Abstract
Background
Malignant Pleural Mesothelioma (MPM) is an aggressive disease related to asbestos exposure, with no effective therapeutic options.
Methods
We undertook unsupervised analyses of RNA-sequencing data of 284 MPMs, with no assumption of discreteness. Using immunohistochemistry, we performed an orthogonal validation on a subset of 103 samples and a biological replication in an independent series of 77 samples.
Findings
A continuum of molecular profiles explained the prognosis of the disease better than any discrete model. The immune and vascular pathways were the major sources of molecular variation, with strong differences in the expression of immune checkpoints and pro-angiogenic genes; the extrema of this continuum had specific molecular profiles: a “hot” bad-prognosis profile, with high lymphocyte infiltration and high expression of immune checkpoints and pro-angiogenic genes; a “cold” bad-prognosis profile, with low lymphocyte infiltration and high expression of pro-angiogenic genes; and a “VEGFR2+/VISTA+” better-prognosis profile, with high expression of immune checkpoint VISTA and pro-angiogenic gene VEGFR2. We validated the gene expression levels at the protein level for a subset of five selected genes belonging to the immune and vascular pathways (CD8A, PDL1, VEGFR3, VEGFR2, and VISTA), in the validation series, and replicated the molecular profiles as well as their prognostic value in the replication series.
Interpretation
The prognosis of MPM is best explained by a continuous model, which extremes show specific expression patterns of genes involved in angiogenesis and immune response.
Keywords: Pleural mesothelioma, Immunotherapy, Angiogenesis, MESOMICS project, French MESOBANK
Research in context.
Evidence before this study
We searched for publically available genomic data of MPM in public databases (European Genome-Phenome Archive, dbGaP, TCGA data portal; last accessed in August 2019) associated with a peer-reviewed article, requiring a minimum of 20 samples with RNA-sequencing data per study. We found two studies that matched our criteria, one by Bueno and colleagues including 211 samples and one by the TCGA consortium including 73 samples. The two studies proposed discrete molecular classifications of MPM that partially match the current histological classification. These two studies were based on the implicit assumption that MPM is subdivided into discrete entities, potentially preventing the discovery of some important aspects of the MPM molecular variation.
Added value of this stud
In this study we characterized the molecular variation of MPM without any assumption of discreteness, to analyse the molecular pathways underlying this variation, and to identify novel candidate markers that could serve both for classification and treatment of this disease. We provide a model that explains the prognosis of MPM better than previous discrete models, both based on histology and molecular data. The continuous model also enabled to identify the immune and vascular pathways as the major sources of molecular variation in MPM. Finally, we provide a validated panel of five proteins that is sufficient to characterize the molecular profile of MPM.
Implications of all the available evidence
Our findings provide novel insights into the combined importance of angiogenesis and the immune response in MPM prognosis and progression and inform future tumour classifications. In addition, the five-protein panel that we provide could be used in the clinic to characterize tumours and inform clinical management and treatment strategies.
Alt-text: Unlabelled Box
1. Introduction
Malignant Pleural Mesothelioma (MPM) is a deadly disease, with most patients dying within 2 years of diagnosis. MPM is related to asbestos exposure, with a long latency between the exposure and the development of the disease [1]. Based on the 2015 WHO classification, there are three major MPM histopathological types, with different prognoses: epithelioid, biphasic, and sarcomatoid [2]. The sarcomatoid component is the marker with the highest prognostic value; however, in the recent IASLC-EURACAN multidisciplinary workshop on mesothelioma classification held in Lyon on the 6–7th July 2018 [3], pathologists agreed that a more precise definition of what constitutes sarcomatoid features, in addition to a more multidisciplinary classification, would be needed to improve diagnosis reproducibility (currently with a kappa of 0.45) [4].
The histopathological classification also has a role in the clinical decision-making but, ultimately, MPM becomes refractory to all conventional treatment modalities, including surgery, chemotherapy, and radiotherapy. Alternative therapeutic options have been evaluated with limited success; for example, although strong pre-clinical data support the role of angiogenesis in MPM, the available phase-II and phase-III clinical trials testing for anti-angiogenic drugs have only shown modest activity [5], [6]. Similarly, preliminary data from ongoing clinical trials suggested that immunotherapy might be a promising approach for this disease [7]; however, PD(L)1 expression measured by immunohistochemistry turned out to be a poor predictive marker of response to PD(L)1 inhibitors, while concerns have been raised about potential toxicities of immunotherapies in patients with mesothelioma [5], [8], [9], [10]. In addition, the outcome of patients treated with systemic agents may be variable across histopathological types: while chemotherapy seems to be less effective in sarcomatoid tumours, antiangiogenic agents and immune checkpoint inhibitors are associated with a lower survival benefit in the epithelioid type. A recent study has highlighted the enormous heterogeneity of the microenvironment of MPM, suggesting that a combination of immunotherapies might be more effective than single-agent approaches [11].
Large-scale genomic studies aiming at characterizing MPM have provided new insights into its classification. Bueno and colleagues [12] proposed a four-class molecular subdivision based on transcriptomic data, consisting of an “Epithelioid” group enriched for epithelioid tumours, a “Biphasic-E” group enriched for biphasic and epithelioid tumours, a “Biphasic-S” group enriched for biphasic and sarcomatoid tumours, and a “Sarcomatoid” group enriched for sarcomatoid tumours. Similarly, Hmeljak and colleagues [13] provided a subdivision into molecular groups based on integrated genomic, transcriptomic, and epigenomic data. Nevertheless, these attempts at classifications were all based on the implicit assumption that MPM is subdivided into discrete entities, potentially preventing the discovery of some important aspects of the MPM molecular variation. Thus, in this study we characterized the molecular variation of MPM without any assumption of discreteness, to analyse the molecular pathways underlying this variation, and to identify novel candidate markers that could serve both for classification and treatment of this disease.
2. Materials and methods
2.1. Ethics
All methods were carried out in accordance with relevant guidelines and regulations. This study is part of a larger study -MESOMICS project- aiming at the comprehensive molecular characterization of malignant pleural mesothelioma, approved by the IARC Ethical Committee (Project No. 15–17). The samples used in this study belong to the French MESOBANK [14], which guidelines include obtaining the informed consent from all subjects.
2.2. Molecular data
We combined the RNA-sequencing (RNA-seq) datasets from Bueno and colleagues [12] (n = 211) and the TCGA [13] (n = 73). Note that we excluded from the full TCGA-MESO cohort of 86 samples, the samples that were excluded in the final report (13 samples). Additionally, we conducted immunohistochemistry (IHC) on two datasets: (1) Tissue MicroArrays (TMAs) of a subset of 106 samples from the Bueno and colleagues study [12], which acts as an orthogonal technical validation; and (2) an independent cohort of 77 samples from the French MESOBANK, which acts as a replication of our results. TMAs were done from 106 cases of MPM, three cores per sample of 0.6 mm of diameter each were used to make six recipient blocks of TMA. The replication dataset of 77 samples come from the French MESOBANK, a multi-centric virtual and exhaustive repository of national data, biological samples, and standardized operational procedures for mesothelioma. This database contains histopathological data for more than 10,000 specimens [14]. The 77 samples were selected from three groups: a long-survival epithelioid group (survival >30months), a short-survival epithelioid group (survival <10 months), and a sarcomatoid group (survival <10 months). The samples from the three groups were matched for age (≤6 years difference) and sex, were all chemo-naive at the time of sample collection but all underwent cisplatin and/or pemetrexed chemotherapy afterwards. Although not matched, we confirmed that smoking status and asbestos exposure were balanced between the three groups (Fisher's exact tests p > 0.05).
2.3. Pathological review and clinical data
Tumour grade, infiltration, and the presence of necrosis were assessed for all 284 samples from digital H&E slides of FFPE. The slides from the TCGA cohorts were visualized from the cancer digital slide archive (http://cancer.digitalslidearchive.net/, accessed in January and February 2018). The histopathological types based on the 2015-WHO classification (epithelioid, biphasic, sarcomatoid) and the clinical information (sex, age, survival, asbestos exposure, pre-treatment, surgery) were retrieved from the supplementary tables of the corresponding manuscripts. The 77 samples from the French MESOBANK replication cohort have all undergone a Central Pathological Review (French standardized procedure of certification of mesothelioma) and contained clinical information on the same variables. For the replication cohort, we also assessed the epithelioid histopathological characteristics (patterns and stromal characteristics), which we subdivided into three subtypes, based on the recent IASCL-EURACAN interdisciplinary meeting recommendations: [3] good-prognosis (regrouping the acinar and papillary subtypes, and samples with abundant myxoid stroma), intermediate-prognosis (trabecular subtype), and bad-prognosis (solid subtype). We confirmed that the epithelioid subtypes were balanced between the long- and short-survival groups (Table 1; Fisher's exact tests p = 1).
Table 1.
Replication series baseline table.
| Characteristics | Epithelioid long survival (n = 26) | Epithelioid short survival (n = 25) | Sarcomatoid (n = 26) |
|---|---|---|---|
| Discrete variables | No. (%) | No. (%) | No. (%) |
| Sex | |||
| Male | 20 (77) | 19 (76) | 20 (77) |
| Female | 6 (23) | 6 (24) | 6 (23) |
| Smoking status | |||
| Never | 6 (30) | 9 (45) | 4 (29) |
| Former | 12 (60) | 8 (40) | 8 (57) |
| Current | 2 (10) | 3 (15) | 2 (14) |
| Asbestos exposure | |||
| No | 5 (19) | 9 (36) | 2 (8) |
| Possible | 0 (0) | 1 (4) | 1 (4) |
| Probable | 4 (15) | 3 (12) | 2 (8) |
| Yes | 17 (65) | 12 (48) | 20 (80) |
| Survival censor (dead) | 26 (100) | 25 (100) | 26 (100) |
| Epithelioid subtypea | |||
| Good-prognosis | 7 (6 acinar, 1 papillary) | 7 (3 acinar, 2 myxoid stroma, 2 papillary) | NA |
| Intermediate-prognosis | 3 | 3 | NA |
| Bad-prognosis | 15 | 14 | NA |
| Continuous variables | Mean (sd) | Mean (sd) | Mean (sd) |
| Age at diagnostic (years) | 74.9 (7.3) | 74.4 (7.3) | 74.8 (6.8) |
| Survival time (months) | 42.2 (5.2) | 5.8 (2.8) | 4.7 (2.4) |
Good-prognosis subtypes: acinar, papillary, myxoid stroma; intermediate-prognosis subtype: trabecular; bad-prognosis subtype: solid.
2.4. Immunohistochemistry
For the 77 French MESOBANK samples, FFPE tissue sections were previously deparaffinised. All the TMA spots and the MESOBANK samples were stained with the CD8 (ROCHE, cl SP57 Rabbit), PDL1 (ROCHE, cl SP263 Rabbit), VEGFR2 (Cell Signaling, cl 55B11 Rabbit), VEGFR3 (R&D, Polyclonal goat), and VISTA (Cell Signaling, cl D1L2G) assays using UltraView Universal DAB Detection Kit (Ventana Medical Systems) and Amplification Kit (Ventana Medical Systems - Roche) on Benchmark ULTRA (Roche, Ventana Meylan, France) individually. For CD8, PDL1 and VISTA, because the available antibodies were all membranous for tumour cells, no dual stainings were performed. For CD8, the percentage of tumour infiltrating lymphocytes (TILS) exhibiting a staining were reported. For PDL1, the percentages of TILS cytoplasmic/membranous staining and tumour cells exhibiting a membranous staining were separately reported. For all other markers, the percentages of tumour cells exhibiting a membranous staining were reported. CD8 and VEGFR3 percentages have been reported as five levels of protein expression: 0%, 25%, 50%, 75% and 100%, instead of a continuous quantification as reported for all other markers, due to the lack of resolution. For VEGFR3 we optimized the membranous staining by adapting the dilution to the best staining of the internal control (vessels), which could explain these difficulties. The percentage of all markers was only reported when the average number of tumour cells was more than 50%. When the slide or staining global quality did not allow the protein level evaluation, the percentage of the marker was not reported. For these reasons, only a subset of 103 out of the 106 samples initially planned was included in the technical validation cohort by IHC on TMA. Among the 77 MESOBANK samples, there were three samples with missing data for CD8 expression, three for PDL1 expression in the tumour, four for PDL1 in TILS, seven for VEGFR2, three for VEGFR3, and eight for VISTA. The positive controls of the five antibodies used are reported in Fig. S1. All IHC photos have been scanned at 20× magnification. All the IHC slides have been read and scored by FGS.
2.5. RNA-seq data processing
The 284 raw reads files were processed in three steps (bioinformatic workflow freely available at https://github.com/IARCbioinfo/RNAseq-nf): [15], [16], [17] (i) reads were scanned for Illumina adapter sequence using software Trim Galore v0.4.2; (ii) reads were mapped to reference genome GRCh38 (gencode version 24) using software STAR v2.5.2b; and (iii) reads were counted for each gene of the comprehensive gencode gene annotation file using software htseq v0.8.0. We quantified the proportion of cells that belong to different immune cell types from the RNA-seq data using software quanTIseq [18] (Table S1). In a nutshell, quanTIseq performs a supervised deconvolution based on the expression signature of a reference panel of blood-derived immune cells from ten different types. Among all the genes from the transcriptome, the quanTIseq authors selected a panel using machine-learning techniques so as to maximize specificity and discriminative power (see Additional File 1 from Finotello et al. 2017 for the source datasets used and the full list of genes included) [18].
2.6. Low-dimensional summary of expression data
The raw read counts of the 284 samples were normalized using the variance stabilization transform (R package DESeq2 v1.18.1) [19]. The genes that displayed the largest variance (7145 genes representing 50% of the total variance; Table S2) were then mean-centered and selected to compute a low-dimensional summary using Principal Component Analysis (PCA; Table S1). Indeed, the visualization of the expression levels across samples for 7145 genes theoretically requires a plot in 7145 dimensions, which would be impossible to interpret. PCA overcomes this issue by reducing the high-dimensional data to a few independent dimensions, each representing groups of genes with correlated expression levels. One limitation of PCA is that it assumes linear relationships between the low-dimensional representation and original variables (gene expression, in our case). Nevertheless, contrary to alternative nonlinear techniques, this linear relationship results in interpretable dimensions [20]. In other words, PCA makes data easy to explore and visualize by reducing the dimensionality of a data set consisting of many variables correlated with each other, while retaining as much as possible the variation present in the original dataset. Note that we report the first two dimensions of the PCA in the results section, because the other main dimensions (from three to seven) explain each <5% of the gene expression variation among the 7145 selected genes, they were not significantly associated with the histopathological type (ANOVA q > 0.05), and none except Dimension 5 were associated with survival (Wald test p > 0.05; Table S3). Importantly, the samples with pre-treatment (31 samples with pre-surgical chemotherapy from the Bueno et al. cohort) did not have significant associations with any of the seven axes. Also note that all 199 samples included in the survival studies underwent similar treatments (chemotherapy plus surgery). Correlation circles were constructed from the correlations of variables on each dimension.
We performed a five-gene PCA following the same protocol, but using only a subset of five genes highly correlated with PCA Dimensions 1 and 2, for which IHC-validated antibodies were available (CD8A, VEGFR2, VEGFR3, PDL1, and VISTA). We compared the PCA performed on all genes (hereafter simply denoted “PCA”) to that performed on the reduced gene set (denoted “five-gene PCA”) using the Kabsch algorithm, which finds the rotation that minimizes the deviation between two sets of points.
PCAs were independently conducted on the validation and replication IHC datasets, after mean-centering expression levels. We used hierarchical clustering on the replication IHC dataset to ensure that the protein expression profiles of the short- and long-survival epithelioid sets were not biased by misclassifications. Indeed, if misclassifications—which are common in MPM, in particular biphasic samples misclassified into epithelioids and sarcomatoids—disproportionately happened between sarcomatoids and short-term survival epithelioids in our series, we would expect the protein expression of the epithelioid short-survival set to be biased toward that of the sarcomatoid set. Using hierarchical clustering analysis on the five-protein expression data (Fig. S2), we found that most sarcomatoid and epithelioid samples had distinct expression patterns, and that there were similar numbers of short- and long-survival epithelioids that had an expression profile in-between that of epithelioid and sarcomatoid samples (four and two samples, respectively, possibly misclassified biphasics). This indicates that misclassifications should impact the two epithelioid sets similarly and thus should not induce a bias in the comparisons between epithelioid sets.
2.7. Interpretation of the PCA dimensions
We tested the association between each dimension and clinical and histopathological variables using linear regression, with sex, age, histopathological type, asbestos exposure, smoking status, necrosis, and grade, as explanatory variables (Table S3). Because each dimension of the PCA summarizes sets of genes, we can infer the main biological processes that correspond to each dimension by looking at the biological functions of these sets of genes. To do so, we computed Gene-Set Enrichment Analyses (GSEA) on hallmarks of cancer gene sets (gene sets in Table S4; results in Table S5) from the study of Kiefer and colleagues [21], after removing the duplicated genes from some hallmarks, using the Principal Component Gene-Set Enrichment method [22]. PCGSE uses t-tests to compare the mean correlation coefficient of non-hallmark genes with that of genes from a focal hallmark. In addition, to test the robustness of GSEA the results to the choice of database, we performed GSEA on the top correlated genes with Dimensions 1 and 2 using the STRING database [23] v11 (Table S6). From the top 300 genes correlated with Dimension 1, 66 Biological Process GO terms were enriched with vascular pathways, with the top 10 including: “regulation of vasculature development” (GO:1901342), “positive regulation of angiogenesis” (GO:0045766), and “artery development” (GO:0060840). Interestingly, the “cell adhesion” (GO:0007155) and “regulation of cell migration” (GO:0030334) GO terms were also in this top 10, suggesting that molecular pathways involved in the epithelial mesenchymal transition are also captured by Dimension 1. We performed the same analysis with the top 300 genes correlated with Dimension 2 coordinates and found 312 Biological Process GO terms enriched for a large majority of pathways related to the immune system. In fact, all nine most strongly associated pathways were directly linked to the immune system. All the results are presented in Table S6.
2.8. Survival analysis
Median survival times were estimated using the Kaplan-Meier nonparametric estimator. Survival predictions were tested using Cox proportional hazards models (R package survival v. 2.42-6) [24]. Goodness of fit was assessed using three diagnoses (following Bradburn and colleagues [25]): (i) to assess the proportional hazards assumption, we computed the Schoenfeld residuals test for each variable, using rank transformation for survival time (function cox.zph from package survival); (ii) to assess the leverage of each observation, we computed the change in regression coefficients when removing each observation (function ggdiagnostics with option “dfbeta” from package survminer); and (iii) to assess the general goodness of fit of the model, we plotted the deviance residuals as a function of linear predictions (function ggdiagnostics with option “deviance” from package survminer). We also assessed the functional forms for continuous variables using plots of martingale residuals against the values of the focal variable; because the percentage of sarcomatoid, PC1 and PC2 presented non-linear functional forms, we modelled them using smoothing splines with four degrees of freedom. Diagnostics and functional forms were assessed using R package survminer, v. 0.4.3.
We compared model fits using the time-dependant Area Under the ROC Curve (AUC) and its integral (iAUC; Section 3.3 of Chambless and Diao, 2006; R package survAUC, v. 1.0–5) [26], computed using leave-one-out cross-validation [27] (Table S7). Time-dependent AUC estimates the ability of a model to predict patients with a survival higher or lower than a given threshold, and iAUC integrates the results of time-dependent AUC over the threshold value, providing an interpretation similar to that of classical AUC.
To assess the ability of the PCA to predict survival, we used the first seven dimensions of the PCA as continuous explanatory variables, and included smoking, and asbestos exposure in the model, and used sex as a stratification variable (Table S3). The use of a stratification variable enables to adjust for the effect of sex, which is in our case a nuisance factor that is not investigated. To assess the ability of specific genes to predict survival, we used the expression of each gene as an explanatory variable, also including age and asbestos exposure as covariables, and sex as a stratification variable (Table S8). All models used the attained age scale, which provides a control for age effects without needing to fit an additional age parameter compatible with the proportional hazards assumption [28]. Because of the high proportion of missing smoking status information and the lack of significant association between this variable and the two first PCs, smoking was not included as a covariable in the model.
2.9. Differential protein expression analysis from IHC data
We performed differential gene-expression analyses from RNA-seq data on the discovery cohort (data from Bueno and colleagues and the TCGA) using univariate independent two-sample Wilcoxon U tests between a long-survival epithelioid group (47 samples from the cohort with survival >30 months), a short-survival epithelioid group (58 samples with survival <10 months), and a sarcomatoid group (eight samples with survival <10 months) as defined in Section 2.2 of Materials and Methods; because we had no a priori hypotheses about the direction of the effects of the groups on gene expression, we used two-sided tests in the discovery cohort. For the replication cohort, we conducted both univariable tests of differential protein expression between the matched sets (short-survival epithelioids, long-survival epithelioids, and sarcomatoids; paired two-sample Wilcoxon T-tests) and between epithelioid subtypes (subtypes and stromal variants; Kruskal-Wallis tests). Note that among the 77 samples of the replication cohort, due to missing data, the sample sizes were 74, 74, 73, 70, 74, and 69, respectively for CD8, PDL1 expression in the tumour, PDL1 in TILS, VEGFR2, VEGFR3, and VISTA. We used nonparametric tests because IHC measures are discrete and thus violate the normality assumption of linear models (linear regression and ANOVA), and we used similar tests in the discovery cohort to maximize the homogeneity between the statistical treatment of discovery and replication cohorts. We conducted Bivariable tests including both sets and epithelioid subtypes using conditional logistic regression. Because the replication cohort was used to confirm the hypotheses generated in the discovery cohort, including the direction of the effects, we performed one-sided tests. See results in Table S9.
2.10. Multiple testing corrections
Whenever multiple tests were performed, we computed q-values—p-values adjusted for a controlled false discovery rate—using the Benjamini-Hochberg procedure [29]. For the remainder of the text, “q” will denote such adjusted p-values and “p” will denote regular p-values.
2.11. Data sharing
TCGA RNA-seq data are available from the GDC portal (TCGA-MESO cohort) and the RNA-seq data from the Bueno and colleagues cohort are available from the European Genome-phenome Archive, EGA:EGAS00001001563. An interactive version of the PCA in Fig. 1a is available for further exploration in https://tumormap.ucsc.edu/ under the project MESOMICS.
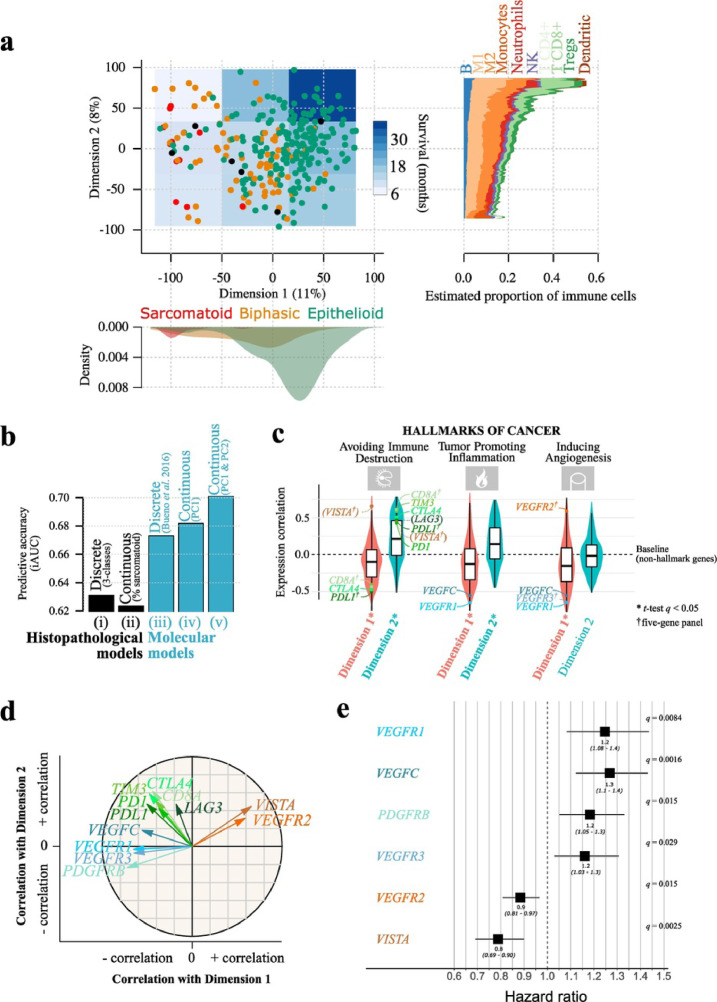
Fig. 1.
Malignant Pleural Mesothelioma expression profiles follow a continuum model.
a) Two-dimensional summary of 284 transcriptomes using Principal Component Analysis (PCA). Point colours represent the three histopathological types, and the overlayed blue-coloured rectangles represent the survival in nine regions; the filled shapes on the bottom panel correspond to the density of samples from each histopathological type on Dimension 1, and the filled shapes on the right panel correspond to the RNA-seq-estimated mean proportion of immune cells from 10 cell types, in each sample, as a function of Dimension 2 coordinates, computed using a moving average with a window size of 30 Dimension 2 units. b) Integral AUC (iAUC) of five Cox proportional hazards survival models: (i) a model based on the three histological types; (ii) a model based on the percentage of sarcomatoid; (iii) a model based on the four molecular clusters from the study of Bueno and colleagues [12]; (iv) a model based on the coordinates of samples on Dimension 1; (v) a model based on the coordinates of samples on Dimensions 1 and 2. c) Gene-Set Enrichment Analysis (GSEA) of the genes most correlated with Dimensions 1 and 2, based on the hallmarks of cancer gene sets; violin plots and boxplots represent the distribution of Pearson correlation coefficients between gene expression and Dimensions 1 (red) or 2 (blue); genes in parenthesis are not part of the current hallmark annotation; only the three hallmarks with the highest correlation are represented; see Fig. S9 for the results of all hallmarks. In the boxplot representation, centre line represents the median and box bounds represent the inter-quartile range (IQR). The whiskers span a 1.5-fold IQR or the highest and lowest observation values if they extend no further than the 1.5-fold IQR. d) Correlation circle of the Principal Component Analysis (PCA) from panel (a) for 12 genes of interest. Arrow lengths and direction correspond to the strength and sign of the correlation between the variable and Dimensions 1 and 2. e) Forest plot of hazard ratios for overall survival with age, sex and asbestos exposure as covariables. The black boxes represent estimated hazard ratios and whiskers represent the associated 95% confidence intervals. Wald test q-values are shown on the right. Only the markers significantly associated with survival are represented (Wald test q < 0.05); see Table S8 for the results of all genes. Data used in (a) and (c) correspond to the n = 211 samples from the study of Bueno and colleagues [12] and the n = 73 transcriptomes from the TCGA MESO cohort [13]. Data used in (b) correspond to the n = 199 samples from the Bueno cohort [12] with RNA-seq data and available percentage of sarcomatoid component. Data used in (e) correspond to n = 205 samples from the Bueno cohort [12] and n = 59 samples from the TCGA MESO cohort [13] with RNA-seq data and available asbestos exposure annotations. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Results
3.1. A continuous molecular classification of MPM
We computed a two-dimensional visual summary of the gene expression variation of 284 MPMs using an unsupervised analysis (Principal Component Analysis, PCA) (see Methods) (Fig. 1a, left panel; Table S1). Each dimension of the PCA summarizes the expression of sets of correlated genes, with the two first dimensions explaining respectively, 11% and 8% of the molecular variation of the most variable genes (7145 genes representing 50% of the total variance; Table S2, see Methods). The first dimension was significantly associated with the reported histopathological type (ANOVA q = 3.0 × 10−20; Fig. 1a, bottom panel; Table S3), with sarcomatoid samples mainly on the left (lowest coordinates on Dimension 1), biphasic samples mainly in the middle (intermediate coordinates on Dimension 1), and epithelioid samples mainly on the right (greatest coordinates on Dimension 1; Fig. 1a, left panel). Nevertheless, samples did not form discrete clusters, and rather conformed to a continuum of expression profiles (see density along Dimension 1 in Fig. S3). This first dimension was also significantly correlated with the percentage of sarcomatoid component in the tumour estimated by the pathologist from the H&E stain (r = −0.74, Pearson correlation test p = 8.4 × 10−36; Fig. S4), presence of necrosis (ANOVA q = 1.1 × 10−2; Table S3), and grade (ANOVA q = 2.5 × 10−2; Table S3), with samples on the left presenting high sarcomatoid component, high grade and necrosis.
The prognostic value of the histopathological classification is well known. In the PCA, samples in the top-right region (greatest coordinates on Dimensions 1 and 2) presented the best survival (Kaplan-Meier median estimate of 36 months, dark blue rectangle in the PCA; Fig. 1a, left panel), and samples on the left, the worst (Kapan-Meier median estimate of 10 months, light blue rectangles; see Table S7 for statistical tests and Fig. S5 for Kaplan-Meier curves). In order to compare the ability of histopathological and molecular data to predict survival, and to compare the relative benefit of using discrete versus continuous variables to predict survival, we compared five survival models: (i) a model based on the three histopathological types (epithelioid, biphasic, and sarcomatoid); (ii) a model based on the sarcomatoid content estimated by the pathologist (continuous phenotypic variable); (iii) a model based on the four molecular groups described by Bueno and colleagues (Epithelioid, Biphasic-E, Biphasic-S, and Sarcomatoid) [12]; (iv) a model based on the one-dimensional summary of gene expression data (using Dimension 1 as a continuous variable); and (v), a model based on the two-dimensional summary of gene expression data (using the two dimensions of the PCA as continuous variables) (Fig. 1b). We found that the models based on molecular (expression) data outperformed the models based on histopathology (iAUC of 0.63, 0.62, 0.67, 0.68, and 0.70, respectively for models i-v), with the continuous molecular models, and in particular, the one based on both dimensions providing the most accurate survival predictions (Fig. 1b). In particular, the continuous molecular model based on PC1 and PC2 provided better predictions for long-term survivors (more than 15% increase in AUC for survival greater than two years; Fig. S6). See Fig. S7 for diagnostics of the goodness of fit for each model, and Fig. S8 for assessments of the functional form of continuous variables. All tests of the proportional hazards assumption (Schoenfeld tests) were non-significant, and no trends were observed in any plot, suggesting adequate models (Fig. S7). One observation—a sarcomatoid tumour with large associated survival (~four years)—displayed a large leverage in most models (Fig. S7b); this observation had a particularly large leverage on the estimate of the sarcomatoid coefficient of model (i) from Fig. 1b (−0.59 change when the sample is removed), because of the very small number of samples in the sarcomatoid group.
Because each dimension of the PCA summarizes the expression of a large group of genes (1793 and 986 genes with an absolute correlation greater than 0.5 with Dimensions 1 and 2, respectively), we used gene-set enrichment analysis (GSEA) on the hallmarks of cancer—10 biological capabilities acquired during the development of tumours— [30] in order to reveal the cellular and molecular processes underlying the two dimensions of the PCA, and to inform their link with survival. We found that Dimension 1 was associated with hallmark “inducing angiogenesis", with samples on the left of the PCA (Fig. 1a, left panel) presenting higher expression levels of genes from this hallmark (negative association with Dimension 1, Fig. 1c; t-test q = 1.5 × 10−7). Dimension 2 was associated with “avoiding immune destruction” and “tumour-promoting inflammation”, with samples at the top of the PCA presenting higher expression levels of genes from these hallmarks (positive associations with Dimension 2; t-test q = 1.7 × 10−101 and q = 7.3 × 10−28, respectively; Fig. 1c; Fig. S9). Of note, genes from eight out of the 10 hallmarks presented a higher expression level in samples on the left of the PCA (t-tests q < 0.05; Fig. S9), which is in line with the worse prognosis of these samples (light blue areas).
Genes from the angiopoietins-tie pathway—which is critical for tumour angiogenesis—and CD31 (PECAM1 in the gencode annotation; which is also a marker of angiogenesis) [31], [32] behave similarly to the “inducing angiogenesis” hallmark. Indeed, many of the genes in the angiopoietins-tie axis (including ANGPTL1, ANGPTL4-5, ANGPTL7, ANGPT1-2, ANGPT4, and TIE1) belong to the genes with the largest molecular variation across samples (Table S2). In addition, many of these genes also contribute to the “inducing angiogenesis” (e.g., ANGPT1-2 and ANGPT4, ANGPTL3-4, and CD31) and the “tumour promoting inflammation” hallmarks (ANGPT1-2 and ANGPT4 and CD31; Table S3). Finally, ANGPT1-7, TIE1, and CD31 expression are all significantly correlated with Dimension 1, supporting our claim that this first dimension represents an angiogenesis axis (Table S5). We show in Table S6 that the association of vascularization with Dimension 1, and immune processes with Dimension 2 are robust to the choice of gene sets, by using GO terms instead of the hallmarks of cancer (see Methods). These results provide a biological interpretation of the dimensions, where Dimension 1 corresponds to an “angiogenesis” axis and Dimension 2 corresponds to an “immune response" and “inflammation” axis.
To assess the importance of tumour infiltrating lymphocytes in driving the gene expression differences across samples captured by the second dimension, we quantified the proportion of immune cells per sample using RNA-seq data [18]. We found that the estimated proportions of B cells, macrophages M2, CD8+ T cells, CD4+ regulatory T cells, and dendritic cells were significantly associated with this second dimension (permutation test q < 0.05; Fig. S10a). In particular, the proportion of CD8+ T cells presented the strongest variation across samples, with samples enriched for these cells being overrepresented in the top-left region of the PCA (Fig. 1a, right panel; Fig. S10). Concordantly, we found that for both Bueno and colleagues [12] and TCGA [13] cohorts, the amount of tumour infiltration estimated by the pathologists from the H&E stains was significantly correlated with the amount estimated from the matched expression data (Pearson correlation tests p = 9.3 × 10−7 and p = 2.8 × 10−4, respectively; Fig. S11).
3.2. Potential markers for classification and therapy
We then focused on finding candidate markers that could have dual roles: classification—accurately representing the continuum of molecular profiles of MPM—and therapy—being associated with possible therapeutic options. Among the 85 “inducing angiogenesis” genes significantly correlated with the first dimension, we found all the members of the vascular endothelial growth factor receptor (VEGFR) family of genes—FLT1 (VEGFR1), KDR (VEGFR2), and FLT4 (VEGFR3)—as well as the VEGFR3 and PDGFRB ligands VEGFC and PDGFB, respectively. Indeed, samples on the right of the PCA presented higher expression of VEGFR2 (Pearson correlation with Dimension 1: r = 0.59, q = 2.9 × 10−26; Fig. 1c–d) and samples on the left presented higher expression of genes PDGFRB, VEGFR1, VEGFR3, and VEGFC (respective correlations with Dimension 1: r = −0.72, −0.65, −0.65, and −0.56, and q = 3.4 × 10−44, 1.2 × 10−33, 5.7 × 10−33, and 9.4 × 10−23; Fig. 1c–d; Table S5). Genes VEGFR1 and VEGFC were also in the “tumour promoting inflammation” hallmark, highlighting their dual pro-angiogenic and pro-inflammatory role (Fig. 1c). The region with the largest amount of CD8+ T cells included samples with a low-survival profile (top-left region of the PCA), also harbouring overexpression of genes from the “avoiding immune destruction” hallmark (Fig. 1c). To gain some insights into this observation, we further investigated the 458 genes from the “avoiding immune destruction” hallmark showing significant correlations with the two dimensions and we found, among them, the CD8+ T-cell marker CD8A, and the immune checkpoints (IC) CTLA4, TIM3 (HAVCR2), PD1 (PDCD1), and PDL1 (CD274, Fig. 1c–d; Table S5). Similarly, other well-known ICs that were not annotated in the hallmarks of cancer list from Kiefer and colleagues [21], such as VISTA (C10orf54) and LAG3 [33], were also significantly correlated with Dimensions 1 and 2 (q < 0.05; Table S5). Interestingly, these data point to an immunosuppressive environment in these samples. The expression levels of six (PDGFRB, VEGFR1, VEGFR2, VEGFR3, VEGFC,VISTA) out of the 12 pro-angiogenic and IC genes above-mentioned, were individually associated with survival differences across samples (Fig. 1e). These associations still held significant for VISTA, VEGFR1 and VEGFC (at the 10% false discovery rate threshold) when restricting the analyses to epithelioid samples (Table S8), suggesting that this association is not only driven by histopathological types. See Fig. S12 for diagnostics of the goodness of fit for each survival model. All tests of the proportional hazards assumption (Schoenfeld tests) were non-significant, and no trends were observed in any plot, suggesting adequate models.
3.3. Technical orthogonal validation and independent replication
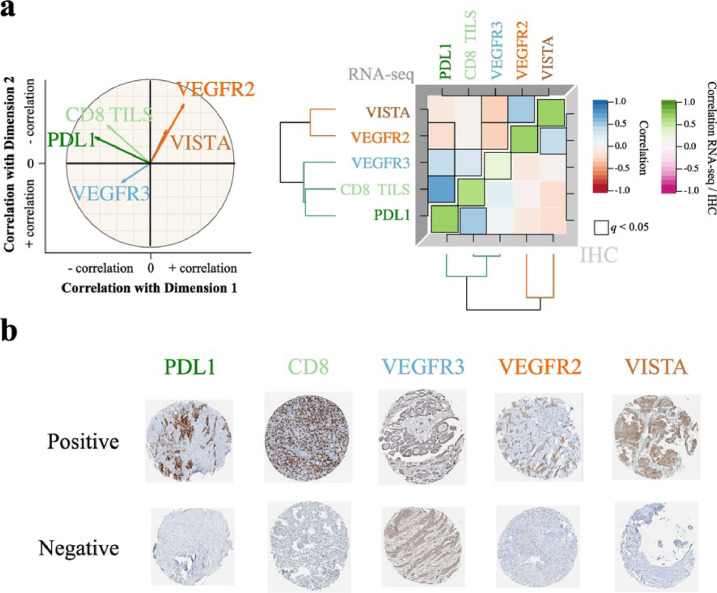
We validated the gene expression levels (from RNA-seq data) at the protein level using immunohistochemistry (IHC) on tissue microarrays (TMAs) generated for a subset of 103 out of the 284 samples included in this study. For this technical orthogonal validation, we selected five genes (out of the 12 genes above-mentioned) for which IHC-validated antibodies were available: CD8, PDL1, VEGFR3, VEGFR2 and VISTA. In order to test if these five genes provided a good approximation of the behaviour of the entire transcriptome, we computed a two-dimensional visual summary of these five genes (five-gene PCA; Fig. S13a) that we contrasted with the two-dimensional summary from the main PCA (Fig. 1a, left panel), based on the entire transcriptome (hereafter simply denoted as PCA). We found that the five-gene PCA provided a good approximation of the PCA: the first two dimensions of the five-gene PCA were significantly correlated with those of the PCA (Pearson correlation test p ≤ 6.2 × 10−59; Fig. S13b-c). In addition, the overall structure (direction and strength) of the correlations between the protein levels and the two dimensions matched that identified using the whole transcriptome (Fig. 2a, left panel versus Fig. 1d; Table S5). The protein levels of the five genes were significantly positively correlated with the gene-expression levels (green diagonal in Fig. 2a, right panel). Interestingly, despite the background detected for the VEGFR3 marker (see Methods), we still found a significant positive correlation with the gene expression. We also validated the correlation structure observed at the RNA-seq level—positive correlations between PDL1 and CD8, and between VEGFR2 and VISTA (upper triangular part in Fig. 2a, right panel)—at the protein expression level (lower triangular part in Fig. 2a, right panel). These observations further support the value of the five markers in explaining the continuum model of MPM, and support the existence of two dimensions in the protein expression of MPM—one associated with angiogenesis and the other one associated with the immune response. Examples of positive and negative samples for the above–mentioned markers are shown in Fig. 2b.
Fig. 2.
Technical validation of a five-gene panel on 103 MPMs.
a) Left panel: correlation circle of the Principal Component Analysis (PCA) based on the RNA-seq expression of the five-gene panel. Arrow lengths and direction correspond to the strength and sign of the correlation between the variable and Dimensions 1 and 2. Data used correspond to the n = 211 samples from the study of Bueno and colleagues [12] and the n = 73 transcriptomes from the TCGA MESO cohort [13]. Right panel: correlation matrix of the five-gene panel expression (upper triangle), of their protein expression (lower triangle), and correlation between expression from RNA-seq data and protein expression from IHC data (green diagonal). Colours correspond to the magnitude and sign of the correlations and statistically significant correlations are surrounded by a black box; dendrograms represent hierarchical clustering of gene or protein expression levels. Data used correspond to the n = 103 samples from the Bueno cohort [12] with RNA-seq data and with Tissue MicroArray (TMA) IHC staining data. b) Tissue MicroArray (TMA) IHC staining from the technical validation series corresponding to n = 103 samples from the Bueno cohort [12], with 0.6 mm core diameter at 5.2× magnification, for the five-gene panel, representing the positive and negative references of the tested protein expression. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The two main findings of this study [(i) the existence of two dimensions of variation in protein expression in MPM respectively linked to angiogenesis and the immune response, and (ii) the prognostic value of the above-mentioned five-marker panel] were independently replicated in a series of 77 additional MPMs from the French MESOBANK [14], on which we performed IHC for the five-gene panel and estimated the percentage of tumour infiltration from the H&E slides. This series was composed of three age- and sex-matched sets of 26 samples each, based on their histopathological type and survival characteristics: a short-survival epithelioid group, a long-survival epithelioid group, and a sarcomatoid group (Table 1). For the epithelioid samples, we also obtained a balanced representation of the different histopathological categories (epithelioid subtypes and stromal characteristics) across the matched sets (Table 1; see details in Methods); in addition, using hierarchical clustering analyses of the IHC data, we confirmed that histopathological type misclassifications are unlikely to bias the analyses (Fig. S2; see details in Methods). The protein levels in this independent series of samples allowed reproducing the two-dimensional summary resulting from the entire transcriptome (denoted IHC PCA and PCA, respectively; Fig. 3a). Indeed, as in the PCA (Fig. 3a, upper panel), a first dimension in the IHC PCA mainly separated sarcomatoid samples from epithelioid samples (Fig. 3a, bottom panel), and was negatively correlated with the expression of PDL1 and CD8 and positively correlated with the expression of VISTA and VEGFR2 (Fig. 3b). A second dimension was mostly orthogonal to the histopathological types, and positively correlated with the expression of PDL1, CD8, VEGFR2, and VISTA, and negatively correlated with the expression of VEGFR3 (Fig. 3b). Concordantly, the correlation structure of the protein expression in this replication series matched that of the discovery series based on RNA-seq data (Fig. 3b versus Fig. 2a).
Fig. 3.
Diagnostic and prognostic value of the expression level of the five-protein panel in the replication series of 77 MPMs, determined by immunohistochemistry.
a) Top panel: two-dimensional summary of the gene expression in the discovery cohort (n = 284) (PCA; subset of Fig. 1a). Bottom panel: two-dimensional summary of the protein expression of the five genes in the replication cohort (n = 77) (IHC PCA). Point colours correspond to the three sample sets from Table 1. b) Top panel: correlation circle of the IHC PCA (n = 77) from (a) bottom panel, where arrow lengths and direction correspond to the strength and sign of the correlation between the variable and Dimensions 1 and 2. Bottom panel: correlation matrix of the protein expression of the 77 MPMs from the replication cohort, where colours correspond to the magnitude and sign of the correlations; the dendrogram represents a hierarchical clustering of protein expression. Significant correlations are surrounded by a black box. c) Left panel: gene expression levels (normalized read counts) in the discovery cohort between long-survival epithelioid, long-survival epithelioid, and sarcomatoid groups, resulting in n = 82 samples from the study of Bueno and colleagues [12] and the n = 31 transcriptomes from the TCGA MESO cohort [13], for the three sets; each row presents violin plots and boxplots for a gene, with stars representing the significance level of pairwise comparisons between groups (q-values from two-sided independent Wilcoxon U tests). Right panel: Protein expression levels (% of cells where the protein is expressed) in the replication cohort, for the three sets; each row presents violin plots and boxplots for a protein, with stars representing the significance level of pairwise comparisons between groups (q-values from one-sided paired Wilcoxon T-tests). Sample sizes were 74, 74, 73, 70, 74, and 69, for CD8, PDL1 in the tumour, PDL1 in TILS, VEGFR2, VEGFR3, and VISTA, respectively. In the boxplot representation, centre line represents the median and box bounds represent the inter-quartile range (IQR). The whiskers span a 1.5-fold IQR or the highest and lowest observation values if they extend no further than the 1.5-fold IQR. d) PDL1 immunohistochemistry of two MPM cases from the replication cohort (left panel: short-survival epithelioid sample; right panel: sarcomatoid sample), both PDL1+ and PDL1 TILS+. Upper panels: Hematoxylin Eosin Saffron (HE) stain at 7× magnification, where white and black arrows show tumour cells and TILS, respectively. Lower panels: corresponding staining with PDL1 rabbit monoclonal antibody (cl SP263; VENTANA) at 7× magnification, where white and black arrows show positive staining of tumour cells and TILS, respectively. e) Protein expression level of VISTA in the replication cohort when considering epithelioid subtypes, independently of the sample set (upper panel) and in addition to the sample set (bottom panel). Data used correspond to n = 63 samples from the replication cohort with available data for all protein markers. In the boxplot representation, centre line represents the median and box bounds represent the inter-quartile range (IQR). The whiskers span a 1.5-fold IQR or the highest and lowest observation values if they extend no further than the 1.5-fold IQR.
In this independent series, we also validated the prognostic value of the markers. Indeed, the second dimension (IHC PCA Dimension 2) was associated with survival, with the region of high expression of VISTA and VEGFR2 (top-right region in Fig. 3a, bottom panel) enriched for long-survival epithelioids (median survival of 35 months). In terms of distinguishing long- and short-survival epithelioids, VISTA seems to be a promising individual marker since both gene expression and protein levels were significantly different between the two groups as shown in the discovery and replication series, respectively (Wilcoxon tests q < 0.01; Fig. 3c). In addition, the IHC also allowed differentiating the expression of PDL1 in tumour cells and in lymphocytes (Fig. 3d); the fact that the tumour cells express PDL1 further supports the suggested immunosuppressive phenotype. VISTA was the only protein with significant expression differences between epithelioid subtypes (Kruskal-Wallis test q = 0.066; Fig. 3e left panel; Table S9). Surprisingly, despite the overall good-prognosis of VISTA expression that we identified, VISTA was overexpressed in the epithelioid subtypes usually associated with intermediate-prognosis (trabecular) and bad-prognosis (solid), compared to those usually associated with good-prognosis (acinar, papillary, and myxoid stroma; Fig. S14). In fact, stratifying by epithelioid subtype revealed larger VISTA expression differences between the short- and long-survival sets (Fig. 3e, right panel; Fig. S14).
4. Discussion
The molecular profile and the prognosis of malignant pleural mesothelioma (MPM) appears to be better explained by a continuous model, with strong differences in the expression of pro-angiogenic and immune checkpoint (IC) genes across samples, pointing to the immune and vascular systems as the major sources of variation. This continuous model can be thought as a refinement of the four-class molecular subdivision from Bueno and colleagues [12] as follows: firstly, we showed that its continuity better captures the molecular variation and provides better prediction than a discrete classification; and secondly, we showed that the continuous model captured a second dimension that was independent of the histopathological classification and that was also associated with survival. This second dimension was not captured by previous molecular classification studies [12], [13], presumably because of their focus on discrete groups correlated with histopathological types. Importantly, we find that this two-dimensional continuous model enables in particular better predictions of long-term survival, which is coherent with the identification of a region of better prognosis (36 months) for high values of both Dimensions 1 and 2. The discovery of a two-dimensional summary of molecular variation uncovered important associations with the 10 currently accepted hallmarks of cancer. In particular, genes of eight hallmarks showed general upregulation in the region enriched for sarcomatoid and biphasic tumours, including the hallmark “activating invasion motility” that encompasses pathways involved in the epithelial-mesenchymal transition, which is known to play an important role in MPM [34], [35]. This could explain the increased aggressiveness reported for these two tumour types, as well as the diverse responses to anti-angiogenic agents and immunotherapies. In addition, among genes from these eight hallmarks that are significantly correlated with the first dimension and upregulated in the region with sarcomatoid and biphasic tumours, there are two well-known indexes used in the clinic: MKI67, which is a well-known proliferation index and which is in the “sustaining proliferative signaling” hallmark, and CASP3, which is a well-known apoptotic index which is in the “evading growth suppressors", “deregulating cellular energetics", and “resisting cell death” hallmarks (Table S4). Because our results for these two genes only correspond to gene expression estimates from RNA-seq data, additional studies are warrantied to confirm the correlation between gene and protein expression in this group of tumours.
At the extremes of the above-mentioned two dimensions, we could define three molecular profiles with prognostic and therapeutic implications (Fig. 4). The first profile (hot/IC+/Angio+) would correspond to “hot” tumours (highly infiltrated with T lymphocytes), enriched for non-epithelioid types (biphasic and sarcomatoid), and characterized by the high expression of pro-angiogenic genes (VEGFR1, VEGFR3, and PDGFRB) and ICs (PD(L)1, CTLA4, TIM3, and LAG3). Patients developing tumours with this profile are expected to show a short median survival (7 months). These characteristics are in line with published data suggesting that PD(L)1 expression by immunohistochemistry is correlated with non-epithelioid histology and poor survival [36].
Fig. 4.
Characteristics of the three Malignant Pleural Mesothelioma transcriptomic profiles.
The schematic position of samples harbouring a given profile in the two-dimensional summary from Fig. 1a (n = 284) is represented in the bottom right panel. For each profile, the hallmarks of cancer generally upregulated are indicated by pictograms in the upper left part, the histological type composition is represented by a pie chart in the upper right part, the proportion of tumour infiltrating lymphocytes estimated from the RNA-seq data (Figs. 1 and S10) is represented by a bar plot in the bottom left part, and the expression of representative genes is represented by a radar plot in the bottom right part. Tissue MicroArray (TMA) IHC staining from the technical validation series, with 0.6 mm core diameter at 5.2× magnification, for the five-gene panel, are presented above each panel.
The second profile (VEGFR2+/VISTA+) would correspond to tumours with high expression levels of VEGFR2 and VISTA, enriched for the epithelioid type. Patients carrying tumours with this profile are expected to show the best median survival (36 months). Despite its suggested immunosuppressive role [37], VISTA expression in tumour cells has been associated with increased tumour-infiltrating lymphocytes, PD-1, a favourable immune microenvironment, and with better overall survival in hepatocellular carcinoma [38] and non-small cell lung cancer [39]. Although also associated with better survival, we found that VISTA expression is associated with VEGFR2 expression, further supporting the possible interaction of these two pathways in MPM. Interestingly, VISTA was the only protein with significant expression differences between epithelioid subtypes, suggesting a potential diagnostic value.
The third and last profile (cold/Angio+) would be represented by “cold” tumours (devoid of immune effector cells) enriched for the non-epithelioid types, and with high expression of pro-angiogenic genes (VEGFR1, VEGFR3, and PDGFRB). Patients with tumours with this profile are expected to show a bad survival (median of 10 months). Of note, when stratifying the analysis by tumour type, we also found that patients with epithelioid tumours of the first and third profiles have a worse survival (median of 10 and 17 months) than those with the second profile (median of 27 months). Tumours in this group also show high levels of VEGFC. Upon activation by VEGFRC, VEGFR3 has a role in lymphangiogenesis, which is an important feature in MPM [40]. It has been shown in cellular models that activation of VEGFR3 on natural killer cells by VEGFC can lead to immunosuppression and that the treatment with the VEGFR3-selective tyrosine-kinase inhibitor MAZ51 counterbalanced this effect [41]. It has also been proven by immunohistochemistry that VEGFR3 is expressed in MPM of different histopathological types, supporting its putative role as a potential therapeutic target in this disease [42].
MPM being refractory to chemotherapy and radiotherapy, there is an urgent need to identify novel and promising candidate therapeutic options as well as the best candidates for those options, especially for the sarcomatoid and biphasic types. Considering the known role of the VEGF/VEGFR axis and the immune response as driving forces in MPM [11], [43], drugs against these pathways have been developed to treat this disease; unfortunately, anti-angiogenic therapies for mesothelioma patients have shown modest activity in clinical trials [6], and recent data from ongoing clinical trials pointed that, while immunotherapy remains promising in the treatment of a subset of mesothelioma patients, better predictive markers of response are needed [44]. Several recent reviews have nicely summarized how the tumour-associated blood and lymphatic vasculature play an important role in avoiding tumour destruction, as well as the therapeutic opportunities to overcome this immune blockage [45], [46], [47], [48], pointing to combinations of anti-angiogenic drugs and immunotherapy as promising options for the management of many cancers. In this study we found a role for the immune and vascular systems in MPM that might not only have a prognostic value, but also allow stratification of patients for the most relevant therapeutic options.
Contrary to already published studies, on which the authors have made an implicit assumption of discreteness by focusing their analyses on (discrete) histopathological types, or on (discrete) molecular clusters (identified using consensuscluster+ or iCluster+), in this study we have made no such assumption. This agnostic characterization of the molecular diversity of these tumours allowed observing an inherent continuity of the tumour phenotypes in MPM that helped uncover clinically relevant pathway interactions that have not been identified in the published studies, presumably because of this implicit assumption of discreteness. Overall, the role of angiogenesis and the heterogeneous microenvironment of MPM could be used as Achilles' heel for this disease; however, the success of future treatments will strongly rely on a deep understanding of the biology of the disease and the interactions that may occur between the most frequently altered pathways.
Funding sources
This work has been funded by the French National Cancer Institute (INCa, PRT-K-15-039 to LFC), and the Ligue Nationale contre le Cancer (LNCC 2017 to LFC). LM has fellowships from the LNCC. The funding sources had no involvement in study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
Declaration of Competing Interests
The authors declare no conflict of interest related to the work presented here. Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organisation, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer/World Health Organisation.
Author contributions
LFC, FGS, and MF conceived and designed the study, and supervised key aspects of the study. NA and LM performed the computational and statistical analyses. CG, MB, FTB, CB, JPLR, GP, NR, DD, JCP, MCC, AS, EW, LW, SL, JMV, AG, CG, CS, CB, VH, PH, JM, VTM, ECT, JM, IR, HB, SL, and RB contributed with samples and the corresponding histopathological, epidemiological, and clinical data. NL, SB, DF, KA, and JDM helped with logistics. JYB, CC, NG, and JDM gave scientific input. NA, LM, JDM, MF and LFC wrote the manuscript, which was reviewed and commented by all the co-authors.
Acknowledgements
We thank the editor and two anonymous reviewers for their comments. We thank the patients donating their tumour specimens. The results shown here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. This study is part of the MESOMICS project and the Rare Cancers Genomics initiative (www.rarecancersgenomics.com).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.09.003.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- 1.Lacourt A., Leveque E., Guichard E., Gilg Soit Ilg A., Sylvestre M.P., Leffondre K. Dose-time-response association between occupational asbestos exposure and pleural mesothelioma. Occup Environ Med. 2017;74:691–697. doi: 10.1136/oemed-2016-104133. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. WHO . 4th ed. 2015. Classification of tumours of the lung, pleura, thymus and heart. [DOI] [PubMed] [Google Scholar]
- 3.Nicholson A.G., Sauter J.L., Nowak A., Kindler H., Gill R., Remy-Jardin M. EURACAN/IASLC proposals for updating the histologic classification of pleural mesothelioma: towards a more multidisciplinary approach. J Thorac Oncol (In Press) 2019 doi: 10.1016/j.jtho.2019.08.2506. [DOI] [PubMed] [Google Scholar]
- 4.Galateau Salle F., Le Stang N., Nicholson A.G., Pissaloux D., Churg A., Klebe S. New insights on diagnostic reproducibility of biphasic mesotheliomas: a multi-institutional evaluation by the international mesothelioma panel from the MESOPATH reference center. J Thorac Oncol: Off Pub Int Assoc Study Lung Cancer. 2018;13:1189–1203. doi: 10.1016/j.jtho.2018.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zalcman G., Mazieres J., Margery J., Greillier L., Audigier-Valette C., Moro-Sibilot D. Bevacizumab for newly diagnosed pleural mesothelioma in the Mesothelioma Avastin Cisplatin Pemetrexed Study (MAPS): a randomised, controlled, open-label, phase 3 trial. Lancet (London, England) 2016;387:1405–1414. doi: 10.1016/S0140-6736(15)01238-6. [DOI] [PubMed] [Google Scholar]
- 6.Zucali P.A. Target therapy: new drugs or new combinations of drugs in malignant pleural mesothelioma. J Thorac Dis. 2018;10:S311–s21. doi: 10.21037/jtd.2017.10.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kindler H.L., Ismaila N., Armato S.G., 3rd, Bueno R., Hesdorffer M., Jahan T. Treatment of malignant pleural mesothelioma: American Society of Clinical Oncology clinical practice guideline. J Clin Oncol. 2018;36:1343–1373. doi: 10.1200/JCO.2017.76.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Gooijer C.J., Baas P. Treat it or leave it: immuno-oncology in mesothelioma observed by the eyes of Argus. J Thorac Oncol. 2018;13:1619–1622. doi: 10.1016/j.jtho.2018.08.2024. [DOI] [PubMed] [Google Scholar]
- 9.European Society for Medical Oncology . ESMO; 2017. Combination immunotherapy in second/third line extends mesothelioma survival to 15 months. http://www.esmo.org/Press-Office/Press-Releases/Combination-Immunotherapy-in-Second-third-Line-Extends-Mesothelioma-Survival-to-15-Months. Topic: lung and other thoracic tumours. [Google Scholar]
- 10.Yap T.A., Aerts J.G., Popat S., Fennell D.A. Novel insights into mesothelioma biology and implications for therapy. Nat Rev Cancer. 2017;17:475–488. doi: 10.1038/nrc.2017.42. [DOI] [PubMed] [Google Scholar]
- 11.Thapa B., Salcedo A., Lin X., Walkiewicz M., Murone C., Ameratunga M. The immune microenvironment, genome-wide copy number aberrations, and survival in mesothelioma. J Thorac Oncol: Off Pub Int Assoc Study Lung Cancer. 2017;12:850–859. doi: 10.1016/j.jtho.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Bueno R., Stawiski E.W., Goldstein L.D., Durinck S., De Rienzo A., Modrusan Z. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. 2016;48:407–416. doi: 10.1038/ng.3520. [DOI] [PubMed] [Google Scholar]
- 13.Hmeljak J., Sanchez-Vega F., Hoadley K.A., Shih J., Stewart C., Heiman D. Integrative molecular characterization of malignant pleural mesothelioma. Cancer Discov. 2018;8:1548–1565. doi: 10.1158/2159-8290.CD-18-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galateau-Salle F., Gilg Soit Ilg A., Le Stang N., Brochard P., Pairon J.C., Astoul P. The French mesothelioma network from 1998 to 2013. Ann Pathol. 2014;34:51–63. doi: 10.1016/j.annpat.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 15.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galore Krueger F.Trim. 2015. A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. [Google Scholar]
- 18.Finotello F., Mayer C., Plattner C., Laschober G., Rieder D., Hackl H. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019;11:34. doi: 10.1186/s13073-019-0638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McInnes L., Healy J., Melville J.U.M.A.P. Uniform manifold approximation and projection for dimension reduction. 2018. https://arxiv.org/abs/1802.03426 arXivorg. 2018/02/09 ed: eprint arXiv:1802.03426.
- 21.Kiefer J., Nasser S., Graf J., Kodira C., Ginty F., Newberg L. Hallmarks of cancer gene set annotation. 2017. https://figshare.com/articles/Hallmarks_of_Cancer_Gene_Set_Annotation/4794025/1
- 22.Frost H.R., Li Z., Moore J.H. Principal component gene set enrichment (PCGSE) BioData Min. 2015;8:25. doi: 10.1186/s13040-015-0059-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–d13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Therneau T. A package for survival analysis in S. version 2.38. 2015. https://cran.r-project.org/web/packages/survival/citation.html
- 25.Bradburn M.J., Clark T.G., Love S.B., Altman D.G. Survival analysis part III: multivariate data analysis -- choosing a model and assessing its adequacy and fit. Br J Cancer. 2003;89:605–611. doi: 10.1038/sj.bjc.6601120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chambless L.E., Diao G. Estimation of time-dependent area under the ROC curve for long-term risk prediction. Stat Med. 2006;25:3474–3486. doi: 10.1002/sim.2299. [DOI] [PubMed] [Google Scholar]
- 27.Potapov S., Adler W., Schmid M. 2012. survAUC: Estimators of prediction accuracy for time-to-event data.https://CRAN.R-project.org/package=survAUC R package version 10-5. 2012-09-04 ed. [Google Scholar]
- 28.Griffin B.A., Anderson G.L., Shih R.A., Whitsel E.A. Use of alternative time scales in cox proportional hazard models: implications for time-varying environmental exposures. Stat Med. 2012;31:3320–3327. doi: 10.1002/sim.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Methodol. 1995;57:289–300. [Google Scholar]
- 30.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Magkouta S., Kollintza A., Moschos C., Spella M., Skianis I., Pappas A. Role of angiopoietins in mesothelioma progression. Cytokine. 2019;118:99–106. doi: 10.1016/j.cyto.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 32.Magkouta S., Pappas A., Pateras I.S., Kollintza A., Moschos C., Vazakidou M.E. Targeting Tie-2/angiopoietin axis in experimental mesothelioma confers differential responses and raises predictive implications. Oncotarget. 2018;9:21783–21796. doi: 10.18632/oncotarget.25004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenkins R.W., Barbie D.A., Flaherty K.T. Mechanisms of resistance to immune checkpoint inhibitors. Br J Cancer. 2018;118:9–16. doi: 10.1038/bjc.2017.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fassina A., Cappellesso R., Guzzardo V., Dalla Via L., Piccolo S., Ventura L. Epithelial-mesenchymal transition in malignant mesothelioma. Mod Pathol: Off J U S Can Acad Pathol, Inc. 2012;25:86–99. doi: 10.1038/modpathol.2011.144. [DOI] [PubMed] [Google Scholar]
- 35.Schramm A., Opitz I., Thies S., Seifert B., Moch H., Weder W. Prognostic significance of epithelial-mesenchymal transition in malignant pleural mesothelioma. Eur J Cardiothorac Surg: Off J Eur Assoc Cardiothorac Surg. 2010;37:566–572. doi: 10.1016/j.ejcts.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 36.Combaz-Lair C., Galateau-Salle F., McLeer-Florin A., Le Stang N., David-Boudet L., Duruisseaux M. Immune biomarkers PD-1/PD-L1 and TLR3 in malignant pleural mesotheliomas. Hum Pathol. 2016;52:9–18. doi: 10.1016/j.humpath.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 37.Lines J.L., Pantazi E., Mak J., Sempere L.F., Wang L., O'Connell S. VISTA is an immune checkpoint molecule for human T cells. Cancer Res. 2014;74:1924–1932. doi: 10.1158/0008-5472.CAN-13-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang M., Pang H.J., Zhao W., Li Y.F., Yan L.X., Dong Z.Y. VISTA expression associated with CD8 confers a favorable immune microenvironment and better overall survival in hepatocellular carcinoma. BMC Cancer. 2018;18:511. doi: 10.1186/s12885-018-4435-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villarroel-Espindola F., Yu X., Datar I., Mani N., Sanmamed M., Velcheti V. Spatially resolved and quantitative analysis of VISTA/PD-1H as a novel immunotherapy target in human non-small cell lung cancer. Clin Cancer Res: Off J Am Assoc Cancer Res. 2018;24:1562–1573. doi: 10.1158/1078-0432.CCR-17-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohta Y., Shridhar V., Bright R.K., Kalemkerian G.P., Du W., Carbone M. VEGF and VEGF type C play an important role in angiogenesis and lymphangiogenesis in human malignant mesothelioma tumours. Br J Cancer. 1999;81:54–61. doi: 10.1038/sj.bjc.6690650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee J.Y., Park S., Min W.S., Kim H.J. Restoration of natural killer cell cytotoxicity by VEGFR-3 inhibition in myelogenous leukemia. Cancer Lett. 2014;354:281–289. doi: 10.1016/j.canlet.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 42.Filho A.L., Baltazar F., Bedrossian C., Michael C., Schmitt F.C. Immunohistochemical expression and distribution of VEGFR-3 in malignant mesothelioma. Diagn Cytopathol. 2007;35:786–791. doi: 10.1002/dc.20767. [DOI] [PubMed] [Google Scholar]
- 43.Carbone M., Kanodia S., Chao A., Miller A., Wali A., Weissman D. Consensus report of the 2015 Weinman international conference on mesothelioma. J Thorac Oncol: Off Pub Int Assoc Study Lung Cancer. 2016;11:1246–1262. doi: 10.1016/j.jtho.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abstract Book . (iMig) TIMIG, editor. 14th International Conference of the International Mesothelioma Interest Group (iMig) The International Mesothelioma Interest Group (iMig); Ottawa, Canada: 2018. http://www.imig2018.org/wp-content/uploads/2018/04/iMig2018_abstractbook.pdf [Google Scholar]
- 45.Fukumura D., Kloepper J., Amoozgar Z., Duda D.G., Jain R.K. Enhancing cancer immunotherapy using antiangiogenics: opportunities and challenges. Nat Rev Clin Oncol. 2018;15:325–340. doi: 10.1038/nrclinonc.2018.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang Y., Kim B.Y.S., Chan C.K., Hahn S.M., Weissman I.L., Jiang W. Improving immune-vascular crosstalk for cancer immunotherapy. Nat Rev Immunol. 2018;18:195–203. doi: 10.1038/nri.2017.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khan K.A., Kerbel R.S. Improving immunotherapy outcomes with anti-angiogenic treatments and vice versa. Nat Rev Clin Oncol. 2018;15:310–324. doi: 10.1038/nrclinonc.2018.9. [DOI] [PubMed] [Google Scholar]
- 48.Schaaf M.B., Garg A.D., Agostinis P. Defining the role of the tumor vasculature in antitumor immunity and immunotherapy. Cell Death Dis. 2018;9:115. doi: 10.1038/s41419-017-0061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2