Abstract
Background
The dysbiosis of gut microbiota has been implicated in the pathogenesis of inflammatory bowel diseases; however, the underlying mechanisms have not yet been elucidated. Heavily glycosylated mucin establishes a first-line barrier against pathogens and serves as a niche for microbial growth.
Methods
To elucidate relationships among dysbiosis, abnormal mucin utilisation, and microbial metabolic dysfunction, we analysed short-chain fatty acids (SCFAs) and mucin components in stool samples of 40 healthy subjects, 49 ulcerative colitis (UC) patients, and 44 Crohn's disease (CD) patients from Japan.
Findings
Levels of n-butyrate were significantly lower in stools of both CD and UC patients than in stools of healthy subjects. Correlation analysis identified seven bacterial species positively correlated with n-butyrate levels; the major n-butyrate producer, Faecalibacterium prausnitzii, was particularly underrepresented in CD patients, but not in UC patients. In UC patients, there were inverse correlations between mucin O-glycan levels and the production of SCFAs, such as n-butyrate, suggesting that mucin O-glycans serve as an endogenous fermentation substrate for n-butyrate production. Indeed, mucin-fed rodents exhibited enhanced n-butyrate production, leading to the expansion of RORgt+Treg cells and IgA-producing cells in colonic lamina propria. Microbial utilisation of mucin-associated O-glycans was significantly reduced in n-butyrate-deficient UC patients.
Interpretation
Mucin O-glycans facilitate symbiosynthesis of n-butyrate by gut microbiota. Abnormal mucin utilisation may lead to reduced n-butyrate production in UC patients.
Fund
Japan Society for the Promotion of Science, Health Labour Sciences Research Grant, AMED-Crest, AMED, Yakult Foundation, Keio Gijuku Academic Development Funds, The Aashi Grass Foundation, and The Canon Foundation.
Keywords: Microbiota, Butyrate, Mucin, Inflammatory bowel disease
Research in context.
Evidence before this study
The dysbiosis of gut microbiota has been implicated in the pathogenesis of inflammatory bowel diseases (IBD). Prior research has suggested abnormalities in mucosal immunity, microbiota composition, and metabolite production in IBD; however, the pathological correlations among these factors were unclear.
Added value of this study
Mucus-associated microbes utilise mucin O-glycans as an endogenous fermentation source to produce mainly n-butyrate, which fuels host cells and contributes to the development of the intestinal immune system by inducing expansion of IgA+ plasma cells and regulatory T cells. The mucin O-glycan–microbiota–n-butyrate axis is severely affected in ulcerative colitis (UC)-associated microbiota. This abnormality may be a significant cause of the reduced n-butyrate production in UC patients.
Implications of all the available evidence
The intestinal microenvironments of Crohn's disease (CD) and UC patients display several different features. n-Butyrate producers, represented by Faecalibacterium prausnitzii, are nearly absent in CD patients; conversely, the gut microbiota of UC patients retains normal levels of F. prausnitzii. However, mucin O-glycan utilisation is compromised in the intestinal microbiota of UC patients, which eventually leads to reduction of n-butyrate production. These findings provide a new model of symbiotic interaction that maintains gut immune homeostasis and offer a new perspective on the pathogenesis of UC.
Alt-text: Unlabelled Box
1. Introduction
Inflammatory bowel diseases (IBDs), such as ulcerative colitis (UC) and Crohn's disease (CD), are recurrent inflammatory disorders caused by both genetic and environmental factors [1,2]. Accumulating evidence has demonstrated that abnormal gut microbiota composition, termed dysbiosis, plays a role in the pathogenesis and/or exacerbation of IBDs in Caucasian patients [3,4]. In IBD patients, the gut microbiota is characterised by diminished microbial diversity, with underrepresentation of Firmicutes and overrepresentation of Proteobacteria [3,[5], [6], [7], [8]]. These characteristics are evident in the gut microbiota of CD patients, and the severity of dysbiosis in rectal mucosa-associated microbiota correlates well with disease score [7]. Changes in gut microbiota composition are less obvious in UC patients than in CD patients, and the associations of the microbial changes with IBD pathogenesis have not yet been elucidated [6,8]. Animal experiments have demonstrated that the gut microbiota contributes to the development of the host intestinal immune system under physiological conditions by inducing the maturation of gut-associated lymphoid tissues, as well as the differentiation of Th17 and regulatory T cells [9]. In contrast, the gut microbiota drives intestinal inflammation under dysbiosis [10,11]. Similarly, the transplantation of microbiota from CD patients into Il10-deficient mice has been shown to accelerate colitis [12]; however, the underlying mechanisms by which dysbiosis promotes the inflammatory response have not yet been fully elucidated.
Short-chain fatty acids (SCFAs) (e.g., acetate, propionate, and n-butyrate) are produced by intestinal microbiota via the microbial fermentation of indigestible carbohydrates (i.e., soluble dietary fibres, oligosaccharides, and resistant starches [RS]) in the colon and caecum of humans [13] and rodents [14], respectively. SCFAs serve as nutrients for the colonic epithelium [15]; they also enhance mucosal barrier function by maintaining epithelial integrity [16], increasing mucin production [17], and triggering the IgA response [18]. Furthermore, n-butyrate exhibits anti-inflammatory effects through the suppression of NF-κB signalling in the colonic epithelium [19]. It has previously been reported that n-butyrate facilitates regulatory T cell differentiation in the colonic lamina propria [20,21]. n-Butyrate is mainly produced by bacterial species classified under Clostridium cluster IV or XIVa. In IBD patients, these butyrate producers, including Faecalibacterium prausnitzii (Clostridium cluster IV) and Lachnospiraceae (Clostridium cluster XIVa) [22,23], are markedly underrepresented, relative to the numbers present in healthy subjects [[24], [25], [26]]. SCFA mixture or n-butyrate enemas have been shown to effectively reduce the inflammatory symptoms of UC on the colonic mucosa [27,28]. These observations suggest that reduced n-butyrate production in IBD patients may exacerbate gut inflammation.
The luminal secretion of mucins by goblet cells establishes a mucus layer which functions as a physicochemical barrier, thus preventing microbial attachment to the epithelial surface [29]. Mucins primarily consist of the heavily glycosylated MUC2 protein, which confers viscosity to mucus to restrict bacterial motility. The stratified inner layer of mucus is impenetrable to intestinal bacteria [30], whereas the outer layer of mucus serves as a habitat for certain bacterial species which utilise mucin as an energy resource [31,32]. Experimental observations using mice devoid of Muc2 or its O-glycans have demonstrated that mucin barrier dysfunction enhances bacterial translocation into the lamina propria, resulting in chronic inflammation of the colon [33,34]. The same phenomenon has been shown for UC, as the colonic mucus of UC patients is less protective than that of healthy subjects; therefore, commensal bacteria often penetrate into the inner mucus layer [35].
The aforementioned abnormalities in mucin and SCFA production due to dysbiosis may be implicated in the development of IBD; however, the relevance of these major pathological events remains obscure. Therefore, we explored the relationships among intestinal microbiota, SCFA levels, and mucin profiles of Japanese IBD patients. We observed reduced n-butyrate levels in the stools of both CD and UC patients; however, this may have resulted from distinct aetiologies. Considerable reductions in the numbers of n-butyrate-producing bacteria diminished n-butyrate synthesis in CD patients, whereas reduced mucin O-glycan utilisation by gut microbiota was observed in UC patients lacking n-butyrate. Herein, we demonstrated that mucin O-glycan is utilised by intestinal microbiota as an endogenous fermentation source to produce n-butyrate, and that this pathway is likely affected in UC.
2. Materials and methods
2.1. Sample collection
To analyse organic acid concentrations, microbiota compositions, and mucin components, the stool samples of 44 healthy subjects, 40 CD patients, and 49 UC patients from Japan were collected at the Department of Gastroenterology and Hepatology in Osaka University Hospital (Study 1; Table 1, S1, S2). In a separate study, the stool samples of 11 healthy subjects and 10 UC patients were collected to analyse mucinase activity (Study 2; Table 2). A validated food frequency questionnaire was used to investigate nutrient intake [36]. Patients were diagnosed with CD or UC based on endoscopic, radiological, histological, and clinical criteria provided by the Council for International Organizations of Medical Sciences of the World Health Organization and the International Organization for the Study of Inflammatory Bowel Disease [[37], [38], [39]]. Clinical disease activity was assessed via clinical disease activity and Crohn's disease Activity Index in UC and CD, respectively [40,41]. Endoscopic disease activity was recorded by ileocolonoscopy, then assessed using Matt's score and modified Rutgeert's score in UC and CD, respectively [42,43]. Endoscopic remission was defined as a Matt's score of 1 in UC and a modified Rutgeert's score of 0 or 1 in CD. All participants provided written informed consent to participate after receiving verbal and written information regarding the study. The study protocol was approved by the ethics committees of Osaka University (#13165–2), The University of Tokyo (#25-42-1122), Keio University (#150421–1), Shizuoka University (#14–11), and NIBIOHN (#72), prior to subject inclusion.
Table 1.
Inflammatory bowel disease patient information (Study 1)
| Healthy |
Ulcerative colitis |
Crohn's disease |
|
|---|---|---|---|
| n = 44 | n = 49 | n = 40 | |
| Median age, years (S.D.) | 33 (9·1) | 46 (13·8) | 40 (13·6) |
| Male (%) | 41 (95·3%) | 27 (55·1%) | 29 (72·5%) |
| Disease duration, years (S.D.) | – | 12·4 (10·5) | 13·4 (11·7) |
| Endoscopic diagnosis | |||
| Active stage | – | 25 (51·0%) | 12 (30·0%) |
| Remission stage | – | 24 (49·0%) | 28 (70·0%) |
| Clinical diagnosis | |||
| Active stage | – | 7 (14·3%) | 10 (25·0%) |
| Remission stage | – | 42 (85·7%) | 30 (75·0%) |
S.D., standard deviation.
Information regarding patient number, age, sex, disease duration, and disease severity based on endoscopic and clinical diagnoses.
Table 2.
Ulcerative colitis patient information (Study 2).
| Healthy |
Ulcerative colitis |
|
|---|---|---|
| n = 11 | n = 10 | |
| Mean age, years (S.D.) | 32·0 (12·7) | 42·8 (14·9) |
| Male (%) | 11 (100%) | 6 (60%) |
| Disease duration, years (S.D.) | – | 10.3 (7·5) |
| Endoscopic diagnosis | ||
| Active stage | – | 7 (70%) |
| Remission stage | – | 3 (30%) |
| Clinical diagnosis | ||
| Active stage | – | 2 (20%) |
| Remission stage | – | 8 (80%) |
S.D., standard deviation.
Information regarding patient number, age, sex, disease duration, and disease severity based on endoscopic and clinical diagnoses.
2.2. Metagenomic 16S rRNA sequencing
Approximately 200 mg of each stool sample was transferred into a 2-mL tube containing 0.1 mm zirconia/silica beads (BioSpec Products, Bartlesville, OK, USA) and 3.0 mm zirconia beads (Biomedical Sciences, Tokyo, Japan). The stool samples were homogenised at 1500 rpm for 20 min with Shake Master Neo (Biomedical Sciences, Tokyo, Japan) after adding the Inhibit EX buffer from the QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany). Genomic DNA was extracted using the kit in accordance with the manufacturer's instructions and then re-suspended in 10 mM Tris-HCl buffer at 5 ng/μL. A library of 16S rRNA genes was prepared in accordance with the protocol described in an Illumina technical note [44]. Briefly, each DNA sample was amplified by polymerase chain reaction (PCR) using KAPA HiFi HS ReadyMix (KAPA Biosystems, Wilmington, MA, USA) and primers specific for variable regions 3 and 4 of the 16S rRNA gene. The PCR products were purified using Agencourt AMPure XP Beads (Beckman Coulter, Brea, CA, USA) and appended by PCR using the Nextera XT index kit (Illumina, San Diego, CA, USA). The libraries were further purified using Agencourt AMPure XP Beads, diluted to 4 nM with 10 mM Tris-HCl buffer, and pooled. The pooled samples were sequenced using the MiSeq system (Illumina) with a 2 × 300-base pair protocol. All sequences analysed in this study were deposited in the DNA Data Bank of Japan (DDBJ)/GenBank/European Molecular Biology Laboratory (EMBL) database under the accession number DRA006094.
2.3. Microbiota composition analysis
We used the join_paired_ends.py QIIME script [45] and the fastq-join method to join paired-end reads, and trimmed sequencing adaptor sequences using cutadapt [46]. We converted the FASTQ files into FASTA files and removed chimeric reads using the identify_chimeric_seqs.py and filter_fasta.py QIIME scripts with usearch61 [47]. Next, we concatenated the FASTA files of individual samples into one FASTA file and used the pick_open_reference_otus.py QIIME script to pick operational taxonomic units (OTUs). We assigned taxonomy using the assign_taxonomy.py QIIME script with the RDP classifier and the Greengenes reference database, clustered at 97% identity. Subsequently, we created an OTU table using the make_otu_table.py QIIME script and removed OTUs lower than 0·005% using the filter_otus_from_otu_table.py script. We subsampled to a depth of 10,000 reads per sample and performed diversity analysis using the core_diversity_analyses.py QIIME script. To analyse the human stool microbiota, we assigned taxonomy to each OTU by similarity searching against the publicly available 16S (RDP ver. 10·27 and CORE update 2 September 2012) and NCBI genome databases using the GLSEARCH program, as described previously [48]. Phylum-, class-, genus-, and species-level assignments were performed at 70%, 90%, 94%, and 97% sequence identity thresholds, respectively. To analyse the caecal microbiota of the mice, taxonomic assignments were performed using the summarize_taxa.py QIIME script with the Greengenes reference database, clustered at 97%, as there were abundant OTUs which lacked sequence homology with any species in the NCBI database. Diversity analyses of the faecal microbiota (α- and β-diversity) were performed using the Vegan package in R [49]. The phenotype of the microbiome was predicted using BugBase [50] with the default parameters.
2.4. Organic acid analysis
The levels of organic acids (formate, acetate, propionate, i-butyrate, n-butyrate, i-valerate, n-valerate, succinate, and lactate) in human stools and rat caecal contents were measured based on the internal standard method with a high-performance liquid chromatography (HPLC) system (LC-6A; Shimadzu, Kyoto, Japan) equipped with a Shim-pack SCR-102H column (internal diameter, 8 mm; length, 30 cm; Shimadzu) and an electroconductivity detector (CDD-6A; Shimadzu) in Shizuoka University, as described previously [51]. Approximately 200 mg of human stool sample or 300 mg of rat caecal contents were homogenised in 2 mL of 10 mmol/L sodium hydroxide solution containing 0·5 g/L crotonic acid as an internal standard, then centrifuged at 10,000 ×g for 15 min. The supernatant obtained was subjected to HPLC analysis. The detection limit for organic acids was 0·1 μmol/g.
Levels of organic acids in mouse caecal contents were measured using a gas chromatography-mass spectrometer (GC–MS) in Keio University, in accordance with the modified methods of Moreau et al. [52]. Approximately 50 mg of mouse caecal contents was homogenised in 9 volumes of H2O (w/w). After centrifugation (10,000 ×g at 4 °C for 15 min), 200 μL of the supernatant was spiked with 10 μL of 1 mM 2-ethyl butyric acid as an internal standard and 20 μL of 20% (w/v) 5-sulfosalicylic acid solution for deproteinisation. After centrifugation (10,000 ×g at 4 °C for 15 min), 200 μL of the supernatant was acidified using 10 μL of 37% HCl, and organic acids were isolated via two rounds of extraction with 1 mL diethyl ether. Next, 500 μL of organic supernatant was mixed with 50 μL of N-tert-butyldimethylsilyl-N-methyltrifluoracetamide (Sigma-Aldrich Co., St. Louis, MO, USA) in a new glass vial and incubated at room temperature for 24 h to allow derivatisation. The derivatised samples were run through a JMS-Q1500GC GC–MS System (JEOL Ltd., Tokyo, Japan) equipped with an HP-5 capillary column (60 m × 0·25 mm × 0·25 μm, Agilent Technologies, Inc., Santa Clara, CA, USA). Pure helium (99·9999%) was used as a carrier gas and delivered at a flow rate of 1·2 mL/min. The following temperature program was used: 60 °C (3 min), 60–120 °C (5 °C/min), 120–290 °C (20 °C/min), and 290 °C (3 min). Organic acid concentrations were determined by comparing their peak areas with those of the standards. Acetate was obtained from Nacalai Tesque, Inc., (Kyoto, Japan); propionate, n-butyrate, succinate, and lactate were obtained from Wako Pure Chemical Industries, Ltd., (Osaka, Japan); and i-butyrate, i-valerate and n-valerate were obtained from Kanto Chemical Co., Inc. (Tokyo, Japan).
2.5. Mucin component analysis
The mucin fraction was isolated as previously described [53]. The sugar compositions were determined using HPLC, in accordance with a previously reported method [54]. In this analysis, approximately 69% and 76% of the stool glycans were occupied by mucin-derived sugars (i.e., fucose, galactose, GalNAc, GlacNAc, and sialic acid) in healthy subjects and UC patients with dietary fibre-derived sugars (xylan, arabinose, ribose, and mannose) as minor components (Fig. S1, Table S3). After appropriate dilution, the O-linked oligosaccharide chains were analysed as previously described [55]. Standard solutions of N-acetylgalactosamine (Sigma-Aldrich) were used to calculate the amounts of oligosaccharide chains released from the mucins during the procedure. Approximately 20 mg of the mucin fraction was completely dried, resuspended in 200 mL of 4 mol/L HCl, and then hydrolysed at 100 °C for 4 h in a heating block. Sulfate levels in the mucin fraction were determined using the method of Harrison and Packer [56]. Solutions of 0·79, 1·59, 3·18, 6·38, and 12·8 mmol sulfate/L (Multi-anion standard solution-1; Wako Pure Chemical Industries) were used as standards. Sialic acid levels were measured as previously described [57]. Briefly, the mucin fractions were hydrolysed in 50 mM HCl at 80 °C for 1 h, and the N-acetylneuraminic acid (NeuAc) and N-glycolylneuraminic acid (NeuGc) released were labelled with 1,2-diamino-4,5-methylenedioxybenzene (DMB) using a commercial kit (Takara, Shiga, Japan). The DMB-labelled sialic acids were analysed by HPLC equipped with a TSK-ODS80Ts column (Tosoh, Tokyo, Japan) and a fluorescence detector (RF-10AXL; Shimadzu).
2.6. Preparation of mucin-containing diets
Semi-purified porcine stomach mucin (Sigma-Aldrich) was suspended in phosphate-buffered saline and filtered to remove contaminants. Ethanol was added to the filtrate to prepare a 60% ethanol (w/w) solution as described previously [53]. Glycosylated mucin was precipitated at −30 °C and collected by centrifugation. This treatment was repeated twice to further purify the mucin. The purified mucin was added to the AIN73-formula diet as a substitute for corn starch.
2.7. Animal experiments
All animal experiments were performed in accordance with protocols approved by the Animal Use Committees of Keio University and Shizuoka University. The rats and mice were maintained in accordance with the guidelines for the care of laboratory animals of Shizuoka University and Keio University, respectively. Male Wister rats were purchased from the Shizuoka Laboratory Animal Center and housed in individual wire screen-bottomed, stainless steel cages at 23 ± 2 °C in a room with controlled lighting (light from 8:00 a.m. to 8:00 p.m.).
C57BL/6J male mice were purchased from CLEA Japan, Inc. (Tokyo, Japan). The mice were fed an AIN93 diet (Oriental Yeast, Tokyo, Japan) for 7 days, then either a diet containing 1·5% mucin or an AIN76-formula control diet for 3 weeks.
2.8. Mucinase activity analysis
Mucinase activity was analysed as described previously [58]. Briefly, faecal samples were mixed with acetate buffer (pH 5·5) at a ratio of 1:400 (w/v) and used for the mucinase assay with porcine stomach mucin as a substrate, in accordance with a previously described method [59]. Our preliminary experiment exhibited that pH 5.5 is optimal pH for the assessment of mucinase activity (Fig. S2). Faecal mucinase activity was expressed as the amount of liberated sugars per minute per protein contained in faecal homogenates or faecal weights. To calculate mucinase activity relative to protein content, the amount of protein in faecal homogenate was determined by Lowry's method [60] using bovine serum albumin standards.
2.9. In vitro fermentation analysis
We employed the use of a previously described in vitro fermentation system [61]. Briefly, caecal contents were obtained from rats fed an AIN93G diet for 7 days and diluted 50-fold in saline. Then, 110 mL of diluted caecal contents was incubated in a jar fermenter at 37 °C under an anaerobic atmosphere, with gentle stirring and pH-controlled (pH > 5·2) conditions. After preincubation overnight, 3·3 g of porcine stomach mucin was added to the culture. To monitor the organic acid contents, 4 mL of each sample was collected at 0, 24, and 48 h after mucin supplementation.
2.10. Preparation of colonic lamina propria cells
Colonic lymphocytes were prepared in accordance with the methods of Weigmann et al [62]. In brief, colonic tissues were treated with Hank's Balanced Salt Solution (Nacalai Tesque) containing 1 mM dithiothreitol and 20 mM EDTA (Nacalai Tesque) at 37 °C for 20 min to remove epithelial cells. The tissues were then minced and dissociated with RPMI 1640 medium (Nacalai Tesque) containing 0·5 mg/mL collagenase (Wako Pure Chemical Industries), 0·5 mg/mL DNase I (Merck, Darmstadt, Germany), 2% fetal calf serum (FCS, MP Biomedicals, Santa Ana, CA, USA), 100 U/mL penicillin, 100 μg/mL streptomycin, and 12·5 mM HEPES (pH 7·2) at 37 °C for 30 min to obtain a single-cell suspension. The suspension was filtered, washed with 2% FCS in RPMI 1640, and separated using a Percoll gradient.
2.11. Flow cytometry
Cell staining was performed as previously described [63]. In brief, colonic lymphocytes were incubated with anti-mouse CD16/CD32 antibodies (93; BioLegend, Inc., San Diego, CA, USA) to block their Fc receptors, then stained using antibodies conjugated with fluorescein isothiocyanate, phycoerythrin, PerCP-Cyanine5·5, redFluor 710, eFluor 450, Brilliant Violet 510, or Brilliant Ultraviolet 737. Anti-CD45R/B220 (RA3-6B2), anti-IgA (C10-3), anti-RORγt (Q31-378), and anti-CD3e (145-2C11) antibodies were obtained from BD Biosciences (San Jose, CA, USA); anti-CD3 (17A2) and anti-CD45R/B220 (RA3) antibodies were obtained from Tonbo Biosciences (San Diego, CA, USA); an anti-CD45 (30-F11) antibody was obtained from BioLegend; and anti-Foxp3 (FJK-16s) and anti-CD4 (RM4-5) antibodies were obtained from Thermo Fisher Scientific (Waltham, MA, USA). 7-Aminoactinomycin D (BioLegend) was added to the cell suspension to label dead cells. To visualise intracellular Foxp3 and RORγt, the cells were stained for surface antigens, fixed, and permeabilised using a Foxp3/Transcription Factor Staining Buffer Set (Thermo Fisher Scientific). Permeabilised cells were then stained with anti-Foxp3 and anti-RORγt antibodies. Dead cells were detected using the Fixable Viability Stain 780 (BD Biosciences) for intracellular staining. Stained cells were analysed using an LSR II Flow Cytometer (BD Biosciences) and FlowJo software ver. 10·4·2 (BD Biosciences).
2.12. Statistical analysis
To analyse organic acid and mucin component levels, differences between two or more groups were analysed by Student's t-test or by analysis of variance followed by Tukey's multiple comparison test, respectively. When variances were not homogeneous, the data were analysed by the Wilcoxon rank-sum test or by the Kruskal–Wallis test followed by Dunn's multiple comparison test. All correlation analyses were performed using Spearman's correlation test with Benjamini–Hochberg false discovery rate (FDR) correction in the multtest R software package [64]. Comparisons of bacterial taxon abundance were performed by LEfSe [65] using the default parameters, or using the Kruskal–Wallis test followed by Dunn's multiple comparison test in the dunn.test R software package. The biomarkers for each group identified using LEfSe were graphically annotated on phylogenetic trees using GraPhlAn [66]. With the exception of LEfSe, these statistical tests were performed in R ver. 3·3·0. The correlation network was visualised using Cytoscape ver. 3·3·0. Sample size was determined based on the available patients or animals, but not on any statistical methods. The experiments were not randomised, and the investigators were not blinded to allocation during experiments or outcome assessment.
3. Results
3.1. Reduced n-butyrate in stools of IBD patients
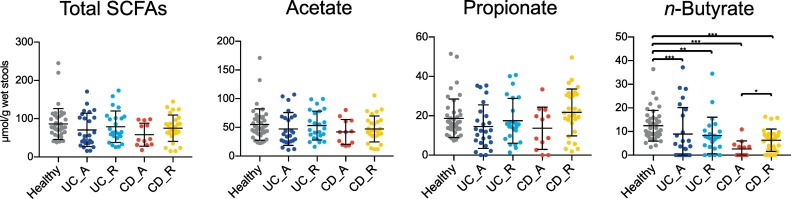
To characterise the altered metabolic functions of the intestinal microbiota in IBD patients, we initially analysed the concentrations of various organic acids, including SCFAs, in stool samples. The disease statuses of UC and CD patients were classified as active or in remission, based on endoscopic assessment (Tables S1 and S2). We observed that faecal n-butyrate levels were significantly lower in UC and CD patients than in healthy subjects (Fig. 1). Notably, a substantial number of CD and UC patients lacked n-butyrate in their stools; in particular, 50% of patients with active CD had undetectable levels of n-butyrate (Table S4). Thus, reduced stool n-butyrate levels reflect disease status in CD. Moreover, n-butyrate concentration was not affected by any other factors, including the site of disease development (i.e., right or left colon in UC, small or large intestine in CD), treatment with probiotics (lactic acid bacteria and Clostridium butyricum), 5-azathiopurine, and anti-TNF antibodies (Fig. S3; Tables S5 and S6).
Fig. 1.
Faecal organic acid concentrations in inflammatory bowel disease patients and healthy subjects.
Faecal organic acid concentrations were analysed in healthy subjects, patients with active (UC-A) or remissive (UC-R) ulcerative colitis (UC), and patients with active (CD-A) or remissive (CD-R) Crohn's disease (CD). Data represent the mean ± standard deviation (n = 12–43/group). ⁎p < 0·05, ⁎⁎p < 0·01, and ⁎⁎⁎p < 0·001 (analysis of variance followed by Tukey's multiple comparison test or the Kruskal–Wallis test followed by Dunn's multiple comparison test).
The stool concentrations of propionate, another beneficial metabolite [67], were also slightly reduced in patients with active UC and CD, although these differences were not statistically significant. n-Valerate was detected in 16·3% of healthy subjects, but not in UC patients (Fig. S4A), whereas elevated levels of succinate were observed in small numbers of UC and CD patients. The amounts of other organic acids, including acetate, formate, and lactate, were similar among groups. Correlation analysis of individual organic acids revealed that levels of n-butyrate were positively correlated with levels of propionate and acetate, but not with levels of formate, lactate, or succinate (Fig. S4B). Overall, these observations support the hypothesis that reduced levels of anti-inflammatory SCFAs, especially n-butyrate, are involved in the development and/or exacerbation of IBDs.
3.2. Characterisation of microbiota compositions in IBD patients
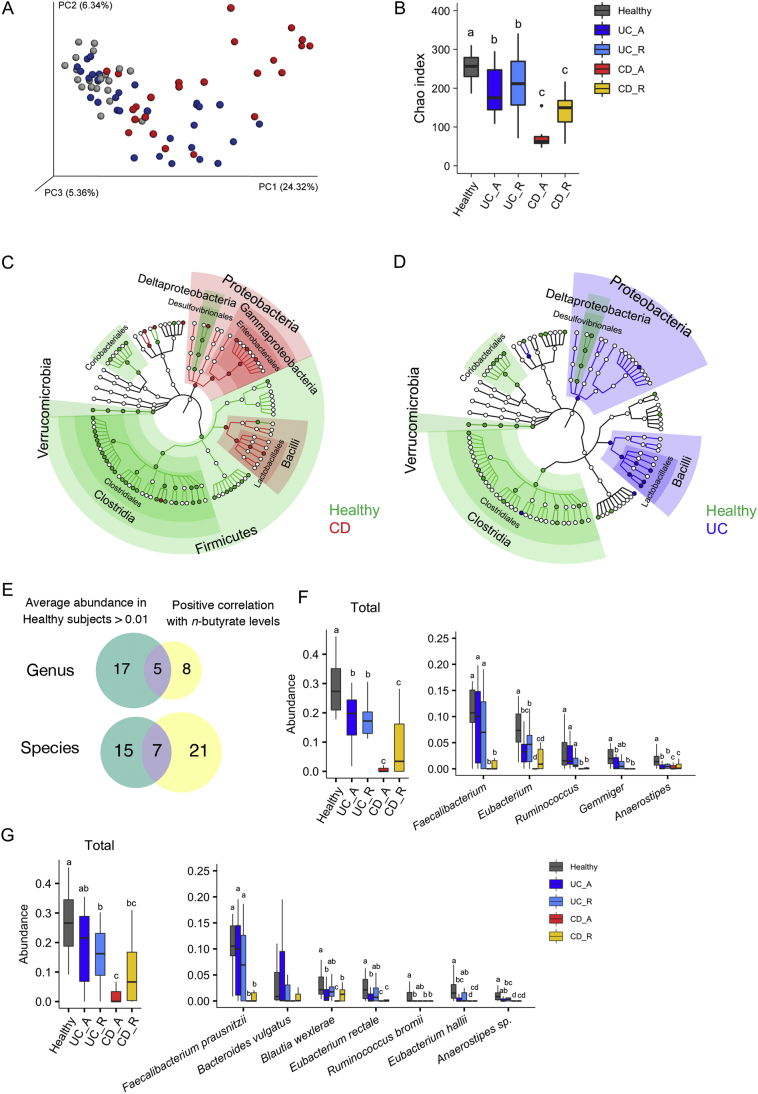
We performed 16S rRNA sequencing on stool samples to characterise the microbiota compositions of the IBD patients. The resulting data were subjected to principal coordinate analysis (PCoA) based on unweighted UniFrac (Fig. 2A). The microbiota compositions of the healthy subjects were grouped within one region, whereas those of UC and CD patients were scattered, indicating that the microbiota compositions of UC and CD patients were distinct from those of the healthy subjects (adonis based on unweighted UniFrac versus healthy microbiota; UC: R2 = 0·074 and p = 0·001; CD: R2 = 0·18 and p = 0·001). Furthermore, microbiota compositions differed significantly between the active and remission phases of both diseases (Fig. S5). Consistent with the findings in previous studies of Caucasian IBD patients [6,7], α-diversity was significantly reduced in Japanese UC and CD patients, particularly during the active stages of disease (Fig. 2B).
Fig. 2.
Microbial signatures of inflammatory bowel disease patients and healthy subjects.
(A) Principal coordinate analysis of unweighted UniFrac distances among the microbiota of inflammatory bowel disease (IBD) patients and healthy subjects. (B) Species richness (Chao 1) of the microbiota of IBD patients and healthy subjects. (C, D) Discriminating taxa between the microbiota of (C) Crohn's disease (CD) or (D) ulcerative colitis (UC) patients and healthy subjects, as determined by LEfSe [38] analysis. Discriminating taxa for the indicated groups are annotated on phylogenetic trees. (E) Venn diagram showing criteria for butyrate-associated bacteria. (F, G) Relative abundances of butyrate-associated bacteria at the genus (F) and species (G) levels. The left panels represent the total abundances of butyrate-associated bacteria, and the right panels indicate the abundances of individual bacterial genera or species. Each boxplot represents the median, interquartile range (IQR), and the lowest and highest values within 1·5 IQRs of the first and third quartiles (n = 7–23/group). Outliers are not shown. Different letters over boxplots indicate significant differences (p < 0·05; Kruskal–Wallis test followed by Dunn's multiple test).
The microbiota compositions of CD patients were characterised by underrepresentation of Firmicutes and Verrucomicrobia (including only Akkermansia muciniphila), and the overrepresentation of Proteobacteria at the phylum level (Fig. 2C and S6A, B). Proteobacteria was also slightly elevated in UC patients. At the class level, patients with active CD exhibited a reduced level of Clostridia and reciprocal increases in the levels of Gammaproteobacteria and Bacilli (Fig. S6C); furthermore, BugBase [50] predicted the outgrowth of facultative anaerobes and the reduction of anaerobes in patients with active CD (Fig. S7). Similar trends were observed to a lesser extent regarding the microbiota compositions of CD patients in remission, active UC patients, and UC patients in remission (Fig. 2A–D, S6 and S7).
3.3. Reduced numbers of butyrate-associated bacteria in IBD patients
To identify the bacteria associated with faecal n-butyrate levels in IBD patients, we performed correlation analyses between the abundance of bacterial taxa and faecal metabolites. We found that the abundances of 12 genera and 28 species were positively correlated with n-butyrate levels (Spearman correlation analysis, FDR < 0·05; Table S7 and S8), whereas the abundances of only a few genera were correlated with acetate, n-valerate, or i-butyrate levels. Other metabolites analysed here did not show significant correlations with any specific genera or species. Among the n-butyrate-associated bacteria, five bacterial genera and seven species occupied 1% or more of the microbial communities of healthy subjects, suggesting that these bacteria may play central roles in n-butyrate biosynthetic pathways (Fig. 2E). Therefore, we considered these to be ‘butyrate-associated bacteria’ (Fig. 2F and G), which included the well-known n-butyrate producers, F. prausnitzii and Eubacterium rectale [22]. Consistent with the findings regarding stool n-butyrate concentration, the overall abundance of butyrate-associated bacteria was considerably lower in CD patients (Fig. 2F and G). Notably, F. prausnitzii, which was the most abundant n-butyrate producer in the microbiota of healthy individuals, was nearly absent in the microbiota of active CD patients (Fig. 2G, right). In contrast, the abundance of F. prausnitzii was not significantly reduced in active UC patients or in UC patients in remission (Fig. 2F and G). Minor populations of n-butyrate-associated bacteria (i.e., Blautia wexlerae, Eubacterium rectale, Ruminococcus bromii, and E. hallii) were underrepresented in active UC patients (Fig. 2G, right).
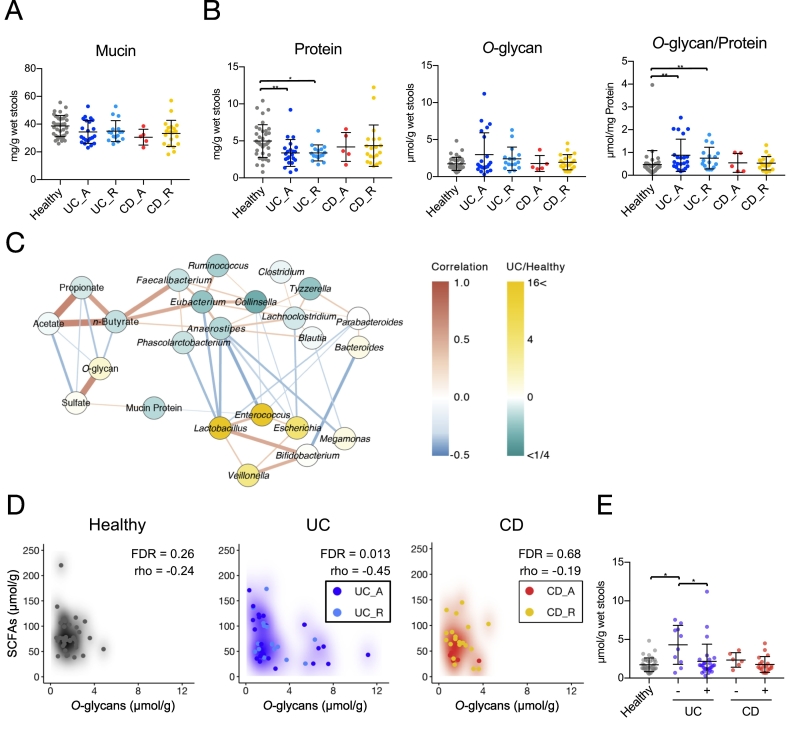
3.4. An increase in O-glycan ratios in stool mucin of UC patients
Mucin O-glycans form core structures, consisting of N-acetylgalactosamine, galactose, and N-acetylglucosamine; these structures are elongated and terminated by fucose, sialic acids, and sulfate residues in the human colon [68]. We hypothesised that the qualities and quantities of mucin components may be affected in IBD patients. Therefore, we explored the amounts of these components in stool mucin (Fig. 3). The protein content of the mucin fraction was significantly lower in UC patients, which may reflect the previously reported downregulation of Muc2 expression [69,70]. Nevertheless, mucin O-glycan levels were slightly higher in these patients than in healthy subjects (Fig. 3B). Consequently, the mucin ratio of O-glycans to protein was significantly higher in UC patients, which may be attributed to the reduced utilisation of mucin O-glycans by UC-associated microbiota. This abnormality was characteristic of UC, but not of CD. Notably, the composition of mucin O-glycan components was mostly intact in UC and CD patients (Table S9), with the exception of NeuGc. Although NeuGc is not synthesised by human cells, diet-derived exogenous NeuGc is occasionally absorbed by host cells and incorporated into endogenous glycoproteins, including mucin [71]. NeuGc was detected more frequently in UC patients than in healthy subjects (Table S10).
Fig. 3.
Stool mucin components of inflammatory bowel disease patients and healthy subjects.
(A, B) Levels of (A) mucin and (B) mucin-associated protein and O-glycan were analysed in the stools of inflammatory bowel disease (IBD) patients and healthy subjects. Data represent the mean ± standard deviation (n = 5–38/group). *p < 0·05 and **p < 0·01 (analysis of variance followed by Tukey's multiple comparison test or the Kruskal–Wallis test followed by Dunn's multiple comparison test). (C) Correlation network of the levels of short-chain fatty acids (SCFAs), mucin components, and bacteria in stool samples. Nodes represent SCFAs, mucin components, or bacterial genera with average levels higher than 1·0%. Node colours reflect average levels in ulcerative colitis (UC) patients relative to those in healthy subjects. Red and blue lines indicate positive (Spearman's correlation coefficient > 0·3) and negative (correlation coefficient < −0·3) correlations, respectively. Edge thickness represents the strength of the correlation. Line colour intensity reflects the extent of the correlation. (D) Scatter density plots of faecal n-butyrate levels versus mucin O-glycan levels for individual disease groups. Increasing intensity of each colour (white to black, blue, or red) reflects scatter plot density. Spearman coefficients (rho) and FDRs are shown. (E) UC and Crohn's disease (CD) patients were classified tinto two groups each, based on faecal n-butyrate detection, to compare faecal levels of mucin O-glycans. Patients with n-butyrate concentrations below 0·1 μmol/g were classified as ‘-’, while the remaining patients were classified as ‘+’. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.5. Impaired mucin O-glycan utilisation may reduce n-butyrate production in UC patients
We generated correlation networks (rho >0·3 or rho < −0·3) of bacterial genera, mucin components, and SCFAs. Correlation network analysis highlighted positive correlations between n-butyrate concentration and butyrate-associated bacteria (i.e. Faecalibacterium, Eubacterium, and Anaerostipes) (Fig. 3C). The butyrate-associated bacteria were inversely correlated with levels of Escherichia, Enterococcus, and Lactobacillus, which were overrepresented in UC patients (Fig. S9). Notably, network analysis revealed inverse correlations between the levels of mucin O-glycan and the levels of each SCFA (Fig. 3C). Of the three groups, inverse correlations between mucin O-glycan and the SCFAs were most prominent in UC patients (total SCFAs: rho = −0·45, FDR = 0·014; acetate: rho = −0·44, FDR = 0·014; propionate: rho = −0·43, FDR = 0·014; n-butyrate: rho = −0·35, FDR = 0·035; Fig. 3D and S10). These data suggest that intestinal microbiota could utilise mucin O-glycans as an endogenous fermentation substrate to produce SCFAs, such as n-butyrate; this was further supported by the reduced utilisation of mucin O-glycans in n-butyrate-deficient UC patients (Fig. 3D). Thus, we hypothesised that mucin O-glycan utilisation is impaired in patients with UC, which could eventually affect n-butyrate production.
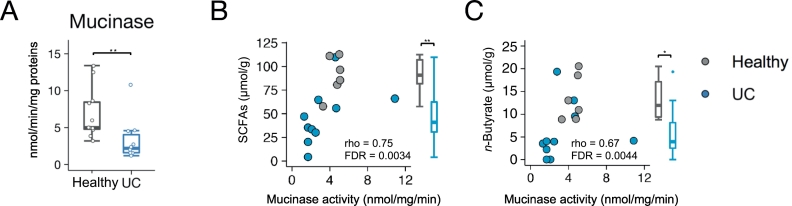
To confirm this hypothesis, we assessed mucin glycan degradation (mucinase activity) in fresh stool samples of newly recruited UC patients and healthy subjects (Fig. 4). We observed a clear reduction in stool mucinase activity in UC patients, compared with the activity in healthy subjects. Mucinase activity exhibited a positive correlation with the stool concentrations of total SCFAs and butyrate, both of which were significantly lower in UC patients (Fig. 4B and S11). The reduction in SCFA levels was not attributed to the dietary habits of UC patients, as there were no significant differences in nutrient intake (including soluble fibre intake) between UC patients and healthy subjects (Fig. S12 and Table S11). Taken together, these data suggest that abnormal mucin O-glycan utilisation may cause reduced n-butyrate production in UC.
Fig. 4.
Stool mucinase activity in ulcerative colitis patients and healthy subjects.
(A) Stool samples of ulcerative colitis (UC) patients and healthy subjects were analysed for mucinase activity. Boxplots represent the median, interquartile range (IQR), and the lowest and highest values within 1·5 IQRs of the first and third quartiles (n = 10–11/group). (B, C) Scatter plots of mucinase activity and (B) total short-chain fatty acids (SCFAs) and (C) n-butyrate concentrations. Spearman's correlation coefficient (rho) and false discovery rate (FDR) are shown. Box plots represent stool concentrations of total SCFAs (B) and butyrate (C) in the two groups (n = 6–10/group). ⁎p < 0·05 and ⁎⁎p < 0·01 (Wilcoxon rank-sum test).
3.6. Mucin supplementation facilitates SCFA production
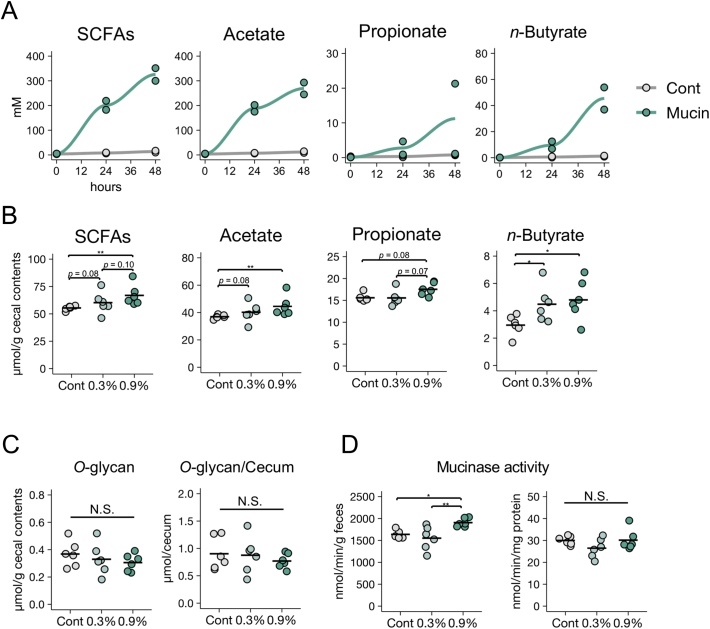
To directly investigate whether gut microbiota utilise mucin as a fermentation source to produce SCFAs, we cultured rat caecal microbiota in the presence or absence of mucin, using an in vitro fermentation system [61]. The caecum is a major site of microbial fermentation in rodents [51]. Within 48 h, mucin supplementation had increased the concentrations of SCFAs, including n-butyrate, in the culture media (Fig. 5A). Thus, we confirmed that the microbial fermentation of mucin O-glycans generates SCFAs.
Fig. 5.
Effects of mucin supplementation on short-chain fatty acid production by gut microbiota.
(A) Rat caecal contents were cultured in an in vitro fermentation system in the presence or absence of porcine gastric mucin and short-chain fatty acid (SCFA) concentrations were measured at the indicated time points. Data are representative of two independent experiments with similar results. (B–D) Caecal SCFA levels (B), O-glycan levels (C), and mucinase activity (D) in the stools of rats fed a control diet or diets containing 0·3% or 0·9% porcine stomach mucin. Data represent the mean ± standard deviation (n = 5–6/group). ⁎p < 0·05 and ⁎⁎p < 0·01 (analysis of variance followed by Dunnett's multiple comparison test or the Kruskal–Wallis test followed by Dunn's multiple comparison test, compared with the control group).
We further corroborated the importance of mucin O-glycans as a fermentation substrate in vivo. We fed rats synthetic diets containing 0·3% or 0·9% (w/w) purified mucins for 2 weeks. The 0·9% mucin diet significantly increased caecal concentrations of acetate and n-butyrate (Fig. 5B), and slightly increased the caecal concentration of propionate. Importantly, the levels of mucin O-glycans in the caecum were similar among all groups (Fig. 5C), indicating that the exogenous mucin O-glycans were efficiently consumed via intestinal microbial fermentation. Furthermore, mucin administration led to increased microbial mucinase activity, relative to faecal weight, in a dose-dependent manner (Fig. 5D). Thus, O-glycan utilisation may be a key determinant for maintenance of SCFA production.
3.7. The mucin–SCFA axis contributes to the development of the gut immune system
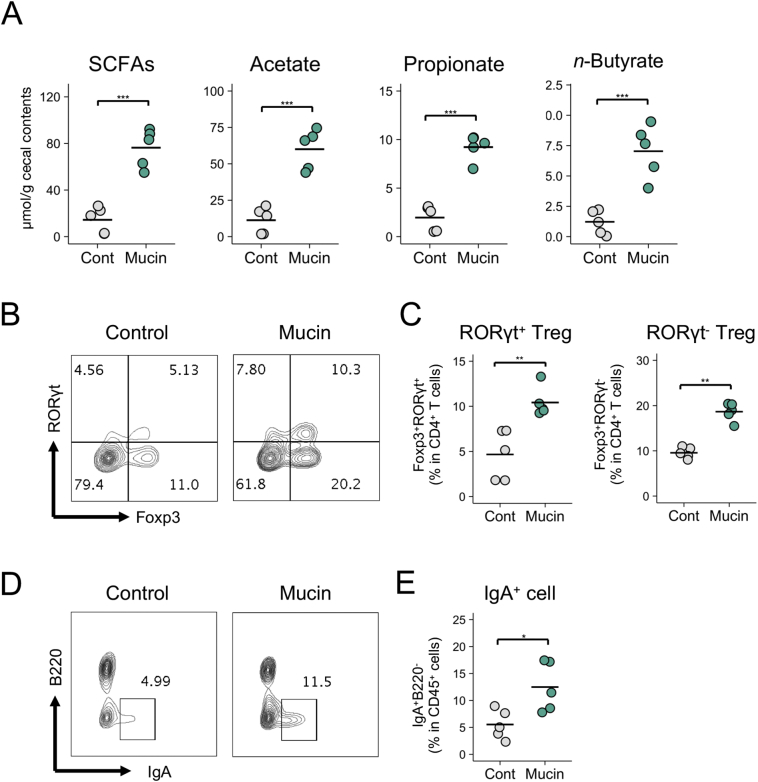
Finally, we investigated the biological significance of the mucin-dependent symbiosynthetic pathway by feeding mice a diet containing 1·5% mucin for 3 weeks. The microbiota composition of the mucin-fed mice was considerably different from that of the controls, characterised by higher α-diversity and increased abundances of Allobaculum, unclassified Bacteroidales S24–7, and Akkermansia (Fig. S13). Consistent with the observations made in the rats in this study, the administration of mucin to mice increased the caecal concentrations of SCFAs, such as n-butyrate (Fig. 6A). Gut microbiota-derived butyrate is known to induce the differentiation of peripherally generated Treg (pTreg) cells, while SCFAs are known to facilitate the development of B220−IgA+ plasma cells in the colonic lamina propria [18,20,21]. Consistent with the findings of these prior reports, the frequencies of RORγt+Foxp3+ pTreg cells in the mucin-fed mice were twice as high as in the control mice (Fig. 6B and C). Notably, the mucin diet also increased RORγt−Foxp3+ thymus-derived Treg (tTreg) cells, suggesting that the colonic migration and/or proliferation of tTreg cells was also enhanced in the mucin-fed group. Furthermore, the mucin-containing diet increased the number of IgA+ plasma cells in the colon (Fig. 6D and E). Based on these observations, mucin serves as a mucosal barrier and contributes to the development of the intestinal immune system by facilitating microbial fermentation to increase luminal SCFA concentrations (Fig. S14).
Fig. 6.
Immune modulation by a mucin-containing diet.
(A–E) Caecal SCFA levels (A), frequencies of Foxp3+RORγt+ cells and Foxp3+RORγt− cells within CD4+ T cells (B and C), and frequencies of IgA+B220− cells within CD45+ cells (D and E) in the colonic mucosa of mice fed a control diet or a diet containing 1·5% porcine stomach mucin for 3 weeks. Representative flow cytometry plots are shown in B and D. Data represent the mean ± standard deviation (n = 5/group). ⁎p < 0·05, ⁎⁎p < 0·01, and ⁎⁎⁎p < 0·001 (two-tailed Student's t-test).
4. Discussion
In this study, we confirmed that Japanese IBD patients exhibit symptoms of dysbiosis, and that the intestinal microenvironments of CD and UC patients display several distinct features. The microbiota composition was less diverse in CD patients than in UC patients (Fig. 2B), whereas microbiota composition during the active stage of CD was characterised by expansions of Gammaproteobacteria and Bacilli with underrepresentation of Clostridia (Fig. 2C). Consequently, the frequencies of anaerobes were markedly reduced (Fig. S7), and n-butyrate producers (mainly F. prausnitzii) were nearly absent in active CD patients (Fig. 2G). Diminished microbial diversity combined with underrepresentation of n-butyrate producers has also been reported in Caucasian CD patients [7,8,24,26]; therefore, these microbial alterations are regarded as hallmarks of CD and are closely associated with disease severity. N-butyrate producers, such as F. prausnitzii and E. rectale, have been identified based on their in vitro metabolic activity [7,[24], [25], [26]]; however, the contributions of these bacteria to n-butyrate production in the human intestine have remained unclear. Our integrated analysis demonstrated that the abundances of these bacteria were positively correlated with concentrations of faecal n-butyrate (Fig. 2G), suggesting that underrepresentation of n-butyrate producers may be the main cause of reduced n-butyrate production in active CD. During CD remission, the abundances of n-butyrate-associated bacteria were slightly higher than in active CD, thus increasing the faecal concentration of n-butyrate.
Conversely, the gut microbiota of UC patients showed only a slight reduction in microbial diversity and retained normal levels of F. prausnitzii, even during the active stage (Fig. 2B and G). Nevertheless, some UC patients were deficient in faecal n-butyrate. To elucidate the underlying mechanism, we analysed the mucin components which serve as nutrient sources for gut microbiota. In particular, we focused on the levels and ratios of O-glycans which were not degraded by gut microbiota, as indexes of mucin O-glycan utilisation by gut microbiota. The ratios of mucin O-glycans in stool were significantly higher in UC patients than in healthy subjects. These observations imply that mucin O-glycan utilisation is compromised in the intestinal microbiota of UC patients. Indeed, lower mucinase activity was detected in stool samples from UC patients than in samples from healthy subjects. Mucin O-glycan utilisation is regarded as a key component in the colony-forming abilities of certain bacterial species on the mucosal surface [72,73]. Additionally, we propose that mucin O-glycans may be utilised as an endogenous fermentation source to produce SCFAs. Several lines of evidence support this hypothesis. First, there was an inverse correlation between the levels of mucin O-glycans and SCFAs in the stool samples of UC patients, whereas mucinase activity was positively correlated with the concentrations of SCFAs. Second, it has been reported that substantial levels of SCFAs are generated in the large intestines of rodents fed synthetic diets without dietary fibre and RS [51,74], indicative of the presence of endogenous fermentation sources in the gut. Intestinal microbiota may utilise mucin as a major fermentation source in the absence of fermentable dietary ingredients. We confirmed this hypothesis by showing that the exogenous administration of purified mucin facilitated the microbial production of SCFAs, both in vitro and in vivo. The mucin-containing diet increased the proportions of Allobaculum, unclassified Bacteroidales S24–7, and Akkermansia in the caecal microbiota of mice, suggesting that these bacteria may utilise mucin as an energy source. Indeed, Akkermansia and certain Bacteroides spp. have been reported to degrade mucin [75,76]. Furthermore, mucinase activity was increased in the caecal contents of mucin-fed rats. Allobacullum species are known to produce butyrate; however, Allobacullum species mainly consume mono- or disaccharides [77]. These results suggest that Allobacullum may utilise the mono- or disaccharides produced from mucin O-glycans by the other two bacteria. Notably, the abundances of Lachnospiraceae and Ruminococcaceae, the major butyrate producers [22,23], were unchanged in mucin-fed mice. Therefore, the administration of exogenous mucin is likely to have altered microbiota composition by promoting the outgrowth of mucin utilisers and enhancing the production of SCFAs, including butyrate. Third, previous studies using an in vitro gut model demonstrated that Firmicutes dominated the gut mucus microbiota, with Clostridium cluster XIVa species constituting approximately 60% of all mucus-colonising bacteria [78]. 16S rRNA gene analysis of mucosa-associated bacteria showed that Firmicutes species were more abundant in the mucus region, and that Clostridium cluster XIVa (Lachnospiraceae and Ruminococcaceae) were the predominant families [79]. Certain gut bacteria, such as A. muciniphila, are capable of utilising mucin O-glycans, but not dietary polysaccharides [76]. Mucus-associated bacteria express mucin glycan utilisation-associated genes at high levels, indicating that these bacteria utilise mucin glycans [32]. Therefore, we presume that certain bacteria in the mucus utilise mucin O-glycans for n-butyrate production, regardless of the presence of dietary fibres. Consequently, we propose a new model of symbiotic interaction in which host cell-derived mucin O-glycans are efficiently metabolised by intestinal microbiota under physiological conditions to produce n-butyrate, which is then utilised as an energy source by colonic epithelial cells and contributes to the development of the colonic immune system. This ‘symbiosynthesis’, based on the mucin O-glycan–n-butyrate axis may play a significant role in establishing host–microbe symbiosis. In UC, the symbiosynthetic pathway appears impaired due to dysbiosis, at least partly contributing to the reduced production of n-butyrate which may be involved in the pathogenesis of UC.
Furthermore, we demonstrated the immunomodulatory functions of mucin in the colon. We confirmed that exogenous mucin expanded Treg cell and IgA+ plasma cell populations, indicating that it has a significant role in the development of the immune system. Given that n-butyrate induces the differentiation of colonic pTreg cells and IgA+ plasma cells [18,20], we speculate that the intake of exogenous mucin increased these immune cell populations by enhancing n-butyrate biosynthesis. Interestingly, mucin administration also induced the expansion of tTreg cell populations, suggesting that mucin facilitates the differentiation, migration, and/or proliferation of tTreg cells. Increased propionate levels may be responsible for this event [80,81]. Although these mechanisms are not yet fully elucidated, these findings emphasise the importance of mucin as a fermentation source in the maintenance of colonic immune homeostasis.
Correlation analysis identified seven species as butyrate-associated bacteria. Among these was the well-documented n-butyrate producer, F. prausnitzii [22]. E. rectale and E. hallii, which belong to the Clostridium cluster XIVa, also produce n-butyrate [22,23]. Notably, these Eubacterium spp. have been shown to accumulate in the mucus layer [78,82] and their numbers were significantly lower in patients with active UC in this study (Fig. 2G). This may reflect the impaired mucin O-glycan–n-butyrate axis in UC. The genus Anaerostipes, another member of Clostridium cluster XIVa, includes several butyrate producers, such as A. caccae and A. hadrus [83,84]. The remaining three species, R. bromii, B. wexlerae, and B. vulgatus, may function as n-butyrate facilitators, as part of a bacterial metabolic web. For example, B. vulgatus has an abundance of genes associated with polysaccharide utilisation [85], whereas R. bromii and B. wexlerae, classified as Clostridium cluster IV and XIVa species, respectively, are known as amylolytic bacteria. R. bromii plays a central role in the degradation and utilisation of RS in the human colon [86], while B. wexlerae utilises RS during fermentation in vitro [87]. The digestion of large-molecule polysaccharides by these bacterial strains may provide oligo- or monosaccharides as fermentation sources for n-butyrate producers. The numbers of R. bromii and B. wexlerae bacteria were significantly lower in active UC patients; therefore, we speculate that the microbial digestion of polysaccharides and RS might be reduced in these patients. This perspective may offer a new therapeutic strategy to restore n-butyrate production in UC patients, by activating the polysaccharide/RS-digestion pathway using prebiotics and/or probiotics [88,89].
In conclusion, we observed that n-butyrate biosynthesis was reduced in both CD and UC patients; however, the underlying mechanisms differed between the two diseases (Fig. S14). In CD patients, n-butyrate levels were reduced by the loss of the majority of n-butyrate producers, including F. prausnitzii. Conversely, UC-associated microbiota utilised mucin O-glycans poorly, which eventually led to reduced n-butyrate production. Although further studies are needed to clarify the precise molecular mechanisms involved in the mucin O-glycan-dependent n-butyrate pathway, our findings offer a new perspective regarding the pathogenesis of UC and the development of intestinal dysbiosis.
Funding sources
This study was supported by grants from the Japan Society for the Promotion of Science (#16H01369, 17KT0055, and 18H04680 to KH), Health Labour Sciences Research Grant (KH and JK), AMED-Crest (#16gm0000000h0101, 17gm1010004h0102, and 18gm1010004h0103 to KH and JK), AMED (#18ek0109303h0001 to KH and JK), Yakult Foundation (KH), Keio Gijuku Academic Development Funds (KH), The Aashi Grass Foundation, and The Canon Foundation (JK).
Author contributions
K.H., T.M., J.K., and H.I. conceived the study. T.Y., Y.Fu., Y.Fr., and K.H. performed the microbiome analysis experiments. H.S. and T.G. analysed mucin and metabolite levels and performed the rat experiments. R.N., H.H., and M.F. performed the in vitro fermentation analysis. T.Y., M.H., and Y.K. performed the mouse experiments. T.Y. and H.S. analysed the data with M.H. and H.O. T.Y. performed the bioinformatic analysis with support from R.A., W.S. and M.H. H.I. collected clinical samples. T.Y., S.H., T.M., and K.H. interpreted the data. T.Y. and K.H. wrote the manuscript.
Declaration of Competing Interest
The authors have declared that no competing interests exist.
Acknowledgements
We would like to thank Yun-Gi Kim for their comments and suggestions, and Koichiro Suzuki for their technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.09.008.
Contributor Information
Tatsuya Morita, Email: morita.tatsuya@shizuoka.ac.jp.
Koji Hase, Email: hase-kj@pha.keio.ac.jp.
Appendix A. Supplementary data
Supplementary Tables
Supplementary Figures
References
- 1.Khor B., Gardet A., Xavier R.J. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wlodarska M., Kostic A.D., Xavier R.J. An integrative view of microbiome-host interactions in inflammatory bowel diseases. Cell Host Microbe. 2015;17:577–591. doi: 10.1016/j.chom.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honda K., Littman D.R. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamada N., Seo S.-U.U., Chen G.Y., Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 5.Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin J., Li R., Raes J., Arumugam M., Burgdorf K., Manichanh C. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gevers D., Kugathasan S., Denson L.A., Vázquez-Baeza Y., Treuren W., Ren B. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halfvarson J., Brislawn C.J., Lamendella R., Vázquez-Baeza Y., Walters W.A., Bramer L.M. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. 2017;2 doi: 10.1038/nmicrobiol.2017.4. nmicrobiol20174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honda K., Littman D.R. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 10.Garrett W.S., Lord G.M., Punit S., Lugo-Villarino G., Mazmanian S.K., Ito S. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elinav E., Strowig T., Kau A.L., Henao-Mejia J., Thaiss C.A., Booth C.J. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagao-Kitamoto H., Shreiner A.B., Gillilland M.G., Kitamoto S., Ishii C., Hirayama A. Functional characterization of inflammatory bowel disease-associated gut dysbiosis in gnotobiotic mice. Cell Mol Gastroenterol Hepatol. 2016;2:468–481. doi: 10.1016/j.jcmgh.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cummings J. Fermentation in the human large intestine: evidence and implications for health. Lancet. 1983;1:1206–1209. doi: 10.1016/s0140-6736(83)92478-9. [DOI] [PubMed] [Google Scholar]
- 14.Høverstad T., Midtvedt T. Short-chain fatty acids in germfree mice and rats. J. Nutr. 1986;116:1772–1776. doi: 10.1093/jn/116.9.1772. [DOI] [PubMed] [Google Scholar]
- 15.Cummings J., Pomare E., Branch W., Naylor C., Macfarlane G. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly C.J., Zheng L., Campbell E.L., Saeedi B., Scholz C.C., Bayless A.J. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 2015;17:662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaudier E., Rival M., Buisine M.-P., Robineau I., Hoebler C. Butyrate enemas upregulate Muc genes expression but decrease adherent mucus thickness in mice colon. Physiol Res. 2008;58:111–119. doi: 10.33549/physiolres.931271. [DOI] [PubMed] [Google Scholar]
- 18.Kim M., Qie Y., Park J., Kim C.H. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe. 2016;20:202–214. doi: 10.1016/j.chom.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin L., Laevsky G., Giardina C. Butyrate suppression of colonocyte NF-kappa B activation and cellular proteasome activity. J. Biol. Chem. 2001;276:44641–44646. doi: 10.1074/jbc.M105170200. [DOI] [PubMed] [Google Scholar]
- 20.Furusawa Y., Obata Y., Fukuda S., Endo T.A., Nakato G., Takahashi D. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 21.Arpaia N., Campbell C., Fan X., Dikiy S., van der Veeken J., de Roos P. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504 doi: 10.1038/nature12726. nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barcenilla A., Pryde S., Martin J., Duncan S., Stewart C., Henderson C. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 2000;66:1654–1661. doi: 10.1128/aem.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louis P., Duncan S.H., McCrae S.I., Millar J., Jackson M.S., Flint H.J. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J. Bacteriol. 2004;186:2099–2106. doi: 10.1128/JB.186.7.2099-2106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank D.N., Amand A.L., Feldman R.A., Boedeker E.C., Harpaz N., Pace N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujimoto T., Imaeda H., Takahashi K., Kasumi E., Bamba S., Fujiyama Y. Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn's disease. J. Gastroenterol. Hepatol. 2013;28:613–619. doi: 10.1111/jgh.12073. [DOI] [PubMed] [Google Scholar]
- 26.Machiels K., Joossens M., Sabino J., Preter V., Arijs I., Eeckhaut V. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 27.Harig J., Soergel K., Komorowski R., Wood C. Treatment of diversion colitis with short-chain-fatty acid irrigation. N. Engl. J. Med. 1989;320:23–28. doi: 10.1056/NEJM198901053200105. [DOI] [PubMed] [Google Scholar]
- 28.Scheppach W., Sommer H., Kirchner T., Paganelli G., Bartram P., Christl S. Effect of butyrate enemas on the colonic mucosa in distal ulcerative colitis. Gastroenterology. 1992;103:51–56. doi: 10.1016/0016-5085(92)91094-k. [DOI] [PubMed] [Google Scholar]
- 29.McGuckin M.A., Lindén S.K., Sutton P., Florin T.H. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9:265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- 30.Johansson M.E., Phillipson M., Petersson J., Velcich A., Holm L., Hansson G.C. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansson M.E., Larsson J.M., Hansson G.C. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host–microbial interactions. Proc Natl Acad Sci. 2011;108:4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H., Limenitakis J.P., Fuhrer T., Geuking M.B., Lawson M.A., Wyss M. The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun. 2015;6 doi: 10.1038/ncomms9292. ncomms9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.der Sluis M., Koning B., Bruijn A., Velcich A., Meijerink J., Goudoever J.B. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 34.Sommer F., Adam N., Johansson M.E., Xia L., Hansson G.C., Bäckhed F. Altered mucus glycosylation in core 1 O-glycan-deficient mice affects microbiota composition and intestinal architecture. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0085254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johansson M.E., Gustafsson J.K., Holmén-Larsson J., Jabbar K.S., Xia L., Xu H. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi K., Yoshimura Y., Kaimoto T., Kunii D., Komatsu T., Yamamoto S. Validation of a food frequency questionnaire based on food groups for estimating individual nutrient intake. Jpn J Nutr Diet. 2001;59:221–232. [Google Scholar]
- 37.Lennard-Jones J. Classification of inflammatory bowel disease. Scand J Gastroenterol Suppl. 1989;170:2–6. doi: 10.3109/00365528909091339. [discussion 16-9] [DOI] [PubMed] [Google Scholar]
- 38.Podolsky D. Inflammatory bowel disease (1) N. Engl. J. Med. 1991;325:928–937. doi: 10.1056/NEJM199109263251306. [DOI] [PubMed] [Google Scholar]
- 39.Podolsky D. Inflammatory bowel disease (2) N. Engl. J. Med. 1991;325:1008–1016. doi: 10.1056/NEJM199110033251406. [DOI] [PubMed] [Google Scholar]
- 40.Rachmilewitz D. Coated mesalazine (5-aminosalicylic acid) versus sulphasalazine in the treatment of active ulcerative colitis: a randomised trial. Br. Med. J. 1989;298:82. doi: 10.1136/bmj.298.6666.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Best W.R., Becktel J.M., Singleton J.W., Kern F. Development of a Crohn's Disease Activity Index National Cooperative Crohn’s Disease Study. Gastroenterology. 1976;70:439–444. [PubMed] [Google Scholar]
- 42.Matts S. The value of rectal biopsy in the diagnosis of ulcerative colitis. Q. J. Med. 1961;30:393–407. [PubMed] [Google Scholar]
- 43.Imaeda H., Bamba S., Takahashi K., Fujimoto T., Ban H., Tsujikawa T. Relationship between serum infliximab trough levels and endoscopic activities in patients with Crohn's disease under scheduled maintenance treatment. J. Gastroenterol. 2014;49:674–682. doi: 10.1007/s00535-013-0829-7. [DOI] [PubMed] [Google Scholar]
- 44.Illumina . 2017. 16S Metagenomic Sequencing Library Preparation; pp. 1–28.http://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf [Google Scholar]
- 45.Caporaso G.J., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7 doi: 10.1038/nmeth.f.303. nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011:10–12. [Google Scholar]
- 47.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 48.Tsuda A., Suda W., Morita H., Takanashi K., Takagi A., Koga Y. Influence of proton-pump inhibitors on the luminal microbiota in the gastrointestinal tract. Clin. Transl. Gastroenterol. 2015;6 doi: 10.1038/ctg.2015.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dixon P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003;14:927–930. [Google Scholar]
- 50.Ward T., Larson J., Meulemans J., Hillmann B., Lynch J., Sidiropoulos D. BugBase predicts organism level microbiome phenotypes. BioRxiv. 2017;133462 [Google Scholar]
- 51.Morita T., Kasaoka S., Ohhashi A., Ikai M., Numasaki Y., Kiriyama S. Resistant proteins alter cecal short-chain fatty acid profiles in rats fed high amylose cornstarch. J. Nutr. 1998;128:1156–1164. doi: 10.1093/jn/128.7.1156. [DOI] [PubMed] [Google Scholar]
- 52.Cobo E., Kissoon-Singh V., Moreau F., Chadee K. Colonic MUC2 mucin regulates the expression and antimicrobial activity of β-defensin 2. Mucosal Immunol. 2015;8:1360–1372. doi: 10.1038/mi.2015.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanabe H., Sugiyama K., Matsuda T., Kiriyama S., Morita T. Small intestinal mucins are secreted in proportion to the settling volume in water of dietary indigestible components in rats. J. Nutr. 2005;135:2431–2437. doi: 10.1093/jn/135.10.2431. [DOI] [PubMed] [Google Scholar]
- 54.Yasuno S., Kokubo K., Kamei M. New method for determining the sugar composition of glycoproteins, glycolipids, and oligosaccharides by high-performance liquid chromatography. Biosci Biotechnol Biochem. 2014;63:1353–1359. doi: 10.1271/bbb.63.1353. [DOI] [PubMed] [Google Scholar]
- 55.Bovee-Oudenhoven I., Termont D., Heidt P., der Meer V.R. Increasing the intestinal resistance of rats to the invasive pathogen Salmonella enteritidis: additive effects of dietary lactulose and calcium. Gut. 1997;40:497–504. doi: 10.1136/gut.40.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harrison M.J., Packer N.H. Glycoprotein methods and protocols. Methods Mol. Biol. 2000;125:211–216. doi: 10.1385/1-59259-048-9:211. [DOI] [PubMed] [Google Scholar]
- 57.Ogata M., Hidari K., Murata T., Shimada S., Kozaki W., Park E.Y. Chemoenzymatic synthesis of sialoglycopolypeptides as glycomimetics to block infection by avian and human influenza viruses. Bioconjug. Chem. 2009;20:538–549. doi: 10.1021/bc800460p. [DOI] [PubMed] [Google Scholar]
- 58.Komura M., Fukuta T., Genda T., Hino S., Aoe S., Kawagishi H. A short-term ingestion of fructo-oligosaccharides increases immunoglobulin A and mucin concentrations in the rat cecum, but the effects are attenuated with the prolonged ingestion. Biosci. Biotechnol. Biochem. 2014;78:1592–1602. doi: 10.1080/09168451.2014.925782. [DOI] [PubMed] [Google Scholar]
- 59.Shiau S.-Y., Chang G.W. Effects of dietary fiber on fecal mucinase and β-glucuronidase activity in rats. J. Nutr. 1983;113:138–144. doi: 10.1093/jn/113.1.138. [DOI] [PubMed] [Google Scholar]
- 60.Lowry O., Rosebrough N., Farr A., Randall R. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 61.Han K.-H., Koyashi Y., Nakamura Y., Shi K., Aritsuka T., Kiyoshi O. Comparison of the effects of longer chain Inulins with different degrees of polymerization on colonic fermentation in a mixed culture of swine fecal bacteria. J Nutr Sci Vitaminol. 2014;60:206–212. doi: 10.3177/jnsv.60.206. [DOI] [PubMed] [Google Scholar]
- 62.Weigmann B., Tubbe I., Seidel D., Nicolaev A., Becker C., Neurath M.F. Isolation and subsequent analysis of murine lamina propria mononuclear cells from colonic tissue. Nat Protoc. 2007;2 doi: 10.1038/nprot.2007.315. nprot.2007.315. [DOI] [PubMed] [Google Scholar]
- 63.Suzuki K., Yamada T., Yamazaki K., Hirota M., Ishihara N., Sakamoto M. Intestinal epithelial cell-specific deletion of α-mannosidase II ameliorates experimental colitis. Cell Struct. Funct. 2018;43:25–39. doi: 10.1247/csf.17022. [DOI] [PubMed] [Google Scholar]
- 64.Pollard K., Dudoit S., van der Laan M. Bioinformatics and computational biology solutions using R and bioconductor. Springer. 2005:249–271. [Google Scholar]
- 65.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:1–18. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Asnicar F., Weingart G., Tickle T.L., Huttenhower C., Segata N. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ. 2015;3 doi: 10.7717/peerj.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamada T., Takahashi D., Hase K. The diet-microbiota-metabolite axis regulates the host physiology. J. Biochem. 2016;160:1–10. doi: 10.1093/jb/mvw022. [DOI] [PubMed] [Google Scholar]
- 68.Larsson J.M., Karlsson H., Sjövall H., Hansson G.C. A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology. 2009;19:756–766. doi: 10.1093/glycob/cwp048. [DOI] [PubMed] [Google Scholar]
- 69.Tytgat K., van der Wal J., Einerhand A., Büller H., Dekker J. Quantitative analysis of MUC2 synthesis in ulcerative colitis. Biochem Biophys Res Commun. 1996;224:397–405. doi: 10.1006/bbrc.1996.1039. [DOI] [PubMed] [Google Scholar]
- 70.Dorofeyev A., Vasilenko I., Rassokhina O., Kondratiuk R. Mucosal barrier in ulcerative colitis and Crohn's disease. Gastroenterol. Res. Pract. 2013;2013:431231. doi: 10.1155/2013/431231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tangvoranuntakul P., Gagneux P., Diaz S., Bardor M., Varki N., Varki A. Human uptake and incorporation of an immunogenic nonhuman dietary sialic acid. Proc Natl Acad Sci. 2003;100:12045–12050. doi: 10.1073/pnas.2131556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tailford L.E., Crost E.H., Kavanaugh D., Juge N. Mucin glycan foraging in the human gut microbiome. Front Genet. 2015;6:81. doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ouwerkerk J.P., de Vos W.M., Belzer C. Glycobiome: bacteria and mucus at the epithelial interface. Best Pract Res Clin Gastroenterol. 2013;27:25–38. doi: 10.1016/j.bpg.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 74.Thorburn A.N., McKenzie C.I., Shen S., Stanley D., Macia L., Mason L.J. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 2015;6:7320. doi: 10.1038/ncomms8320. [DOI] [PubMed] [Google Scholar]
- 75.Derrien M., Vaughan E.E., Plugge C.M., de Vos W.M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 76.Desai M.S., Ekatz A., Koropatkin N.M., Kamada N., Hickey C.A., Wolter M. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167:1339–1353. doi: 10.1016/j.cell.2016.10.043. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Greetham H.L., Gibson G.R., Giffard C., Hippe H., Merkhoffer B., Steiner U. Allobaculum stercoricanis gen. nov., sp. nov., isolated from canine feces. Anaerobe. 2004;10:301–307. doi: 10.1016/j.anaerobe.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 78.den Abbeele P., Belzer C., Goossens M., Kleerebezem M., Vos W.M., Thas O. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013;7:949. doi: 10.1038/ismej.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nava G.M., Friedrichsen H.J., Stappenbeck T.S. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 2011;5:627. doi: 10.1038/ismej.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Obata Y., Furusawa Y., Endo T.A., Sharif J., Takahashi D., Atarashi K. The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nat Immunol. 2014;15:571–579. doi: 10.1038/ni.2886. [DOI] [PubMed] [Google Scholar]
- 81.Smith P.M., Howitt M.R., Panikov N., Michaud M., Gallini C., Bohlooly-Y M. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Louis P., Flint H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 83.Schwiertz A., Hold G.L., Duncan S.H., Gruhl B., Collins M.D., Lawson P.A. Anaerostipes caccae gen. Nov., sp. nov., a new saccharolytic, acetate-utilising, butyrate-producing bacterium from human faeces. Syst. Appl. Microbiol. 2002;25:46–51. doi: 10.1078/0723-2020-00096. [DOI] [PubMed] [Google Scholar]
- 84.Allen-Vercoe E., Daigneault M., White A., Panaccione R., Duncan S.H., Flint H.J. Anaerostipes hadrus comb. nov., a dominant species within the human colonic microbiota; reclassification of Eubacterium hadrum Moore et al. 1976. Anaerobe. 2012;18:523–529. doi: 10.1016/j.anaerobe.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 85.Kaoutari A., Armougom F., Gordon J.I., Raoult D., Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11 doi: 10.1038/nrmicro3050. nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 86.Ze X., Duncan S.H., Louis P., Flint H.J. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012;6:1535. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang J., Martínez I., Walter J., Keshavarzian A., Rose D.J. In vitro characterization of the impact of selected dietary fibers on fecal microbiota composition and short chain fatty acid production. Anaerobe. 2013;23:74–81. doi: 10.1016/j.anaerobe.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 88.Walker A.W., Ince J., Duncan S.H., Webster L.M., Holtrop G., Ze X. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Scott K.P., Antoine J.-M., Midtvedt T., van Hemert S. Manipulating the gut microbiota to maintain health and treat disease. Microb Ecol Health D. 2015;26:25877. doi: 10.3402/mehd.v26.25877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables
Supplementary Figures