zxkzw4nz68.1 (Mendeley data).
Fig. 1.
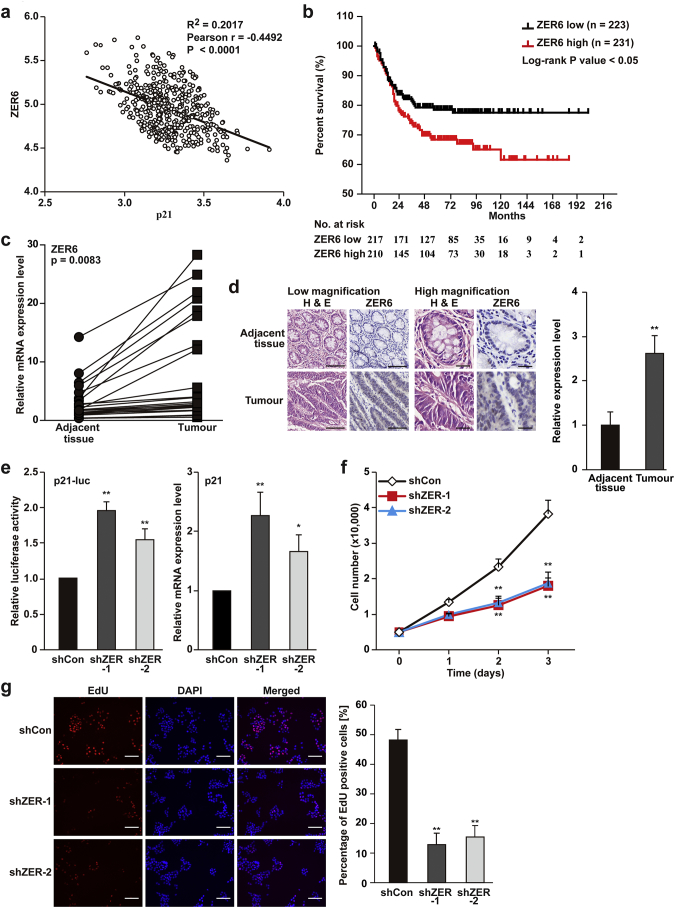
ZER6 shows negative correlation with p21 in tumour cells. (a–b) Bioinformatics analysis using GEO data set GSE39582 (n = 454): (a) correlation analysis of ZER6 and p21 expression in colon carcinoma specimens; and (b) Kaplan-Meier survival plots of colon carcinoma patients with either low (black) or high (red) ZER6 expression. (c) ZER6 mRNA expression level in clinical human colon carcinoma and corresponding adjacent tissue samples (n = 23), as determined by quantitative RT-PCR (qPCR). (d) Immunohistochemistry staining showing the expression levels and localisation of ZER6 in clinical human colon carcinoma and adjacent tissues samples. Representative of low-magnification (left; scale bars, 100 μm) and high magnification (right; scale bars, 25 μm) images, as well as quantification results were shown. (e) Activity of p21 reporter (left) and p21 mRNA expression level (right) in HCT116 cells transfected with shRNA expression vectors targeting different sites of ZER6, as determined by dual luciferase assay and qPCR, respectively. (f) Total cell number of ZER6-silenced HCT116 cells at the indicated time points. (g) Number of proliferative ZER6-silenced HCT116 cells, as determined by the EdU incorporation assay. Representative images (left) and the percentage of EdU-positive cells to DAPI positive cells (right) were shown. Cells transfected with control vector (shCon) were used as control. β-Actin was used for qPCR normalisation. Quantitative data were expressed as mean ± SEM of three independent experiments. ⁎P < .05; ⁎⁎P < .01 (ANOVA, unless otherwise indicated). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 2.
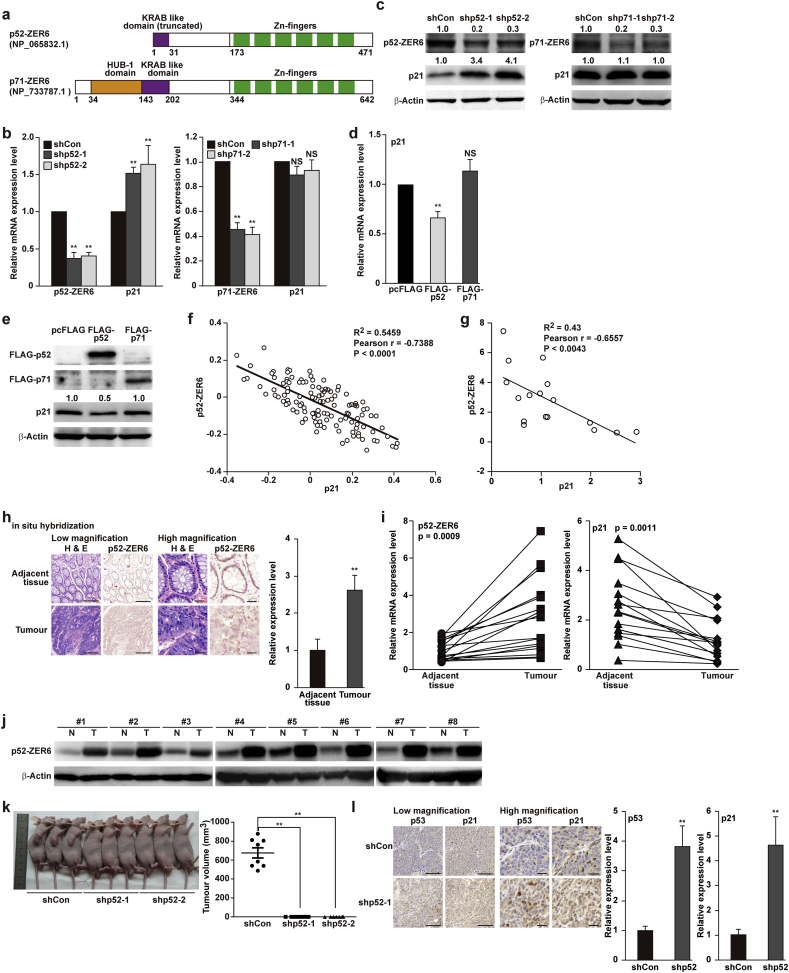
p52-ZER6 negatively regulates p21 and correlated with tumourigenesis. (a) Schematic diagram of the amino acid sequences of ZER6 isoforms. (b–c) p21 mRNA (b) and protein (c) expression levels in p52-ZER6- or p71-ZER6-silenced HCT116 cells, as determined using qPCR and western blotting, respectively. (d–e) p21 mRNA (d) and protein (e) expression levels in HCT116 cells overexpressing p52-ZER6 or p71-ZER6, as determined using qPCR and western blotting, respectively. (f) Correlation analysis of p52-ZER6 and p21 in colon carcinoma specimens (GEO data set: GSE42284, n = 120). (g) Correlation analysis between the mRNA expression levels of p52-ZER6 and p21 in clinical human colon carcinoma tissue, as determined using qPCR. (h) in situ hybridization showing the expression level of p52-ZER6 in clinical human colon carcinoma and adjacent tissues samples. Representative of low magnification (left; scale bars, 200 μm) and high magnification (right; scale bars, 25 μm) images, as well as quantification results were shown. Nuclei were stained with hematoxylin. (i–j) mRNA (i; n = 17) and protein (j; n = 8) expression levels of p52-ZER6 and p21 in clinical human colon carcinoma and corresponding adjacent tissue samples, as determined using qPCR and western blotting, respectively. (k) Representative morphological images (left) and tumour volume (right) of the BALB/c-nu/nu mice transplanted subcutaneously with HCT116/shCon, HCT116/shp52–1 and HCT116/shp52–2 stable cell lines at 14 days post-transplantation. (l) Immunohistochemistry staining showing the expression levels of p53 and p21 in tissue sections of xenografted tumours in BALB/c-nu/nu mice transplanted with HCT116/shCon (sacrificed 14 days post-transplantation) or HCT116/shp52–1 (sacrificed 28 days post-transplantation) stable cell lines. Representative of low-magnification (left; scale bars, 100 μm) and high magnification (right; scale bars, 25 μm) images, as well as quantification results were shown. Cells transfected with control vector (shCon or pcFLAG) were used as control. β-Actin was used for qPCR normalisation and as western blotting loading control. Quantitative data were expressed as mean ± SEM of three independent experiments. ⁎⁎P < .01; NS: not significant (ANOVA); N: adjacent normal tissue; T: tumour tissue.
3. Results
3.1. ZER6 is a novel p21 regulator
Through high-throughput screening, we previously identified ZER6 as a candidate for the control of p21, a key cell cycle regulator [11]. To elucidate the role of ZER6 in tumourigenesis, we firstly performed bioinformatics analysis using the NCBI GEO Database [26], and found a negative correlation between ZER6 and p21 expression, as well as between ZER6 level and patient survival in colon carcinoma (Fig. 1a and b). Furthermore, the results of qPCR and immunohistochemistry revealed that ZER6 expression was up-regulated in human colon carcinoma lesions compared to normal adjacent tissues (Fig. 1c and d).
p21 is a negative regulator of cell proliferation, and its expression is frequently aberrant in tumours [27,28]. To further confirm the link between ZER6 and p21, we constructed two shRNA expression vectors targeting different sites of ZER6 and confirmed that they efficiently suppressed ZER6 expression (Fig. S1). As shown in Fig. 1e, ZER6 silencing significantly enhanced p21 promoter activity and mRNA expression in human HCT116 colon carcinoma cells. Furthermore, ZER6 silencing robustly reduced HCT116 cell numbers (Fig. 1f) and cell proliferation as indicated by fewer EdU-positive cells (Fig. 1g). These findings identified ZER6 as a novel regulator of p21 transcription, with a possible role in tumour cell proliferation and tumourigenesis.
3.2. p52-ZER6 is critical for tumourigenesis
The two isoforms of ZER6, p52-ZER6 and p71-ZER6, have distinct N-termini and 5′-untranslated regions in their proteins and mRNAs, respectively (Figs. 2a and S2a) [16,29]. To investigate how these two isoforms regulated p21, we constructed shRNA vectors specific for p52-ZER6 and p71-ZER6, and examined their effect on p21 expression. Schematic diagrams with the locations of shRNA targets and primers used for qPCR are shown in Fig. S2b and c. The specificity of shRNAs targeting p52-ZER6 and p71-ZER6 were confirmed, as shRNA against p52-ZER6 had no significant effect on p71-ZER6 mRNA and protein, and vice versa (Fig. S2d and e). Intriguingly, only p52-ZER6 silencing, but not p71-ZER6 silencing, significantly enhanced p21 transcriptional activity (Fig. S2f) and expression (Fig. 2b and c). Similarly, p21 expression was suppressed only in HCT116 cells overexpressing p52-ZER6, whereas p71-ZER6 overexpression failed to yield any significant result (Fig. 2d and e).
Furthermore, analysis of the NCBI GEO Database [30,31] revealed an inverse expression pattern between p52-ZER6 and p21 in colon carcinoma and hepatocellular carcinoma (Figs. 2f and S3a), which was further confirmed in clinical colon carcinoma tissues (Fig. 2g). Moreover, in situ hybridisation revealed higher p52-ZER6 expression in clinical colon carcinoma tissues (Fig. 2h), which was further confirmed by qPCR and western blot analysis (Fig. 2i and j). Next, we performed a xenograft experiment by transplanting HCT116/shp52–1 or HCT116/shp52–2 stable cell lines (Fig. S3b and c). At the time of sacrifice (14 days post-transplantation), the size of xenografted tumours formed by control HCT116/shCon stable cell lines reached almost 2.0 cm, which corresponded to the maximum allowed by the ethics committee of the laboratory animal facility where the experiment was performed. In contrast, no obvious tumour formation could be detected in mice transplanted with HCT116/shp52 cell lines at the same time (Fig. 2k). Indeed, only a single very small tumour was formed in one mouse from each group transplanted with HCT116/shp52–1 and HCT116/shp52–2 cells 28 days after transplantation. Concomitantly, immunohistochemistry of tissue sections from xenografted tumours showed enhanced p53 and p21 expression in tumour initiated by p52-ZER6-silenced cells (Fig. 2l). Together, these results suggested that p52-ZER6 was crucial for colorectal tumourigenesis.
3.3. The p52-ZER6/p21 pathway regulates cell cycle and tumour cell proliferation
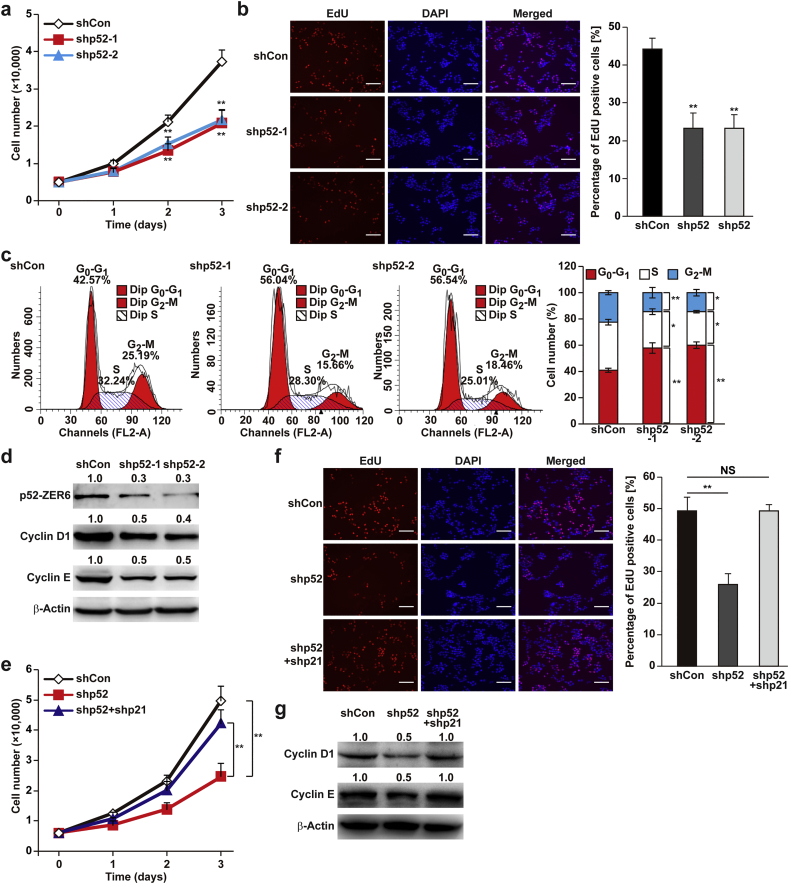
p21 is a master regulator of the cell cycle; thus, next, we investigated the role of p52-ZER6 in cell cycle and proliferation. p52-ZER6 silencing largely reduced total cell number and the percentage of EdU-positive cells (Fig. 3a and b), while its overexpression significantly induced them (Fig. S4a and b). Furthermore, p52-ZER6 silencing grossly increased the percentage of G0-G1 cells (Fig. 3c). Concomitantly, it down-regulated the expression of G1 phases cyclins D1 and E (Fig. 3d).
Fig. 3.
p52-ZER6 regulates cell cycle progression and cell proliferation. (a) Total cell number of p52-ZER6-silenced HCT116 cells at the indicated time points. (b) Number of proliferative p52-ZER6-silenced HCT116 cells, as determined by the EdU incorporation assay. Representative images (left) and the percentage of EdU-positive cells to DAPI positive cells (right) were shown. (c) Different cell cycle phases of p52-ZER6-silenced HCT116 cells. Cells were stained with propidium iodide, and the percentages were determined by flow cytometry. Representative images are shown (left), and the average percentage of cells in each cell cycle phase from three independent experiments was calculated (right). (d) Protein expression levels of G1 cyclins in p52-ZER6-silenced HCT116 cells. (e) Total cell number of p52-ZER6, p21-double knockdown HCT116 cells at the indicated time points. (f) Number of proliferative p52-ZER6, p21-double knockdown HCT116 cells, as determined by the EdU incorporation assay. Representative images (left) and the percentage of EdU-positive cells to DAPI positive cells (right) were shown. (g) Protein expression levels of G1 cyclins in p52-ZER6, p21-double knockdown HCT116 cells. Cells transfected with control vector (shCon) were used as control. β-Actin was used as western blotting loading control. Quantitative data were expressed as mean ± SEM of three independent experiments. ⁎P < .05; ⁎⁎P < .01; NS: not significant (ANOVA).
To elucidate the role of p21 in p52-ZER6-regulated cell proliferation, we suppressed the increase in p21 caused by p52-ZER6 silencing by knocking down both p52-ZER6 and p21 using shRNA expression vectors (Fig. S5a and b). Silencing of both genes significantly restored the total cell number and that of proliferative cells (Fig. 3e and f). Thus, p52-ZER6 silencing suppressed tumour cell proliferation by enhancing p21 expression, which was further confirmed by increased expression of cyclins D1 and E in double-knockdown cells (Fig. 3g).
3.4. p52-ZER6 enhances p53 ubiquitination
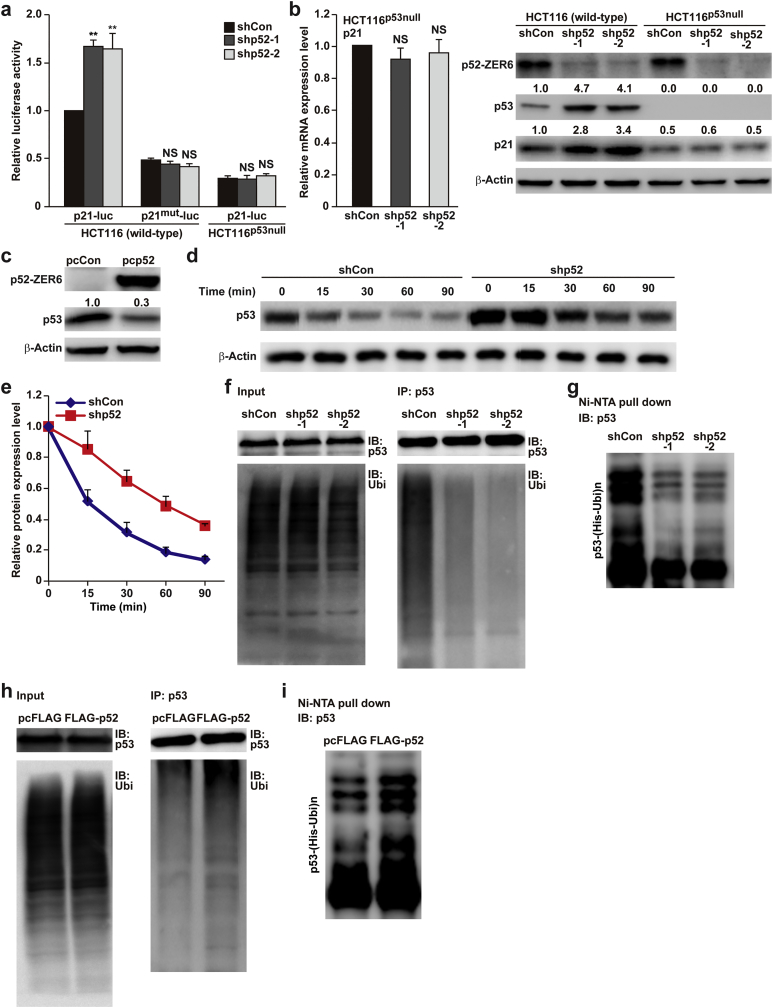
Given that p53 is a transcriptional activator of p21, we next analysed the activity of a wild-type p21 reporter (p21-luc) and a p21 reporter without p53 binding site (p21mut-luc) [11] in p52-ZER6-silenced wild-type HCT116 cells. Whereas p52-ZER6 silencing robustly increased wild-type p21 activity, it failed to affect that of p21mut-luc (Fig. 4a). Furthermore, p52-ZER6 silencing in HCT116p53null cells (Fig. S6a) failed to significantly affect the activity of the wild-type p21 reporter (Fig. 4a). Concomitantly, p52-ZER6 silencing did not affect the expression levels of p21 as well as cyclin D1 and cyclin E in HCT116p53null cells (Figs. 4b and S6b). Even though p21 expression could also be regulated in a p53-independent manner [32], these results indicated that p53 was necessary for p52-ZER6-dependent regulation of p21 transcription.
Fig. 4.
p52-ZER6 suppresses p21 expression by promoting p53 ubiquitination and enhancing its instability. (a) Activities of wild-type p21-luc and p21mut-luc in the indicated cells, as analysed by dual luciferase assay. Wild-type HCT116 cells co-transfected with shCon and p21-luc were used as control. (b) p21 mRNA (left) and protein (right) expression levels in p52-ZER6-silenced HCT116p53null cells, as determined using qPCR and western blotting, respectively. (c) p53 protein expression level in p52-ZER6-overexpressed HCT116 cells, as analysed using western blotting. (d–e) Degradation rates of p53 in HCT116/shCon and HCT116/shp52 stable cell lines. The levels of p53 protein at the indicated time points after the addition of cycloheximide (final concentration: 200 μg/ml) were analysed using western blotting (d), and the half-life of p53 protein was determined based on the quantification of western blotting results (e). (f–i) p53 ubiquitination level in p52-ZER6-silenced (f, g) or p52-ZER6-overexpressed (h, i) HCT116 cells were analysed using anti-ubiquitin immunoblotting of cell lysates immunoprecipitated with anti-p53 antibody (f, h), or using in vivo ubiquitination assay conducted using Ni-NTA pull-down under denaturing condition followed by immunoblotting (g, i). Cells were treated with MG132 to inhibit proteasomal degradation. For inputs of immunoprecipitation assay, 40 μg of corresponding samples were loaded. Ubi, ubiquitin; IP: immunoprecipitation; IB: immunoblotting. Cells transfected with shCon or pcFLAG were used as controls. β-Actin was used for qPCR normalisation and as western blotting loading control. ⁎⁎P < .01; NS: not significant (ANOVA).
Furthermore, our results showed that p52-ZER6-silencing induced p53 accumulation in HCT116 cells (Fig. 4b); while p52-ZER6 overexpression suppressed p53 accumulation (Fig. 4c), suggesting a regulatory effect of p52-ZER6 on p53 expression. Given that transcriptional inhibition using actinomycin D, as well as inhibition of de novo protein synthesis using cycloheximide did not alter the effect of p52-ZER6 silencing on p53 accumulation (Fig. S6c), we hypothesised that p52-ZER6 regulated p53 at a post-translational stage.
Ubiquitin-mediated degradation through the proteasomal pathway has been well-known as the canonical mechanism for regulating p53 protein homeostasis. Time-dependent protein degradation rate assays revealed that p52-ZER6 silencing substantially decreased the p53 degradation rate (Fig. 4d); concomitantly, p52-ZER6 silencing prolonged p53 protein half-life from about 15 min to about 45 min (Fig. 4e), indicating that p52-ZER6 might be involved in p53 protein stability. Furthermore, p52-ZER6 silencing robustly suppressed p53 ubiquitination (Fig. 4f and g), whereas its overexpression promoted it (Fig. 4h and i). Together, these results suggested that p52-ZER6 regulated the p53/p21 pathway by promoting p53 protein ubiquitination and degradation. Moreover, increase of p53 and p21 proteins was observed in p52-ZER6-silenced human HepG2 hepatocellular carcinoma and U2OS sarcoma cell lines (Fig. S6d), suggesting that regulation of the p53/p21 axis by p52-ZER6 might be common in cancers.
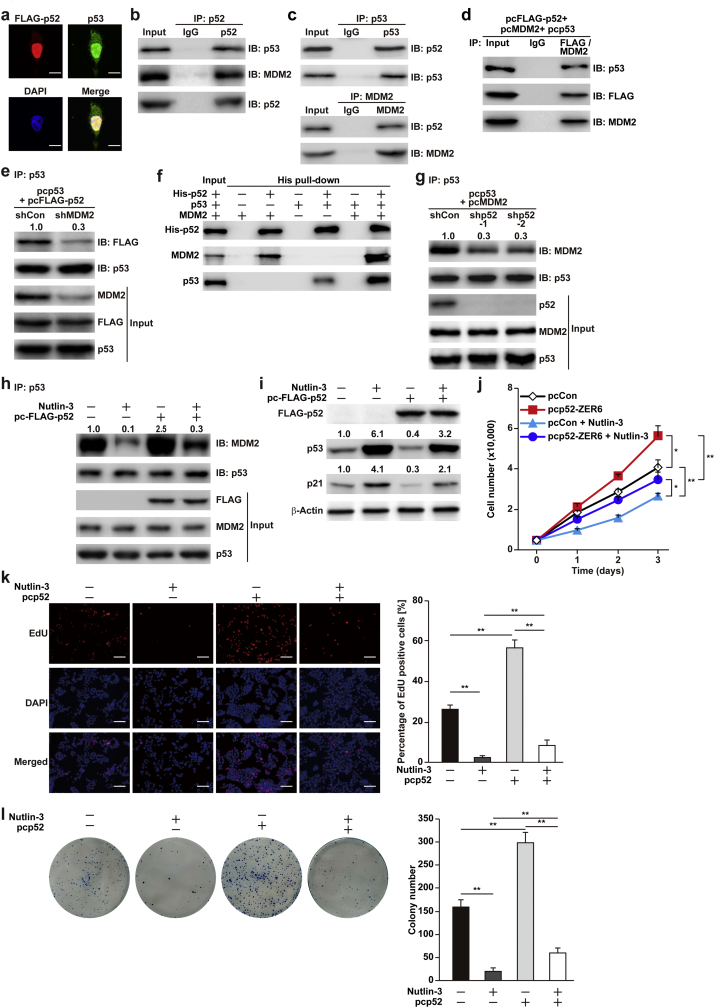
3.5. p52-ZER6 destabilises p53 by enhancing the MDM2-p53 interaction
Next, we sought to determine whether p52-ZER6 regulated p53 stability directly. We found that p52-ZER6 co-localised with p53 in the nucleus (Fig. 5a), where p53 ubiquitination occurs [33]. Immunoprecipitation of endogenous p52-ZER6 protein using anti-p52-ZER6 antibody demonstrated the existence of a physical interaction between p52-ZER6 and p53, as well as with MDM2, an E3 ligase that promotes p53 ubiquitination (Fig. 5b). These results were further confirmed by immunoprecipitation using anti-p53 or anti-MDM2 antibodies (Fig. 5c). Moreover, the results were consistent with immunoprecipitation in HCT116 cells overexpressing p52-ZER6 and p53 or p52-ZER6 and MDM2 (Fig. S7a and b). Furthermore, through sequential immunoprecipitation using anti-FLAG and anti-MDM2 antibodies, we detected p53 in the p52-ZER6/MDM2 complex (Fig. 5d), revealing that p52-ZER6, MDM2, and p53 could form at least a tertiary protein complex. Moreover, MDM2 silencing (Fig. S7c) reduced the amount of p52-ZER6 co-precipitated with p53 (Fig. 5e); while overexpression of p52-ZER6 in MDM2-silenced cells failed to suppress the expression of p53 and p21 (Fig. S7d). These results indicated that MDM2 was required for the interaction between p53 and p52-ZER6, as well as for the regulation of p52-ZER6 on p53. However, MDM2 expression in p52-ZER6-silenced HCT116 cells did not show any significant changes (Fig. S7e). The bindings between p52-ZER6, p53 and MDM2 proteins were further confirmed by in vitro analysis: a His pull-down assay clearly revealed the physical interactions between p52-ZER6, MDM2, and p53 (Fig. 5f). Interestingly, the His pull-down assay also demonstrated that binding between p52-ZER6 and MDM2 or p53 increased robustly when all three proteins were present.
Fig. 5.
p52-ZER6 enhances the integrity of MDM2/p53 protein complex. (a) Co-localisation of p52-ZER6 and p53 in HCT116 cells, as determined by immunofluorescence staining. Scale bars, 10 μm. (b–c) Physical interactions between endogenous p52-ZER6 and p53 or MDM2 proteins in HCT116 cells, as determined by anti-p53 or anti-MDM2 immunoblotting of cell lysate immunoprecipitated with anti-ZER6 antibody (b), and vice versa (c). (d) Physical interaction between p52-ZER6, MDM2 and p53 in HCT116 cells transfected with pcFLAG-p52, pcMDM2 and pcp53, as detected by sequential immunoprecipitation using anti-FLAG and anti-MDM2 antibodies consecutively. (e) Binding capacity of p52-ZER6 to p53 in MDM2-silenced HCT116 cells. Cells were treated with MG132 to inhibit proteasomal degradation. Cell lysates were immunoprecipitated against anti-p53 antibody. The presence of p52-ZER6 was detected by immunoblotting. (f) Physical interactions between p52-ZER6 (His-p52), MDM2 and p53, as determined by an in vitro His pull-down assay. (g) Binding capacity of MDM2 to p53 in p52-ZER6-silenced HCT116 cells. Cells were treated with MG132 to inhibit proteasomal degradation. Cell lysates were immunoprecipitated against anti-p53 antibody. The presence of MDM2 was detected by immunoblotting. (h) Binding capacity of MDM2 to p53 in HCT116 cells overexpressing p52-ZER6 and treated with nutlin-3 (final concentration: 5 μM). Cells were treated with MG132 to inhibit proteasomal degradation. Cell lysates were immunoprecipitated against anti-p53 antibody. The presence of MDM2 was detected by immunoblotting. (i) p53 and p21 protein accumulation in HCT116 cells overexpressing p52-ZER6 and treated with nutlin-3 (final concentration: 5 μM), as examined using western blotting. (j) Total cell number of p52-ZER6-overexpressed HCT116 cells treated with nutlin-3 at the indicated time points. (k–l) Proliferation (k) and colony formation potential (l) of p52-ZER6-overexpressed HCT116 cells treated with nutlin-3, as determined by the EdU incorporation assay and colony formation assay, respectively. Representative images (left) and quantification results (right) were shown. For inputs of immunoprecipitation assay and His-pull down assay, 40 μg and 40 ng of corresponding samples were loaded, respectively. IP: immunoprecipitation; IB: immunoblotting. Cells transfected with shCon or pcFLAG were used as controls. β-Actin was used as western blotting loading control.
We next questioned whether p52-ZER6 affected MDM2 and p53 binding. As shown in Fig. 5g, the amount of MDM2 co-precipitated together with p53 decreased when p52-ZER6 was silenced, indicating that p52-ZER6 enhanced MDM2-induced p53 ubiquitination by strengthening the MDM2-p53 interaction. The nutlin family is a class of cis-imidazoline analogues that inhibits MDM2-p53 binding [34]. As shown in Fig. 5h and i, nutlin-3 grossly suppressed the enhanced MDM2/p53 complex integrity resulting from p52-ZER6 overexpression, concomitantly restoring p53 and p21 protein accumulation. Furthermore, these results showed that the effect of nutlin-3 on MDM2-p53 binding and p53 accumulation was much weaker in cells with high p52-ZER6 expression. We next examined the role of MDM2/p53 axis in the tumourigenesis potential of HCT116 cells overexpressing p52-ZER6. As shown in Fig. 5j and k, while overexpression of p52-ZER6 enhanced the total HCT116 cell numbers as well as the number of proliferative cells, blocking the binding between p53 and MDM2 proteins by nutlin-3 significantly suppressed this effect. Furthermore, p52-ZER6 enhanced the colony formation potential of HCT116 cells, while addition of nutlin-3 suppressed it (Fig. 5l). Together, our results suggested that p52-ZER6 regulated p53 expression at a post-translational level by enhancing its binding to MDM2, leading to the promotion of p53 ubiquitination and proteasomal degradation, and subsequently, increased tumourigenesis potential.
3.6. tKRAB is the p52-ZER6 domain responsible for regulating the MDM2/p53 axis
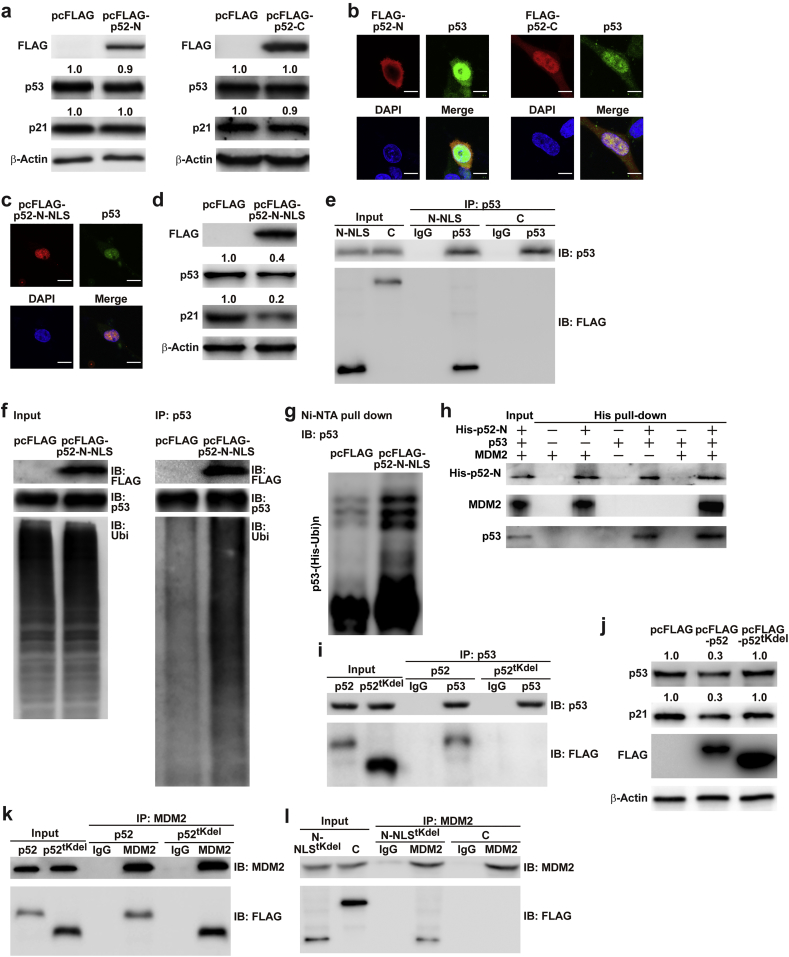
The N-terminus of p52-ZER6 consists of a tKRAB domain and a 142-amino acid region, whereas its C-terminus consists of zinc-finger domains. To investigate the function of each terminus in regulating p53 accumulation, we overexpressed FLAG-conjugated p52-ZER6 N-terminal and FLAG-conjugated p52-ZER6 C-terminal regions (Fig. S8a, FLAG-p52-N and FLAG-p52-C, respectively). Surprisingly, none of them affected the levels of p53 and its downstream target p21 (Fig. 6a). Indeed, whereas full-length p52-ZER6 and FLAG-p52-C co-localised with p53 in the nucleus (Figs. 5a and 6b), FLAG-p52-N localised in the cytoplasm (Fig. 6b). To induce FLAG-p52-N to enter the nucleus, we inserted a nuclear localisation signal (NLS) at the C-terminus of FLAG-p52-N (FLAG-p52-N-NLS, Figs. S8a and Fig. 6c), and found that FLAG-p52-N-NLS overexpression suppressed p53 and p21 accumulation (Fig. 6d). Concomitantly, an immunoprecipitation assay revealed that only the N-terminus, but not the C-terminus of p52-ZER6 interacted physically with p53 (Fig. 6e), and was sufficient to promote p53 ubiquitination (Fig. 6f and g). Furthermore, an in vitro His pull-down assay showed that the N-terminus of p52-ZER6 was sufficient for bindings to both p53 and MDM2 (Fig. 6h).
Fig. 6.
p52-ZER6 N-terminus binds and regulates MDM2/p53 complex integrity. (a) p53 protein expression level in HCT116 cells overexpressing N-terminus (left) or C-terminus (right) of p52-ZER6. (b–c) Co-localisation of p52-ZER6 N- or C-terminal fragments (FLAG-p52-N and FLAG-p52-C, respectively; b); or NLS-fused p52-ZER6 N-terminal fragment (FLAG-p52-N-NLS; c) and p53 in HCT116 cells, as determined by immunofluorescence staining. Scale bars, 10 μm. (d) p53 and p21 protein expression levels in HCT116 cells overexpressing FLAG-p52-N-NLS, as analysed using western blotting. (e) Physical interaction between p52-ZER6 fragments and p53 in HCT116 cells transfected with FLAG-p52-N-NLS or FLAG-p52-C overexpression vectors and pcp53, as determined by immunoprecipitation. Cell lysates were immunoprecipitated against IgG or anti-p53 antibody. The presence of p52-ZER6 fragments were detected by immunoblotting with anti-FLAG antibody. (f–g) p53 ubiquitination levels in HCT116 cells overexpressing FLAG-p52-N-NLS were analysed using anti-ubiquitin immunoblotting of cell lysates immunoprecipitated with anti-p53 antibody (f), or using in vivo ubiquitination assay conducted using Ni-NTA pull-down under denaturing condition followed by immunoblotting (g). Cells were treated with MG132 to inhibit proteasomal degradation. (h) Physical interactions between the N-terminus of p52-ZER6 (His-p52-N), MDM2 and p53, as determined by an in vitro His pull-down assay. (i) Physical interaction between p53 and p52-ZER6 without tKRAB domain (FLAG-p52tKdel) in HCT116 cells overexpressing p53 and FLAG-p52tKdel. Cell lysates were immunoprecipitated against anti-p53 antibody. The presence of p52-ZER6 fragments were detected by immunoblotting with anti-FLAG antibody. (j) p53 and p21 protein expression levels in HCT116 cells overexpressing FLAG-p52tKdel, as examined using western blotting. (k) Physical interaction between MDM2 and FLAG-p52tKdel in HCT116 cells. Cell lysates were immunoprecipitated against anti-MDM2 antibody. The presence of p52-ZER6 fragments were detected by immunoblotting with anti-FLAG antibody. (l) Physical interactions between MDM2 and FLAG-p52-C or NLS-fused N-terminal of p52-ZER6 lacking tKRAB domain (FLAG-N-p52-NLStKdel) in HCT116 cells. Cell lysates were immunoprecipitated against anti-MDM2 antibody. The presence of p52-ZER6 fragments was detected by immunoblotting with anti-FLAG antibody. For inputs of immunoprecipitation assay, 40 μg of corresponding samples were loaded. For input of His pull-down assay, 40 ng of corresponding sample was loaded. Ubi, ubiquitin; IP: immunoprecipitation; IB: immunoblotting. Cells transfected with pcFLAG were used as controls. β-Actin was used as western blotting loading control.
To discern the function of each part of the N-terminus, we constructed a vector overexpressing p52-ZER6 without the tKRAB domain (FLAG-p52tKdel, Fig. S8b). Deletion of the tKRAB domain disrupted the physical interaction between p52-ZER6 and p53 (Fig. 6i), while diminishing the role of p52-ZER6 in suppressing p53 and p21 proteins accumulation (Fig. 6j). Concomitantly, overexpression of FLAG-p52tKdel failed to induce HCT116 cell proliferation to the same extent as with full-length p52-ZER6 (Fig. S8c and d). This finding indicated that, while the role of a p53-independent pathway needs further investigation, our results pointed to a critical role of p53 in p52-ZER6-dependent regulation of tumour proliferation.
Intriguingly, deletion of the tKRAB domain did not abolish the interaction between p52-ZER6 and MDM2 (Fig. 6k). Indeed, MDM2 could be immunoprecipitated with the N-terminus of p52-ZER6 lacking the tKRAB domain (FLAG-p52-N-NLStKdel, Fig. S8b), but not with the C-terminus of p52-ZER6 (Fig. 6l), suggesting that the region between tKRAB and the zinc-finger domains (hereafter named MDM2-binding region, MBR) was essential for the interaction between p52-ZER6 and MDM2. These results clearly demonstrated that the N-terminus of p52-ZER6 was specifically responsible for MDM2/p53 complex integrity through bindings of tKRAB to p53 and MBR to MDM2; whereas the C-terminus was crucial for its localisation to the nucleus.
3.7. The HUB-1 domain inhibits p52-ZER6 regulation of p53
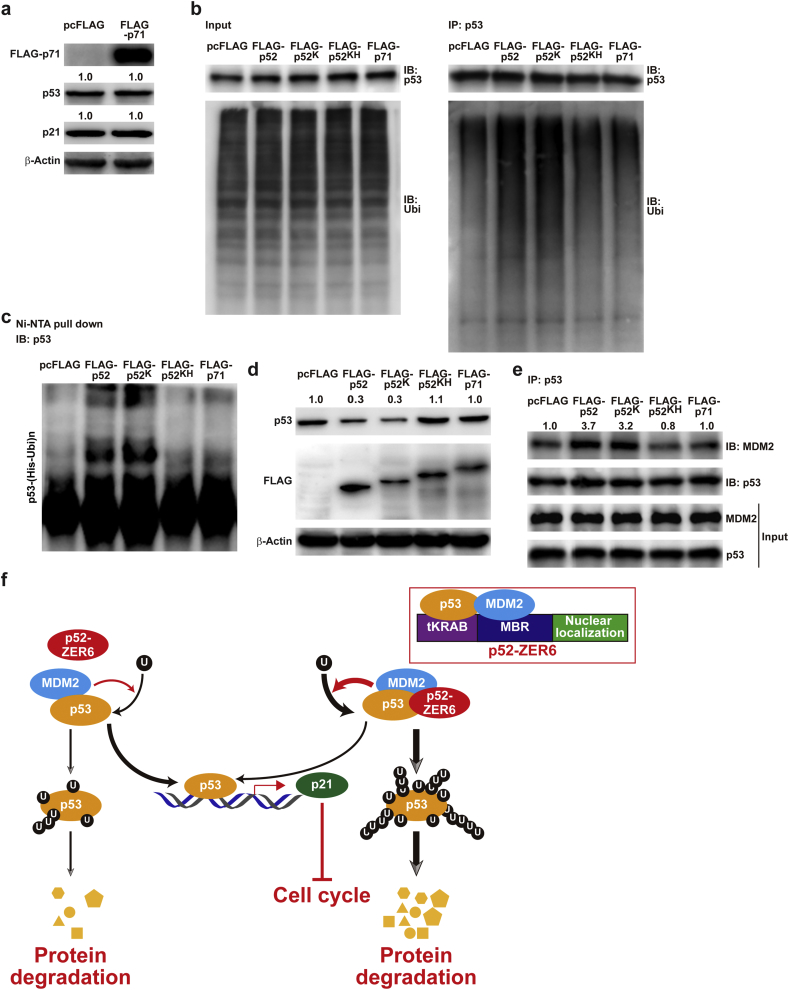
As shown in Fig. 2a, even though p71-ZER6 possessed a full-length KRAB domain, it did not affect the transcriptional activity and expression of p21 nor p53 protein accumulation (Figs. S2f, 2c, and 7a). These findings prompted us to investigate whether differences in the N-termini were responsible for the distinct effect of p52-ZER6 and p71-ZER6 isoforms on p53.
Fig. 7.
HUB-1 domain blocks p52-ZER6 regulation on the integrity of MDM2/p53 complex. (a) p53 and p21 protein expression levels in HCT116 cells overexpressing p71-ZER6, as analysed using western blotting. (b–c) p53 ubiquitination levels in HCT116 cells overexpressing FLAG-p52, FLAG-p52 added with full-length KRAB domain (FLAG-p52K), FLAG-p52 added with full-length KRAB and HUB-1 domains (FLAG-p52KH), and FLAG-p71 were analysed by anti-ubiquitin immunoblotting of cell lysates immunoprecipitated with anti-p53 antibody (b), or using in vivo ubiquitination assay conducted using Ni-NTA pull-down under denaturing condition followed by immunoblotting (c). Cells were treated with MG132 to inhibit proteasomal degradation. (d) p53 protein expression level in HCT116 cells transfected with indicated vectors, as determined by western blotting. (e) Binding capacity of MDM2 to p53 in HCT116 cells overexpressing FLAG-p52, FLAG-p52K, FLAG-p52KH, and FLAG-p71; as determined by immunoprecipitation. Cell lysates were immunoprecipitated with anti-p53 antibody. The presence of MDM2 was detected by immunoblotting. Cells transfected with pcFLAG were used as controls. β-Actin was used as western blotting loading control. Ubi, ubiquitin; IP: immunoprecipitation; IB: immunoblotting. For inputs of immunoprecipitation assay, 40 μg of corresponding samples were loaded. (f) Schematic diagram showing the molecular mechanism of p52-ZER6 regulation on MDM2/p53/p21 axis; U: ubiquitin.
We thus constructed overexpression vectors carrying intermediate fragments between p52-ZER6 and p71-ZER6: FLAG-conjugated p52-ZER6 with additional 29 amino acids to form a full KRAB domain (FLAG-p52K), or with additional 138 amino acids to form full KRAB and HUB-1 domains (FLAG-p52KH) (Fig. S9). Ubiquitination assays revealed that overexpression of p52-ZER6 and FLAG-p52K grossly induced p53 ubiquitination and concomitantly, promoted p53 protein degradation in HCT116 cells; however, overexpression of FLAG-p52KH as well as p71-ZER6 did not affect them (Fig. 7b–d). To determine the underlying mechanism, we evaluated the amount of MDM2 that interacted with p53 in HCT116 cells overexpressing Flag-p52, Flag-p52K, Flag-p52KH, and Flag-p71 (Fig. 7e). Accordingly, whereas p52-ZER6 and FLAG-p52K increased the amount of MDM2 interacting with p53, FLAG-p52KH and p71-ZER6 failed to exert a similar effect, suggesting that the additional amino acids found in the N-terminus of p71-ZER6 prevented enhanced MDM2/p53 complex integrity, leading to a distinct effect on p53 than with p52-ZER6.
Together, these findings point to p52-ZER6 as a novel oncogene and p53-regulating factor, which promotes p53 ubiquitination and destabilisation by enhancing the integrity of the MDM2/p53 complex (Fig. 7f). Moreover, our results reveal the structural difference between p52-ZER6 and p71-ZER6 as a source of their functional disparity in regulating the MDM2/p53 pathway.
4. Discussion
Ubiquitination-induced proteasomal degradation of p53 is key for maintaining p53 protein homeostasis, whose disruption is a molecular hallmark of tumours [10]. In spite of it being known for 40 years [35,36], regulation of p53 has not been totally unravelled [11,37]. Here we describe for the first time the aberrant up-regulation of an isoform of ZER6, p52-ZER6, in tumour tissue, as well as its critical role in regulating p53 protein stability, and subsequently, tumourigenesis. Conroy et al. reported that only p52-ZER6, but not p71-ZER6, bound in a ligand-dependent fashion to oestrogen receptor alpha in the presence of 17β-estradiol, leading to the inhibition of p52-ZER6 transactivational activity; and predicted that the presence of the HUB-1 domain in p71-ZER6 blocked this binding [16]. Despite prediction of genes' transcription being regulated through a ZER6-binding element [16], no ZER6 target has been identified at present. Thus, the biological and pathological functions of ZER6 isoforms remain completely uncharacterised. Herein, we identified p52-ZER6 as a novel regulator of the MDM2/p53 pathway: p52-ZER6 binds to the MDM2/p53 complex and is crucial for its integrity, thereby enhancing p53 ubiquitination and degradation. Concomitantly, p53 accumulation is enhanced in p52-ZER6-silenced cells, leading to inhibition of cell cycle progression, cell proliferation, and subsequently, tumourigenesis. Furthermore, recent studies have shown that p53 is critical for tumour angiogenesis, metabolism, and stemness [[37], [38], [39], [40], [41]]. Indeed, xenograft experiments show that p52-ZER6 silencing drastically repressed tumourigenesis, indicating that p52-ZER6 might also affect other aspects of tumourigenesis, which require further investigation. Nevertheless, our study is the first to describe the function of p52-ZER6 in promoting tumour cell proliferation as well as tumourigenesis, as well as its molecular mechanism in regulating the MDM2/p53 axis.
Abnormal MDM2/p53 regulation is common in tumours [42] and is closely related to aberrant p53 expression. Indeed, while p53 gene mutation could be found in around 50% of tumour patients, p53 expression is frequently down-regulated in patient with wild-type p53 gene, reflecting the importance of altered p53 regulation in tumourigenesis. Previous studies have revealed that MDM2 self-ubiquitination affects p53 ubiquitination [11,43]. Here we demonstrate another aspect in the MDM2-mediated regulation on p53, as p52-ZER6 binds to the MDM2/p53 complex through the N-terminal tKRAB domain and MBR, critically affecting its integrity. p52-ZER6 silencing significantly disrupts the binding between MDM2 and p53, reduces p53 ubiquitination, and consequently enhances its stability. It is noteworthy that binding of p52-ZER6 to p53 and MDM2 is strengthened when all three proteins are present, suggesting that they affect each other. Furthermore, in spite of being the core functional region of p52-ZER6, the N-terminus itself is insufficient for promoting p53 ubiquitination, as its C-terminus is indispensable for co-localising p52-ZER6 with p53 in the nucleus. On the other hand, although it functions as a nuclear localisation signal, C-terminus itself could not bind with p53 and MDM2, and thus could not regulate p53 protein stability. C-terminus of p52-ZER6 contains six C2H2-type zinc finger domains, which could function as transcriptional factor. Indeed, while its specific target genes currently have not been reported, Conroy et al. showed that p52-ZER6 itself possesses transactivational activity [16]. Thus, besides its function in enhancing MDM2-p53 binding, it is possible that p52-ZER6 could also regulate tumourigenesis by functioning as a transcriptional regulator.
Targeting the binding between MDM2 and p53 to increase p53 accumulation in tumour cells has become an attractive strategy for cancer therapy [44]. Several compounds, including nutlins, have shown promising results as potential anti-cancer drugs by interfering with MDM2-p53 binding [[44], [45], [46]]. However, our results show that high expression of p52-ZER6 might reduce the ability of nutlin to interfere with MDM2-p53 binding and subsequent p53 accumulation. These results suggests that the therapeutic effect of this kind of drugs might be weaker in clinical tumour patients with high p52-ZER6 expression. Thus, the expression level of p52-ZER6 might be a potential biomarker for determining patient suitability for treatment targeting MDM2-p53 binding.
Nearly one-third of all zinc-finger proteins contain an N-terminal KRAB domain [47]. KRAB-type zinc-finger proteins are involved in various biological events [48]; and could act as both oncogenes and tumour suppressors [[49], [50], [51], [52]]. They bind to co-repressor proteins and inhibit gene transcription [48,49], and could regulate p53 transactivational activity and attenuating its acetylation [47,51]. However, to our knowledge, there is no reports regarding the function of tKRAB domain at present. Here we show that both p52-ZER6, which possesses tKRAB domain, as well as an artificial construct in which N-terminal amino acids of a full-length KRAB has been added to the p52-ZER6, could destabilises p53 protein. Furthermore, the tKRAB domain in p52-ZER6 is essential for destabilising p53. However, whether the tKRAB domain is sufficient for the function of other zinc-finger proteins, and whether such function is analogous to that of full-length KRAB domains remain to be determined. Furthermore, these facts raise another intriguing question: what kind of natural selection pressure led to loss of the N-terminal region of the KRAB domain while maintaining its function? Overall, our findings widen our understanding of the KRAB domain, whether it is full-length or truncated.
Intriguingly, while p52-ZER6 affects MDM2-p53 binding and enhances p53 degradation, p71-ZER6 does not significantly affect p53 accumulation, most likely due to the additional 109 amino acids homologous to the HUB-1 domain. The latter prevents p52-ZER6 from enhancing the binding between MDM2 and p53. In spite of being functionally critical for suppressing human T-cell lymphotrophic virus type I long terminal repeat-mediated expression [53,54], the function of proteins with the HUB-1 domain remains poorly characterised. Although a detailed investigation is needed, it is possible that the HUB-1 domain folds back and binds to the tKRAB domain, thus masking the p53 binding site. Furthermore, it is also possible that the HUB-1 domain causes p71-ZER6 to bind to other proteins, preventing it from destabilising p53. Interestingly, alteration of p71-ZER6 expression level also regulates the tumourigenesis potential, as demonstrated by its proliferative and colony formation potentials (Fig. S10a–c). Given that p71-ZER6 could not suppress p53 protein accumulation, this result suggests that different from that of p52-ZER6, p71-ZER6 might be involved in tumourigenesis through a p53-independent pathway.
In summary, we identify p52-ZER6 as a novel oncogene responsible for promoting tumour progression by enhancing the integrity of the MDM2/p53 complex. This leads to repression of p53 accumulation, subsequently promoting cell cycle progression and proliferation. Given the widespread importance of p53, p52-ZER6 might have a far-reaching impact, both in tumourigenesis and other physiological events. Finally, our findings provide new insights on the regulation of the MDM2/p53 axis and the function of KRAB-type domains, as well as the potential of targeting p52-ZER6 for cancer therapy.
Acknowledgments
Acknowledgments
We thank Dr. Bert Vogelstein (Johns Hopkins University School of Medicine) for kindly providing wild-type and p53-null HCT116 cell lines.
Funding sources
This work was supported by grants from the National Natural Science Foundation of China (31871367, 81872273, 11832008); the Natural Science Foundation of Chongqing (cstc2018jcyjAX0411, cstc2018jcyjAX0374); and the Fundamental Research Funds for the Central Universities (2019CDQYSW010).
Declaration of competing interests
Patents related to the results of this study have been filed with Chinese patent application Nos. 201910626899.0 and 201910626900.X. All of authors declare no conflicts of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.08.070.
Appendix A. Supplementary data
Supplementary material
References
- 1.Aubrey B.J., Kelly G.L., Janic A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018;25:104–113. doi: 10.1038/cdd.2017.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaiser A.M., Attardi L.D. Deconstructing networks of p53-mediated tumor suppression in vivo. Cell Death Differ. 2018;25:93–103. doi: 10.1038/cdd.2017.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Epping M.T., Meijer L.A., Krijgsman O. TSPYL5 suppresses p53 levels and function by physical interaction with USP7. Nat Cell Biol. 2011;13:102–108. doi: 10.1038/ncb2142. [DOI] [PubMed] [Google Scholar]
- 4.Han Y., Wu P., Wang Z. Ubiquinol-cytochrome C reductase core protein II promotes tumorigenesis by facilitating p53 degradation. EBioMedicine. 2019;40:92–105. doi: 10.1016/j.ebiom.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieging K.T., Mello S.S., Attardi L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vousden K.H., Lane D.P. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 7.Miyamoto T., Lo P.H.Y., Saichi N. Argininosuccinate synthase 1 is an intrinsic Akt repressor transactivated by p53. Sci Adv. 2017;3 doi: 10.1126/sciadv.1603204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mello S.S., Attardi L.D. Deciphering p53 signaling in tumor suppression. Curr Opin Cell Biol. 2018;51:65–72. doi: 10.1016/j.ceb.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bublik D.R., Bursac S., Sheffer M. Regulatory module involving FGF13, miR-504, and p53 regulates ribosomal biogenesis and supports cancer cell survival. Proc Natl Acad Sci U S A. 2017;114:E496–E505. doi: 10.1073/pnas.1614876114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wade M., Li Y.C., Wahl G.M. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C., Wu S., Ji H. Identification of XBP1-u as a novel regulator of the MDM2/p53 axis using an shRNA library. Sci Adv. 2017;3 doi: 10.1126/sciadv.1701383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.el-Deiry W.S., Tokino T., Velculescu V.E. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 13.Harper J.W., Adami G.R., Wei N. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 14.Abbas T., Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Feng H., Gu H. The p53-PUMA axis suppresses iPSC generation. Nat Commun. 2013;4:2174. doi: 10.1038/ncomms3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conroy A.T., Sharma M., Holtz A.E. A novel zinc finger transcription factor with two isoforms that are differentially repressed by estrogen receptor-alpha. J Biol Chem. 2002;277:9326–9334. doi: 10.1074/jbc.M107702200. [DOI] [PubMed] [Google Scholar]
- 17.Haupt Y., Maya R., Kazaz A. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 18.Fang S., Jensen J.P., Ludwig R.L. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 19.Kussie P.H., Gorina S., Marechal V. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 20.Kwon D.H., Eom G.H., Ko J.H. MDM2 E3 ligase-mediated ubiquitination and degradation of HDAC1 in vascular calcification. Nat Commun. 2016;7 doi: 10.1038/ncomms10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyagishi M., Taira K. Strategies for generation of an siRNA expression library directed against the human genome. Oligonucleotides. 2003;13:325–333. doi: 10.1089/154545703322617005. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y., Wu S., Huang C. Yin Yang 1 promotes the Warburg effect and tumorigenesis via glucose transporter GLUT3. Cancer Sci. 2018;109:2423–2434. doi: 10.1111/cas.13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu S., Wang H., Li Y. Transcription factor YY1 promotes cell proliferation by directly activating the pentose phosphate pathway. Cancer Res. 2018;78:4549–4562. doi: 10.1158/0008-5472.CAN-17-4047. [DOI] [PubMed] [Google Scholar]
- 24.Okamoto N., Yasukawa M., Nguyen C. Maintenance of tumor initiating cells of defined genetic composition by nucleostemin. Proc Natl Acad Sci U S A. 2011;108:20388–20393. doi: 10.1073/pnas.1015171108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun X.X., Challagundla K.B., Dai M.S. Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J. 2012;31:576–592. doi: 10.1038/emboj.2011.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marisa L., de Reynies A., Duval A. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10 doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Deiry W.S. p21(WAF1) mediates cell-cycle inhibition, relevant to cancer suppression and therapy. Cancer Res. 2016;76:5189–5191. doi: 10.1158/0008-5472.CAN-16-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Y.Q., Hu Z.Y., Mangala L.S. MYC targeted long noncoding RNA DANCR promotes cancer in part by reducing p21 levels. Cancer Res. 2018;78:64–74. doi: 10.1158/0008-5472.CAN-17-0815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stabach P.R., Thiyagarajan M.M., Weigel R.J. Expression of ZER6 in ER alpha-positive breast cancer. J Surg Res. 2005;126:86–91. doi: 10.1016/j.jss.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Roepman P., Schlicker A., Tabernero J. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int J Cancer. 2014;134:552–562. doi: 10.1002/ijc.28387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamb J.R., Zhang C., Xie T. Predictive genes in adjacent normal tissue are preferentially altered by sCNV during tumorigenesis in liver cancer and may rate limiting. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macleod K.F., Sherry N., Hannon G. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev. 1995;9:935–944. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- 33.Lee J.T., Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010;17:86–92. doi: 10.1038/cdd.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vassilev L.T., Vu B.T., Graves B. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 35.Lane D.P., Crawford L.V. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 36.Linzer D.I., Levine A.J. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 37.Ni T., Li X.Y., Lu N. Snail1-dependent p53 repression regulates expansion and activity of tumour-initiating cells in breast cancer. Nat Cell Biol. 2016;18:1221–1232. doi: 10.1038/ncb3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang H., He X., Wang S. A microtubule-associated zinc finger protein, BuGZ, regulates mitotic chromosome alignment by ensuring Bub3 stability and kinetochore targeting. Dev Cell. 2014;28:268–281. doi: 10.1016/j.devcel.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu K., Lee J., Kim J.Y. Mitophagy controls the activities of tumor suppressor p53 to regulate hepatic cancer stem cells. Mol Cell. 2017;68:281–292. doi: 10.1016/j.molcel.2017.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mello S.S., Valente L.J., Raj N. A p53 super-tumor suppressor reveals a tumor suppressive p53-Ptpn14-yap Axis in pancreatic cancer. Cancer Cell. 2017;32:460–473. doi: 10.1016/j.ccell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamakuchi M., Lotterman C.D., Bao C. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci U S A. 2010;107:6334–6339. doi: 10.1073/pnas.0911082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashcroft M., Vousden K.H. Regulation of p53 stability. Oncogene. 1999;18:7637–7643. doi: 10.1038/sj.onc.1203012. [DOI] [PubMed] [Google Scholar]
- 43.Leslie P.L., Ke H., Zhang Y. The MDM2 RING domain and central acidic domain play distinct roles in MDM2 protein homodimerization and MDM2-MDMX protein heterodimerization. J Biol Chem. 2015;290:12941–12950. doi: 10.1074/jbc.M115.644435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shangary S., Wang S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu Rev Pharmacol Toxicol. 2009;49:223–241. doi: 10.1146/annurev.pharmtox.48.113006.094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hai J., Sakashita S., Allo G. Inhibiting MDM2-p53 interaction suppresses tumor growth in patient-derived non-small cell lung cancer xenograft models. J Thorac Oncol. 2015;10:1172–1180. doi: 10.1097/JTO.0000000000000584. [DOI] [PubMed] [Google Scholar]
- 46.Mohammad R.M., Wu J., Azmi A.S. An MDM2 antagonist (MI-319) restores p53 functions and increases the life span of orally treated follicular lymphoma bearing animals. Mol Cancer. 2009;8:115. doi: 10.1186/1476-4598-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian C., Xing G., Xie P. KRAB-type zinc-finger protein Apak specifically regulates p53-dependent apoptosis. Nat Cell Biol. 2009;11:580–591. doi: 10.1038/ncb1864. [DOI] [PubMed] [Google Scholar]
- 48.Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4 doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi W.I., Kim M.Y., Jeon B.N. Role of promyelocytic leukemia zinc finger (PLZF) in cell proliferation and cyclin-dependent kinase inhibitor 1A (p21WAF/CDKN1A) gene repression. J Biol Chem. 2014;289:18625–18640. doi: 10.1074/jbc.M113.538751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu R.Z., Peng G., Dai H. ZNF668 functions as a tumor suppressor by regulating p53 stability and function in breast cancer. Cancer Res. 2011;71:6524–6534. doi: 10.1158/0008-5472.CAN-11-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeon B.N., Kim M.K., Choi W.I. KR-POK interacts with p53 and represses its ability to activate transcription of p21WAF1/CDKN1A. Cancer Res. 2012;72:1137–1148. doi: 10.1158/0008-5472.CAN-11-2433. [DOI] [PubMed] [Google Scholar]
- 52.Shahi P., Wang C.Y., Lawson D.A. ZNF503/Zpo2 drives aggressive breast cancer progression by down-regulation of GATA3 expression. Proc Natl Acad Sci U S A. 2017;114:3169–3174. doi: 10.1073/pnas.1701690114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okumura K., Sakaguchi G., Naito K. HUB1, a novel Kruppel type zinc finger protein, represses the human T cell leukemia virus type I long terminal repeat-mediated expression. Nucleic Acids Res. 1997;25:5025–5032. doi: 10.1093/nar/25.24.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Itoh M., Watanabe M., Yamada Y. HUB1 is an autoantigen frequently eliciting humoral immune response in patients with adult T cell leukemia. Int J Oncol. 1999;14:703–708. doi: 10.3892/ijo.14.4.703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material